Abstract
We introduce a portable biochemical analysis platform for rapid field deployment of nucleic acid-based diagnostics using consumer-class quadcopter drones. This approach exploits the ability to isothermally perform the polymerase chain reaction (PCR) with a single heater, enabling the system to be operated using standard 5 V USB sources that power mobile devices (via battery, solar, or hand crank action). Time-resolved fluorescence detection and quantification is achieved using a smartphone camera and integrated image analysis app. Standard sample preparation is enabled by leveraging the drone’s motors as centrifuges via 3D printed snap-on attachments. These advancements make it possible to build a complete DNA/RNA analysis system at a cost of ∼$50 ($US). Our instrument is rugged and versatile, enabling pinpoint deployment of sophisticated diagnostics to distributed field sites. This capability is demonstrated by successful in-flight replication of Staphylococcus aureus and λ-phage DNA targets in under 20 min. The ability to perform rapid in-flight assays with smartphone connectivity eliminates delays between sample collection and analysis so that test results can be delivered in minutes, suggesting new possibilities for drone-based systems to function in broader and more sophisticated roles beyond cargo transport and imaging.
The recent Ebola outbreak has exposed some of the key limitations facing current infectious disease management strategies, particularly when applied in remote underdeveloped areas. Existing approaches are highly resource intensive, relying on dispatching specially trained personnel to isolated locations where biological samples are collected and returned to dedicated laboratories for analysis. These inefficient channels become overwhelmed when field sites are not connected by a modern transportation infrastructure, introducing a considerable time lag between sample collection, diagnosis, and implementation of countermeasures. The resulting bandwidth of information flow is often extremely narrow, delaying treatment of infected individuals and making it challenging for medical authorities to proactively formulate effective courses of action. A need therefore exists for inexpensive and robust tools that can be broadly deployed to accelerate diagnosis, enable pinpoint delivery of therapeutics, and provide real-time data to better inform decision making.1−10
Nucleic acid-based approaches like the polymerase chain reaction (PCR) are considered diagnostic gold standards in terms of both sensitivity and specificity, but PCR is largely ruled out for portable deployment owing to an unfavorable combination of (i) excessive electrical power requirements associated with repeated heating and cooling of reagents during thermocycling and (ii) complexities involved with inexpensively implementing fluorescence-based product detection. Here, we show how convective thermocycling can be leveraged to overcome these limitations, laying the foundation for a new generation of simple ruggedized PCR-based diagnostic platforms (Figure 1). The same natural convection phenomena at play in ordinary motion lamps (e.g., Lava Lamps) enable the PCR to be isothermally executed using a single heater maintained at the reaction’s denaturing temperature. This inherently simple design dramatically reduces electrical power consumption and can be readily interfaced with a fluorescence detection system that exploits the versatility and connectivity of ordinary smartphones.11−13 These breakthroughs make it possible to construct ultraportable, rapid, and quantitatively accurate PCR-based nucleic acid analysis systems for ∼$50 ($US) enabling conventional laboratory protocols to be followed with little or no modification (Figure S1, Table S1).
Figure 1.
Lab-on-a-drone. (A) Convective thermocycling enables the PCR to be actuated isothermally using a single heater. (B) The instrument can be assembled for ∼$50 ($US) using readily available components. (C) Fluorescence detection of reaction products is achieved using an ordinary smartphone camera. (D) The entire assembly is incredibly lightweight, enabling deployment on consumer-class quadcopter drones. (E) Ruggedization is demonstrated by performing in-flight PCR as a drone payload. Successful in-flight replication of two different DNA targets is achieved (lane M, FlashGel DNA marker; lane 1, 147 bp S. aureus target (16 min in-flight reaction time); lane 2, 237 bp target from a λ-phage DNA template (18 min in-flight reaction time)). Tamb ∼ 23 °C.
Since its inception a little over a decade ago,14 convective thermocycling has remained an intriguing avenue to enable rapid PCR, but a crucial roadblock to practical implementation of this approach has been the inherent interdependence between the internal flow field and the reactor geometry (expressed in terms of the height, h, and diameter, d, of a cylindrically shaped configuration, Figure 1A). The spatial temperature gradient established between the top (cool) and bottom (hot) surfaces of the reactor not only actuates the denaturing, annealing, and extension steps necessary to perform the PCR but also supplies the driving force to physically transport reagents between these reaction zones. It has previously been assumed that this interplay implies a need to custom design reactor geometries to match the individual thermal requirements of each PCR assay to be performed (e.g., multiplex analysis involving different primers with different annealing temperatures), and that robustness is constrained by a need to maintain a specific orientation with respect to the gravitational driving force. We recently developed a 3D coupled flow-reaction model that contradicts this view, revealing an unexpectedly broad design space dominated by chaotic advection where reaction rates are greatly accelerated and remain essentially unchanged over virtually the entire range of realistic PCR conditions.15,16 Any reactor geometry selected within this regime is therefore universally functional (i.e., analogous to standardized PCR tubes and plates), making it possible to execute a 30 cycle PCR in 10–20 min regardless of temperature set point and spatial orientation.
Experimental Section
Convective Thermocycling
Cylindrical reactors were constructed from 1 in. diameter polycarbonate rod stock (Amazon Supply) by cutting them to lengths and machining holes to produce reactors of desired height (10 mm) and diameter (2.5 mm) yielding an ∼20 μL reactor volume. Smartphone-enabled real time PCR experiments were performed to replicate a 237 base pair target from a λ-phage DNA template using a SYBR Green PCR master mix (Table S2). Sensitivity to initial copy number (Figure 2F), ambient temperature (Figure 3C), and inclination angle of the convective reactor (Figure 3E) were established without fluorescence detection by replicating a 237 base pair target from λ-phage DNA template with KOD polymerase enzyme (Table S3). Analysis of Ebola (150 bp target from positive control template, Table S4) and Staphylococcus aureus (147 bp target from purified genomic DNA, Table S5) was performed using test kits supplied with the respective samples. Convective PCR runs for Ebola and Staphylococcus aureus detection were performed using a preincubated hot start reagent mixture.
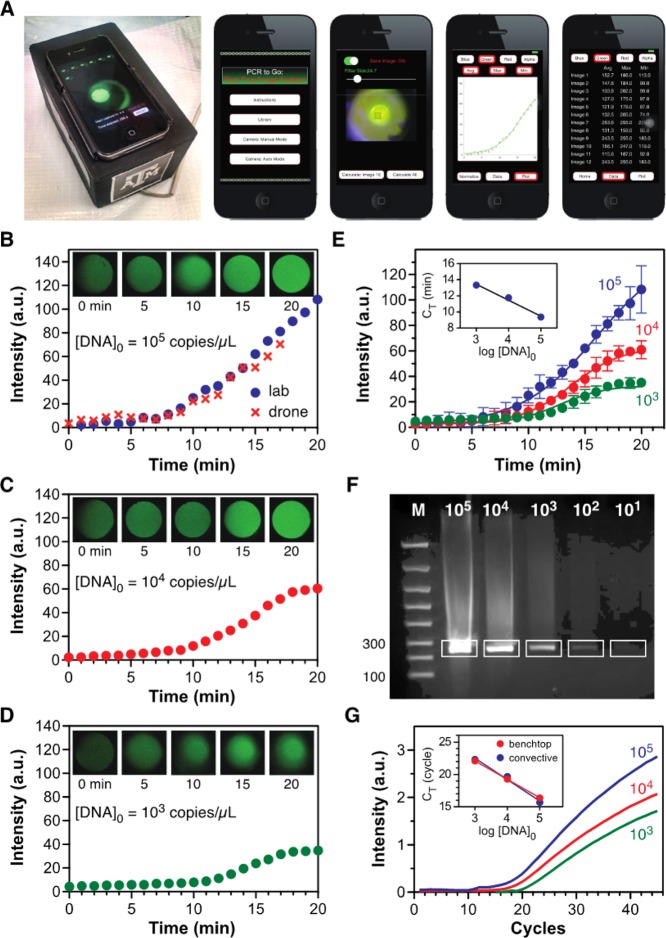
Figure 2.
Quantitative smartphone-based fluorescence detection. (A) The PCR to Go analysis app enables smartphone-based image acquisition, processing, and data analysis. (B–D) Measured fluorescence as a function of initial template copy number (237 bp target from a λ-phage DNA template; images acquired by the smartphone camera are shown at the top of each plot). The data in (B) also show that comparable results are obtained when analysis is performed in-flight as a drone payload. (E) Quantification is achieved by applying sigmoidal fits to these data (mean ± sd of 3 replicates) and using reaction times when fluorescence exceeds a threshold value of 20 units to construct a standard curve (inset, CT = 9.4, 11.8, and 13.3 min for [DNA]0 = 105, 104, and 103 copies/μL, respectively), whose slope yields a doubling time of 35.8 s. (F) Convective thermocycling (25 min reaction time) achieves sensitivity in the 101 to 102 copy/μL range (lane M, FlashGel DNA marker, remaining lanes correspond to initial DNA copy numbers indicated on the gel image). (G) Smartphone-based fluorescence analysis yields quantification comparable to a benchtop real time PCR instrument, with nearly identical standard curves.
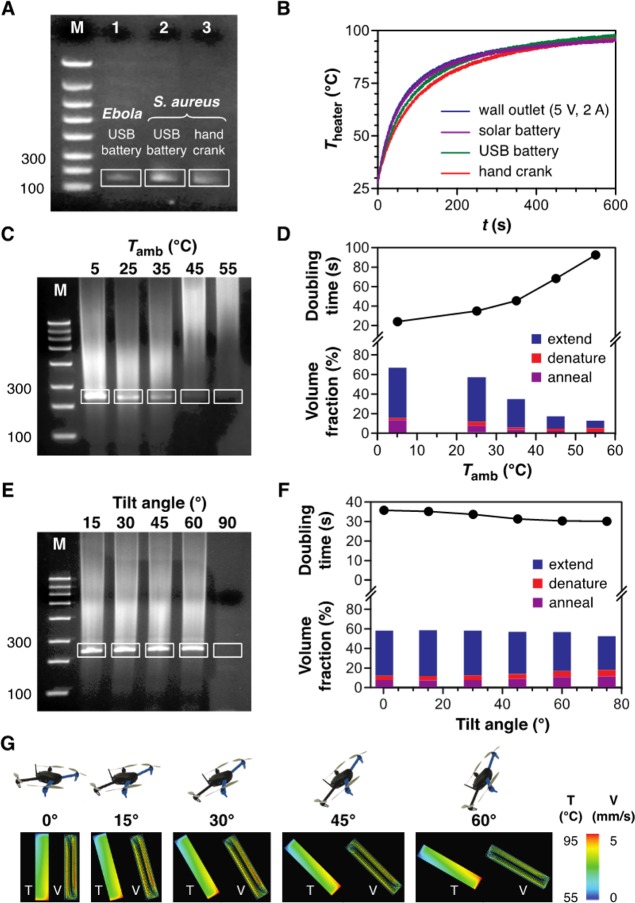
Figure 3.
A versatile and rugged PCR analysis platform. (A) Convective thermocycling enables replication of a 150 bp Ebola target (lane 1) and a 147 bp S. aureus target (lanes 2, 3) in 20 min (lane M, FlashGel DNA marker) using USB battery and hand crank power sources. (B) Comparable heater performance is achieved via on- or off-grid electrical power sources. Experiments (C) and reaction model simulations (D) display consistent performance over a broad ambient temperature range. Experiments (E) and simulations (F) show that product yields are insensitive to the reactor’s orientation with respect to the vertical direction up to tilt angles of at least 60°. (G) Simulations verify that favorable flow fields are maintained at these orientations (V and T denote velocity magnitude and temperature profiles). Gels in (C) and (E) show replication of a 237 bp target from a λ-phage DNA template in 25 min (lane M, FlashGel DNA marker; some smearing in the product bands is evident because a hot start enzyme protocol was not employed in these tests). Plots in (D) and (F) show doubling time (above) and fractional reactor volume maintained at each PCR temperature condition (below).
Prior to reagent loading, convective reactors were rinsed with a 10 mg/mL aqueous solution of bovine serum albumin (Cat. no. A2153; Sigma-Aldrich) followed by Rain-X Anti-Fog (ITW Global Brands) and dried to minimize sidewall adsorption and enhance surface wettability. After loading reagents, reactors were placed on the preheated (95 °C) ceramic heater surface of the portable PCR device and secured using adhesive tape (Scotch Magic Tape, 3M). The PCR to Go iPhone app was run in continuous mode and set to capture images of the top reactor surface at 1 min intervals. After incubation for a desired time, convective reactors were removed and the post PCR products were pipetted out for subsequent analysis by agarose gel electrophoresis using a Lonza FlashGel system (1 μL of FlashGel loading dye (Cat. no. 50462) and 4 μL of DNA samples). Samples were run at 280 V for 8 min with a FlashGel DNA marker (100 bp–4 kb, Cat. no. 50473; scale (bp): 100, 200, 300, 500, 800, 1250, 2000, and 4000).
Computational Modeling and Reactor Design
Detailed formulation of the computational fluid dynamics and the PCR kinetic model have been described previously.15 Briefly, the finite volume solver of STAR-CCM+ (CD-adapco) was used to simultaneously evaluate the continuity, 3D Navier-Stokes, and energy conservation equations with the Boussinesq approximation. The resulting convective flow field was coupled with a PCR kinetic model yielding time-resolved evolution of the target DNA concentration. A doubling time (time required for the concentration of target DNA molecules to double) was then extracted as a metric for the global DNA replication rate. For the single heater setup, a convective heat loss boundary condition was imposed at the top reactor surface (Q = hc (Ttop – Tamb); hc is the convective heat transfer coefficient (1000 W/(m2 K)); Ttop is the top boundary temperature; Tamb is the ambient temperature). There was no appreciable change in the velocity and temperature fields when the adiabatic side walls were replaced with conductive acrylic walls. Fluid properties were cast as temperature dependent field functions. Geometries for 3D simulation were created and meshed using Gambit (ANSYS). Nonuniform hexahedral grids were generated, and grid independent solution was verified.
Circuit Design
The isothermal heater, blue excitation LED, and LCD screen were operated via a Arduino UNO R3 microcontroller (ATmega328 - assembled) (circuit diagram shown in Figure S3, parts list and cost analysis provided in Table S6). Two 10 Ω ceramic wire wound resistors (Mouser Electronics) arranged in parallel functioned as heaters by converting electric current to heat. The heaters were encapsulated using poly(dimethylsiloxane) (PDMS) to provide insulation, allowing a 95 °C set point to be reached in ∼5 min (Figure 3B). One of the digital pins (D3) in the microcontroller was assigned to provide the heater’s 5 V input through a 100 μF capacitor. A temperature probe (tmp35) sensed the temperature of the heater, and the signal was sent to one of the analog pins in the microcontroller (A5). This analog reading was converted into temperature which was continuously monitored to operate the heaters isothermally at 95 °C by actuating the pulse width modulation (PWM) pin of the ATMEGA microprocessor. The PWM pin outputs 100% of its signal until the temperature of the heater reaches the set point and reduces to 85% thereafter. If the temperature exceeds 97 °C, the PWM signal reduces to zero. In our testing, this simple scheme was sufficient to maintain the heater at 95 ± 2 °C. The 16 MHz ceramic resonator (Mouser Electronics) with two 22 pF capacitors (Mouser Electronics) connected to the microcontroller generated a clock signal to control timing in the circuit. A 5 mW blue LED was connected to a low-dropout (LDO) regulator (TLV2217-33) with two capacitors (10 and 47 μF; Mouser Electronics) and a 100 Ω resistor (Mouser Electronics) to provide a constant 3.3 V via one of the digital pins in the microcontroller (D4). A LCD (Nokia 5110) was connected to 6 digital pins (D6, D9, D10, D11, D12, D13) in the microcontroller and was used to display and monitor real-time relevant information such as the heater temperature and blue LED status. The Arduino program was written and compiled in Arduino 1.0.6 integrated development environment (IDE) and was loaded into the microcontroller through an FTDI chip (Sparkfun). The final circuit and all the components were soldered on a printed circuit board which was housed in a plastic case designed using FreeCAD and printed using a MakerGear M2 3D printer. The device can be powered by any 5 V USB-compatible electrical source. Testing in Figure 3A,B was performed using the following power sources: (1) a standard USB wall outlet, (2) a USB battery pack (Vinsic Tulip, 5 V, 1 A), (3) a hand crank generator (SOScharger; hand crank power was manually applied during the entire heating and reaction time), and (4) a solar-rechargeable USB battery pack (Solar Juice; solar power was used to charge the battery pack prior to testing). The convective thermocycler assembly weighs less than 300 g (Table S1).
In-Flight PCR as a Quadcopter Drone Payload
In-flight PCR was performed using a 3D Robotics IRIS+ quadcopter drone ($799 retail price in fall 2014, available for under $500 as of December 2015). The IRIS+ is a consumer-class drone that provides ∼25 min of flight time over programmed paths with an operation radius of 5–10 miles. We routinely obtained flight times of 15–18 min with the convective thermocycler and smartphone payload. The thermocycling instrument was preheated for 3 to 5 min prior to mounting the PCR reactor onto the heater base using adhesive tape (Scotch Magic Tape, 3M). The assembled instrument was fastened to the drone using an elastic Velcro strap. A 3200 mA h portable external battery charger (Vinsic Tulip, 5 V, 1 A) was used to power the convective PCR device, while the drone was powered by its internal high capacity rechargeable battery. After landing, the postflight PCR products were collected for further analysis. We also obtained successful results using a DJI Phantom 2 quadcopter drone, but with reduced flight times of 10–12 min owing to its lower payload capacity.
Drone-Based Centrifuge
Drone-based centrifugation experiments were performed by removing the quadcopter propellers and replacing them with 3D printed centrifuge rotors designed to fit the motor shaft threading. A low filament infill percentage was selected when optimizing 3D prints of these parts to ensure that the rotors remained lightweight (comparable to the propeller blade mass) and maintained a circularly symmetric mass distribution to help reduce additional wear on the drone motors during centrifugation. The throttle/yaw stick on the drone controller (left control stick on the IRIS+ quadcopter) was gradually moved from the fully down position to the fully up position to change the drone motors’ rotational speed. An optical tachometer (Model 4059 Traceable Touchless/Contact Digital Tachometer, Traceable Products) was used to measure rotational speeds attained by the centrifuge attachment at each throttle setting. Rotational speeds (rpm) were converted to relative centrifugal force (RCF) units via RCF (× g) = 1.12·R·(rpm/1000)2, where R was taken as the radius from the motor shaft to the tip of a spin column tube (40 mm). The drone motors were then activated to carry out centrifugation involving spin column-based nucleic acid extraction/purification and separation/enrichment of nanosized colloidal suspensions. 350 nm fluorescent magnetic particles (Cat. no. FCM-02556-2, Spherotech, Inc.) and 40 nm colloidal gold particles (Cat. no. 15707-20, Ted Pella, Inc.) were centrifuged for 5 and 10 min, respectively, using the drone at a throttle setting of 10 000 rpm (4500g). The drone centrifuge was operated in a shrouded enclosure, and appropriate personal safety protection measures were taken to prevent injury while the rotors were armed.
Nucleic acid extraction was demonstrated using the positive control DENV-1-4 of the CDC DENV-1-4 real-time PCR assay for detection and serotype identification of Dengue virus (Cat. no. KK0128, US Centers for Disease Control and Prevention (CDC)). This positive control contains heat-inactivated Dengue virus serotype DENV-1 Haw, DENV-2 NGC, DENV-3 H87, and DENV-4 H241 in human serum. Total nucleic acid extraction was performed using a QIAamp DNA Mini Kit (Cat. no. 51304, QIAGEN) using 200 μL of DENV-1-4 positive spiked serum in each extraction. The nucleic acid was eluted with 200 μL of Qiagen Buffer AE to keep the concentration factor of 1, following the centrifugation protocol in Table S7. Spin times were increased in the drone-based protocol to compensate for the lower RCF values than prescribed in the standard protocol.
To test the quality and efficiency of nucleic acid extraction performed with the drone and with the benchtop centrifuge, reverse transcription PCR was performed using primers and TaqMan-based hydrolysis probe provided in the CDC DENV-1-4 real-time PCR assay kit. The PCR reaction mixture was based on the SuperScript III Platinum One-Step qRT-PCR kit (Cat. no. 11732-020, Life Technologies), with forward and reverse primers (D3-F and D3-R) and probe (D3-Texas Red TaqMan probe). A 5 μL aliquot of the purified RNA sample was used as template for the RT-PCR (25 μL reaction volume). The reactions were performed in a benchtop qPCR machine (CFX96 Real-Time System, Bio-Rad). The temperature cycling protocol involved cDNA synthesis at 50 °C for 30 min, denaturation at 95 °C for 2 min followed by 45 cycles of denaturation at 95 °C for 15 s, and annealing/extension at 60 °C for 1 min.
iPhone-Based Fluorescence Analysis
The PCR to Go application was written and developed in Xcode 5.0 using objective C. The core graphics and AVFoundation frameworks were incorporated from Apple’s iOS software development kit (SDK) to access and control advanced iPhone camera features such as exposure time and focal length. This enabled images to be accessed via either the user’s photo library or directly from the camera. A clip-on magnifying lens (mini currency detector microscope, www.amazon.com) provided a 10× optical zoom to the camera, and the PCR to Go app interface applied an additional 6× digital zoom via the SDK’s core graphics affine transformation framework, allowing the user to access a maximum of 60× zoom without degradation of image quality. The focus and exposure are locked before each image acquisition session ensuring constant lighting.
The app interface enables a sequence of images to be automatically captured at regular time intervals. User touch gestures are incorporated to select a desired region of interest for image analysis. The selected analysis area is converted into bitmap image format stored as a mutable data set containing 4 bits per pixel (one for each red, blue, green and alpha pixel values) corresponding to RGBA color space. The average, maximum, and minimum RGBA pixel values for each image are calculated and can be either tabulated on the screen or exported via email for further manipulation. The average green pixel values were used for all fluorescence intensity analysis.
A gamma correction was applied to the intensity values via Icorrected = (Ioriginal/255)γ, where Ioriginal is the pixel intensity acquired from the smartphone camera, and a value of γ = 2.2 was used.17 Background fluorescence was removed by subtracting the average intensity values over the analysis area in the first image of the sequence (i.e., the pre-PCR sample) from all subsequent images, yielding a common baseline. Quantification was performed by applying a sigmoidal fit to the fluorescence intensity versus time data18,19 as follows
| 1 |
where Imax and Io represent the maximum and background fluorescence intensities, respectively. The quantities t1/2 and k express the time required for intensity to reach half of the maximum value and slope of the curve, respectively, and were used as fitting parameters. Critical threshold reaction times (Figure 2E, inset) were determined from the point where the sigmoidal fit exceeded a threshold value of 20 fluorescence units.
Results and Discussion
In-Flight PCR
We leveraged this approach by first selecting a cylindrically shaped reactor geometry whose internal flow field is dominated by chaotic advection and then tuning its height h such that passive heat loss to the surrounding environment automatically imposes the desired annealing conditions at the top surface when the bottom is held at the denaturing temperature (Figure 1A). In this way, the PCR is actuated isothermally by maintaining a single heater at a constant temperature, drastically reducing electrical consumption to a level that can be supplied by standard 5 V USB sources that power ordinary consumer mobile devices. Instrument design is also greatly simplified, with thermal management achievable by exploiting Joule heating from off-the-shelf ceramic resistors regulated by a straightforward feedback loop programmed using an Arduino-based microcontroller (Figure 1B). A 3D printed enclosure and interchangeable smartphone cradle ensures alignment of the camera and optical components independent of the specific mobile device (Figure 1C). The instrument is incredibly lightweight (<300 g, not including the smartphone, Table S1), making pinpoint deployment into remote field locations feasible using ordinary consumer-class quadcopter drones (Figure 1D). Unprecedented ruggedness (i.e., ability to perform outside of an idealized lab environment) is achievable, as evident by the ability to perform PCR across different target/template systems during flight as a drone payload. This capability is demonstrated by successful in-flight replication of a 147 bp Staphylococcus aureus and 237 bp λ-phage DNA targets in under 20 min (Figure 1E, Movie S1).
Smartphone-Based Fluorescence Detection and Quantification
We next explored the ability to perform quantitatively accurate in-flight product analysis. This capability, combined with smartphone connectivity, challenges the existing view of drones (i.e., primarily focused on operations involving cargo transport and imaging20,21) by introducing the potential to function as versatile mobile analytical platforms (Figure S1). However, fluorescence-based approaches are challenging to implement in a portable format due to limitations on size, cost, and ruggedness. We addressed this by drawing on previous advancements in exploiting the CMOS-based imaging sensor embedded in ordinary smartphone cameras to record fluorescence generated by a conventional SYBR Green reagent chemistry.22 The reactor is illuminated from the side by a low-power blue LED, while the smartphone camera is used to view the top surface of the reactor from above with the aid of an inexpensive mini-microscope and band-pass emission filter (Figure 1A, Experimental Section). Images are acquired using a dedicated smartphone app that allows the user to adjust camera exposure, focus, and the time interval between successive snapshots (Figure 2A).23 The collected images are gamma corrected and split into 8-bit RGBA channels, after which the green channel intensity is analyzed within a user-defined region of interest. Analysis can be performed either in an end-point mode to obtain yes/no results or in a time-resolved manner enabling reaction progress to be continuously monitored.
Sensitivity and quantification were evaluated by replicating a 237 bp target sequence from a λ-phage DNA template over a series of initial copy numbers (Figure 2B–D). Comparable results are obtained regardless of whether fluorescence analysis is performed in the lab or in-flight as a drone payload (Figure 2B). A standard two parameter sigmoidal fit was applied to the intensity data extracted from each sequence of images, enabling the reaction time when fluorescence exceeds a threshold level to be obtained (Figure 2E). This time scale is analogous to the critical cycle number in conventional real-time PCR. These data can be used to construct a standard curve whose slope yields an effective doubling time of 35.8 s (Figure 2E, inset), in good agreement with the value of 35.2 s predicted by computational simulations. Increased variability in the fluorescence data is observed at the 19 and 20 min time points under the highest initial DNA starting concentration, partially obscuring transition to the plateau phase of replication. This variability is a consequence of the smartphone-based imaging approach, where saturation of the fluorescence signal is approached at high starting concentrations. The convective thermocycling approach is not subject to inherent sensitivity limitations beyond those encountered in conventional instruments and can be interfaced with more sophisticated detection approaches to extend quantification down to the 10–100 copies/μL range (Figure 2F). As a comparison, we evaluated the same series of reactions in a benchtop real-time PCR instrument (LightCycler 96, Roche) and converted the convective data in Figure 2E (expressed in units of reaction time) to equivalent cycle numbers by assuming that one conventional replication cycle occurs during the 35.8 s doubling time. Both approaches agree remarkably well, yielding virtually identical standard curves (Figure 2G).
Our adaptation of smartphone-based fluorescence detection enables quantitative analysis at template concentrations down to 1000 copies/μL, a level sufficient for many diagnostic scenarios (i.e., depending on the severity of an infection) and particularly impressive considering the simplified instrument format. It is also envisioned that in practice each PCR reactor will include a calibration target on the surface and multiple reactor cells to enable parallel control reactions to ensure consistent results. In the context of Ebola, viral loads on the order of 105 to 106 RNA copies/mL characterize many acute nonfatal infection scenarios.24 In developing countries lacking an extensive healthcare infrastructure, patients often display acute symptoms by the time they are able to visit a clinic, suggesting that the technology proposed here can potentially help to bridge the gap between current-generation rapid diagnostic tests and the PCR gold standard typically not feasible for broad implementation.25 Other diagnostic scenarios can potentially benefit from this level of performance, including viral load monitoring of HIV infection (generally involving quantification in the range of 103–107 copies/mL). There is interest in expanding availability of viral load testing given its importance for diagnostics, monitoring of treatment efficacy, and assessment of/adherence to treatments. We also note that these detection limits are dictated solely by our choice of LED illumination and smartphone camera-based fluorescence detection and, therefore, can be improved if it makes sense to do so within the context of a specific diagnostic scenario or application need.
Design for Portability
Versatility and ruggedization are also essential considerations in the context of portable field operation. The conventional USB interface makes it possible to employ a variety of power supplies suitable for use in remote settings, including battery pack, hand crank, and solar chargers. Consistent reaction yield (Figure 3A) and heater performance (Figure 3B) are achievable regardless of electrical power source. We also evaluated the effect of ambient environmental temperature and the reactor’s orientation with respect to the vertical direction. Ambient temperature conditions impact thermocycling performance by virtue of their influence on the thermal gradient established within the reactor. Experiments and computational simulations both reveal that our system operates consistently across ambient temperatures ranging from 5 to 35 °C (Figure 3C,D; “volume fraction” refers to the percentage of the reactor volume at annealing, extension, and denaturing temperatures, respectively). Reactions proceed most efficiently at 5 °C, with performance dropping above 35 °C where it becomes difficult to maintain the spatially distinct temperature zones required to actuate each stage of the reaction. Ambient temperature conditions can also affect fluorescence detection via the dependence of LED emission intensity, which decreases with increasing junction temperature, but these effects are of small magnitude over the range of interest here (<3% of the room temperature luminosity for junction temperatures not exceeding 30 °C according to the LED manufacturer’s datasheet) and can be straightforwardly addressed by incorporating a circuit design that increases the drive current with increasing temperature. Regarding the potential impact of temperature change with altitude, we maintained altitudes of 50–100 ft during the flight tests employed here, where a general rule of thumb is to expect a 3.5–5 °F temperature decrease per 1000 ft increase in elevation (depending on environmental factors such as relative humidity). Therefore, we do not anticipate significant temperature changes to occur within the envisioned operational envelopes.
The reactor’s orientation with respect to the vertical direction would also intuitively be viewed as an important consideration given the gravitational origin of the driving force for convective transport. However, coordinated experiments and simulations both confirm that this is not the case, revealing no appreciable change in reaction performance as the reactor’s tilt angle is increased up to at least 60° (Figure 3E–G). This invariance with spatial orientation is a reflection of the chaotic advection regime’s inherent robustness, where consistent flow phenomena are predicted over orders of magnitude in the thermal driving force.15 Computational simulations indicate that favorable flow fields can be maintained at tilt angles approaching ∼80°, beyond which the onset of secondary circulatory features becomes evident (Figure S2).
Drone-Based Centrifuge
Sample preparation remains a critical bottleneck in most portable diagnostic workflows, in large part because benchtop centrifugation processes are challenging to miniaturize. The quadcopter platform uniquely addresses this need because its on-board rotors inherently deliver sufficient power to function as laboratory centrifuges. We explored this possibility by 3D printing centrifuge rotors compatible with standard spin column-based extraction/purification kits (Figure 4A). These attachments mount directly onto the motor shaft and leverage the same self-tightening threading built into standard rotor blades (diagonally opposed rotors spin in the same direction for balanced operation). The quadcopter’s throttle control enables rotational speeds up to 10 000 rpm to be accessed, generating centrifugal forces comparable to benchtop instruments (Figure 4B). The resulting platform is incredibly versatile, maintaining compatibility with commercial spin column preparation kits (Figure 4C) and is robust enough to enable separation and enrichment of nanosized colloidal materials from suspension (Figure 4D,E) with performance comparable to conventional benchtop systems (Movie S2). Practical utility of the drone-based centrifuge platform is further underscored by leveraging it to perform kit-based extraction of Dengue viral RNA in human serum, yielding results on par with those obtained from samples processed using a benchtop centrifuge (Figure 4F). This flexibility enables virtually any standard sample preparation protocol to be deployed with minimal modification to established laboratory practices. Notably, drone run times between battery charges increase by over 50% when the unit is used as a centrifuge (as compared with flight operation) and can be further extended by programming the controller to activate only a single motor. The 10 000 rpm rotation speed also delivers sufficient power to enable more complex lab-on-a-CD analysis operations to be performed on a drone-based platform.26,27
Figure 4.
Drone-based sample preparation. (A) The quadcopter blades are replaced with 3D printed rotor attachments to enable standard centrifuge-based workflows. (B) These attachments transform the quadcopter into a centrifuge capable of rotation speeds ω up to 10 000 rpm (below, mean ± sd of three replicates, error bars smaller than the plotted symbols), yielding performance comparable to benchtop instruments (above). (C–E) Images depicting samples before (left) and after (right) drone-based centrifugation. (C) Colored buffer enables visualization of elution through a standard spin column kit centrifuged for 2 min at 10 000 rpm. (D) A 350 nm magnetic nanoparticle suspension centrifuged for 5 min at 10 000 rpm. (E) A 40 nm gold colloid suspension centrifuged for 10 min at 10 000 rpm. (F) Real-time PCR plot of Dengue virus serotype 3 amplification using templates extracted by drone-based and benchtop centrifuges. The quantity of RNA extracted in both platforms is comparable, as evident by quantification cycle (Cq) values of 31.8 and 31.6 obtained using the drone-based platform (two replicates) whereas Cq = 32.5 was obtained when the protocol was performed using a benchtop centrifuge.
Conclusions
A performance/portability/cost comparison between our instrument and current commercially available systems is presented as the Supplementary Note. Here, we briefly consider notable options beginning with a recently reported novel approach leveraging a solar focusing lens to establish spatially distinct temperature zones needed to perform continuous flow PCR.28 Although heating occurs passively, electrical power is required to circulate reagents through the microfluidic network using a syringe pump, post-PCR detection is achieved off-chip using a benchtop microscope, and effective operation is contingent upon availability of sufficient ambient sunlight. The suitcase-sized POCKIT system (GeneReach) is a commercial convective thermocycler but is of limited utility for portable diagnostics due to lack of an integrated battery and limited end point fluorescence detection. The Palm PCR instrument (Ahram Biosystems), a competing convection-based device, offers battery power but lacks integrated fluorescence detection. Others have demonstrated capillary-based convective real time PCR with integrated CCD-based detection,29,30 but these instruments are not portable and require a desktop computer for operation and data analysis. The integrated sample preparation capability enabled by our drone-based centrifuge platform is also a significant advancement. The rotor design in Figure 4 delivers a relative centrifugal force (RCF) of ∼4500g at 10 000 rpm, surpassing the capabilities of previously developed systems aimed at performing blood plasma separations in resource-limited settings via mechanisms based on hand operated egg beaters (RCF = 280g at 1200 rpm),31 bicycle wheels (RCF = 240g at 750 rpm),32 and salad spinners (RCF = 31g at 602 rpm).33
Our system delivers a rugged inexpensive biochemical analysis platform ideally suited for field deployment (Figure S1). The ∼$50 ($US) hardware cost (not including the smartphone, Table S6) and compatibility with consumer-class quadcopter drones (whose prices continue to rapidly decrease) breaks existing price barriers by several orders of magnitude (Supplementary Note, Figure S4), while the simplified imaging and device agnostic design makes it possible to leverage a growing low-cost smartphone marketplace to deliver affordable fluorescence detection. In addition to ongoing initiatives aimed at producing affordable camera-equipped smartphones for developing country markets, there is already a huge market for second hand smartphones that may be obsolete in first-world markets but remain well equipped for diagnostic applications in resource-limited settings.22 Smartphone-based analysis also introduces extraordinary connectivity, making it possible to tap into existing communication networks for real-time delivery of analysis results.34 Drones are inherently capable of autonomous operation, enabling targeted establishment of pop-up hubs for programmed payload exchange, battery charging, and routine maintenance. The distribution of these nodes can dynamically evolve so that cargo can be directed to specific locations based on real-time data, enabling access to very remote settings. These capabilities suggest new potential for drone-based systems to function in broader and more sophisticated roles beyond cargo transport and imaging. Areas for future improvement include development and packaging of assay kits for reagent delivery in parallel with instrument deployment and broader quantification of product yields and detection limits.
The United States clinical laboratory improvement amendments (CLIA) classify clinical diagnostic tests as high, moderate, or waived complexity based upon the nature of the test performed.35,36 PCR-based diagnostics are currently classified as high complexity due to prerequisite operational training and sophisticated instrumentation, thereby making them expensive and impractical for mass distribution in portable applications. The versatile platform introduced here offers potential to enable PCR to be classified in the moderate or waived complexity categories, opening the door for a new generation of fast, accurate, and affordable diagnostic tools impacting a host of new scenarios where rapid field-deployable analysis is needed but not yet widely available (e.g., emergency response, agricultural biosurveillance, and veterinary field care scenarios).
Acknowledgments
We thank Magda Lagoudas in the Dwight Look College of Engineering at Texas A&M for support of the undergraduate research team via the AggiE-Challenge program. We gratefully acknowledge Dr. Arul Jayaraman and Rani Menon for providing access to and assistance with the benchtop real-time PCR instrument and Dr. Maria King for helpful discussions regarding the same. AI Biosciences, Inc. acknowledges funding from the US National Institutes of Health under grant number 1R43AI107984. The logo for Texas A&M University is copyrighted, and permission was obtained for the use of the logo in this publication.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.analchem.5b04153.
Author Contributions
¶ Y.B., M. Carpio, J.C., M. Coen, D.C., J.H., J.J., A.K., R.L., S.L., A.M., A. Pangelinan, N.P., and L.S. contributed equally to this work as members of an undergraduate AggiE-Challenge research team and are listed alphabetically.
The authors declare no competing financial interest.
Supplementary Material
References
- Yager P.; Edwards T.; Fu E.; Helton K.; Nelson K.; Tam M. R.; Weigl B. H. Nature 2006, 442, 412–418. 10.1038/nature05064. [DOI] [PubMed] [Google Scholar]
- Boppart S. A.; Richards-Kortum R. Sci. Transl. Med. 2014, 6, 253rv2. 10.1126/scitranslmed.3009725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin C. D.; Linder V.; Sia S. K. Lab Chip 2007, 7, 41–57. 10.1039/B611455E. [DOI] [PubMed] [Google Scholar]
- Chin C. D.; Laksanasopin T.; Cheung Y. K.; Steinmiller D.; Linder V.; Parsa H.; Wang J.; Moore H.; Rouse R.; Umviligihozo G.; et al. Nat. Med. 2011, 17, 1015–1019. 10.1038/nm.2408. [DOI] [PubMed] [Google Scholar]
- Easley C. J.; Karlinsey J. M.; Bienvenue J. M.; Legendre L. A.; Roper M. G.; Feldman S. H.; Hughes M. A.; Hewlett E. L.; Merkel T. J.; Ferrance J. P.; et al. Proc. Natl. Acad. Sci. U. S. A. 2006, 103, 19272–19277. 10.1073/pnas.0604663103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez A. W.; Phillips S. T.; Whitesides G. M.; Carrilho E. Anal. Chem. 2010, 82, 3–10. 10.1021/ac9013989. [DOI] [PubMed] [Google Scholar]
- Sorger P. K. Nat. Biotechnol. 2008, 26, 1345–1346. 10.1038/nbt1208-1345. [DOI] [PubMed] [Google Scholar]
- Richards-Kortum R.; Oden M. Science 2013, 342, 1055–1057. 10.1126/science.1243473. [DOI] [PubMed] [Google Scholar]
- Lee W. G.; Kim Y.-G.; Chung B. G.; Demirci U.; Khademhosseini A. Adv. Drug Delivery Rev. 2010, 62, 449–457. 10.1016/j.addr.2009.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A. A.; Hennek J. W.; Smith B. S.; Kumar S.; Beattie P.; Jain S.; Rolland J. P.; Stossel T. P.; Chunda-Liyoka C.; Whitesides G. M. Angew. Chem., Int. Ed. 2015, 54, 5836–5853. 10.1002/anie.201411741. [DOI] [PubMed] [Google Scholar]
- Koydemir H. C.; Gorocs Z.; Tseng D.; Cortazar B.; Feng S.; Chan R. Y. L.; Burbano J.; McLeod E.; Ozcan A. Lab Chip 2015, 15, 1284–1293. 10.1039/C4LC01358A. [DOI] [PubMed] [Google Scholar]
- Wei Q.; Luo W.; Chiang S.; Kappel T.; Mejia C.; Tseng D.; Chan R. Y. L.; Yan E.; Qi H.; Shabbir F.; Ozkan H.; Feng S.; Ozcan A. ACS Nano 2014, 8, 12725–12733. 10.1021/nn505821y. [DOI] [PubMed] [Google Scholar]
- Laksanasopin T.; Guo T. W.; Nayak S.; Sridhara A. A.; Xie S.; Olowookere O. O.; Cadinu P.; Meng F.; Chee N. H.; Kim J. Sci. Transl. Med. 2015, 7, 273re271. [DOI] [PubMed] [Google Scholar]
- Krishnan M.; Ugaz V. M.; Burns M. A. Science 2002, 298, 793. 10.1126/science.298.5594.793. [DOI] [PubMed] [Google Scholar]
- Priye A.; Hassan Y. A.; Ugaz V. M. Anal. Chem. 2013, 85, 10536–10541. 10.1021/ac402611s. [DOI] [PubMed] [Google Scholar]
- Muddu R.; Hassan Y. A.; Ugaz V. M. Angew. Chem., Int. Ed. 2011, 50, 3048–3052. 10.1002/anie.201004217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skandarajah A.; Reber C. D.; Switz N. A.; Fletcher D. A. PLoS One 2014, 9, e96906. 10.1371/journal.pone.0096906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W.; Saint D. A. Biochem. Biophys. Res. Commun. 2002, 294, 347–353. 10.1016/S0006-291X(02)00478-3. [DOI] [PubMed] [Google Scholar]
- Rutledge R. Nucleic Acids Res. 2004, 32, e178. 10.1093/nar/gnh177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornace K. M.; Drakeley C. J.; William T.; Espino F.; Cox J. Trends Parasitol. 2014, 30, 514–519. 10.1016/j.pt.2014.09.001. [DOI] [PubMed] [Google Scholar]
- Schiffman R. Science 2014, 344, 459. 10.1126/science.344.6183.459. [DOI] [PubMed] [Google Scholar]
- Ozcan A. Lab Chip 2014, 14, 3187–3194. 10.1039/C4LC00010B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priye A.; Ugaz V. M. Apple iOS App Store; https://itunes.apple.com/us/app/pcr-to-go/id909227041?mt=8, 2014.
- Towner J. S.; Rollin P. E.; Bausch D. G.; Sanchez A.; Crary S. M.; Vincent M.; Lee W. F.; Spiropoulou C. F.; Ksiazek T. G.; Lukwiya M.; Kaducu F.; Downing R.; Nichol S. T. J. Virol. 2004, 78, 4330–4341. 10.1128/JVI.78.8.4330-4341.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nouvellet P.; Garske T.; Mills H. L.; Nedjati-Gilani G.; Hinsley W.; Blake I. M.; Van Kerkhove M. D.; Cori A.; Dorigatti I.; Jombart T.; Riley S.; Fraser C.; Donnelly C. A.; Ferguson N. M. Nature 2015, 528, S109–S116. 10.1038/nature16041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madou M.; Zoval J.; Jia G.; Kido H.; Kim J.; Kim N. Annu. Rev. Biomed. Eng. 2006, 8, 601–628. 10.1146/annurev.bioeng.8.061505.095758. [DOI] [PubMed] [Google Scholar]
- Ducrée J.; Haeberle S.; Lutz S.; Pausch S.; Von Stetten F.; Zengerle R. J. Micromech. Microeng. 2007, 17, S103. 10.1088/0960-1317/17/7/S07. [DOI] [Google Scholar]
- Jiang L.; Mancuso M.; Lu Z.; Akar G.; Cesarman E.; Erickson D. Sci. Rep. 2014, 4, 4137. 10.1038/srep04137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou W. P.; Chen P. H. Jr; Miao M.; Kuo L. S.; Yeh S. H.; Chen P. J. BioTechniques 2011, 50, 52–57. 10.2144/000113589. [DOI] [PubMed] [Google Scholar]
- Hsieh Y.-F.; Lee D.-S.; Chen P.-H.; Liao S.-K.; Yeh S.-H.; Chen P.-J.; Yang A.-S. Sens. Actuators, B 2013, 183, 434–440. 10.1016/j.snb.2013.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong A. P.; Gupta M.; Shevkoplyas S. S.; Whitesides G. M. Lab Chip 2008, 8, 2032–2037. 10.1039/b809830c. [DOI] [PubMed] [Google Scholar]
- Stinson L. In Wired Online, December 10, 2014, http://www.wired.com/2014/12/clever-bike-powered-centrifuge-developing-countries.
- Brown J.; Theis L.; Kerr L.; Zakhidova N.; O’Connor K.; Uthman M.; Oden Z. M.; Richards-Kortum R. Am. J. Trop. Med. Hyg. 2011, 85, 327–332. 10.4269/ajtmh.2011.10-0399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns M. A. Science 2002, 296, 1818–1819. 10.1126/science.1073562. [DOI] [PubMed] [Google Scholar]
- Ehrmeyer S. S.; Laessig R. H. Clin. Chem. Lab. Med. 2007, 45, 766–773. 10.1515/CCLM.2007.164. [DOI] [PubMed] [Google Scholar]
- Niemz A.; Ferguson T. M.; Boyle D. S. Trends Biotechnol. 2011, 29, 240–250. 10.1016/j.tibtech.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.