Abstract
Staphylococcus aureus is a crucial human pathogen expressing various immune-evasion proteins that interact with the host-cell molecules. Clumping factor A (ClfA) is a microbial surface protein that promotes S. aureus binding to fibrinogen, and is associated with septic arthritis and infective endocarditis. In order to identify the major human serum proteins that bind the ClfA, we utilized recombinant ClfA region A in a plate-based assay. SDS-PAGE analysis of the bound proteins yielded five prominent bands, which were analysed by MS yielding apolipoprotein E (ApoE) as the predominant protein. ClfA-sufficient S. aureus bound purified ApoE by more than one log greater than an isogenic ClfA-deficient mutant. An immunodot-blot assay yielded a linearity model for ClfA binding to human ApoE with a stoichiometric-binding ratio of 1.702 at maximal Pearson's correlation coefficient (0.927). These data suggest that ApoE could be a major and novel binding target for the S. aureus virulence factor ClfA. Thus, ClfA recruitment of serum ApoE to the S. aureus surface may sequester ApoE and blunt its host defence function against S. aureus-invasive infections to humans. In this context, compounds that can block or suppress ClfA binding to ApoE might be utilized as prophylactic or therapeutic agents.
Introduction
Staphylococcus aureus is one of the primary causes of community and nosocomial bacterial infections in humans (Rosenthal et al., 2010). S. aureus accounts for more superficial and invasive infections than any other bacteria, leading to considerable morbidity and mortality worldwide (Hakim et al., 2007; Haupt et al., 2008). A number of S. aureus surface proteins enable the microbe to adhere to and invade the host cells, as well as to evade immune defence mechanisms (Bur et al., 2013). Clumping factor A (ClfA) is categorized as a member of the microbial surface component recognizing adhesive matrix molecules (MSCRAMM) family and has been shown to be a major virulence factor in many animal models, including being the most crucial cell-surface-anchored virulence factor in S. aureus-induced sepsis (McAdow et al., 2011). ClfA mediates adhesion of S. aureus to host tissues by binding to extracellular matrix proteins such as fibrinogen (McCormack et al., 2014), and it participates in impeding macrophage phagocytosis, as well as in stimulating platelet aggregation and fibrin clot formation (Kerrigan et al., 2008; Palmqvist et al., 2004). Our laboratory has previously shown that ClfA can recruit the plasma complement regulatory protein factor I, resulting in inhibition of complement-mediated opsonophagocytosis (Hair et al., 2008, 2010).
Plasma apolipoproteins (Apos) are central therapeutic targets as they play a vital role in macrophage biology and metabolic disorders (Sigel et al., 2012). Interestingly, human nasal fluid contains all major lipid classes as well as ApoA–I, which contribute in the inherent antibacterial activity of human mucosal defence (Do Carmo et al., 2008). Moreover, the host immune response was enhanced by high-density lipoprotein components and phagocytic activity was markedly improved by ApoA in particular (Tanaka et al., 2010). Some Apos can interfere with the S. aureus switch from colonizing to invasive pathogen by antagonizing the S. aureus quorum-sensing system that upregulates genes required for invasive infection. This antagonism is mediated by the binding of ApoB to S. aureus autoinducing pheromone, preventing attachment of this pheromone to the bacterial cell and subsequently suppressing signal transduction through its receptor (Peterson et al., 2008). Sera of familially hypercholesterolaemic patients, with elevated ApoB, could suppress lipoteichoic acid (LTA)-induced inflammation in humans through its binding to LTA of S. aureus (Sigel et al., 2012). As a part of innate immune responses to infection, human serum ApoB binds and masks the quorum-signalling molecule of S. aureus, auto-inducing peptide (AIP), leading to transcriptional downregulation of agr expression in S. aureus (James et al., 2013). Human serum ApoA and ApoC directly inhibit the growth of coagulase-negative staphylococci in vitro and promote innate defence mechanisms of normal and immunocompromised individuals (Tada et al., 1993). Although ApoD may not play a role in S. aureus infection, it has neuroprotective function against human coronavirus-induced encephalitis (Do Carmo et al., 2008). Moreover, serum ApoH can bind staphylococcal Sbi cell-surface protein, which is expressed at diverse levels between different S. aureus strains (Zhang et al., 2011).
ApoE is a major cholesterol carrier that has diverse functions in regulating lipid transport, glucose metabolism, mitochondrial function, neuronal signalling and modulating neuroinflammation, as well as possessing a role in the innate immune response (Liu et al., 2013). Although ApoE binds to several cell-surface receptors (Liu et al., 2013), its interaction with cell-surface proteins of S. aureus, particularly with ClfA, has not been reported so far. The current study identifies ApoE as a novel ligand for the S. aureus surface protein ClfA and evaluates the extent to which ApoE binds to ClfA on the molecular level as well as on the microbial cell level.
Methods
S. aureus strains.
S. aureus wild-type ClfA-sufficient Newman strain (ClfA+), as well as the isogenic S. aureus ClfA-deficient strain (ClfA−) with the genotype clfA2 : : Tn917 (McDevitt et al., 1994), were used in whole-cell binding assays as well as in cell-wall-binding experiments. For preservation of strains, a single colony of each strain was inoculated in 5 ml tryptic soy broth (TSB; MP Biomedicals). After incubation for 24 h at 37 °C, glycerol was added at a final concentration of 15 % (v/v) to TSB cultures and fractioned as 1 ml aliquots into sterile polypropylene cryotubes (Sigma-Aldrich), which were stored at −80 °C (Arciola et al., 2001). Before each experiment, the tested strains were grown to mid-exponential phase (OD600 1.0–1.5) at 37 °C in Columbia broth supplemented with 2 % (w/v) NaCl.
Serum and plasma.
Normal human serum and plasma were made as described previously (Cunnion et al., 2001) from the blood of healthy human volunteers in agreement with an Institutional Review Board standard protocol of Eastern Virginia Medical School (EVMS IRB 02-06-EX-0216). The serum was pooled, aliquoted and frozen at −80 °C.
Proteins, antibodies and chemicals.
Fraction V heat shock-treated BSA was obtained from Fisher Scientific. Recombinant clumping factor A (rClfA) with a molecular mass of 80 kDa was generated in Escherichia coli and purified as previously described (Hair et al., 2010). Human ApoE (34 kDa), Goat anti-ApoE IgG and anti-goat IgG horseradish peroxidase (HRP) conjugate antibodies were purchased from EMD Millipore Chemicals. Unless otherwise stated, chemicals and buffers’ ingredients were obtained from Sigma-Aldrich.
Plasma proteins binding to rClfA.
The bottom of an Immulon 2HB plate (ImmunoChemistry Technologies) was coated with rClfA (20 µg ml−1), as previously described (Hair et al., 2010). The plate was blocked with 3 % (w/v) BSA, washed, then 20 % (v/v) human plasma was added (in 60 % DGVBS2+ buffer) and incubated. The wells were then washed and stripped with 2 % (w/v) SDS. The stripped extracts were run on a 4–20 % SDS-PAGE gel. The SDS-PAGE gel was stained for all proteins using SYPRO Ruby, according to the manufacturer’s instructions.
MS identification of human proteins.
Protein bands were excised from SYPRO Ruby stained SDS-PAGE gels and processed for liquid chromatography electrospray ionization tandem mass spectrometry (LC-ES-MS-MS) as previously described (Hair et al., 2013). The acquired data were processed and the proteins were identified using Mascot Daemon client application (Matrix Science) software using an indexed bacterial subset database of the non-redundant proteins database from ExPASy/Swiss-Prot. Statistical analysis of strength of peptide identification was performed as previously described (Hair et al., 2013).
Combined ELISA Western blot analysis (CEWA) assay of serum ApoE binding to rClfA.
This previously described method of CEWA was adapted for assessment of ApoE binding to rClfA (Sharp et al., 2012). Briefly, Immulon 2HB plates were coated with 20 µg S. aureus rClfA ml−1 in carbonate buffer (50 µl per well) and incubated at 4 °C for 24 h. After incubation, the wells were rinsed three times with PBS-Tween 20 (PBST) solution. Blocking solution (3 %, w/v, BSA in PBST) was applied (50 µl per well) overnight at 4 °C. All the wells were then washed three times with PBST and incubated with 20 % (v/v) human serum in PBS (100 µl per well) at room temperature for 1 h. The wells were washed three times with PBST and stripped of retained proteins with 25 µl of 2 % (w/v) SDS buffer. Control wells contained 3 % (w/v) BSA in PBS.
SDS-PAGE and Western blotting.
Solutions of stripped proteins were mixed with 4× reducing sample buffer, heated at 95 °C for 5 min, loaded onto 4–20 % Tris/HCl mini SDS-PAGE gels (Bio-Rad Laboratories) and electrophoresed 200 V for 45 min (Faghri et al., 2012). PageRuler Plus prestained protein ladder (Thermo Scientific) with a size range of 10–250 kDa, MagicMark XP Western protein standard (Invitrogen) with a size range of 20–220 kDa and human ApoE (EMD Millipore Chemicals) were used in each experiment. After electrophoresis, the proteins were blotted to PVDF membrane (EMD Millipore) as described elsewhere (Walsh et al., 2004) and the membrane was then incubated on a rotary shaker (100 r.p.m.) at room temperature for 1 h with goat anti-ApoE antibody. After washing the PVDF membrane three times with Tris-buffered saline with 0.05 % Tween 20 (TBST) solution, the membrane was incubated with mouse anti-goat IgG antibody HRP conjugate at room temperature on a rotary shaker (100 r.p.m.) for 30 min, followed by rinsing with TBST three times. The developer solution Immun-Star HRP chemiluminescent kit (Bio-Rad Laboratories) was applied to the membrane according to the manufacturer's instructions and a chemiluminescence image was captured at 20 s exposure time with the VersaDoc imaging system (Bio-Rad Laboratories); optical densitometry analysis was performed using Quantity One® analysis Software (Bio-Rad).
Whole-cell binding assay.
This assay was performed as described elsewhere (Miajlovic et al., 2010) with some modifications. Briefly, Immulon 2HB plates were coated with 20 µg purified human ApoE ml−1 in carbonate buffer (50 µl per well), followed by incubation at 4 °C for 24 h. After coating, the wells were rinsed twice with 3 % (w/v) BSA in PBS solution and the surfaces were then blocked with the same solution at 37 °C for 1 h. Bacterial suspensions [100 µl per well of 108 c.f.u. ml−1 in 3 % (w/v) BSA in PBS] were transferred to ApoE-coated wells and the plates were incubated for 2 h at room temperature on a rotary shaker (100 r.p.m.). After incubation, the wells were washed twice with PBS and the bound cells were mechanically stripped in PBS (100 µl per well). The harvested cells were tenfold serially diluted in PBS, plated onto Mueller–Hinton II agar (BD) and incubated at 37 °C for 18–20 h (Masago et al., 2008). Colony counting was performed, as described by Kuwahara et al. (2010).
ApoE binding to S. aureus cell wall.
Based on a published described method (Friberg et al., 2008) with some modifications, S. aureus strains were grown to mid-exponential phase (OD600 1.0–1.5) at 37 °C in Columbia broth supplemented with 2 % (w/v) NaCl. The cells were harvested by centrifugation at 10 844 g for 3 min, washed twice with sterile PBS and resuspended in PBS containing 0.5 % (w/v) BSA (PBS-BSA) to match 0.5 McFarland turbidity standard (Thermo Scientific Remel), which is equivalent to 1.5×108 c.f.u. ml−1. Cell suspensions were incubated with ApoE (25 µg ml−1) for 2 h at 37 °C, followed by centrifugation at 10 844 g for 3 min and washing twice with sterile PBS-BSA. A formerly described method (Sharp & Cunnion, 2011) was employed for preparation of cell-wall extracts. In brief, bacterial cells were suspended in 600 µl digestion buffer (30 %, w/v raffinose in 0.05 M Tris pH 7.5 with 0.145 M NaCl) containing 10 µg lysostaphin (Sigma-Aldrich), 10 µg DNase (Sigma-Aldrich) and 100 µl protease inhibitor (Roche Diagnostics). The cell suspensions were incubated at 37 °C for 1 h, with rotational mixing. The protoplasts were settled down by centrifugation at 8500 r.p.m. for 10 min and cell-wall proteins were recovered in the supernatant. Cell-wall-bound ApoE was assessed as described under SDS-PAGE and Western blotting, and pure human ApoE was used as a control.
Immunodot-blot assay.
For establishment of an ApoE calibration curve, the immunodot-blot technique (Dhaliwal et al., 2009) was implemented. In short, methanol-treated PVDF membrane was loaded with escalating amounts of purified ApoE (0.32–3.2 µg), followed by air-drying at room temperature. After drying, the membrane with blocked with 3 % (w/v) BSA in Tris-buffered saline (TBS) at room temperature on a rotary shaker (100 r.p.m.) for 1 h followed by rinsing three times with TBST solution. The membrane was incubated with goat anti-ApoE antibody for 1 h on a rotary shaker (100 r.p.m.) at room temperature and then rinsed three times with TBST. Mouse anti-goat IgG antibody HRP conjugate was incubated with the membrane at room temperature on a rotary shaker (100 r.p.m.) for 45 min followed by rinsing with TBST three times. The developer solution Immun-Star HRP chemiluminescent kit (Bio-Rad) was applied to the membrane and chemiluminescence was captured at 20 s exposure time with a VersaDoc imaging system (Bio-Rad). Density of the chemiluminescence signals was quantified and expressed as counts (CNT) mm−2 using Quantity One® analysis software (Bio-Rad). Conversely, the PVDF membrane was loaded with increasing amounts of rClfA (0.32–2.6 µg) followed by air-drying, and BSA (0.32–2.6 µg) was used as a control. The membrane was incubated with blocking solution (TBS–BSA) at room temperature for 1 h with shaking. Diluted human serum (20 %, v/v, human serum in TBS–BSA) was incubated with the membrane for 1 h at room temperature on a rotary shaker (100 r.p.m.) followed by rinsing with TBST three times. The membrane was incubated with goat anti-ApoE antibody and subsequently with mouse anti-goat IgG antibody HRP conjugate as described above.
Statistical analysis.
Quantitative assays were performed in triplicate and means (±sd) of the data were represented graphically. Results were analysed with the Mann–Whitney non-parametric test. The linearity model of protein interaction and Pearson’s correlation coefficients of rClfA binding to ApoE were determined at the two-tailed significance level. Data output of analyses with calculated P values of ≤0.05 were considered statistically significant. Statistical analyses in the current study were performed using spss, version 18.0 (SPSS).
Results
Plasma proteins bound to rClfA
In order to identify human plasma proteins that would bind to the A region of ClfA, we utilized rClfA, incubating human plasma with surface-bound rClfA in an ELISA plate. After washing and extraction of bound proteins, the supernatants were analysed by SDS-PAGE with total protein staining by SYPRO Ruby (Fig. 1). The five predominant bands that could be easily visualized on a UV viewer were excised and processed for MS-based protein identification. The number of unique peptides identified and their respective proteins in each band are shown in Table 1. The number of peptides identified includes variants of the peptides that have different post-translational modifications or slightly different retention times. Thus, excluding the minor variants of the 44 peptides identified for ApoE, yielded nine significant peptides (Table 2). The ApoE peptides were identified with high statistical significance (i.e. peptide score >30 and expect score <0.05). The tandem MS data of these peptides showed continuous series of both b and y fragmentation ions, which further validated their identities. These peptides provide a coverage map of 38 %, demonstrating good coverage and a high degree of confidence in the identification (Fig. 2). These data strongly suggest that ApoE could be a novel ligand for ClfA.
Fig. 1.
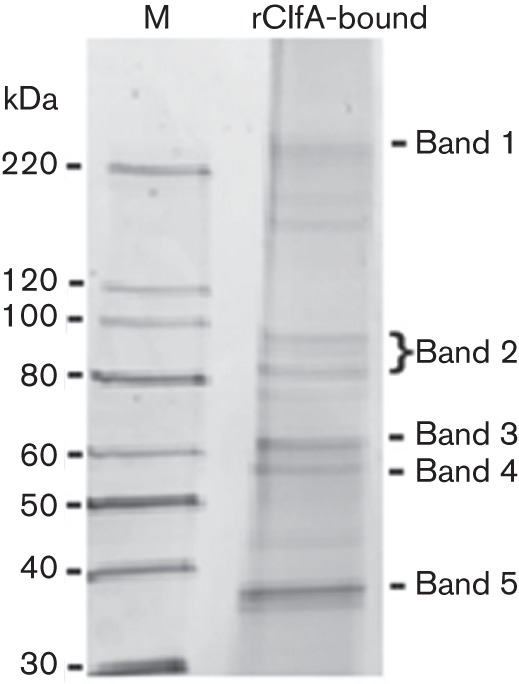
Plasma proteins binding to surface-coupled rClfA in an ELISA well. Extracted proteins were analysed by SDS-PAGE and the predominant bands (bands 1–5) were excised for MS-based identification. M, Molecular mass marker.
Table 1. MS identification of the predominant peptides and corresponding protein for each band.
| Band no. | Predominant protein | No. of peptides |
| 1 | Clumping factor A | 4 |
| 2 | Complement C3 | 1 |
| 3 | Complement C3 | 2 |
| 4 | Albumin | 10 |
| 5 | Apolipoprotein E | 44 |
Table 2. MS identification of the predominant apolipoprotein E peptides.
| M r (expected) | M r (calculated) | Peptide score | Expected score | Peptide |
| 967.8622 | 967.5451 | 52 | 0.0016 | R.LGPLVEQGR.V |
| 1033.0740 | 1032.5352 | 45 | 0.01 | R.LQAEAFQAR.L |
| 1214.2628 | 1213.6778 | 90 | 3.4×10−07 | R.LSKELQAAQAR.L |
| 1312.7996 | 1312.7099 | 73 | 9.6×10−06 | R.AKLEEQAQQIR.L |
| 1496.7558 | 1496.7947 | 84 | 7.4×10−07 | R.AATVGSLAGQPLQER.A |
| 1619.6998 | 1619.7904 | 125 | 6.6×10−11 | K.VQAAVGTSAAPVPSDN.H |
| 1663.2076 | 1662.7883 | 94 | 1.3×10−07 | R.GEVQAMLGQSTEELR.V |
| 1684.8298 | 1684.8744 | 78 | 2.6×10−06 | R.DRLDEVKEQVAEVR.A |
| 2092.6094 | 2092.0324 | 94 | 1.5×10−07 | K.AYKSELEEQLTPVAEETR.A |
Fig. 2.
Peptide map of apolipoprotein E (ApoE). Peptides identified by MS are in bold and underlined.
Serum ApoE binding to surface-coupled rClfA
In order to confirm that serum ApoE binds to ClfA, we assayed serum ApoE binding to rClfA in a CEWA. Briefly, rClfA was coupled to Immulon 2HB plates and then incubated with human serum. After washing, retained proteins were stripped and assayed by Western blot analysis with probing for ApoE. Wells coated with rClfA showed a marked binding of ApoE from diluted human serum (20 %, v/v, human serum in PBS) to S. aureus rClfA, whereas serum ApoE binding to BSA (control) could not be detected. In these experiments, standard human ApoE was used to confirm the protein identity of the human serum ApoE and both bands appeared at the expected molecular mass of 34 kDa under reducing SDS-PAGE conditions, as shown in Fig. 3.
Fig. 3.
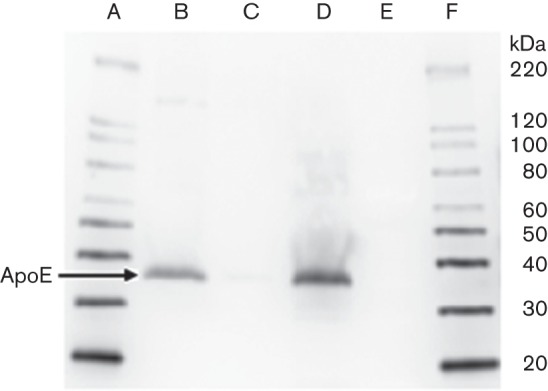
CEWA assay of serum ApoE binding to S. aureus rClfA. Sequence of lanes on Western blot: A, MagicMark XP Western protein standard; B, rClfA eluted test wells of Immulon 2HB plates showing ApoE binding; C, BSA eluted control wells of Immulon 2HB plates showing no ApoE binding; D, purified human ApoE used as an identity control; E, PageRuler Plus prestained protein ladder for SDS-PAGE monitoring; F, MagicMark XP Western protein standard.
Immunodot-blot assay
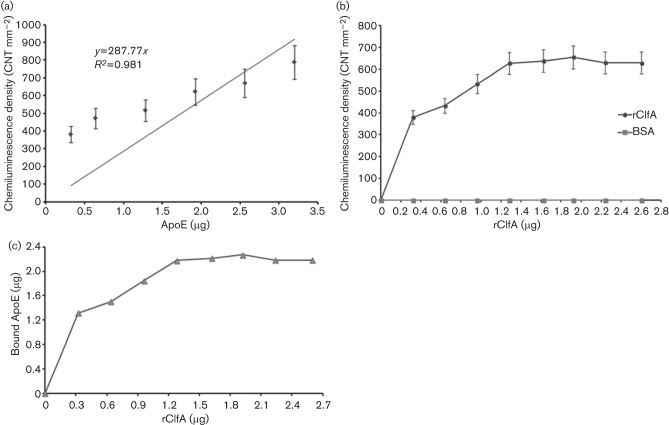
To establish an ApoE calibration curve, the immunodot-blot technique was employed by blotting increasing amounts of ApoE (0.32–3.2 µg) on PVDF membrane, followed by probing of ApoE. The constructed calibration curve shows a good linearity (R 2 = 0.981) between ApoE (µg) and density of the chemiluminescence (CNT mm−2) as shown in Fig. 4(a). To quantitatively characterize the binding relationship of serum ApoE to rClfA, escalating amounts of rClfA (0.32–2.6 µg) was dot blotted onto the membrane, which was then incubated with human serum, and rinsed. Subsequently, the membrane was incubated with goat anti-ApoE antibody followed by probing for ApoE (Fig. 4b). Negligible ApoE bound to BSA (control) and the binding of ApoE to rClfA demonstrated two phases of binding kinetics: (a) a linear phase with a rClfA concentration range of 0–1.28 µg, and (b) a plateau phase with a rClfA concentration range of 1.28–2.60 µg. By correlating optical densitometry values with the ApoE calibration curve, the amounts of ApoE (µg) that bound to rClfA could be determined as shown in Fig. 4(c). The linearity model of the ApoE in human serum binding to rClfA (0–2.6 µg) demonstrated a statistically significant (P = 0.024) positive correlation with a stoichiometric binding ratio (rClfA : ApoE) of 1.702 at the highest Pearson's correlation coefficient of 0.927, as shown in Table 3.
Fig. 4.
(a) Calibration curve of human ApoE (0.32–3.2 µg) dot blotted on PVDF membrane and probed for ApoE. (b) Serum ApoE binding to dot blotted rClfA. Density of the chemiluminescence signals of ApoE in diluted human serum (20 %, v/v, human serum in TBS–BSA) that bound to escalating concentrations of rClfA (0.32–2.6 µg) on PVDF membrane (BSA was used as a control). (c) Calculated ApoE (µg) in diluted human serum (20 %, v/v, human serum in TBS–BSA) that bound to escalating concentrations of rClfA (0.32–2.6 µg) on PVDF membrane using immunodot-blot assay.
Table 3. Linearity model of protein interaction depending on rClfA (0–2.6 µg) binding to ApoE in diluted human serum (20 %, v/v, human serum in TBS–BSA) as determined by spss, version 18.0 (SPSS).
Calculations in the linearity model were based on the chemiluminescence densities obtained from immunodot-blot assays of serum ApoE binding to rClfA. Quantification of the chemiluminescence densities to the corresponding ApoE (µg) were performed using the ApoE calibration curve.
| Concentration range of rClfA (µg) | Pearson's correlation coefficient | P value (two-tailed) | Stoichiometric ratio (rClfA : ApoE) |
| 0–0.64 | 0.917 | 0.262 | 2.351 |
| 0–0.96 | 0.915 | 0.085 | 1.929 |
| 0–1.28 | 0.927 | 0.024 | 1.702 |
| 0–1.62 | 0.908 | 0.012 | 1.368 |
| 0–1.92 | 0.891 | 0.007 | 1.185 |
| 0–2.24 | 0.853 | 0.007 | 0.976 |
| 0–2.60 | 0.815 | 0.007 | 0.840 |
ApoE binding to S. aureus cell wall
To further examine the binding capability of human ApoE to bacterial cell surface, mid-exponential phase ClfA-sufficient wild-type S. aureus (ClfA+) and the isogenic ClfA-deficient mutant (ClfA−) were incubated with human ApoE (25 µg ml−1), washed and cell-wall extracts were prepared. Western-blot analysis of the cell-wall preparation for the wild-type (ClfA+) showed the presence of human ApoE with the characteristic band at 34 kDa. However, binding of human ApoE to the cell wall of the ClfA-deficient mutant (ClfA−) could not be detected under the same experimental conditions (Fig. 5).
Fig. 5.
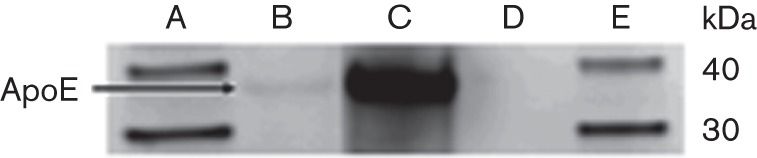
Purified human ApoE binding to mid-exponential S. aureus wild-type (ClfA+) and the isogenic ClfA-deficient mutant (ClfA−) after incubation for 2 h at 37 °C. Cell-wall proteins were solubilized from the washed bacteria and analysed via Western blotting and probing of ApoE. Sequence of lanes on Western blot: A, MagicMark XP Western protein standard; B, ApoE in the wild-type ClfA+ strain cell-wall preparation; C, purified human ApoE (EMD Millipore Chemicals) used as an internal control; D, absence of ApoE binding to cell wall of the ClfA-deficient mutant; E, MagicMark XP Western protein standard.
Whole-cell binding assay
In order to evaluate the extent to which S. aureus surface expression of ClfA contributed to ApoE binding, we performed a whole-cell binding assay. Wild-type ClfA-expressing (ClfA+) S. aureus Newman strain was compared with an isogenic ClfA-deficient (ClfA–) mutant. The wild-type S. aureus (ClfA+) and the ClfA-deficient strain (ClfA–) in suspension were incubated with ApoE-coated Immulon 2HB plates. After washing, the bound cells were then striped and colony counting was performed by the agar plating technique. The results depicted in Fig. 6 indicate statistically significant (P = 0.05; as determined by Mann–Whitney non-parametric test) lower binding of the ClfA-deficient strain (ClfA−) as compared to that of the wild-type (ClfA+).
Fig. 6.
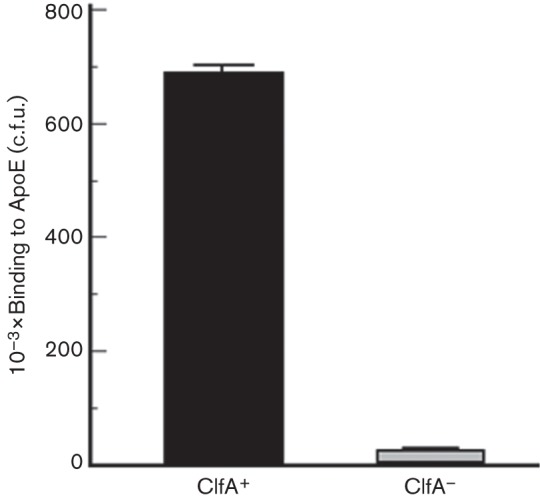
Whole-cell binding of S. aureus wild-type (ClfA+) and the isogenic ClfA-deficient mutant (ClfA−) to human-ApoE-coated Immulon 2HB plates. Columns indicate the means of colony counting after bacterial cell stripping from the ApoE-coated plates and are expressed as c.f.u. Error bars indicate ses.
Discussion
S. aureus is a premier human pathogen that causes life-threatening diseases and has a complex pathogenesis (Hawkins et al., 2012), including attachment and evasion of immune defences (Sharp et al., 2012). The current study identifies ApoE as a potentially new ligand for the S. aureus surface-expressed virulence factor ClfA. S. aureus strains express ClfA, a conserved surface protein, initially described for its ability to promote the precipitation (clumping reaction) of staphylococci in vitro via its binding to the soluble fibrinogen (McAdow et al., 2011). ClfA has been clearly demonstrated to be a major virulence factor in many animal models, including animal models of endocarditis and septic arthritis (Hu et al., 2009; Josefsson et al., 2001; Moreillon et al., 1995). However, the extents to which ClfA-enhanced virulence is mediated by its interaction with fibrinogen (McAdow et al., 2011), complement regulator factor I (Hair et al., 2008, 2010) or other serum proteins remain unresolved.
Plasma Apos and lipids participate in host innate immunity against different pathogens (Singh et al., 1999; Zhang et al., 2011). ApoE possesses multiple immune regulatory functions, including modulation of macrophage functions, suppression of T cell proliferation, upregulation of the production of platelet nitric oxide and facilitation of lipid antigen presentation by CD1 molecules to natural killer T cells. Moreover, inflammatory cytokines can either upregulate or downregulate the production of ApoE in various tissue types, highlighting the complex roles of ApoE and cytokines in various diseases (Zhang et al., 2011). ApoE is believed to mediate its non-lipid associated functions through binding to cell-surface receptors (Jofre-Monseny et al., 2008). Earlier studies that employed ApoE-deficient mice revealed increased susceptibility to Klebsiella pneumoniae infection (de Bont et al., 1999), as well as elevated mortality by Listeria monocytogenes, as compared to the wild-type mice (Roselaar & Daugherty, 1998). Furthermore, serum ApoE-deficient mice demonstrated an inability to neutralize LPS and demonstrated considerably enhanced lethality by LPS-induced endotoxaemia as compared to the wild-type mice (Van Oosten et al., 2001). ApoE deficiency was also associated with delayed emergence of adaptive immunity to tuberculosis and ApoE-deficient mice were extremely susceptible to tuberculosis (Martens et al., 2008). A peptide analogue of the receptor-binding region of ApoE, ApoE dimer tandem repeat peptide (ApoE dp), could reproduce some of the biological properties of ApoE and demonstrated antimicrobial activities against S. aureus, Pseudomonas aeruginosa, herpes simplex viruses, as well as human immunodeficiency virus. These biological activities were attributed in part to the prevention of early stages of microbial attachment to the mammalian cells (Dobson et al., 2006). Moreover, a novel peptide derivative of human ApoE (ApoE dpL-W) showed broad-spectrum antibacterial activity and noticeable antimicrobial activity against emerging S. aureus biofilms (Forbes et al., 2013).
In the current study, we identified, using MS, ApoE as the major plasma protein to bind surface-coupled recombinant ClfA. To confirm and elucidate the binding of human ApoE to ClfA, we utilized purified and serum ApoE, and evaluated their binding to rClfA through CEWA and dot blot assays. Furthermore, an isogenic ClfA-deficient S. aureus strain revealed statistically significant (P = 0.05) decreased binding to human serum ApoE compared to the wild-type ClfA-sufficient strain. Our Western blot analysis revealed that purified human ApoE bound to the bacterial cell surface of the wild-type ClfA-sufficient strain (ClfA+), but no binding was detected with the isogenic ClfA-deficient strain (ClfA−). Together, these data suggest that ApoE may be a novel ligand for ClfA.
Previous studies, described above, suggested that ApoE plays a role in host defences against many pathogens, although the precise mechanisms remain unclear. Thus, ClfA recruitment of ApoE to the S. aureus surface may sequester ApoE and blunt its host defence functions. Our lab has previously demonstrated that a secreted or shed form of ClfA is elaborated from the S. aureus surface (Hair et al., 2008), which could potentially bind ApoE remote from the bacterial surface. In these scenarios, ClfA sequestration of ApoE could mediate a host defence evasion mechanism for S. aureus. The previous study with ApoE-derived peptides suggested that ApoE may possess an anti-biofilm effect on S. aureus as a host defence mechanism, potentially modulated by binding ClfA (Forbes et al., 2013). Alternatively, it is also possible that ApoE binding to the virulence factor ClfA could potentially mask the virulence factor from interacting with other known ligands, fibrinogen or factor I; thus, moderating ClfA-mediated virulence. However, if ApoE binding to ClfA were a major mechanism of immunological control of S. aureus, then we would expect that ClfA-deficient strains should demonstrate increased virulence. The opposite is true: ClfA-deficient strains are markedly less virulent, suggesting that ApoE binding to ClfA is unlikely to contribute to immunological control of S. aureus. Thus, it appears more likely that S. aureus may sequester ApoE via ClfA as an immune evasion tactic or as a means to increase virulence via another mechanism.
Conclusion
In the current study, we identified a novel ligand, ApoE, for the known S. aureus virulence factor ClfA. The binding of ApoE to ClfA was demonstrated on both molecular and cellular levels. Future studies will focus on elucidating whether ClfA binding of ApoE enhances or inhibits S. aureus virulence and the mechanism by which this occurs. Additionally, in vivo studies utilizing ApoE-knockout animal models and ClfA-deficient S. aureus strains are required to address the in-depth mechanism and the impact of ClfA–ApoE interaction on S. aureus pathogenesis and cell signalling.
Acknowledgements
This work was supported by a grant from the National Institutes of Health (NIH) (R21AI082398) to K. M. Cunnion. S. aureus Newman, the isogenic ClfA-deficient mutant, and the E. coli expressing rClfA were kindly provided by Dr Timothy Foster and Dr Joan Geoghegan. Access to the mass spectrometers of the Leroy T. Canoles Jr Cancer Research Center was kindly provided by Dr O. John Semmes.
References
- Arciola C. R., Baldassarri L., Montanaro L. ( 2001. ). Presence of icaA and icaD genes and slime production in a collection of staphylococcal strains from catheter-associated infections. J Clin Microbiol 39, 2151–2156. 10.1128/JCM.39.6.2151-2156.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bur S., Preissner K. T., Herrmann M., Bischoff M. ( 2013. ). The Staphylococcus aureus extracellular adherence protein promotes bacterial internalization by keratinocytes independent of fibronectin-binding proteins. J Invest Dermatol 133, 2004–2012. 10.1038/jid.2013.87 [DOI] [PubMed] [Google Scholar]
- Cunnion K. M., Lee J. C., Frank M. M. ( 2001. ). Capsule production and growth phase influence binding of complement to Staphylococcus aureus . Infect Immun 69, 6796–6803. 10.1128/IAI.69.11.6796-6803.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bont N., Netea M. G., Demacker P. N., Verschueren I., Kullberg B. J., van Dijk K. W., van der Meer J. W., Stalenhoef A. F. ( 1999. ). Apolipoprotein E knock-out mice are highly susceptible to endotoxemia and Klebsiella pneumoniae infection. J Lipid Res 40, 680–685. [PubMed] [Google Scholar]
- Dhaliwal W., Kelly P., Bajaj-Elliott M. ( 2009. ). Differential effects of staphylococcal enterotoxin B-mediated immune activation on intestinal defensins. Clin Exp Immunol 156, 263–270. 10.1111/j.1365-2249.2008.03808.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Do Carmo S., Jacomy H., Talbot P. J., Rassart E. ( 2008. ). Neuroprotective effect of apolipoprotein D against human coronavirus OC43-induced encephalitis in mice. J Neurosci 28, 10330–10338. 10.1523/JNEUROSCI.2644-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobson C. B., Sales S. D., Hoggard P., Wozniak M. A., Crutcher K. A. ( 2006. ). The receptor-binding region of human apolipoprotein E has direct anti-infective activity. J Infect Dis 193, 442–450. 10.1086/499280 [DOI] [PubMed] [Google Scholar]
- Faghri J., Shahbazzadeh D., Pooshang Bagheri K., Moghim S., Ghasemian Safaei H., Nasr Esfahani B., Fazeli H., Yazdani R., Mirmohammad Sadeghi H. ( 2012. ). Two dimensional structural analysis and expression of a new Staphylococcus aureus adhesin based fusion protein. Iran J Basic Med Sci 15, 725–738. [PMC free article] [PubMed] [Google Scholar]
- Forbes S., McBain A. J., Felton-Smith S., Jowitt T. A., Birchenough H. L., Dobson C. B. ( 2013. ). Comparative surface antimicrobial properties of synthetic biocides and novel human apolipoprotein E derived antimicrobial peptides. Biomaterials 34, 5453–5464. 10.1016/j.biomaterials.2013.03.087 [DOI] [PubMed] [Google Scholar]
- Friberg N., Carlson P., Kentala E., Mattila P. S., Kuusela P., Meri S., Jarva H. ( 2008. ). Factor H binding as a complement evasion mechanism for an anaerobic pathogen, Fusobacterium necrophorum . J Immunol 181, 8624–8632. 10.4049/jimmunol.181.12.8624 [DOI] [PubMed] [Google Scholar]
- Hair P. S., Ward M. D., Semmes O. J., Foster T. J., Cunnion K. M. ( 2008. ). Staphylococcus aureus clumping factor A binds to complement regulator factor I and increases factor I cleavage of C3b. J Infect Dis 198, 125–133. 10.1086/588825 [DOI] [PubMed] [Google Scholar]
- Hair P. S., Echague C. G., Sholl A. M., Watkins J. A., Geoghegan J. A., Foster T. J., Cunnion K. M. ( 2010. ). Clumping factor A interaction with complement factor I increases C3b cleavage on the bacterial surface of Staphylococcus aureus and decreases complement-mediated phagocytosis. Infect Immun 78, 1717–1727. 10.1128/IAI.01065-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hair P. S., Foley C. K., Krishna N. K., Nyalwidhe J. O., Geoghegan J. A., Foster T. J., Cunnion K. M. ( 2013. ). Complement regulator C4BP binds to Staphylococcus aureus surface proteins SdrE and Bbp inhibiting bacterial opsonization and killing. Results Immunol 3, 114–121. 10.1016/j.rinim.2013.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakim H., Mylotte J. M., Faden H. ( 2007. ). Morbidity and mortality of staphylococcal bacteremia in children. Am J Infect Control 35, 102–105. 10.1016/j.ajic.2006.09.016 [DOI] [PubMed] [Google Scholar]
- Haupt K., Reuter M., van den Elsen J., Burman J., Hälbich S., Richter J., Skerka C., Zipfel P. F. ( 2008. ). The Staphylococcus aureus protein Sbi acts as a complement inhibitor and forms a tripartite complex with host complement factor H and C3b. PLoS Pathog 4, e1000250. 10.1371/journal.ppat.1000250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins J., Kodali S., Matsuka Y. V., McNeil L. K., Mininni T., Scully I. L., Vernachio J. H., Severina E., Girgenti D., et al. ( 2012. ). A recombinant clumping factor A-containing vaccine induces functional antibodies to Staphylococcus aureus that are not observed after natural exposure. Clin Vaccine Immunol 19, 1641–1650. 10.1128/CVI.00354-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu D. L., Narita K., Hyodo M., Hayakawa Y., Nakane A., Karaolis D. K. ( 2009. ). c-di-GMP as a vaccine adjuvant enhances protection against systemic methicillin-resistant Staphylococcus aureus (MRSA) infection. Vaccine 27, 4867–4873. 10.1016/j.vaccine.2009.04.053 [DOI] [PubMed] [Google Scholar]
- James E. H., Edwards A. M., Wigneshweraraj S. ( 2013. ). Transcriptional downregulation of agr expression in Staphylococcus aureus during growth in human serum can be overcome by constitutively active mutant forms of the sensor kinase AgrC. FEMS Microbiol Lett 349, 153–162. 10.1111/1574-6968.12309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jofre-Monseny L., Minihane A. M., Rimbach G. ( 2008. ). Impact of apoE genotype on oxidative stress, inflammation and disease risk. Mol Nutr Food Res 52, 131–145. 10.1002/mnfr.200700322 [DOI] [PubMed] [Google Scholar]
- Josefsson E., Hartford O., O’Brien L., Patti J. M., Foster T. ( 2001. ). Protection against experimental Staphylococcus aureus arthritis by vaccination with clumping factor A, a novel virulence determinant. J Infect Dis 184, 1572–1580. 10.1086/324430 [DOI] [PubMed] [Google Scholar]
- Kerrigan S. W., Clarke N., Loughman A., Meade G., Foster T. J., Cox D. ( 2008. ). Molecular basis for Staphylococcus aureus-mediated platelet aggregate formation under arterial shear in vitro. Arterioscler Thromb Vasc Biol 28, 335–340. 10.1161/ATVBAHA.107.152058 [DOI] [PubMed] [Google Scholar]
- Kuwahara T., Kaneda S., Shimono K., Inoue Y. ( 2010. ). Growth of microorganisms in total parenteral nutrition solutions without lipid. Int J Med Sci 7, 43–47. 10.7150/ijms.7.43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C. C., Kanekiyo T., Xu H., Bu G. ( 2013. ). Apolipoprotein E and Alzheimer disease: risk, mechanisms and therapy. Nat Rev Neurol 9, 106–118. 10.1038/nrneurol.2012.263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens G. W., Arikan M. C., Lee J., Ren F., Vallerskog T., Kornfeld H. ( 2008. ). Hypercholesterolemia impairs immunity to tuberculosis. Infect Immun 76, 3464–3472. 10.1128/IAI.00037-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masago Y., Shibata T., Rose J. B. ( 2008. ). Bacteriophage P22 and Staphylococcus aureus attenuation on nonporous fomites as determined by plate assay and quantitative PCR. Appl Environ Microbiol 74, 5838–5840. 10.1128/AEM.00352-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAdow M., Kim H. K., Dedent A. C., Hendrickx A. P., Schneewind O., Missiakas D. M. ( 2011. ). Preventing Staphylococcus aureus sepsis through the inhibition of its agglutination in blood. PLoS Pathog 7, e1002307. 10.1371/journal.ppat.1002307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormack N., Foster T. J., Geoghegan J. A. ( 2014. ). A short sequence within subdomain N1 of region A of the Staphylococcus aureus MSCRAMM clumping factor A is required for export and surface display. Microbiology 160, 659–670. 10.1099/mic.0.074724-0 [DOI] [PubMed] [Google Scholar]
- McDevitt D., Francois P., Vaudaux P., Foster T. J. ( 1994. ). Molecular characterization of the clumping factor (fibrinogen receptor) of Staphylococcus aureus . Mol Microbiol 11, 237–248. 10.1111/j.1365-2958.1994.tb00304.x [DOI] [PubMed] [Google Scholar]
- Miajlovic H., Fallon P. G., Irvine A. D., Foster T. J. ( 2010. ). Effect of filaggrin breakdown products on growth of and protein expression by Staphylococcus aureus . J Allergy Clin Immunol 126, 1184–1190. 10.1016/j.jaci.2010.09.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreillon P., Entenza J. M., Francioli P., McDevitt D., Foster T. J., François P., Vaudaux P. ( 1995. ). Role of Staphylococcus aureus coagulase and clumping factor in pathogenesis of experimental endocarditis. Infect Immun 63, 4738–4743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmqvist N., Patti J. M., Tarkowski A., Josefsson E. ( 2004. ). Expression of staphylococcal clumping factor A impedes macrophage phagocytosis. Microbes Infect 6, 188–195. 10.1016/j.micinf.2003.11.005 [DOI] [PubMed] [Google Scholar]
- Peterson M. M., Mack J. L., Hall P. R., Alsup A. A., Alexander S. M., Sully E. K., Sawires Y. S., Cheung A. L., Otto M., Gresham H. D. ( 2008. ). Apolipoprotein B is an innate barrier against invasive Staphylococcus aureus infection. Cell Host Microbe 4, 555–566. 10.1016/j.chom.2008.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roselaar S. E., Daugherty A. ( 1998. ). Apolipoprotein E-deficient mice have impaired innate immune responses to Listeria monocytogenes in vivo. J Lipid Res 39, 1740–1743. [PubMed] [Google Scholar]
- Rosenthal V. D., Maki D. G., Jamulitrat S., Medeiros E. A., Todi S. K., Gomez D. Y., Leblebicioglu H., Abu Khader I., Miranda Novales M. G., et al. ( 2010. ). International Nosocomial Infection Control Consortium (INICC) report, data summary for 2003-2008, issued June 2009. Am J Infect Control 38, 95–104, e2. 10.1016/j.ajic.2009.12.004 [DOI] [PubMed] [Google Scholar]
- Sharp J. A., Cunnion K. M. ( 2011. ). Disruption of the alternative pathway convertase occurs at the staphylococcal surface via the acquisition of factor H by Staphylococcus aureus . Mol Immunol 48, 683–690. 10.1016/j.molimm.2010.11.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp J. A., Echague C. G., Hair P. S., Ward M. D., Nyalwidhe J. O., Geoghegan J. A., Foster T. J., Cunnion K. M. ( 2012. ). Staphylococcus aureus surface protein SdrE binds complement regulator factor H as an immune evasion tactic. PLoS ONE 7, e38407. 10.1371/journal.pone.0038407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigel S., Bunk S., Meergans T., Doninger B., Stich K., Stulnig T., Derfler K., Hoffmann J., Deininger S., et al. ( 2012. ). Apolipoprotein B100 is a suppressor of Staphylococcus aureus-induced innate immune responses in humans and mice. Eur J Immunol 42, 2983–2989. 10.1002/eji.201242564 [DOI] [PubMed] [Google Scholar]
- Singh I. P., Chopra A. K., Coppenhaver D. H., Ananatharamaiah G. M., Baron S. ( 1999. ). Lipoproteins account for part of the broad non-specific antiviral activity of human serum. Antiviral Res 42, 211–218. 10.1016/S0166-3542(99)00032-7 [DOI] [PubMed] [Google Scholar]
- Tada N., Sakamoto T., Kagami A., Mochizuki K., Kurosaka K. ( 1993. ). Antimicrobial activity of lipoprotein particles containing apolipoprotein Al. Mol Cell Biochem 119, 171–178. 10.1007/BF00926868 [DOI] [PubMed] [Google Scholar]
- Tanaka N., Abe-Dohmae S., Iwamoto N., Fitzgerald M. L., Yokoyama S. ( 2010. ). Helical apolipoproteins of high-density lipoprotein enhance phagocytosis by stabilizing ATP-binding cassette transporter A7. J Lipid Res 51, 2591–2599. 10.1194/jlr.M006049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Oosten M., Rensen P. C., Van Amersfoort E. S., Van Eck M., Van Dam A. M., Breve J. J., Vogel T., Panet A., Van Berkel T. J., Kuiper J. ( 2001. ). Apolipoprotein E protects against bacterial lipopolysaccharide-induced lethality. A new therapeutic approach to treat gram-negative sepsis. J Biol Chem 276, 8820–8824. 10.1074/jbc.M009915200 [DOI] [PubMed] [Google Scholar]
- Walsh E. J., O’Brien L. M., Liang X., Hook M., Foster T. J. ( 2004. ). Clumping factor B, a fibrinogen-binding MSCRAMM (microbial surface components recognizing adhesive matrix molecules) adhesin of Staphylococcus aureus, also binds to the tail region of type I cytokeratin 10. J Biol Chem 279, 50691–50699. 10.1074/jbc.M408713200 [DOI] [PubMed] [Google Scholar]
- Zhang H., Wu L.-M., Wu J. ( 2011. ). Cross-talk between apolipoprotein E and cytokines. Mediators Inflamm 2011, 949072. 10.1155/2011/949072 [DOI] [PMC free article] [PubMed] [Google Scholar]