Abstract
Given their free-ranging habits, feral swine could serve as reservoirs or spatially dynamic ‘mixing vessels’ for influenza A virus (IAV). To better understand virus shedding patterns and antibody response dynamics in the context of IAV surveillance amongst feral swine, we used IAV of feral swine origin to perform infection experiments. The virus was highly infectious and transmissible in feral swine, and virus shedding patterns and antibody response dynamics were similar to those in domestic swine. In the virus-inoculated and sentinel groups, virus shedding lasted ≤ 6 and ≤ 9 days, respectively. Antibody titres in inoculated swine peaked at 1 : 840 on day 11 post-inoculation (p.i.), remained there until 21 days p.i. and dropped to < 1 : 220 at 42 days p.i. Genomic sequencing identified changes in wildtype (WT) viruses and isolates from sentinel swine, most notably an amino acid divergence in nucleoprotein position 473. Using data from cell culture as a benchmark, sensitivity and specificity of a matrix gene-based quantitative reverse transcription-PCR method using nasal swab samples for detection of IAV in feral swine were 78.9 and 78.1 %, respectively. Using data from haemagglutination inhibition assays as a benchmark, sensitivity and specificity of an ELISA for detection of IAV-specific antibody were 95.4 and 95.0 %, respectively. Serological surveillance from 2009 to 2014 showed that ∼7.58 % of feral swine in the USA were positive for IAV. Our findings confirm the susceptibility of IAV infection and the high transmission ability of IAV amongst feral swine, and also suggest the need for continued surveillance of IAVs in feral swine populations.
Introduction
Influenza viruses (family Orthomyxoviridae) are classified into types A, B and C (Alexander & Brown, 2000; Mahy, 1997). Influenza A virus (IAV), an enveloped RNA virus, contains eight negative-sense ssRNA genome segments. The IAVs can cause seasonal epidemics, affecting one or many countries, as well as pandemics. IAVs have been recovered from at least 105 wild bird species of 26 different families (Olsen et al., 2006a), and species living in wetland and aquatic environments (e.g. Anseriformes and Charadriiformes spp.) constitute the major natural IAV reservoir (Webster et al., 1992). However, in addition to circulating amongst avian species, IAVs also circulate amongst a wide spectrum of other host species, including humans, swine, equines, canines and marine mammals (Keawcharoen et al., 2004; Peiris et al., 2007; Sun et al., 2011; Webster et al., 1992).
Amongst the natural hosts of IAVs, swine have been shown to be susceptible to many IAV subtypes (Kida et al., 1994). In domestic swine, IAVs can cause respiratory diseases characterized by fever, lethargy, sneezing, coughing, difficulty breathing and decreased appetite, which usually lead to weight loss. For the past decade, IAV subtypes H1N1, H1N2 and H3N2 have been the predominant strains circulating amongst the domestic swine population in the USA (Vincent et al., 2008). Antigenic characterization revealed that the circulating H1N1 IAVs formed four genetic clusters: swH1α (classic H1N1), swH1β (reassortant H1N1-like), swH1γ (H1N2-like) and swH1δ (human-like H1). Viruses within cluster swH1δ can be further classified into two subclusters: swH1δ1 (human-like H1N2) and swH1δ2 (human-like H1N1) (Vincent et al., 2006, 2009). The 2009 pandemic influenza A(H1N1)pdm09 virus is a classic subtype H1N1-origin swine virus, but it differs genetically from the four genetic clusters identified from the USA (Lorusso et al., 2011). Antigenic characterization showed variations amongst viruses in the subtype H1N1 clusters (Lorusso et al., 2011).
Similar to H1 IAVs, the H3N2 subtypes in the US swine population are also genetically and antigenically diverse. Four genetic clusters of H3N2 subtype IAVs (clusters I–IV) have been identified (Hause et al., 2010; Olsen et al., 2006b; Richt et al., 2003). Cluster IV, which has become predominant amongst the US swine population, has further evolved into two antigenic clusters: H3N2-α and H3N2-β (Feng et al., 2014). Many of these H3N2 genetic clusters are currently co-circulating in swine populations and frequent reassortments of these IAVs have occurred. In 2011, a predominant H3N2 genotype containing a matrix gene from influenza A(H1N1)pdm09 virus led to the emergence of an H3N2 variant virus that caused disease in humans (Bowman et al., 2012; Nelson et al., 2012; Shu et al., 2012); this variant IAV is antigenically similar to H3N2-β viruses (Feng et al., 2014).
In addition to the prevalent H1 and H3 IAVs, other haemagglutinin subtype viruses, such as H1N1, H4N6, H5N1, H6N6 and H9N2, have been transiently detected in swine (Choi et al., 2005; Guan et al., 1996; Olsen, 2002; Peiris et al., 2001; Zhang et al., 2011). As swine have the avian-like NeuAc-2,3-α-Gal receptors and the human-like NeuAc-2,6-α-Gal receptors in their respiratory tracts, they have been proposed as a ‘mixing vessel’ for the generation of IAV reassortants (Scholtissek, 1994).
In the USA, there are ∼5 million feral swine across >40 states and the number is increasing (Bevins et al., 2014; Fogarty, 2007). Contacts between feral and domestic swine provide the opportunities for bi-directional transmission for pathogens such as IAVs (Wyckoff et al., 2009). Also, because feral swine and wild birds are sympatric throughout their ranges, direct contact (e.g. feral swine scavenging and predation on wild birds) and indirect contact (e.g. use of common resources like water and forage) provide the opportunity for virus transmission amongst these species (Bevins et al., 2014). The free-ranging nature of feral swine and their direct and indirect interactions with other IAV hosts position them as ideal, spatially dynamic ‘mixing vessels’. Previous studies have recovered subtype H3N2 and influenza A(H1N1)pdm09 viruses from feral swine (Clavijo et al., 2012). Results from serological surveillance during October 2011–September 2012 documented that 9.15 % of serum samples from feral swine in 31 states were positive for IAV exposure, of which >60 % were positive for H3 subtype viruses (Feng et al., 2014). These findings indicate that IAVs are widely present in feral swine, and IAVs circulating amongst feral swine are antigenically and genetically similar to those circulating in domestic swine.
Surveillance for IAV infection is more challenging in feral swine than in domestic swine because multiple haemagglutinin subtypes can be present in feral swine and little is known about IAV infection dynamics in these animals. Current methods for IAV diagnosis and surveillance in feral swine were adapted from protocols designed for use with domestic swine or poultry. To improve these methods and to evaluate protocols for IAV diagnosis and surveillance in feral swine, knowledge of virus shedding patterns and antibody response dynamics is needed. Furthermore, it is known that feral swine can acquire IAV from domestic swine (Bevins et al., 2014; Fogarty, 2007), but it is not clear whether IAVs can be easily transmitted amongst feral swine.
To better understand IAV shedding patterns, antibody response dynamics and the potential for IAV transmission amongst feral swine, we performed infection and reinfection experiments using a subtype H3N2 IAV of feral swine origin. Other goals were to investigate whether feral swine with low IAV antibody titres could infect and shed homologous viruses, and to evaluate the protocol for influenza diagnosis and surveillance in feral swine.
Results
H3N2 IAV is highly infectious and transmissible in feral swine
We determined virus shedding patterns and antibody response dynamics in 12 feral swine by performing infection experiments with a subtype H3N2 IAV of feral swine origin that was highly infectious and transmissible amongst these animals. The swine were divided into treatment (n = 8) and sentinel (n = 4) groups, and housed at one or two animals per pen (a total of six adjoining pens for the treatment group and three for the sentinel group); pens for treatment and sentinel groups were 1.2 m apart (Fig. 1). Swine in the treatment group began shedding virus 1 day post-inoculation (p.i.) and continued to shed virus until 6 days p.i. Virus titres in nasal wash and nasal swab samples from the treatment group ranged from 100.575 to 101.878 TCID50 ml− 1 and peaked at 5 days p.i. In the four sentinel swine, virus shedding was detectable 1 day post-exposure (p.e.) to the IAV-inoculated swine. Virus titres in sentinel swine peaked at 8 days p.e. (Fig. 2) and shedding was sustained for up to 9 days (mean titres 101.532–104.673 TCID50 ml− 1). No obvious clinical signs of infection (e.g. cough, fever, weight loss) were observed in the inoculated or sentinel swine. To investigate whether feral swine with low IAV antibody titres could infect and shed homologous viruses, we reinoculated swine in the treatment group and inoculated swine in the sentinel group with subtype H3N2 IAV on day 103 p.i. However, the antibody in the treatment swine quickly rose and we were unable to determine virus titres in samples from the treatment group.
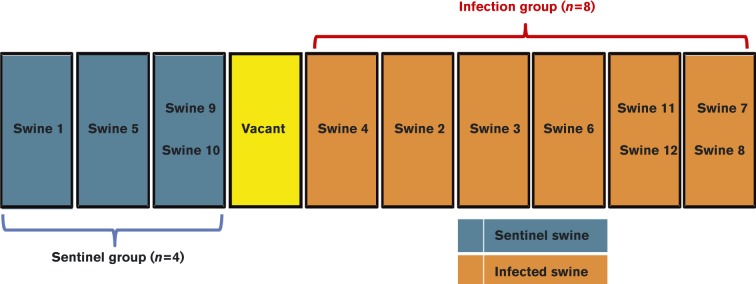
Fig. 1.
Physical layout of pens housing swine in a study of the virus shedding patterns and antibody response dynamics during IAV infection in feral swine. An empty pen was located between six pens housing a total of eight swine in the treatment group and three pens housing a total of four swine in the sentinel group. Swine in the treatment group were intranasally inoculated twice (days 0 and 103) with 106 TCID50 influenza A/swine/Texas/A01104013/2012(H3N2) virus; swine in the sentinel group were intranasally inoculated once (day 103) with 106 TCID50 influenza A/swine/Texas/A01104013/2012(H3N2) virus.
Fig. 2.
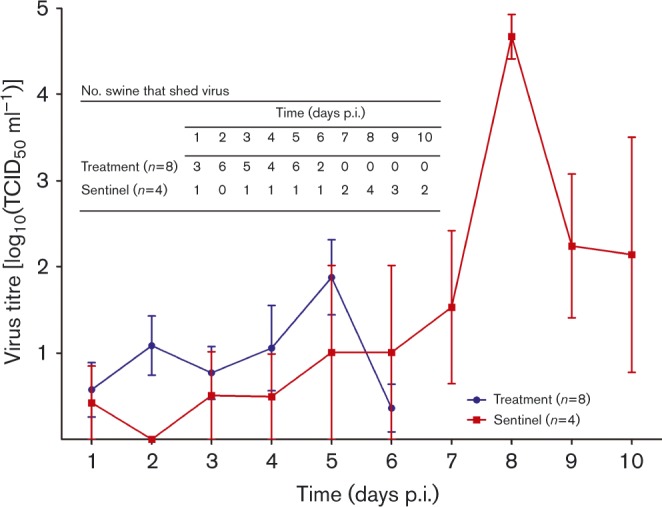
Mean titres of influenza viruses recovered from nasal wash or nasal swab samples of feral swine following nasal inoculation of influenza virus. Swine in the infection group were inoculated with 106 TCID50 influenza A/swine/Texas/A01104013/2012(H3N2) virus (in 1 ml volume). Swine in the sentinel group were inoculated with 1 ml PBS. Virus titres were measured in nasal wash or swab fluids collected on indicated days following titration in Madin–Darby canine kidney (MDCK) cells; ending titres are expressed as mean ± sd. The inset table shows the number of swine that shed virus on the various days after virus inoculation or exposure.
Antibody response dynamics in feral swine after inoculation with H3N2 IAV
Feral swine in the treatment group seroconverted at 8 days p.i.; the mean haemagglutination inhibition (HI) titre was 1 : 450 (Fig. 3). The mean HI titres peaked at 1 : 840 at 11 days p.i. and remained at that level until 21 days p.i. before gradually declining to < 1 : 220 at 42 days p.i.; titres then continued to decline to 1 : 130 at 93 days p.i. Before swine in the treatment group were reinoculated with H3N2 IAV at 103 days p.i., the mean HI titre was 1 : 120.
Fig. 3.
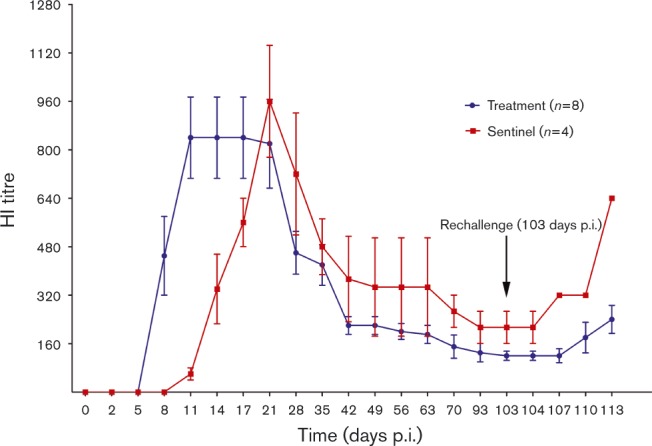
Mean HI titres of IAVs in serum samples collected from feral swine following nasal inoculation of influenza virus. Swine in the infection group were inoculated twice (days 0 and 103) with 106 TCID50 influenza A/swine/Texas/A01104013/2012(H3N2) virus (in 1 ml volume). Swine in the sentinel group were inoculated once with 1 ml PBS (day 0) and once (day 103) with 106 TCID50 influenza A/swine/Texas/A01104013/2012(H3N2) virus (in 1 ml volume). HI titres were measured in serum samples obtained on indicated days following inoculation. HI assays were conducted by using 0.5 % turkey red blood cells. Titres are expressed as mean ± SD from the results in eight swine in the treatment group or four swine in sentinel group.
Feral swine in the sentinel group seroconverted (mean HI titre 1 : 60) at 11 days p.e.; HI titres in these swine peaked at 1 : 960 at 21 days p.e. and gradually declined to 1 : 373 at 42 days p.e. Before swine in the sentinel group were inoculated with H3N2 IAV at 103 days p.e., the mean HI titre was 1 : 213.
After swine in the treatment group were reinoculated and the sentinel group was inoculated with IAV, the antibody titre increased in both groups of swine. At 113 days p.i. (10 days after reinoculation), HI titres in the treatment group swine increased to 1 : 240, whereas titres in the sentinel swine group increased to 1 : 640 (Fig. 3).
During 21–113 days p.i./p.e., antibody titres in sentinel swine were significantly higher (P < 0.001) than those in inoculated swine.
Amino acid polymorphisms in IAVs recovered from experimentally infected feral swine
To explore the mechanism for the discrepancy in antibody levels between sentinel and treated swine, we recovered viruses from the sentinel swine and compared them with the WT virus that was used to inoculate swine in the treatment group. The growth kinetics study demonstrated that, compared with virus isolates from the sentinel swine, WT virus replicated better in Madin–Darby canine kidney (MDCK) cells (Fig. 4). Titres of WT virus were 0.5- to 1.5-fold higher (P < 0.05) than titres for sentinel swine test isolate P10D8 at 24, 48 and 72 h p.i., and for sentinel swine test isolate P5D8 at 24 h p.i. Further sequencing of these isolates identified amino acid and nucleic acid polymorphisms in PB2, PB1, PA, HA, NP, NA, M1, and NS1, (Tables 1, S1 and S2, available in the online Supplementary Material). Overall, these polymorphisms suggested that the original cell-adapted virus inoculum adapted within the infected swine and that this adaptation may underlie the increase in HI titre levels.
Fig. 4.
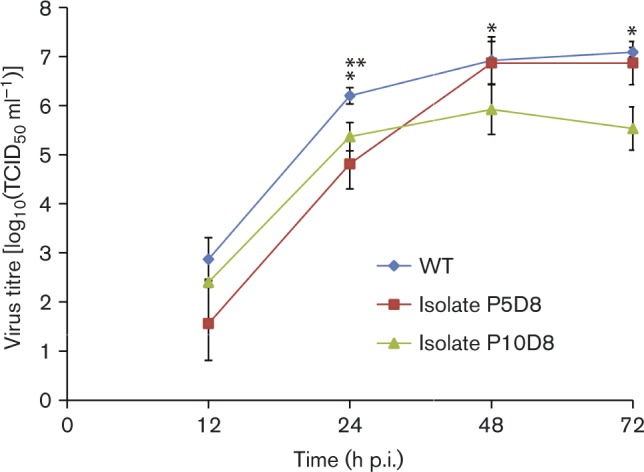
Growth kinetics of influenza virus isolates and WT virus characterized in MDCK cells. MDCK cells were infected with individual viruses at m.o.i. 0.001. Virus titres were measured in MDCK cells; ending titres are expressed as mean ± sd from three independent experiments. *WT virus is significantly higher than that of isolate P10D8 (P < 0.05); **WT virus is significantly higher than that of isolate P5D8 (P < 0.05).
Table 1. Amino acid polymorphisms amongst sequences of WT virus used for inoculation and amongst virus isolates recovered from sentinel swine in a study of the dynamics of virus shedding and antibody responses during IAV infection in feral swine.
| Gene | Position* | Amino acid† (by virus‡) | |||
|---|---|---|---|---|---|
| P1D8 | P5D8 | P10D8 | WT | ||
| HA | 443 | L/H (137/65) | L | L | L |
| NA | 346 | G | G/S (279/211) | G | G |
| PB1 | 59 | T | T | T/K (2041/575) | T |
| NP | 473 | S | S | S/G (2223/232) | S/G (459/275) |
| NS1 | 157 | V | V | I/V (803/6400) | I/V (1747/1605) |
Numbering of the residues was from the first amino acid in the methionine start site of each gene of the influenza viruses.
Numbers in parentheses represent number of supporting reads.
Isolates P1D8, P5D8 and P10D8 were isolated from sentinel swine numbers 1, 5 and 10, respectively, 8 days after swine in the treatment group were infected. WT virus was influenza strain A/swine/Texas/A01104013/2012(H3N2) that was used to inoculate swine in the treatment group.
Sensitivity of nasal swab specimens for diagnosing IAV infection in feral swine
To assess the value of a matrix gene-based quantitative reverse transcription (qRT)-PCR method using nasal swab samples for determining IAV infection, we compared qRT-PCR and TCID50 results for 141 swab samples collected 1–14 days p.i. (Tables 2 and 3). The qRT-PCR results were positive for 55 samples and cell cultures were positive for 41 samples; 32 of the samples had positive results from both the qRT-PCR and cell culture methods. The sensitivity and specificity of the qRT-PCR method using nasal swab samples for diagnosis of IAV infection in feral swine was 78.90 and 78.05 %, respectively.
Table 2. Results of qRT-PCR of nasal swab samples from feral swine after inoculation with influenza A/swine/Texas/A01104013/2012(H3N2) virus and from feral swine after exposure to infected swine .
nc, no samples were collected.
| Group, swine no.* | C t value (day p.i./p.e.) | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | |
| Inoculation | |||||||||||||||
| 2 | 39.5 | 29.2 | 22.1 | 26.4 | 22.3 | 27.7 | 28.8 | 34.1 | 38.2 | 39.5 | 0 | 0 | 0 | 0 | 0 |
| 3 | 0 | 33.5 | nc | 24.4 | nc | 30.2 | nc | 37.1 | nc | 0 | nc | 0 | nc | 0 | nc |
| 4 | 0 | 34.1 | 29.2 | 26.0 | 28.3 | 27.2 | 28.8 | 33.8 | 36.8 | 39.2 | 0 | 33.3 | 0 | 37.1 | 34.1 |
| 6 | 0 | 35.5 | nc | 30.2 | nc | 26.9 | nc | 32.7 | nc | 0 | nc | 0 | nc | 0 | nc |
| 7 | 0 | 36.0 | nc | 30.0 | nc | 28.3 | nc | 40.0 | nc | 0 | nc | 0 | nc | 0 | nc |
| 8 | 0 | 0 | nc | 31.1 | nc | 32.8 | nc | 38.6 | nc | 0 | nc | 0 | nc | 0 | nc |
| 11 | 0 | 36.9 | 30.4 | 27.7 | 28.6 | 28.5 | 33.9 | 35.6 | 40.6 | 0 | 0 | 0 | 0 | 0 | 0 |
| 12 | 0 | 34.3 | 29.2 | 28.4 | 28.4 | 29.0 | 31.1 | 34.4 | 36.6 | 0 | 0 | 0 | 0 | 0 | 0 |
| Sentinel | |||||||||||||||
| 1 | 0 | 0 | 0 | 0 | 0 | 39.5 | 40.0 | 29.8 | 26.0 | 26.2 | 30.3 | 35.5 | 37.2 | 0 | 0 |
| 5 | 0 | 0 | 0 | 0 | 33.3 | 24.1 | 26.3 | 25.9 | 23.9 | 28.1 | 31.8 | 34.9 | 35.5 | 39.1 | 37.9 |
| 9 | 0 | 0 | nc | 0 | nc | 37.8 | nc | 34.4 | nc | 32.8 | nc | 39.5 | nc | 0 | nc |
| 10 | 0 | 40.9 | nc | 0 | nc | 0 | nc | 36.8 | nc | 33.1 | nc | 32.9 | nc | 33.5 | nc |
A feral swine index was used to randomly assign swine to the treatment or sentinel group. Swine in both groups were housed in pens in the same barn (Fig. 1). The following swine were housed together: swine 7 and 8, swine 9 and 10, and swine 11 and 12.
Evaluation of current protocols for using an AI MultiS-Screen Ab Test kit to detect IAV in feral swine
To evaluate current protocols for IAV surveillance in the feral swine population, we used an AI MultiS-Screen Ab Test kit (see Methods) to determine the receiver operating characteristic curve. Results showed that when the threshold of the sample-to-negative control (S/N) ratio increased, the sensitivity of the AI MultiS-Screen Ab Test kit also increased, but specificity decreased (Fig. 5). When the S/N threshold was 0.50, sensitivity and specificity of the AI MultiS-Screen Ab assay were 81.44 and 100 %, respectively. When the S/N threshold was increased to 0.681 (i.e. the value currently used by the US Department of Agriculture as the standard for testing feral swine samples for IAV exposure), sensitivity and specificity of the AI MultiS-Screen Ab assay increased to 95.36 and 95.00 %, respectively. If specificity and sensitivity were combined, an S/N value of 0.681 would be reasonable for the diagnosis of IAV in feral swine. By applying the S/N threshold of 0.681, we determined that 7.58 % (585) of 7714 serum samples collected from feral swine during 2009–2014 were positive for IAV (US Department of Agriculture, unpublished data); this finding confirms the prevalence of IAV infection in feral swine. Our results also demonstrated that the 0.681 threshold currently used in field surveillance programs is effective in identifying IAV-positive serum samples from feral swine.
Fig. 5.
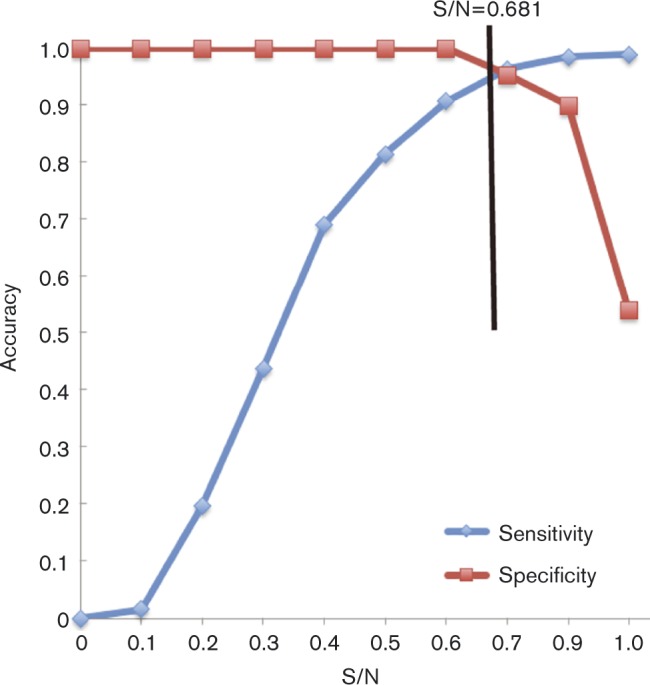
Receiver operating characteristic curves showing sensitivity and specificity of ELISAs for detection of IAV in experimentally infected feral swine and in sentinel swine exposed to infected swine. The threshold of 0.681 for the S/N was used by the US Department of Agriculture and this cut-off is indicated by a black straight line.
Discussion
Under laboratory conditions, enzootic swine-origin subtype H1N1, H3N2 and H1N2 IAVs have been shown to replicate in the respiratory tracts of domestic swine, and virus shedding lasts for 5–6 days (De Vleeschauwer et al., 2009a; Van Reeth et al., 2009). However, infection with avian-origin subtype H1N1, H4N1, H5N1 and H7N1 IAVs results in virus shedding for 3–6 days (De Vleeschauwer et al., 2009b). Furthermore, influenza A(H1N1)pdm09 virus infection in domestic swine can lead to virus shedding for up to 10 days (Bragstad et al., 2013). Thus, the duration of virus shedding in IAV-infected domestic swine depends on the virus subtype and strain. In this study, we showed that feral swine infected with the H3N2 IAV of feral swine origin can shed virus for up to 10 days.
After infection, no obvious clinical signs were observed in feral swine of either group; domestic swine infected with IAVs show only mild respiratory distress or, sometimes, no clinical signs (Bragstad et al., 2013; Ferrari et al., 2010). The virus titration results suggested low shedding in both treatment and sentinel swine. However, the high HI titres suggested that both groups were successfully infected with H3N2 virus. It should be noted that this H3N2 IAV replicated poorly in MDCK cells and thus virus titration in MDCK cells could generate biases in the virus shedding data.
Results showed that virus titres in the sentinel group were higher than those in the treatment group. This finding is consistent with results from another swine experiment with influenza strain A/swine/Flanders/1/98(H3N2) (De Vleeschauwer et al., 2009b). In that study, nasal shedding in inoculated pigs peaked at 1 day p.i. [titre of ∼105 EID50 (100 mg nasal secretions)–1, where EID50 is the 50 % egg infectious dose] and shedding continued for ∼7 days; however, nasal shedding in contact pigs peaked at 6 days p.e. [titre of 106 EID50 (100 mg nasal secretions)–1]. Other reports showed evidence of higher replication capability and/or pathogenesis by IAV after passage in a host, suggesting possible adaptation of the viruses to host species (Mase et al., 2006; Wei et al., 2014). Thus, it is likely that before being transmitted from treatment group swine to sentinel swine, the WT viruses acquired improved replication efficiency and infection capability during passage in the treatment group swine. Of note, the viruses inoculated into the treatment group swine had been passaged twice in MDCK cells. Furthermore, the data obtained from genomic sequencing identified a few changes between the isolates recovered from the sentinel swine and the WT viruses inoculated into the treatment swine: changes were noted in non-protein-coding sequences, and in amino acid and nucleotide sequences in the protein-coding sequences (Tables 1, S1 and S2). Notably, there was amino acid divergence in position 473 in the nucleoprotein. Amino acid at position 473 (in the peptide of 473–481) in the nucleoprotein was relatively conserved in subtype H1N1 and H3N2 IAVs, and was reported to stimulate the host to produce IFN-γ (Wahl et al., 2009). These findings are of interest because the non-coding regions and some coding regions of genome segments affect viral RNA synthesis and packing (Liang et al., 2005; Muster et al., 1991; Ng et al., 2008; Sun et al., 2015; Watanabe et al., 2003; Zheng et al., 1996). Further experiments are needed to validate the roles of sequence changes in the discrepancy seen in patterns of virus shedding and antibody response dynamics between the swine in the treatment and sentinel groups.
It is likely that seroconversion of sentinel swine occurred after they were exposed to contaminated fomites or aerosolized virus disseminated through the ventilation system. This supposition is supported by the fact that one sentinel swine began shedding virus at 1 day p.e., even though virus detection by qRT-PCR was negative for this swine until 5 days p.e. (Table 2). Although it is possible that infectious aerosols (e.g. those created during pen washing) or contact with infectious fomites could have led to virus transmission amongst swine (Allerson et al., 2013; Tellier, 2006), the sentinel and treatment groups were separated by 1.2 m, and our study protocol included procedures to avoid the unintended transmission of virus as a result of routine cleaning and feeding (e.g. sentinel group pens were cleaned before treatment group pens and the sentinel group was fed before the treatment group). Regardless, the findings in this study demonstrate that subtype H3N2 IAV can be easily transmitted amongst feral swine.
Isolation of enzootic subtype H3N2 and H1N1 IAVs from feral swine suggested that there have been multiple introductions of IAVs from domestic swine to feral swine (Bevins et al., 2014; Fogarty, 2007). It would be of interest to know whether these introduced H3N2 IAVs can be maintained in the feral swine population. In domestic swine, subtype H3N2 IAV can persist in the population through chronic infection of swine or periodic reintroduction of the virus (Kyriakis et al., 2013; Wallace, 1979). As feral swine can live ∼8 years (versus < 1 year for most domestic swine) and are free-ranging (unlike domestic swine), they have more opportunities to be exposed to other feral swine, domestic swine and ponds contaminated by avian influenza-infected wild birds, including migratory waterfowl. Our findings suggest that H3N2 virus of feral swine origin can be transmitted easily amongst feral swine. Thus, feral swine could facilitate antigenic drift or shift, which could lead to the generation of novel IAVs, further complicating our ability to elucidate the complex ecology of IAVs.
Swine in the treatment group seroconverted (HI titres ≥ 1 : 40) at 8 days p.i. and swine in the sentinel group seroconverted 3 days later. These results were similar to those in a study of domestic swine experimentally infected with H1 IAV, in which seroconversion occurred at 7–8 days p.i. (Lee et al., 1995; Panyasing et al., 2013). In addition, another study showed that antibody titres in IAV-inoculated domestic swine were lower than those in domestic swine infected through contact transmission (De Vleeschauwer et al., 2009b). Our finding that mean HI titres at different time points were higher in the sentinel group than in the treatment group were also consistent with findings in these previous studies. Unfortunately, we were unable to detect viruses in nasal wash samples from swine that were reinoculated with H3N2 IAV at 103 days p.i. because of a spike in HI antibody titre levels.
Although next-generation sequencing detected a few amino acid polymorphisms across the genomes of WT virus and virus isolates recovered from sentinel swine, it is still not clear whether these polymorphisms led to the discrepancy in antibody levels between sentinel and treated swine. Further experiments using IAV reassortants with a specific mutation are needed to identify any specific polymorphisms responsible for such a difference.
A previous study in feral swine documented a very low rate of virus recovery: amongst 1983 nasal swab samples from feral swine, nine were matrix gene qRT-PCR-positive and only one IAV strain was isolated (Feng et al., 2014). Our study demonstrated that nasal swab samples from feral swine provide adequate virus for the detection of IAV. The low rates of IAV detection and isolation in the previous study (Feng et al., 2014) were probably due to the short duration of virus shedding. Nasal swab samples from younger, immunologically naive feral swine would more likely to lead to a higher qRT-PCR detection and virus isolation success rate.
Comparison of virus titres for swine with only nasal swab samples and those with alternating nasal wash and nasal swab samples demonstrated that the virus titres in the former group were slightly higher than those in the latter group (Table 3). It is interesting that, once virus shedding was detected, the swine with only swab samples had consistent shedding patterns, whereas those with alternating nasal wash and nasal swab samples did not. This might have occurred because nasal washes likely cleared some viral particles from the nasal cavity.
Table 3. Virus titration results for nasal wash and nasal swab samples from feral swine after inoculation with influenza A/swine/Texas/A01104013/2012(H3N2) virus and from feral swine after exposure to infected swine.
| Group, swine no.* | Titre [log10(TCID50 ml− 1)]† (day p.i./p.e.) | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | |
| Inoculation | ||||||||||||||
| 2 | 2.199 | 2.199 | 0.699 | 3.699 | 2.199 | 2.199 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 3 | 1.699 | 2.366 | 0.699 | 0 | 3.032 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 4 | 0 | 0.699 | 0.699 | 0.699 | 3.199 | 0.699 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 6 | 0.699 | 0.699 | 0 | 0 | 2.199 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 7 | 0 | 0.699 | 0 | 0 | 2.366 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 8 | 0 | 0 | 0 | 2.366 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 11 | 0 | 0 | 2.366 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 12 | 0 | 2.032 | 1.699 | 1.699 | 2.032 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Sentinel | ||||||||||||||
| 1 | 0 | 0 | 0 | 0 | 0 | 0 | 2.93 | 5.199 | 4.032 | 2.866 | 0 | 0 | 0 | 0 |
| 5 | 1.699 | 0 | 2.032 | 1.98 | 4.032 | 4.032 | 3.199 | 5.032 | 2.468 | 0 | 0 | 0 | 0 | 0 |
| 9 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4.199 | 0 | 0 | 0 | 0 | 0 | 0 |
| 10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4.262 | 2.468 | 5.699 | 0 | 0 | 0 | 0 |
A feral swine index was used to randomly assign swine to the treatment group or the sentinel group. Swine in the treatment group and the sentinel group were housed in pens in the same barn (Fig. 1). The following swine were housed together: swine 7 and 8, swine 9 and 10, and swine 11 and 12.
Virus titres were measured in MDCK cells; the titres in italics were for nasal wash samples, and the others for nasal swab samples.
Serological analyses demonstrated that ELISAs are less sensitive than HI assays for the detection of IAV in feral swine. However, by optimizing the S/N threshold, both the sensitivity and specificity of ELISAs can be improved to >95 % with a S/N threshold of 0.681. With this threshold, the serological surveillance from 2009 to 2014 showed that ∼7.58 % of feral swine in the USA were positive for IAV, confirming the prevalence of IAV infection in feral swine. These findings suggest the need for continued surveillance to monitor the distribution, and the genomic and antigenic diversities of IAVs in feral swine populations.
Methods
Virus and cells
We used influenza A/swine/Texas/A01104013/2012(H3N2) virus, a WT strain of feral swine origin (Feng et al., 2014), to infect feral swine. MDCK cells (American Type Culture Collection) were maintained at 37 °C with 5 % CO2 in Dulbecco's modified Eagle's medium (Gibco-BRL) supplemented with 10 % FBS (Atlanta Biologicals) and penicillin/streptomycin (Gibco-BRL). Virus amplification was performed in MDCK cells at 37 °C with 5 % CO2 in Opti-MEM I Reduced Serum Medium (Life Technologies) supplemented with TPCK (l-1-tosylamido-2-phenylethyl chloromethyl ketone)-treated trypsin (1.5 μg ml− 1).
RNA extraction, PCR, qRT-PCR and genomic sequencing
Viral RNA was extracted by using an RNeasy Plus Mini kit (Qiagen). The full-length cDNA for eight influenza gene segments was amplified by using SuperScript One-Step RT-PCR (Invitrogen) with previously described primers (Hoffmann et al., 2001; Zhou et al., 2009). To address the possibility of mutations changing the growth phenotype of viruses, the full genome of isolates from sentinel swine and WT virus used for inoculation were amplified by using a method described previously (Zhou et al., 2009). Amplified viral DNA products were quantified by using a High Sensitivity DNA kit on an Agilent 2100 Bioanalyzer (Agilent Technologies). An equal amount of each sample was used with an Illumina Nextera DNA Sample Preparation kit (Illumina) to prepare a sequencing library. Library samples were further quantified, normalized and then pooled together. Pooled library samples were sequenced by using a MiSeq Reagent kit version 2 (500 cycles) on a MiSeq sequencer (Illumina) according to the sequencing protocol suggested by the manufacturer.
Feral swine
A total of 12 feral swine (body weight 16–22 kg) were trapped in a rural area of Starkville, MS, USA, by using corral traps constructed of 4.9 × 1.5 m utility panels with 10.2 cm2 mesh and steel T-posts. Animals were transported in an air-conditioned, enclosed trailer to the Wildlife Services, National Wildlife Research Center, Mississippi Field Station in Mississippi State, MS, USA. The captured swine were quarantined for 1 week, and tested for exposure to brucellosis, pseudorabies and IAV by ELISA; all test results were negative. The swine were then housed in 1.2 × 2.7 m pens with solid concrete floors in an enclosed, air-conditioned building (Fig. 1). Once a day, the swine were fed a 16 % crude protein commercial swine ration equal to 4 % of their body weight.
Animal experiments
The 12 feral swine were each assigned to one of two groups: a treatment group (n = 8) or a sentinel group (n = 4). Animals in the treatment group were housed in six adjoining pens (one or two swine per pen); swine in the sentinel group were housed in three adjoining pens (one or two swine per pen), separated from those in the treatment group by 1.2 m (Fig. 1).
Prior to virus inoculation and sampling, animals were anaesthetized by using a syringe pole to inject TKX (4.4 mg Telazol kg− 1, 2.2 mg ketamine kg− 1 and 2.2 mg xylazine kg− 1) (Ko et al., 1993) at 0.044 ml (kg body weight)− 1 behind the ear, in the loin or in the ham. Animals in the treatment group were intranasally inoculated with 106 TCID50 influenza A/swine/Texas/A01104013/2012(H3N2) in 1 ml volume (Fig. 1). To investigate whether feral swine with low IAV antibody titres could infect and shed homologous viruses, we reinoculated swine in the treatment group at 103 days p.i. and we intranasally inoculated swine in the sentinel group with 106 TCID50 (in 1 ml volume) of the same virus. We originally planned to administer these inoculations at 90 days p.i., but at that time, the US government had shut down all non-essential government work. Thus, we had to postpone the reinfection experiments until 103 days p.i., when we were permitted to continue our experimental activities.
Nasal swab samples were collected from half of the swine (four treatment and two sentinel swine) daily from 1 to 14 days p.i. and from 104 to 113 days p.i., and stored in 15 ml tubes with 2 ml PBS. In the remaining swine (four treatment and two sentinel swine), nasal swab or nasal wash samples were collected on alternating days from 1 to 14 days p.i. and from 104 to 113 days p.i. Nasal washes were performed in both nostrils by using 2 ml PBS. qRT-PCR was used to determine virus loads in nasal swab samples, and viruses in both nasal wash and nasal swab samples were titrated by TCID50. Blood samples (5 ml) were obtained from swine for serological assays at 0, 2, 5, 8, 11, 14, 17, 21, 28, 35, 42, 49, 56, 63, 70, 93, 103, 104, 107, 110, and 113 days p.i.
Two swine per day (one treatment and one sentinel) were euthanized on 104–106 days p.i. and necropsies were performed. Euthanasia was performed by administration of barbiturates solution [1 ml (4.5 kg body weight)–1] in fully anaesthetized swine [0.044 ml TKX (kg body weight)− 1]. The remaining six swine were euthanized at 113 days p.i. and necropsies were performed.
Swine were monitored daily for subjective signs of influenza infection (e.g. lethargy, nasal discharge, coughing and dyspnoea) and objective signs (e.g. body temperature) until 14 days p.i. Body weight was measured weekly.
Serological assays
To determine the antibody response dynamics in feral swine after IAV infection, we conducted a HI assay, using 0.5 % turkey red blood cells, as described previously (Sun et al., 2013). ELISAs were performed by using an AI MultiS-Screen Ab Test kit (IDEXX).
Growth kinetics of viruses in MDCK cells
To characterize the growth kinetics of influenza strain A/swine/Texas/A01104013/2012(H3N2), we inoculated MDCK cells with the virus at m.o.i. 0.001. After being incubated for 1 h at 37 °C, the inoculum was removed and the cells were washed three times in PBS. The cells were then incubated (37 °C in 5 % CO2) in Opti-MEM I (Gibco) containing TPCK-treated trypsin (1.5 μg ml− 1). At 12, 24, 48 and 72 h p.i., supernatant was collected from three of the 12 wells (1 ml per well) and then titrated, by TCID50, in MDCK cells. TCID50 was calculated by using the Reed–Muench method (Reed & Muench, 1938).
Data analyses
Student's t-test was used to determine whether serological responses in the treatment group differed from responses in sentinel swine. Specificity and sensitivity of ELISA kits were calculated. Results from HI assays were viewed as true positive and true negative. We similarly measured the specificity and sensitivity of the nasal swab sample-based qRT-PCR method for influenza diagnosis. Cell culture results (TCID50) for nasal swab samples were used as true positive and true negative. The data from the growth curve of the WT virus and the isolates recovered from the sentinel swine were analysed by using a one-way repeated-measure ANOVA followed by Turkey and Duncan's multiple comparison test.
The MiSeq sequence reads were matched to the reference genome by using the Bowtie2 alignment program (Langmead & Salzberg, 2012). Single nucleotide polymorphisms were analysed by using VarScan version 2.3.6 (Koboldt et al., 2012). A minimum of 10-fold mapping coverage and a quality score of 30 were required for a given nucleotide position. A minor variant was determined only if it was supported by at least 20 % of reads at a given position.
Biosafety protocols for laboratory and animal experiments
The laboratory analysis and inoculation trials in feral swine were carried out under Biosafety Level 2 conditions, with investigators wearing appropriate personal protective equipment, in compliance with the Institutional Animal Care and Use Committee of US Department of Agriculture-approved protocols, and Biosafety in Microbiological and Biomedical Laboratories, as well as Risk Group Classification for Infectious Agents.
Acknowledgements
This work was supported by the Wildlife Services, National Wildlife Research Center, US Department of Agriculture (13-7428-0961-CA and 14-7428-1041-CA), and partially funded by the National Institutes of Health (P20GM103646). Funding for virus sequencing was supported by the Global Emerging Infection Systems, a Division of the Armed Forces Health Surveillance Center. The opinions or assertions contained herein are the private views of the authors and are not to be construed as reflecting the official views of the US Army or the US Department of Defense.
Supplementary Data
Supplementary Data
References
- Alexander D.J., Brown I.H. (2000). Recent zoonoses caused by influenza A viruses Rev Sci Tech 19 197–225. [DOI] [PubMed] [Google Scholar]
- Allerson M.W., Cardona C.J., Torremorell M. (2013). Indirect transmission of influenza A virus between pig populations under two different biosecurity settings PLoS One 8 e67293 10.1371/journal.pone.0067293 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevins S.N., Pedersen K., Lutman M.W., Gidlewski T., DeLiberto T.J. (2014). Consequences associated with the recent expansion of nonnative feral swine Bioscience 64 291–299 10.1093/biosci/biu015 . [DOI] [Google Scholar]
- Bowman A.S., Nolting J.M., Nelson S.W., Slemons R.D. (2012). Subclinical influenza virus A infections in pigs exhibited at agricultural fairs, Ohio, USA, 2009–2011 Emerg Infect Dis 18 1945–1950 10.3201/eid1812.121116 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bragstad K., Vinner L., Hansen M.S., Nielsen J., Fomsgaard A. (2013). A polyvalent influenza A DNA vaccine induces heterologous immunity and protects pigs against pandemic A(H1N1)pdm09 virus infection Vaccine 31 2281–2288 10.1016/j.vaccine.2013.02.061 . [DOI] [PubMed] [Google Scholar]
- Choi Y.K., Nguyen T.D., Ozaki H., Webby R.J., Puthavathana P., Buranathal C., Chaisingh A., Auewarakul P., Hanh N.T., other authors (2005). Studies of H5N1 influenza virus infection of pigs by using viruses isolated in Vietnam and Thailand in 2004 J Virol 79 10821–10825 10.1128/JVI.79.16.10821-10825.2005 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clavijo A., Nikooienejad A., Esfahani M.S., Metz R.P., Schwartz S., Atashpaz-Gargari E., Deliberto T.J., Lutman M.W., Pedersen K., other authors (2012). Identification and analysis of the first 2009 pandemic H1N1 influenza virus from U.S. feral swine Zoonoses Public Health 60 327–335. [DOI] [PubMed] [Google Scholar]
- De Vleeschauwer A., Atanasova K., Van Borm S., van den Berg T., Rasmussen T.B., Uttenthal A., Van Reeth K. (2009a). Comparative pathogenesis of an avian H5N2 and a swine H1N1 influenza virus in pigs PLoS One 4 e6662 10.1371/journal.pone.0006662 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vleeschauwer A., Van Poucke S., Braeckmans D., Van Doorsselaere J., Van Reeth K. (2009b). Efficient transmission of swine-adapted but not wholly avian influenza viruses among pigs and from pigs to ferrets J Infect Dis 200 1884–1892 10.1086/648475 . [DOI] [PubMed] [Google Scholar]
- Feng Z., Baroch J.A., Long L.P., Xu Y., Cunningham F.L., Pedersen K., Lutman M.W., Schmit B.S., Bowman A.S., other authors (2014). Influenza A subtype H3 viruses in feral swine, United States, 2011–2012 Emerg Infect Dis 20 843–846 10.3201/eid2005.131578 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari M., Borghetti P., Foni E., Robotti C., Di Lecce R., Corradi A., Petrini S., Bottarelli E. (2010). Pathogenesis and subsequent cross-protection of influenza virus infection in pigs sustained by an H1N2 strain Zoonoses Public Health 57 273–280 10.1111/j.1863-2378.2009.01239.x . [DOI] [PubMed] [Google Scholar]
- Fogarty E. (2007). National distribution of and stakeholder attitudes toward feral hogs Thesis, Mississippi State University, Mississippi State, MS, USA..
- Guan Y., Shortridge K.F., Krauss S., Li P.H., Kawaoka Y., Webster R.G. (1996). Emergence of avian H1N1 influenza viruses in pigs in China J Virol 70 8041–8046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hause B.M., Oleson T.A., Bey R.F., Stine D.L., Simonson R.R. (2010). Antigenic categorization of contemporary H3N2 Swine influenza virus isolates using a high-throughput serum neutralization assay J Vet Diagn Invest 22 352–359 10.1177/104063871002200302 . [DOI] [PubMed] [Google Scholar]
- Hoffmann E., Stech J., Guan Y., Webster R.G., Perez D.R. (2001). Universal primer set for the full-length amplification of all influenza A viruses Arch Virol 146 2275–2289 10.1007/s007050170002 . [DOI] [PubMed] [Google Scholar]
- Keawcharoen J., Oraveerakul K., Kuiken T., Fouchier R.A., Amonsin A., Payungporn S., Noppornpanth S., Wattanodorn S., Theambooniers A., other authors (2004). Avian influenza H5N1 in tigers and leopards Emerg Infect Dis 10 2189–2191 10.3201/eid1012.040759 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kida H., Ito T., Yasuda J., Shimizu Y., Itakura C., Shortridge K.F., Kawaoka Y., Webster R.G. (1994). Potential for transmission of avian influenza viruses to pigs J Gen Virol 75 2183–2188 10.1099/0022-1317-75-9-2183 . [DOI] [PubMed] [Google Scholar]
- Ko J.C., Williams B.L., Smith V.L., McGrath C.J., Jacobson J.D. (1993). Comparison of Telazol Telazol-ketamine, Telazol-xylazine, and Telazol-ketamine-xylazine as chemical restraint and anesthetic induction combination in swine Lab Anim Sci 43 476–480. [PubMed] [Google Scholar]
- Koboldt D.C., Zhang Q., Larson D.E., Shen D., McLellan M.D., Lin L., Miller C.A., Mardis E.R., Ding L., Wilson R.K. (2012). VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing Genome Res 22 568–576 10.1101/gr.129684.111 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyriakis C.S., Rose N., Foni E., Maldonado J., Loeffen W.L., Madec F., Simon G., Van Reeth K. (2013). Influenza A virus infection dynamics in swine farms in Belgium, France, Italy and Spain, 2006–2008 Vet Microbiol 162 543–550 10.1016/j.vetmic.2012.11.014 . [DOI] [PubMed] [Google Scholar]
- Langmead B., Salzberg S.L. (2012). Fast gapped-read alignment with Bowtie 2 Nat Methods 9 357–359 10.1038/nmeth.1923 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B.W., Bey R.F., Baarsch M.J., Larson M.E. (1995). Class specific antibody response to influenza A H1N1 infection in swine Vet Microbiol 43 241–250 10.1016/0378-1135(94)00099-I . [DOI] [PubMed] [Google Scholar]
- Liang Y., Hong Y., Parslow T.G. (2005). cis-Acting packaging signals in the influenza virus PB1, PB2, and PA genomic RNA segments J Virol 79 10348–10355 10.1128/JVI.79.16.10348-10355.2005 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorusso A., Vincent A.L., Harland M.L., Alt D., Bayles D.O., Swenson S.L., Gramer M.R., Russell C.A., Smith D.J., other authors (2011). Genetic and antigenic characterization of H1 influenza viruses from United States swine from 2008 J Gen Virol 92 919–930 10.1099/vir.0.027557-0 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahy B. (1997). Influenza A virus (FLUA). In A Dictionary of Virology 2nd edn., pp. 170–171. Edited by Mahy B. W. J. San Diego, CA: Academic Press. [Google Scholar]
- Mase M., Tanimura N., Imada T., Okamatsu M., Tsukamoto K., Yamaguchi S. (2006). Recent H5N1 avian influenza A virus increases rapidly in virulence to mice after a single passage in mice J Gen Virol 87 3655–3659 10.1099/vir.0.81843-0 . [DOI] [PubMed] [Google Scholar]
- Muster T., Subbarao E.K., Enami M., Murphy B.R., Palese P. (1991). An influenza A virus containing influenza B virus 5′ and 3′ noncoding regions on the neuraminidase gene is attenuated in mice Proc Natl Acad Sci U S A 88 5177–5181 10.1073/pnas.88.12.5177 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson M.I., Vincent A.L., Kitikoon P., Holmes E.C., Gramer M.R. (2012). Evolution of novel reassortant A/H3N2 influenza viruses in North American swine and humans, 2009–2011 J Virol 86 8872–8878 10.1128/JVI.00259-12 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng S.S., Li O.T., Cheung T.K., Malik Peiris J.S., Poon L.L. (2008). Heterologous influenza vRNA segments with identical non-coding sequences stimulate viral RNA replication in trans Virol J 5 2 10.1186/1743-422X-5-2 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen C.W. (2002). The emergence of novel swine influenza viruses in North America Virus Res 85 199–210 10.1016/S0168-1702(02)00027-8 . [DOI] [PubMed] [Google Scholar]
- Olsen B., Munster V.J., Wallensten A., Waldenström J., Osterhaus A.D., Fouchier R.A. (2006a). Global patterns of influenza a virus in wild birds Science 312 384–388 10.1126/science.1122438 . [DOI] [PubMed] [Google Scholar]
- Olsen C.W., Karasin A.I., Carman S., Li Y., Bastien N., Ojkic D., Alves D., Charbonneau G., Henning B.M., other authors (2006b). Triple reassortant H3N2 influenza A viruses, Canada, 2005 Emerg Infect Dis 12 1132–1135 10.3201/eid1207.060268 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panyasing Y., Goodell C.K., Giménez-Lirola L., Kittawornrat A., Wang C., Schwartz K.J., Zimmerman J.J. (2013). Kinetics of influenza A virus nucleoprotein antibody (IgM, IgA, and IgG) in serum and oral fluid specimens from pigs infected under experimental conditions Vaccine 31 6210–6215 10.1016/j.vaccine.2013.10.040 . [DOI] [PubMed] [Google Scholar]
- Peiris J.S., Guan Y., Markwell D., Ghose P., Webster R.G., Shortridge K.F. (2001). Cocirculation of avian H9N2 and contemporary “human” H3N2 influenza A viruses in pigs in southeastern China: potential for genetic reassortment? J Virol 75 9679–9686 10.1128/JVI.75.20.9679-9686.2001 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiris J.S., de Jong M.D., Guan Y. (2007). Avian influenza virus (H5N1): a threat to human health Clin Microbiol Rev 20 243–267 10.1128/CMR.00037-06 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed L.J., Muench H. (1938). A simple method of estimating fifty percent endpoints Am J Epidemiol 27 5. [Google Scholar]
- Richt J.A., Lager K.M., Janke B.H., Woods R.D., Webster R.G., Webby R.J. (2003). Pathogenic and antigenic properties of phylogenetically distinct reassortant H3N2 swine influenza viruses cocirculating in the United States J Clin Microbiol 41 3198–3205 10.1128/JCM.41.7.3198-3205.2003 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholtissek C. (1994). Source for influenza pandemics Eur J Epidemiol 10 455–458 10.1007/BF01719674 . [DOI] [PubMed] [Google Scholar]
- Shu B., Garten R., Emery S., Balish A., Cooper L., Sessions W., Deyde V., Smith C., Berman L., other authors (2012). Genetic analysis and antigenic characterization of swine origin influenza viruses isolated from humans in the United States, 1990-2010 Virology 422 151–160 10.1016/j.virol.2011.10.016 . [DOI] [PubMed] [Google Scholar]
- Sun H., Jiao P., Jia B., Xu C., Wei L., Shan F., Luo K., Xin C., Zhang K., Liao M. (2011). Pathogenicity in quails and mice of H5N1 highly pathogenic avian influenza viruses isolated from ducks Vet Microbiol 152 258–265 10.1016/j.vetmic.2011.05.009 . [DOI] [PubMed] [Google Scholar]
- Sun H., Yang J., Zhang T., Long L.P., Jia K., Yang G., Webby R.J., Wan X.F. (2013). Using sequence data to infer the antigenicity of influenza virus MBio 4 e00230–e00213 10.1128/mBio.00230-13 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H., Cui P., Song Y., Qi Y., Li X., Qi W., Xu C., Jiao P., Liao M. (2015). PB2 segment promotes high-pathogenicity of H5N1 avian influenza viruses in mice Front Microbiol 6 73 10.3389/fmicb.2015.00073 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tellier R. (2006). Review of aerosol transmission of influenza A virus Emerg Infect Dis 12 1657–1662 10.3201/eid1211.060426 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Reeth K., Braeckmans D., Cox E., Van Borm S., van den Berg T., Goddeeris B., De Vleeschauwer A. (2009). Prior infection with an H1N1 swine influenza virus partially protects pigs against a low pathogenic H5N1 avian influenza virus Vaccine 27 6330–6339 10.1016/j.vaccine.2009.03.021 . [DOI] [PubMed] [Google Scholar]
- Vincent A.L., Lager K.M., Ma W., Lekcharoensuk P., Gramer M.R., Loiacono C., Richt J.A. (2006). Evaluation of hemagglutinin subtype 1 swine influenza viruses from the United States Vet Microbiol 118 212–222 10.1016/j.vetmic.2006.07.017 . [DOI] [PubMed] [Google Scholar]
- Vincent A.L., Ma W., Lager K.M., Janke B.H., Richt J.A. (2008). Swine influenza viruses a North American perspective Adv Virus Res 72 127–154 10.1016/S0065-3527(08)00403-X . [DOI] [PubMed] [Google Scholar]
- Vincent A.L., Ma W., Lager K.M., Gramer M.R., Richt J.A., Janke B.H. (2009). Characterization of a newly emerged genetic cluster of H1N1 and H1N2 swine influenza virus in the United States Virus Genes 39 176–185 10.1007/s11262-009-0386-6 . [DOI] [PubMed] [Google Scholar]
- Wahl A., Schafer F., Bardet W., Buchli R., Air G.M., Hildebrand W.H. (2009). HLA class I molecules consistently present internal influenza epitopes Proc Natl Acad Sci U S A 106 540–545 10.1073/pnas.0811271106 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace G.D. (1979). Natural history of influenza in swine in Hawaii: swine influenza virus (Hsw1N1) in herds not infected with lungworms Am J Vet Res 40 1159–1164. [PubMed] [Google Scholar]
- Watanabe T., Watanabe S., Noda T., Fujii Y., Kawaoka Y. (2003). Exploitation of nucleic acid packaging signals to generate a novel influenza virus-based vector stably expressing two foreign genes J Virol 77 10575–10583 10.1128/JVI.77.19.10575-10583.2003 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster R.G., Bean W.J., Gorman O.T., Chambers T.M., Kawaoka Y. (1992). Evolution and ecology of influenza A viruses Microbiol Rev 56 152–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei K., Sun H., Sun Z., Sun Y., Kong W., Pu J., Ma G., Yin Y., Yang H., other authors (2014). Influenza A virus acquires enhanced pathogenicity and transmissibility after serial passages in swine J Virol 88 11981–11994 10.1128/JVI.01679-14 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyckoff A.C., Henke S.E., Campbell T.A., Hewitt D.G., VerCauteren K.C. (2009). Feral swine contact with domestic swine: a serologic survey and assessment of potential for disease transmission J Wildl Dis 45 422–429 10.7589/0090-3558-45.2.422 . [DOI] [PubMed] [Google Scholar]
- Zhang G., Kong W., Qi W., Long L.P., Cao Z., Huang L., Qi H., Cao N., Wang W., other authors (2011). Identification of an H6N6 swine influenza virus in southern China Infect Genet Evol 11 1174–1177 10.1016/j.meegid.2011.02.023 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H., Palese P., García-Sastre A. (1996). Nonconserved nucleotides at the 3′ and 5′ ends of an influenza A virus RNA play an important role in viral RNA replication Virology 217 242–251 10.1006/viro.1996.0111 . [DOI] [PubMed] [Google Scholar]
- Zhou B., Donnelly M.E., Scholes D.T., St George K., Hatta M., Kawaoka Y., Wentworth D.E. (2009). Single-reaction genomic amplification accelerates sequencing and vaccine production for classical and Swine origin human influenza a viruses J Virol 83 10309–10313 10.1128/JVI.01109-09 . [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data