Abstract
Polycystic ovary syndrome (PCOS) is a complex disorder affecting approximately 5–10 percent of all women of reproductive age. It is a multi-factorial endocrine disorder, which demonstrates menstrual disturbance, infertility, anovulation, hirsutism, hyper androgenism and others. It has been indicated that differential expression of genes, genetic level variations, and other molecular alterations interplay in PCOS and are the target sites for clinical applications. Therefore, integrating the PCOS-associated genes along with its alteration and underpinning the underlying mechanism might definitely provide valuable information to understand the disease mechanism. We manually curated the information from 234 published literatures, including gene, molecular alteration, details of association, significance of association, ethnicity, age, drug, and other annotated summaries. PCOSDB is an online resource that brings comprehensive information about the disease, and the implication of various genes and its mechanism. We present the curated information from peer reviewed literatures, and organized the information at various levels including differentially expressed genes in PCOS, genetic variations such as polymorphisms, mutations causing PCOS across various ethnicities. We have covered both significant and non-significant associations along with conflicting studies. PCOSDB v1.0 contains 208 gene reports, 427 molecular alterations, and 46 phenotypes associated with PCOS
Background
Polycystic ovary syndrome (PCOS) is considered to be the leading causes of female subfertility and the most frequent endocrine problems in women of reproductive age [1]. PCOS is a complex disorder affecting approximately 5–10% of all women of reproductive age [2]. It is a multifactorial endocrine disorder, which demonstrates menstrual disturbance, infertility, anovulation, hirsutism, and hyperandrogenism [3]. PCOS is characterized by arrested follicular development prior to selection of a dominant follicle. The increase in the secretion of androgens by the ovaries and the adrenal glands is one of the pathological effects observed in PCOS [4]. PCOS is also associated with an increased risk of developing Type 2 diabetes, dyslipidemia, and cardiovascular diseases [5]. Women with PCOS are also at an increased risk of developing gestational diabetes, preterm birth (PTB) and likely to give birth to premature babies [7]. The etiology of the disease has been difficult to determine because of its hetero genousity. The cause of PCOS is still unclear; however, it has been observed that various environmental and genetic factors, such as genetic variations, differential regulation of genes, and affected pathways, may contribute to the pathogenesis of PCOS [5]. We have reviewed the association of differential regulation of genes at various levels, including genes that are upregulated and downregulated in PCOS and the associated effects of dysregulation of genes [6]. The detailed literature study revealed that the differential expression of genes involved in the androgen biosynthesis, angiogenesis, follicular development, and at different stages of the embryonic development, contributes to the various changes at the molecular level [7],[8], including the differential expression of genes and miRNAs in the PCOS and its serious effects,including endometrial receptivity, implantation failure, early pregnancy loss, PTB, insulin resistance, hyper androgenesim in women with PCOS [9],[10]. The genetic variations play an important role in the pathogenesis of PCOS across different ethnicities [11]. The detailed literature study revealed several genes and the genetic variations in PCOS and its critical effects,such as ovary failure, obesity [12], spontaneous abortion [13] and recurrent pregnancy loss [14]. The causal genetic variants were assembled at various levels,including mutation, single nucleotide polymorphism, etc., in PCOS and the associated phenotypic effects. Although several studies have been performed on PCOS, the information is dispersed in the literature, which is the most specific challenge for researchers. Hence, the need to have a comprehensive coverage of evidence-based information on PCOS-associated genes and its molecular mechanism becomes evident. At present, we do not have a database available in the public domain; other alternatives are more on the clinical trials [15], patient information on PCOS [16], pathways and networks [17]. Furthermore, literature-based information on PCOS genes with associated evidence to understand the underlying mechanism becomes crucial for better prognosis and treatment. Therefore,it is clear that integration of the PCOS genes along with literature support is of prime concern. Thus, we developed a database, called PCOSDB (Polycystic Ovary Syndrome Database), with the literature-based structured information of genes and its molecular alterations in PCOS condition. We populated the database with literature-driven information on several susceptible genes in PCOS condition, including significant and non-significant association of variations in PCOS, along with conflicting data has been covered in the database. We have underpinned the critical genetic variations in PCOS across different ethnicities and its associated effects,comprehensively in PCOSDB. The database would help in identifying the candidate genes or biomarkers in the disease condition. More than two hundreds of genes have been covered in PCOSDB. The gene identifiers are hyperlinked to external database, Entrez Gene; the references are linked to PubMed. The database is freely available at http://www.pcosdb.net
Methodology
Article Screening and Strategy:
‘PCOS’ or ‘Polycystic Ovary Syndrome’ AND ‘Gene’ AND ‘Mutation OR Polymorphism OR Variation OR SNP’ were used as keywords in PubMed Medline Database to search for the research papers. Around 1200 references were screened at the abstract level to segregate the false positive papers from the hit list. All potential published studies on candidate genes and PCOS were evaluated. The true positive papers were collected to perform the manual data curation process.
Data extraction:
Manual curation process was adopted to extract the information. All papers were read, and specific information on PCOS, associated genes, mechanism of association, details of the association, significance of association mentioned in the papers were carefully captured according to the authors’ interpretation of the results. Database organization and web interface: PCOSDB is built with Hypertext preprocessor program PHP (http://www.php.net/). The database tables are stored in MySQL Server relational database, a lightweight database management system. MySQL, PHP, and JavaScript technology were preferred as they are open source software. A simple and efficient search tool was developed using Ajax technology. A user-friendly web interface has been designed and implemented for ‘PCOSDB’, which provides interfaces to search, browse, retrieve, and visualize the information freely.
Utility:
The aim of PCOSDB is to provide reliable information on disease gene association. It is a unique catalogue of reliable manually curated database on experimentally associated information on molecular alterations in PCOS. It includes upto- date information on the genes, and all associated genetic variations, dysregulation of genes and miRNAs in PCOS condition.
PCOSDB Web Interface:
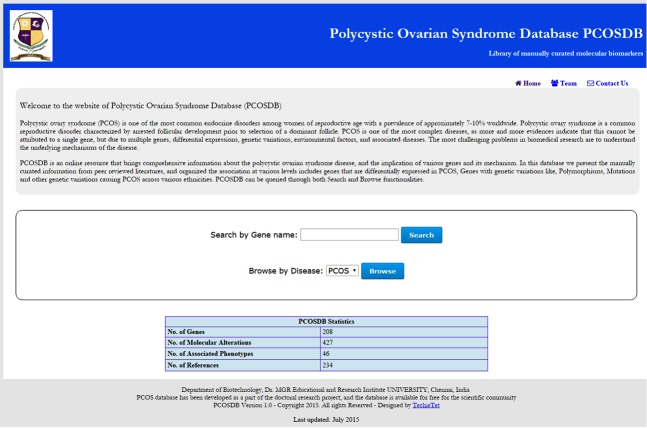
The PCOSDB portal is composed of a database and a web interface. The web interface supports searching and browsing of PCOS data (Figure 1). The web interface offers two entry points: 1. Search view: It allows the user to search a specific gene in the database using gene name or gene symbol. A dropdown menu appears with the potential list of genes, and the user can select the gene of interest. As a result, the user retrieves a gene report (or gene page), which will contain all information, as described in Table 1 along with the literature reference. 2. Browse view: It allows the user to explore the complete list of genes associated with PCOS (Figure 2). From the list of the genes, user can select the gene of interest and the respective gene report (Figure 3)can be accessed, i.e. the results are shown in the same way as when using the Search view. The gene report also provides links to external resources such as Entrez Gene (NCBI) and PubMed for references. Gene reports are accessed via both Search and Browse tool. Gene reports are represented as one page report, covers information about Gene and Disease.
Figure 1.
Web interface of the PCOSDB Basic home page displaying the search and browser tool with PCOSDB data statistics.
Table 1. PCOSDB data fields - a short description of the data fields along with examples.
| Data fields | Content Description | Example |
| GeneID | Entrez Gene identifier | Gene id: 367 |
| Gene Description | Full description of the Gene name | Androgen Receptor |
| Gene Symbol | Official Gene Symbol | AR |
| Gene Aliases | Synonyms and alternative names of the gene | RP11-383C12.1, AIS, DHTR, HUMARA, HYSP1, KD, NR3C4, SBMA, SMAX1, TFM |
| Chromosome Loci | Chromosome locus position | Xq12 |
| Species | Species information | Human |
| Disease | Primary disease name | PCOS, Polycystic Ovary Syndrome Disease, PCOD |
| Associated Diseases | Secondary and associated diseases | Androgen excess; infertility |
| Type of Association | Type of molecular alteration | Polymorphism; Gene expression |
| Details of Association | Details of the molecular alteration | Androgen receptor gene CAG trinucleotide repeats |
| Significance of Association | Significance of molecular alteration | Risk of PCOS development |
| Population | Studied population or ethnicity | Indian and Chinese, Australian, Caucasian |
| Drug Name | It contains drug name | none |
| Additional Information | Annotated comments briefly describing the experimental details or the author's conclusion or results described in the article | Androgens function through the X-linked androgen receptor (AR), studies based on the investigation of the AR encoded by an increasingly polymorphic CAG trinucleotide repeat tract in polycystic ovary syndrome revealed that there is an association between short CAG repeat length and the pathological process of polycystic ovaries in PCOS patients (PMID: 10999852) |
| Contradictory results | Negative correlation, contradictory information related to molecular alterations | A study was conducted to determine the relationship between CAG length variations in AR gene and polycystic ovary syndrome. The results revealed that the CAG length variations in AR gene was not associated with polycystic ovary syndrome (PMID: 23628801) |
| Reference | List of curated references specific to the gene report | Association of the CAG repeats polymorphisms in androgen receptor gene with polycystic ovary syndrome: a systemic review and meta-analysis. Gene. 2013 (PMID: 23628801) |
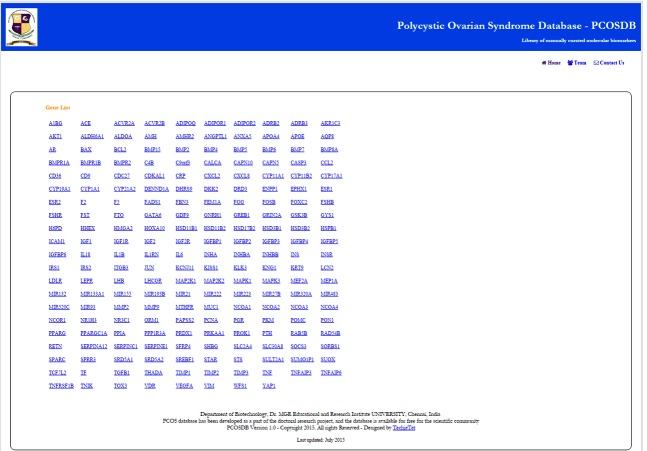
Figure 2.
Web interface of the PCOSDB browser view. List of genes available in the PCOSDB displayed.
Figure 3.
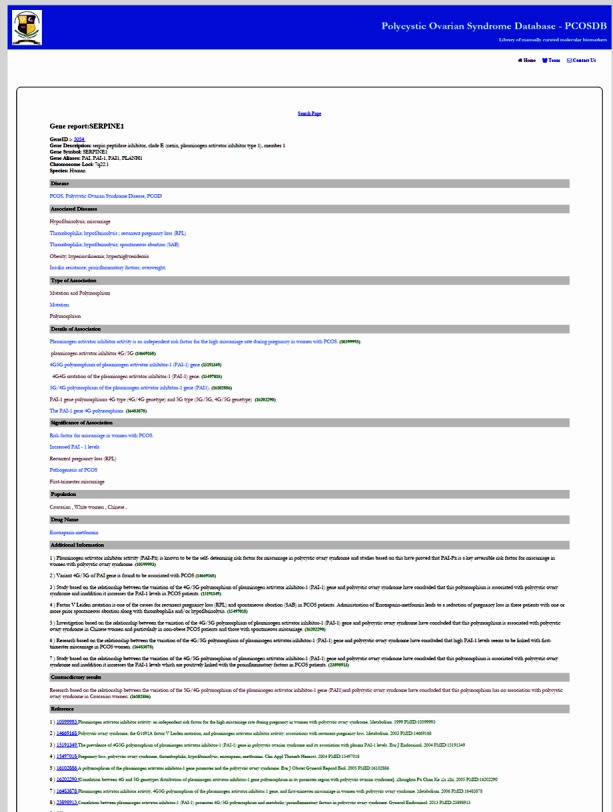
A detailed gene report.
Summary of the information currently available in PCOSDB
PCOSDB.v1 contains 208 PCOS-associated genes, 427 molecular alterations along with detailed annotations, 46 associated phenotypes, curated from 234 references.
Conclusion and future scope
PCOSDB has been developed as a new resource to help the scientific and medical community. Currently, PCOSDB provide useful targets or biomarkers relevant for clinical diagnosis. It helps in accelerating the research as it presents the underlying molecular mechanism of the disease, underpinning the targets. The database content is carefully maintained and updated. Repeated literature searches and curation are planned to allow for identification and periodic update of new data into the database. A module on the integration of UCSC genome browser for genome analysis is planned for future. We plan to streamline the search functionality by accommodating the search based on gene identifiers, disease name. We will also consider the inclusion of data for other related diseases, to broaden the scope of the database to a larger audience.
Competing Interest:
It should be noted that a concurrent database with similar interest is also available elsewhere [18]. Comparison of data between databases is of interest for further development and advancement.
Author’s contributions:
JM performed the research. DM conceived the study, VM assisted on data fields. JM constructed the database and website with the help of UV.
Footnotes
Citation:Jesintha Maryet al. Bioinformation 12(1): 4-8 (2016)
References
- 1.Boomsma CM, et al. Semin Reprod Med. 2008;26:72. doi: 10.1055/s-2007-992927. [DOI] [PubMed] [Google Scholar]
- 2.Dunaif A. Endocr Rev. 1997;18:774. doi: 10.1210/edrv.18.6.0318. [DOI] [PubMed] [Google Scholar]
- 3.Franks S, et al. Hum Reprod Update. 2008;14:367. doi: 10.1093/humupd/dmn015. [DOI] [PubMed] [Google Scholar]
- 4.Escobar-Morreale HF, et al. Trends Endocrinol Metab. 2007;18:266. doi: 10.1016/j.tem.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 5.Lindholm A, et al. Int J Gynaecol Obstet. 2008;102:39. doi: 10.1016/j.ijgo.2008.01.023. [DOI] [PubMed] [Google Scholar]
- 6.Jesintha Mary M, et al. Int J Pharm Bio Sci. 2015;6:893. [Google Scholar]
- 7.Mikola M, et al. Hum Reprod. 2001;16:226. doi: 10.1093/humrep/16.2.226. [DOI] [PubMed] [Google Scholar]
- 8.Jakimiuk AJ, et al. J Clin Endocrinol Metab. 1999;84:2414. doi: 10.1210/jcem.84.7.5863. [DOI] [PubMed] [Google Scholar]
- 9.Gregory CW, et al. J Clin Endocrinol Metab. 2002;87:2960. doi: 10.1210/jcem.87.6.8572. [DOI] [PubMed] [Google Scholar]
- 10.Cermik D, et al. J Clin Endocrinol Metab. 2003;88:238. doi: 10.1210/jc.2002-021072. [DOI] [PubMed] [Google Scholar]
- 11.Jesintha Mary M, et al. Asian J Pharm Clin Res. 2015;8:62. [Google Scholar]
- 12.Movahed Z, et al. Taiwan J Obstet Gynecol. 2015;54:280. doi: 10.1016/j.tjog.2014.05.007. [DOI] [PubMed] [Google Scholar]
- 13.Sun L, et al. J Endocrinol Invest. 2010;33:77. doi: 10.1007/BF03346557. [DOI] [PubMed] [Google Scholar]
- 14.Rogenhofer N, et al. Metabolism. 2013;62:1057. doi: 10.1016/j.metabol.2013.02.005. [DOI] [PubMed] [Google Scholar]
- 15. https://clinicaltrials.gov/ct2/show/record/NCT01338519/ Clinical Trials Database.
- 16. http://www.isrctn.com/ISRCTN70196169/ ISRCTN Registry.
- 17.Mohamed-Hussein ZA, Harun S. Theor Biol Med Model. 2009; 6:18. doi: 10.1186/1742-4682-6-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Joseph S, et al. Nucleic Acids Res. 2016;44(D1):D1032. doi: 10.1093/nar/gkv1146. [DOI] [PMC free article] [PubMed] [Google Scholar]