Abstract
Resection of DNA double-strand break (DSB) ends, which results in 3′ single-stranded tails, is an early event of DSB repair and can be a critical determinant in choice of repair pathways and eventual genome stability. Current techniques for examining resection are restricted to model in vivo systems with defined substrates (i.e., HO-endonuclease targets). We present here a robust assay that can analyze not only the resection of site-specific DSBs which typically have “clean” double-strand ends but also random “dirtyended” DSBs such as those generated by ionizing radiation and chemotherapeutic agents. The assay is based on our finding that yeast chromosomes with single-stranded DNA tails caused by resection are less mobile during pulsed-field gel electrophoresis (PFGE) than those without a tail. In combination with the use of a circular chromosome and enzymatic trimming of single-stranded DNA, resection of random DSBs can be easily detected and analyzed. This mobility-shift assay provides a unique opportunity to examine the mechanisms of resection, early events in DSB repair, as well as factors involved in pathway regulation.
Keywords: DNA, double-strand break repair, resection, pulsed-field gel electrophoresis (PFGE), ionizing radiation, HO endonuclease, I-SceI, mung bean nuclease, telomere
1. Introduction
DNA double-strand breaks (DSBs) are among the most lethal and destabilizing DNA lesions that cells can encounter. They are induced by a variety of factors including ionizing radiation (IR), chemotherapeutic agents, endogenously arising reactive oxygen species, errors during replication such as fork collapse, as well as processing of closely spaced single-strand lesions (1). Two major pathways have been identified to repair DSBs: non-homologous end joining (NHEJ) and homologous recombination (HR). DSB repair via the HR pathway is a multi-stage process using undamaged homologous DNA sequence as a template for accurate repair (2). An early step in this pathway involves resection of DSBs to produce 3′ single-stranded DNA tails that are critical for recombinational repair. The resected tails are utilized in strand invasion processes for priming repair synthesis and serve as a signal for checkpoint activation (3, 4).
Although long studied, mechanisms of resection have remained elusive, especially at the ends of random DSBs. To date, most studies on resection employ in vivo model systems with defined substrates such as DSBs induced by HO endonuclease or I-SceI endonuclease (5). In these studies, the nuclease recognition site is placed at a defined location, and the cut is induced by the expression of a site-specific endonuclease (6–8). A direct approach for addressing resection involves a combination of restriction site analysis and probes to specific sequences at different distances from the DSB. Loss of restriction sites due to resection diminishes Southern blot hybridization signal (9, 10). Resection at a defined DSB can also be detected by using denaturing alkaline gels. In this case, the loss of restriction sites due to resection results in the formation of higher molecular weight bands that could be detected by sequence-specific probes. Finally, formation of ssDNA resection intermediates can be detected by slot blots which take advantage of the ssDNA binding to positively charged nylon membranes (whereas dsDNA cannot bind). The amount of ssDNA formed is determined by hybridization with strand-specific probes (11).
Both HO and I-SceI recognize long nonpalindromic sequences and generate 4-bp staggered cuts with 3′-OH overhangs (12, 13). The DSB ends generated in this way are considered “clean” since they have 5′-P and 3′-OH groups suitable for ligation via end-joining processes or for priming DNA synthesis (14). However, most spontaneous or biologically relevant DSBs caused by environmental and therapeutic reagents such as IR, oxidative stress, and cancer drugs produce a variety of chemically modified termini or even protein–DNA adducts that cannot be directly ligated. These types of DSBs are referred to as “dirty” ends and require end processing by nucleases or other modifying enzymes to enable repair by HR or NHEJ (15). Analyzing the resection and repair of random “dirty” DSBs in vivo has been a challenge in the field. The appearance and repair of these types of DSBs can be determined qualitatively by the appearance of foci of proteins associated with DSB induction, such as H2AX chromatin modification, or foci appearing at various steps in repair (16, 17). However, there are few opportunities to address molecular events associated with random DSBs. Here we present a system capable of detecting resection at randomly induced DSBs as well as uncapped telomeres in addition to events at site-specific DSBs.
1.1. Large DNA Molecules with Single-Stranded Tail(s) Show Slower Mobility on PFGE
Pulsed-field gel electrophoresis (PFGE) is a widely used approach to monitor yeast chromosome changes since it permits very large DNA molecules to be resolved on agarose gels (for, e.g., see (18)). The system that we developed for the detection of resection is based on the finding that large chromosomal DNAs with single-stranded tails have significantly reduced mobility on PFGE. This mobility shift was observed in a study of the fate of radiation-induced DSBs in repair-deficient rad50, rad51, and rad52 mutants of the yeast Saccharomyces cerevisiae (19). The repair was assessed by monitoring the fragmentation and restitution of full-size yeast chromosomes in nocodazole-arrested G2/M haploid yeast. Unexpectedly, rad51 and rad52 mutants showed a decrease in mobility of the smear of the chromosome fragments, initially interpreted as representing a low level of repair. There was no such PFGE mobility shift in the rad50 mutant up to 4 h after irradiation. Further analysis that employed a circular chromosome and in vitro biochemical assays of the broken chromosome, as described below, demonstrated that the PFGE mobility change associated with the smear is due to the presence of single-stranded DNA (19).
1.2. Detecting Resection of Randomly Produced Single DSBs Using Circular Chromosome
The combination of PFGE along with an analysis of changes in circular chromosomes that have been broken provides the opportunity to study events at random DSBs (19). The use of a circular chromosome to detect a single DSB was initially developed in yeast by Game and colleagues (20). The principle of this method is that under most PFGE conditions a circular form of yeast chromosome is unable to move through the agarose matrix and is, therefore, retained in the loading well. However, the circle is converted into a full-length linear molecule by a single DSB, which enables the molecule to enter into the gel and give rise to a single band upon PFGE. The band is detectable either by Southern hybridization or by ethidium bromide. Since any single DSB on the circular chromosome leads to full-length linear DNA molecules of a uniform size, this approach provides the opportunity to address DSBs regardless of where they appear in a circular chromosome.
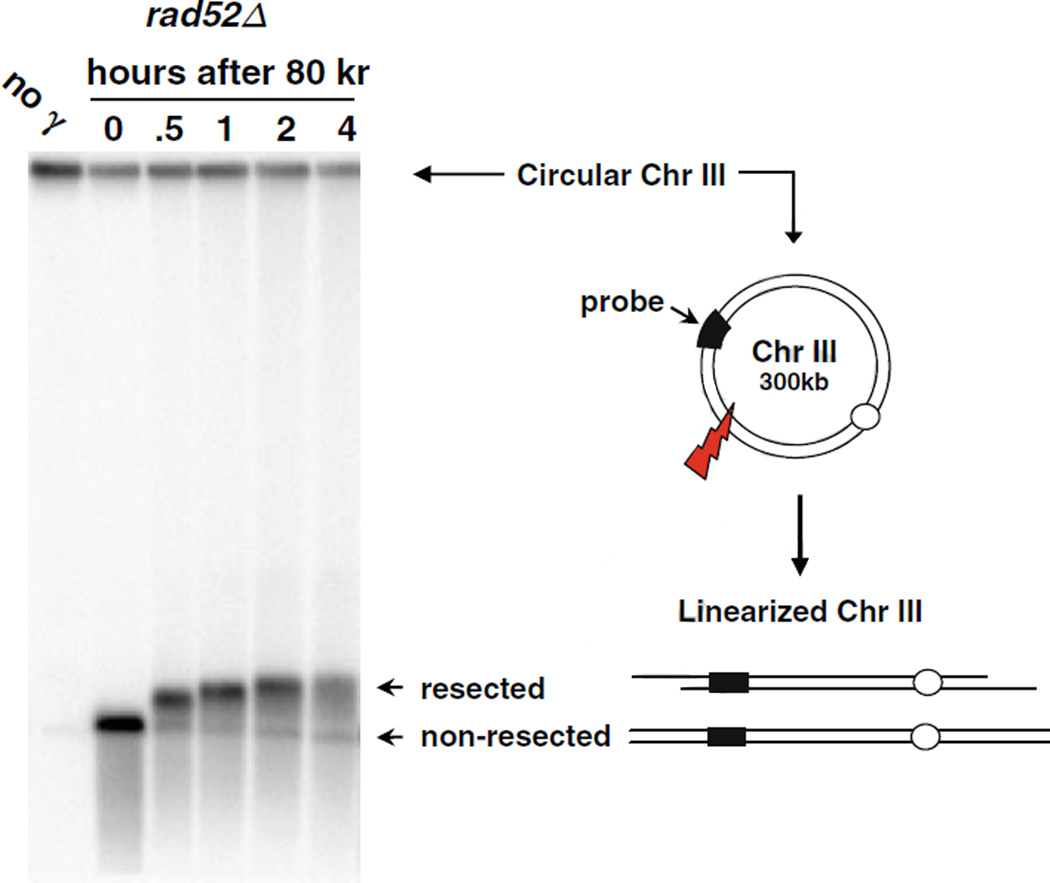
We recently found that the resection of IR-induced DSBs can be readily detected based on the shift in mobility of linearized circular chromosomes that have experienced a single DSB (19). Figure 2.1 shows the “PFGE-shift” of the corresponding linear band that is seen in samples taken at various times after γ-irradiation (IR) of a recombination-deficient rad52 mutant that is unable to repair DSBs. The yeast strain we constructed contained a circularized Chr III (~300 kb) (21). At “0” time after an 80 krad exposure an intense single band was detected (Fig. 2.1). The smear below this band corresponds to Chr III molecules with multiple DSBs. With time after post-irradiation incubation in YPDA, the DNA exhibited a shift that is clearly seen by 30 min after IR with further shift in PFGE mobility at 1 h reaching a plateau of ~430 kb apparent size by 4 h. We found that the increase in apparent size was actually due, paradoxically, to a loss in mass of the chromosomes due to resection, as described in the next section.
Fig. 2.1.
PFGE-shift of circular chromosomes broken by IR-induced random DSBs. Nocodazole-arrested G2/M rad52Δ cells were irradiated with 80 krads, and post-IR incubation was done in YPDA medium. Cells were collected and prepared at the indicated times. Plug preparation and CHEF parameters are described in text. Chr III was detected by a probe targeting the CHA1 gene. The circular (unbroken) form of Chr III is trapped in the well during PFGE. A single random DSB results in full-length linearized Chr III molecules that can migrate out of the well, forming a unique 300 kb band. The PFGE-shifted DNA corresponding to resected DNA reaches a plateau with “apparent” size of 430 kb. (This image is from 19.)
The resection is initiated uniformly and progresses at a comparable speed among the molecules examined based on the fairly sharp PFGE-shifted band at various times after irradiation. This also suggests that resection is not markedly affected by DNA sequence/structures. Nearly all the linearized molecules exhibited a shift by 1 h, independent of dose (19). The shift during post-irradiation incubation appears to occur even if the resected tail is a few hundred nucleotides based on the observation of shift in as little as 7.5 min after IR (19). The reasons for the shift remain to be established. The slower mobility of the resected DNA might be due to extension and contraction of single-stranded DNA (ssDNA) tails during PFGE providing stronger interactions than double-stranded DNA rods. It is also possible that secondary structure in the resected ssDNA contributes to its reduced migration.
This system based around PFGE-shift also has the potential to address the issue of whether resection of the two ends of the same DSB is coordinated or not. We found that the mobility shift of linear lambda DNA molecules with ssDNA tails generated in vitro at both ends moves much slower than molecules of the same length with ssDNA at only one end (19). This property provides a unique opportunity to address resection at both sides of a single randomly induced DSBs in circular molecules, where the two ends of the break are connected by the intervening intact DNA of the rest of the molecule. For example, for rad50 mutants exposed to low IR doses, multiple PFGE-shift bands are detected that appear to be due to one- and two-end events (19). Since DSBs induced in linear chromosomes would result in the two ends becoming separated (the two fragments each bounded by a telomere at one end), it has not been possible until now to address events at both sides of the same DSB.
1.3. Measuring Resection Length Using a ssDNA-Degrading Enzyme
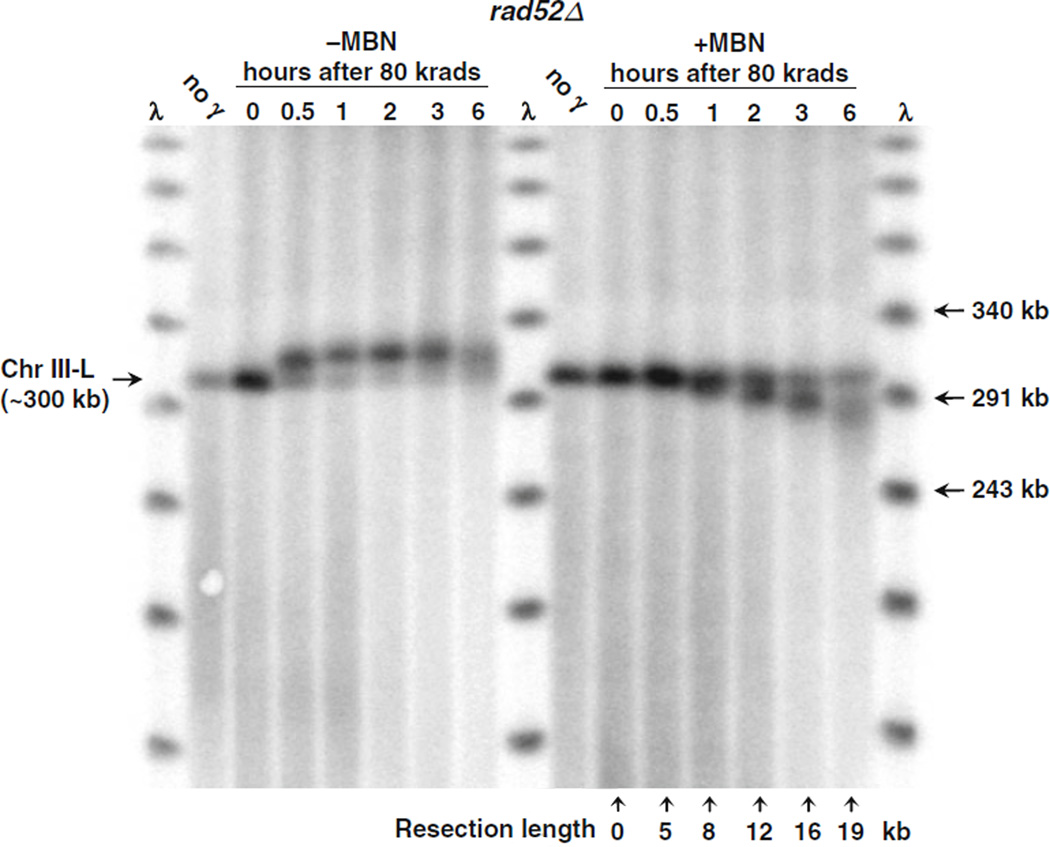
To establish that the PFGE-shift of the linearized molecules was due to resection, chromosomal DNA was treated with mung bean nuclease (MBN) in order to degrade the single-stranded tails. As shown in Fig. 2.2 (using DNA from IR-exposed rad52 cells), MBN treatment of the chromosomal DNAs within the plugs used for PFGE led to a reduction in the apparent MW of the Chr III linear molecules that showed PFGE-shift. This demonstrates that the PFGE-shift in radiation-broken chromosomes is due to the formation of ssDNA resulting from resection at the DSB ends. The mobility of the molecules at “0” time, when no resection is expected to occur, did not change with MBN treatment. The PFGE-shift in combination with MBN provides a sensitive method for measuring resection length and processing rate. In rad52 cells treated with 80 krads, the resection rate was ~2 kb/h per DSB end. The opportunity to follow resection of random DSBs makes it possible to characterize the roles of different genetic components in DSB repair, especially the initial stage which is critical for signaling and repair pathway regulation.
Fig. 2.2.
PFGE-shift DNA is due to resection, based on mung bean nuclease treatment which can also be used to quantitate resection length. PFGE plugs from an experiment involving 80 krads to rad52Δ cells and post-irradiation incubation (such as that described in Fig. 2.1) were treated with MBN (+MBN lanes) or without MBN (−MBN lanes) and run on a CHEF gel. Chromosome bands after Southern blotting were detected by probing for the LEU2 gene (see Note 4). The mung bean nuclease treatment (right half of image) abolished the PFGE-shift seen with untreated plugs (left half of image); the products ran at a faster rate than the unresected monomer in the 0 h lane. The numbers below each lane (right half of image) indicate the molecular weight change compared to the unresected linear Chr III band. The molecular weight of each band was calculated by comparing to positions identified in lambda DNA ladder (first and last lanes). (This image is from 19.)
1.4. Assessing Resection at Site-Specific DSBs and Telomeres
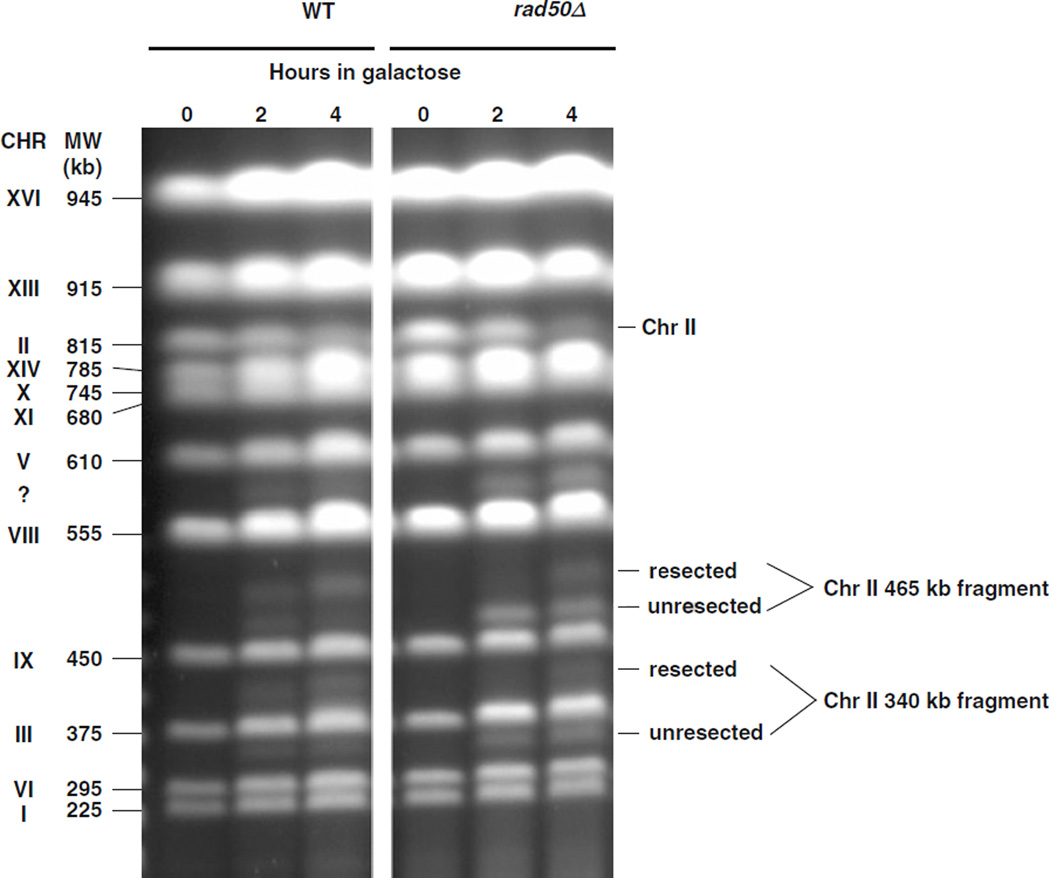
The resection-related PFGE-shift can be detected over a broad range of chromosome sizes that extends from tens of kilobytes (lambda DNA) to large chromosomes over 800 kb (e.g., yeast Chr II). The approach can also be employed to analyze resection at site-specific DSBs. We note that the induction of DSBs by ionizing radiation is “synchronous” in that they are induced simultaneously, unlike the enzymatically induced DSBs. Following induction of a single, DSB induced in a linear Chr III of G2/M yeast by HO endonuclease, we observed PFGE-shift with kinetics similar to those for IR-induced DSBs under somewhat different PFGE conditions (19). The results obtained with an I-SceI-induced DSB in Chr II (Nakai and Resnick, unpublished) using the PFGE procedures described here are presented in Fig. 2.3. Within 2 h after expression of I-SceI, the two expected fragments (340 and 465 kb, respectively) were observed with the wild-type and the rad50 null strains. PFGE-shift was detected in the ethidium bromide stained gels (and confirmed by Southerns) for most of the broken molecules of the WT strain, but for less than half of the molecules in the rad50 mutant.
Fig. 2.3.
PFGE-shift of chromosome fragments generated by an I-SceI site-specific break is detected on ethidium bromide stained gels. The galactose-inducible I-SceI endonuclease that cuts at a specific site engineered into Chr II was induced by transferring cells to galactose (see (23)). Samples were taken at 0, 2, and 4 h and analyzed by PFGE. The I-SceI site is on Chr II (815 kb) and cuts the DNA into 340 and 465 kb fragments. The efficiency of I-SceI cutting was 60% in WT and 80% in a rad50-null mutant at 4 h. Most of the fragments generated in WT cells within 4 h after transferring to galactose were shifted on PFGE (left image). However, for the rad50 mutant, less than half of the molecules were shifted (right half of image). These results are consistent with those described by Westmoreland et al. (19) using an HO endonuclease acting at a different site and demonstrate with the PFGE-shift approach a role for the MRX complex in resection. (We note that in these experiments an unidentified fragment appeared between 555 and 610 kb as shown by the symbol “?” The origin of this cryptic target remains to be determined but the site of cutting is likely highly related to the I-SceI site.) Experimental protocol: The experiment was performed at 30°C. Cells were grown overnight in YPDA medium, resuspended in YEP lactate medium (3.15% lactic acid, pH 5.5), and grown for an additional 18 h. The cells were then transferred to synthetic lactate medium (3.15% lactic acid, pH 5.5) containing 2% galactose. Cells were harvested at 0, 2, and 4 h and plugs were prepared for PFGE as described in the text.
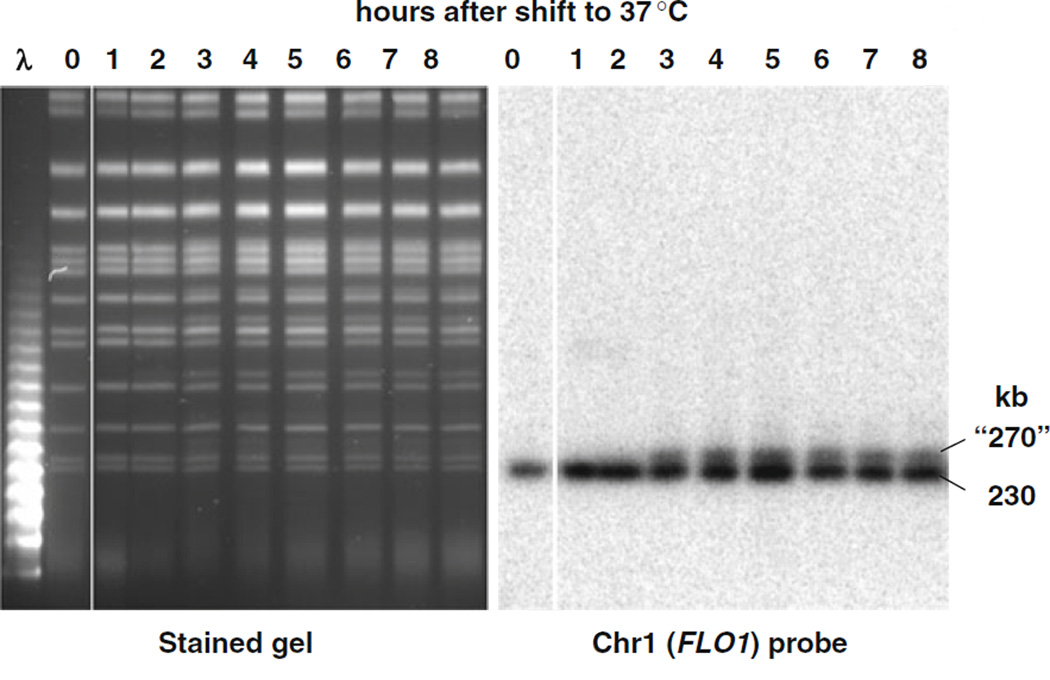
The PFGE-shift phenomenon can also be used to distinguish events at uncapped telomeres of individual chromosomes. Using the temperature-sensitive mutant cdc13-1, which is deficient in telomere capping, we detected resection of telomeres at elevated temperatures as shown in Fig. 2.4. These findings are consistent with those of Maringele and Lydall (22, 23) using a very different approach that involves quantitative amplification of ssDNA (QAOS). Upon PFGE analysis, many chromosomes appeared as doublets. Based on Southern hybridization of Chr I (Fig. 2.4) there was, in fact, a doublet consisting of the original chromosome (230 kb) and an apparently larger version (~270 kb). This shift is considered to be due to the telomeres of this mutant becoming uncapped at 37°C and subject to resection by the repair system that deals with DSBs. Southern analysis of other chromosomes revealed that most (except Chr IV) exhibited a PFGE-shift (19). This approach for detecting resection at telomeres is expected to provide a useful tool for addressing mechanisms that maintain telomeres as well as the impact on genome stability of altered telomere metabolism.
Fig. 2.4.
PFGE-shift of uncapped telomeres. A temperature-sensitive cdc13-1 strain, which is defective for telomere capping, was grown to stationary phase at permissive temperature, 23°C, then diluted 20-fold in fresh YPDA media at the nonpermissive temperature, 37°C, to induce telomere uncapping and subsequent 5′ to 3′ resection. (By 3 h, over 90% of the cells were arrested in G2.) Samples were collected at the indicated times following 37°C incubation. In the subsequent PFGE analysis, novel bands were observed at positions corresponding to molecular weights of ~40 kb above several of the chromosomal bands (left image). The shift in chromosomes was confirmed by Southern blot using a FLO1 probe which is specific to Chr I (right image). This image is from (19). Likewise, shifts in Chromosomes II (813 kb), III (340 kb), V (576 kb), and VIII (565 kb) were also confirmed using chromosome-specific probes (data not shown). PFGE-shifts were not detected for cells incubated at the permissive temperature (data not shown). Although the image shown was obtained with a Beckman Geneline II TAFE system (no longer commercially available), we also have similar unpublished results with cdc13-1 strains using CHEF. The TAFE running parameters were as follows: The first 18 h were run at constant current of 350 mA with 9 h of 60 s pulses, 3 h of 70 s pulses, 3 h of 80 s pulses, and 3 h of 90 s pulses. The remaining 6 h used 300 mA constant current and 4 min pulses.
2. Materials and Methods
2.1. Yeast Strains
All strains used here are haploids, although the approaches can be applied to diploid cells. Construction of strains containing circular Chr III (mwj49, mwj50, and derived yeast mutants) was described in (21). Construction of yeast strains for I-SceI-induced DSBs (KS406 and derived mutants) was described in (24). Construction of strains containing the cdc13-1 ts mutation (DAG760) was described in (25).
2.2. Media and Solutions
2.2.1. Media for Yeast Cultures
YPDA: 1% yeast extract, 2% Bacto Peptone, 2% dextrose, and 60 µg/ml adenine sulfate, autoclave.
YEP lactate: 1% yeast extract, 2% Bacto Peptone, 3.7% lactic acid (pH 5.5), and 60 µg/ml adenine sulfate, autoclave.
Nocodazole stock solution: 10 mg/ml dissolved in DMSO; store at −20°C.
2.2.2. Solutions for PFGE and Southern Blotting
Cell suspension buffer: 10 mM Tris (pH 8.0), 100 mM EDTA, and 2 mM NaCl.
2% low-melting agarose (LMP): 2% low-melting point agarose dissolved and melted in 10 mM Tris–HCl (pH 8.0), 100 mM EDTA.
Zymolyase: 1 mg/ml Zymolyase dissolved in 50% glycerol.
Agarose plug molds: see, for example, Bio-Rad, catalog no. 170–3622.
Proteinase K reaction buffer: 10 mM Tris (pH 8.0), 100 mM EDTA, 1.0% N-lauroyl sarcosine, 1 mg/ml proteinase K.
Plug washing buffer: 10 mMTris, 50mMEDTA (pH 8.0). 7. TBE 10X stock solution: 890 mM Tris base, 890 mM boric acid, 20 mM EDTA, pH 8.0.
TE buffer: 10 mM Tris, pH 7.4, 1 mM EDTA.
Mung bean nuclease (Promega, Madison, WI): stock solution 100 U/µl.
DNA detection: 10 mg/ml ethidium bromide solution or other DNA stains.
Southern blotting solutions. The following are used for Southern blotting: 0.25 N HCl; alkaline solution (0.4 N NaOH and 1.5 M NaCl); neutralizing buffer (0.5 M Tris–HCl and 1.5 M NaCl); 10× SSC (1.5 M NaCl, 0.15 M citrate, pH 7.0); Sigma PerfectHyb Plus hybridization buffer.
2.3. Probe to Detect Yeast Chromosome III
Chr III is detected by Southern blot with probes specifically targeting either the CHA1 gene or the LEU2 gene. The CHA1 probe size is 279 bp, and the following primer pairs were used to amplify this fragment:
CHA1-5′: AACGGCCGTGATCTCTAATC
CHA1-3′: TCCAACGCTTCTTCCAAGTC
The LEU2 probe size is 288 bp, and the following primer pairs were used to amplify:
LEU2-5′: TGTCAGAGAATTAGTGGGAGG
LEU2-3′: ATCATGGCGGCAGAATCAAT
2.4. Equipment and Other Materials
PFGE systems: transverse alternating field electrophoresis (TAFE) (Gene Line II apparatus from Beckman Instruments, Fullerton, CA, or equivalent) or contour-clamped homogeneous electric field (CHEF) (CHEFMapper XA system from Bio-Rad, Hercules, CA, or equivalent).
Southern blotting apparatus and materials: UV crosslinker (Stratagene Stratalinker or equivalent); nylon membrane (Hybond N+, GE Healthcare or equivalent); Stratagene Prime-It RmT Random Primer Labeling Kit; ProbeQuant G-25 or G-50 Micro Columns; hybridization oven and bottles (260 × 40 mm); Whatman 3MM filter paper.
3. Methods
3.1. Cell Culture and Yeast Preparation
3.1.1. G2 Yeast Cell-Cycle Synchronization by Nocodazole
Growth: cells are grown logarithmically under aerobic conditions in liquid YPDA medium at 30°C to a concentration of 5–20 × 106 cells/ml.
Arrest at G2/M with nocodazole: nocodazole is added to a final concentration of 20 µg/ml and an additional 10 µg/ml every 1 h. Cells are incubated for 3 h at 30°C. Most cells are arrested in G2/M as determined microscopically by the presence of large budded cells and verification using flow cytometry.
3.2. Pulsed-Field Gel Electrophoresis (PFGE)
3.2.1. Preparation of Agarose-Embedded DNA (DNA Plug)
Prepare 2% low-melting agarose and keep it warm in a 55°C heat block.
Centrifuge ~1.2 × 108 cells and resuspend in cell suspension buffer at a total volume of 120 µl; add 20 µl Zymolyase (1 µg/µl), vortex and warm up to ~40–50°C using a heat block. Zymolyase should be added immediately prior to imbedding the cells in agarose (see Note 1).
Add 60 µl 2% agarose, quickly mix by gentle but thorough vortexing. Transfer the mixture to plug molds using sterile transfer pipettes (two plugs). Allow the agarose to solidify at room temperature or, to expedite this process, place the molds at 4°C for 10–15 min. (Note: this results in ~6 × 107 G2-arrested cells per 100 µl plug, which is the amount normally used in our experiments.)
Push the solidified agarose plugs into cell suspension buffer in a container such as multi-well tissue culture plate or conical centrifuge tube. Using ~1 ml for two plugs, incubate at 37°C for 1–2 h.
Remove cell suspension buffer and add 1 ml of Proteinase K reaction buffer for two plugs. Incubate the plugs overnight at 37°C without agitation.
Wash the plugs three to four times with plug washing buffer, 1 h for each wash at room temperature with gentle agitation.
Store plugs at 4°C. Depending on the type of DNA lesions induced, the plugs should be stable for a few weeks.
3.2.2. PFGE to Separate Yeast Chromosomes
The following protocol is for the preparation of a CHEF gel. The preparation of TAFE gels is similar and the running parameters for TAFE are provided in Fig. 2.4.
Preparation of gel casting stand with removable end plates (comes with the CHEF Mapper system) and comb. We found that a 3 mm thick preparative well comb (i.e., no teeth) is convenient for placing and organizing plugs during loading.
Melt 1% LE agarose (Seakem, Rockland, ME) in 0.5× TBE and pour into casting stand. While gel is solidifying, prepare 2.2 l 0.5× TBE running buffer and put into CHEF apparatus tank; cool to 14°C.
Take the DNA-containing agarose plug out of buffer; use a clean razor blade to cut out 1/4–1/2 size pieces (a thickness of ~2 mm); load into the bottom of a preparative well. Seal the well containing the plugs using 1% agarose and allow to set ~30 min at room temperature.
Install the gel from the casting stand into the PFGE electrophoresis tank according to CHEF Mapper instructions. Make sure the gel is not able to move or float during the electrophoresis. Equilibrate the gel placed in the tank with 14°C gel running buffer for 10 min before starting electrophoresis.
Run CHEF gel with appropriate conditions to separate the target DNA. For example, the following conditions can be used to separate all yeast chromosomes: 6 V/cm (120 V in the CHEF or DRII Bio-Rad units) at 14°C, 120° switch angle, switch time is ramped from 10 to 90 s over the 24 h run time.
3.3. Southern Blot and Hybridization
After electrophoresis, stain the gel for 60 min to overnight in 0.5× TBE with 1 µg/ml ethidium bromide. Destain in 0.5× TBE for 2–3 h and photograph the gel.
Rinse the gel briefly with water; add 0.25 N HCl to the tray containing the CHEF gel and gently shake for1 45–60 min.
Rinse gel briefly with water; treat with the alkaline solution for 30–60 min.
Neutralize with neutralization buffer for 30 min.
Cut a Hybond N+ membrane, wet first in water, and then soak for 10–15 min in 10× SSC.
Select a suitable method for Southern transfer. For example, use capillary method or a vacuum blotter according to the manufacturer’s instructions.
Rinse the membrane with 10× SSC. Dry the membrane or UV-crosslink with Stratalinker (120 mJ/cm2). Clearly mark the DNA side and top of the membrane.
Before hybridization, wet the membrane with 10× SSC and place it into a hybridization bottle.
Prehybridization: pour 15–20 ml of hybridization solution (e.g., PerfectHyb from Sigma) into the bottle (260 × 40 mm), add 10 µl of denatured salmon sperm DNA (10 mg/ml) per ml of hybridization buffer, and rotate at 68°C for 1 h in a hybridization oven.
Prepare radioactively labeled probe during prehybridization, using 50–100 ng of template DNA (preparation described in 3.4.1). 32P-labeled double-stranded DNA probe can be prepared by random priming using an appropriate commercial kit according to the manufacturer’s instructions (e.g., Stratagene Prime-It RmT Random Primer Labeling Kit). Purify the radiolabeled probe using a gel filtration spin column (e.g., ProbeQuant G-50 or G-25 Micro Columns).
Denature the probe at 100°C for 10 min and quickly cool down in ice. Add denatured probe directly to the hybridization bottle with prehybridization solution. (No need to replace with fresh hybridization solution.) Rotate hybridization bottle at 68°C overnight (16–24 h).
Cold washes: discard the hybridization solution, put the membrane into a tray, add 300–400 ml of 2× SSC/0.1% SDS, and shake at ambient temperature for 30 min.
Stringent washes: add 400 ml of pre-warmed (68°C) 0.1× SSC/0.1% SDS into the tray, shake at 68°C for 20 min, two to three washes.
Wrap the blot with plastic wrap and expose to phosphor screen or film for 1–2 days.
3.4. Detection of DSBs and Resection by PFGE
3.4.1. Detection of Random DSBs Using Circularized Chromosome
Chr III is detected by Southern blot with probes that specifically target either the CHA1 gene or the LEU2 gene.
Amplify the CHA1 or LEU2 sequence by PCR using yeast genomic DNA and purify by agarose gel electrophoresis with an appropriate gel extraction kit. Use the purified DNA as template for secondary PCR with the same primers to prepare a large amount of probe for long-term use. Purify the second PCR product by gel extraction or PCR purification methods; dissolve in TE and store at −20°C.
After Southern transfer of DNA materials onto membrane, use the CHA1 or LEU2 probe for hybridization at a concentration of 5–10 ng probe/ml hybridization buffer (50–100 ng/hybridization tube). Autoradiographs can be analyzed by specific software such as Carestream MI.
3.4.2. Detection of Resection at Single, Random DSBs in Circularized Chromosomes by PFGE-Shift
The following protocol for DSB induction and repair is derived from studies that employed ionizing radiation. The method can be modified to detect random DSBs generated by other sources causing DNA damage such as chemotherapeutic reagents.
Harvest nocodazole-arrested G2 yeast by centrifugation (2,000×g, 2 min), wash once with water, and resuspend in ice-cold water at 5–10 × 107 cells/ml. Save 1.2 × 108 cells (for two DNA plugs as described below) to be used as the unirradiated control for PFGE.
Cell suspensions are kept on ice throughout the entire irradiation process. Irradiate cells at desired doses (we typically use 5–80 krads with a 137Cs irradiator (J. L. Shepherd Model 431, 2.3 krads/min)) in plastic 50 ml tubes and vortex well every 10 krads exposure to assure good aeration.
Following irradiation, collect a volume corresponding to 1.2 × 108 cells, centrifuge and resuspend the pellet in icecold cell suspension buffer. These cells represent the time point “0” of DSB repair.
To address events during post-irradiation incubation, centrifuge the remaining cells and resuspend in YPDA with nocodazole (final concentration is 5–10 × 106 cells/ml) and incubate at 30°C with shaking. Since nocodazole is unstable in aqueous solution, add 10 µg/ml nocodazole every hour during incubation to maintain cells in G2/M.
At designated post-irradiation time points such as 30 min, 1 h, 2 h, collect cells to assess repair events. Cells (1.2 × 108 for two DNA plugs) are centrifuged and resuspended in ice-cold cell suspension buffer for DNA plug preparation as described above.
Run CHEF gel. The following parameters (or modify to other suitable parameters) can be used to detect DSBs (linear Chr III) and resected chromosomes (shifted band of linear Chr III): 6 V/cm, switch angle 120°, switch time 10–90 s with linear ramp, 24 h run time at 14°C with buffer recirculation (See Note 2).
Southern blot and hybridize with CHA1 or LEU2 probe.
3.4.3. Detection of Resection at Site-Specific DSBs Using PFGE-Shift
Procedures similar to those described above can be used to follow events at a site-specific DSB produced by galactose-induced HO endonuclease (19) or I-SceI (Nakai and Resnick, unpublished, also see Fig. 2.3).
3.4.4. Detection of Resection at Telomeres Using PFGE-Shift
The following is an example of how to detect resection associated with unstable telomere ends using the temperature-sensitive yeast mutant cdc13-1 which is defective for telomere capping (DAG760 described in (25)).
Grow cdc13-1 cells to stationary phase for 3 days in YPDA medium at the permissive temperature, 23°C.
Dilute 20-fold into fresh YPDA and incubate at 37°C, a condition resulting in 5′ to 3′ resection at telomeres (22). Within 3 h, greater than 90% of the cells are arrested in G2.
Collect cells at different time points after shifting to 37°C along with control cells kept at 23°C. The cells are processed for PFGE and Southern blot analysis as described above.
3.4.5. Mung Bean Nuclease Digestion of DNA in PFGE Plugs to Identify Resection and Determine Length
Mung bean nuclease can be used to measure resection length. It removes the single-stranded resected ends that develop at DSBs. The nuclease generates blunt ends resulting in linear chromosomal DNAs with reduced length as exhibited by greater PFGE mobility. The resection length can be determined by comparing length after MBN treatment with the length at the time of DSB induction.
For MBN digestion of yeast plugs, cut the plug in half (50 µl) and put into a 96-well multi-well plate. Plugs are equilibrated with three changes (20 min) of 150 µl of TE at room temperature. The other half of the plug is used as a non-MBN control.
Remove the TE buffer and incubate with 40 U/ml of MBN in 150 µl of MBN reaction buffer for 20 min at room temperature with gentle shaking (see Note 3).
Quickly remove the MBN reaction solution and wash four times with ice-cold 50 mM EDTA to stop the reaction.
Preparation of CHEF gel, for sufficient separation of DNA at ~300 kb range, a long gel is preferred (using the 14 cm (width) × 21 cm (length) casting stand from Bio-Rad, see Note 2). Load plugs (with and without MBN treatment) into the CHEF gel, using lambda DNA as the length marker. Run the gel with the following parameters: 6 V/cm (120 V in the CHEF and DRII Bio-Rad apparatuses), 120° switch angle, switch time 6–36 s, linear ramp, 48 h run time at 14°C with buffer recirculation.
Southern blot and hybridize with LEU2 probe (see Note 4). The molecular weights associated with bands can be calculated using Kodak MI (version 4.0) software (Eastman Kodak Co., Rochester, NY) by comparing with positions in marker bands (lambda DNA ladder; New England Biolabs, Beverly, MA).
Acknowledgments
This work was supported by the Intramural Research Program of the NIEHS (NIH, DHHS) under project 1 Z01 ES065073 (MAR).
Footnotes
During plug preparation, the cell suspension after adding Zymolyase should be kept at 40–50°C as briefly as possible in order to minimize inactivation of enzymatic activity and avoid possible damage to DNA.
The 14 cm long by 21 cm wide gel can be used to visualize the PFGE-shift caused by resection. But for measuring resection length with mung bean nuclease, a 21 cm long gel should be used.
Mung bean nuclease should be used at low concentration (40 U/ml) and <30 min incubation to minimize the generation of nonspecific DSBs. The small amount of nonspecific activity at this low concentration does not interfere with the measurement of resection.
For measuring resection length, it is important to include appropriate DNA size standards on the same gel. In general, one needs to use two probes to visualize both the size marker and the chromosome in the autoradiograph of the Southern blot. We have found under our conditions of LEU2 gene amplification, both the lambda DNA ladder (from NEB) and Chromosome III were detectable (see Fig. 2.2).
References
- 1.Ma W, Panduri V, Sterling JF, Van Houten B, Gordenin DA, Resnick MA. The transition of closely opposed lesions to double-strand breaks during long-patch base excision repair is prevented by the coordinated action of DNA polymerase delta and Rad27/Fen1. Mol Cell Biol. 2009;29:1212–1221. doi: 10.1128/MCB.01499-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Krogh BO, Symington LS. Recombination proteins in yeast. Annu Rev Genet. 2004;38:233–271. doi: 10.1146/annurev.genet.38.072902.091500. [DOI] [PubMed] [Google Scholar]
- 3.Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
- 4.Haber JE. Genome Dynamics and Stability. Vol. 3. Berlin/Heidelberg: Springer; 2008. Evolution of models of homologous recombination; pp. 1–64. [Google Scholar]
- 5.Mimitou EP, Symington LS. DNA end resection: many nucleases make light work. DNA Repair (Amst) 2009;8:983–995. doi: 10.1016/j.dnarep.2009.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kramer KM, Brock JA, Bloom K, Moore JK, Haber JE. Two different types of double-strand breaks in Saccharomyces cerevisiae are repaired by similar RAD52-independent, nonhomologous recombination events. Mol Cell Biol. 1994;14:1293–1301. doi: 10.1128/mcb.14.2.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Plessis A, Perrin A, Haber JE, Dujon B. Site-specific recombination determined by I-SceI, a mitochondrial group I intron-encoded endonuclease expressed in the yeast nucleus. Genetics. 1992;130:451–460. doi: 10.1093/genetics/130.3.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haber JE. Transpositions and translocations induced by site-specific double-strand breaks in budding yeast. DNA Repair (Amst) 2006;5:998–1009. doi: 10.1016/j.dnarep.2006.05.025. [DOI] [PubMed] [Google Scholar]
- 9.Ira G, Pellicioli A, Balijja A, Wang X, Fiorani S, Carotenuto W, Liberi G, Bressan D, Wan L, Hollingsworth NM, et al. DNA end resection, homologous recombination and DNA damage checkpoint activation require CDK1. Nature. 2004;431:1011–1017. doi: 10.1038/nature02964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhu Z, Chung WH, Shim EY, Lee SE, Ira G. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell. 2008;134:981–994. doi: 10.1016/j.cell.2008.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sugawara N, Haber JE. Repair of DNA double strand breaks: in vivo biochemistry. Methods Enzymol. 2006;408:416–429. doi: 10.1016/S0076-6879(06)08026-8. [DOI] [PubMed] [Google Scholar]
- 12.Colleaux L, D’Auriol L, Galibert F, Dujon B. Recognition and cleavage site of the intron-encoded omega transposase. Proc Natl Acad Sci USA. 1988;85:6022–6026. doi: 10.1073/pnas.85.16.6022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kostriken R, Heffron F. The product of the HO gene is a nuclease: purification and characterization of the enzyme. Cold Spring Harb Symp Quant Biol. 1984;49:89–96. doi: 10.1101/sqb.1984.049.01.012. [DOI] [PubMed] [Google Scholar]
- 14.Daley JM, Palmbos PL, Wu D, Wilson TE. Nonhomologous end joining in yeast. Annu Rev Genet. 2005;39:431–451. doi: 10.1146/annurev.genet.39.073003.113340. [DOI] [PubMed] [Google Scholar]
- 15.Wyman C, Kanaar R. DNA double-strand break repair: all’s well that ends well. Annu Rev Genet. 2006;40:363–383. doi: 10.1146/annurev.genet.40.110405.090451. [DOI] [PubMed] [Google Scholar]
- 16.Lisby M, Barlow JH, Burgess RC, Rothstein R. Choreography of the DNA damage response: spatiotemporal relationships among checkpoint and repair proteins. Cell. 2004;118:699–713. doi: 10.1016/j.cell.2004.08.015. [DOI] [PubMed] [Google Scholar]
- 17.Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem. 1998;273:5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- 18.Argueso JL, Westmoreland J, Mieczkowski PA, Gawel M, Petes TD, Resnick MA. Double-strand breaks associated with repetitive DNA can reshape the genome. Proc Natl Acad Sci USA. 2008;105:11845–11850. doi: 10.1073/pnas.0804529105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Westmoreland J, Ma W, Yan Y, Van Hulle K, Malkova A, Resnick MA. RAD50 is required for efficient initiation of resection and recombinational repair at random, gamma-induced double-strand break ends. PLoS Genet. 2009;5:e1000656. doi: 10.1371/journal.pgen.1000656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Game JC, Sitney KC, Cook VE, Mortimer RK. Use of a ring chromosome and pulsed-field gels to study interhomolog recombination, double-strand DNA breaks and sister-chromatid exchange in yeast. Genetics. 1989;123:695–713. doi: 10.1093/genetics/123.4.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ma W, Resnick MA, Gordenin DA. Apn1 and Apn2 endonucleases prevent accumulation of repair-associated DNA breaks in budding yeast as revealed by direct chromosomal analysis. Nucleic Acids Res. 2008;36:1836–1846. doi: 10.1093/nar/gkm1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maringele L, Lydall D. EXO1-dependent single-stranded DNA at telomeres activates subsets of DNA damage and spindle checkpoint pathways in budding yeast yku70Delta mutants. Genes Dev. 2002;16:1919–1933. doi: 10.1101/gad.225102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zubko MK, Maringele L, Foster SS, Lydall D. Detecting repair intermediates in vivo: effects of DNA damage response genes on single-stranded DNA accumulation at uncapped telomeres in budding yeast. Methods Enzymol. 2006;409:285–300. doi: 10.1016/S0076-6879(05)09016-6. [DOI] [PubMed] [Google Scholar]
- 24.Lobachev K, Vitriol E, Stemple J, Resnick MA, Bloom K. Chromosome fragmentation after induction of a double-strand break is an active process prevented by the RMX repair complex. Curr Biol. 2004;14:2107–2112. doi: 10.1016/j.cub.2004.11.051. [DOI] [PubMed] [Google Scholar]
- 25.Yang Y, Sterling J, Storici F, Resnick MA, Gordenin DA. Hyper-mutability of damaged single-strand DNA formed at double-strand breaks and uncapped telomeres in yeast Saccharomyces cerevisiae. PLoS Genet. 2008;4:e1000264. doi: 10.1371/journal.pgen.1000264. [DOI] [PMC free article] [PubMed] [Google Scholar]