Abstract
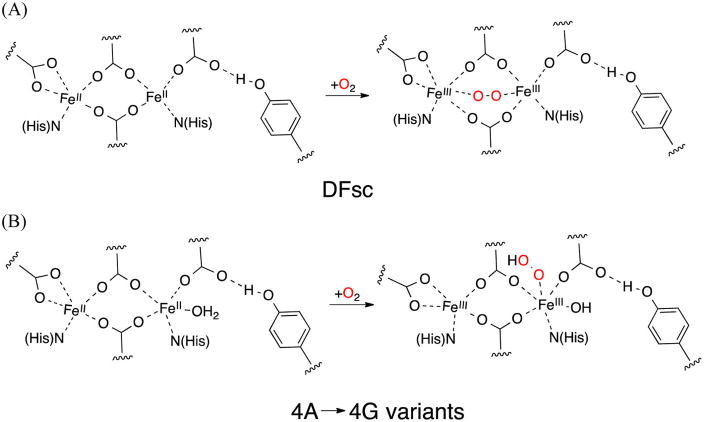
DFsc (single-chain due ferri) proteins allow for modeling binuclear non-heme iron enzymes with a similar fold. Three 4A → 4G variants of DFsc were studied to investigate the effects of (1) increasing the size of the substrate/solvent access channel (G4DFsc), (2) including an additional His residue in the first coordination sphere along with three additional helix-stabilizing mutations [3His-G4DFsc(Mut3)], and (3) the three helix-stabilizing mutations alone [G4DFsc-(Mut3)] on the biferrous structures and their O2 reactivities. Near-infrared circular dichroism and magnetic circular dichroism (MCD) spectroscopy show that the 4A → 4G mutations increase coordination of the diiron site from 4-coordinate/5-coordinate to 5-coordinate/5-coordinate, likely reflecting increased solvent accessibility. While the three helix-stabilizing mutations [G4DFsc(Mut3)] do not affect the coordination number, addition of the third active site His residue [3His-G4DFsc(Mut3)] results in a 5-coordinate/6-coordinate site. Although all 4A → 4G variants have significantly slower pseudo-first-order rates when reacting with excess O2 than DFsc (~2 s−1), G4DFsc and 3His-G4DFsc(Mut3) have rates (~0.02 and ~0.04 s−1) faster than that of G4DFsc(Mut3) (~0.002 s−1). These trends in the rate of O2 reactivity correlate with exchange coupling between the Fe(II) sites and suggest that the two-electron reduction of O2 occurs through end-on binding at one Fe(II) rather than through a peroxy-bridged intermediate. UV–vis absorption and MCD spectroscopies indicate that an Fe(III)Fe(III)-OH species first forms in all three variants but converts into an Fe(III)-μ-OH-Fe(III) species only in the 2-His forms, a process inhibited by the additional active site His ligand that coordinatively saturates one of the iron centers in 3His-G4DFsc(Mut3).
Graphical abstract

Binuclear non-heme iron enzymes are ubiquitous in nature and involved in many important biological processes.1–4 Their diiron active sites are often located in a common ferritin-like 4-helix bundle motif and ligated by varying numbers of carboxylate and histidine residues.1,2,5–10 A 2-histidine/4-carboxylate ligand set is present in the most studied subclasses of these enzymes. Examples of diiron enzymes with the 4-helix bundle motif and 2-histidine/4-carboxylate ligand set include the R2 subunit of ribonucleotide reductases (RR), which react with O2 and an exogenous electron to form a tyrosine radical required for deoxyribonucleotide biosynthesis,4,5 Δ9-desaturase (Δ9D), which catalyzes carbon–carbon double bond formation in stearoyl-bound ACP (acyl carrier protein),4,10 and methane monooxygenase (MMO), which catalyzes the conversion of methane to methanol.7,8,11 The biferrous sites in these enzymes initially reduce O2 by 2e− to form peroxy-bridged biferric species.1,12–19
Although these diiron enzymes have similar protein motifs and binuclear iron cofactors, the reactions that they catalyze are quite varied. Substrate and solvent accessibility to the diiron sites have been suggested to play key roles in this variation in reactivity.20 The higher efficiency of alkene/aromatic monooxygenases (AMOs) in hydroxylating aromatics relative to MMOs may be related to structural differences that increase the level of exposure of the diiron core of AMO to solvent, potentially allowing greater active site accessibility for the aromatic substrates.20,21 Moreover, conformational changes of protein gates have been implicated in allowing small molecular movement between cavities in MMO.20,22 Also, the gating of a hydrophilic pore in toluene/o-xylene monooxygenase (ToMO) by N202 and Q228 residues was proposed to be implicated in protecting activated oxygen intermediates involved in catalysis from being quenched.23
While solvent and substrate accessibility should influence substrate reactivity, spectroscopic characterizations (derived from solution studies that use crystallographic information) of the biferrous enzymes with 2-histidine/4-carboxylate ligand sets indicate differing coordinations of the ligating residues may also influence their O2 activation. RR has a 5-coordinate/4-coordinate biferrous site with two μ-1,3 bridging carboxylates, one histidine bound to each iron ion, one carboxylate bound bidentate to one iron ion, and another carboxylate bound monodendate to the other iron center (Figure 1, left).24 Δ9D is similar to RR, except that both nonbridging carboxylates bind in bidentate fashion to each iron ion, making its diiron site 5-coordinate/5-coordinate (Figure 1, middle).9 MMO also has a 5-coordinate/5-coordinate iron site, but with the nonbridging carboxylates bound monodentate to each iron site and one of the carboxylate bridges bound in a μ-1,1 rather than a μ-1,3 mode (Figure 1, right).7,9,25 While biferrous RR reacts rapidly with O2, addition of component B to MMO and steryl-bound acyl carrier protein to Δ9D is necessary to turn on their reactivity.7,9,24,25 The active site of MMO was found to rearrange into a new 5-coordinate/5-coordinate site upon addition of component B, and the diiron site of Δ9D converted to a 4-coordinate/5-coordinate site upon addition of substrate.9,26 These rearrangements were suggested to be necessary for proper orbital overlap between dioxygen and the two iron centers, which would allow for its two-electron reduction [one electron from each Fe(II)] in the formation of the peroxy-bridged intermediate.
Figure 1.
Spectroscopically derived structures for the biferrous sites of ribonucleotide reductase (left), Δ9-desaturase (middle), and soluble methane monooxygenase (right).7,9,24,25
While many binuclear non-heme iron enzymes have 2-histidine/4-carboxylate ligand sets, other histidine to carboxylate ratios are observed. One such exception is p-aminobenzoate N-oxygenase (AurF), which catalyzes the formation of p-nitro-benzoic acid from p-aminobenzoic acid, a six-electron process that is necessary in the biosynthesis of the antibiotic aureothin.27 Its diiron site, also located in a 4-helix bundle motif, is ligated by an additional histidine, resulting in a 3-histidine/4-carboxylate ligand set. This binuclear iron site reacts with dioxygen to form a long-lived peroxy intermediate with spectroscopic characteristics different from those of MMO, RR, and Δ9D, but similar to those of the peroxy intermediate in ToMO.18
Unlike small molecule models, de novo-designed due ferri binuclear iron proteins offer a scaffold for systematically investigating the effects of altered solvent environments, variations of the primary ligand sphere, and second-shell effects while still maintaining the simplicity of relatively small molecules.28–30 Significant expansion of the number of available DF scaffolds (DF1, DF2, DF2turn, DFtet, and DFsc) has allowed for better modeling of the folding characteristics of native proteins.31 Early studies focused on constructs that self-associate to form homodimeric or heterotetrameric four-helix bundles, while more recent studies have focused on the DFsc family of proteins, which are bacterially expressed as a single-chain protein. The dimetal cofactor in the initial DF protein32 was buried in a site that had very limited access to solvent or substrates. However, in subsequent designs, a substrate access cavity was introduced by a series of large-to-small mutations, which opened a deep cleft between two α-helices in the protein.33–35 Introduction of four small Ala residues created a substrate access cavity, which was expanded in a 4-Gly mutant, resulting in catechol and phenol oxidase activities.36,37
The introduction of a large cavity not only provides a substrate-binding site but also can have a major influence on the ligand environment and reactivity of the dimetal cofactor. For example, in a series of crystallographic structures of di-Mn(II) DF derivatives, the number of external solvent ligands increased from zero to two as the bulk of the residues lining the access cavity was systematically decreased from Leu to Ala to Gly.33–35
In DF, expansion of the substrate access cavity from the 4-Gly substitution, as well as perturbation of residues near the diiron site, leads to large changes in reactivity. The biferrous sites of the 4-Ala version of DF proteins have been spectroscopically determined to model that of RR [4-coordinate/5-coordinate (Figure 1, left)] and rapidly react with O2 at rates similar to those of natural diiron proteins. Moreover, the resulting biferric form oxidizes substrates, as in natural diiron proteins, but not at appreciable rates.38,39 By contrast, the biferrous form of G4DFsc catalyzes the O2-dependent two-electron oxidation of 4-amino-phenol (4AP).40 However, the oxidation of 4-aminophenol by G4DFsc appears to be much slower than in the 4-Ala version of the protein, despite the fact that G4DFsc has a less sterically encumbered binding site. Moreover, when a third His ligand is incorporated near the diiron site in the G4DFsc construct to model AurF, oxidase activity is diminished and arylamine activity, similar to AurF, is observed. This incorporation (I100H) required additional mutations (Mut3 = Y18L, I37N, and L81H) to stabilize the protein (Y18L allows space for His at position 100, while I37N and L81H provide H-bonding).
In this study, we probe the mechanism of O2 reactivity for the 4-Gly variants and explore the structural effects of 4-Gly substitution and 3-His incorporation near the diiron site on O2 reaction rates. We focus on G4DFsc, 3His-G4DFsc(Mut3), and also the new G4DFsc(Mut3) variant. These DF forms allow for investigation of the effects of systematic perturbations that include increased solvent accessibility, first-sphere mutation, and three additional helix-stabilizing residue mutations. Compared to DFsc, G4DFsc allows for increased level of access of solvent to the diiron site. As described above, 3His-G4DFsc(Mut3) contains an additional third His ligand at the active site along with three supporting mutations that result in a hydrogen bond network from H100 to H81 to N37. G4DFsc(Mut3) allows for the study of the effects of the three supporting mutations in the absence of the ligating H100 residue in the active site, and comparisons between G4DFsc(Mut3) and 3His-G4DFsc-(Mut3) allow us to isolate contributions to the active site from the H100 residue alone.
The biferrous active site structures for all three proteins are evaluated using near-IR (NIR) circular dichroism (CD), magnetic circular dichroism (MCD), and variable-temperature variable-field (VTVH) MCD spectroscopies. NIR CD and MCD spectroscopy provide geometric insight (i.e., coordination number) into biferrous sites from the energy positions of the d→d transitions of each Fe(II). VTVH MCD spectroscopy provides information about the spin manifold of the ground state by probing the MCD intensities of the d→d transitions. This allows for determination of spin-Hamiltonian parameters: zero-field splitting (D and E) and magnetic exchange coupling (J) associated with bridging ligands of the binuclear iron site. The O2 reactivity of G4DFsc, G4DFsc(Mut3), and 3His-G4DFsc-(Mut3) is further investigated using stopped flow kinetics. Biferric reaction intermediates and products of the O2 reaction are characterized via UV–vis MCD spectroscopy. This uses their ligand to metal charge transfer transitions to probe the ground states of these oxidized sites. The information obtained from these studies provides a detailed understanding of how solvent exposure and active site ligation impact O2 reactivity in the DFsc proteins and has implications for our understanding of O2 reactivity among natural carboxylate-bridged diiron enzymes.
MATERIALS AND METHODS
Mutagenesis, Expression, and Protein Purification
G4DFsc and 3His-G4DFsc(Mut3) were expressed and purified as described previously.40 To create G4DFsc(Mut3), a single-point mutation (H100I) was introduced via site-directed mutagenesis into the pET-28a vector (Novagen) containing the 3His-G4DFsc(Mut3) gene using PrimeSTAR HS DNA polymerase premix (Takara) and primers from Eurofins Genomics (forward, GAGCCTGGTTCAACAGCACCTGCA-GGATGAGCAG; reverse, CTGCTCATCCTGCAGGTGCT-GTTGAACCAGGCTC). The plasmid containing the mutated gene was isolated from XL1-Blue supercompetent cells (Agilent) using the QIAprep Spin Miniprep Kit (Qiagen), verified by DNA sequencing (Eurofins Genomics), and transformed into Escherichia coli BL21(DE3) competent cells (Novagen) for expression. Cells were lysed using repeated freeze–thaw cycles,41 and the crude lysate was then collected via centrifugation at 10000 rpm for 15 min. The protein was purified by reverse-phase high-performance liquid chromatography as described previously for G4DFsc and 3His-G4DFsc.40
Protein Reconstitution
Lyophilized purified protein was dissolved in 150 mM MOPS/150 mM NaCl buffer at pH 7 and spin filtered to remove any remaining particulate matter. Stock solution concentrations were measured by UV–visible absorption spectroscopy at 280 nm of 20-fold diluted samples in 6 M guanidine hydrochloride using a calculated extinction coefficient of 8250 M−1 cm−1 for G4DFsc and 6990 M−1 cm−1 for 3His-G4DFsc(Mut3) and G4DFsc(Mut3).
Biferrous Protein
Apoprotein in buffer (150 mM MOPS/150 mM NaCl at pH 7) was degassed on a Schlenk line by cycles (at least eight) of vacuum for very short intervals (<2 s) and purging headspace with Ar(g) (99.9%, Praxair) for longer intervals (>1 min) followed by purging sample headspace continuously for at least 2 h. Buffer was degassed by spurging on the Schlenk line for at least 3 h. The anaerobic nature of all solutions was tested by addition of 2 μL to a solution of reduced methyl viologen (0.1–0.5 mg/mL in buffer). FeSO4·6H2O (J. T. Baker) was dissolved in deoxygenated buffer (5–20 mg/200 μL). This solution was diluted and added to protein to obtain the requisite equivalents of Fe(II) ions (usually 1.7–1.8 equiv). The sample was incubated for 15–20 min before use. Deuterated samples were prepared similarly except apoprotein was buffer exchanged with deuterated buffer (150 mM MOPS/150 mM NaCl in D2O at pD 7) prior to being degassed. This included at least eight cycles of dilution to 1/4 concentration with D2O buffer and concentration to 2–3 mM. The voltage associated with the absorption of the CD spectrum of the apoprotein was checked at 1500 nm (~6700 cm−1) to monitor buffer exchange. The concentration was determined by diluting the sample if necessary, taking the absorption at 280 nm with an Agilent 8453 UV–visible spectrophotometer and using the calculated extinction coefficients at 280 nm [8250 M−1 cm−1 for G4DFsc and 6990 M−1 cm−1 for 3His-G4DFsc(Mut3) and G4DFsc-(Mut3)]. MOPS and NaCl were obtained from Sigma-Aldrich. Deuterium oxide (D2O; 99.9% 2H) was obtained from Cambridge Isotope Laboratories (Andover, MA).
Near-IR CD Experiments
Deoxygenated apoprotein (120 μL) was added to an anaerobic quartz cell in a glovebox. For the CD titration for G4DFsc(Mut3), equivalents of FeSO4·6H2O (0, 0.25, 0.5, 0.75, 1, 1.25, 1.5, 1.75, 2, and 3 equiv) were successively added to the solution anaerobically and their CD spectra recorded. For all other preparations, 1.7–1.8 equiv of Fe(II) ions was added to the protein. To determine the H2O2 reactivity of the biferrous site in 3His-G4DFsc(Mut3), the CD spectrum was taken before and after the addition of H2O2 (2 equiv) to biferrous protein. Protein concentrations were 2–3 mM for each experiment. The near-IR (600–1900 nm region) CD spectrum was then taken on a JASCO J730D spectropolarimeter with a cooled InSb detector. A circulator connected to the CD cell holder was used to maintain the anaerobic quartz CD cell at 4–12 °C. Multiple scans (5–20) were averaged.
MCD Experiments
MCD samples were prepared by adding anaerobically D-glycerol (d8-glycerol; 98% 2H from Cambridge Isotope Laboratories) [54% (v/v)] to CD samples (with protein concentrations of 1–2 mM). Possible effects of glycerol on the diiron site were probed by taking a CD spectrum after the addition of D-glycerol (see Figure S2 of the Supporting Information). MCD cells (quartz windows) were loaded with sample anaerobically and immediately submerged in N2(l). Near-IR MCD spectra were recorded at ±7, ±5, ±3, ±1, and 0 T at 2 K and ±7 and 0 T for temperatures of 5, 10, and 20 K. UV–vis MCD spectra were taken at ±7 and 0 T for temperatures ranging from 5 to 80 K. The MCD baseline was corrected by averaging positive and negative fields [e.g., (+7 T − −7 T)/2]. CD and MCD spectra were analyzed by resolving them into the minimum number of required Gaussian bands using a nonlinear least-squares procedure to fit both data sets while allowing for limited sharpening and shifting of peaks in the MCD spectrum (taken at low temperature). Near-IR (600–1900 nm region) MCD was performed on a JASCO J730D spectropolarimeter with a cooled InSb detector, and UV–vis MCD was performed using a JASCO J810 spectropolarimeter equipped with a photomultiplier tube that has an extended S20 photocathode. A He(l)-cooled Oxford Instrument SM4000 7T superconducting magnet was used for field and temperature variation.
Near-IR VTVH MCD Experiments
Data were taken at fields of 0, ±0.35, ±0.7, ±1.4, ±2.1, ±2.8, ±3.5, ±4.2, ±4.7, ±5.6, ±6.3, and ±7.0 T and temperatures of 2, 3, 5, 7.5, 10, 15, 20, and 25 K. MCD intensity was averaged over 100–200 scans for each field at each temperature. The reported data points are baseline-subtracted averages of the measurements for positive and negative field taken at the same temperature (error bars represent the propagation of errors from the standard deviations). The reduced χ2 value was used to judge the goodness of fit for the VTVH MCD data. The VTVH MCD data were fit with doublet and spin projection models as described in Results and Analysis.
O2 Kinetics
All stopped flow experiments were conducted at 4 °C. Deoxygenated protein and buffer were prepared. Two injection ports on the stopped flow instrument were degassed with ~3.0 mM sodium dithionite for ~20 min and kept anaerobic with a gaseous nitrogen flow. All protein samples were loaded on the stopped flow at concentrations of 200–250 μM. O2-saturated buffer was prepared by purging buffer (on ice) with O2 gas for 15 min. O2-saturated buffer was added to the protein solution in a 1:1 ratio to obtain a final protein concentration of 100–125 μM. An Applied Photophysics SX20 stopped flow absorption spectrophotometer with a Hg/Xe Arc lamp, PEEK tubing, and a Fisher Scientific Isotemp 3016 water/ethanol bath for controlling temperature was used.
Biferric Species
For the preparation of species B of 3His-G4DFsc(Mut3), species C of G4DFsc(Mut3), and species C of G4DFsc, biferrous protein was diluted to <50 μM with O2-saturated buffer, allowed to react for at least 2 h at 4–12 °C, and then concentrated. Glycerol was added [54% (v/v)], and the sample was loaded into an MCD cell for a final protein concentration of 300–500 μM. For biferric species B of G4DFsc, biferrous protein was prepared with glycerol [54% (v/v)] and placed into an MCD cell. O2-saturated buffer was prepared on ice and quickly mixed with glycerol [54% (v/v)] before being added to the MCD cell; 100 s after addition, the MCD cell was submerged in liquid nitrogen (final protein concentration of ~200 μM with 50–70% of species B approximated from the kinetic model).
H2O2 Detection
Biferrous protein was prepared. O2-saturated buffer was added to biferrous protein, and the solution was allowed to react at 4–12 °C. A solution containing 2,2′-azinobis(3-ethylbenzothiazoline-6-sulfonic acid) (ABTS) (3 mg/L) and horseradish peroxidase (20 mg/L) (Sigma-Aldrich) was prepared and its UV–vis absorption recorded. An O2-reacted protein solution was added to a solution of ABTS and peroxidase to obtain a final protein concentration of 10 μM, ABTS (3 mg/L), and horseradish peroxidase (20 mg/L) and its UV–vis absorption recorded. UV–vis absorption data were also taken for a reference solution containing hydrogen peroxide (30%, 10 μM), ABTS (3 mg/L), and horseradish peroxidase (20 mg/L).
RESULTS AND ANALYSIS
Protein Design
The single-chain DF protein (DFsc) is a stable and highly water-soluble mimic of natural binuclear non-heme iron proteins, has folding properties similar to those of natural proteins, and can be systematically mutated.38,40 Previous studies have suggested that incorporation of Ala to Gly mutations along the active site channel in DF proteins would decrease steric hindrance of solvent and substrate active site accessibility. One such study40 showed that the perturbation of DFsc with four Ala to Gly mutations (A10G, A14G, A43G, and A47G), G4DFsc, altered protein reactivity. In addition, the incorporation of a third His at the diiron core was found to substantially affect function. To obtain a third His incorporation, three additional mutations were necessary: Y18L to avoid steric clashes with the additional His and I37N and L81H to construct a hydrogen bonding network for stabilization of the diiron core. To extend these studies to form a systematic investigation, a 2His version that included these three extra mutations without H100 was also constructed [G4DFsc(Mut3)]. Figure 2A shows the sequences of the DF forms studied with the 4A → 4G (A10G, A14G, A43G, and A47G) form shown in red, the three mutations, Mut3 (Y18L, I37N, and L81H), shown in green, and the incorporated third His (I100H) shown in purple. These three variants [G4DFsc, G4DFsc(Mut3), and 3His-G4DFsc(Mut3)] allow the study of the effects of solvent accessibility (Figure 2B), construction of a hydrogen bonding network among second-sphere residues (Figure 2C), and incorporation of a third His ligand into the active site (Figure 2D) on the biferrous active site structure and its dioxgyen reactivity.
Figure 2.
Protein design.40 (A) Amino acid sequences for DFsc and variants studied in this work, highlighting the mutations for each protein [A10G, A14G, A43G, and A47G in bold red for G4DFsc; Y18L, I37N, and L81H in bold green for G4DFsc(Mut3); and I100H in bold purple for 3His-G4DFsc(Mut3)]. (B) Surface model of the 4A → 4G mutations (red) demonstrating enlargement of the solvent access channel using the Zn NMR structure of DFsc (Protein Data Bank entry 2HZ8). (C) Zn NMR structure of DFsc (left) and a 3His/Mut3/2A → 2G variant (right, Protein Data Bank 2LFD) in which mutations are colored green. (D) Incorporation of the third His at the active site from Zn NMR structures of DFsc (left) and a 3His/Mut3/2A → 2G variant (right).
Biferrous Spectroscopy
Previous studies showed that the binding of Fe(II) to the apoprotein of G4DFsc and 3His-G4DFsc(Mut3) resulted in near-IR circular dichroism (NIR CD) features that maximized at 2 equiv of Fe(II).40 The maximum limits of the Fe(II) dissociation constants for G4DFsc (<0.1 mM) and 3His-G4DFsc(Mut3) (<0.05 mM) are consistent with that of DFsc (<0.04 mM).38,40 G4DFsc(Mut3) also shows similar behavior where titrating in Fe(II) gives NIR CD signals that saturate at 2 equiv of Fe(II) (Figure S1A of the Supporting Information). A least-squares fit of an averaged Fe(II) titration curve to eq S1 of the Supporting Information gives a dissociation constant of <0.04 mM (Figure S1C of the Supporting Information). These studies show that the 4A → 4G, Mut3, and 3His mutations do not substantially affect the loading of Fe(II) on the apoprotein and result in binuclear iron cores. The energy positions of the NIR CD features for G4DFsc, G4DFsc(Mut3), and 3His-G4DFsc(Mut3) show little perturbation upon addition of glycerol [55% (v/v)] (Figure S2 of the Supporting Information), allowing the use of this glassing agent for low-temperature MCD studies. The CD spectra for G4DFsc and G4DFsc(Mut3) were Gaussian resolved into two bands (Figure 3A,B, top). For G4DFsc, these features are present at 7400 and 9400 cm−1. Gaussian resolution of the NIR CD spectrum of G4DFsc(Mut3) reveals two bands at 7200 and 9200 cm−1. Thus, inclusion of the mutations necessary for the hydrogen bonding network in the DF scaffold (Mut3) does not appear to substantially affect the energy positions of the NIR CD features. Incorporating the third His to form 3His-G4DFsc(Mut3), however, results in a dramatic change in the NIR CD (Figure 3C, top). Gaussian resolution of this spectrum requires three, rather than two, bands at 7400(+), 9200(−), and 10400(+) cm−1.
Figure 3.
CD and MCD of (A) G4DFsc, (B) G4DFsc(Mut3), and (C) 3His-G4DFsc(Mut3). Experimental results are presented in solid blue with the Gaussian-resolved features shown in dashed gray and numbered. The energy maximum for each numbered band is indicated in the graph. For all forms, CD spectra were collected between 4 and 12 °C and MCD spectra were collected at 2 K and 7 T. Protein concentrations were 1–3 mM. Black arrows on the MCD spectra indicate energies at which VTVH MCD data were collected.
The intensity of NIR magnetic CD (NIR MCD) features for G4DFsc, G4DFsc(Mut3), and 3His-G4DFsc(Mut3) increases with a decrease in temperature (Figure S3 of the Supporting Information, top) and increasing field (Figure S3 of the Supporting Information, bottom). Thus, these are C-terms associated with paramagnetic metal sites.1 Their temperature behavior and energy positions indicate that these are Fe(II) d → d transitions. The MCD spectra of both 2His variants [G4DFsc and G4DFsc(Mut3)] Gaussian resolve into two bands (Figure 3A,B, bottom) at 7600 and 9300 cm−1 for G4DFsc and 7600 and 9100 cm−1 for G4DFsc(Mut3). These correlate well to the bands in the corresponding NIR CD where the sharpening and energy shift are consistent with the lower temperature of the MCD spectra (2 K vs 277 K for CD). In addition, the low-temperature (2 K) NIR CD spectrum could be obtained for G4DFsc(Mut3) (Figure S4 of the Supporting Information). This shows the shift and sharpening of the CD features upon cooling, where the CD features at 2 K have the same energies and bandwidths as those in the NIR MCD spectrum (at 2 K). This provides further evidence that the 2His variants have only two ligand-field transitions in the NIR region and are different from DFsc, which has a third low-energy band that is observed in its CD spectrum.38 Thus, incorporation of the 4A → 4G mutations results in two rather than three observable NIR Fe(II) d → d transitions and suggests a change in the coordination of the diiron core.
While initial inspection of the NIR MCD for 3His-G4DFsc-(Mut3) (Figure 3C, bottom) suggests the presence of only two features, the higher-energy MCD band is broader (full width at half-maximum of ~3000 cm−1) than the corresponding CD band (~2000 cm−1 at 277 K). Because electronic transitions must sharpen at lower temperatures, the broadness of the low-temperature MCD feature requires the presence of an additional band. The NIR MCD of 3His-G4DFsc(Mut3) was thus fit with three bands at 7700, 9500, and 10300 cm−1. These Gaussian-resolved features correlate well with those in the CD spectrum. The additional higher-energy band suggests a higher coordination at the biferrous site upon incorporation of the third His.
Variable-temperature variable-field MCD (VTVH MCD) curves of the different MCD features were collected (black arrows, Figure 3, bottom) for G4DFsc, G4DFsc(Mut3), and 3His-G4DFsc(Mut3) (Figure 4). Overlays of the VTVH MCD isotherms are included in Figure 4. In both 2His forms, the VTVH MCD curves for bands 1 and 2 (Figure 4A,B) do not overlay within error, especially the lower-temperature isotherms. This indicates that these two bands arise from different iron ions. For 3His-G4DFsc(Mut3), the VTVH MCD curves for bands 1 and 2 (Figure 4D) do not overlay for the lower-temperature isotherms, while bands 1 and 3 do (Figure 4C). The overlay of bands 1 and 3 suggests that they derive from the same iron center, while band 2 derives from the second iron center.
Figure 4.
Overlay of the VTVH MCD isotherms collected at 2, 3, 5, 7.5, 10, 15, 20, and 25 K. (A) G4DFsc, overlay of bands 1 (gray) and 2 (purple). (B) G4DFsc(Mut3), overlay of bands 1 (gray) and 2 (purple). (C) 3His-G4DFsc(Mut3), overlay of bands 1 (gray) and 3 (purple). (D) 3His-G4DFsc(Mut3), overlay of bands 1 (gray) and 2 (purple).
For an integer spin system associated with a high-spin biferrous site, the VTVH MCD data may be modeled as a series of non-Kramers doublets in which different sublevels of the biferrous spin manifold contribute to the total MCD intensity through their Boltzmann population and field dependence of their wave functions.1,42 Fits using this modeling are provided in Figure 5 (top) with their related ground state energy level diagrams given at the bottom. The VTVH MCD isotherms for G4DFsc (Figure 5A) were fit with a doublet ground state having an effective g parallel value (g||) of 4.0 and a rhombic zero-field spitting (δ) of 3 cm−1 and a singlet excited state at 4.8 cm−1. The effective g|| value indicates an Ms = ±1 ground state that requires an antiferromagnetic exchange-coupled biferrous site. The VTVH MCD curves for G4DFsc(Mut3) (Figure 5B) were fit with a doublet ground state having a g|| of 8.0 and a δ of 0.17 cm−1 and two doublet excited states having a g|| of 4.0, one at 1.74 cm−1 with a δ of 0.02 cm−1 and another at 7.8 cm−1 with a δ of 0.2 cm−1. This fit indicates an Ms = ±2 ground state with an Ms = ±1 excited state. These fits indicate that the differences in VTVH MCD behavior between the 2His forms can be attributed to differences in the ground state spin manifold dictated by their spin-Hamiltonian parameters (vide infra). The VTVH MCD curves of 3His-G4DFsc(Mut3) were also fit with a ground state having a g|| of 8.0 with a δ of 4.0 cm−1 and an excited state having a g|| of 4.0 with a δ of 6.0 cm−1 at 3.0 cm−1. Final doublet fit parameters for these sites are listed in Table 1. The Ms = ±1 or ±2 ground states, as defined for the three G4 variants, require that the two irons be antiferromagnetically coupled and have oppositely signed axial zero-field splitting parameters.
Figure 5.
Doublet fits (top) with the models (bottom): (A) G4DFsc, 9100 cm−1; (B) G4DFsc(Mut3), 9100 cm−1; and (C) 3His-G4DFsc(Mut3), 9200 cm−1. Data are colored purple and fits red.
Table 1.
Summary of the Doublet Fit and Spin-Hamiltonian Parameters for DF Variants
| G4DFsc | G4DFsc(Mut3) | 3His-G4DFsc(Mut3) | |
|---|---|---|---|
| doublet-fit parameters | |||
| g||GS (cm−1) | 4.0 | 8.0 | 8.0 |
| δGS (cm−1) | 3.0 | 0.2 | 4.0 |
| Atot | 1.1 | 0.8 | 2.0 |
| B-term (%Atot) | 6.7 | 15.3 | 0.5 |
| energy (cm−1) | 0.0 | 0.0 | 0.0 |
| g||ES1 (cm−1) | – | 4.0 | 4.0 |
| δES1 (cm−1) | – | 0.0 | 6.0 |
| Atot | – | 0.6 | 3.4 |
| B-term (%Atot) | 2.61 (arb) | −1.2 | 0.2 |
| energy (cm−1) | 4.8 | 1.7 | 3.0 |
| g||ES2 (cm−1) | – | 4.0 | – |
| δES2 (cm−1) | – | 0.2 | – |
| Atot | – | 6.1 | – |
| B-term (%Atot) | – | −3.0 | – |
| energy (cm−1) | – | 7.8 | – |
| −J (cm−1) | 3–4 | 0.2–0.3 | 1–3 |
| spin-Hamiltonian parameters | |||
| D1 (cm−1) | 5–10 (band 2) | 7–8 (band 1) | 10–15 (bands 1 and 3) |
| (E/D)1 | 0.33 | 0.33 | 0.33 |
| D2 (cm−1) | −7 to −14 (band 1) | −2 to −3 (band 2) | −10 to −15 (band 2) |
| (E/D)2 | 0.15 | 0 | 0.33 |
Axial zero-field splitting (D) will spit the Ms sublevels of the S = 2 ground state of a mononuclear high-spin Fe(II) ion.1 The resulting Ms = ±2, ±1, and 0 sublevels are further split and mixed because of rhombic zero-field splitting (E). The magnetic coupling of two high-spin ferrous sites through a superexchange pathway, associated with the presence of a bridging ligand, results in Stot = |S1 + S2|···|S1 − S2| = 4, 3, 2, 1, 0 levels split by 8J, 6J, 4J, and 2J (Hex = −2JŜ1 Ŝ2, where J quantifies the exchange coupling). These combined zero-field and exchange effects on the ground state of a binuclear non-heme iron site are given by eq 1.
| (1) |
Applying eq 1 to an uncoupled basis set |S1,S2,Ms1,Ms2〉 for Fe1 and Fe2 results in a 25 × 25 matrix with the eigenvectors and eigenvalues describing the ground spin state sublevels and their relative energies.1 The results from the VTVH MCD fits to the doublet model can be correlated to specific values of J, D, and E. This is represented by the energy correlation diagram (Figure 6) for oppositely signed D values (−7 and 10 cm−1) with varying J values. Inspection of the resulting spin state sublevels and their energy splittings allows one to obtain ranges of D, E, and J for each biferrous site variant. As presented previously, these ranges can be further narrowed by fitting the VTVH MCD curves directly to an equation that correlates MCD intensity with the spin projection values of a specific Fe(II) ion on the ground sublevels of the coupled system given by the spin Hamiltonian in eq 1.43 The spin projection fits to the different VTVH MCD behaviors of the 2His forms are displayed in Figure 7. Combining the results from the non-Kramers doublet fits and those of the spin projection fits, we obtained spin-Hamiltonian parameters for the 4A → 4G variants. Spin-Hamiltonian and doublet fit parameters for the three 4A → 4G variants are listed in Table 1.
Figure 6.
Energy correlation diagram for D1 = 10 cm−1 and D2 = −7 cm−1 with E/D1 and E/D2 set to 0. G4DFsc, G4DFsc(Mut3) (labeled Mut3), and 3His-G4DFsc(Mut3) (labeled 3His) are indicated on the basis of their J values.
Figure 7.
Spin projection fits of the VTVH MCD data for (A) G4DFsc, (B) G4DFsc(Mut3), and (C) 3His-G4DFsc(Mut3). For G4DFsc and G4DFsc(Mut3), the VTVH MCD of band 1 (7,600 cm−1) is shown on the bottom and band 2 (9100 cm−1) is shown on the top. For 3His-G4DFsc(Mut3), spin projection fits of the VTVH MCD data for band 1 (7500 cm−1, bottom) and band 2 (9200 cm−1, top). For G4DFsc, only the first three isotherms (2, 3, and 5 K) were used because of larger uncertainties at higher energies. For G4DFsc(Mut3), all isotherms were used (2, 3, 5, 7.5, 10, 15, 20, and 25 K). Only the first four isotherms (2, 3, 5, and 7.5K) were used for 3His-G4DFsc(Mut3).
All three forms have similar D and E values for the two irons where G4DFsc has one iron (Fe1) with D1 = 5–10 cm−1 and E/D1 = 0.33 (associated with band 1 in Figure 3A) and the other (Fe2) with D2 = −7 to −10 cm−1 and E/D2 = 0.15 (associated with band 2). For G4DFsc(Mut3), one iron (Fe1) has D1 = 7–8 cm−1 and E/D1 = 0.33 (associated with band 1 in Figure 3B) and the other (Fe2) has D2 = −2 to −3 cm−1 and E/D2 ~ 0 (associated with band 2). 3His-G4DFsc(Mut3) was found to have D1 = 10–15 cm−1 and E/D1 = 0.33 (associated with bands 1 and 3) and D2 = −10 to −15 cm−1 and E/D2 = 0.33 (associated with band 2). All three forms have negative J values, indicating antiferromagnetic exchange coupling, with −J values of 3–4 cm−1 for G4DFsc, 0.2–0.3 cm−1 for G4DFsc(Mut3), and 1–3 cm−1 for 3His-G4DFsc(Mut3) (arrows in Figure 6, bottom). The magnitude and sign of the J values are consistent with two μ-1,3 carboxylate bridging ligands as seen in the Zn NMR structures for DFsc44 and a 2A → 2G 3His variant of DFsc40 (Figure 2D). Changes in the exchange coupling between the two Fe(II) sites in these DF variants predominately reflect perturbations of the carboxylate bridges and their binding to each Fe(II) center that define the superexchange pathways.
The 4A → 4G 2His variants [G4DFsc and G4DFsc(Mut3)] have two features in their CD and MCD spectra (~9400 and ~7400 cm−1), which could arise from either one or two iron sites. However, the intensities of these CD features saturate at 2 equiv of Fe(II). Additionally, the VTVH MCD curves of these bands do not overlay within error, and the non-Kramers doublet fit of the VTVH MCD data in Figure 5A for G4DFsc indicates an |Ms| = 1 ground state. Taken together, these attributes indicate the presence of two inequivalent iron sites. The lack of a higher-energy band (>10000 cm−1) suggests that neither a 6-coordinate center nor a 5-coordinate square pyramidal center is present. The lack of an observable low-energy band (~6000 cm−1) suggests that a 4-coordinate Fe(II) geometry may also be excluded. Five-coordinate trigonal bipyramidal Fe(II) sites show only one transition in this energy region (<10000 cm−1) with the second transition outside the range of the spectrometer (<3800 cm−1) and generally have either negative or rhombic zero-field splittings.1,42 Therefore, G4DFsc and G4DFsc(Mut3) may be described as having two inequivalent 5-coordinate trigonal bipyramidal Fe(II) centers bridged by two carboxylates that provide weak [and even weaker for G4DFsc(Mut3)] anti-ferromagnetic coupling. The coordination environment is different from that of DFsc, which was determined to have a 4-coordinate/5-coordinate biferrous center [similar to that of RR (Figure 1, left)].38 Considering the 4A → 4G mutations in these 2His variants, it is likely that increasing accessibility has resulted in solvent binding to one ferrous center. A notable difference between G4DFsc and G4DFsc(Mut3) is the order of magnitude larger antiferromagnetic exchange −J = 3–4 cm−1 for G4DFsc compared to −J = 0.2–0.3 cm−1 for G4DFsc(Mut3) (Figure 6, arrows), which appears to correlate to their rates of O2 reactivity (vide infra).
The 3His-G4DFsc(Mut3) variant has three bands present in both the MCD and CD spectra (~7700, ~9200, and ~10300 cm−1) and, thus, also requires the presence of two inequivalent iron centers, as indicated by the saturation of the CD features with 2 equiv of Fe(II).2 The VTVH MCD curves for bands 1 (~7700 cm−1) and 3 (~10300 cm−1) overlay within error, while bands 1 and 2 (~9200 cm−1) do not. This suggests that bands 1 and 3 can be attributed to the same iron site (D1 ~ 10–15 cm−1). They are centered at ~10000 cm−1 and split by ~2600 cm−1, which is characteristic of a 6-coordinate Fe(II) center. Because band 2 (D2 ~ −10 to −15 cm−1) in Figure 3C is at <10000 cm−1 with no MCD feature at low energy, this band is attributed to a 5-coordinate trigonal bipyramidal Fe(II) center. These data suggest that the addition of the third His residue near the active site has resulted in its binding to a ferrous center. Finally, we note that the J value of 3His-G4DFsc(Mut3) is also an order of magnitude greater than that of G4DFsc(Mut3). The consequences of these notably different J values for O2 reactivity are considered below.
Dioxygen Reactivity and Spectroscopy of Intermediates
Previous studies of G4DFsc and 3His-G4DFsc(Mut3) demonstrated their O2 reactivity with the appearance of absorption features in the 300–400 nm region.40 It should be noted that, in these previous studies, Fe(II) was added to apoprotein under ambient O2 conditions. To extend this study, we employed stopped flow absorption spectroscopy to investigate the kinetics of the O2 reactions in anaerobically preloaded Fe(II) G4DFsc and 3His-G4DFsc(Mut3), as well as the new G4DFsc(Mut3) variant. All three 4A → 4G variants of DFsc were reacted with O2-saturated buffer at 4 °C (>~0.65 mM O2 calculated from Henry’s law constant for dissolved oxygen in water45 and ~100 μM protein) and monitored via UV–vis absorption spectroscopy. The changes in the stopped flow data are shown in Figure 8. Upon reaction of G4DFsc with O2, two features simultaneously grow in at 330 and 380 nm (Figure 8A). After ~60 s, the 380 nm feature decays while the 330 nm feature remains. In contrast, in the reaction of the biferrous form of G4DFsc(Mut3) with O2, a 330 nm band is the only distinguishable feature that develops. For the biferrous form of 3His-G4DFsc(Mut3), reaction with O2 leads to absorption bands at 330 and 380 nm, but in contrast to G4DFsc, the 380 nm band remains for an extended period of time. Thus, the species formed upon reacting the biferrous 4A → 4G variants with O2 have similar absorption features and can be identified as two types: those with both 330 and 380 nm features (blue spectra in Figure S5 of the Supporting Information) and those with only a 330 nm band and no prominent 380 nm shoulder (red spectra in Figure S5 of the Supporting Information). Neither of these types shows a prominent 520 nm UV–vis absorption band in contrast to DFsc, which forms a tyrosine-bound biferric species upon reacting with O2.46
Figure 8.
Change in stopped flow absorption upon addition of O2. (A) G4DFsc with kinetic fits to Scheme 1 (right). (B) G4DFsc(Mut3) with kinetic fits to Scheme 2 (right). (C) 3His-G4DFsc(Mut3) with kinetic fits to Scheme 3 (right).
The reactivity of G4DFsc can be modeled using kinetic Scheme 1:
Scheme 1.

where species A is the biferrous form, species B is the intermediate with 330 and 380 nm features, and species C is the final product having only the 330 nm band. In Scheme 1, two reaction rates from noncompetitive parallel paths are required to fit the kinetic data (Figure 8, right) to form B. Mathematical representation of Scheme 1 is provided in the Supporting Information. Kinetic fits to this scheme are included in Figure 8A (right). The following kinetic rates were obtained: k1 = 0.02 ± 0.01 s−1, k2 = (0.8 ± 0.3) × 10−3 s−1, and k3 = (2.5 ± 0.5) × 10−3 s−1. The G4DFsc(Mut3) form followed the simpler reaction scheme:
Scheme 2.

The Supporting Information provides the mathematical representation of this scheme. The kinetic rate for this process is k1 = (2 ± 1) × 10−3 s−1 (fit in Figure 8B, right), significantly slower than for G4DFsc. [Note that if G4DFsc(Mut3) goes through species B (vide infra) and k1 represents its rate of formation (i.e., the slow step), it would not be observed if the B to C conversion rate is the same as k3 in G4DFsc.] For 3His-G4DFsc(Mut3), only the formation of species B is observed and its kinetic data again require two rates for its formation (Scheme 3):
Scheme 3.
The fit to the growth of the 330 nm feature is provided in Figure 8C (right), with the fitting details in the Supporting Information. Kinetic rates for this reaction were found to be similar to those of G4DFsc with k1 = 0.04 ± 0.02 s−1 and k2 = (2 ± 1) × 10−4 s−1. Kinetic rates for all forms are summarized in Table 2. Approximately 0.5 equiv of hydrogen peroxide was detected upon the completion of the reaction of O2 with 3His-G4DFsc(Mut3) (Figure S6D of the Supporting Information) [peroxide was detected by its oxidation of 2,2′-azino-bis(3-ethylbenzothiazoline-6-sulfonate) (ABTS) catalyzed by horse-radish peroxidase].47 This provides evidence that formation of species B is a simple two-electron oxidation of the diiron core by O2 with loss of H2O2. The second reaction pathway may thus reflect the released H2O2 reacting with the diiron site (biferric or biferrous). The lack of observable H2O2 after the reaction of G4DFsc with O2 (Figure S6C of the Supporting Information) may then be related to the faster k2 reaction rate in Scheme 1, where the H2O2 would be more rapidly consumed.
Table 2.
Kinetic Parameters of O2 Reactivity for the 4A → 4G DF Variantsa
| kinetic parameter (s−1) | G4DFsc | G4DFsc(Mut3) | 3His-G4DFsc(Mut3) |
|---|---|---|---|
| k1 | 0.02 ± 0.01 | (2 ± 1) × 10−3 | 0.04 ± 0.02 |
| k2 | (0.8 ± 0.3) × 10−3 | – | (2 ± 1) × 10−4 |
| k3 | (2.5 ± 0.5) × 10−3 | – | – |
Amplitudes are 0.5–0.7 for the k1 process and 0.3–0.5 for the k2 process for G4DFsc and 3His-G4DFsc(Mut3).
To help identify the products of the O2 reactions, UV–vis MCD spectra of species B and C for the three 4A → 4G DF variants were collected. In both G4DFsc and 3His-G4DFsc-(Mut3), the intensity of the MCD spectra of species B increased with a decrease in temperature, indicating that the MCD features are C-terms and identifying species B as paramagnetic (Figure 9A,B). For both G4DFsc and G4DFsc(Mut3), the MCD spectra of species C do not change with a decrease in temperature, indicating that the MCD features are B-terms and identifying species C as diamagnetic (Figure 9C,D). Non-heme ferrous sites give rise to NIR CD features. Because no significant NIR CD features are present for species B [3His-G4DFsc(Mut3) (Figure S8 of the Supporting Information)], the paramagnetism is not due to any ferrous-containing sites (i.e., biferrous or mixed valent forms). Thus, species B has a paramagnetic biferric site. (MCD data are correlated with the associated absorption data in Figure S9 of the Supporting Information.)
Figure 9.
UV–vis MCD spectra of the O2 reaction products of 4A → 4G DF variants. The MCD of species B is shown for (A) G4DFsc and (B) 3His-G4DFsc(Mut3), and that of species C is shown for (C) G4DFsc and (D) G4DFsc(Mut3). Fields for all spectra are 7 T. Δε values reflect the total protein concentration. 3His-G4DFsc(Mut3) species B and G4DFsc(Mut3) and G4DFsc species C represent >99% of the protein, and G4DFsc species B represents 50–70% of the protein (determined from kinetic fits). MCD spectra are correlated with the associated absorption data in Figure S9 of the Supporting Information.
The observed loss of H2O2 and the absence of any prominent absorption feature in the 800–500 nm energy region indicate that the absorption changes in the O2 reactions of the 4A → 4G DF variants (Figure 8) are not associated with the formation of the peroxy intermediate that has been observed in a number of binuclear iron enzymes and models.1,16,48–50 Although bridged μ-oxo to ferric charge transfer transitions are often observed in the 400–300 nm region, their extinction coefficients are generally in the range of 6000–12000 M−1 cm−1, greater than those observed for the oxidized forms of the 4A → 4G DF variants (2000–3000 M−1 cm−1), which suggests that neither species B nor species C is a μ-oxo-bridged biferric site.51,52 It should be noted that hydroxo-bridged ferric dimers have absorption features similar to those of species C (and B). Because hydroxo-bridged ferric dimers are antiferromagnetically coupled with −J values of ~20 cm−1,1 these would appear to be diamagnetic at low temperature. The assignment as a OH-bridged biferric species is thus reasonable for species C, but not species B, which is paramagnetic and thus has only the weak exchange coupling as would be associated with carboxylate bridges. The decay of B into C may thus reflect a terminal hydroxide ligand in B that goes on to bridge in C. The loss of the 380 nm band in the UV–vis absorption upon conversion of B to C may then be due to a blue shift of a hydroxide-to-ferric charge transfer transition caused by the formation of the OH bridge.
DISCUSSION
The 4A → 4G variants of DFsc allow for evaluation of the effects of solvent access to the diiron site on both structural and functional levels. Comparing these variants with the original DFsc protein provides insight into binuclear non-heme iron structure and reactivity. DFsc was previously found to contain a 4-coordinate/5-coordinate biferrous core that reasonably modeled the active site of ribonucleotide reductase.38 This site is represented in Figure 10A with a 2-histidine/4-carboxylate ligand set in which the two μ-1,3 carboxylate bridges allow for weak magnetic coupling between the two ferrous ions. The quadruple A to G mutations (A10G, A14G, A43G, and A47G), which were proposed to increase solvent accessibility, resulted in a loss of a low-energy Fe(II) ligand-field transition suggesting an increased level of coordination of the biferrous site to two 5-coordinate trigonal bipyramidal centers. This change (represented in Figure 10B) is likely the result of additional solvent access (4A → 4G) leading to solvent binding to an open coordination site on the 4-coordinate iron center in DFsc. Note that the antiferromagnetic coupling of DFsc and G4DFsc is comparable (−J = 1.5–2.9 cm−1 for DFsc and 3–4 cm−1 for G4DFsc), which reflects similar carboxylate superexchange pathways between the two Fe(II) sites. The relative energy positions (7200–7600 and 9100–9400 cm−1) of the two features in the CD and MCD spectra for G4DFsc appear to be unperturbed when the three helix-stabilizing mutations in G4DFsc(Mut3) are incorporated. Thus, the diiron site in G4DFsc(Mut3) retains coordination similar to that of G4DFsc with two 5-coordinate trigonal bipyramidal iron sites (as in Figure 10B). However, the VTVH-MCD of G4DFsc(Mut3) is different with a spin-sublevel ground state with Ms = ±2 (Ms = ±1 for G4DFsc). This indicates that the magnitude of the magnetic coupling has decreased (from 3–4 to 0.2–0.3 cm−1) (arrows, Figure 6), consistent with the Mut3 mutations causing a perturbation in the orientation of the μ-1,3 carboxylate bridges that further decreases the number of already limited super-exchange pathway(s). While incorporation of the third active site His results in a magnitude and sign of the magnetic coupling indicating μ-1,3 carboxylate bridges similar to those in the 2His forms, the CD and MCD spectra are greatly perturbed. The appearance of a higher-energy feature (~10300 cm−1) indicates that one iron is 6-coordinate and the second is a 5-coordinate trigonal bipyramidal center. This coordination number increase indicates that the third His binds to one of the Fe(II) ions. This agrees well with prior NMR structural data (Protein Data Bank entry 2LFD) that showed H100 bound to a Zn ion in a di-Zn form of a 2A → 2G analogue of 3His-G4DFsc(Mut3). Using the information from the NIR CD, MCD, and VTVH MCD data, as well as the relative position of the additional active site His in the NMR structure, the diiron site of 3His-G4DFsc(Mut3) is represented in Figure 10C.
Figure 10.
Spectroscopically derived models for the biferrous forms of (A) DFsc, (B) the 4A → 4G 2His variants of DFsc, and (C) 3His-G4DFsc(Mut3).
The O2 reaction rate of DFsc is comparable to that of natural binuclear non-heme iron enzymes (2 s−1 compared to ~1 s−1 of E. coli ribonucleotide reductase).46 These similar rates suggest that the 4-coordinate/5-coordinate diiron site of DFsc reduces O2 through a bridged peroxy structure as seen in most binuclear non-heme iron enzymes. The μ-1,3 carboxylate bridges are inefficient superexchange pathways, and thus, bridging of the two iron ions by dioxygen allows for an efficient 2-electron transfer and relatively fast O2 reaction rates. This reactivity is presented in Figure 11A. Allowing solvent exposure to the diiron site with the 4A → 4G mutations significantly decreases the O2 reactivity (0.02 ± 0.01 s−1). The O2 reaction rate of 3His-G4DFsc(Mut3) (0.04 ± 0.02 s−1) is similar to that of the G4DFsc form. One of the iron ions in the 3His form is coordinatively saturated, preventing coordination of O2 between the iron ions and thus precluding the formation of a bridged peroxy species. Therefore, the 2-electron transfer to O2 likely proceeds through a mechanism by which O2 binds in a terminal fashion to only the unsaturated ferrous site. Interestingly, even though the G4DFsc(Mut3) variant has iron coordination similar to that of G4DFsc, its O2 reaction rate is an additional order of magnitude slower [(2 ± 1) × 10−3 s−1].
Figure 11.
Proposed binding and reduction of O2 to form the biferric peroxy intermediate. (A) DFsc likely reacts with O2 to form a bridged species. (B) 4A → 4G variants likely react with O2 to form an end-on species (for the 2His forms, O2 binding may occur at either site).
The effects of the three mutations (Y18L, L81H, and I37N) that differentiate G4DFsc(Mut3) from G4DFsc on the potential of the diiron site are expected to be minimal. In the Zn NMR structure for DFsc, the Y18 residue is oriented parallel (at ~5 Å) to the diiron site (and assumed to remain similarly oriented in G4DFsc), limiting the effects of an induced dipole. The anticipated hydrogen bond between N37 and H81 could increase an induced negative dipole at H81, 7 Å from the diiron site, which would reduce the potential and lead to a more reactive center. However, the O2 reaction rate decreases (rather than increases) in G4DFsc(Mut3). Thus, with dioxygen binding to only one iron center while being reduced by 2 electrons [confirmed for 3His based on the observation of H2O2 (Figure S6 of the Supporting Information)], the efficiency of the superexchange pathway(s) associated with the bridging carboxylate ligands would modulate this reduction rate. The weaker magnetic coupling in the G4DFsc(Mut3) form combined with its slower O2 reaction rate provides evidence for this O2 reduction mechanism. This 2-electron reduction at a single Fe(II) center (represented in Figure 11B) is consistent with the reaction rate of the more coordinately saturated 3His form being similar to that of G4DFsc, where dioxygen can bind to only the 5-coordinate iron.
In the reaction of biferrous DFsc with O2, a tyrosine-bound biferric species was observed in the UV–vis spectrum with the appearance of a 520 nm feature.46 No prominent 520 nm bands appear in the UV–vis absorption in the O2 reactivity of the biferrous form of any of the 4A → 4G variants studied here. The higher coordination of these variants likely protects the ferric sites from tyrosine binding. Instead, a paramagnetic biferric species (species B) is observed in G4DFsc, which decays to a diamagnetic biferric species (species C). Only species C is observed in G4DFsc(Mut3). The absence of an observed species B in the Mut3 form is likely because of the slow initial O2 reaction rate, where conversion of species B to species C is faster. A plausible structure for the 2His species B is a nonbridging hydroxo-bound biferric site (Figure 12A). This species would become diamagnetic upon conversion to a bridging hydroxide biferric site (Figure 12A), which would be consistent with the spectral features of species C. The 6-coordinate iron site in the 3His form should inhibit the formation of species C (Figure 12B), consistent with the observation of only species B for 3His-G4DFsc(Mut3). Antiferromagnetically coupled ferric sites bridged by oxo or hydroxo ligands, such as species C, are common end products in the O2 reaction of many binuclear non-heme iron enzymes, which include the biferric forms of methane monooxygenase, flavin diiron proteins, hemerythrin, myo-inositol oxygenase, ribonucleotide reductase, etc.1,53,54 The results suggest that increasing the coordination of the diiron site may inhibit the formation of this common end product.
Figure 12.
Spectroscopically derived structural models for (A) 2His species B and species C and (B) 3His species B.
In summary, these results offer insight into the effects of solvent exposure on the structure and reactivity of a binuclear non-heme ferrous center. Enlarging the access channel to the 2-histidine/4-carboxylate site allows for solvent coordination. This increase in coordination prevents O2 from binding to both iron ions to form a bridged peroxy species. Electron transfer from the unbound iron ion allowing direct 2-electron reduction must then proceed through the inefficient μ-1,3 carboxylate superexchange pathways to reduce dioxygen by 2 electrons. This results in the loss of H2O2 and a final bound hydroxide ligated to one iron. If the second iron of the biferric site is not coordinatively saturated, a hydroxo-bridged biferric species can form, which is precluded in 3His-G4DFsc(Mut3), resulting in a paramagnetic species with only μ-1,3 bridging carboxylates. These results provide a foundation for investigating the oxygen-dependent catalysis of the 4A → 4G DF proteins. The slow O2 reactivity of the DF variants may then be compared to the relatively faster O2 reactivity of the native enzymes. While the mechanism for O2 reduction by these DF variants parallels that of hemerytherin where O2 binds to one iron and undergoes a 2-electron reduction with the transfer of the second electron proceeding through a superexchange pathway, O2 reduction is facilitated by an efficient μ-hydroxo/μ-oxo bridge in the case of hemerythrin.1 Δ9D, on the other hand, has only inefficient μ-1,3 carboxylate superexchange pathways available, which necessitates the addition of substrate to perturb the diiron site and to create an open coordination position that allows for O2 bridging of the two irons and, thus, the efficient 2-electron reduction of dioxygen.9 However, the DF variants have neither efficient μ-hydroxo bridges nor binding sites for O2 to bridge. They thus demonstrate a mechanism for a slow O2 reactivity of diiron sites where electron transfer occurs through inefficient μ-1,3 carboxylate superexchange pathways.
Supplementary Material
Acknowledgments
Funding
This work was supported by the National Science Foundation (MCB-1404866 to E.I.S. and CHE-1413295 to W.F.D.) and the National Institutes of Health (F32-GM808852 and R15-GM110657 to A.J.R. and GM54616 and GM71628 to W.F.D.).
Footnotes
Notes
The authors declare no competing financial interest.
CD Fe(II) titration of G4DFsc(Mut3); CD of G4DFsc, G4DFsc(Mut3), and 3His-G4DFsc(Mut3) with and without glycerol addition; MCD spectra of G4DFsc, G4DFsc(Mut3), and 3His-G4DFsc(Mut3) at different temperatures and magnetic fields showing C-term behavior; low-temperature CD of G4DFsc(Mut3); UV–vis absorption spectrum of species B and C for G4DFsc, G4DFsc(Mut3), and 3His-G4DFsc(Mut3); modeling for Scheme 4.1; H2O2 detection in 3His-G4DFsc-(Mut3); CD of Fe(II)Fe(II) 3His-G4DFsc(Mut3) reacted with H2O2 CD of species B for 3His-G4DFsc(Mut3); and overlay of UV–vis MCD and Abs for species B and C of G4DFsc, G4DFsc(Mut3), and 3His-G4DFsc(Mut3). The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.biochem.5b00324.
References
- 1.Solomon EI, Brunold TC, Davis MI, Kemsley JN, Lee SK, Lehnert N, Neese F, Skulan AJ, Yang YS, Zhou J. Geometric and electronic structure/function correlations in non-heme iron enzymes. Chem Rev. 2000;100:235–349. doi: 10.1021/cr9900275. [DOI] [PubMed] [Google Scholar]
- 2.Andrews SC. The ferritin-like superfamily: Evolution of the biological iron storeman from a rubrerythrin-like ancestor. Biochim Biophys Acta, Gen Subj. 2010;1800:691–705. doi: 10.1016/j.bbagen.2010.05.010. [DOI] [PubMed] [Google Scholar]
- 3.Theil EC. Ferritin: At the crossroads of iron and oxygen metabolism. J Nutr. 2003;34:1549S–1553S. doi: 10.1093/jn/133.5.1549S. [DOI] [PubMed] [Google Scholar]
- 4.Shao J, Zhou B, Chu B, Yen Y. Ribonucleotide reductase inhibitors and future drug design. Curr Cancer Drug Targets. 2006;6:409–431. doi: 10.2174/156800906777723949. [DOI] [PubMed] [Google Scholar]
- 5.Wei PP, Skulan AJ, Mitić N, Yang YS, Saleh L, Bollinger JM, Jr, Solomon EI. Electronic and spectroscopic studies of the non-heme reduced binuclear iron sites of two ribonucleotide reductase variants: Comparison to reduced methane monooxygenase and contributions to O2 reactivity. J Am Chem Soc. 2004;126:3777–3788. doi: 10.1021/ja0374731. [DOI] [PubMed] [Google Scholar]
- 6.Lindqvist Y, Huang W, Schneider G, Shanklin J. Crystal structure of Δ9 stearoyl-acyl carrier protein desaturase from castor seed and its relationship to other di-iron proteins. EMBO J. 1996;15:4081–4092. [PMC free article] [PubMed] [Google Scholar]
- 7.Whittington DA, Lippard SJ. Crystal structures of the soluble methane monooxygenase hydroxylase from Methylococcus capsulatus (bath) demonstrating geometrical variability at the dinuclear iron active site. J Am Chem Soc. 2001;123:827–838. doi: 10.1021/ja003240n. [DOI] [PubMed] [Google Scholar]
- 8.Mitić N, Schwartz JK, Brazeau BJ, Lipscomb JD, Solomon EI. CD and MCD studies of the effects of component B variant binding on the biferrous active site of methane monooxygenase. Biochemistry. 2008;47:8386–8397. doi: 10.1021/bi800818w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang YS, Broadwater JA, Pulver SC, Fox BG, Solomon EI. Circular dichroism and magnetic circular dichroism studies of the reduced binuclear non-hem iron site of stearoyl-ACP Δ9 desaturase: Substrate binding and comparison to ribonucleotide reductase. J Am Chem Soc. 1999;121:2770–2783. [Google Scholar]
- 10.Stubbe J. Di-iron-tyrosyl radical ribonucleotide reductases. Curr Opin Chem Biol. 2003;7:183–188. doi: 10.1016/s1367-5931(03)00025-5. [DOI] [PubMed] [Google Scholar]
- 11.Lipscomb JD. Biochemistry of the soluble methane monooxygenase. Annu Rev Microbiol. 1994;48:371–399. doi: 10.1146/annurev.mi.48.100194.002103. [DOI] [PubMed] [Google Scholar]
- 12.Liu KE, Wang D, Huynh BH, Edmondson DE, Salifoglou A, Lippard SJ. Spectroscopic detection of intermediates in the reaction of dioxygen with the reduced methane monooxygenase/hydroxylase from Methylococcus capsulatus (bath) J Am Chem Soc. 1994;116:7465–7466. [Google Scholar]
- 13.Tong WH, Chen S, Lloyd SG, Edmondson DE, Huynh BH, Stubbe J. Mechanism of assembly of the diferric cluster-tyrosyl radical cofactor of Escherichia coli ribonucleotide reductase from the diferrous form of the R2 subunit. J Am Chem Soc. 1996;118:2107–2108. [Google Scholar]
- 14.Bollinger JM, Jr, Krebs C, Vicol A, Chen S, Ley BA, Edmondson DE, Huynh BH. Engineering the diiron site of E. coli ribonucleotide reductase protein R2 to accumulate peroxodiiron(III) complex from the methane monooxygenase catalytic cycle. J Am Chem Soc. 1998;120:1094–1095. [Google Scholar]
- 15.Yun D, Garcia-Serres R, Chicalese BM, An YH, Huynh BH, Bollinger JM., Jr (μ-1,2-peroxo)diiron(III/III) complex as a precursor to the diiron(III/IV) intermediate X in the assembly of the iron-radical cofactor of ribonucleotide reductase from mouse. Biochemistry. 2007;46:1925–1932. doi: 10.1021/bi061717n. [DOI] [PubMed] [Google Scholar]
- 16.Broadwater JA, Ai J, Loehr TM, Sanders-Loehr J, Fox BG. Peroxodiferric intermediate of stearoyl-acyl carrier protein Δ9Desaturase: Oxidase reactivity during single turnover and implications for the mechanism of desaturation. Biochemistry. 1998;37:14664–14671. doi: 10.1021/bi981839i. [DOI] [PubMed] [Google Scholar]
- 17.Pereira AS, Small W, Krebs C, Tavares P, Edmondson DE, Theil EC, Huynh BH. Direct spectroscopic and kinetic evidence for the involvement of a peroxodiferric intermediate during the ferroxidase reaction in fast ferritin mineralization. Biochemistry. 1998;37:9871–9876. doi: 10.1021/bi980847w. [DOI] [PubMed] [Google Scholar]
- 18.Korboukh VK, Li N, Barr EW, Bollinger JM, Jr, Krebs C. A long-lived, substrate-hydroxylating peroxodiiron(III/III) intermediate in the amine oxygenase, AurF, from Streptomyces thioluteus. J Am Chem Soc. 2009;131:13608–13609. doi: 10.1021/ja9064969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murray LJ, Garcia-Serres R, Naik S, Huynh BH, Lippard SJ. Dioxygen activation at non-heme diiron centers: Characterization of intermediates in a mutant form of toluene/o-xylene monooxygenase hydroxylase. J Am Chem Soc. 2006;128:7458–7459. doi: 10.1021/ja062762l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sazinsky MH, Lippard SJ. Correlating structure with function in bacterial multicomponent monooxygenases and related diiron proteins. Acc Chem Res. 2006;39:558–566. doi: 10.1021/ar030204v. [DOI] [PubMed] [Google Scholar]
- 21.Sazinsky MH, Bard J, Di Donato A, Lippard SJ. Crystal structure of the toluene/o-xylene monooxygenase hydroxylase from Pseudomonas stutzeri OX1. Insight into the substrate specificity, substrate channeling, and active site tuning of multicomponent monooxygenases. J Biol Chem. 2004;279:30600–30610. doi: 10.1074/jbc.M400710200. [DOI] [PubMed] [Google Scholar]
- 22.Sazinsky MH, Lippard SJ. Product bound structures of the soluble methane monooxygenase hydroxylase from Methylococcus capsulatus (bath): Protein motion in the alpha-subunit. J Am Chem Soc. 2005;127:5814–5825. doi: 10.1021/ja044099b. [DOI] [PubMed] [Google Scholar]
- 23.Liang AD, Wrobel AT, Lippard SJ. A flexible glutamine regulates the catalytic activity of toluene/o-xylene mono-oxygenase. Biochemistry. 2014;53:3585–3592. doi: 10.1021/bi500387y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang YS, Baldwin J, Ley BA, Bollinger JM, Jr, Solomon EI. Spectroscopic and electronic structure description of the reduced binuclear non-heme iron active site in ribonucleotide reductase from E. coli: Comparison to reduced Δ9Desaturase and electronic structure contributions to differences in O2 reactivity. J Am Chem Soc. 2000;122:8495–8510. [Google Scholar]
- 25.Rosenzweig AC, Brandstetter H, Whittington DA, Nordlund P, Lippard SJ, Frederick CA. Crystal structures of the methane monooxygenase hydroxylase from Methyl-ococcus capsulatus (bath): Implications for substrate gating and component interactions. Proteins: Struct, Funct, Genet. 1997;29:141–152. [PubMed] [Google Scholar]
- 26.Pulver SC, Froland WA, Lipscomb JD, Solomon EI. Ligand field circular dichroism and magnetic circular dichroism studies of component B and substrate binding to the hydroxylase component of methane monooxygenase. J Am Chem Soc. 1997;119:387–395. [Google Scholar]
- 27.Choi YS, Zhang H, Brunzelle JS, Nair SK, Zhao H. In vitro reconstitution and crystal structure of p-aminobenzoate N-oxygenase (AurF) involved in aureothin biosynthesis. Proc Natl Acad Sci U S A. 2008;105:6858–6863. doi: 10.1073/pnas.0712073105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.DeGrado WF, Wasserman ZR, Lear JD. Protein design, a minimalist approach. Science. 1989;243:622–628. doi: 10.1126/science.2464850. [DOI] [PubMed] [Google Scholar]
- 29.DeGrado WF, Summa CM, Pavone V, Nastri F, Lombardi A. De novo design and structural characterization of proteins and metalloproteins. Annu Rev Biochem. 1999;68:779–819. doi: 10.1146/annurev.biochem.68.1.779. [DOI] [PubMed] [Google Scholar]
- 30.Bryson JW, Betz SF, Lu HS, Suich DJ, Zhou HX, O’Neil KT, DeGrado WF. Protein design: A hierarchic approach. Science. 1995;270:935–941. doi: 10.1126/science.270.5238.935. [DOI] [PubMed] [Google Scholar]
- 31.Calhoun JR, Nastri F, Maglio O, Pavone V, Lombardi A, DeGrado WF. Artificial diiron proteins: From structure to function. Biopolymers. 2005;80:264–278. doi: 10.1002/bip.20230. [DOI] [PubMed] [Google Scholar]
- 32.Lombardi A, Summa CM, Geremia S, Randaccio L, Pavone V, DeGrado WF. Inaugural article: Retrostructural analysis of metalloproteins: Application to the design of a minimal model for diiron proteins. Proc Natl Acad Sci U S A. 2000;97:6298–305. doi: 10.1073/pnas.97.12.6298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Di Costanzo L, Wade H, Geremia S, Randaccio L, Pavone V, DeGrado WF, Lombardi A. Toward the de novo design of a catalytically active helix bundle: A substrate-accessible carboxylate-bridged dinuclear metal center. J Am Chem Soc. 2001;123:12749–57. doi: 10.1021/ja010506x. [DOI] [PubMed] [Google Scholar]
- 34.Geremia S, Di Costanzo L, Randaccio L, Engel DE, Lombardi A, Nastri F, DeGrado WF. Response of a designed metalloprotein to changes in metal ion coordination, exogenous ligands, and active site volume determined by X-ray crystallography. J Am Chem Soc. 2005;127:17266–76. doi: 10.1021/ja054199x. [DOI] [PubMed] [Google Scholar]
- 35.DeGrado WF, Di Costanzo L, Geremia S, Lombardi A, Pavone V, Randaccio L. Sliding helix and change of coordination geometry in a model di-Mn(II) protein. Angew Chem, Int Ed. 2003;42:417–20. doi: 10.1002/anie.200390127. [DOI] [PubMed] [Google Scholar]
- 36.Kaplan J, DeGrado WF. De novo design of catalytic proteins. Proc Natl Acad Sci U S A. 2004;101:11566–70. doi: 10.1073/pnas.0404387101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Faiella M, Andreozzi C, de Rosales RTM, Pavone V, Maglio O, Nastri F, DeGrado WF, Lombardi A. An artificial di-iron oxo-protein with phenol oxidase activity. Nat Chem Biol. 2009;5:882–884. doi: 10.1038/nchembio.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bell CB, III, Calhoun JR, Bobyr E, Wei P, Hedman B, Hodgson KO, DeGrado WF, Solomon EI. Spectroscopic definition of the biferrous and biferric sites in de novo designed four-helix bundle DFsc peptides: Implications for O2 reactivity of binuclear non-heme iron enzymes. Biochemistry. 2009;48:59–73. doi: 10.1021/bi8016087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wei P, Skulan A, Wade H, DeGrado WF, Solomon EI. Spectroscopic and computational studies of the de novo designed protein DF2t: Correlation to the biferrous active site of ribonucleotide reductase and factors that affect O2 reactivity. J Am Chem Soc. 2005;127:16098–16106. doi: 10.1021/ja053661a. [DOI] [PubMed] [Google Scholar]
- 40.Reig AJ, Pires MM, Snyder RA, Wu Y, Jo H, Kulp DW, Butch SE, Calhoun JR, Szyperski T, Solomon EI, DeGrado WF. Alteration of the oxygen-dependent reactivity of de novo due ferri proteins. Nat Chem. 2012;4:900–906. doi: 10.1038/nchem.1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Johnson BH, Hecht MH. Recombinant proteins can be isolated from E. coli cells by repeated cycles of freezing and thawing. Bio/Technology. 1994;12:1357–1360. doi: 10.1038/nbt1294-1357. [DOI] [PubMed] [Google Scholar]
- 42.Solomon EI, Pavel EG, Loeb KE, Campochiaro C. Magnetic circular dichroism spectroscopy as a probe of the geometric and electronic structure of non-heme ferrous enzymes. Coord Chem Rev. 1995;144:369–460. [Google Scholar]
- 43.Neese F, Solomon EI. MCD C-term signs, saturation behavior, and determination of band polarizations in randomly oriented systems with spin S ≥ 1/2. applications to S = 1/2 and S = 5/2. Inorg Chem. 1999;38:1847–1865. doi: 10.1021/ic981264d. [DOI] [PubMed] [Google Scholar]
- 44.Calhoun JR, Kono H, Lahr S, Wang W, DeGrado WF, Saven JG. Computational design and characterization of a monomeric helical dinuclear metalloprotein. J Mol Biol. 2003;334:1101–1115. doi: 10.1016/j.jmb.2003.10.004. [DOI] [PubMed] [Google Scholar]
- 45.Sander R. Compilation of henry’s law constants, version 3.99. Atmos Chem Phys Discuss. 2014;14:29615–30521. [Google Scholar]
- 46.Calhoun JR, Bell CB, III, Smith TJ, Thamann TJ, DeGrado WF, Solomon EI. Oxygen reactivity of the biferrous site in the de novo designed four helix bundle peptide DFsc: Nature of the “intermediate” and reaction mechanism. J Am Chem Soc. 2008;130:9188–9189. doi: 10.1021/ja801657y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kadnikova EN, Kostic NM. Oxidation of ABTS by hydrogen peroxide catalyzed by horseradish peroxidase encapsulated into sol–gel glass. J Mol Catal B: Enzym. 2002;18:39–48. [Google Scholar]
- 48.Bou-Abdallah F, Papaefthymiou GC, Scheswohl DM, Stanga SD, Arosio P, Chasteen ND. μ-1,2-peroxobridged di-iron(III) dimer formation in human H-chain ferritin. Biochem J. 2002;364:57–63. doi: 10.1042/bj3640057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Broadwater JA, Achim C, Munck E, Fox BG. Mössbauer studies of the formation and reactivity of a quasi-stable peroxo intermediate of stearoyl-acyl carrier protein Δ9Desaturase. Biochemistry. 1999;38:12197–204. doi: 10.1021/bi9914199. [DOI] [PubMed] [Google Scholar]
- 50.Jensen KP, Bell CB, III, Clay MD, Solomon EI. Peroxo-type intermediates in class I ribonucleotide reductase and related binuclear non-heme iron enzymes. J Am Chem Soc. 2009;131:12155–12171. doi: 10.1021/ja809983g. [DOI] [PubMed] [Google Scholar]
- 51.Brown CA, Remar GJ, Musselman RL, Solomon EI. Spectroscopic and electronic-structure studies of met-hemerythrin model complexes - a description of the ferric-oxo dimer bond. Inorg Chem. 1995;34:688–717. [Google Scholar]
- 52.Kurtz DM., Jr Oxo- and hydroxo-bridged diiron complexes: A chemical perspective on a biological unit. Chem Rev. 1990;90:585–606. [Google Scholar]
- 53.Brown PM, Caradoc-Davies TT, Dickson JM, Cooper GJ, Loomes KM, Baker EN. Crystal structure of a substrate complex of myo-inositol oxygenase, a di-iron oxygenase with a key role in inositol metabolism. Proc Natl Acad Sci U S A. 2006;103:15032–15037. doi: 10.1073/pnas.0605143103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Silaghi-Dumitrescu R, Kurtz DM, Jr, Ljungdahl LG, Lanzilotta WN. X-ray crystal structures of Moorella thermoacetica FprA. Novel diiron site structure and mechanistic insights into a scavenging nitric oxide reductase. Biochemistry. 2005;44:6492–6501. doi: 10.1021/bi0473049. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.