Abstract
Cell-surface glycans are a diverse class of macromolecules that participate in many key biological processes, including cell-cell communication, development, and disease progression. Thus, the ability to modulate the structures of glycans on cell surfaces provides a powerful means not only to understand fundamental processes but also to direct activity and elicit desired cellular responses. Here, we describe methods to sculpt glycans on cell surfaces and highlight recent successes in which artificially engineered glycans have been employed to control biological outcomes such as the immune response and stem cell fate.
Introduction
Cell-surface glycans participate in many important processes throughout the lifespan of an organism, ranging from cell migration and tissue patterning to the immune response, disease progression, and cell death (Fuster and Esko, 2005; Häcker et al., 2005; Lichtenstein and Rabinovich, 2013; van Kooyk and Rabinovich, 2008). At a molecular level, glycans are often the first points of contact between cells, and they function by facilitating a variety of interactions both in cis (on the same cell) and in trans (on different cells). The glycan covering that surrounds the cell surface, termed the glycocalyx, can both promote and hinder the binding of canonical protein ligands to their cell-surface receptors, as well as mediate ligand-independent receptor clustering and activation (Bishop et al., 2007; Coles et al., 2011; Haines and Irvine, 2003; Rogers et al., 2011). Indeed, the integral roles of cell-surface glycans in regulating cellular signaling events are only beginning to be understood.
The ability to modulate and re-engineer the diverse structures of glycans at the cell surface provides a powerful means to elucidate the molecular mechanisms that underlie glycan-mediated signaling events and their downstream cellular consequences. From a mechanistic standpoint, systematically altering glycan structures provides insights into structure-function relationships and the importance of individual structures in glycan-mediated processes. From an engineering standpoint, the ability to remodel glycan architectures on cell surfaces offers a novel approach to manipulate cellular physiology and phenotypic outcomes. Here, we will describe the methods available to tailor the structures of glycans on cells and provide notable examples of how remodeling of cell-surface glycans have led to both new biological insights and novel cellular functions.
Genetic Approaches
Genetic manipulation of glycosyltransferases (GTs) or other genes involved in glycan biosynthesis provides a powerful method to perturb specific glycan subpopulations in cells and organisms. A variety of approaches have been developed, including gene deletion or knockout, gene knockdown by RNAi, and gene overexpression (Figure 1). Genetic methods offer excellent spatial and temporal control, enabling the precise manipulation of specific genes in a cell-specific and inducible manner. However, as GTs typically operate on multiple protein substrates and assemble a variety of glycan structures, it is difficult to study the impact of a single glycan on an individual protein of interest. Moreover, genetic approaches usually subtract from existing glycan structures rather than add new chemical functionalities. Finally, knocking out GTs can lead to developmental defects or embryonic lethality, which can hinder the identification of functions in the adult organism. Nonetheless, genetic approaches have provided invaluable information on the functional importance of GTs and protein glycosylation in vivo.
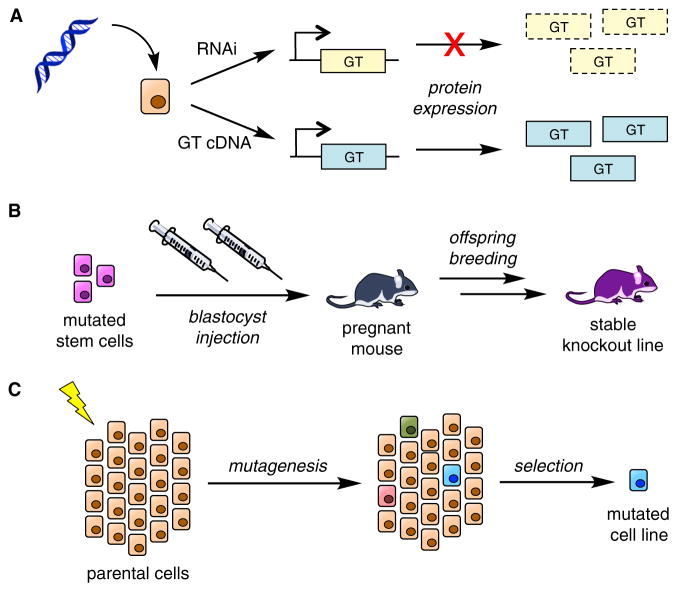
Figure 1. Genetic Approaches.
(A) Transient transfection of cells in vitro with either shRNA or siRNA (RNAi) targeting a gene causes a decrease in expression of the protein. Transfection with the cDNA of a protein leads to its over-expression.
(B) Mouse knockout models are traditionally produced by first stably disrupting gene expression in mouse embryonic stem cells by homologous recombination. Mutated stem cells are injected into blastocysts, which are then implanted into mice to produce mosaic offspring. Mice are further bred to produce a stable knockout line.
(C) To generate stable CHO knockout cell lines, parental cells were treated with a mutagenic agent (e.g., ionizing radiation) and then selected for resistance to cytotoxic lectins, which bind to specific carbohydrate epitopes now missing on the mutated cells.
The power of genetic approaches is exemplified by elegant studies on the Notch signaling pathway. Notch signaling is essential for proper development, and dysregulation of the pathway leads to various human diseases, including congenital disorders and cancer (Andersson et al., 2011; Kopan and Ilagan, 2009). Glycosylation of the extracellular domain of the Notch receptor has emerged as an important mechanism for the regulation of Notch activity (Figure 2A). Early studies identified a β-1,3-N-acetylglucosaminyltransferase called Fringe that modifies O-linked Fuc residues on Notch (Brückner et al., 2000; Moloney et al., 2000). Ectopic expression of a catalytically inactive, mutant form of Fringe in Drosophila demonstrated that the GT activity of Fringe was required for certain Notch ligand-receptor interactions and proper wing formation in vivo (Moloney et al., 2000). Other GTs have also been shown to be critical for Notch signaling. For example, protein O-fucosyltransferase 1 (Pofut1 in mammals or Ofut1 in Drosophila) catalyzes the addition of O-Fuc to Notch, generating the acceptor substrate for Fringe (Wang et al., 2001). RNAi-mediated knockdown of Ofut1 showed that the enzyme is required for both Fringe-dependent and Fringe-independent Notch function in Drosophila (Okajima and Irvine, 2002). In mice, genetic deletion of Pofut1 results in embryonic lethality with phenotypic defects in cardiovascular and neurologic development, consistent with loss of all Notch paralog signaling (Shi and Stanley, 2003). In addition, inducible inactivation of Pofut1 in the immune-responsive cells and hematopoietic tissues of adult mice revealed a key role for O-Fuc glycans on Notch in the regulation of hematopoietic homeostasis (Yao et al., 2011). Together, these genetic studies demonstrate the essential nature of glycosylation for Notch receptor function.
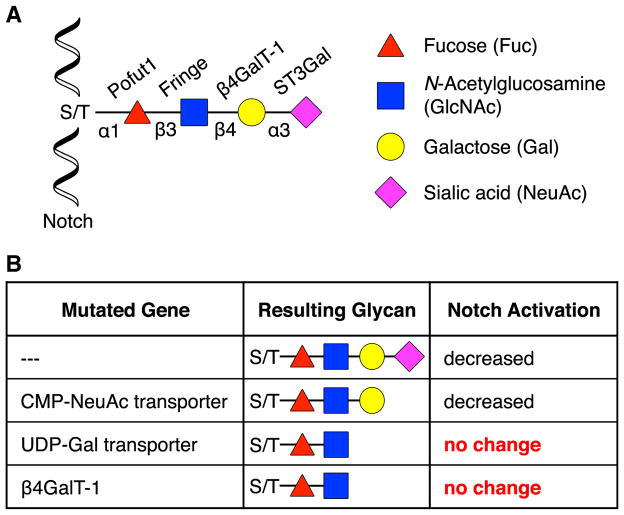
Figure 2. Genetic Studies of Notch.
(A) Structure of the O-fucose glycan found on mammalian Notch.
(B) The O-fucose glycan of Notch causes a decrease in Jagged1-mediated stimulation. By eliminating specific monosaccharides on the Notch glycan using genetic methods, it was found that a trisaccharide was the minimum structure necessary to decrease Jagged1 activity.
Mass spectrometry analysis indicated that the O-Fuc glycans on mammalian Notch exist as a mixture of di-, tri-, and tetrasaccharides of NeuAcα(2–3)Galβ(1–4)GlcNAcβ(1–3)Fuc (Moloney et al., 2000). To decipher the minimum structure necessary for activity, a library of mutant Chinese hamster ovary (CHO) cell lines deficient in GTs and activated nucleotide (NXP)-sugar transporters was employed (Figures 1C and 2B) (Chen et al., 2001). Expression of Fringe in parental CHO cells led to a decrease in Jagged1 stimulation of Notch, presumably due to the loss of Jagged1-Notch complexation mediated by the O-Fuc glycan. However, the Lec8 cell line, which contains mutations in the uridine diphosphate (UDP)-Gal Golgi transporter that result in defective protein galactosylation, showed no change in Notch activation upon Fringe expression, suggesting that the Gal moiety was required to disrupt the Jagged1-Notch interaction. Conversely, the Lec2 cell line containing an inactive cytidine monophosphate (CMP)-NeuAc Golgi transporter that prevents protein sialylation showed the same response to Jagged1 stimulation as the parental cell line, indicating that the NeuAc residue was unnecessary. Of the six mammalian β-1,4-galactosyltransferase (β4GalT) enzymes that transfer Gal to GlcNAc in CHO cells, only β4GalT-1 was required for Notch stimulation, providing the identity of a key enzyme involved in synthesizing the minimum trisaccharide. Thus, genetic approaches not only illustrate the necessity of specific glycans for activity in physiological systems but also can be systematically used to determine their structure and the enzymes required for their biosynthesis.
Another example of the power of genetic approaches to reveal important developmental functions of glycans involves studies on heparan sulfate (HS) glycosaminoglycans (GAGs). HS binds to diffusible chemical signals known as morphogens and aids in establishing gradients of these secreted proteins to relay positional information in developing organisms (Figure 3A) (Häcker et al., 2005). Phenotypic screens in Drosophila showed that mutations in tout-velu (ttv), a homolog of the mammalian HS polymerase Ext1, caused pattern defects in the wing imaginal disc similar to the loss of Hedgehog (Hh) signaling (Bellaiche et al., 1998; The et al., 1999). The morphogen Hh specifies anterior-posterior orientation during wing development by diffusing from the posterior compartment of the imaginal disc to anterior cells in a graded fashion (Tabata and Kornberg, 1994). To elucidate the importance of ttv, mosaic knockout clones were produced in which defined regions of the Drosophila embryo lacked ttv (Bellaiche et al., 1998). Loss of ttv in the posterior compartment where Hh is secreted showed no effect on the distance of Hh-mediated activation into the anterior compartment. However, anterior ttv−/− clones showed a severe spatial attenuation of active Hh signaling in the anterior compartment. Defective distribution of Hh signaling was also observed in mutant clones of the Ext2 homolog sister-of-tout-velu (sotv) and the Extl3 homolog brother-of-tout-velu (botv) (Han et al., 2004), suggesting that the HS produced by these enzymes is necessary for Hh binding, gradient formation, and signal transduction. Furthermore, wingless (Wg) and decapentaplegic (dpp) morphogen signaling were shown to be disrupted by altered HS biosynthesis and display (Baeg et al., 2004; Belenkaya et al., 2004; Kirkpatrick et al., 2004), providing evidence for a general mechanism by which HS mediates morphogen gradients.
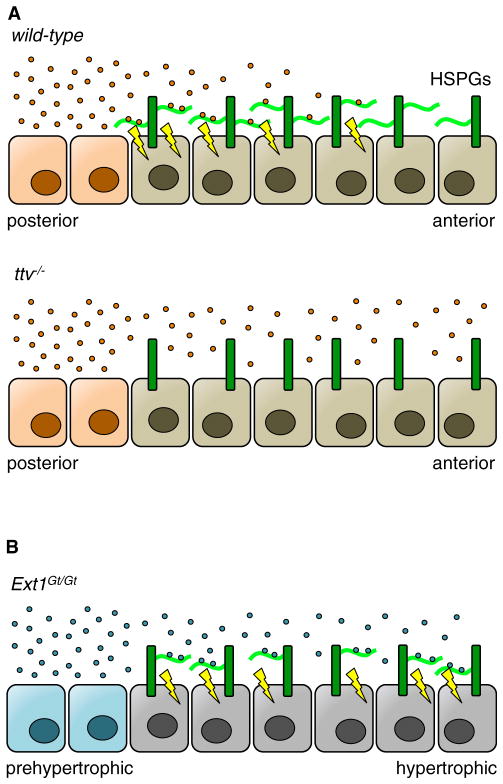
Figure 3. Genetic Studies Provide Insights into How HS Glycans Control Morphogen Gradients.
(A) In wild-type Drosophila, Hedgehog ligands (Hh, orange) are secreted from posterior cells of the imaginal wing disc and diffuse to anterior cells. HS chains (light green) on HS proteoglycans (HSPGs, dark green) displayed on the cell surface trap these ligands for cellular signaling and prevent further diffusion. However, the lack of HS in ttv−/− Drosophila prevents Hh recruitment to the cell surface and downstream signaling.
(B) Mice with a hypomorphic mutation in Ext1 (Ext1Gt/Gt) produced less HS on the cell surface. When Indian hedgehog ligands (Ihh, blue) were secreted by prehypertrophic chondrocytes, the reduced amount of cell-surface HS allowed Ihh to diffuse further into the hypertrophic region. Unlike full knockout as seen in the ttv−/− Drosophila, chondrocytes with a hypomorphic mutation in Ext1 maintained enough cell-surface HS to transduce signaling, but not enough to impede diffusion.
Interestingly, ostensibly conflicting data were obtained from mouse models (Koziel et al., 2004). Here, hypomorphic mutation of Ext1, which severely limits HS production, led to an extended tissue distribution of Indian hedgehog (Ihh), the mouse homolog of Hh, during endochondral ossification (Figure 3B). Using haploinsufficiency studies in which one or both alleles of Ext1 were replaced with the hypomorphic variant, lower production of HS correlated with higher rates of proliferation in a region of chondrocytes distal from Ihh secretion, implying increased Ihh diffusion. In vitro growth of embryonic mouse forelimbs with exogenous GAGs showed that Ihh diffusion was inversely proportional to HS concentration, suggesting that HS retarded the diffusion of Ihh, unlike the results seen for ttv in Drosophila. It has been proposed that the disparity in these results is due to differences in the genetic approaches used (Häcker et al., 2005). As HS is required for ligand-receptor binding, the use of a null allele for ttv prohibits all HS biosynthesis and Hh-mediated signaling. In contrast, the hypomorphic Ext1 allele produces enough HS to elicit Ihh-protein complexation, and the lower overall amount of HS may facilitate further morphogen diffusion. Thus, complete ablation of HS in Drosophila decreases Hh signaling due to a loss of ligand-receptor binding, but the decrease of HS in mice increases Ihh signaling by allowing greater diffusion of Ihh. The interpretation of these results highlights that careful consideration is necessary in selecting genetic models due to the multi-faceted nature of glycan-protein interactions in vivo.
The examination of naturally occurring mutations in human genes provides a complementary approach to establish the molecular determinants that underlie development and disease-related phenotypes. Human genetic mutations that lead to abnormal glycosylation have been well documented and span a wide clinical spectrum, impacting nearly every organ system in the body. Advances in next-generation DNA sequencing technologies have helped to propel the discovery of >100 such congenital disorders of glycosylation (CDGs), providing unique opportunities to understand the significance of glycosylation in human physiology. As CDGs have been well reviewed (Freeze, 2013; Hennet and Cabalzar, 2015), we highlight only a few representative examples.
Similar to classical forward genetics, genetic mapping of disease-related mutations has led to the discovery of novel genes responsible for a single set of pathological symptoms. Hereditary multiple exostoses (HME), a condition characterized by abnormal growths at the ends of long bones, has been associated with loss-of-function mutations in the Ext1, Ext2, and Ext3 genes by chromosomal linkage analysis (Cook et al., 1993; Le Merrer et al., 1994; Wu et al., 1994). These genes were later determined to encode GTs responsible for HS biosynthesis (McCormick et al., 1998). Missense mutations that were mapped for Ext1 were shown to cause a deficiency in HS production. As described above, decreased levels of HS results in increased activation of Ihh (Koziel et al., 2004). This signaling dysregulation triggers uncontrolled cellular growth and leads to cartilaginous abscesses and benign tumors in endochondrial bone. Thus, this particular example highlights how the mutational profile of a disease enhanced an understanding of the molecular basis of exostoses formation.
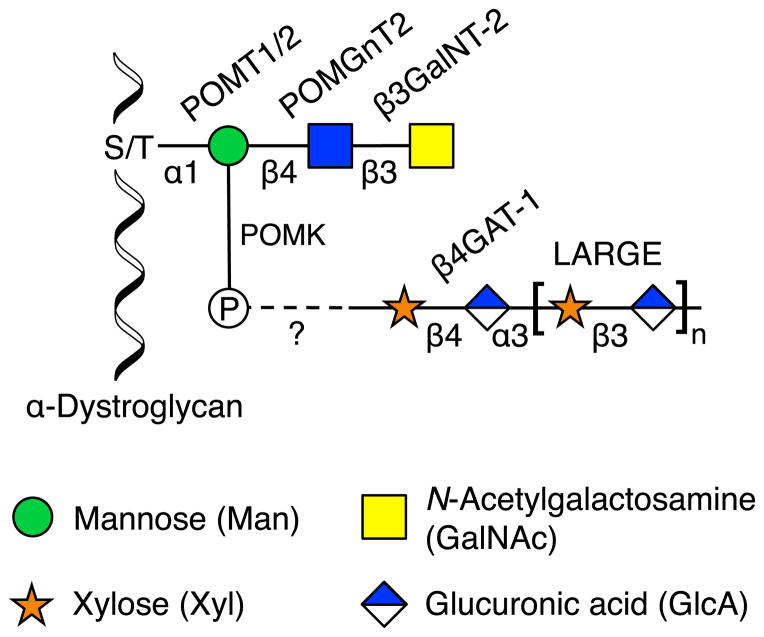
Genetic mapping of distinct but related disease states identified multiple important genes that are components of the same glycosylation pathway. Mutations in at least ten enzymes have been linked to congenital muscular dystrophies (Wells, 2013), which are commonly associated with defective O-glycosylation of α-dystroglycan, an integral component of muscle fibers (Figure 4). The O-glycans of α-dystroglycan mediate interactions with extracellular matrix proteins such as laminin (Yoshida-Moriguchi et al., 2010) and facilitate the formation of compact basement membranes and mature neuromuscular junctions (Goddeeris et al., 2013). Loss of these glycans disrupts extracellular matrix organization and predisposes muscular tissue to dystrophy. Walker-Warburg syndrome and muscle-eye-brain disease, two similar types of muscular dystrophy typified by anomalies of the brain and eye, were linked to a number of proteins that produce O-mannosyl glycans, including protein O-mannosyl-transferase 1 and 2 (POMT1/2) (Beltran-Valero de Bernabé et al., 2002; van Reeuwijk et al., 2005), protein O-mannosyl N-acetylglucosaminyltransferase 1 and 2 (POMGnT1/2) (Manzini et al., 2012; Yoshida et al., 2001), β-1,3-N-acetylgalactosaminyltransferase 2 (β3GalNT2) (Stevens et al., 2013), and protein O-mannosyl kinase (POMK) (Yoshida-Moriguchi et al., 2013). Merosin-deficient congenital muscular dystrophy 1D (MDC1D) was associated with mutations in like-acetylglucosaminyltransferase (LARGE) (Longman et al., 2003), which transfers GlcAβ(1–3)Xylα(1–3) repeating units onto the O-mannosyl glycan (Goddeeris et al., 2013). Finally, patients with Fukuyama-type congenital muscular dystrophy, a frequent disorder in Japan that presents as brain micropolygria due to improper neuronal migration, contained mutations in a novel gene called fukutin (FKTN) (Kobayashi et al., 1998). Although its exact function remains unknown, FKTN was postulated to be a glycosyltransferase and was found to be required for LARGE activity (Ohtsuka et al., 2015). Thus, an examination of these and other α-dystroglycanopathies converged on genes involved in the synthesis of the α-dystroglycan O-mannosyl glycan, linking the biosynthetic pathway of a complex glycan to a suite of developmental disorders.
Figure 4. O-Mannosyl Glycan of α-Dystroglycan.
The laminin-binding glycan of α-dystroglycan contains a core trisaccharide produced by POMT1/2, POMGnT2, and β3GalNT-2. The 6-OH position of mannose is phosphorylated by POMK, on which an unknown structure is added. β4GAT-1 makes an acceptor substrate for LARGE, which adds repeating GlcAβ(1–3)Xylα(1–3) units. The required roles of other associated enzymes such as FKTN are still unknown.
Next-generation sequencing has allowed for both targeted and untargeted screens to identify genes involved in the etiology of glycosylation-related diseases. Targeted sequencing of 25 known CDG-related genes uncovered no observable mutations in a patient with widespread mental and physical developmental delays (Jones et al., 2012). Subsequent untargeted whole-exome sequencing against roughly 22,000 genes revealed two mutations that caused premature truncation of DDOST, a subunit of the oligosaccharyltransferase (OST) complex. The DDOST mutations impaired cell-surface expression of intracellular adhesion molecule 1 (ICAM-1), whose trafficking to the cell surface depends on proper N-glycosylation. Complementation with wild-type DDOST partially restored ICAM-1 trafficking, indicating that the DDOST mutations were indeed pathogenic and that their clinical manifestations were likely due to widespread defects in N-glycosylation. Untargeted genetic approaches have also revealed unanticipated genes important for proper glycosylation. Microarray and sequencing analysis of genes within autozygous regions of two glycosylation-deficient siblings with multiple forms of dysplasia uncovered mutations in TMEM165 (Foulquier et al., 2012). This protein has no known function but is implicated in maintaining calcium levels within the Golgi (Demaegd et al., 2013), suggesting that proper glycosylation relies on not only GTs and transporters but also homeostatic regulation of Golgi pH and ion concentrations. The identification of DDOST and TMEM165 mutations by these methods illustrates the ability of next-generation sequencing to reveal pathologies caused by glycosylation defects and potentially expose novel regulatory mechanisms of glycosylation as targets for therapeutic intervention.
Thus, genetic methods provide a powerful means to understand the importance of glycosylation in development and disease. However, knockout, knockdown, and overexpression approaches result in loss of glycan structures or enhanced expression of only natural structures. A complementary approach is the use of chemical biology methods that enable expansion past the confines of naturally occurring glycans through incorporation of non-natural sugars. These non-natural labeling approaches have facilitated the imaging, tracking, and identification of glycan structures. Beyond advancing the study of glycans, such approaches provide a novel way to control cellular physiology and function by altering the glycan structures presented on the cell surface. Below, we highlight a few notable examples whereby cell-surface glycan engineering has been exploited to interrogate and control glycan-mediated biological processes.
Metabolic Oligosaccharide Engineering
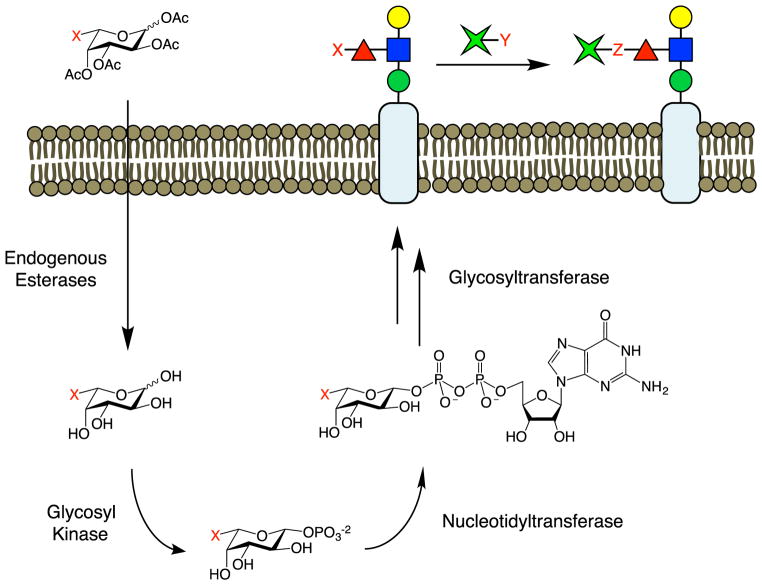
Metabolic oligosaccharide engineering (MOE) is a widely used method for modifying cellular glycans with non-natural sugars (Dube and Bertozzi, 2003; Laughlin and Bertozzi, 2009a; Rouhanifard et al., 2013). In this method, non-natural monosaccharide analogs are taken up by cells into biosynthetic salvage pathways, converted into activated nucleotide phosphate donors, and incorporated by GTs into specific glycan structures (Figure 5). A range of non-natural sugar analogs based on Fuc, NeuAc, GalNAc, and GlcNAc have been successfully installed in a variety of cells and organisms such as zebrafish and Caenorhabditis elegans. In principle, the analog is inserted into all glycans containing the monosaccharide of interest, which allows for the simultaneous detection of multiple glycan structures. However, this also precludes the selective detection of specific structures, such as motifs that differ by neighboring glycans or by glycosidic linkage. As the non-natural analogs compete with natural substrates, their incorporation into cellular glycans is sub-stoichiometric, which minimizes perturbation to the system but also reduces detection sensitivity. The analogs must also contain relatively small non-natural functionalities to allow efficient recognition by enzymes and incorporation into glycans. Nonetheless, a number of chemical groups, including inert alkyl chains (Keppler et al., 1995), as well as reactive groups such as ketones, thiols, azides, alkynes, alkenes, isonitriles, and cyclopropenes have been installed (Cole et al., 2013; Hang et al., 2003; Hsu et al., 2007; Laughlin et al., 2008; Mahal et al., 1997; Sampathkumar et al., 2006; Späte et al., 2014; Stairs et al., 2013). Reactive groups have enabled the covalent attachment of a variety of small-molecule reporters (e.g., biotin, fluorescent dyes, crosslinking agents) via bioorthogonal chemistry (McKay and Finn, 2014; Sletten and Bertozzi, 2009). Consequently, MOE provides a facile platform for endowing glycan structures with new chemical functionalities.
Figure 5. Metabolic Oligosaccharide Engineering.
MOE requires the passage of peracetylated glycans across the cell membrane and into the cell, where they are deacetylated by endogenous esterases. The monosaccharides are phosphorylated and activated with a nucleotide phosphate group. Glycosyltransferases then incorporate the non-natural sugars into nascent glycans. The functional handle (X) installed by the biosynthetic machinery can be reacted with other groups (Y) via bioorthogonal chemistry (X + Y produce Z) for detection, imaging, or introduction of novel biological activity.
A beautiful example is the application of MOE to image cell-surface glycans in developing zebrafish (Laughlin et al., 2008). By incubation of embryos with peracetylated N-azidoacetylgalactosamine (Ac4GalNAz), followed by two different strained cyclooctyne-conjugated fluorophores, newly synthesized glycans were distinguished from older glycans and tracked in vivo. Interestingly, distinct regions of the embryo containing only newly synthesized glycans were observed, suggesting that the production of glycoproteins is highly choreographed during differentiation. To image glycosylation at earlier stages of development, N-azidoacetylsialic acid (NeuAz/SiaNAz) was microinjected into zebrafish embryos to bypass diffusion into the cell and label sialic acid-containing glycans (Dehnert et al., 2012). Dual-labeling experiments were also coducted in C. elegans with Ac4GalNAz to distinguish regions of active glycan production during both larval and adult stages of development (Laughlin and Bertozzi, 2009b). Thus, MOE provides a powerful approach to visualize the spatiotemporal dynamics of glycan biosynthesis in live organisms.
In addition to imaging applications, MOE has been used to identify the binding partners of lectins such as the sialic acid-binding immunoglobulin-like lectin (Siglec) CD22 (Han et al., 2005). CD22 functions as a negative regulator of B cell receptor activation by establishing a minimum signaling threshold to prevent aberrant immune reactions (Nitschke et al., 1997). Although it was known that cis interactions of CD22 with glycoproteins on the same cell modulated CD22 activity, the identity of the glycoprotein(s) involved was unknown. Cells were incubated with NeuAc containing an aryl-azide moiety (9-AAz-NeuAc) to photo-chemically crosslink NeuAc-bearing glycoproteins to their cis-binding partners. Surprisingly, the only proteins pulled down with CD22 were other CD22 receptors, which formed self-regulating, homomultimeric complexes. To find the trans-binding partners of CD22, the soluble ectodomain of CD22 was photo-chemically crosslinked to cells displaying 9-AAz-NeuAc-containing glycans (Ramya et al., 2010). The B cell receptor IgM was discovered to be the preferred trans-binding partner. Notably, IgM did not appear to crosslink to CD22 when presented in cis, underscoring the ability of this MOE approach to dissect components of different CD22 binding complexes.
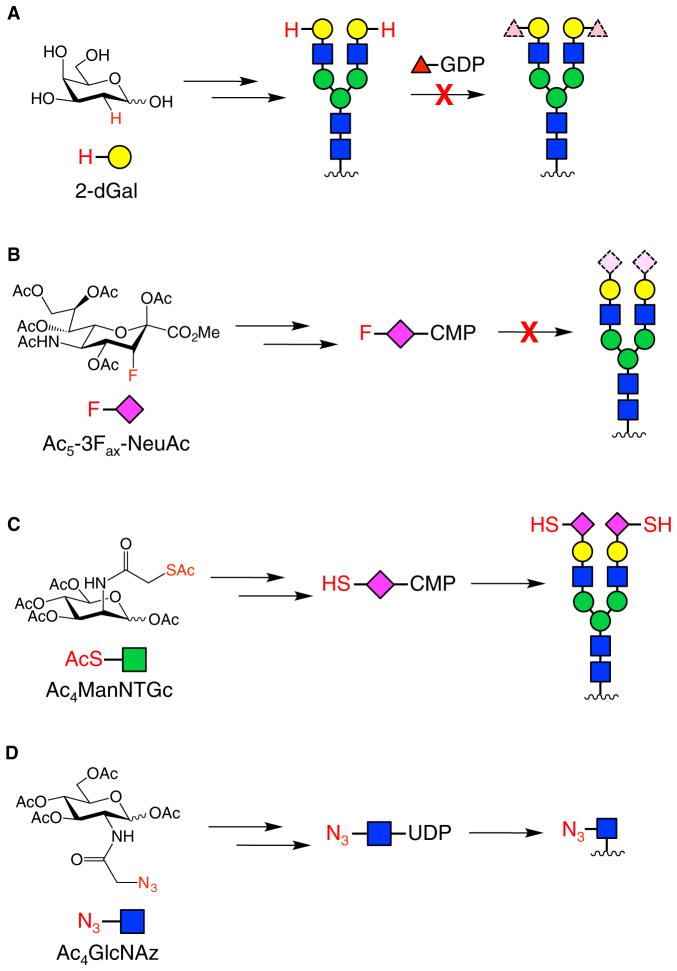
The ability of MOE to remodel glycan structures at the cell surface provides a powerful means to modulate cellular responses. For example, Fucα(1–2)Gal-containing glycans have been implicated in cognitive processes such as learning and memory. To study these glycans, 2-deoxygalactose (2-dGal) was used to prevent the attachment of Fuc to the 2-position of Gal in the Fucα(1–2)Gal structure (Figure 6A). Intracerebral injection of 2-dGal into day-old chicks induced reversible amnesia and interfered with the maintenance of long-term potentiation (Bullock et al., 1990; Krug et al., 1991; Rose and Jork, 1987), an electro-physiological model of learning and memory. The amnesic effect was not observed with other monosaccharides, and memory formation was rescued by co-injection with Gal but not Fuc (Rose and Jork, 1987). At the cellular level, 2-dGal decreased neurite outgrowth of embryonic cortical neurons and induced collapse of synapses, while 3-deoxygalactose (3-dGal) and 6-deoxygalactose (6-dGal) had no effect (Kalovidouris et al., 2005; Murrey et al., 2006). Treatment with Gal reversed the inhibitory effects of 2-dGal on neurite outgrowth, presumably by enabling re-synthesis of Fucα(1–2)Gal-containing glycans (Kalovidouris et al., 2005). Interestingly, Fucα(1–2)Gal glycans were enriched on synaptic proteins such as synapsin Ia and Ib (Murrey et al., 2006), which regulate neurotransmitter release and synapse formation. 2-dGal disrupted the fucosylation of synapsin and promoted its degradation. Thus, the alteration of Fucα(1–2)Gal glycans on cell surfaces using non-natural sugars has provided insights into the functions of these glycans in the nervous system, suggesting that they play roles in the regulation of important synaptic proteins and morphological changes that underlie synaptic plasticity.
Figure 6. Non-Natural Monosaccharides Used for MOE.
(A) 2-dGal was incorporated into cell-surface glycans to prevent attachment of Fuc in neurons. A representative glycan containing terminal Fucα(1–2)Gal is shown.
(B) Fluorinated analogs of Fuc and NeuAc were metabolized to their respective nucleotide phosphate donors and used to inhibit native GTs and affect leukocyte rolling velocity.
(C) Ac5ManNTGc was converted to a thiol-containing analog of NeuAc and incorporated into cell-surface glycans to alter cell-cell adhesion and stem cell differentiation.
(D) Ac4GlcNAz was selectively incorporated into cell-surface glycans of Helicobacter pylori, allowing the installation of a functional handle into unknown glycan structures to elicit an immune response against the bacteria.
An alternative to inhibiting protein glycosylation with deoxy sugars is the use of fluorinated sugar analogs, which can serve as inhibitors of GTs. Peracetylated and fluorinated Fuc and NeuAc analogs were used to block protein fucosylation and sialylation, respectively, through a two-pronged mechanism (Figure 6B) (Rillahan et al., 2012). First, the fluorinated monosaccharide inhibitors were converted by salvage pathways to the corresponding activated nucleotide sugar donors. Once activated, the inhibitors bound to GTs but were not transferred to nascent glycan structures, preventing protein fucosylation or sialylation. The resulting accumulation of sugar donors also stimulated feedback inhibition loops that suppressed de novo biosynthesis of Fuc or NeuAc. This approach was utilized to inhibit the production of Lewis X (LeX) and sialyl LeX antigens, resulting in reduced binding to E- and P-selectins and enhanced leukocyte rolling, a process important for fighting infections.
The remodeling of cell-surface glycans with reactive chemical handles provides another powerful means to modulate cellular responses. For example, the incorporation of a thiol group into NeuAc-containing glycoproteins promoted spontaneous cell-cell clustering of Jurkat T cell lymphoma cells, as well as lineage-specific differentiation of human embryoid body-derived (hEBD) stem cells (Figure 6C) (Sampathkumar et al., 2006). The hEBD cells showed a decrease in proliferation, along with an increase in β-catenin expression and morphological changes consistent with differentiation to a neural cell type. Moreover, Jurkat cells decorated with thiol-containing NeuAc could be immobilized onto gold or maleimide-functionalized glass surfaces without compromising cell viability. Thus, the remodeling of cell surfaces with non-natural sugars can both change their adhesive properties and artificially induce cellular signaling processes.
In another notable example, MOE was used to trigger an artificial immune response against Helicobacter pylori, a bacterium responsible for stomach ulcers (Kaewsapsak et al., 2013). Although tetraacetyl N-azidoacetylglucosamine (Ac4GlcNAz) is incorporated primarily into cytosolic glycoproteins in mammalian cells, H. pylori robustly processed the sugar for cell-surface display (Figure 6D). The azido-functionalized glycoproteins on the cell surface were subsequently conjugated to an immunostimulant, 2,4-dinitrophenol (DNP), via Staudinger ligation. After labeling, the bacteria were treated with anti-DNP antibodies, leading to a nearly two-fold increase in bacterial cell death, as measured by an antibody-dependent cell-mediated cytotoxicity assay. This method highlights how the differential metabolic utilization of glycans can potentially be exploited to selectively target and elicit an immune response against pathogenic bacteria.
Overall, MOE provides a versatile approach to control the glycan architecture on cell surfaces through the incorporation of non-natural monosaccharides. In addition to facilitating the detection and study of glycoconjugates, MOE can be exploited to modulate cellular processes and possibly engineer new activities through chain termination, installation of reactive groups, and functionalization with bioactive compounds.
Chemoenzymatic Labeling
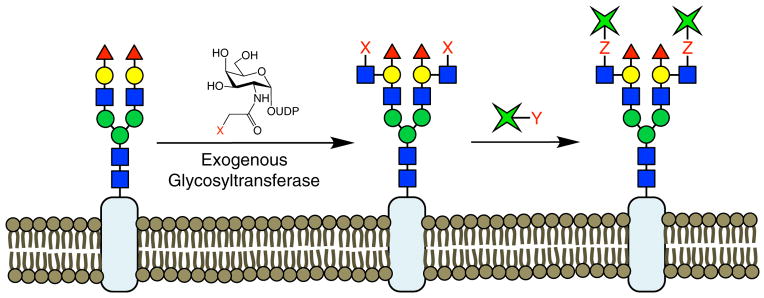
With MOE, the non-natural analog in principle can be incorporated into all glycans containing the monosaccharide of interest. However, determining the precise glycan structures labeled by the non-natural sugar and demonstrating labeling specificity can be challenging in practice and can complicate efforts to study glycan function (Boyce et al., 2011; Chuh et al., 2014). The chemoenzymatic labeling (CL) of glycans represents a complementary strategy to remodel glycans with non-natural functionalities. In this approach, an exogenous GT transfers a non-natural sugar that contains a reactive group onto the glycan structure of interest (Figure 7). Subsequent reaction allows for the attachment of a variety of chemical reporters as readouts for the glycan of interest. The specificity of the exogenous GT can be determined using glycan microarrays, and this approach can be used to distinguish and selectively label disaccharide or higher-order motifs of specific sugar composition and glycosidic linkage, such as Fucα(1–2)Gal and N-acetyllactosamine (LacNAc). Another advantage of this approach is that the chemo-enzymatic reactions often proceed quantitatively, which leads to highly sensitive detection. Finally, this system can be employed in biological contexts where feeding cells non-natural sugar analogs is not possible, such as the ex vivo detection of disease biomarkers. Although identifying a suitable GT that recognizes the motif of interest is no small task and may limit the overall scope of the approach, CL has been used both to provide novel insights into glycan function and to control cellular processes.
Figure 7. Chemoenzymatic Labeling.
In CL, cells are treated with a purified, exogenous GT and an appropriate nucleotide phosphate donor to stoichiometrically attach non-natural sugars onto glycans of defined structure. The functional handle (X) can be further elaborated using bioorthogonal chemistry to detect and study glycans or to endow cells with new biological properties.
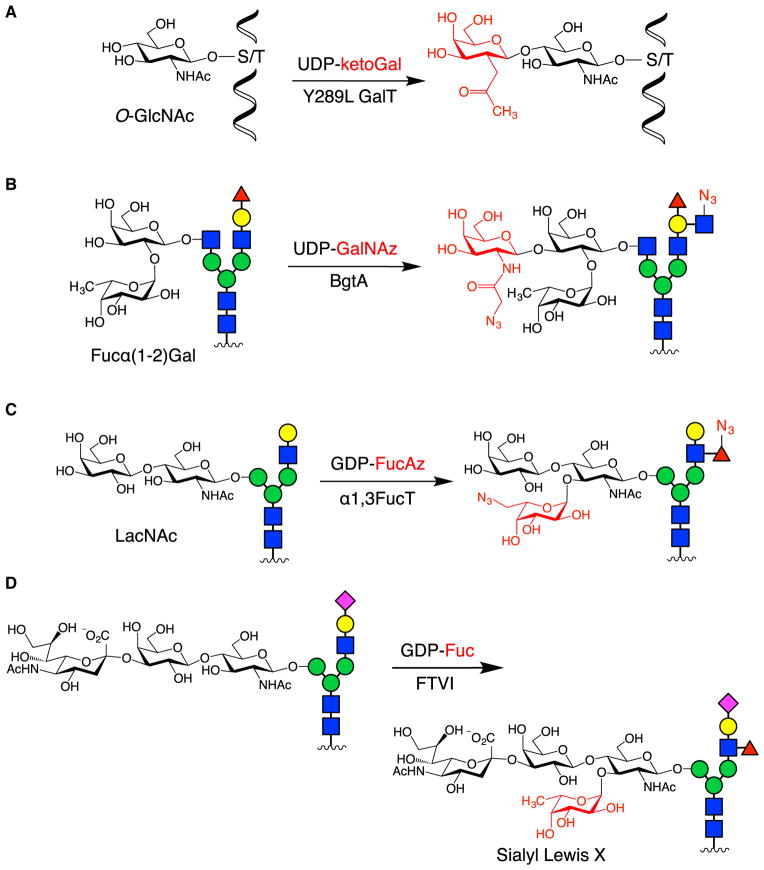
A powerful example is the application of CL to study the function of O-linked GlcNAc glycosylation on cAMP response element-binding protein (CREB), a crucial transcription factor involved in neuronal development, plasticity, and long-term memory formation (Lonze and Ginty, 2002). Whereas traditional methods such as wheat germ agglutinin lectin and O-GlcNAc antibodies failed to detect O-GlcNAc glycosylated CREB, CL using an engineered β-1,4-galactosyltransferase enzyme and UDP-2-acetonyl-2-deoxygalactose (UDP-ketoGal) allowed for its bio-tinylation and subsequent rapid, sensitive detection (Figure 8A) (Khidekel et al., 2003; Rexach et al., 2012). To determine the stoichiometry of glycosylation, O-GlcNAc glycosylated proteins from neuronal lysates were labeled with a 2,000-Da polyethylene glycol mass tag (Rexach et al., 2010), which shifted the molecular weight of the glycosylated proteins. Immunoblotting for CREB allowed for quantification of the glycosylated species, demonstrating that nearly 50% of the CREB population was O-GlcNAc-modified in neurons (Rexach et al., 2012). Furthermore, expression of CREB mutants in which glycosylation sites were mutated to alanine, followed by measurement of their glycosylation stoichiometry, enabled determination of the major glycosylation site in neurons, which was also found to be the site induced by neuronal depolarization. Identification of this key site facilitated in-depth functional studies and revealed that glycosylation at this site repressed both basal and activity-induced CREB-mediated transcription, regulating both neuronal growth and long-term memory.
Figure 8. Examples of CL.
(A) The O-GlcNAc modification was detected and quantified by labeling with ketoGal, followed by installation of a biotin or PEG mass tag.
(B) The Fucα(1–2)Gal epitope was selectively labeled using BgtA to install GalNAc derivatives with functional handles for downstream detection.
(C) α1,3FucT from H. pylori modified LacNAc moieties with Fuc derivatives for flow cytometry and imaging analysis.
(D) Human FTVI was used to functionalize the N-glycans of CD44 on MSCs, thereby producing SLeX moieties to help target MSCs to bone marrow tissue.
In another example, a CL approach was developed to detect the potential biomarker Fucα(1–2)Gal, which is upregulated in inflammatory diseases and cancer (Figure 8B) (Chaubard et al., 2012). Terminal Fucα(1–2)Gal residues on complex cell-surface glycans were labeled using a bacterial homolog of the human blood group A antigen GT (BgtA) and UDP-GalNAz or UDP-keto-Gal. Notably, no mutagenesis of the BgtA active site was required to transfer the non-natural sugars, highlighting the substrate promiscuity of prokaryotic GTs. Glycan microarray data demonstrated that BgtA specifically labeled glycans containing a terminal Fucα(1–2)Gal disaccharide motif, in contrast to MOE, which would have labeled all fucosylated glycans. Moreover, BgtA tolerated a variety of distal architectures appended to the reducing end of Gal, labeling diverse N- and O-glycan structures with a terminal Fucα(1–2)Gal moiety, including Globo H and Fuc-GM1. Fluorescent labeling of Fucα(1–2)Gal glycans and flow cytometry analysis demonstrated a significant increase in Fucα(1–2)Gal expression on prostate cancer cells compared with normal prostate epithelial cells. Thus, this CL method may provide a novel tool to discriminate cancerous cells from normal cells and to study glycans associated with tumor pathogenesis.
CL has also been applied to detect and image LacNAc, a universal component of the antennae of hybrid- and complex-type N-glycans (Zheng et al., 2011). Here, α-1,3-fucosyltransferase (FucT) from H. pylori was combined with GDP-Fuc derivatives to label LacNAc on the surface of cell lines, primary spleenocytes, and zebrafish embryos (Figure 8C). To demonstrate its efficacy at labeling cells ex vivo, T and B cells isolated from mice were treated in situ with exogenous FucT and GDP-6-azidofucose (GDP-FucAz), followed by reaction with strained cyclooctyne-conjugated biotin and incubation with fluorescently labeled streptavidin. Flow cytometry analysis of the labeled cell population revealed that activated immune cells produced higher fluorescence signal than their naïve counterparts, suggesting that immune cell activation increased cell-surface LacNAc. When applied to zebrafish, only a short incubation with the non-natural sugar analog and exogenous enzyme were needed to produce robust labeling, thereby obviating the need for microinjection and minimizing perturbation to the developing animal. Moreover, FucT tolerated fluorescein-labeled GDP-Fuc as a substrate, which avoided the subsequent dye functionalization step.
In addition to glycan detection, the modification of cell-surface glycans by CL has been exploited to control biological activity. Although the use of mesenchymal stem cells (MSCs) to regrow tissue and cure skeletal diseases holds great potential (Horwitz et al., 2002), injected MSCs often do not migrate efficiently and selectively to the tissues of interest. To provide a homing target for circulating MSCs, the CD44 glycan on MSCs was modified ex vivo by human-derived α-1,3-fucosyltransferase (FTVI) to produce the SLeX glycan motif (Figure 8D) (Sackstein et al., 2008). This motif binds to E-selectin receptors that are specifically expressed in bone marrow tissue. Engineered HCELL+ MSCs showed much stronger sheer-resistant adhesive interactions with E-selectin-expressing endothelial cells compared with un-modified MSCs, suggesting that the Fuc modification of CD44 glycans endowed cells with new adherent properties. Furthermore, HCELL+ MSCs that were injected intravenously into mice exhibited much greater levels of homing to bone marrow tissue, highlighting the ability of CL to engineer cellular functions in vivo.
Overall, CL provides a complementary alternative to MOE for labeling and modifying intracellular and cell-surface glycans. Both methods have strengths in particular biological systems and can alter the properties of cells. In the future, mining of prokaryotic genomes may reveal many novel GT candidates, underscoring the opportunity to expand the collection of GTs and chemical diversity of glycan architectures remodeled by CL.
De novo Glycan Display
The methods described thus far involve the modification of natural glycans on cell surfaces by manipulating the biosynthetic machinery used for glycan production. An alternative approach, which we term de novo glycan display, focuses on inserting defined glycan or glycomimetic structures directly into plasma membranes using approaches such as lipid insertion, liposomal fusion, or protein conjugation (Frame et al., 2007; Huang et al., 2014; Hudak et al., 2013; Paszek et al., 2014; Pulsipher et al., 2014, 2015). De novo display methods provide excellent control over the glycan structure of interest and enable the presentation of both small and large glycans at the cell surface. Moreover, this approach is not constrained by the substrate specificity of natural enzymes, thus expanding the scope of glycan engineering to complex structures unattainable by other methods. However, exogenous sugars are typically displayed alongside the native glycan population, which could obscure the biological effects of the newly added carbohydrates. To address this complication, de novo glycan display methods can be used in combination with genetic depletion approaches to minimize the contributions of interfering endogenous glycans (Huang et al., 2014). The versatility of the technique also allows for the display of a wide range of carbohydrate-based structures, including glycomimetics such as synthetic glycopolymers, glycans appended to simplified proteins, or even the glycan component of glycoproteins alone (Huang et al., 2014; Hudak et al., 2013; Paszek et al., 2014; Pulsipher et al., 2014, 2015). Despite the use of minimalist structures and possible interference from endogenous carbohydrates, de novo glycan display has provided remarkable insights into a variety of biological systems.
One of the first examples of de novo glycan display examined the effects of altering blood group antigens on erythrocyte cell surfaces (Frame et al., 2007). Blood type is determined by the presence of specific glycolipid structures, the O-, A- and B-type antigens, on red blood cells. Synthetic glycolipid mimics that contained each antigen type were incubated with O-type erythrocytes (which lack A- and B-type antigens) to allow for lipid insertion of the glycomimetics in the membrane. The newly remodeled blood cells displaying the artificial A- and B-type glycolipids acquired a strong immune response to anti-A and anti-B antibodies, respectively. This proof-of-concept approach demonstrated that de novo glycan display through the passive insertion of lipid-anchored glycans into membranes could be used to control biological responses.
More recently, the immunoevasive properties of tumorigenic cells were examined through membrane insertion of synthetic glycopolymers. Many cancer cells develop mechanisms such as increased surface sialylation to avoid detection by natural killer (NK) cells in the innate immune response (Büll et al., 2014; Fuster and Esko, 2005). Using glycan-functionalized polymers containing a terminal phospholipid, different carbohydrates were anchored to the surface of cancer cells (Figure 9A) (Hudak et al., 2013). Cells were then co-incubated with NK cells to determine if the exogenous glycans recapitulated the naturally observed avoidance of NK-mediated cell death. Only sialylated NeuAc-containing polymers significantly attenuated cytotoxicity, and the polymer-displaying cells also were found to prevent NK degranulation, a key process in cell killing. Notably, the NeuAc polymers protected bone marrow-derived hematopoietic stem cells against NK-mediated cytotoxicity, suggesting that de novo glycan display of such polymers could help prevent cellular rejection during transplantation.
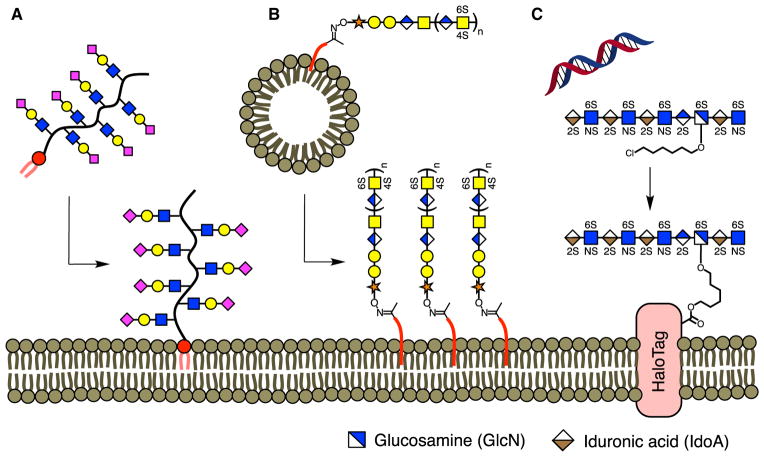
Figure 9. De novo Display of Cell-Surface Glycans.
(A) NeuAc-containing glycans and HS GAGs were chemically appended to a ketone-functionalized polymer and inserted into the plasma membrane using a phospholipid tail.
(B) CS GAGs were displayed on the cell surface by functionalization onto ketone-containing liposomes.
(C) Genetic incorporation of the HaloTag protein allowed for the immobilization of HS GAGs modified with a chlorohexyl linker.
The remodeling of cell surfaces with lipid-anchored glycopolymers has also provided key insights into the mechanobiology of integrin signaling. Integrins are cell-surface receptors that regulate cancer cell migration, invasion, and proliferation (Guo and Giancotti, 2004). Computational modeling studies predicted that the glycocalyx promotes integrin receptor clustering and enhances integrin signaling through mechanical interactions between bulky carbohydrates and transmembrane proteins (Paszek et al., 2009). To test this hypothesis, synthetic mucin-like glycopolymers of different lengths were conjugated to a phospholipid tail and inserted into the membranes of mammary epithelial cells (Paszek et al., 2014). Cells displaying longer glycopolymers (80 nm) showed segregated regions of clustered integrins, whereas cells decorated with shorter glycopolymers (3 and 30 nm) showed no effect. The larger glycopolymers were found to expand the distance between the cell surface and the extracellular matrix by 19 nm, as determined by scanning angle interference microscopy. This larger spacing enhanced the clustering of integrins into fewer, larger focal adhesions, promoting the formation of active signaling nodes on the plasma membrane by physically restructuring the cell surface. Corroborating these data, overexpression of mucin glycoprotein MUC1 recapitulated the overall heightened membrane and isolated regions of focal adhesion formation. Thus, de novo glycan display provided a facile method to modulate systematically the chemical composition and physical parameters of the glycocalyx.
De novo glycan display has also provided a physiologically relevant model for studying developing neurons in the hippocampus. Neuronal growth and synapse formation are highly regulated processes in the hippocampus, a brain region critical for the consolidation of short- and long-term memory. Studies suggested that chondroitin sulfate glycosaminoglycans (CS GAGs) containing certain sulfation motifs can stimulate the outgrowth of embryonic hippocampal neurons by recruiting growth factors to the cell surface and facilitating activation of their cognate cell-surface receptors (Clement et al., 1999; Gama et al., 2006; Rogers et al., 2011). However, evidence for this mechanism was primarily based on the growth-promoting effects of CS GAGs adhered to cell culture plates or other artificial substrata. A liposome-based approach was developed to recapitulate the natural display of CS GAGs on neuronal cell surfaces and probe the role of sulfation (Pulsipher et al., 2014). Liposomes were formed from commercially available lipids in the presence of 2-dodecanone, which provided a reactive ketone handle for covalent attachment of aminooxy-functionalized CS polysaccharides (Figure 9B). Membrane fusion led to presentation of CS GAGs enriched in specific sulfation motifs on the surface of embryonic hippocampal neurons. Notably, only neurons displaying the disulfated CS-E motif showed significantly greater neurite outgrowth in response to nerve growth factor (NGF) compared with neurons lacking lipid-anchored CS GAGs. The extent of neurite outgrowth could be finely tuned by controlling the amount of CS-E delivered to the cell surface. As further support for the mechanism, the presence of CS-E led to enhanced activation of NGF-mediated signaling pathways, suggesting that CS-E facilitates surface recruitment of NGF. Thus, these studies demonstrated the proof-of-principle that de novo glycan display of CS GAGs can be used to control neuronal signaling pathways and downstream cellular responses.
GAG-mediated cell signaling plays a critical role not only in neurons but also in stem cell development. For example, previous reports have suggested that HS sulfation levels influence stem cell fate by regulating growth factor-mediated signaling events that determine both pluripotent self-renewal and differentiation (Forsberg et al., 2012; Kraushaar et al., 2012). To explore whether artificial cell-surface glycans could be used to direct this process, synthetic phospholipid-anchored ketone polymers were functionalized with different aminooxy-tagged HS disaccharides produced by enzymatic digestion (Huang et al., 2014). Passive insertion of glycomimetics containing both 2-O- and 6-O-sulfation, but not other sulfation motifs, activated fibroblast growth factor 2 (FGF2) signaling pathways involved in stem cell differentiation. Furthermore, stem cells decorated with the active HS glycopolymers formed neural rosettes, suggesting the formation of intermediate neuroprogenitor cells.
A major challenge of de novo glycan display methods is that the presentation of glycans at the cell surface is often short-lived. Lipid-anchored glycans typically exhibit a membrane half-life of about 6–8 hr (Huang et al., 2014; Hudak et al., 2013; Pulsipher et al., 2014). To achieve long-lived display, a genetically encoded, transmembrane-anchored HaloTag protein was exploited (Figure 9C) (Pulsipher et al., 2015). The HaloTag protein is a bacterial alkyl dehalogenase that forms a covalent bond with chloroalkyl ligands (Los et al., 2008), which allows for attachment of HS GAGs and other molecules. Notably, fluorescent probes anchored to HaloTag proteins persisted on the cell surface for over 8 days. Moreover, stem cells decorated with highly sulfated HS on HaloTag proteins underwent an accelerated loss of pluripotency and differentiation into mature neuronal cells. Homogeneous neuronal cell populations could be valuable as cell replacement therapies for neuro-degenerative disorders such as Parkinson’s disease (Freed et al., 2001; Mendez et al., 2008). This method constitutes the first example of long-term glycan display, significantly broadening the potential application of de novo display to a wide range of biological processes.
De novo glycan display provides a powerful, versatile means to direct key signaling events and biological outcomes such as the immune response, cancer proliferation, and stem cell differentiation. The technique is modular by design, providing a general platform to test numerous glycan structures and presentation architectures that best suit the biological system of interest. Moreover, the method allows for the controlled display of large polysaccharides, which are inaccessible to other forms of glycan engineering. De novo glycan display approaches have provided novel insights into the roles of specific carbohydrates in biology and have the potential to direct biomedically important processes such as tissue transplantations or stem cell fate.
Outlook
Considerable progress has been made toward developing novel methods to remodel the structures of glycans at cell surfaces. These studies have broadly demonstrated the ability to modulate signaling pathways and important downstream biological events by controlling glycan-protein interactions. To date, the majority of glycan engineering techniques have been applied in vitro. The dearth of in vivo experiments underscores the need to expand these approaches to complex living systems. The ability to perturb glycans in a tissue-specific manner or at an organism-wide level will present new opportunities to unravel the intricacies of glycan-mediated processes that cannot be fully recapitulated in vitro.
Excitingly, recent advances in other fields promise to fuel the future development and expansion of glycan engineering technologies. With the advent of genome editing methods such as CRISPR/Cas9 (Hsu et al., 2014), scientists now can more rapidly knock out genes and screen for enzymatic function, and perhaps even more impressively, knock in exogenous or evolved enzymes with new activities. These advances should greatly facilitate the identification of still unknown GTs and glycosylation-associated proteins critically involved in development and disease. Furthermore, the integration of facile genetic editing with tissue-specific or temporally regulated expression should enable the production of animal models that possess modified glycan structures during specific stages of development, growth, and disease.
Such technologies should also accelerate the expansion of MOE and CL to a wider range of non-natural glycan structures. Genetic knockins or targeted delivery of agents could produce specific cell populations and even entire organisms capable of incorporating a diverse assortment of non-natural sugars inaccessible to the natural glycosylation machinery, possibly allowing multiple glycan populations to be monitored in real time using orthogonal non-natural functionalities. Furthermore, tagged glycans could be purified from living tissues to ascertain the differences in individual glycoprotein structures over time. With the increase in non-natural sugars that could be simultaneously incorporated, a complementary wider set of functional handles would provide further power to control cellular function and endow cells with novel properties.
Ongoing developments in DNA sequencing and bioinformatics have dramatically increased the number of sequenced and annotated microbial genomes (Markowitz et al., 2012), which could provide novel enzymes involved in glycosylation and expand the chemical diversity of glycan architectures remodeled by CL. As CL is uniquely suited for biomarker detection, these new GTs could also provide a suite of tools to monitor human pathology and perhaps predict the onset and progression of disease states.
Finally, recent advances in de novo glycan display, particularly the expansion of its temporal limits, have opened an exciting new frontier to explore the structure-function relationships of carbohydrates. These methods now present unparalleled opportunities to engineer novel signaling pathways and functions into cells. Liposomes decorated with tissue-specific ligands could be used to deliver exogenous glycans to specific cell populations in vivo. Similarly, protein anchors like the HaloTag protein could be incorporated through genetic engineering to direct glycan display in model organisms. The attachment of glycans to various protein architectures could help mimic the natural presentation of sugars as glycoproteins and dissect the importance of different protein-carbohydrate combinations. Together, the successes in glycan engineering underscore the powerful integration of chemistry and biology and its ability to drive technological innovations and new discoveries.
Acknowledgments
This research was supported by the NIH (R01-GM093627 and R01-GM084724) and a National Science Foundation Graduate Research Fellowship (DGE-1144469).
References
- Andersson ER, Sandberg R, Lendahl U. Notch signaling: simplicity in design, versatility in function. Development. 2011;138:3593–3612. doi: 10.1242/dev.063610. [DOI] [PubMed] [Google Scholar]
- Baeg GH, Selva EM, Goodman RM, Dasgupta R, Perrimon N. The wingless morphogen gradient is established by the cooperative action of Frizzled and heparan sulfate proteoglycan receptors. Dev Biol. 2004;276:89–100. doi: 10.1016/j.ydbio.2004.08.023. [DOI] [PubMed] [Google Scholar]
- Belenkaya TY, Han C, Yan D, Opoka RJ, Khodoun M, Liu HZ, Lin XH. Drosophila Dpp morphogen movement is independent of dynamin-mediated endocytosis but regulated by the glypican members of heparan sulfate proteoglycans. Cell. 2004;119:231–244. doi: 10.1016/j.cell.2004.09.031. [DOI] [PubMed] [Google Scholar]
- Bellaiche Y, The I, Perrimon N. Tout-velu is a Drosophila homologue of the putative tumour suppressor EXT-1 and is needed for Hh diffusion. Nature. 1998;394:85–88. doi: 10.1038/27932. [DOI] [PubMed] [Google Scholar]
- Beltran-Valero de Bernabé D, Currier S, Steinbrecher A, Celli J, van Beusekom E, van der Zwaag B, Kayserili H, Merlini L, Chitayat D, Dobyns WB, et al. Mutations in the O-mannosyltransferase gene POMT1 give rise to the severe neuronal migration disorder Walker-Warburg syndrome. Am J Hum Genet. 2002;71:1033–1043. doi: 10.1086/342975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop JR, Schuksz M, Esko JD. Heparan sulphate proteoglycans fine-tune mammalian physiology. Nature. 2007;446:1030–1037. doi: 10.1038/nature05817. [DOI] [PubMed] [Google Scholar]
- Boyce M, Carrico IS, Ganguli AS, Yu SH, Hangauer MJ, Hubbard SC, Kohler JJ, Bertozzi CR. Metabolic cross-talk allows labeling of O-linked beta-N-acetylglucosamine-modified proteins via the N-acetyl-galactosamine salvage pathway. Proc Natl Acad Sci USA. 2011;108:3141–3146. doi: 10.1073/pnas.1010045108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brückner K, Perez L, Clausen H, Cohen S. Glycosyltransferase activity of Fringe modulates Notch-Delta interactions. Nature. 2000;406:411–415. doi: 10.1038/35019075. [DOI] [PubMed] [Google Scholar]
- Büll C, den Brok MH, Adema GJ. Sweet escape: sialic acids in tumor immune evasion. Biochim Biophys Acta. 2014;1846:238–246. doi: 10.1016/j.bbcan.2014.07.005. [DOI] [PubMed] [Google Scholar]
- Bullock S, Potter J, Rose SPR. Effects of the amnesic agent 2-deoxygalactose on incorporation of fucose into chick brain glycoproteins. J Neurochem. 1990;54:135–142. doi: 10.1111/j.1471-4159.1990.tb13293.x. [DOI] [PubMed] [Google Scholar]
- Chaubard JL, Krishnamurthy C, Yi W, Smith DF, Hsieh-Wilson LC. Chemoenzymatic probes for detecting and imaging fucose-α(1-2)-galactose glycan biomarkers. J Am Chem Soc. 2012;134:4489–4492. doi: 10.1021/ja211312u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Moloney DJ, Stanley P. Fringe modulation of Jagged1-induced Notch signaling requires the action of β4galactosyltransferase-1. Proc Natl Acad Sci USA. 2001;98:13716–13721. doi: 10.1073/pnas.241398098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuh KN, Zaro BW, Piller F, Piller V, Pratt MR. Changes in metabolic chemical reporter structure yield a selective probe of O-GlcNAc modification. J Am Chem Soc. 2014;136:12283–12295. doi: 10.1021/ja504063c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clement AM, Sugahara K, Faissner A. Chondroitin sulfate E promotes neurite outgrowth of rat embryonic day 18 hippocampal neurons. Neurosci Lett. 1999;269:125–128. doi: 10.1016/s0304-3940(99)00432-2. [DOI] [PubMed] [Google Scholar]
- Cole CM, Yang J, Seckute J, Devaraj NK. Fluorescent live-cell imaging of metabolically incorporated unnatural cyclopropene-mannosamine derivatives. Chembiochem. 2013;14:205–208. doi: 10.1002/cbic.201200719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coles CH, Shen Y, Tenney AP, Siebold C, Sutton GC, Lu W, Gallagher JT, Jones EY, Flanagan JG, Aricescu AR. Proteoglycan-specific molecular switch for RPTPσ clustering and neuronal extension. Science. 2011;332:484–488. doi: 10.1126/science.1200840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook A, Raskind W, Blanton SH, Pauli RM, Gregg RG, Francomano CA, Puffenberger E, Conrad EU, Schmale G, Schellenberg G, et al. Genetic heterogeneity in families with hereditary multiple exostoses. Am J Hum Genet. 1993;53:71–79. [PMC free article] [PubMed] [Google Scholar]
- Dehnert KW, Baskin JM, Laughlin ST, Beahm BJ, Naidu NN, Amacher SL, Bertozzi CR. Imaging the sialome during zebrafish development with copper-free click chemistry. Chembiochem. 2012;13:353–357. doi: 10.1002/cbic.201100649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demaegd D, Foulquier F, Colinet AS, Gremillon L, Legrand D, Mariot P, Peiter E, Van Schaftingen E, Matthijs G, Morsomme P. Newly characterized Golgi-localized family of proteins is involved in calcium and pH homeostasis in yeast and human cells. Proc Natl Acad Sci USA. 2013;110:6859–6864. doi: 10.1073/pnas.1219871110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dube DH, Bertozzi CR. Metabolic oligosaccharide engineering as a tool for glycobiology. Curr Opin Chem Biol. 2003;7:616–625. doi: 10.1016/j.cbpa.2003.08.006. [DOI] [PubMed] [Google Scholar]
- Forsberg M, Holmborn K, Kundu S, Dagalv A, Kjellén L, Forsberg-Nilsson K. Undersulfation of heparan sulfate restricts differentiation potential of mouse embryonic stem cells. J Biol Chem. 2012;287:10853–10862. doi: 10.1074/jbc.M111.337030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foulquier F, Amyere M, Jaeken J, Zeevaert R, Schollen E, Race V, Bammens R, Morelle W, Rosnoblet C, Legrand D, et al. TMEM165 deficiency causes a congenital disorder of glycosylation. Am J Hum Genet. 2012;91:15–26. doi: 10.1016/j.ajhg.2012.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frame T, Carroll T, Korchagina E, Bovin N, Henry S. Synthetic glycolipid modification of red blood cell membranes. Transfusion. 2007;47:876–882. doi: 10.1111/j.1537-2995.2007.01204.x. [DOI] [PubMed] [Google Scholar]
- Freed CR, Greene PE, Breeze RE, Tsai WY, DuMouchel W, Kao R, Dillon S, Winfield H, Culver S, Trojanowski JQ, et al. Transplantation of embryonic dopamine neurons for severe Parkinson’s Disease. N Engl J Med. 2001;344:710–719. doi: 10.1056/NEJM200103083441002. [DOI] [PubMed] [Google Scholar]
- Freeze HH. Understanding human glycosylation disorders: biochemistry leads the charge. J Biol Chem. 2013;288:6936–6945. doi: 10.1074/jbc.R112.429274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuster MM, Esko JD. The sweet and sour of cancer: glycans as novel therapeutic targets. Nat Rev Cancer. 2005;5:526–542. doi: 10.1038/nrc1649. [DOI] [PubMed] [Google Scholar]
- Gama CI, Tully SE, Sotogaku N, Clark PM, Rawat M, Vaidehi N, Goddard WA, III, Nishi A, Hsieh-Wilson LC. Sulfation patterns of glycosaminoglycans encode molecular recognition and activity. Nat Chem Biol. 2006;2:467–473. doi: 10.1038/nchembio810. [DOI] [PubMed] [Google Scholar]
- Goddeeris MM, Wu B, Venzke D, Yoshida-Moriguchi T, Saito F, Matsumura K, Moore SA, Campbell KP. LARGE glycans on dystroglycan function as a tunable matrix scaffold to prevent dystrophy. Nature. 2013;503:136–140. doi: 10.1038/nature12605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W, Giancotti FG. Integrin signalling during tumour progression. Nat Rev Mol Cell Biol. 2004;5:816–826. doi: 10.1038/nrm1490. [DOI] [PubMed] [Google Scholar]
- Häcker U, Nybakken K, Perrimon N. Heparan sulphate proteoglycans: the sweet side of development. Nat Rev Mol Cell Biol. 2005;6:530–541. doi: 10.1038/nrm1681. [DOI] [PubMed] [Google Scholar]
- Haines N, Irvine KD. Glycosylation regulates Notch signalling. Nat Rev Mol Cell Biol. 2003;4:786–797. doi: 10.1038/nrm1228. [DOI] [PubMed] [Google Scholar]
- Han C, Belenkaya TY, Khodoun M, Tauchi M, Lin X, Lin X. Distinct and collaborative roles of Drosophila EXT family proteins in morphogen signalling and gradient formation. Development. 2004;131:1563–1575. doi: 10.1242/dev.01051. [DOI] [PubMed] [Google Scholar]
- Han S, Collins BE, Bengtson P, Paulson JC. Homomultimeric complexes of CD22 in B cells revealed by protein-glycan cross-linking. Nat Chem Biol. 2005;1:93–97. doi: 10.1038/nchembio713. [DOI] [PubMed] [Google Scholar]
- Hang HC, Yu C, Kato DL, Bertozzi CR. A metabolic labeling approach toward proteomic analysis of mucin-type O-linked glycosylation. Proc Natl Acad Sci USA. 2003;100:14846–14851. doi: 10.1073/pnas.2335201100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennet T, Cabalzar J. Congenital disorders of glycosylation: a concise chart of glycocalyx dysfunction. Trends Biochem Sci. 2015;40:377–384. doi: 10.1016/j.tibs.2015.03.002. [DOI] [PubMed] [Google Scholar]
- Horwitz EM, Gordon PL, Koo WKK, Marx JC, Neel MD, McNall RY, Muul L, Hofmann T. Isolated allogeneic bone marrow-derived mesenchymal cells engraft and stimulate growth in children with osteogenesis imperfecta: implications for cell therapy of bone. Proc Natl Acad Sci USA. 2002;99:8932–8937. doi: 10.1073/pnas.132252399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu TL, Hanson SR, Kishikawa K, Wang SK, Sawa M, Wong CH. Alkynyl sugar analogs for the labeling and visualization of glycoconjugates in cells. Proc Natl Acad Sci USA. 2007;104:2614–2619. doi: 10.1073/pnas.0611307104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang ML, Smith RAA, Trieger GW, Godula K. Glycocalyx remodeling with proteoglycan mimetics promotes neural specification in embryonic stem cells. J Am Chem Soc. 2014;136:10565–10568. doi: 10.1021/ja505012a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudak JE, Canham SM, Bertozzi CR. Glycocalyx engineering reveals a Siglec-based mechanism for NK cell immunoevasion. Nat Chem Biol. 2013;10:69–75. doi: 10.1038/nchembio.1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones MA, Ng BG, Bhide S, Chin E, Rhodenizer D, He P, Losfeld ME, He M, Raymond K, Berry G, et al. DDOST mutations identified by whole-exome sequencing are implicated in congenital disorders of glycosylation. Am J Hum Genet. 2012;90:363–368. doi: 10.1016/j.ajhg.2011.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaewsapsak P, Esonu O, Dube DH. Recruiting the host’s immune system to target Helicobacter pylori’s surface glycans. Chembiochem. 2013;14:721–726. doi: 10.1002/cbic.201300006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalovidouris SA, Gama CI, Lee LW, Hsieh-Wilson LC. A role for fucose-α(1–2)-galactose carbohydrates in neuronal growth. J Am Chem Soc. 2005;127:1340–1341. doi: 10.1021/ja044631v. [DOI] [PubMed] [Google Scholar]
- Keppler OT, Stehling P, Herrmann M, Kayser H, Grunow D, Reutter W, Pawlita M. Biosynthetic modulation of sialic acid-dependent virus-receptor interactions of two primate polyoma viruses. J Biol Chem. 1995;270:1308–1314. doi: 10.1074/jbc.270.3.1308. [DOI] [PubMed] [Google Scholar]
- Khidekel N, Arndt S, Lamarre-Vincent N, Lippert A, Poulin-Kerstien KG, Ramakrishnan B, Qasba PK, Hsieh-Wilson LC. A chemoenzymatic approach toward the rapid and sensitive detection of O-GlcNAc post-translational modifications. J Am Chem Soc. 2003;125:16162–16163. doi: 10.1021/ja038545r. [DOI] [PubMed] [Google Scholar]
- Kirkpatrick CA, Dimitroff BD, Rawson JM, Selleck SB. Spatial regulation of wingless morphogen distribution and signaling by dally-like protein. Dev Cell. 2004;7:513–523. doi: 10.1016/j.devcel.2004.08.004. [DOI] [PubMed] [Google Scholar]
- Kobayashi K, Nakahori Y, Miyake M, Matsumura K, Kondo-Iida E, Nomura Y, Segawa M, Yoshioka M, Saito K, Osawa K, et al. An ancient retrotransposal insertion causes Fukuyama-type congenital muscular dystrophy. Nature. 1998;394:388–392. doi: 10.1038/28653. [DOI] [PubMed] [Google Scholar]
- Kopan R, Ilagan MXG. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137:216–233. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koziel L, Kunath M, Kelly OG, Vortkamp A. Ext1-dependent heparan sulfate regulates the range of Ihh signaling during endochondral ossification. Dev Cell. 2004;6:801–813. doi: 10.1016/j.devcel.2004.05.009. [DOI] [PubMed] [Google Scholar]
- Kraushaar DC, Rai S, Condac E, Nairn A, Zhang S, Yamaguchi Y, Moremen K, Dalton S, Wang L. Heparan sulfate facilitates FGF and BMP signaling to drive mesoderm differentiation of mouse embryonic stem cells. J Biol Chem. 2012;287:22691–22700. doi: 10.1074/jbc.M112.368241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krug M, Jork R, Reymann K, Wagner M, Matthies H. The amnesic substance 2-deoxy-D-galactose suppresses the maintenance of hippocampal LTP. Brain Res. 1991;540:237–242. doi: 10.1016/0006-8993(91)90513-u. [DOI] [PubMed] [Google Scholar]
- Laughlin ST, Bertozzi CR. Imaging the glycome. Proc Natl Acad Sci USA. 2009a;106:12–17. doi: 10.1073/pnas.0811481106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laughlin ST, Bertozzi CR. In vivo imaging of Caenorhabditis elegans glycans. ACS Chem Biol. 2009b;4:1068–1072. doi: 10.1021/cb900254y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laughlin ST, Baskin JM, Amacher SL, Bertozzi CR. In vivo imaging of membrane-associated glycans in developing zebrafish. Science. 2008;320:664–667. doi: 10.1126/science.1155106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Merrer M, Legeai-Mallet L, Jeannin PM, Horsthemke B, Schinzel A, Plauchu H, Toutain A, Archard F, Munnich A, Maroteaux P. A gene for hereditary multiple exostoses maps to chromosome 19p. Hum Mol Genet. 1994;3:717–722. doi: 10.1093/hmg/3.5.717. [DOI] [PubMed] [Google Scholar]
- Lichtenstein RG, Rabinovich GA. Glycobiology of cell death: when glycans and lectins govern cell fate. Cell Death Differ. 2013;20:976–986. doi: 10.1038/cdd.2013.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longman C, Brockington M, Torelli S, Jimenez-Mallebrera C, Kennedy C, Khalil N, Feng L, Saran RK, Voit T, Merlini L, et al. Mutations in the human LARGE gene cause MDC1D, a novel form of congenital muscular dystrophy with severe mental retardation and abnormal glycosylation of α-dystroglycan. Hum Mol Genet. 2003;12:2853–2861. doi: 10.1093/hmg/ddg307. [DOI] [PubMed] [Google Scholar]
- Lonze BE, Ginty DD. Function and regulation of CREB family transcription factors in the nervous system. Neuron. 2002;35:605–623. doi: 10.1016/s0896-6273(02)00828-0. [DOI] [PubMed] [Google Scholar]
- Los GV, Encell LP, McDougall MG, Hartzell DD, Karassina N, Zimprich C, Wood MG, Learish R, Ohana RF, Urh M, et al. HaloTag: a novel protein labeling technology for cell imaging and protein analysis. ACS Chem Biol. 2008;3:373–382. doi: 10.1021/cb800025k. [DOI] [PubMed] [Google Scholar]
- Mahal LK, Yarema KJ, Bertozzi CR. Engineering chemical reactivity on cell surfaces through oligosaccharide biosynthesis. Science. 1997;276:1125–1128. doi: 10.1126/science.276.5315.1125. [DOI] [PubMed] [Google Scholar]
- Manzini MC, Tambunan DE, Hill RS, Yu TW, Maynard TM, Heinzen EL, Shianna KV, Stevens CR, Partlow JN, Barry BJ, et al. Exome sequencing and functional validation in zebrafish identify GTDC2 mutations as a cause of Walker-Warburg syndrome. Am J Hum Genet. 2012;91:541–547. doi: 10.1016/j.ajhg.2012.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markowitz VM, Chen IMA, Palaniappan K, Chu K, Szeto E, Grechkin Y, Ratner A, Jacob B, Huang J, Williams P, et al. IMG: the Integrated Microbial Genomes database and comparative analysis system. Nucleic Acids Res. 2012;40:D115–D122. doi: 10.1093/nar/gkr1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick C, Leduc Y, Martindale D, Mattison K, Esford LE, Dyer AP, Tufaro F. The putative tumour suppressor EXT1 alters the expression of cell-surface heparan sulfate. Nat Genet. 1998;19:158–161. doi: 10.1038/514. [DOI] [PubMed] [Google Scholar]
- McKay CS, Finn MG. Click chemistry in complex mixtures: bio-orthogonal bioconjugation. Chem Biol. 2014;21:1075–1101. doi: 10.1016/j.chembiol.2014.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez I, Viñuela A, Astradsson A, Mukhida K, Hallett P, Robertson H, Tierney T, Holness R, Dagher A, Trojanowski JQ, et al. Dopamine neurons implanted into people with Parkinson’s disease survive without pathology for 14 years. Nat Med. 2008;14:507–509. doi: 10.1038/nm1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moloney DJ, Panin VM, Johnston SH, Chen JH, Shao L, Wilson R, Wang Y, Stanley P, Irvine KD, Haltiwanger RS, et al. Fringe is a glycosyltransferase that modifies Notch. Nature. 2000;406:369–375. doi: 10.1038/35019000. [DOI] [PubMed] [Google Scholar]
- Murrey HE, Gama CI, Kalovidouris SA, Luo WI, Driggers EM, Porton B, Hsieh-Wilson LC. Protein fucosylation regulates synapsin la/lb expression and neuronal morphology in primary hippocampal neurons. Proc Natl Acad Sci USA. 2006;103:21–26. doi: 10.1073/pnas.0503381102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitschke L, Carsetti R, Ocker B, Köhler G, Lamers MC. CD22 is a negative regulator of B-cell receptor signalling. Curr Biol. 1997;7:133–143. doi: 10.1016/s0960-9822(06)00057-1. [DOI] [PubMed] [Google Scholar]
- Ohtsuka Y, Kanagawa M, Yu CC, Ito C, Chiyo T, Kobayashi K, Okada T, Takeda S, Toda T. Fukutin is prerequisite to ameliorate muscular dystrophic phenotype by myofiber-selective LARGE expression. Sci Rep. 2015;5:8316. doi: 10.1038/srep08316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okajima T, Irvine KD. Regulation of Notch signaling by O-linked fucose. Cell. 2002;111:893–904. doi: 10.1016/s0092-8674(02)01114-5. [DOI] [PubMed] [Google Scholar]
- Paszek MJ, Boettiger D, Weaver VM, Hammer DA. Integrin clustering is driven by mechanical resistance from the glycocalyx and the substrate. PLoS Comput Biol. 2009;5:e1000604. doi: 10.1371/journal.pcbi.1000604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paszek MJ, DuFort CC, Rossier O, Bainer R, Mouw JK, Godula K, Hudak JE, Lakins JN, Wijekoon AC, Cassereau L, et al. The cancer glycocalyx mechanically primes integrin-mediated growth and survival. Nature. 2014;511:319–325. doi: 10.1038/nature13535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulsipher A, Griffin ME, Stone SE, Brown JM, Hsieh-Wilson LC. Directing neuronal signaling through cell-surface glycan engineering. J Am Chem Soc. 2014;136:6794–6797. doi: 10.1021/ja5005174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulsipher A, Griffin ME, Stone SE, Hsieh-Wilson LC. Long-lived engineering of glycans to direct stem cell fate. Angew Chem Int Ed Engl. 2015;54:1466–1470. doi: 10.1002/anie.201409258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramya TNC, Weerapana E, Liao L, Zeng Y, Tateno H, Liao L, Yates JR, III, Cravatt BF, Paulson JC. In situ trans ligands of CD22 identified by glycan-protein photocross-linking-enabled proteomics. Mol Cell Proteomics. 2010;9:1339–1351. doi: 10.1074/mcp.M900461-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rexach JE, Rogers CJ, Yu SH, Tao J, Sun YE, Hsieh-Wilson LC. Quantification of O-glycosylation stoichiometry and dynamics using resolvable mass tags. Nat Chem Biol. 2010;6:645–651. doi: 10.1038/nchembio.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rexach JE, Clark PM, Mason DE, Neve RL, Peters EC, Hsieh-Wilson LC. Dynamic O-GlcNAc modification regulates CREB-mediated gene expression and memory formation. Nat Chem Biol. 2012;8:253–261. doi: 10.1038/nchembio.770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rillahan CD, Antonopoulos A, Lefort CT, Sonon R, Azadi P, Ley K, Dell A, Haslam SM, Paulson JC. Global metabolic inhibitors of sialyl-and fucosyltransferases remodel the glycome. Nat Chem Biol. 2012;8:661–668. doi: 10.1038/nchembio.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers CJ, Clark PM, Tully SE, Abrol R, Garcia KC, Goddard WA, III, Hsieh-Wilson LC. Elucidating glycosaminoglycan-protein-protein interactions using carbohydrate microarray and computational approaches. Proc Natl Acad Sci USA. 2011;108:9747–9752. doi: 10.1073/pnas.1102962108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose SPR, Jork R. Long-term memory formation in chicks is blocked by 2-deoxygalactose, a fucose analog. Behav Neural Biol. 1987;48:246–258. doi: 10.1016/s0163-1047(87)90808-9. [DOI] [PubMed] [Google Scholar]
- Rouhanifard SH, Nordstrøm LU, Zheng T, Wu P. Chemical probing of glycans in cells and organisms. Chem Soc Rev. 2013;42:4284–4296. doi: 10.1039/c2cs35416k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sackstein R, Merzaban JS, Cain DW, Dagia NM, Spencer JA, Lin CP, Wohlgemuth R. Ex vivo glycan engineering of CD44 programs human multipotent mesenchymal stromal cell trafficking to bone. Nat Med. 2008;14:181–187. doi: 10.1038/nm1703. [DOI] [PubMed] [Google Scholar]
- Sampathkumar SG, Li AV, Jones MB, Sun Z, Yarema KJ. Metabolic installation of thiols into sialic acid modulates adhesion and stem cell biology. Nat Chem Biol. 2006;2:149–152. doi: 10.1038/nchembio770. [DOI] [PubMed] [Google Scholar]
- Shi SL, Stanley P. Protein O-fucosyltransferase 1 is an essential component of Notch signaling pathways. Proc Natl Acad Sci USA. 2003;100:5234–5239. doi: 10.1073/pnas.0831126100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sletten EM, Bertozzi CR. Bioorthogonal chemistry: fishing for selectivity in a sea of functionality. Angew Chem Int Ed Engl. 2009;48:6974–6998. doi: 10.1002/anie.200900942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Späte AK, Schart VF, Schöllkopf S, Niederwieser A, Wittmann V. Terminal alkenes as versatile chemical reporter groups for metabolic oligosaccharide engineering. Chemistry. 2014;20:16502–16508. doi: 10.1002/chem.201404716. [DOI] [PubMed] [Google Scholar]
- Stairs S, Neves AA, Stöckman H, Wainman YA, Ireland-Zecchini H, Brindle KM, Leeper FJ. Metabolic glycan imaging by isonitrile-tetrazine click chemistry. Chembiochem. 2013;14:1063–1067. doi: 10.1002/cbic.201300130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens E, Carss KJ, Cirak S, Foley AR, Torelli S, Willer T, Tambunan DE, Yau S, Brodd L, Sewry CA, et al. Mutations in B3GALNT2 cause congenital muscular dystrophy and hypoglycosylation of α-dystroglycan. Am J Hum Genet. 2013;92:354–365. doi: 10.1016/j.ajhg.2013.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabata T, Kornberg TB. Hedgehog is a signaling protein with a key role in patterning Drosophila imaginal discs. Cell. 1994;76:89–102. doi: 10.1016/0092-8674(94)90175-9. [DOI] [PubMed] [Google Scholar]
- The I, Bellaiche Y, Perrimon N. Hedgehog movement is regulated through tout velu-dependent synthesis of a heparan sulfate proteoglycan. Mol Cell. 1999;4:633–639. doi: 10.1016/s1097-2765(00)80214-2. [DOI] [PubMed] [Google Scholar]
- van Kooyk Y, Rabinovich GA. Protein-glycan interactions in the control of innate and adaptive immune responses. Nat Immunol. 2008;9:593–601. doi: 10.1038/ni.f.203. [DOI] [PubMed] [Google Scholar]
- van Reeuwijk J, Janssen M, van den Elzen C, Beltran-Valero de Bernabé D, Sabatelli P, Merlini L, Boon M, Scheffer H, Brockington M, Mutoni F, et al. POMT2 mutations cause α-dystroglycan hypoglycosylation and Walker-Warburg syndrome. J Med Genet. 2005;42:907–912. doi: 10.1136/jmg.2005.031963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Shao L, Shi S, Harris RJ, Spellman MW, Stanley P, Haltiwanger RS. Modification of epidermal growth factor-like repeats with O-fucose: molecular cloning and expression of a novel GDP-fucose protein O-fucosyltransferase. J Biol Chem. 2001;276:40338–40345. doi: 10.1074/jbc.M107849200. [DOI] [PubMed] [Google Scholar]
- Wells L. The O-mannosylation pathway: glycosyltransferases and proteins implicated in congenital muscular dystrophy. J Biol Chem. 2013;288:6930–6935. doi: 10.1074/jbc.R112.438978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu YQ, Heutink P, de Vries BBA, Sandkuijl LA, van den Ouweland AMW, Neirmeijer MF, Galjaard H, Reyniers E, Willems PJ, Halley DJJ. Assignment of a second locus for multiple exostoses to the pericentromeric region of chromosome 11. Hum Mol Genet. 1994;3:167–171. doi: 10.1093/hmg/3.1.167. [DOI] [PubMed] [Google Scholar]
- Yao D, Huang Y, Huang X, Wang W, Yan Q, Wei L, Xin W, Gerson S, Stanley P, Lowe JB, et al. Protein O-fucosyltransferase 1 (Pofut1) regulates lymphoid and myeloid homeostasis through modulation of Notch receptor ligand interactions. Blood. 2011;117:5652–5662. doi: 10.1182/blood-2010-12-326074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida A, Kobayashi K, Manya H, Taniguchi K, Kano H, Mizuno M, Inazu T, Mitsuhashi H, Takahashi S, Takeuchi M, et al. Muscular dystrophy and neuronal migration disorder caused by mutations in a glycosyl-transferase, POMGnT1. Dev Cell. 2001;1:717–724. doi: 10.1016/s1534-5807(01)00070-3. [DOI] [PubMed] [Google Scholar]
- Yoshida-Moriguchi T, Yu L, Stalnaker SH, Davis S, Kunz S, Madson M, Oldstone MBA, Schachter H, Wells L, Campbell KP. O-Mannosyl phosphorylation of alpha-dystroglycan is required for laminin binding. Science. 2010;327:88–92. doi: 10.1126/science.1180512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida-Moriguchi T, Willer T, Anderson ME, Venzke D, Whyte T, Muntoni F, Lee H, Nelson SF, Yu L, Campbell KP. SGK196 is a glycosylation-specific O-mannose kinase required for dystroglycan function. Science. 2013;341:896–899. doi: 10.1126/science.1239951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng T, Jiang H, Gros M, Soriano del Amo D, Sundaram S, Lauvau G, Marlow F, Liu Y, Stanley P, Wu P. Tracking N-acetyllactosamine on cell-surface glycans in vivo. Angew Chem Int Ed Engl. 2011;50:4113–4118. doi: 10.1002/anie.201100265. [DOI] [PMC free article] [PubMed] [Google Scholar]