Summary
Salmonella enterica catabolizes ethanolamine inside a compartment known as the metabolosome. The ethanolamine utilization (eut) operon of this bacterium encodes all functions needed for the assembly and function of this structure. To date, the roles of the EutQ and EutP were not known. Herein we show that both proteins have acetate kinase activity, and that EutQ is required during anoxic growth of S. enterica on ethanolamine and tetrathionate. EutP and EutQ-dependent ATP synthesis occurred when enzymes were incubated with ADP, Mg(II) ions and acetyl-phosphate. EutQ and EutP also synthesized acetyl-phosphate from ATP and acetate. Although EutP had acetate kinase activity, ΔeutP strains lacked discernable phenotypes under the conditions where ΔeutQ strains displayed clear phenotypes. The kinetic parameters indicate that EutP is a faster enzyme than EutQ. Our evidence supports the conclusions that EutQ and EutP represent novel classes of acetate kinases. We propose that EutQ is necessary to drive flux through the pathway under physiological conditions, preventing a buildup of acetaldehyde. We also suggest that ATP generated by these enzymes may be used as a substrate for EutT, the ATP-dependent corrinoid adenosyltransferase, and for the EutA ethanolamine ammonia-lyase reactivase.
Keywords: bacterial metabolism, ethanolamine catabolism, acetate kinases, metabolosome, bacterial microcompartments, acetate activation, substrate-level phosphorylation
Introduction
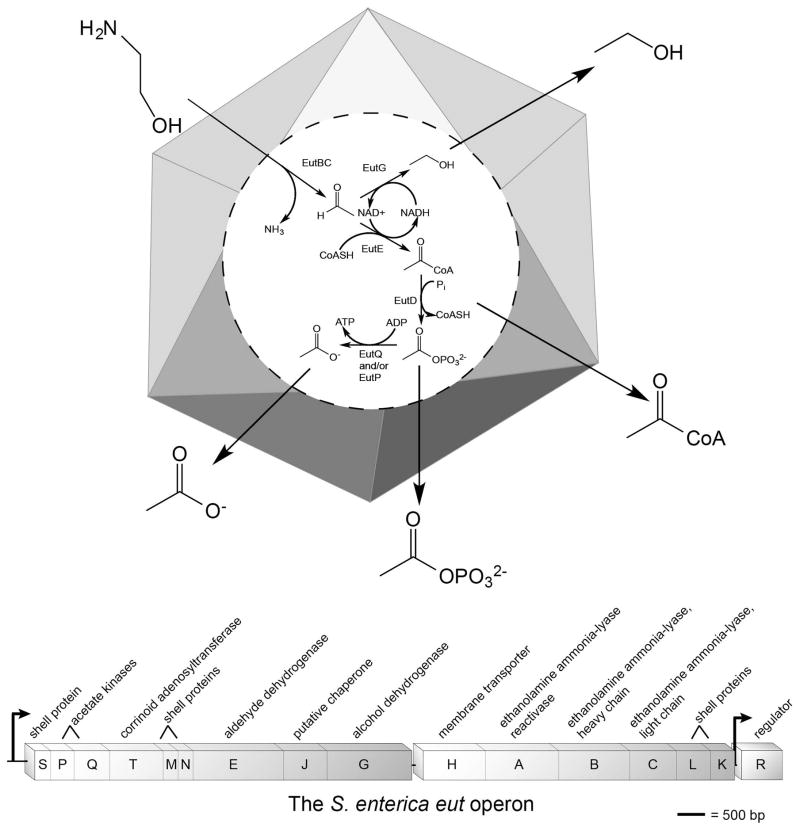
Salmonella enterica serovar Typhimurium strain LT2 (hereafter S. Typhimurium) is a Gamma-proteobacterium that can generate energy via fermentation or oxidative phosphorylation. The latter mechanism can use molecular oxygen or, under anoxic conditions, this bacterium can respire to diverse terminal electron acceptors, such as nitrate, fumarate, timethylamine-N-oxide, dimethyl sulfoxide, and tetrathionate (Escalante-Semerena & Roth, 1987, Barrett & Clark, 1987). S. Typhimurium can use ethanolamine as a carbon, nitrogen and energy source (Chang & Chang, 1975, Roof & Roth, 1988b, Roof & Roth, 1989), and the breakdown of this two-carbon amino alcohol occurs within a metabolosome, also known as a bacterial microcompartment (BMC1 or MCP) (Bobik, 2006, Cheng et al., 2008, Yeates et al., 2011) (Fig. 1A). The metabolosome is a proteinaceous polyhedron that is assembled by proteins encoded by the 17-gene ethanolamine utilization (eut) operon (Fig. 1B), whose expression is induced by the presence of ethanolamine and coenzyme B12 (Kofoid et al., 1999, Buan et al., 2004, Held et al., 2013, Roof & Roth, 1988a). It is thought that the metabolosome serves several functions: i) to prevent cell damage by containing the acetaldehyde product of ethanolamine ammonia-lyase (Brinsmade et al., 2005); ii) to prevent carbon loss from volatilization of acetaldehyde (Penrod & Roth, 2006); and iii) to conserve and recycle cofactors used in ethanolamine catabolism (Huseby & Roth, 2013). All of the proteins encoded by the eut operon have assigned functions except for EutQ, EutP and EutJ.
Figure 1. The ethanolamine catabolic pathway.
(A) The ethanolamine metabolosome, made of EutSMNLK, is represented as a polyhedral shell. The scheme for the breakdown of ethanolamine is shown as occurring within the shell; products of the pathway are shown outside the shell. (B) The ethanolamine utilization (eut) operon of S. Typhimurium.
In the intestine, ethanolamine is derived from phosphatidylethanolamine (Larson et al., 1983, Fung & Proulx, 1969), a major component of eukaryotic membranes (Cotton, 1972). Although ethanolamine is an abundant source of carbon and nitrogen in the intestinal environment occupied by S. Typhimurium, not all microbes present in it can ferment or respire ethanolamine. Most microbes require molecular oxygen to respire aminoalcohol, but S. Typhimurium sidesteps this need by using tetrathionate as terminal electron acceptor (Thiennimitr et al., 2012, Price-Carter et al., 2001). When S. Typhimurium triggers intestinal inflammation and white blood cells are recruited to the infection site, the reactive oxygen species released generate tetrathionate from thiosulfate present in the gut (Winter et al., 2010). Access to tetrathionate gives S. Typhimurium a competitive advantage over other intestinal microbes (Thiennimitr et al., 2012). Generation of tetrathionate via the inflammatory response is therefore important for S. Typhimurium pathogenesis (Baumler et al., 2011).
Previous studies did not assign roles to the proteins encoded by eutP, eutQ, and eutJ. Strains carrying deletions in these genes grew as well as the wild-type strain on ethanolamine as a carbon and energy source (Kofoid et al., 1999, Stojiljkovic et al., 1995, Buan & Escalante-Semerena, 2006). This paper reports the functions of EutP and Eut, but not the function of EutJ (a putative DnaK-like chaperone). Relevant to the work reported here is the observation by Penrod and Roth (Penrod & Roth, 2006), who showed that eutP and eutQ strains excreted more acetaldehyde than the wild-type strain when grown on glycerol with ethanolamine as a nitrogen source. From a structural standpoint, EutQ belongs to the cupin superfamily (SSF51182, PF06249), which includes a wide variety of enzymes such as carbohydrate-binding isomerases, flavonoid-binding dioxygenases and nuclear factors (Dunwell et al., 2004). The structure of EutP has not been reported, but bioinformatics analyses show that EutP is a member of the P-loop containing nucleoside triphosphate hydrolase superfamily (SSF52504, PF10662). In light of scant experimental and bioinformatics data, the functions of these proteins remained unknown.
Given that S. Typhimurium respires ethanolamine to tetrathionate under anoxic conditions, we decided to investigate possible functions of EutP and EutQ proteins during anoxic growth of this bacterium with ethanolamine as the sole source of carbon and energy and tetrathionate as terminal electron acceptor. Our data show that EutQ is required under such conditions, yet it is dispensable when S. Typhimurium is growing on ethanolamine in the presence of oxygen. Results of in vitro activity assays and complementation studies support the conclusion that EutQ is an acetate kinase. The physiological roles of EutP and EutQ under oxic and anoxic conditions are discussed.
Results
Efforts in our laboratory looking for growth defects caused by the absence of the EutP or EutQ protein of S. Typhimurium during growth on ethanolamine under oxic conditions were not fruitful. However, when we shifted our attention to anoxic growth conditions, it became clear that, at least the EutQ function was needed for growth of this bacterium on ethanolamine and tetrathionate.
Phenotypes of eutP and eutQ strains growing under anoxic conditions on ethanolamine and tetrathionate
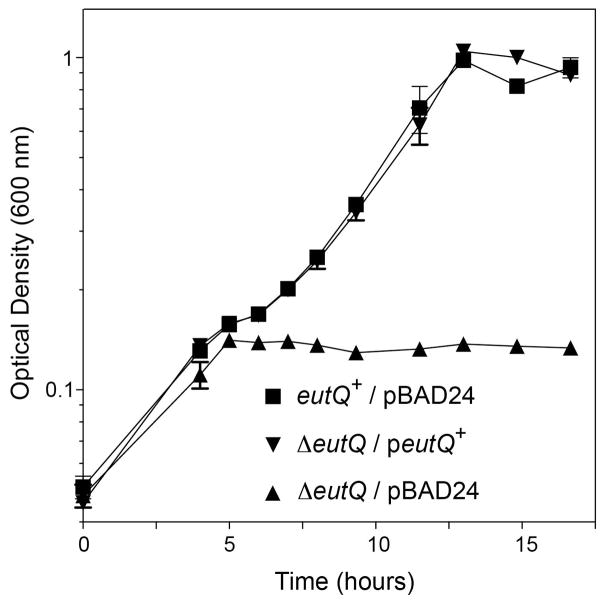
To determine whether or not eutP and eutQ strains had a growth phenotype on ethanolamine as the sole source of carbon and energy under anoxic conditions, we grew each strain on ethanolamine (30 mM) as carbon and energy source and tetrathionate (40 mM) as terminal electron acceptor. After consuming the small amount of glycerol (0.5 mM) present in the medium to stimulate growth, the eutQ strain stopped growing at an OD600 ~0.2 (Fig. 2, triangles), whereas the eutQ+ strain grew to an OD600 ~1 (Fig. 2, squares). Induction of a plasmid-encoded eutQ+ allele using low concentrations of inducer (arabinose, 0.05 mM) restored growth (Fig. 2, inverted triangles). In contrast, the eutP strain grew as well as the eutP+ strain under the same conditions (Fig. S1).
Figure 2. Growth phenotype of a ΔeutQ strain, and correction of it by ectopic expression of eutQ+.
Strains expressing eutQ+ from a plasmid under anoxic growth conditions in minimal medium supplemented with ethanolamine (30 mM), NH4Cl (30 mM), tetrathionate (40 mM) and L(+)-arabinose (0.05 mM). Growth rates were calculated for each culture during the logarithmic phase of growth. Oxidized tetrathionate can precipitate, making standard doubling times based on optical density difficult to calculate. Therefore, we normalized the mean percent change of optical density of each strain (triplicate) to the linear portion of the log phase in lieu of a standard doubling time calculation (mean ΔOD% • h−1): eutQ+/pBAD24, 14.0; ΔeutQ/pBAD24, no growth; ΔeutQ/peutQ+, 13.5.
Excess EutQ has a negative effect on growth on ethanolamine and tetrathionate, but the effect is avoided when the medium is supplemented with pantothenate
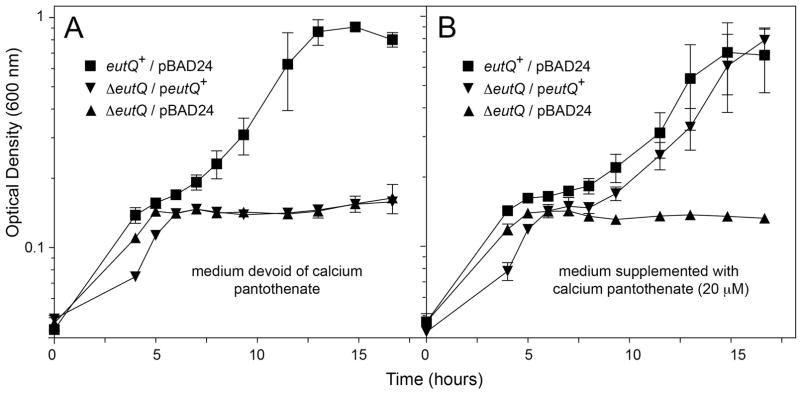
When we increased the expression of plasmid-borne eutQ+ we observed a dramatic negative effect on the ability of S. Typhimurium to grow on ethanolamine and tetrathionate. As shown in figure 3A, the growth behavior of the ΔeutQ/peutQ+ strain in ethanolamine/tetrathionate medium containing 1 mM arabinose was indistinguishable from that of the ΔeutQ strain carrying the empty cloning vector pBAD24 (compare triangles vs inverted triangles). Surprisingly, this effect was avoided by the addition of pantothenate, a precursor to coenzyme A (CoA) (Fig. 3B, inverted triangles). Our interpretation of these results is discussed below.
Figure 3. Elevated expression of eutQ+ is deleterious for growth, but the effect is counteracted by exogenous pantothenate.
A. Strains expressing eutQ+ from a plasmid under anoxic growth conditions in minimal medium supplemented with ethanolamine (30 mM), NH4Cl (30 mM), tetrathionate (40 mM) and L(+)-arabinose (1 mM). Growth rates were calculated for each culture during the logarithmic phase of growth (mean ΔOD% • h−1): eutQ+/pBAD24, 14; ΔeutQ/pBAD24, 2; ΔeutQ/peutQ+, 1. B. Same conditions as in panel A, but with the addition of calcium pantothenate (20 μM). Growth rates: eutQ+/pBAD24, 15; ΔeutQ/pBAD24, 1; ΔeutQ/peutQ+, 13.
Inhibitory levels of acetaldehyde are responsible for the EutQ phenotype
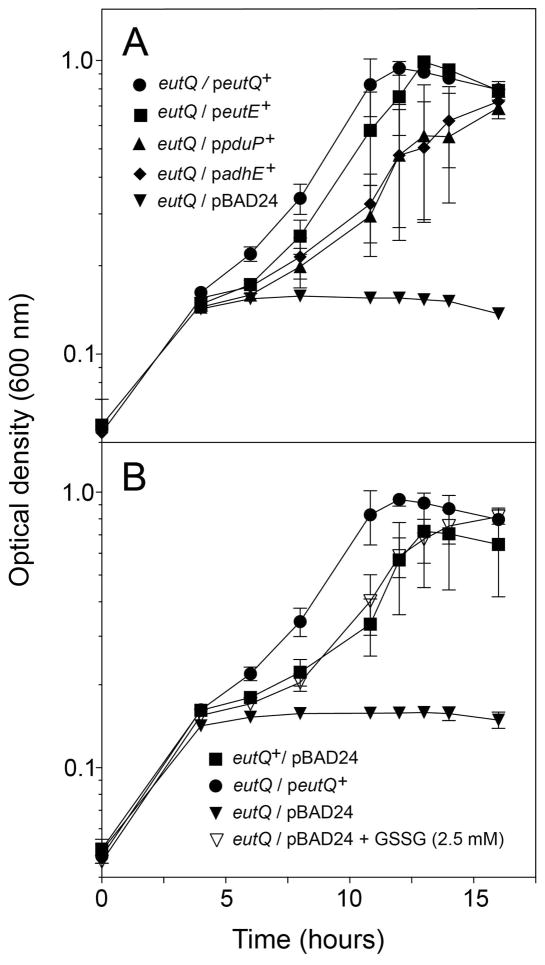
As mentioned above, Penrod and Roth reported that a S. Typhimurium ΔeutQ strain released acetaldehyde into the medium during growth on glycerol glycerol and ethanolamine (Penrod & Roth, 2006). Germane to this observation are results from previous work from our laboratory, which implicated acetaldehyde as a strong inhibitor of S. Typhimurium growth on ethanolamine (Brinsmade et al., 2005). The correlation between aldehyde accumulation and growth inhibition prompted us to directly test the idea that acetaldehyde caused the EutQ phenotype. For this purpose we introduced into a ΔeutQ strain individual plasmids carrying wild-type alleles of genes encoding bona fide acetaldehyde dehydrogenases, such as S. Typhimurium EutE (Zhu et al., 2011), S. Typhimurium PduP (Leal et al., 2003), and S. Typhimurium AdhE (Burton & Stadtman, 1953). Data presented in figure 4A show that eutQ strains that synthesized acetaldehyde dehydrogenases grew at similar rates and reached full density. In contrast, the control ΔeutQ strain carrying the empty cloning vector failed to grow (Fig. 4A, inverted triangles). We obtained additional experimental evidence to support the hypothesis that acetaldehyde accumulation was responsible for the EutQ phenotype. The experiment we performed was based on the previously reported beneficial effect of glutathione on cells growing on ethanolamine (Rondon et al., 1995, Brinsmade et al., 2005). As seen in figure 4B (open triangles), the addition of glutathione restored growth of the alluded strain at a rate similar to that of the ΔeutQ/peutQ+ strain, and allowed the culture to reach full density.
Figure 4. The EutQ phenotype is caused by acetaldehyde, and can be corrected by aldehyde dehydrogenases or glutathione.
Strains were grown anoxically on minimal medium supplemented with ethanolamine (30 mM), NH4Cl (30 mM), and tetrathionate (40 mM) at 37°C. A. In all cases, ectopic gene expression was induced using arabinose (0.5 mM), except for eutQ+, which was induced using a 10-fold lower concentration of arabinose (0.05 mM). Growth rates were calculated at the log phase of the cultures (mean ΔOD% • h−1): ΔeutQ/peutQ+, 15; ΔeutQ/peutE+, 16; ΔeutQ/ppduP+, 9; ΔeutQ/padhE+, 9; ΔeutQ/pBAD24, no growth. B. All strains were induced with 0.05 mM arabinose. ΔeutQ/pBAD24 was supplemented with oxidized glutathione (GSSG, 2.5 mM) where indicated. Growth rates: eutQ+/pBAD24, 12; ΔeutQ/peutQ+,16; ΔeutQ/pBAD24, 1; ΔeutQ/pBAD24 + GSSG, 14.
In vivo evidence suggests that EutQ has acetate kinase activity
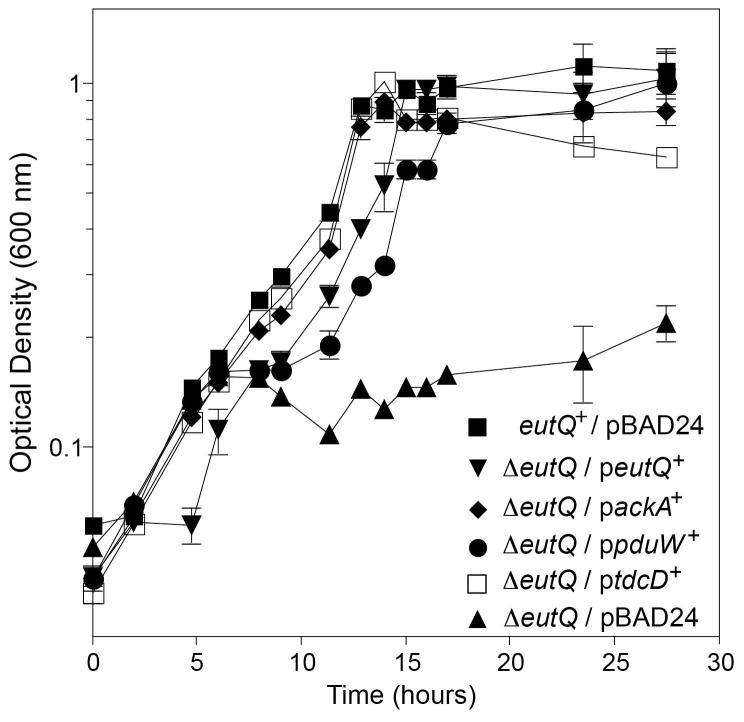
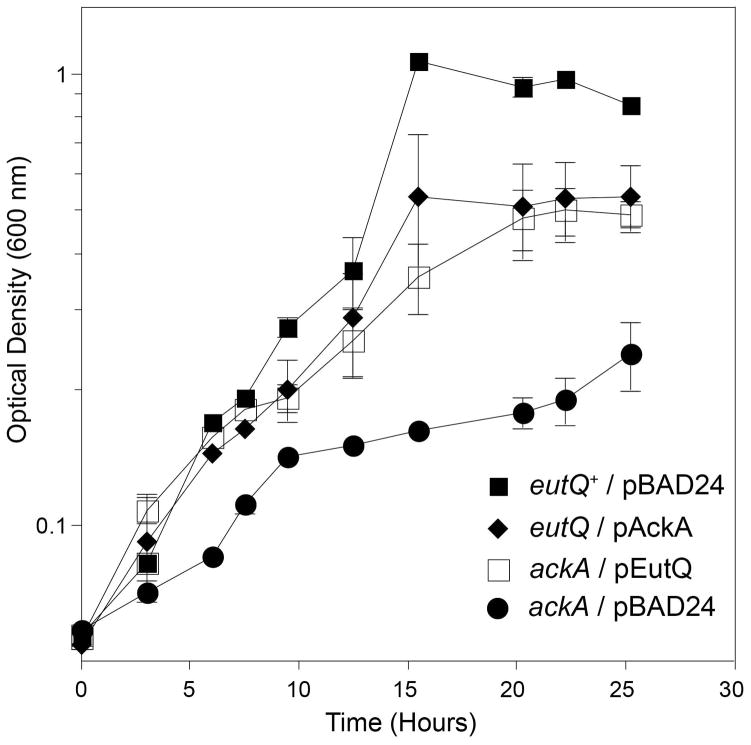
Although the above results could be interpreted to mean that EutQ had acetaldehyde dehydrogenase activity, we considered that such a possibility was unlikely, given the fact that EutE has been identified as the acetaldehyde dehydrogenase encoded by the eut operon (Zhu et al., 2011). Instead, we hypothesized that the synthesis of acetyl-CoA from acetaldehyde and CoA was, for some reason, an unfavorable reaction and that EutQ played a key role in pulling the reaction forward with the eventual release of acetate. Given that EutD is a phosphotransacetylase (Brinsmade & Escalante-Semerena, 2004), we posited that EutQ had acetate kinase activity. To test this idea we introduced into the ΔeutQ strain wild-type alleles encoding the housekeeping acetate kinase AckA (EC 2.7.2.1), or propionate/acetate kinases (EC 2.7.2.15) such as PduW and TdcD (Wolfe, 2005, Palacios et al., 2003, Hesslinger et al., 1998, Bobik et al., 1999). Remarkably, synthesis of AckA, PduW or TdcD restored anoxic growth of the eutQ strain on ethanolamine and tetrathionate (Fig. 5). These results strongly suggested that EutQ had acetate kinase activity. To obtain additional in vivo evidence to support this conclusion we performed a reciprocal experiment where EutQ would be expected to compensate for the absence of the AckA acetate kinase. A ΔackA strain had a significant growth defect on ethanolamine and tetrathionate, which was efficiently corrected by ectopically expressed eutQ+ or ackA+ (Fig. 6, squares, diamonds). Our interpretation of these results is presented in the Discussion section.
Figure 5. Complementation of the EutQ phenotype by acetate kinases.
Strains were grown anaerobically on minimal medium supplemented with ethanolamine (30 mM), NH4Cl (30 mM), tetrathionate (40 mM) and L(+)-arabinose (0.05 mM) at 37°C. Growth rates were calculated for each culture during the logarithmic phase of growth (mean ΔOD% • h−1): eutQ+/pBAD24, 12; ΔeutQ/peutQ+, 14; ΔeutQ/packA+, 15; ΔeutQ/ppduW+, 18; ΔeutQ/ptdcD+, 16; ΔeutQ/peutQ+, 14; ΔeutQ/pBAD24, 1.
Figure 6. Complementation of the AckA phenotype on 30 mM ethanolamine by EutQ.
Strains were grown anaerobically on minimal medium supplemented with ethanolamine (30 mM), tetrathionate (40 mM), and L(+)-arabinose (0.05 mM). Growth rates were calculated for each culture during the logarithmic phase of growth (mean ΔOD% • h−1): eutQ+/pBAD24, 11; ΔeutQ/packA+, 11; ΔackA/pBAD24 6; ΔackA/peutQ+, 9.
EutQ and EutP have acetate kinase activity in vitro
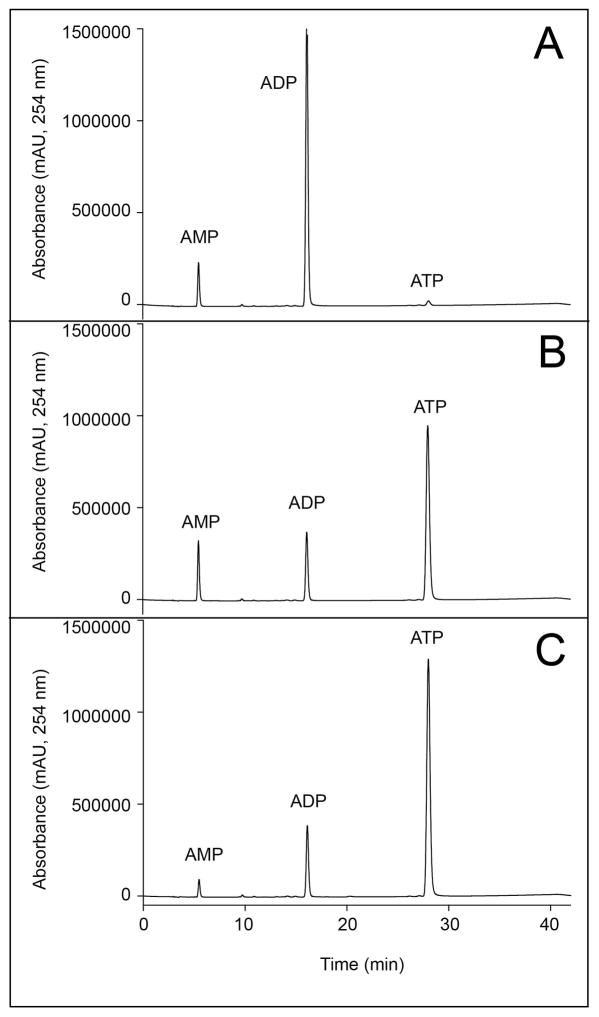
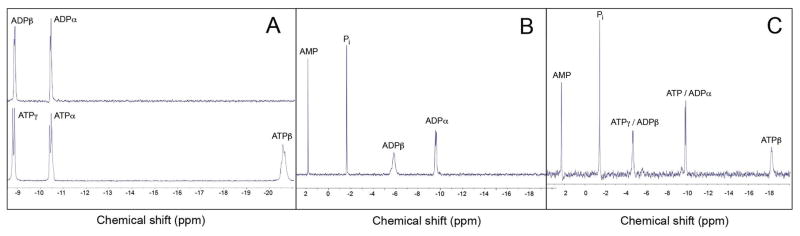
Highly purified preparations of EutP and EutQ (Fig. S2) were assayed for acetate kinase activity in vitro using HPLC to resolve substrates from products. Due to the lability of AcP and the difficulty in detecting low amounts of this compound by HPLC, we performed the EutQ reaction in the opposite direction, providing AcP and ADP as substrates for the enzyme to make ATP (Fig. 7). Panels B, C of figure 7 show that EutP and EutQ synthesized ATP from AcP and ADP, relative to the results obtained in the absence of either enzyme (panel A). The presence of AMP in the no-enzyme control indicates that it might have been an impurity of the stock solution. The formation of ATP was confirmed using 31P-NMR spectroscopy (Fig. 8). Fig. 8A shows the chemical shifts of phosphates associated with commercially available ADP and ATP. Panel B shows the spectrum of a reaction mixture devoid of enzyme, and panel C shows the spectrum of the sample containing EutQ, which contained all the peaks expected for ATP. Similar results were obtained with EutP (data not shown).
Figure 7. EutQ and EutP generate ATP in vitro via phosphorylation of ADP.
All reaction mixtures contained AcP and ADP (1 mM each), and were incubated for 1 hour at 37°C. A. No-enzyme control; B. EutQ (0.7 μM). C) EutP (0.7 μM). Peaks were labeled in comparison to authentic standards. All reactions were monitored at 254 nm.
Figure 8. EutQ dependent synthesis of ATP from ADP and acetyl-phosphate monitored by 31P-NMR spectroscopy.
Reaction mixtures (1 ml final volume) containing EutQ enzyme (3.5 μg), ADP (5 mM), AcP (5 mM), and MgCl2 (5 mM) were prepared for 31P-NMR analysis as described under Experimental procedures. Panel A shows proton-decoupled 31P-NMR spectra of commercially available ADP (top) and ADP (bottom), both at 10 mM. B. No-enzyme control; C. Products of the EutQ reaction. Note that the shifts of ATP and ADP are shifted in the positive direction because of complexation with the Mg2+ ions in the EutQ reaction, a shift that has been described in the literature (Bock, 1980, Szabo, 2008).
EutQ and EutP were also assayed for acetate kinase activity in a continuous spectrophotometric assay that measured the oxidation of NADH. The reaction rate was dependent on the concentration of protein (Figure S3), and both enzymes failed to use the non-hydrolyzable ATP analogue, AMP-PCP, as substrate (data not shown). To test for the presence of divalent ions, the EutQ acetate kinase was dialyzed against buffer A containing EDTA (2 mM), but lacking imidazole (three buffer changes, 4°C for 2 h each). EutQ dialyzed under such conditions did not lose any activity compared to untreated enzyme (data not shown). EutQ used guanosine triphosphate (GTP) as substrate (870 μM/min/mg), but did it at a 45% slower rate than the same reaction containing an equimolar ATP concentration (1300 μM/min/mg). Neither EutQ nor EutP displayed acetate kinase activity when cytidine triphosphate (CTP) or thymidine triphosphate (TTP) was used in lieu of ATP.
The eutP gene is immediately upstream of eutQ in the eut operon (Fig. 1B), and its 3′ end overlaps with the start codon of eutQ. Although we have been unable to find a phenotype for ΔeutP strains under conditions in which a ΔeutQ strain has a phenotype, we considered the possibility that EutP and EutQ may interact. Surprisingly, EutP demonstrated acetate kinase activity independently of EutQ, and generated ATP from ADP and acetyl-phosphate, and acetyl-phosphate from ATP and acetate (Figs. 8, 9). Similarly to EutQ, EutP did not have activity with AMP-PCP, CTP, or TTP, but had a 28% lower specific activity with GTP (1750 μmol/min/mg) compared to ATP (2400 μmol/min/mg). Dialysis of EutP against EDTA did not have an effect on its specific activity, and its kinetic parameters indicated that EutP was a slightly faster enzyme than EutQ, with nearly identical Km values, and kcat values that were approximately 30–40% higher than those for EutQ for either acetate or ATP (Table 2).
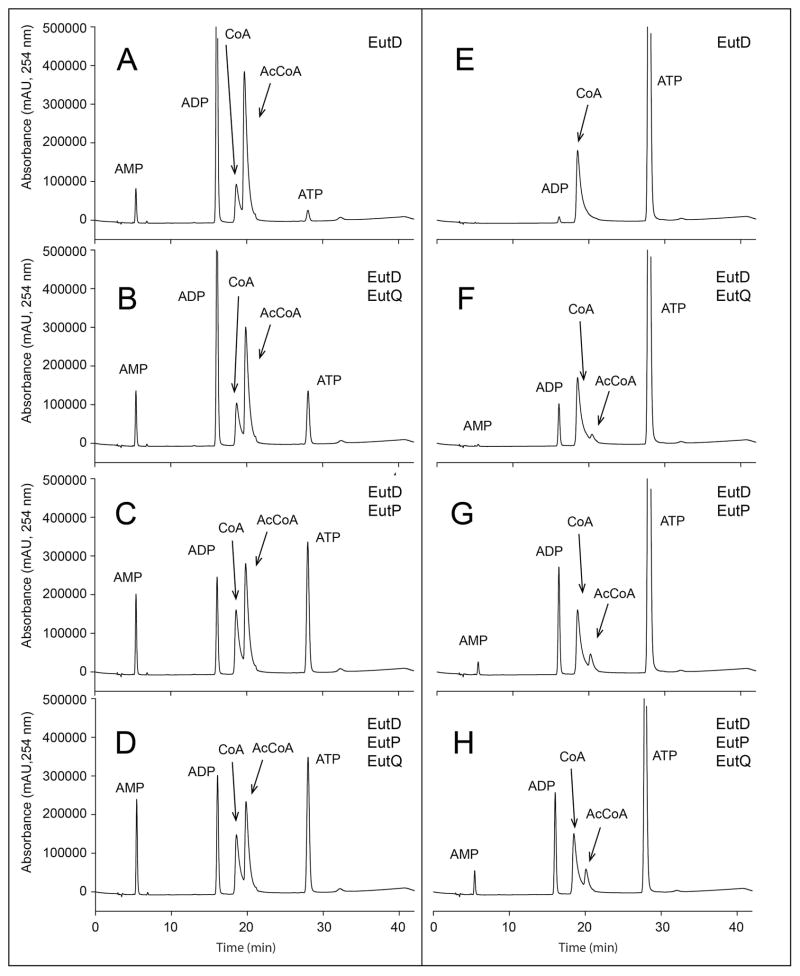
Figure 9. Use of HPLC to monitor the reversibility of the EutP- and EutQ-catalyzed reactions coupled to the EutD phosphotransacetylase.
Reaction mixtures included enzymes shown in the upper right of each panel. Panels A–D. In these experiments all reaction mixtures contained Pi + ADP + AcCoA. Under these conditions bona fide acetate kinases synthesized ATP at the expense of AcP generated by EutD. Panels E–H. In these experiments EutQ and/or EutP converted acetate to acetyl-phosphate at the expense of ATP. EutD was added to the reaction mixture for the in situ generation of AcCoA from AcP + CoA. All reactions were monitored at 254 nm, and all proteins used in these experiments were >95% homogeneous (Fig. S2).
Table 2.
Kinetic constants for EutQ and EutP
| Enzyme | Substrate: | Km (mM) | kcat (s−1) | kcat/Km (s−1 mM−1) |
|---|---|---|---|---|
| EutQ | Acetate | 0.7 ± 0.2 | 318 | 454 ± 101 |
| ATP | 0.5 ± 0.1 | 434 | 868 ± 145 | |
| EutP | Acetate | 0.7 ±0.2 | 545 | 778 ± 173 |
| ATP | 0.5 ± 0.2 | 657 | 1314 ± 377 |
Data were obtained using the lactate dehydrogenase/pyruvate kinase coupled assay described under Experimental procedures. Assays were run in triplicate. Values (means ± standard deviations) were calculated using nonlinear regression in Prism4 (GraphPad Software, v. 4.0a, 2003)
The EutP- and EutQ-catalyzed reactions can be coupled to a physiologically relevant phosphotransacetylase
EutP and EutQ can make AcP from acetate and ATP, or can catalyze the substrate-level phosphorylation of ADP using AcP as the phosphate donor. These conclusions were reached on the basis of data presented in figure 9. The reaction mixtures used to obtain the results shown in panels A–D used the phosphotransacetylase EutD to generate AcP from AcCoA and ortho-phosphate. When EutQ, EutP or both were present in the reaction mixture, ATP was formed at the expense of AcCoA (panels B–D); in the absence of EutP or EutQ ATP was not formed (panel A). The reaction was driven in reverse, with AcP generated by EutQ or EutP. The resulting AcP was used by EutD to yield AcCoA from CoA present in the mixture (panels F–H); again, in the absence of EutQ or EutP, AcCoA was not formed (panel E). Notably, the amount of acetyl-CoA converted to CoA by EutD increased upon co-incubation of EutQ (by 15%) or EutP (by 44%).
Kinetic analysis of EutQ and EutP
Kinetic parameters of EutQ and EutP were determined using the lactate dehydrogenase/pyruvate kinase coupled assay describe under Experimental procedures. Our results differ slightly from published values for the housekeeping S. enterica acetate kinase AckA (Chittori et al., 2012). In comparison, EutQ had a ten-fold higher Km for ATP (0.70 mM) than that of AckA (0.07 mM), but a two-fold lower Km for acetate (0.7 mM) than that of AckA (1.2 mM).
As mentioned above, EutQ belongs to the cupin superfamily of proteins, and many members of this superfamily bind transition metals to catalyze a variety of reactions. However, EutQ belongs to a class of cupins that lack a metal-binding motif (Dunwell et al., 2004). In place of one of the conserved histidines of the metal binding motif is an aspartate residue (D175), which has been proposed to form an acidic pair with a nearby glutamate (E173) (Fig. S4). These two residues are located deep within a negatively charged cleft, as seen in the three-dimensional crystal structure of EutQ from Clostridium difficile (PDB 4AXO) (Pitts et al., 2012). This cleft was proposed to be the active site, and remarkably, the authors modeled an acetyl moiety deep within the site, demonstrating the feasibility of this enzyme binding acetate (Pitts et al., 2012). However, changing either of these two residues to alanine did not impact the ability of the EutQ variants to restore growth of the ΔeutQ strain on ethanolamine and tetrathionate. Changing both of the residues to alanine resulted in a slight growth defect, compared to complementation by the eutQ+ allele (Fig. S5)
EutP and EutQ have no similarity to known acetate kinases
Comparisons of the EutP (159 aa) and EutQ (229 aa) sequences to the S. Typhimurium AckA enzyme (400 aa) (Figs. S6B, C) clearly show marked differences in size and sequence, placing EutP and EutQ in a different class of acetate kinases. In spite of these differences, there are several residues conserved in EutP, EutQ and AckA. Methanosarcina thermophila AckA acetate kinase was crystallized with a variety of nucleotide and acetyl substrates (PDB 1G99, 1TUY, 1TUU) (Ingram-Smith et al., 2005, Gorrell et al., 2005, Miles et al., 2002, Gorrell & Ferry, 2007) and from its comparison to the S. Typhimurium AckA, we identified residues able to make polar contact with the substrates (Fig. S5A). From this analysis, it appears that ADP-binding residues D285 and N337 of S. Typhimurium AckA align with E147 and N170 of EutQ, and the acetyl-binding residue H94 aligns to H14 of EutQ. These residues were identified using ClustalW2, which is not ideal for alignment of only two proteins. The more appropriate algorithm, BLAST, did not identify significant similarity between EutQ and AckA (E value: 1.3, coverage: 11%, identity: 35%). Future site-directed mutagenesis studies will determine if these residues contribute to EutQ function.
Discussion
EutQ is required for ethanolamine catabolism in S. Typhimurium under ecologically relevant conditions
Our data (Fig. 2) show for the first time that EutQ activity is required for growth S. Typhimurium on ethanolamine in an anoxic environment where the electron acceptor tetrathionate is present. Surprisingly, the absence of EutQ did not affect growth of this bacterium on ethanolamine in an oxic environment, a behavior that is discussed below under a separate context. The conditional need for EutQ highlights the importance of ethanolamine respiration to tetrathionate in the anoxic environment in the intestine of its host. In addition, the need for EutQ also raises questions regarding metabolite flux through the pathway and about the mechanisms of import and egress of enzyme substrates and products between the metabolosome lumen and the cytosol by as-yet-unidentified means.
EutQ and EutP are novel acetate kinases
In vivo and in vitro evidence reported here (Figs. 5–9) strongly support the conclusion that EutQ and EutP have acetate kinase activity. Unfortunately, the available crystal structure of the S. Typhimurium EutQ apo-protein (PDB 2PYT) does not provide insights into the identity of the active site. Interestingly, the investigators that reported the crystal structure of the Clostridum difficile EutQ homologue did model an acetyl moiety in it, without knowing that the enzyme was an acetate kinase (Pitts et al., 2012). This putative binding site for acetate, however, is not yet supported by experimental evidence. The authors proposed that the active site of EutQ was located deep within a negative cleft of each beta-barrel in the cupin-like structure, and that a conserved aspartate (D175) and glutamate (E173) residue were important for activity (Pitts et al., 2012) (Fig. S3). In our hands, S. Typhimurium EutQ acetate kinase activity did not require divalent metal cofactors, as dialysis with EDTA did not affect its activity. Consistent with the idea that EutQ is not a metalloprotein, we note that the His residues involved in metal binding among cupins are not conserved. Changing either D175 or E173 to alanine did not affect the ability of the resulting EutQ variants to restore growth of a ΔeutQ strain on ethanolamine/tetrathionate under anoxic conditions, which suggests that these residues do not play a significant role in substrate binding or catalysis.
Do EutP and EutQ work together or independently, and are they needed for tetrathionate respiration?
Bioinformatics information available from databases suggests that EutP binds ATP near its N terminus (Protein Feature View, RSCB PDB). Our data show that EutP has acetate kinase in vitro activity, and its kinetic parameters indicate that EutP is a slightly more efficient enzyme than EutQ, including a ~40% higher kcat with respect to ATP than EutQ (Table 2). From a physiologic standpoint it is not clear why S. Typhimurium has retained two genes encoding acetate kinases within the eut operon. One possibility could be that EutP and EutQ may be monomers of an acetate-kinase hetero-oligomer. If this were the case, the lack of phenotypes for ΔeutP strains (Fig. S1) would indicate that EutQ is the critical monomer of such a putative hetero-oligomer. As mentioned above, EutQ has been proposed to have an acetate-binding site, and EutP contains a nucleotide-binding P-loop. These two domains may interact to enhance acetate kinase activity. It is also possible that in addition to its enzymatic activity, EutQ may interact with other Eut proteins, including EutD or the proteins comprising the shell of the metabolosome, and that such roles are critical to the function of the putative hetero-oligomer. Another possibility is that eutP is transcribed very minimally compared to eutQ under the conditions tested. At present, data to support these ideas are lacking.
The presence of EutQ and EutP is not linked to the ability of S. Typhimurium to use tetrathionate as an electron acceptor. Notably, the genome of many ethanolamine-catabolizing organisms such as Klebsiella pneumoniae, Escherichia coli and Shigella flexneri encode copies of eutQ and eutP, yet these bacteria cannot respire to tetrathionate (Barrett & Clark, 1987). Therefore, EutQ and EutP are not directly involved in tetrathionate respiration in S. Typhimurium. A requirement for eutQ or eutP has yet to be tested in other organisms.
Are EutP and EutQ functions required in other ethanolamine catabolizing bacteria?
Across organisms with eut operons, the eutP and eutQ genes are not always adjacent, and in some cases one, the other or both are missing. For example, the Nocardioides, Listeriaceae, and Entercoccaceae eut operons only have eutQ, and the Clostridiaciae and Fusobacterium nucleatum do not have eutP and eutQ located next to one another (Tsoy et al., 2009). Numerous other genomes do not have either eutP or eutQ, although these organisms often have minimal eut operons lacking the shell and most other eut proteins except for EutBC, the small and large subunits of the ethanolamine ammonia-lyase central to ethanolamine catabolism.
Despite the lack of a phenotype for ΔeutP strains, there may be a link between EutP activity and pathogenesis. Of the 24 strains encoding eutP, 54% are known to cause food poisoning (Tsoy et al., 2009). Of the 24 strains carrying eutQ, 38% are pathogenic. In comparison, of the 85 strains carrying eutBC, the critical genes for ethanolamine catabolism, only 17% can cause disease. Although it is unclear if all these genes are active, the high percentage of pathogenic ethanolamine-respiring organisms encoding eutP may indicate a function for EutP activity in virulence.
Acetate kinase activity may be important for increasing flux through the EutD phosphotransacetylase
The growth inhibition caused by overexpression of eutQ+ was intriguing, but even more so was the relief of such inhibition by exogenous pantothenate (Fig. 3). Although the toxicity could be due to EutQ aggregation caused by non-physiological protein levels casued by overexpression, such an argument would not explain the effect of pantothenate. One possible explanation for both observations is that high levels of EutQ deprive the cell of AcCoA needed for anabolic purposes by increasing the conversion of AcCoA to AcP by EutD. We propose that pantothenate reverses this problem by increasing the pool of free CoA, which in turn is used by the CoA-consuming acetaldehyde dehydrogenase (EutE) enzyme to generate excess AcCoA, some of which can be diverted to central metabolism and promote cell growth. This idea is supported by results from in vitro experiments in which EutD was coupled to EutQ and EutP. EutQ, EutP, or both in combination enhanced the EutD-catalyzed conversion of AcCoA to CoA (Fig. 9E–H).
The above hypothesis can also help us explain the increased excretion of acetaldehyde in ΔeutQ strains. If in fact EutQ and EutP increase flux through the EutD enzyme as proposed above, their absence apparently results in less active EutD, which in turn would lead to less active EutE aldehyde dehydrogenase. With less flux through EutE, the acetaldehyde produced by ethanolamine ammonia-lyase (EutBC) would accumulate. This hypothesis is consistent with an earlier observation that eutQ and eutP mutant strains excrete more acetaldehyde than the wild-type strain (Penrod & Roth, 2006). If this hypothesis were correct, expressing aldehyde dehydrogenases in eutQ strains should correct the phenotype, a prediction that was experimentally validated (Fig. 4). An alternative hypothesis suggesting that EutQ activity is more important for energy charge maintenance is discussed below.
EutQ may play a role in increasing the energy charge and ATP recycling inside the metabolosome
The standard oxidation/reduction potential of tetrathionate/thiosulfate is low, +170 mV, relative to that of the oxygen/water couple (+815 mV) (Kapralek, 1972, Thauer et al., 1977). When the cell respires ethanolamine to tetrathionate a ΔG = −94.6 kJ/mol are available for ATP synthesis, compared to a ΔG = −219 kJ/mol available when molecular oxygen is the electron acceptor. This substantial difference in free energy likely results in a lower energy charge when tetrathionate is the electron acceptor, hence placing a greater importance on ATP synthesis via substrate-level phosphorylation by EutP and EutQ.
EutQ may be important for energy generation via substrate-level phosphorylation, which would explain why ΔeutQ strains and ΔackA strains grow poorly on ethanolamine/tetrathionate. Complementation by alternative acetate kinases in a ΔeutQ strain could be due to the restoration of ATP levels, which supports the substrate-level phosphorylation hypothesis. However, if EutQ activity is only important for generation of ATP, why is overexpression of EutQ toxic? and why can this toxicity be rescued by pantothenate? We suggest that if EutQ is necessary for substrate-level phosphorylation, and to ensure flux through EutD, as proposed above.
It is also possible that EutQ is required to recycle ATP within the metabolosome, although it is not known for certain if EutQ is localized inside or outside the compartment. Cofactor recycling within the metabolosome is an important aspect of the functionality of metabolosomes. Several enzymes have been shown to be involved in cofactor recycling within both the eut and pdu metabolosomes of S. Typhimurium. For example, EutD, EutE and EutG are needed to recycle CoA and NAD+ within the eut metabolosome, while PduP, PduQ and PduL fulfill the same role in the pdu metabolosome (Huseby & Roth, 2013, Liu et al., 2015, Cheng et al., 2012). The cobalamin-recycling enzyme EutT (Johnson et al., 2004, Sheppard et al., 2004, Buan & Escalante-Semerena, 2006, Mera et al., 2007, Moore et al., 2014) and the ethanolamine ammonia-lyase reactivase EutA (Aaron et al., 2007, Mori et al., 2004) are ATP-consuming enzymes, hence ATP is either generated inside, is transported into the metabolosome or both. Notably, ATP is the only co-substrate whose regeneration inside the metabolosome has not been accounted for. On the basis of our data, we propose that EutQ, and possibly EutP, fulfill this role. EutA is the only Eut enzyme that generates ADP as reaction product, but whether EutA is inside or outside of the metabolosome remains an open question. If EutA were located inside the metabolosome, there would be a need for ATP-recycling inside the structure or ATP flux into it.
If cofactor recycling is a role of EutQ, then why do ΔeutQ strains grow on ethanolamine under oxic conditions? One explanation would be that the flow of ATP into the metabolosome increases during oxic growth on ethanolamine due to the high-energy charge generated under such conditions.
Other ideas for why oxic growth conditions may enable a ΔeutQ strain to tolerate higher acetaldehyde concentrations
If the absence of EutQ slows the activity of EutE as proposed above, under oxic growth conditions a strain lacking EutQ may tolerate the resulting increased concentration of acetaldehyde for several reasons. First, the concentration of the detoxifying tripeptide glutathione (GSH) is four times higher during oxic growth than under anoxic growth (Fahey et al., 1978). Additionally, during oxic growth other proteins known to consume acetaldehyde, such as aldehyde dehydrogenase (AdhE) may be present at higher levels. Such a combination of detoxifying factors would prevent acetaldehyde from damaging proteins and DNA.
Why do cytosolic acetate kinases compensate for the absence of EutQ?
It is unlikely that the acetate/propionate kinases AckA, PduW or TdcD ever enter the metabolosome, yet they can compensate for the absence of EutQ during growth of a ΔeutQ strain on ethanolamine/tetrathionate (Fig. 5). One intepretation is that EutQ is located outside the metabolosome, and that AcP diffuses outside the shell and can be equally accessed by any acetate kinase. However, if this hypothesis is correct, why is there a need for specialized eut acetate kinases at all? We suggest it is more likely that EutQ, and perhaps EutP, are located within the metabolosome. Inside the metabolosome, they can interact immediately with their upstream product, which is likely AcP from the EutD phosphotransacetylase. Our interpretation for the complementation by alternative acetate kinases in a ΔeutQ strain is that AcP generated by EutD inside the metabolosome accumulates without being consumed by EutQ, and escapes into the cytosol through pores proposed to be small molecule transporters (Chowdhury et al., 2015). Once in the cytosol, AcP is consumed by AckA, PduW or TdcD, reducing the concentration of AcP inside the metabolosome with the concomitant acceleration of AcCoA and acetaldehyde by EutD and EutE, respectively. Additionally, the ATP produced by these enzymes may increase the energy charge of the bacterium to a level sufficient to overcome growth arrest. Future work will address these possibilities.
Conclusion
This work provides the first experimental evidence that the S. Typhimurium EutQ and EutP proteins have acetate kinase activity. Bioinformatics analyses support the conclusion that EutP and EutQ belong to new classes of acetate kinases. We conclude that acetaldehyde accumulation is responsible for the observed lack of growth of ΔeutQ strains under growth conditions that lower the energy charge of the cell and the concentration of aldehyde quenching biomolecules. We also suggest that substrate-level phosphoylation via the EutQ acetate kinase plays a significant role in cellular growth during ethanolamine respiration. Our findings are significant, not only because we have identified new classes of acetate kinases, but because of the need for these enzymes during respiration of ethanolamine to tetrathionate, a condition known to be critical to S. Typhimurium in the context of the disease-causing capabilities of this bacterium. In vivo evidence supporting a role for the EutP enzyme in ethanolamine catabolism is yet to be obtained.
Experimental procedures
Construction of expression and complementation vectors
To generate a recombinant construct of EutQ with a cleavable N-terminal hexahistidine (H6) tag, the S. Typhimurium eutQ+ allele was PCR amplified from strain JE6583 (Table 1). The primers used for the amplification were 5′-NNGCTCTTCNTTCGTGAAAAAACTTATCACAGCTAACGA-3′ (forward), and 5′-NNGCTCTTCNTTATCATACGGATTGCCAGTTTG-3′ (reverse). The PCR fragment and the vector pTEV18 were cut with restriction enzyme BspQI and ligated, yielding plasmid pEUT112. The latter directed the synthesis of a recombinant H6-EutQ whose tag was removed using recombinant tobacco etch virus (rTEV) protease (Blommel & Fox, 2007). Tagless EutQ contained two residues (Gly-Thr) upstream of the N-terminal Met residue; these residues did not affect enzyme function. This cloning strategy was also used to generate pTEV18 constructs containing eutP+ (plasmid pEUT110) and eutP+ eutQ+ (plasmid pEUT129). For eutP+, the following primers were used: 5′-NNGCTCTTCNTTCATGAAACGTATTGCTTTTGTCG-3′ (forward), and 5′-NNGCTCTTCNTTATTAGCTGTGATAAGTTTTTTCACCTG-3′ (reverse). Plasmid pEUT129 was generated using the eutP+ forward primer and the eutQ+ reverse primer. eutQ alleles encoding variant EutQ proteins were generated using the QuikChange XL site-directed mutagenesis kit (Stratagene). The pEUT112 and the pEUT56 (lab collection) plasmids were used as templates for the PCR-based site-directed mutagenesis according to the manufacturer’s instructions. Mutations in the eutQ sequence were confirmed by DNA sequencing using BigDye® protocols (ABI-Prism). DNA sequences were determined at the Georgia Genomics Facility at the University of Georgia-Athens.
Table 1.
Strains and plasmids used over the course of this study
| Strains | Genotype | Cloning vector | Plasmid | Protein encoded | Source, reference |
|---|---|---|---|---|---|
| E. coli strains | |||||
| JE3892 | BL21 (λDE3) | Lab collection | |||
| JE8833 | XL10 Gold | pBAD30 | pEUT56 | EutQWT | Lab collection |
| JE19891 (DH5α) | fhuA2 Δ(argF-lacZ)U169 phoA glnV44 Φ80 Δ(lacZ)M15 gyrA96 recA1 relA1 endA1 thi-1 hsdR17 | pBAD24 | pACK7 | AckAWT | Lab collection |
| JE8066 | DH5α | pADH3 | AdhEWT | Lab collection | |
| JE8111 | DH5α | pBAD30 | pPDU18 | PduPWT | Lab collection |
| JE7994 | DH5α | pEUT30 | EutEWT | Lab collection | |
| S. Typhimurium strains1 | |||||
| JE6583 | metE205 ara-9 | Lab collection | |||
| Derivatives of strain JE6583 | |||||
| JE6814 | prpC114::MudJ2 | pTDCD1 | TdcDWT | Lab collection | |
| JE8749 | Δack103 | Lab collection | |||
| JE8816 | ΔeutQ1183 | Lab collection | |||
| JE21128 | Δack103 | pBAD24 | Control | This work | |
| JE21130 | Δack103 | pBAD30 | pEUT56 | This work | |
| JE5304 | hsdSA29 hsdSB121 hsdL6 metA22 metE551 trpC2 ilv-452 rpsL120 xyl-404 galE719 H1-b H2-e, n,x) fla-66 nm | Formerly JR501 (Ryu & Hartin, 1990) | |||
| Derivatives of JE5304 | |||||
| JE20867 | pEUT110 | EutPWT | This work | ||
| JE20869 | pTEV18 | pEUT112 | EutQWT | This work | |
| JE5912 | pBAD30 | pPDU1 | PduWWT | Lab collection | |
| Derivatives of JE8816 | |||||
| JE14190 | pBAD24 | Control | None | Lab collection | |
| JE14116 | pBAD24 | pEUT56 | EutQWT | This work | |
| JE21131 | pBAD24 | pACK7 | AckAWT | This work | |
| JE21167 | pBAD30 | pPDU1 | PduWWT | This work | |
| JE21168 | pBAD18s | pTDCD1 | TdcDWT | This work | |
| JE21381 | pTAC-85 | pADH3 | AdhEWT | Lab colection | |
| JE21382 | pBAD30 | pPDU18 | PduPWT | Lab collection | |
| JE21383 | pBAD30 | pEUT30 | EutEWT | Lab collection |
S. Typhimurium strains were derivatives of S. enterica sv Typhimurium strain LT2
Protein overproduction and purification
Plasmids expressing eutP+ or eutQ+ were transformed into Escherichia coli BL21(λDE3) using a described heat shock protocol (Maniatis et al., 1982). Overnight cultures were inoculated 1:100 (v/v) into 2L of Terrific Broth (http://openwetware.org/wiki/Terrific_Broth) supplemented with ampicillin (100 μg/mL). Cultures were grown to an OD600 ~0.7 at 37°C with shaking. Protein expression was induced with isopropyl-β-D-1 thiogalactopyranoside (IPTG, 0.5 mM) and shaken overnight at 10°C. Cell pellets were harvested at 12,000 × g using a Beckman/Coulter Avanti J-25I centrifuge equipped with a JLA-16.250 rotor. Cell pellets were frozen at −80°C until used. Cell pellets were re-suspended in buffer A (4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES, 50 mM, pH 7) containing sodium chloride (NaCl, 500 mM), imidazole (70 mM) and tris-(2-carboxyethyl)phosphine (TCEP, 0.25 mM)), at a ratio of 5 mL per gram of cell paste. Cells were broken using a French pressure cell under 103 MPa and 4°C. To ensure cell breakage, the procedure was performed twice. Cellular debris was removed from solution by centrifugation (45,000 × g for 45 min) followed by filtration through a 0.45-μm-pore filter.
His-tagged proteins were separated from crude cell-free extracts using an AKTApurifier fast protein liquid chromatograph equipped with a 5-mL HisTrap column (GE Healthcare). The column was washed with 40 mL of buffer A, and tagged protein was eluted using a 50-mL linear gradient to 100% buffer B (buffer A containing 500 mM imidazole). H7-rTEV protease (1 mg/mL) was added to the elutate at a 1:50 ratio (v/v) to cleave the H6-tag from the protein of interest. H7-rTEV was prepared as described (Rocco et al., 2008). The tagged protein/protease mixture was dialyzed three times into buffer A (1L each), once for 1 h, once overnight, and once for 3 h, all at 4°C.
The protein of interest was loaded onto a fresh HisTrap column and eluted with 25 mL of buffer A to remove the cleaved His-tag and H7-rTEV. Tagless protein was dialyzed into buffer C (buffer A with no imidazole and 10%(v/v) glycerol) thrice as described above. Proteins were flash-frozen in liquid N2 and stored at −80°C until use. Protein purity was assessed by sodium dodecyl-sulfate polyacrylamide gel electrophoresis (SDS-PAGE) (Laemmli, 1970), followed by staining with Coomassie Brilliant Blue R (Sigma) (Sasse, 1991).
In vivo assessment of function
To assess the role of EutQ in vivo, a strain (JE8816, metE205 ara-9 ΔeutQ1182) was tested for phenotypes when grown on minimal medium. No-carbon essential (NCE) minimal medium (Vogel & Bonner, 1956, Berkowitz et al., 1968) was supplemented with ethanolamine (30 mM), methionine (0.5 mM), NH4Cl (30 mM), MgSO4 (1 mM) L-(+)-arabinose (500 μM), glycerol (0.5 mM), and Wolfe’s trace minerals (Balch & Wolfe, 1976). The small amount of glycerol added to all cultures greatly reduced the length of the lag phase. The growth rate in the early stages of growth observed in all curves was due to the use of glycerol as the source of carbon and energy. All reported growth rates were calculated after glycerol depletion. When growth curves were performed under anoxic conditions, tetrathionate (40 mM) was supplied as electron acceptor. Anoxic medium was degassed in an anoxic chamber (Coy) for 24 h, and stirred for 1 h to remove all oxygen prior to inoculation. Growth under the stated conditions was monitored for 24 h or less due to precipitation of sulfur granules after prolonged growth (Price-Carter et al., 2005). Plasmids used for in vivo analyses were constructed as described above and in Table 1. Plasmids carrying eutQ wild type or mutant alleles were under the control of the arabinose-inducible ParaBAD promoter in plasmid pBAD24 (Guzman et al., 1995).
Spectrophotometric acetate kinase assay
To determine the kinetic parameters of homogeneous EutQ, we used a modified version of a described acetate kinase coupled assay (Bergmeyer et al., 1984). Briefly, the assay measures the rate at which an acetate kinase phosphorylates acetate at the expense of ATP. The resulting ADP is consumed by pyruvate kinase, which reacts it with PEP to make ATP and pyruvate. Pyruvate is reduced to lactate by lactic dehydrogenase at the expense of NADH. NADH consumption was monitored spectrophotometrically. Each reaction contained HEPES buffer (84 mM, pH adjusted at 25°C), sodium acetate (NaOAc, made fresh, variable concentration), MgCl2 (6.6 mM), NADH (1.1 mM), pyruvate kinase/lactate dehydrogenase (1–1.6/1.6–2.3 U, respectively), myokinase (0.66 U), phophoenolpyruvate (PEP, 1.9 mM, made fresh) and ATP (variable concentration). Reactions were set up in a 96-well plate and initiated by the addition of EutQ or EutP (2 μM). The reaction was monitored at 340 nm on a Spectramax Plus (Molecular Devices). Reactions were performed at 40°C.
HPLC determination of EutQ and EutP reaction products
EutP and EutQ can make AcP at the expense of acetate and ATP yielding ADP as a byproduct. The reaction can be reversed by providing ADP and AcP to the enzymes. We used HPLC to show that the acetate kinase activities of EutP and EutQ are reversible. For this purpose, the chromatograph was equipped with a Partisil-10 SAX column (250 × 4.6 mm, Phenomenex) and an NH2 SecurityGuard cartridge (Phenomenex). A gradient of potassium phosphate (monobasic, 25°C) from 0.05 (pH 4.9) to 1 M (pH 4.0) was applied to the column over a 35-min period at a flow rate of 1 mL/min at 25°C. Reaction products were detected at 254 nm. Prior to HPLC analysis, reaction mixtures were incubated at 37°C for 1 h (when using AcP and ADP as substrates) or 2 h (when coupled to EutD), and filtered through Spin-X columns (Costar, 0.45 μm). To test the back reaction (i.e. AcP + ADP
 acetate + ATP), we mixed EutP or EutQ (0.7 μM each), ADP (1 mM), AcP (1mM) and MgCl2 (1 mM) in PIPES buffer (20 mM, pH7) in a 500 μl final volume. In some cases we used the EutD phosphotransacetylase to generate AcP in situ from AcCoA and ortho-phosphate (Pi). In such cases, the reaction mixtures contained EutD (0.7 μM), AcCoA (0.5 mM), and KH2PO4 (1 mM) in lieu of AcP. The forward reaction (i.e acetate + ATP
acetate + ATP), we mixed EutP or EutQ (0.7 μM each), ADP (1 mM), AcP (1mM) and MgCl2 (1 mM) in PIPES buffer (20 mM, pH7) in a 500 μl final volume. In some cases we used the EutD phosphotransacetylase to generate AcP in situ from AcCoA and ortho-phosphate (Pi). In such cases, the reaction mixtures contained EutD (0.7 μM), AcCoA (0.5 mM), and KH2PO4 (1 mM) in lieu of AcP. The forward reaction (i.e acetate + ATP
 AcP + ADP) was demonstrated by coupling the production of AcP by EutP or EutQ to the synthesis of AcCoA by EutD. For this purpose, the reaction mixtures contained EutD (0.7 μM), EutP or EutQ (0.7 μM), ATP (1 mM), acetate (1 mM), MgCl2 (1 mM), and coenzyme A (0.5 mM).
AcP + ADP) was demonstrated by coupling the production of AcP by EutP or EutQ to the synthesis of AcCoA by EutD. For this purpose, the reaction mixtures contained EutD (0.7 μM), EutP or EutQ (0.7 μM), ATP (1 mM), acetate (1 mM), MgCl2 (1 mM), and coenzyme A (0.5 mM).
31P-NMR spectroscopy
For 31P-NMR analysis, reaction mixtures were prepared as described for HPLC analysis. Reaction mixtures (500 μl) were brought up to a final volume of 600 μL in D2O (17%, v/v). Proton-decoupled 31P-NMR spectra were obtained using a Varian Unity Inova500 500 mHz spectrometer in the Chemical Sciences Magnetic Resonance Facility fo the University of Georgia. A total of 1024 scans were performed per sample. Chemical shifts were referenced to H3PO4 (85%) set to 0.0 ppm.
Supplementary Material
Acknowledgments
This works was supported by USPHS grant R37 GM040313 to J.C.E.-S. We would like to thank Dongtao Cui at the University of Georgia Chemical Sciences Magnetic Resonance facility for her technical assistance.
Footnotes
The authors do not have any conflict of interest to declare.
References
- Aaron M, Charbon G, Lam H, Schwarz H, Vollmer W, Jacobs-Wagner C. The tubulin homologue FtsZ contributes to cell elongation by guiding cell wall precursor synthesis in Caulobacter crescentus. Mol Microbiol. 2007;64:938–952. doi: 10.1111/j.1365-2958.2007.05720.x. [DOI] [PubMed] [Google Scholar]
- Balch WE, Wolfe RS. New approach to the cultivation of methanogenic bacteria: 2-mercaptoethanesulfonic acid (HS-CoM)-dependent growth of Methanobacterium ruminantium in a pressurized atmosphere. Appl Environ Microbiol. 1976;32:781–791. doi: 10.1128/aem.32.6.781-791.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett EL, Clark MA. Tetrathionate reduction and production of hydrogen sulfide from thiosulfate. Microbiological reviews. 1987;51:192. doi: 10.1128/mr.51.2.192-205.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumler AJ, Winter SE, Thiennimitr P, Casadesus J. Intestinal and chronic infections: Salmonella lifestyles in hostile environments. Environ Microbiol Rep. 2011;3:508–517. doi: 10.1111/j.1758-2229.2011.00242.x. [DOI] [PubMed] [Google Scholar]
- Bergmeyer J, Grassl M, Berger R. Principles of enzymatic analysis. In: Bergmeyer HU, editor. Methods of enzymatic analysis. Weinheim/Bergstrasse; Verlag Chemie: 1984. [Google Scholar]
- Berkowitz D, Hushon JM, Whitfield HJ, Jr, Roth J, Ames BN. Procedure for identifying nonsense mutations. J Bacteriol. 1968;96:215–220. doi: 10.1128/jb.96.1.215-220.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blommel PG, Fox BG. A combined approach to improving large-scale production of tobacco etch virus protease. Protein Expr Purif. 2007;55:53–68. doi: 10.1016/j.pep.2007.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobik TA. Polyhedral organelles compartmenting bacterial metabolic processes. Appl Microbiol Biotechnol. 2006;70:517–525. doi: 10.1007/s00253-005-0295-0. [DOI] [PubMed] [Google Scholar]
- Bobik TA, Havemann GD, Busch RJ, Williams DS, Aldrich HC. The propanediol utilization (pdu) operon of Salmonella enterica serovar Typhimurium LT2 includes genes necessary for formation of polyhedral organelles involved in coenzyme B12-dependent 1, 2-propanediol degradation. J Bacteriol. 1999;181:5967–5975. doi: 10.1128/jb.181.19.5967-5975.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock JL. The binding of metal ions to ATP: a proton and phosphorus nmr investigation of diamagnetic metal--ATP complexes. J Inorg Biochem. 1980;12:119–310. doi: 10.1016/s0162-0134(00)80123-3. [DOI] [PubMed] [Google Scholar]
- Brinsmade SR, Escalante-Semerena JC. The eutD gene of Salmonella enterica encodes a protein with phosphotransacetylase enzyme activity. J Bacteriol. 2004;186:1890–1892. doi: 10.1128/JB.186.6.1890-1892.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinsmade SR, Paldon T, Escalante-Semerena JC. Minimal functions and physiological conditions required for growth of Salmonella enterica on ethanolamine in the absence of the metabolosome. J Bacteriol. 2005;187:8039–8046. doi: 10.1128/JB.187.23.8039-8046.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buan NR, Escalante-Semerena JC. Purification and initial biochemical characterization of ATP:Cob(I)alamin adenosyltransferase (EutT) enzyme of Salmonella enterica. J Biol Chem. 2006;281:16971–16977. doi: 10.1074/jbc.M603069200. [DOI] [PubMed] [Google Scholar]
- Buan NR, Suh SJ, Escalante-Semerena JC. The eutT gene of Salmonella enterica encodes an oxygen-labile, metal-containing ATP:corrinoid adenosyltransferase enzyme. J Bacteriol. 2004;186:5708–5714. doi: 10.1128/JB.186.17.5708-5714.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton RM, Stadtman ER. The oxidation of acetaldehyde to acetyl coenzyme A. J Biol Chem. 1953;202:873–890. [PubMed] [Google Scholar]
- Chang GW, Chang JT. Evidence for the B12-dependent enzyme ethanolamine deaminase in Salmonella. Nature. 1975;254:150–151. doi: 10.1038/254150a0. [DOI] [PubMed] [Google Scholar]
- Cheng S, Fan C, Sinha S, Bobik TA. The PduQ enzyme is an alcohol dehydrogenase used to recycle NAD+ internally within the Pdu microcompartment of Salmonella enterica. PLoS One. 2012;7:e47144. doi: 10.1371/journal.pone.0047144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng S, Liu Y, Crowley CS, Yeates TO, Bobik TA. Bacterial microcompartments: their properties and paradoxes. Bioessays. 2008;30:1084–1095. doi: 10.1002/bies.20830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chittori S, Savithri HS, Murthy MR. Structural and mechanistic investigations on Salmonella typhimurium acetate kinase (AckA): identification of a putative ligand binding pocket at the dimeric interface. BMC Struct Biol. 2012;12:24. doi: 10.1186/1472-6807-12-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury C, Chun S, Pang A, Sawaya MR, Sinha S, Yeates TO, Bobik TA. Selective molecular transport through the protein shell of a bacterial microcompartment organelle. Proc Natl Acad Sci U S A. 2015;112:2990–2995. doi: 10.1073/pnas.1423672112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotton PB. Non-dietary lipid in the intestinal lumen. Gut. 1972;13:675–681. doi: 10.1136/gut.13.9.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunwell JM, Purvis A, Khuri S. Cupins: the most functionally diverse protein superfamily? Phytochemistry. 2004;65:7–17. doi: 10.1016/j.phytochem.2003.08.016. [DOI] [PubMed] [Google Scholar]
- Escalante-Semerena JC, Roth JR. Regulation of cobalamin biosynthetic operons in Salmonella typhimurium. J Bacteriol. 1987;169:2251–2258. doi: 10.1128/jb.169.5.2251-2258.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahey RC, Brown WC, Adams WB, Worsham MB. Occurrence of glutathione in bacteria. J Bacteriol. 1978;133:1126–1129. doi: 10.1128/jb.133.3.1126-1129.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung CK, Proulx P. Metabolism of phosphoglycerides in E. coli. 3. The presence of phospholipase A. Can J Biochem. 1969;47:3713. [PubMed] [Google Scholar]
- Gorrell A, Ferry JG. Investigation of the Methanosarcina thermophila acetate kinase mechanism by fluorescence quenching. Biochemistry. 2007;46:14170–14176. doi: 10.1021/bi701292a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorrell A, Lawrence SH, Ferry JG. Structural and kinetic analyses of arginine residues in the active site of the acetate kinase from Methanosarcina thermophila. J Biol Chem. 2005;280:10731–10742. doi: 10.1074/jbc.M412118200. [DOI] [PubMed] [Google Scholar]
- Guzman LM, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Held M, Quin MB, Schmidt-Dannert C. Eut bacterial microcompartments: insights into their function, structure, and bioengineering applications. J Mol Microbiol Biotechnol. 2013;23:308–320. doi: 10.1159/000351343. [DOI] [PubMed] [Google Scholar]
- Hesslinger C, Fairhurst SA, Sawers G. Novel keto acid formate-lyase and propionate kinase enzymes are components of an anaerobic pathway in Escherichia coli that degrades L- threonine to propionate. Mol Microbiol. 1998;27:477–492. doi: 10.1046/j.1365-2958.1998.00696.x. [DOI] [PubMed] [Google Scholar]
- Huseby DL, Roth JR. Evidence that a metabolic microcompartment contains and recycles private cofactor pools. J Bacteriol. 2013;195:2864–2879. doi: 10.1128/JB.02179-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingram-Smith C, Gorrell A, Lawrence SH, Iyer P, Smith K, Ferry JG. Characterization of the acetate binding pocket in the Methanosarcina thermophila acetate kinase. J Bacteriol. 2005;187:2386–2394. doi: 10.1128/JB.187.7.2386-2394.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson CL, Buszko ML, Bobik TA. Purification and initial characterization of the Salmonella enterica PduO ATP:Cob(I)alamin adenosyltransferase. J Bacteriol. 2004;186:7881–7887. doi: 10.1128/JB.186.23.7881-7887.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapralek F. The physiological role of tetrathionate respiration in growing Citrobacter. J Gen Microbiol. 1972;71:133–139. doi: 10.1099/00221287-71-1-133. [DOI] [PubMed] [Google Scholar]
- Kofoid E, Rappleye C, Stojiljkovic I, Roth J. The 17-gene ethanolamine (eut) operon of Salmonella typhimurium encodes five homologues of carboxysome shell proteins. J Bacteriol. 1999;181:5317–5329. doi: 10.1128/jb.181.17.5317-5329.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Larson TJ, Ehrmann M, Boos W. Periplasmic glycerophosphodiester phosphodiesterase of Escherichia coli, a new enzyme of the glp regulon. J Biol Chem. 1983;258:5428–5432. [PubMed] [Google Scholar]
- Leal NA, Havemann GD, Bobik TA. PduP is a coenzyme-A-acylating propionaldehyde dehydrogenase associated with the polyhedral bodies involved in B12-dependent 1,2-propanediol degradation by Salmonella enterica serovar Typhimurium LT2. Arch Microbiol. 2003;180:353–361. doi: 10.1007/s00203-003-0601-0. [DOI] [PubMed] [Google Scholar]
- Liu Y, Jorda J, Yeates TO, Bobik TA. The PduL phosphotransacylase is used to recycle coenzyme A within the Pdu microcompartment. J Bacteriol. 2015 doi: 10.1128/JB.00056-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T, Fritsch EF, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory; 1982. Introduction of plasmid and bacteriophage lambda into Escherichia coli; pp. 250–251. [Google Scholar]
- Mera PE, Maurice MS, Rayment I, Escalante-Semerena JC. Structural and functional analyses of the human-type corrinoid adenosyltransferase (PduO) from Lactobacillus reuteri. Biochemistry. 2007;46:13829–13836. doi: 10.1021/bi701622j. [DOI] [PubMed] [Google Scholar]
- Miles RD, Gorrell A, Ferry JG. Evidence for a transition state analog, MgADP-aluminum fluoride-acetate, in acetate kinase from Methanosarcina thermophila. J Biol Chem. 2002;277:22547–22552. doi: 10.1074/jbc.M105921200. [DOI] [PubMed] [Google Scholar]
- Moore TC, Mera PE, Escalante-Semerena JC. the EutT enzyme of Salmonella enterica is a unique ATP:Cob(I)alamin adenosyltransferase metalloprotein that requires ferrous ions for maximal activity. J Bacteriol. 2014;196:903–910. doi: 10.1128/JB.01304-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori K, Bando R, Hieda N, Toraya T. Identification of a reactivating factor for adenosylcobalamin-dependent ethanolamine ammonia lyase. J Bacteriol. 2004;186:6845–6854. doi: 10.1128/JB.186.20.6845-6854.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios S, V, Starai J, Escalante-Semerena JC. Propionyl coenzyme A is a common intermediate in the 1,2-propanediol and propionate catabolic pathways needed for expression of the prpBCDE operon during growth of Salmonella enterica on 1,2-propanediol. J Bacteriol. 2003;185:2802–2810. doi: 10.1128/JB.185.9.2802-2810.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penrod JT, Roth JR. Conserving a volatile metabolite: a role for carboxysome-like organelles in Salmonella enterica. J Bacteriol. 2006;188:2865–2874. doi: 10.1128/JB.188.8.2865-2874.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitts AC, Tuck LR, Faulds-Pain A, Lewis RJ, Marles-Wright J. Structural insight into the Clostridium difficile ethanolamine utilisation microcompartment. PLOS One. 2012;7:e48360. doi: 10.1371/journal.pone.0048360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price-Carter M, Fazzio TG, Vallbona EI, Roth JR. Polyphosphate kinase protects Salmonella enterica from weak organic acid stress. J Bacteriol. 2005;187:3088–3099. doi: 10.1128/JB.187.9.3088-3099.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price-Carter M, Tingey J, Bobik TA, Roth JR. The alternative electron acceptor tetrathionate supports B12-dependent anaerobic growth of Salmonella enterica serovar Typhimurium on ethanolamine or 1,2-propanediol. J Bacteriol. 2001;183:2463–2475. doi: 10.1128/JB.183.8.2463-2475.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocco CJ, Dennison KL, Klenchin VA, Rayment I, Escalante-Semerena JC. Construction and use of new cloning vectors for the rapid isolation of recombinant proteins from Escherichia coli. Plasmid. 2008;59:231–237. doi: 10.1016/j.plasmid.2008.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rondon MR, Kazmierczak R, Escalante-Semerena JC. Glutathione is required for maximal transcription of the cobalamin biosynthetic and 1,2-propanediol utilization (cob/pdu) regulon and for the catabolism of ethanolamine, 1,2-propanediol, and propionate in Salmonella typhimurium LT2. J Bacteriol. 1995;177:5434–5439. doi: 10.1128/jb.177.19.5434-5439.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roof DM, Roth JR. Ethanolamine utilization in Salmonella typhimurium. J Bacteriol. 1988a;170:3855–3863. doi: 10.1128/jb.170.9.3855-3863.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roof DM, Roth JR. Ethanolamine utilization in Salmonella typhimurium. J Bacteriol. 1988b;170:3855–3863. doi: 10.1128/jb.170.9.3855-3863.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roof DM, Roth JR. Functions required for vitamin B12-dependent ethanolamine utilization in Salmonella typhimurium. J Bacteriol. 1989;171:3316–3323. doi: 10.1128/jb.171.6.3316-3323.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu J, Hartin RJ. Quick transformation in Salmonella typhimurium LT2. Biotechniques. 1990;8:43–45. [PubMed] [Google Scholar]
- Sasse J. Detection of proteins. In: Ausubel FA, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K, editors. Current Protocols in Molecular Biology. New York: Wiley Interscience; 1991. pp. 10.16.11–10.16.18. [Google Scholar]
- Sheppard DE, Penrod JT, Bobik T, Kofoid E, Roth JR. Evidence that a B12-adenosyl transferase is encoded within the ethanolamine operon of Salmonella enterica. J Bacteriol. 2004;186:7635–7644. doi: 10.1128/JB.186.22.7635-7644.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stojiljkovic I, Baumler AJ, Heffron F. Ethanolamine utilization in Salmonella typhimurium: nucleotide sequence, protein expression, and mutational analysis of the cchA cchB eutE eutJ eutG eutH gene cluster. J Bacteriol. 1995;177:1357–1366. doi: 10.1128/jb.177.5.1357-1366.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo Z. Multinuclear NMR studies of the interaction of metal ions with adenine-nucleotides. Coordin Chem Rev. 2008;252:2362–2380. [Google Scholar]
- Thauer RK, Jungermann K, Decker K. Energy conservation in chemotrophic anaerobic bacteria. Bacteriol Rev. 1977;41:100–180. doi: 10.1128/br.41.1.100-180.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiennimitr P, Winter SE, Baumler AJ. Salmonella, the host and its microbiota. Curr Opin Microbiol. 2012;15:108–114. doi: 10.1016/j.mib.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsoy O, Ravcheev D, Mushegian A. Comparative genomics of ethanolamine utilization. J Bacteriol. 2009;191:7157–7164. doi: 10.1128/JB.00838-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel HJ, Bonner DM. Acetylornithinase of Escherichia coli: partial purification and some properties. J Biol Chem. 1956;218:97–106. [PubMed] [Google Scholar]
- Winter SE, Thiennimitr P, Winter MG, Butler BP, Huseby DL, Crawford RW, Russell JM, Bevins CL, Adams LG, Tsolis RM, Roth JR, Baumler AJ. Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature. 2010;467:426–4269. doi: 10.1038/nature09415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe AJ. The acetate switch. Microbiol Mol Biol Rev. 2005;69:12–50. doi: 10.1128/MMBR.69.1.12-50.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeates TO, Thompson MC, Bobik TA. The protein shells of bacterial microcompartment organelles. Curr Opin Struct Biol. 2011;21:223–231. doi: 10.1016/j.sbi.2011.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Gonzalez R, Bobik TA. Coproduction of acetaldehyde and hydrogen during glucose fermentation by Escherichia coli. Appl Environ Microbiol. 2011;77:6441–6450. doi: 10.1128/AEM.05358-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.