Abstract
Here we describe protocols to culture adult zebrafish hearts as explants, and to study regeneration of epicardial tissue ex vivo. Uninjured or injured adult hearts are excised, washed, and cultured in an incubator with gentle agitation. Preparation of heart explants is accomplished within 2 hours, and explants can be maintained in culture for 30 days or longer. Dynamic behaviors of epicardial cells are monitored through live imaging of fluorescent transgenes in the explant, using stereofluorescence microscopy. We also describe ex vivo procedures for genetic ablation of the epicardium, cell proliferation assays, tissue grafts, and bead grafts. Basic cell culture and surgical skills are required to carry out these protocols. As opposed to existing protocols for culturing isolated zebrafish epicardial cells on matrices, procedures described here maintain epicardial cells on an intact cardiac surface, better enabling in vivo cell behaviors. Our protocols complement and extend in vivo studies of heart regeneration.
Keywords: heart regeneration, epicardium, explant culture, genetic ablation, live imaging
INTRODUCTION
Zebrafish provide a valuable model system to study regeneration of cardiac tissue, as injured hearts can regenerate large portions of lost muscle through the proliferation of spared cardiomyocytes. Non-muscle supporting cells like epicardium, endocardium, vasculature, nerves, and inflammatory cells play critical roles during heart regeneration1-6. The epicardium, a mesothelial layer covering the surface of all vertebrate hearts, is required for normal muscle regeneration after cardiac injury in zebrafish4, 5, 7-10. Studies in zebrafish and mammals have led to many proposed roles for the epicardium: in vascularization of new muscle, as a source of mitogens, as a contributor to muscle survival, and as a cellular target for direct reprogramming strategies11-17.
Tissue regeneration is by definition an in vivo phenomenon. However, regeneration studies in adult zebrafish suffer from their long animal generation times (~3 months) and the dearth of molecular genetic tools for inducibly manipulating adult regenerative events. Moreover, the heart as an internal organ is technically challenging to access for live imaging of regenerative events. To address these challenges, we have developed an explant system that enables culturing of adult hearts for several weeks, while maintaining much of the morphology and contractility of the heart7. With this method and molecular genetic tools for genetic ablation and live monitoring of epicardial cells, we defined and dissected regenerative capacity of adult zebrafish epicardial tissue in a recent research report7. Additionally, this explant culture system can be adapted for tissue grafting and chemical screening approaches. Here, we describe in detail this protocol for explant culture, and important applications.
Development of the protocol
Our original methods for culturing heart explants incubated extracted hearts for 7 days with Liebovit's L-15 medium supplemented with 10% fetal calf serum and antibiotics10. With this method, heart explants tended to attach to the dish bottom, altering ventricular shape and challenging live visualization of epicardial cell behaviors on the cardiac surface. Spontaneous internal infarcts were formed within ~7 days inside the explants, suggesting poorly maintained cardiac muscles. An additional protocol exists to culture isolated epicardial cells from adult zebrafish on a fibrin matrix18. With this method, epicardial cells migrate into the fibrin gel from explanted chunks of ventricle apex. This method is advantageous for cellular and molecular analysis; however it is a more artificial environment than the cardiac surface. Several other methods exist for culturing mammalian cardiac slices for weeks while maintaining contractile activity19-22; yet, these systems are not applicable for studies of the epicardium.
To visualize and manipulate epicardial cells on an intact cardiac surface for extended imaging experiments, we refined our explant culture protocol with the following specific modifications. a) Instead of non-carbonate buffered L-15 medium, we used carbonate buffered DMEM medium, which is ideal for a CO2 incubator. We also tested DMEM/F12 and IMDM medium without noticing apparent differences. We ultimately chose to use DMEM medium, as it is inexpensive and sufficient for the experiments described here. b) We added 1% non-essential amino acids (NEAA) and 0.1% 2-mercaptoethanol, used previously for long-term heart slice cultures19. NEAA increases cell growth and viability, while 2-mercaptoethanol is a reducing agent that than can limit oxygen radical levels. c) We used a cocktail of antibiotics - Primocin - for the first few days of primary culture to reduce the incidence of contamination18. We also tested a combination of penicillin-streptomycin and fungizone, which was inadequate in reducing contamination. d) Most importantly, we agitated the culture plate at 150 rpm in an orbital shaker while culturing, which allows the explant to remain suspended in the culture and prevents ventricular adherence and deformation. Agitating manually every 12 h without a shaker was typically not sufficient to prevent attachment of explants to the dish bottom. These alterations in methodology allow the heart surface to remain largely intact during culture. With the refined protocol, we were able to longitudinally monitor epicardial cell migration and proliferation on the surface of a beating heart for at least one month. This protocol is suitable for studies that focus on the epicardium and epicardial regeneration. We have not tested pacing of the explanted hearts, different energy substrates and application of hyperoxygenation, which have the potential to further optimize the protocol.
Overview of the procedure
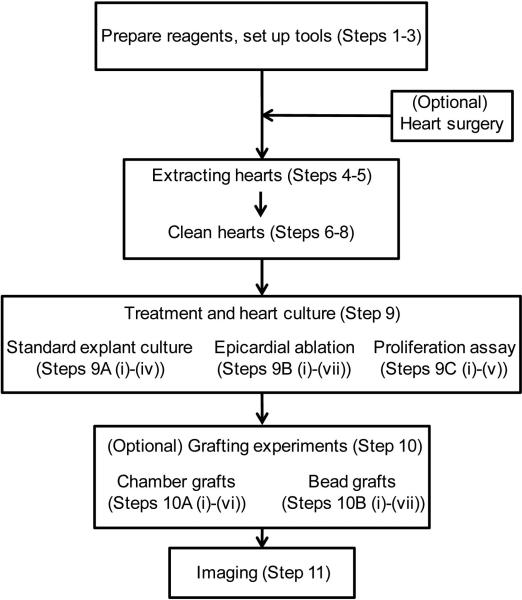
The overall experimental scheme is shown in Fig. 1. In this protocol, we will describe explant culture of intact hearts and ex vivo epicardial ablation. To ablate epicardial cells, we generated a BAC line by fusing a bacterial Nitroreductase (NTR) to an mCherry fluorescent reporter gene cassette, driven by tcf21 regulatory sequences, which are epicardial and epicardial-derived-cell-specific in the heart (tcf21:mCherry-NTR)5, 7. After treatment with metronidazole (Mtz), which is processed by NTR into a cytotoxin, this line induced cell death specifically of epicardial and epicardial-derived cells23, 24. We typically use a tcf21:nucEGFP BAC reporter strain to mark epicardial cell nuclei5. In optional procedures (Box 1-3), we describe an assay for proliferation by epicardial cells in culture by incorporation of the nucleotide analog 5-ethynyl-2’-deoxyuridine (EdU) (Box 1); the engraftment of a freshly collected outflow tract to the base of an epicardially-ablated ventricle (Box 2); and embedding of beads with Hh proteins onto the ventricular base (Box 3).
Figure 1.
Flowchart of experimental procedures.
Box 1. Proliferation assay TIMING 5-8 h.
Following Steps 1-11. To detect proliferation, EdU should be added to the culture medium at a concentration of 25 μM.
-
a1.
Warm DMEM culture medium to 28°C and add EdU to 25 μM. Change medium of the explants with EdU medium and return to the incubator. A one-hour incorporation will efficiently label cells undergoing DNA synthesis. If explants will not be fixed immediately after the 1 h treatment, the medium with EdU should be filter sterilized before use. If planning to wash out EdU and keep culturing, pipette out EdU medium and wash twice with PBS as described in Step 12, and then change to fresh medium without EdU.
-
a2.
Wash hearts with PBS twice and fix hearts in 4% paraformaldehyde. EdU staining can be done as described7.
CRITICAL STEP PBS washes are important; otherwise unincorporated EdU will produce high background.
Box 3. Bead grafts TIMING about 26 h.
Following Steps 1-12. Here we graft beads soaked with Hh proteins onto the ventricular base as an example.
CRITICAL STEP Steps c1-c6 can be performed in a biological hood using sterile tools.
-
c1.
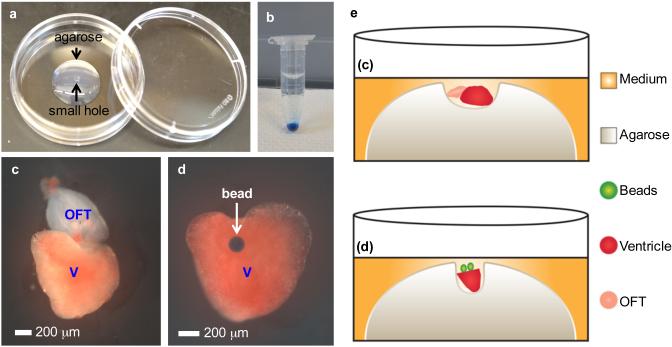
Prepare culture dishes with low melting agarose. Make a round hole in the middle of the agarose as described above (Fig. 3a; Supplementary Video 2).
-
c2.
Affi-Gel Blue beads were prepared by thoroughly washing the beads in PBS for 5 times in an EP tube. During each wash, allow beads to settle by gravity, and then gently remove PBS without disturbing beads at the bottom (Fig. 3b). After the last washing, remove as much PBS as possible by using a 20 μl pipette tip.
CRITICAL STEP Beads must be washed thoroughly to remove preservative.
-
c3.
Recombinant mouse Sonic Hedgehog (C25II), N-terminus protein was reconstituted at 100 μg/ml in phosphate buffered saline (PBS) containing 0.1% bovine serum albumin (BSA). Then 20 μl Shh solution is added to the beads and incubated for 2 hours at room temperature while rotating on a shaker (GeneMate GyroMixer). A solution with the same concentration of BSA protein is used as the control. The bead solutions can be stored at 4°C.
-
c4.
Transfer ventricular explants to the agarose, and gently place the ventricle into the hole with the ventricular base on top, using forceps.
-
c5.
Tilt the dish and pipette out extra medium before adding beads. Use a 20 μl pipette tip to pipette beads and apply to the base of ventricular explants. Add medium carefully to cover only the explant and beads without allowing the beads to float (Fig. 3d, e). Transfer the dish to the incubator.
CRITICAL STEP Two to three beads can be applied to one site. Not all beads will be successfully grafted.
CRITICAL STEP Do not add too much medium, as this may cause the beads to float.
CRITICAL STEP Gently place the dish in the incubator, and keep still until the release of explants from the agarose.
-
c6.
After 24 hours, release the ventricles from the agarose with the attached beads and transfer to a new dish with culture medium and culture while shaking. Images are taken every day as described in Step 13.
TROUBLESHOOTING
Box 2. Chamber grafts TIMING 25-50 h.
Following Steps 1-12. Here, we graft freshly collected outflow tracts to an epicardially ablated ventricle as an example. The ventricle was treated with 1 mM Mtz for 24 h and cultured for 2 more days before engraftment.
CRITICAL STEP Steps b1-b5 should be performed in a biological hood with sterile tools.
-
b1.
Prepare culture dishes with low melting agarose. Under a dissecting scope, construct a hole in the middle of the agarose by using forceps, just large enough to hold the ventricle (Fig. 3a).
CRITICAL STEP Several holes with different sizes can be made in case heart size varies among your animals.
-
b2.
Collect outflow tracts by following Steps 4-8.
-
b3.
Transfer ventricular explants on top of the agarose by using a 1 ml pipette tip with a cut off end (or a plastic transfer pipette). Gently place the ventricle into the hole with the ventricular base postioned laterally, using forceps. Then, transfer an outflow tract to the same hole, modifying the size of the hole to ensure the tissues contact each other at their bases (Fig. 3c, e; Supplementary Video 1).
CRITICAL STEP Ensure the size of the hole is appropriate - not too small to squeeze chambers.
-
b4.
Add sufficient medium carefully to the dish to cover the agarose with explants, without agitating the explants. Culture in the incubator without shaking.
CRITICAL STEP Gently place the dish in the incubator, and prevent agitation until explants are released from the agarose in subsequent steps.
-
b5.
After 24-48 h of culture, release the grafted explants from the agarose by disrupting the agarose with forceps and flushing gently with culture medium. Transfer the explants to a new culture dish with fresh medium and culture while shaking. Images are taken every day as described in Step 13.
TROUBLESHOOTING
Comparison with existing methods
Our original ex vivo explantation method was developed to study epicardial and endocardial cell gene expression after the formation of spontaneous internal infarcts10. This method is suitable only for short-term culture (7 days). Also, as heart explants attach to the culture dish and cardiac muscle undergoes rapid necrosis, there is considerable opportunity for artifacts. In vitro primary cell culture of isolated epicardial cells is effective to characterize epicardial cell-extracellular matrix interactions, and can be expanded to tests of cell-cell interactions by co-culture18. Although a more artificial environment than the cardiac surface, dissociated cell culture has certain advantages for cellular and molecular studies. including the capacities for gene transfection and live imaging. The refined protocol we describe here combines the advantages of these two systems. It is suitable for cellular and molecular analyses while largely maintaining the cardiac surface environment (described in the next section). Other methods for culturing cardiac slices are sufficient for testing cardiac muscle behaviors but are not applicable for studies of the epicardium19-22.
Applications of the method
Recently, by generating transgenic tools to ablate the epicardium in zebrafish, we found that the epicardium is required for normal regeneration of heart muscle after injury. Moreover, with the aid of this ex vivo explant culture system, we identified unexpected features by which the epicardium itself regenerates after its depletion. The procedures we describe have considerable advantages for investigating the cellular and molecular mechanisms of epicardial cell gene regulation, proliferation, migration, and overall regeneration in real time while largely recapitulating in vivo environment. The heart explants are also suitable for assays of gene expression by in situ hybridization. We include procedures here that enable the study of tissue-tissue interactions, including tissue and bead grafting, which can potentially be expanded to include other tissues. A clear application for these methods is chemical screening; for instance, one can test hundreds or thousands of compounds for influences on epicardial cell behaviors while on the cardiac surface. Those factors that induce epicardial cell proliferation or the activation of markers of tissue regeneration have the to potential to influence the efficacy of heart regeneration.
Limitations
The protocol described here has been successfully used in studying epicardial cell behaviors, but it is not suitable for detailed studies of cardiomyocytes. We detect very limited proliferation of cardiomyocytes during culture, possibly due to missing ingredients or suboptimal environmental conditions. Rates of contraction decrease during culture, and hearts occasionally show internal infarcts. Further development of the protocol is required to improve its applicability for ex vivo studies of cardiac muscle. Most applications can be completed in 2 weeks, during which the hearts appear grossly normal in morphology and beating. For long-term culture of more than 2 weeks, the heart shape becomes slightly altered. Shrinking is observable, and the pyramidal ventricle appears more rounded, likely secondary to the shaking of the culture. After grafting of cardiac chambers, the ventricular shape may change slightly due to pressure exerted from the agarose used in the procedure. However, no apparent difference in epicardial cell behavior (such as proliferation and migration) was observed in ventricles of different shapes.
Experimental design
Partial ventricular resection surgeries and inducible epicardial ablation
Depending on the experimental purpose, partial resection can be performed by removing 10-20% of the ventricular apex as described previously, before explant culture1. Epicardial ablation can be performed before or during explant culture. Hearts can be collected at any time following in vivo manipulations. Examples can be found in our recent publication7. Hearts after resection or ablation injury will be adhesive and may be difficult to separate from pericardial material. In this report, we will describe explant culture of intact hearts and those undergoing ex vivo epicardial ablation. As heart sizes, surgery size and positions, and ablation effects are variable among different fish and clutches, at least 10 explants per treatment are required for each experiment. Five sham-treated (uninjured or unablated) explants should be included each time as a system control for culture procedures. At least two independent replicates are required for conclusions. If treatments will be applied during explant culture, such as chemical treatment and bead embedding, uninjured explants and/or explants with surgery or ablation should be randomized into different groups for each experiment. Finally, only clutchmates (or hearts collected from clutchmates) will be used for each experiment.
Choice of culture dish
While shaking, explants will gather in the middle of the culture dish or well. If epicardial cells are ablated or heart surgeries are performed prior to culturing, explants are adhesive and will become attached to each other when cultured as multiples. In this situation, one heart per well is recommended and multi-well plates are economical. On the other hand, small wells of 24- or 48- well plate are not conducive for live imaging. As the samples must remain covered while imaging, only the center of these small wells can provide an undistorted view. The 12-well plate is the best choice for most experiments. Uninjured hearts can be cultured together in larger wells or petri dishes.
Proliferation assay
There are various ways to assay indices of proliferation in cells on the cardiac cell surface. To mark cells undergoing DNA synthesis, we use the nucleotide analog EdU. A 1-hour period of EdU incorporation at 25 μM is sufficient to label a large number of cells. Phosphorylated Histone H3 is a marker of cells in G2/M phase, recognized by several commercially available antibodies. We choose EdU as an example here. Epicardial cell nuclei are best identified by the tcf21:nucEGFP strain5. Two independent replicates and 10 explants per treatment are required for quantification of proliferation indices.
Chemical treatment
Small molecule compounds can be included in the incubation protocols to examine effects on epicardial proliferation and regeneration7. It is important to maintain sterile medium, accomplished either by filter sterilization of the medium with diluted drug, or by keeping the drug stocks sterile. For each chemical compound, two independent replicates with at least 10 explants of uninjured or epicardially ablated hearts per replicate are treated, and 10 more vehicle treated explants of hearts from the same clutch of zebrafish served as a control. Medium should be changed every other day with fresh chemicals. For uninjured hearts, effects on epicaridial proliferation are assessed by quantification of cell numbers using the tcf21:nucEGFP reporter at 2-6 days of culture. For effects on epicardial regeneration after ablation, chemicals are added at 2 days post Mtz treatment, and explants from the tcf21:nucEGFP strain are imaged every day for up to 14 days to assess epicardial proliferation and migration.
Tissue grafts
We have successfully engrafted the bulbous arteriosus (cardiac outflow tract) or ventricular apex to the base or apex of ventricles that have had their epicardium genetically depleted7. Here we describe the engraftment of freshly collected outflow tract to the base of an epicardially-ablated ventricle. In this experiment, about 50% of the explants will be tightly grafted together after 24 h contact in agarose. For 10 successful grafts, at least 20 attempts are required. As the grafted positions vary across grafts, we prefer to make conclusions based on 3 independent replicate experiments. The protocol should also be applicable to other tissue combinations.
Bead grafts
Agarose beads can be used to test factor function by controlled protein release. Here we will embed beads soaked with Hh proteins onto the ventricular base as an example. Beads soaked with BSA protein serve as a control. At least 50% of ventricles successfully engraft and maintain beads during culture. Three independent replicates with at least 20 attempts per replicate are required for each experiment. Heparin acrylic beads can also be used25.
MATERIALS
Zebrafish
Adult zebrafish of the Ekkwill and Ekkwill/AB strains were used, maintained as described1. Animals between 4 and 12 months of both sexes are generally used. Transgenic lines used in this study were Tg(tcf21:mCherry-NTR)pd108 (ref. 7), Tg(tcf21:nucEGFP)pd41 (ref. 26), Tg(cmlc2:actin3-EGFP)sd10 (ref. 27) and Tg(fli1a:EGFP)y1 (ref. 28). All transgenic strains were analyzed as hemizygotes. Animal procedures were approved by the Institutional Animal Care & Use Committee at Duke.
REAGENTS
Fish water (aquarium water from the zebrafish facility)
Tricaine (Sigma, cat. no. A5040) CAUTION Avoid inhalation and direct contact with eyes and skin.
DMEM cell culture medium (Invitrogen, cat. no. 11965)
FBS (Thermo Scientific, cat. no. SH30071.03)
Primocin (InvivoGen, cat. no. ant-pm-2)
PBS (Corning, cat. no. 46-013-CM)
Ethanol (VWR, cat. no. 89125-172)
Metronidazole (Mtz, Sigma, cat. no. M1547) CAUTION Mtz is a carcinogen. Avoid contact with skin and eyes. Avoid inhalation. Wear gloves and eye protection.
L-Glutamine (Invitrogen, cat. no. 25030-081)
MEM-NEAA (Invitrogen, cat. no. 11140-050)
Penicillin-streptomycin (Invitrogen, cat. no. 15140-122)
Low-melting agarose (Fisher Scientific, cat. no. BP1360-100)
Affi-Gel Blue beads (Bio-Rad, cat. no. 1537301)
5-ethynyl-2’-deoxyuridine (EdU, Life Technologies, cat. no. A10044) CAUTION Avoid contact with skin and eyes. Avoid inhalation. Wear gloves and eye protection.
Recombinant mouse Sonic Hedgehog (C25II), N-terminus protein (R & D Systems, cat. no. 464-SH-025)
Bovine serum albumin (BSA, VWR, cat. no. 97061-420)
Paraformaldehyde (Fisher Scientific, cat. no. O4042-500) CAUTION Avoid contact with skin and eyes. Avoid inhalation. Wear gloves and eye protection.
Fluoromount G mounting medium (Fisher Scientific, cat. no. 0100-01)
EQUIPMENT
Finger bowls (90 × 50 mm; VWR, cat. no. 89000-288)
1.7 ml Eppendorf tubes (EP tubes, Genesee Scientific, cat. no. 24-281)
12-well plates (Corning, cat. no. 3737)
60 mm Petri dishes (Corning, cat. no. 351007)
Millex-GP Filter units (0.22μm, Millipore, cat. no. SLGP033RB)
Syringes (BD, cat. no. 302831)
Sterile pipette tips (Corning, cat. no. 4135, 4139 and 4140)
Sterile disposable plastic transfer pipettes (VWR, cat. no. 414004-016)
Cover slips (Fisher Scientific, cat. no. 12-544-E)
Stainless steel microforceps (Dumont, cat. no. 5-inox-H)
Stainless steel microscissors (World Precision Instruments, cat no. 14124)
Sponges (1.5 × 5 × 3 cm) with a single center groove cut using scissors (0.5 × 2.5 cm)
Plastic spoons, heavy duty (Staples)
Falcon tubes (15 ml, 50 ml, Corning, cat. no. 430790, 430828)
Dissecting microscope (Leica)
Water bath (Fisher Scientific, ISOTEMP 150)
125 ml flasks (Corning)
Biological safety cabinet with laminar flow and UV light (Labconco, Class II, type A2)
Cell culture incubator (28°C, 5% CO2, humidified; Fisher Scientific, Model 3530)
Orbital shaker (Troemner, cat. no. 12620-938)
GeneMate GyroMixer, Variable Speed (BioExpress, cat. no. R-3200-1XL)
Leica MZ05FA stereofluorescence microscope with camera
REAGENT SETUP
Tricaine stock solution
The stock solution contains 15 mM ethyl 3-aminobenzoate methanesulfonate (tricaine) and 20 mM Tris-HCl, pH 7.4. Store this stock solution at 4°C for as long as one month, and at −20°C for longer term storage. To anaesthetize adult zebrafish, dilute 4.2 ml of the stock solution into 100 ml of fish water.
Culture medium
Culture medium consists of DMEM plus 10% fetal bovine serum, 1% MEM-NEAA (non-essential amino acids), 100 U/ml penicillin, 100 μg/ml streptomycin, and 50 μM 2-mercaptoethanol. Primocin™ was added just before use to prohibit microbial contaminants during the first few days primary culture. Sterile medium can be stored at 4°C for up to one month without effects on culture results. CAUTION Medium and PBS must be sterile filtered.
Ablation medium
For epicardial ablation, freshly prepare 1 mM Mtz solution in culture medium for use. First make 10-20 ml of a 10 mM Mtz solution in a 50 ml Falcon tube by vortexing and pipetting. Then dilute 1:10 to the desired volume, and filter sterilize with a 0.22 μm filter unit.
CAUTION As Mtz is difficult to dissolve, ensure it is fully dissolved before use.
Low-melting agarose
Keep a separate bottle of agarose powder for culture use only, to reduce chance of contamination. To make 30 ml 1% low-melting agarose, weigh 0.3 g of powder and put into autoclaved 125 ml flask and add 30 ml sterile PBS. Microwave for several minutes until agarose is fully dissolved. Store the agarose in a 37°C water bath until use.
CRITICAL Use sterile PBS and flask.
PROCEDURE
Preparation TIMING 1 h
-
1.
Prepare reagents as described above. Warm DMEM culture medium and PBS to 28°C (or room temperature, 25 °C), and prepare ablation medium if required. Place the orbital shaker in the incubator.
-
2.
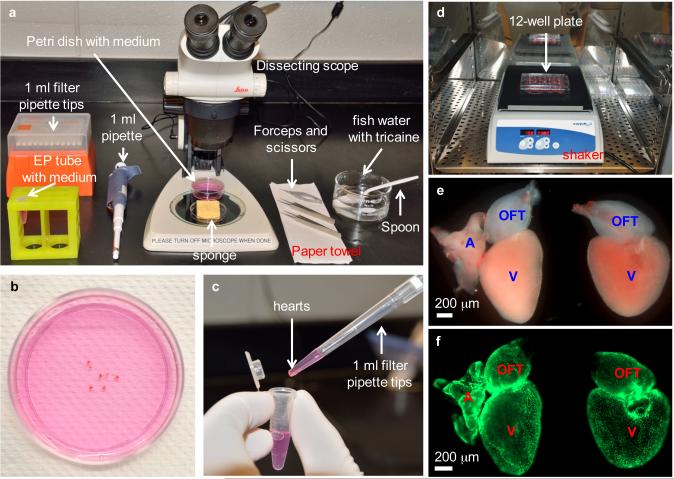
Sterilize forceps and scissors by immersing in 70% ethanol. Clean bench and dissecting scope stage with 70% ethanol. Clean sponge with ethanol and squeeze off residual ethanol prior to moistening with fish water (Fig. 2a).
Figure 2.
Heart dissection and explant culture. (a) Equipment setup for heart dissection, including dissecting scope, forceps, scissors, pipette and filter tips, grooved sponge, 60 mm Petri dish with culture medium, 1.5 ml EP tube with culture medium, finger bowel with diluted Tricaine in aquarium water, spoon and paper towel. (b) Collected hearts are placed in a 60 mm Petri dish to remove blood clots and extra tissues. (c) Cleaned hearts are transferred to an EP tube by using a 1 ml pipette tip with the end cut off. (d) Washed heart explants are transferred to a 12-well culture plate, and the plate is placed on a shaker in a CO2 incubator, shaking at 150 rpm. (e) Close-up view of two heart explants with (left) or without (right) outflow tract in a culture plate. Hearts are from tcf21:nucEGFP zebrafish. A, atrium; V, ventricle; OFT, outflow tract. (f) Fluorescent view of explants in (e), showing nuclear EGFP signals. Work with zebrafish was approved by the Institutional Animal Care & Use Committee at Duke.
CAUTION A sponge that is used repeatedly could accumulate contaminating microorganisms. Washing with EtOH will reduce the chance of contamination.
CRITICAL STEP Steps 2–7 must be performed on a clean lab bench using sterile tools and filter pipette tips.
-
3.
Prepare a 60 mm petri dish with 5 ml culture medium, and EP tubes with 1 ml culture medium per tube. All samples will be placed in one tube (Fig. 2a).
Extracting hearts TIMING 2-5 min per heart
-
4.
Euthanize the fish with Tricaine in a glass bowl until gill movements end, and there is no response to a tail pinch. Use a plastic spoon to transfer the fish, dry briefly on a paper towel, and then place fish ventral side up onto the groove of moist sponge under a dissecting microscope.
CRITICAL STEP Animal procedures must be performed in accordance with institutional guidelines.
-
5.
Open the ventral wall of the fish with a longitudinal cut at the gill area. Locate the outflow tract. Grab the distal end of outflow tract with forceps and gently pull the heart out of the chest cavity. Sever the ventral aorta and sinus venosus with forceps. Place the heart into the 60 mm Petri dish with culture medium (Fig. 2b). A video demonstrating a similar heart extraction procedure can be found in a previous protocol18.
CRITICAL STEP If more then one heart is processed, store hearts in a Petri dish with medium at room temperature. Keep the lid of the culture dish closed.
Clean hearts TIMING 5 min
-
6.
Carefully remove blood clots on the heart surface by using forceps while holding the end of the outflow tract with the other forceps. Remove the atrium or outflow tract if required by experiment.
CRITICAL STEP Steps 6-10: Be careful not to damage the epicardium, which may influence experiments.
-
7.
After all hearts are dissected, cut off the tip of a 1 ml pipette tip, and use this (or a sterile plastic transfer pipette) to transfer hearts to the EP tube containing 1 ml culture medium (Fig. 2c).
CRITICAL STEP All explant transfer procedures should be performed in this way.
-
8.
In a biological safety hood, wash hearts three times, using 1 ml PBS per wash.
CRITICAL STEP Steps 8–12 must be performed in a biological safety hood using sterile pipette tips and PBS.
Heart culture and epicardial ablation TIMING 5 min
-
9.
Add medium to 12-well plate using 1 ml per well. If planning to ablate epicardium, use the ablation medium.
CRITICAL STEP Medium must be warmed before use. Ablation medium must be freshly prepared just before dissecting.
-
10.
Transfer heart to culture dishes with 1 ml pipette tips.
-
11.
Place culture plate onto the horizontal shaker in a 28°C incubator and shake at 150 rpm (Fig 2d). Culture medium should be changed every other day. If ablation medium is used, replace ablation medium with normal culture medium after 24 h.
CRITICAL STEP Ensure the shaker is working properly throughout the culture period.
TROUBLESHOOTING
Box 1. Proliferation assay
Release of Mtz treatment TIMING 30 min
-
12.
Warm PBS and culture medium to 28°C in water bath. After 24 h culture in ablation medium with 1 mM Mtz, gently remove medium with a 1 ml tip. Ensure not to touch the hearts, which may injure the epicardium. Add 1 ml PBS to the well, gently shake a few times and then pipette out the PBS. Repeat washing with 1 ml PBS and then add 1 ml fresh medium without Mtz to each well. Return to the incubator and continue culturing.
CAUTION Follow institutional guidelines to dispose of medium with Mtz.
TROUBLESHOOTING
Box 2. Chamber grafts
Box 3. Bead grafts
Imaging TIMING up to 30 days
-
13.
Images can be acquired every day using a Leica M205FA stereofluorescence or other similar dissecting scope (Fig. 2e, f). We typically image two sides of a heart, the atrial side and the opposite side.
CRITICAL While taking images, keep the lids of culture plates closed.
TROUBLESHOOTING
TROUBLESHOOTING
| Step | Problem | Possible reason | Solution |
|---|---|---|---|
| 11-13, b5, c6 | Contamination with bacteria | Bench is not clean | Clean bench with 70% ethanol or perform procedures in a cell culture hood |
| Tools are not sterile | Sterilize tools with 70% ethanol, use sterile pipette tips or filter tips | ||
| Reagents are not sterile | Filter sterilize reagents | ||
| Plate lid is open during culture or imaging | Keep lid closed while culturing and imaging | ||
| Primocin is not added | Add Primocin for the first few days of culture | ||
| 11 | Heart explant is attached to dish bottom | Culture plate is not shaking properly | Make sure the shaker is always working at 150 rpm during culture |
| 13 | Absent or weak epicardial ablation effect | Mtz is not fully dissolved | Ensure Mtz is fully dissolved when preparing 10 mM solution |
| b5 | Chamber engraft fails | Chambers are not contacted closely | Use smaller hole to hold chambers tightly |
| c6 | Beads floating | Too much medium | Add less medium |
TIMING
Steps 1-3, Preparation: 1 h
Steps 4-5, Extracting hearts: 2-5 min per heart
Steps 6-8, Clean hearts: 5 min
Steps 9-11, Heart culture and epicardial ablation: 5 min
Step 12, Release Mtz treatment: 30 min
Step 13, Imaging: up to 30 days
Optional Steps a1-a3: Proliferation assay: 5-8 h
Optional Steps b1-b5: Chamber engraftment: 25-50 h
Optional Steps c1-c5: Beads embedding: about 26 h
ANTICIPATED RESULTS
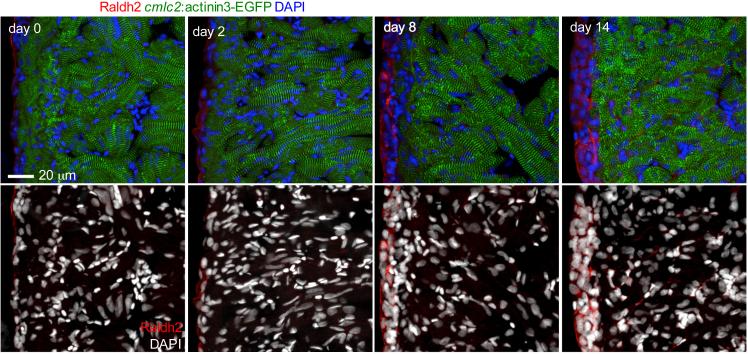
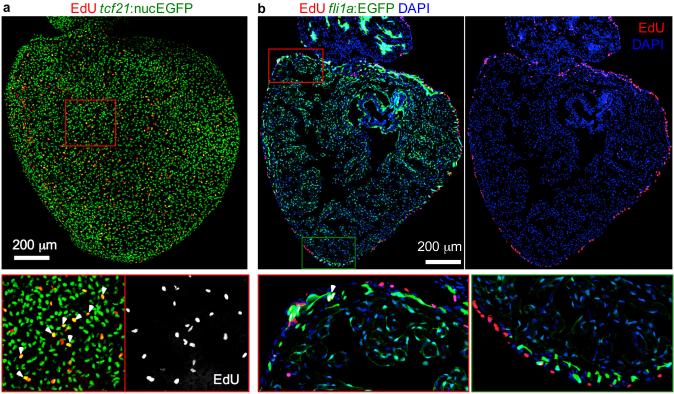
During culture, cardiac contraction will gradually slow, although explants can continue beating for more than one month (ref. 7, Supplementary video 1)7. Sarcomeric structure, visualized using a transgenic cmlc2:actinin3-EGFP line, is largely maintained during the first 2 weeks of culture, and muscle necrosis is rare (Fig. 4). The heart shape will also change gradually, shrink slightly and become rounded. Despite these minor changes, epicardial proliferation and migration are not affected. Epicardial cells will proliferate spontaneously without any treatment, with cell density increasing during the first 2 days, as assessed by staining with an antibody against Raldh2, and EdU incorporation and staining (Fig. 4; Fig. 5 and ref. 7)7. Proliferating endocardial cells were very rare, and were not detected in most hearts after 24h EdU incorporation (day 2-3) and assessment at day 3 by using a fli1a:EGFP reporter line (Fig. 5b). Some EGFP+/EdU+ endothelial cells were detected, but a nuclear marker of endothelial cells is required for further confirmation.
Figure 4.
Sarcomere morphology and expansion of epicardial cells during explant culture. Cultured hearts from cmlc2: actinin3-EGFP fish were fixed at the indicated time point. (Top panels) Heart sections were stained with an anti-Raldh2 antibody to denote epicardial and endocardial cells (red) and DAPI for DNA (blue). Sarcomeric structure (green) was largely maintained in the first 2 weeks, and the epicardial cell population expanded. Cardiac muscle compresses somewhat at day 14, which likely causes tissue shrinkage. (Bottom panels) DAPI (white) staining showed largely normal myocardial nuclei. Necrosis was rare within 14 days of culture.
Figure 5.
Spontaneous proliferation of epicardial cells during explant culture. (a) Spontaneous proliferation of epicardial cells after 48 h in culture. This heart explant from a tcf21:nucEGFP fish was treated with 25 μM EdU for 1 h at 47-48 h of culture. The framed region is enlarged to show details. Arrowheads indicate EGFP+/EdU+ nuclei. (b) Section of a 72 h heart explant from a fli1a:EGFP fish treated with 25 μM EdU for 24 h at 48-72 h of culture. Most EdU+ cells were in the ventricular wall. EGFP+/EdU+ endocardial cells were very rare. There are some EGFP+/EdU+ endothelial cells (arrowhead).
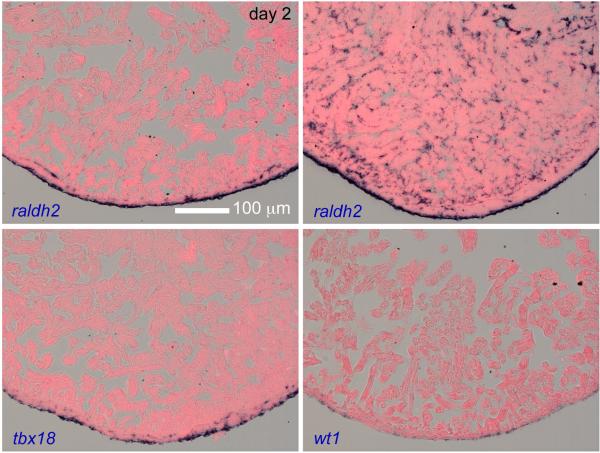
The cultured explants are suitable for in situ hybridization (Fig. 6), and we detected epicardial induction of raldh2, tbx18 and wt1 in uninjured hearts during explant culture. For most explants, raldh2 was only detected in epicardial cells, while some explants displayed endocardial expression.
Figure 6.
In situ hybridization on sections of ventricular explants after 2 days of culture. raldh2, tbx18 and wt1 were induced during explant culture. For most explants, raldh2 was only detected in the wall (top left), while some explants showed endocardial expression (top right), suggestive of endocardial activation by injury.
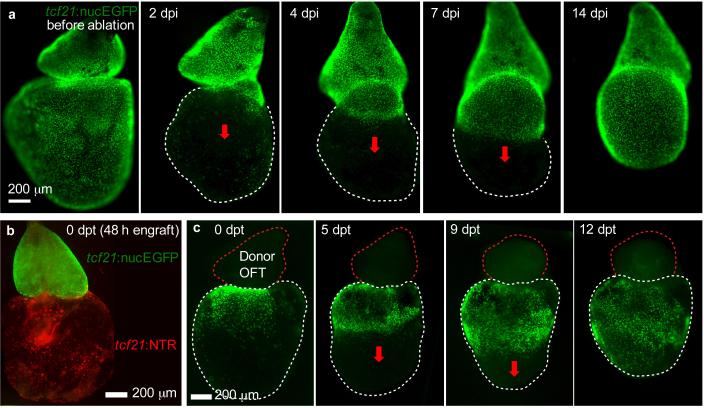
With Mtz treatment, epicardial cell death can be observed from day 2 (1 day post Mtz incubation, dpi) with debris protruding from the heart surface. Full ablation can be achieved at day 3 (2 dpi) with a significant portion of the ventral surface demonstrating absent tcf21:nucEGFP fluorescent. Epicardial regeneration is visualized by daily imaging; epicardial cells located at the ventricular base proliferate and migrate toward the ventricular apex (Fig. 7a). It takes approximately 2 weeks for epicardial cells to completely repopulate the ventricular surface (Fig. 7a and ref. 7, Fig. 2). In chamber engraftment experiments, about 50% of the explants will be tightly connected after 24 h contact in agarose (Fig. 7b, c). The ratio will be higher after 48 h contact in agarose. The ventricular epicardium loses regenerative capacity after removal of the outflow tract; however, a fresh donor outflow grafted to the ventricular base can rescue the defect (Fig. 7c and ref. 7, Fig. 3). In bead-grafting experiments, some ventricles lose the beads after release from agarose; these samples must be discarded. Ventricles also lose beads during 1-week culture and also must be discarded. At least 50% of ventricles successfully engraft and maintain beads during culture. Beads soaked with Hh protein can induce epicardial cell regeneration within the host ventricle after outflow tract removal (ref. 7, Fig. 4), implicating this factor in mechanisms of regeneration.
Figure 7.
Ex vivo epicardial regeneration and chamber engraftment. (a) Ventricular epicardium regenerates in a base-to-apex direction (arrows). Hearts from tcf21:NTR; tcf21:nucEGFP adults were incubated with 1 mM Mtz for 24 h. Whole mount images were taken daily. dpi, days post Mtz incubation. (b) A freshly collected outflow tract from a tcf21:nucEGFP fish (green) was grafted to the base of a ventricle from tcf21:NTR fish (red), by 48 h contact in agarose. The ventricle was treated with 1 mM Mtz for 24 h and cultured for 2 more days before engraftment. (c) A non-transgenic donor outflow tract (OFT) was transplanted to the base of an epicardially ablated ventricle. Base-to-apex epicardial regeneration (arrows) was observed from host tissue. dpt, days post-transplantation.
Figure 3.
Heart chamber and bead grafts. (a) A 35 mm Petri dish with a drop of 1% low-melting agarose in the center. A small hole is made at the top of the gel using forceps. (b) Washed Affi-Gel Blue beads settle to the bottom of EP tube. (c) A ventricle and an outflow tract from two hearts are placed together closely in the small hole at the top of the agarose gel. V, ventricle; OFT, outflow tract. (d) A ventricular explant inside the small hole is grafted with a bead at the base. (e) Cartoon of sectional view of the experimental samples in (c) and (d).
In summary, we describe in detail an explant culture system that maintains aspects of cardiac physiology and recapitulates in vivo observations of epicardial regeneration, while better representing such observations than cultured primary cells7. At the same time, cellular and molecular studies are convenient and efficient.
Supplementary Material
ACKNOWLEDGMENTS
We thank J. Wang and S. Singh for generating transgenic fish; J. Burris, N. Lee, A. Dunlap, S. Davies, T. Thoren, and N. Benkaci for fish care; A.L. Dickson for artwork; and A. Shoffner for comments on the manuscript. This work was funded by a postdoctoral fellowship from the American Heart Association to J.C. and grants from NIH (R01 HL081674 and R01 HL131319) to K.D.P.
Footnotes
Key reference:
Wang, J., Cao, J., Dickson, A.L. & Poss, K.D. Epicardial regeneration is guided by cardiac outflow tract and Hedgehog signalling. Nature 522, 226-230 (2015).
AUTHOR CONTRIBUTIONS
J.C. and K.D.P. designed the study and wrote the manuscript. J.C. performed all experiments.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
REFERENCES
- 1.Poss KD, Wilson LG, Keating MT. Heart regeneration in zebrafish. Science. 2002;298:2188–2190. doi: 10.1126/science.1077857. [DOI] [PubMed] [Google Scholar]
- 2.Jopling C, et al. Zebrafish heart regeneration occurs by cardiomyocyte dedifferentiation and proliferation. Nature. 2010;464:606–609. doi: 10.1038/nature08899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kikuchi K, et al. Primary contribution to zebrafish heart regeneration by gata4(+) cardiomyocytes. Nature. 2010;464:601–605. doi: 10.1038/nature08804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lepilina A, et al. A dynamic epicardial injury response supports progenitor cell activity during zebrafish heart regeneration. Cell. 2006;127:607–619. doi: 10.1016/j.cell.2006.08.052. [DOI] [PubMed] [Google Scholar]
- 5.Kikuchi K, et al. tcf21+ epicardial cells adopt non-myocardial fates during zebrafish heart development and regeneration. Development. 2011;138:2895–2902. doi: 10.1242/dev.067041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mahmoud AI, et al. Nerves Regulate Cardiomyocyte Proliferation and Heart Regeneration. Dev. Cell. 2015;34:387–399. doi: 10.1016/j.devcel.2015.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang J, Cao J, Dickson AL, Poss KD. Epicardial regeneration is guided by cardiac outflow tract and Hedgehog signalling. Nature. 2015;522:226–230. doi: 10.1038/nature14325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gemberling M, Karra R, Dickson AL, Poss KD. Nrg1 is an injury-induced cardiomyocyte mitogen for the endogenous heart regeneration program in zebrafish. Elife. 2015;4 doi: 10.7554/eLife.05871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang J, Karra R, Dickson AL, Poss KD. Fibronectin is deposited by injury-activated epicardial cells and is necessary for zebrafish heart regeneration. Dev. Biol. 2013;382:427–435. doi: 10.1016/j.ydbio.2013.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kikuchi K, et al. Retinoic acid production by endocardium and epicardium is an injury response essential for zebrafish heart regeneration. Dev. Cell. 2011;20:397–404. doi: 10.1016/j.devcel.2011.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou B, et al. Adult mouse epicardium modulates myocardial injury by secreting paracrine factors. J. Clin. Invest. 2011;121:1894–1904. doi: 10.1172/JCI45529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang GN, et al. C/EBP transcription factors mediate epicardial activation during heart development and injury. Science. 2012;338:1599–1603. doi: 10.1126/science.1229765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smart N, et al. De novo cardiomyocytes from within the activated adult heart after injury. Nature. 2011;474:640–644. doi: 10.1038/nature10188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smart N, et al. Thymosin beta4 induces adult epicardial progenitor mobilization and neovascularization. Nature. 2007;445:177–182. doi: 10.1038/nature05383. [DOI] [PubMed] [Google Scholar]
- 15.Ieda M, et al. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell. 2010;142:375–386. doi: 10.1016/j.cell.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qian L, et al. In vivo reprogramming of murine cardiac fibroblasts into induced cardiomyocytes. Nature. 2012;485:593–598. doi: 10.1038/nature11044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Song K, et al. Heart repair by reprogramming non-myocytes with cardiac transcription factors. Nature. 2012;485:599–604. doi: 10.1038/nature11139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim J, Rubin N, Huang Y, Tuan TL, Lien CL. In vitro culture of epicardial cells from adult zebrafish heart on a fibrin matrix. Nat. Protoc. 2012;7:247–255. doi: 10.1038/nprot.2011.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Habeler W, et al. An in vitro beating heart model for long-term assessment of experimental therapeutics. Cardiovasc. Res. 2009;81:253–259. doi: 10.1093/cvr/cvn299. [DOI] [PubMed] [Google Scholar]
- 20.Brandenburger M, et al. Organotypic slice culture from human adult ventricular myocardium. Cardiovasc. Res. 2012;93:50–59. doi: 10.1093/cvr/cvr259. [DOI] [PubMed] [Google Scholar]
- 21.Stuckmann I, Evans S, Lassar AB. Erythropoietin and retinoic acid, secreted from the epicardium, are required for cardiac myocyte proliferation. Dev. Biol. 2003;255:334–349. doi: 10.1016/s0012-1606(02)00078-7. [DOI] [PubMed] [Google Scholar]
- 22.Pillekamp F, et al. Force measurements of human embryonic stem cell-derived cardiomyocytes in an in vitro transplantation model. Stem Cells. 2007;25:174–180. doi: 10.1634/stemcells.2006-0094. [DOI] [PubMed] [Google Scholar]
- 23.Singh SP, Holdway JE, Poss KD. Regeneration of amputated zebrafish fin rays from de novo osteoblasts. Dev. Cell. 2012;22:879–886. doi: 10.1016/j.devcel.2012.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Curado S, et al. Conditional targeted cell ablation in zebrafish: a new tool for regeneration studies. Dev. Dyn. 2007;236:1025–1035. doi: 10.1002/dvdy.21100. [DOI] [PubMed] [Google Scholar]
- 25.Brito JM, Teillet MA, Le Douarin NM. An early role for sonic hedgehog from foregut endoderm in jaw development: ensuring neural crest cell survival. Proc. Natl. Acad. Sci. U S A. 2006;103:11607–11612. doi: 10.1073/pnas.0604751103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang J, et al. The regenerative capacity of zebrafish reverses cardiac failure caused by genetic cardiomyocyte depletion. Development. 2011;138:3421–3430. doi: 10.1242/dev.068601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin YF, Swinburne I, Yelon D. Multiple influences of blood flow on cardiomyocyte hypertrophy in the embryonic zebrafish heart. Dev. Biol. 2012;362:242–253. doi: 10.1016/j.ydbio.2011.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lawson ND, Weinstein BM. In vivo imaging of embryonic vascular development using transgenic zebrafish. Dev. Biol. 2002;248:307–318. doi: 10.1006/dbio.2002.0711. [DOI] [PubMed] [Google Scholar]
- 29.Gupta V, Poss KD. Clonally dominant cardiomyocytes direct heart morphogenesis. Nature. 2012;484:479–484. doi: 10.1038/nature11045. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.