Abstract
Begomovirus-associated epidemics currently threaten tomato production worldwide due to the emergence of highly pathogenic virus species and the proliferation of a whitefly B biotype vector that is adapted to tomato. To generate an efficient defence against begomovirus, we modulated the activity of the immune defence receptor nuclear shuttle protein (NSP)-interacting kinase (NIK) in tomato plants; NIK is a virulence target of the begomovirus NSP during infection. Mutation of T474 within the kinase activation loop promoted the constitutive activation of NIK-mediated defences, resulting in the down-regulation of translation-related genes and the suppression of global translation. Consistent with these findings, transgenic lines harbouring an activating mutation (T474D) were tolerant to the tomato-infecting begomoviruses ToYSV and ToSRV. This phenotype was associated with reduced loading of coat protein viral mRNA in actively translating polysomes, lower infection efficiency and reduced accumulation of viral DNA in systemic leaves. Our results also add some relevant insights into the mechanism underlying the NIK-mediated defence. We observed that the mock-inoculated T474D-overexpressing lines showed a constitutively infected wild-type transcriptome, indicating that the activation of the NIK-mediated signalling pathway triggers a typical response to begomovirus infection. In addition, the gain-of-function mutant T474D could sustain an activated NIK-mediated antiviral response in the absence of the virus, further confirming that phosphorylation of Thr-474 is the crucial event that leads to the activation of the kinase.
Keywords: NIK, NSP-interacting kinase, nuclear shuttle protein, tomato-infecting begomoviruses, begomovirus, leucine-rich repeats receptor-like kinase
Introduction
Begomoviruses (whitefly-transmitted geminiviruses) cause severe diseases of high economic impact in a variety of agriculturally relevant crops in tropical and subtropical areas (Rojas et al., 2005). Current climate changes are expected to further alter the whitefly distribution across the globe, posing a major threat to agriculture worldwide. The threat is particularly strongly for tomato plants, which are inflicted by a variety of emergent species of tomato-infecting begomoviruses. Previous attempts to develop pathogen-derived resistance using begomovirus DNA sequences in transgenic plants have failed to achieve immunity, resistance or tolerance, even though a delay of infection and/or attenuation of symptoms were frequently observed (Day et al., 1991; Hashmi et al., 2011; Hong and Stanley, 1996; Kunik et al., 1994; Lin et al., 2012; Noris et al., 1996; Stanley et al., 1991). The only exception was a single common bean transgenic line expressing a siRNA that targets the replication protein from BGMV (bean golden mosaic virus), which has been shown to be immune to this begomovirus (Aragão and Faria, 2009). However, it has been extremely difficult to engineer broad-spectrum resistance against tomato-infecting begomoviruses through a similar RNA silencing strategy (Lucioli et al., 2008). One explanation for the failure of siRNA tomato transgenic lines to resist begomovirus infection is the emergence of new species of tomato-infecting begomoviruses that evolve rapidly through recombination or pseudo-recombination, which produces divergent genome sequences, giving the virus an advantage over its host’s recognition system (Albuquerque et al., 2012; Castillo-Urquiza et al., 2008; Galvão et al., 2003). More recently, transgenic tomato lines expressing peptide aptamers, which bind efficiently to and inhibit the begomovirus replication protein (Rep), have been shown to display enhanced tolerance to Tomato yellow leaf curl virus or Tomato mottle virus (Reyes et al., 2013). Thus, the Rep-binding peptide octamers may serve as an efficient strategy for engineering transgenic tomato plants that are resistant to diverse begomoviruses. Likewise, expression of the single-stranded DNA binding protein virE2 from Agrobacterium in tobacco has been shown to reduce Mungbean yellow mosaic virus DNA accumulation, although the spectrum of the resistance has not been established (Sunitha et al., 2011).
Begomoviruses are single-stranded DNA viruses with a monopartite or bipartite genome configuration. For the bipartite begomoviruses, the proteins required for replication (Rep and REn), transactivation of viral genes (TrAP), the suppression of RNAi defence functions (TrAP and AC4) and encapsidation of viral DNA (CP) are encoded by the DNA-A component, whereas the nuclear shuttle protein (NSP) and intercellular movement protein (MP) are encoded by DNA-B (Rojas et al., 2005). NSP facilitates the traffic of viral DNA from the nucleus to the cytoplasm and acts in concert with MP to move the viral DNA to the adjacent, uninfected cells. The mechanistic model for viral DNA intracellular trafficking holds that NSP binds to newly replicated viral DNA in the nuclei of infected cells and utilizes the nuclear export machinery to move the viral DNA to the cytoplasm (Carvalho et al., 2008a,b; Gafni and Epel, 2002). Consistent with this model, NSP contains a HIVRev-like or TFIIA-like leucine-rich nuclear export signal (NES) that can be functionally replaced by TFIIIA NES in both nuclear export and infectivity (Ward and Lazarowitz, 1999). NSP is found within the nuclei of transfected plant, insect and yeast cells, but is relocated to the cell periphery when co-expressed with viral movement protein (MP, Carvalho et al., 2008b; Sanderfoot and Lazarowitz, 1995, 1996; Sanderfoot et al., 1996; Zhang et al.,2001). The fundamental role of NSP in virus movement predicts that this viral protein may interact with host factors in different subcellular compartments. Accordingly, NSP has been shown to interact with an Arabidopsis thaliana nuclear acetylase, designated nuclear shuttle protein interactor (AtNSI) and a cytosolic GTPase that facilitates the release of the viral DNA-NSP complex from the nuclear pores to the cytosol, designated NIG (NSP-interacting GTPase). NSP also interacts with plasma membrane receptor-like kinases, designated NsAKs (NSP-activating kinases) from Arabidopsis and NIKs (NSP-interacting kinases) from tomato, soya bean and Arabidopsis (Carvalho et al., 2008b; Florentino et al., 2006; Mariano et al., 2004; McGarry et al., 2003). In Arabidopsis, NSP interacts with three members of the LRR-receptor-like kinase (RLK) family, NIK1, NIK2 and NIK3, which have been shown to be authentic serine/threonine kinases with biochemical properties consistent with a receptor signalling function (Fontes et al., 2004).
NIK was recently discovered as a component of the antiviral plant immune system (Carvalho et al., 2008c; Fontes et al., 2004). The viral NSP binds to the kinase domain of NIK to suppress its activity and increase begomovirus pathogenicity (Fontes et al., 2004). The current model for NIK activation holds that upon begomovirus infection, NIK oligomerizes and transphosphorylates the kinase domain on a key threonine residue at position 474 (T474; Carvalho et al., 2008c; Rocha et al., 2008; Santos et al., 2009). This phosphorylation-dependent activation of NIK leads to the phosphorylation of a downstream component, the ribosomal protein L10A (RPL10), which in turn translocates to the nucleus, where it interacts with LIMYB to fully down-regulate translation machinery-related genes, leading to host translation suppression that affects the translation of begomovirus mRNAs (Carvalho et al., 2008c; Zorzatto et al., 2015). To counteract this mechanism, the viral NSP binds to the kinase domain of NIK and prevents phosphorylation of T474, leading to the suppression of the kinase activity and establishing an environment that is more favourable to begomovirus infection. Accordingly, the loss of NIK function enhances the susceptibility of NIK null alleles to begomovirus infection, whereas the overexpression of Arabidopsis (At) NIK1 in begomovirus-infected tobacco leaves titrates the virally produced NSP inhibitor and the molar excess of NIK overcomes NSP-mediated inhibition (Santos et al., 2010). Likewise, enhanced accumulation of AtNIK1 in tomato plants attenuates begomovirus infection. However, the effectiveness of the NIK-mediated signalling pathway against begomovirus infection is limited because the viral NSP functions as a NIK suppressor and because activation of the pathway seems to be dependent on the onset of infection.
To increase the effectiveness of the NIK-mediated pathway against virus infection, we prepared a gain-of-function AtNIK1 mutant (T474D) by replacing a threonine residue at position 474 with an aspartic acid residue (a phosphomimic). We have previously shown that this mutation leads to hyperactivation of the kinase activity, with a 1.5-fold increase in substrate phosphorylation activity and an enhanced capacity to relocate RPL10 to the nucleus (Santos et al., 2009). In this study, we used the gain-of-function mutant from Arabidopsis to generate an antiviral strategy to fight tomato-infecting begomoviruses and to gain insights into the activation of the NIK-mediated defence response. We showed that the antiviral signalling activity of a constitutively activated NIK receptor from the cruciferous plant Arabidopsis thaliana is retained after its transfer to the solanaceous plant tomato (Solanum lycopersicum), making the tomato transgenic lines more resistant to different species of begomovirus.
Results
NSP-NIK complex formation is barely detected in vivo by BiFC
Proteins that shuttle between the nucleus and the cytoplasm are concentrated within the nucleus because the rate of nuclear import exceeds that of export. In fact, NSPs from several begomoviruses ectopically expressed in transfected cells are nuclear localized (Carvalho et al., 2008b; Sanderfoot and Lazarowitz, 1995; Sanderfoot et al., 1996). Likewise, the ectopically expressed NSPs from CaLCuV, ToYSV and ToSRV colocalized with the nuclear marker AtWWP1 in the nuclei of N. benthamiana epidermal cells (Figure S1). This preferential nuclear localization of NSP when transiently expressed in leaf cells compromises the use of the BiFC (bimolecular fluorescence complementation) assay and co-immunoprecipitation of ectopically expressed proteins in efficiently detecting interactions between NSP and membrane proteins in planta. Accordingly, our attempts to examine interactions between NSP from CaLCuV and NIK in vivo by BiFC resulted in a very low frequency of transfected cells displaying any reconstituted fluorescence on the plasma membrane (Figure 1a), although it was greater than the background levels (right panels). In addition, the formation of an NSP-NIK1 complex occurred in vivo independently of the orientation of the NSP or NIK1 fusions (N-terminus or C-terminus of YFP; Figure 1a). These results confirmed that NSP interacts with NIK in vivo, but they also demonstrated that the detection of NIK-NSP complex formation in planta clearly depends on the saturation of the nuclear import machinery by high expression of NSP, which is limited when driven by the BiFC vectors. Therefore, the efficiency of detection of NSP-NIK interactions in planta by transient expression of NSP is extremely dependent on the opportunistic low concentration of NSP in the cell periphery, which very often is below the level of detection.
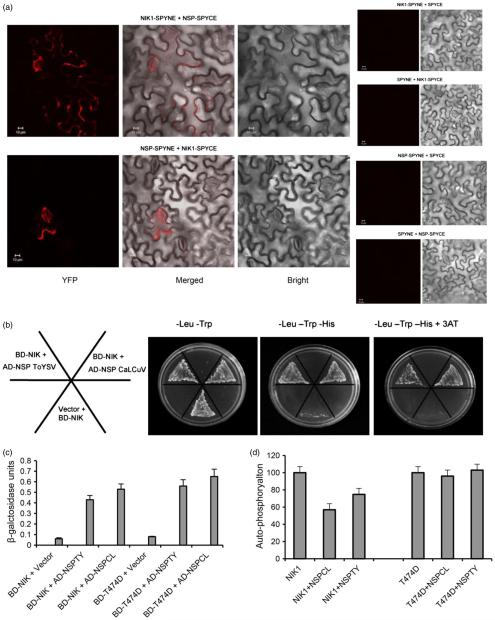
Figure 1.
T474D, a hyperactive AtNIK1 mutant, bypasses NSP inhibition and yet binds to ToYSV NSP. (a) In vivo interaction between NSP from CaLCuV and AtNIK1 by BiFC analysis. Fluorescence (YFP), bright and merged confocal images were taken of epidermal cells of tobacco leaves co-expressing NIK1-YFPC (NIK1-SPYCE) +NSP-YFPN (NSP-SPYNE) or NIK1YFPN (NIK1-SPYNE) + NSP-YFPC (NSP-SPYCE) fusion proteins in the presence of HC-Pro suppressor, 2 h after agro-infiltration with the indicated DNA constructs. Negative controls are shown on the right. Scale bars = 10 μm. (b) and (c) Interactions of ToYSV NSP with AtNIK1 and T474D. NSPs from CaLCuV (NSPCL, control) and ToYSV (NSPTY) were expressed in yeast as GAL4 activation domain (AD) fusions (AD-NSP ToYSV and AD-NSP CaLCuV), and the kinase domains of AtNIK1 (NIK) and T474D were expressed as GAL4 binding domain (BD) fusions (BD-NIK). Interactions between the tested proteins were examined by monitoring His prototrophy (b) and confirmed by measuring the activity (mean ± SD, n = 3) of the β-galactosidase reporter enzyme corresponding to the second reporter gene β-Gal (c). Error bars represent the confidence interval (α = 0.05) of three technical replicates. (d) ToYSV NSP inhibits AtNIK1 autophosphorylation, but does not suppress T474D kinase activity. The kinase domain of AtNIK1 (NIK1) or T474D was expressed as a GST fusion and incubated with [γ-32P]ATP in the presence or absence of GST-NSP. Phosphorylated proteins were quantified by counting the scintillation of the excised protein bands. Autophosphorylation activity in the presence of NSP was expressed as the percentage of the total activity of NIK1 or T474D alone. Error bars represent the confidence interval (α = 0.05) of three technical replicates from two independent experiments.
The hyperactive AtNIK1 mutant T474D bypasses NSP inhibition from tomato-infecting begomoviruses
Because of the low detection level of NSP interactions with plasma membrane proteins in transient expression assays, we assayed whether NSP from a tomato-infecting begomovirus interacts with the kinase domain of NIK from Arabidopsis through a yeast two-hybrid assay, which has been shown to efficiently detect interactions between host and begomovirus proteins (Carvalho et al., 2008b; Florentino et al., 2006; Fontes et al., 2004; Mariano et al., 2004). We showed that, similar to the NSP from the Arabidopsis-infecting begomovirus CaLCuV (Cabbage leaf curly virus), which interacts with the NIK immune receptors in yeast and in planta (Fontes et al., 2004; Figure 1a), NSP from the tomato-infecting begomovirus ToYSV (Tomato yellow spot virus) interacted stably with the kinase domain of NIK from Arabidopsis in yeast (Figure 1b). Yeast transformed with the empty vector (pAD-GAL4) and BD-NIK was used as a negative control, and yeast co-transformed with BD-NIK and AD-NSPCaLCuV was used as a positive control (Fontes et al., 2004). These interactions were further confirmed by monitoring β-galactosidase activity in yeast protein extracts (Figure 1c). Replacing T474 with aspartate did not prevent NSP binding (Figure 1b) but impaired the NSP-mediated inactivation of kinase activity (Figure 1d). These results suggest that the hyperactive NIK T474D mutant is a more effective target for engineering resistance against begomovirus.
Activation of the NIK-mediated signalling pathway triggers a typical response to begomovirus infection
We prepared tomato plants expressing different amounts of T474D (AtNIK1 mutant), which exhibit enhanced accumulation of the protein (Figure S2). In the first generation, some transgenic lines displayed shorter roots and stunted growth compared to wild type, but this phenotype did not persist over the subsequent generations. In fact, the development, overall physiological performance and horticultural traits of T474D-2-, T474D-5- and T474D-6-overexpressing lines were similar to those of wild-type plants and AtNIK1-expressing transgenic lines (NIK1-4; 15) for more than three generations (from R1 to R3) under normal greenhouse conditions. During the vegetative phase, the transgenic lines and wild-type control displayed similar plant height and biomass accumulation, as measured by shoot and root dry weight (Figures S3a–d). The net CO2 assimilation rate (A), transpiration rate (E), stomatal conductance to water vapour (gs) ration and internal to ambient CO2 concentration ratio (Ci/Ca) of fully expanded leaves did not differ significantly among the transgenic lines and wild-type control (Figure S3e–h). Fully ripened tomatoes were harvested and analysed for colour, morphology, total soluble solids (brix) and nutritional quality (Figure S4). The ripe fruits of the transgenic lines were bright red (Figure S4a) and displayed an overall shape (Figure S4a), colour (Figure S4d), size (Figure S4e) and nutritional value (Figure S4c, f, g) similar to those of the wild-type control. The fruit yield (Figure S4b, i) and the overall development of the reproductive phase (Figure S4h, i) of the transgenic lines were similar to those of wild-type plants grown under normal greenhouse conditions.
To examine whether the virus infection alone triggered NIK-mediated defence signalling, the transgenic lines T474D-6 and NIK1-4 were challenged with infectious clones of the begomovirus ToYSV. Using RNA deep sequencing, we compared the virus-induced and NIK1 or T474D mutant transcriptomes. The hierarchical clustering via the multiscale bootstrap resampling method was employed to obtain clusters from normalized treatments data sets (Figure 2a). These global comparisons of the expressed sequences among the mock-treated and infected wild type (WT), NIK and T474D lines showed that the transcriptomes of the infected wild-type and mock-inoculated T474D lines were more closely related, as these samples clustered together. These transcriptomes differed greatly from the NIK mock-inoculated transcriptome, confirming that the gain-of-function T474D mutant may be activated in a constitutive manner that allows it to support a sustained NIK-mediated response. In addition, a sustained NIK-mediated response may lead to a ‘priming’ state that further enhances the response to begomovirus infection in T474D- and NIK-overexpressing leaves; the virus-induced transcriptomes of these samples cluster together and differ from the T474D mock-inoculated transcriptome (Figure 2a). The values of the approximately unbiased P-value (au) and the bootstrap probability (bp) were significant using a threshold of 0.05. Therefore, this analysis clearly uncovered a strong similarity between the general gene expression profile of the mock T474D and infected WT plants, suggesting that activation of the NIK-mediated signalling pathway in tomato triggers a typical response to begomovirus infection similar to that in Arabidopsis (Zorzatto et al., 2015). Consistent with this finding, upon trimming the mock T474D–mock WT differentially expressed (DE) genes off of all treatments, the effect of viral infection seemed to be titrated off, and each T474D mock-, NIK mock- and WT mock-inoculated treatment grouped together with the infected counterpart (Figure 2b). Taken together, these results suggest that the gain-of-function mutant T474D can sustain an activated NIK-mediated antiviral response in the absence of the virus and that the activation of the NIK-mediated signalling pathway triggers a typical response to begomovirus infection. However, the T474D-induced infection response represents only a subset of the transgenic response because a residual effect of the transgene promoted the separation of the genotypes in the cluster analysis.
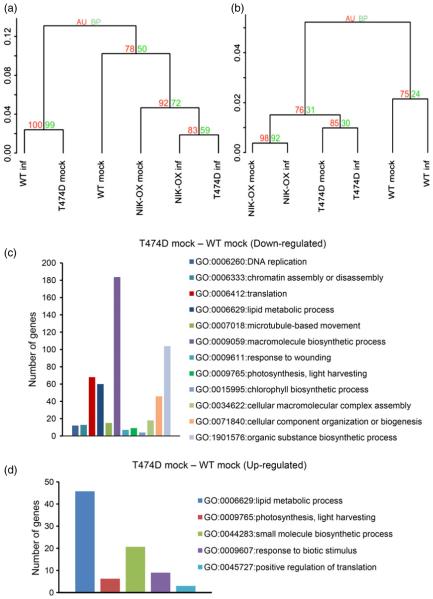
Figure 2.
Transcriptomic analyses of mock-inoculated and infected (inf) T474D, NIK-OX and wild-type lines (a) Mock-inoculated T474D-overexpressing lines display a constitutive infected wild-type (WT) transcriptome. Clustering analysis was performed using the R package pvclust (hierarchical clustering with P-values via multiscale bootstrap resampling) using the TMM normalization method and the Ward agglomerative clustering method. The dendrogram provides two types of P-values: AU (approximately unbiased – red, from multiscale bootstrap resampling) and BP (bootstrap probability – green, normal bootstrap resampling). The AU P-value comes from multiscale bootstrap resampling, and the BP value represents normal bootstrap resampling. These P-values were calculated by Multiscale Bootstrap Resampling using the R-cran package pvclust with a cut-off of 0.05. These P-values show the significance of the proximity of each GE experiment profile. NIK-OX indicates NIK-overexpressing lines. (b) Elimination of the mock T474D–mock WT DE genes from the raw data minimizes the gene expression variation as a result of virus infection. The hierarchical clustering of the gene expression (GE) data was performed as described in Figure 2, except that the DE genes of mock T474D–mock WT were removed from the raw data from all treatments prior to the clustering analysis. The red numbers indicate unbiased P-values (au), and the green ones indicate bootstrap probabilities. (c) and (d) Bar graphs of the enriched down- (c) and up-regulated (d) categories based on biological process from the gene ontology (GO) database. Using at least four DGE methods, the enriched GO categories were determined by at least three DGE methods and included more than three genes (Table S1).
Constitutive activation of AtNIK (T474D) in tomato plants causes a general down-regulation of translation machinery-related genes and impairs translation
To determine the global gene expression variation of the infected WT and the mock-inoculated overexpressing NIK and T474D tomato plants, we analysed the following combinations: infected WT–mock WT, mock NIK-OX–mock WT and mock T474D–mock WT, using the differential gene expression (DGE) methods edgeR/TMM, edgeR/TC, DESeq and baySeq. The differentially expressed (DE) genes were stored using SQL tables at the PostgreSQL relational database (http://inctipp.bioagro.ufv.br/tomatodb/), which listed the corresponding log2FC (fold change) and P-value corrected by FDR (q-value) for all DE genes. We performed a gene enrichment analysis using the GSEA methods based on biological process from the GO data. Many enriched categories were found using P-value cut-off of <0.05 (http://tomatodb.inctipp.ufv.br). Because of the poor annotation of the tomato genome regarding some GO categories, we changed the P-value cut-off to <0.01 and only accepted the GO categories that had been labelled by at least three DGE methods (Table S1). A strong bias arose from some poorly annotated GO categories in the enrichment analysis, which led to the recognition of some significant groups in spite of their very low number of genes (P-value <0.01). Thus, we only considered the groups with more than three genes (Table S1). The enriched categories for the contrast mock T474D–mock WT, which included the highest number of enriched GO categories, are shown in Figure 2. Among all the analysed combinations, the enriched category GO:0006412 (translation) from the mock T474D–mock WT combination displayed the most significant P-value (average P-value 4.69e-07; Table S1). This category is represented by components of the translational machinery, including ribosomal genes and other components of protein synthesis, such as translational initiation and elongation factors and molecular chaperones. To determine whether down-regulation of the translation machinery-related genes directly resulted from T474D expression, we plotted all the annotated genes attributed to GO:0006412 (translation) in a smear-plot as red dots (Figure S5). In the T474D mock–WT mock combination, the translation-related genes (red dots) tended to be down-regulated, because their log2FC values were clearly concentrated below zero. In contrast, this trend was not shared by the down-regulated GO:0006629: lipid metabolic process-enriched category in the mock T474D–mock WT combination; these genes were highly dispersed without any tendency for up- or down-regulated profiles (Figure S5, see red dots). These results indicate that merging the DGE data as the average of at least three DGE methods may have underestimated the number of translation-related DE genes in the T474D mock–WT mock combination. Constitutive activation of AtNIK1 in Arabidopsis has also been shown to promote the down-regulation of translation-related genes, which is reflected by the suppression of global translation (Zorzatto et al., 2015).
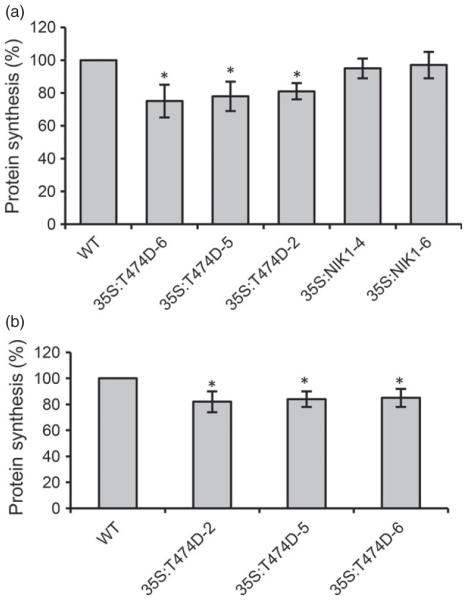
To confirm that protein synthesis was impaired by the constitutive activation of AtNIK in the T474D tomato lines, we labelled leaf proteins in vivo with [35S]Met in control plants and T474D overexpression lines (Figure 3a). There was a significant decrease (25% in T474D-5, 22.5% in T474D-6 and 19.6% in T474D-2; P < 0.05) in the amount of newly synthesized protein in T474D-overexpressing leaves compared with the amounts found in wild-type and NIK-overexpressing leaves. We observed a slight variation in the T474D-mediated inhibition of translation during development, as the reduction of translation was 18.1% (T474D-2), 16.8% (T474D-5) and 15.5% (T474D-6) when the incorporation of [35S]Met into total proteins was measured in 28-day-old leaves (Figure 3b). Our data indicated that the gain-of-function mutant T474D from Arabidopsis functions similarly in tomato plants as it causes a general down-regulation of translation machinery-related genes and affects translation in transgenic tomato lines. This suppression of translation might underlie at least part of the molecular mechanism involved in NIK-mediated antiviral defences.
Figure 3.
Ectopic expression of the T474 mutant receptor down-regulates global protein synthesis in leaves of 10 days (a) old and 28 days old (b) tomato plants. Equal fresh weights of tomato leaves (300 mg) were incubated with 50 μg/mL chloramphenicol and 20 μCi of [35S]methionine for 3 h at room temperature. Incorporation of [35S]Met into protein was measured in the TCA-precipitated total protein (mean ± SD, n = 3, P < 0.05) from wild-type and T474D transgenic lines. Asterisks indicate significant differences from the wild-type control (P < 0.05, Student’s t-test).
Constitutive activation of NIK confers broad-spectrum tolerance to tomato-infecting begomoviruses
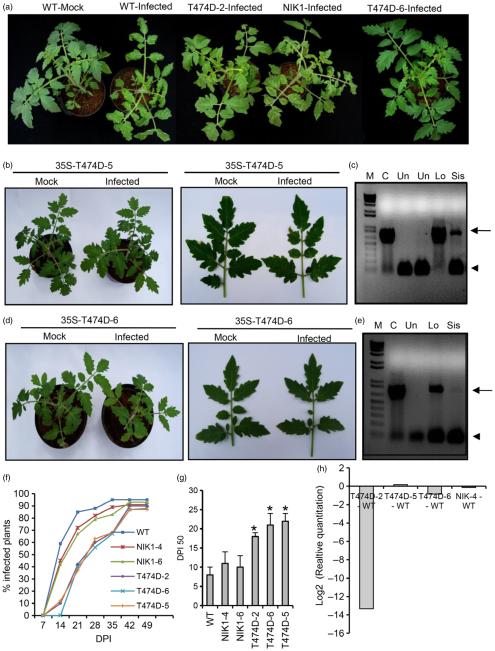
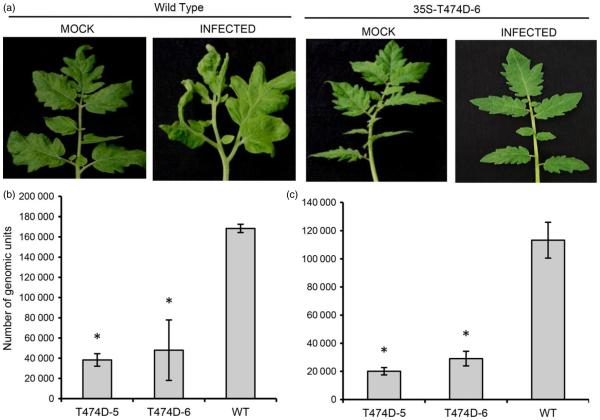
We next examined whether the constitutive activation of NIK was effective at controlling begomovirus infection. To this end, four independent T474D-overexpressing transgenic lines (T474D-9, T474D-6, T474D-5 and T47D-2), a wild-type (untransformed) line and the NIK-overexpressing lines NIK1-4 and NIK1-6 (Carvalho et al., 2008c) were inoculated with tandemly repeated ToYSV DNA-A and DNA-B (Andrade et al., 2006) using biolistic delivery, and the plants were assayed for symptoms of infection and the accumulation of viral DNA, as detected by PCR and qPCR. The wild-type plants displayed typical symptoms of ToYSV infection, such as leaf curling and yellow spots all over the leaves (>10 spots/cm2; Figure 4a). Consistent with a previous observation (Carvalho et al., 2008c), the NIK-overexpressing line NIK1-4 displayed attenuated symptoms (less accentuated leaf distortion and a lower number of yellow spots per leaf area, <6 spots/cm2). The symptoms in the T474D-overexpressing lines, however, were even more attenuated, with few spots per leaf area (varying among the lines) and no visible leaf curling (see T474D-2 and T474D-5 lines, Figure 4a, b). The T474D-6 transgenic line displayed typical tolerance to begomoviruses, as it did not develop symptoms (Figure 4a, d), but we detected viral DNA accumulation in both inoculated and systemically infected leaves (Figure 4e). The symptomless ToYSV infections of the T474D-6 line were associated with a delayed course of infection (Figure 4f), a lower rate of infection (DPI50, days postinoculation to infect 50% of plants; Figure 4g), and a lower accumulation of viral DNA in the systemically infected leaves, as shown by qPCR (Figure 4h).
Figure 4.
Ectopic expression of T474D in tomato confers tolerance to ToYSV infection. (a) Ectopic expression of T474D in tomato plants attenuates the development of symptoms upon ToYSV infection. Tandemly repeated ToYSV DNA-A and DNA-B sequences were introduced into the indicated lines by biolistic inoculation. Photographs were taken at 21 days postinoculation (DPI). (b) Symptoms associated with ToYSV infection in the 35S-T474D-5 line. Photographs were taken at 21 DPI. (c) Viral DNA accumulation in the infected leaves of the T474-5 line. Total DNA was isolated from infected plants at 21 DPI, and PCR was performed with viral DNA-B-specific primers and actin-specific primers (as an internal control) in the same reaction. Lo indicates inoculated and Sys denotes systemic leaves. ‘Un’ indicates mock-inoculated leaves. The gel shows representative samples of WT and the 35S::T474D-5 transgenic line. The upper band (1.2 kb, arrow) is the amplified genomic fragment from actin, and the lower band (0.5 kb, arrowhead) is the viral DNA fragment. (d) and (e) The line 35S-T474D-6 displayed tolerance to ToYSV infection. The T474D-6 line was infected with ToYSV, and photographs were taken at 21 DPI. Viral DNA accumulation was detected by PCR in inoculated (‘Lo’) and systemic (‘Sys’) leaves. (f) The course of infection was delayed in T474D lines. The indicated lines were infected with ToYSV by the biolistic method, and the course of infection was monitored by PCR amplification of viral DNA. In each experiment (three biological replicates), 20 plants of each line were inoculated. Values represent the percentage of systemically infected plants at different DPI. (g) Infection efficiency in T474D-overexpressing lines. The infection efficiency is expressed as the DPI required to infect 50% of the plants (mean ± SD of three replicates). Asterisks indicate significantly different means (P < 0.05, Student’s t-test). (h) Viral DNA accumulation in T474D-overexpressing lines, as determined by quantitative PCR at 28 DPI. The fold variation (±SD, n = 3 biological replicates) is shown as log2-scaled copy units of the viral genome. Viral DNA accumulation was determined in systemic leaves at 28 DPI.
Likewise, in the T474D-2 and T474D-5 lines, the progress and rate of infection were delayed compared with those of the wild-type control lines and the NIK1-overexpressing lines (Figure 4f, g). In the case of ToYSV, which showed high levels of accumulation in all samples analysed, both the T474D-2 and T474D-6 lines displayed lower viral DNA accumulation levels in the systemically infected leaves, although the high dispersion of the data among the samples prevented us from ascertaining the statistical significance of these findings (Figure 4h). Collectively, these results indicate that the T474D-overexpressing lines are more tolerant to ToYSV infection as compared to AtNIK1-overexpressing lines and wild-type lines.
Tomatoes are usually infected by a variety of rapidly evolving species of tomato-infecting begomoviruses and engineering broad-spectrum resistance to these viruses in crops constitutes a relevant trait to achieve durable resistance against begomoviruses. These transgenic lines were also challenged with the tomato-infecting begomovirus ToSRV (Tomato severe rugose virus), which displays a highly divergent genomic sequence from ToYSV sequence (Albuquerque et al., 2012). ToSRV infection caused severe leaf distortion in wild-type leaves but not in the T474D-6 overexpressing line (Figure 5a). In these T474D-overexpressing lines, the viral DNA accumulation in the systemic leaves from ToSRV infections was significantly lower at 14 and 21 DPI (P < 0.05; Figure 5b, c). These results indicate that the ectopic expression of T474D in tomato confers tolerance to heterologous species of tomato-infecting begomoviruses, which are phylogenetically separated within the two major groups of begomoviruses found in Brazil (Albuquerque et al., 2012). The performance of the T474D-overexpressing lines upon begomovirus infection further confirmed that the T474D mutant protein could mount a sustained NIK-mediated antiviral defence in the absence of viral infection. The constitutive activation of T474D and its ability to bypass viral NSP inhibition likely account for the tolerance to begomovirus infection displayed by the transgenic lines.
Figure 5.
The T474D-6 overexpressing line is also tolerant to ToSRV infection. (a) Symptoms associated with ToSRV infection in the 35S-T474D-6 line. The T474D-6 line was infected with ToSRV, and photographs were taken at 21 DPI. (b) and (c) Viral DNA accumulation in the T474D-6- and T474-5-overexpressing lines at 14 DPI (b) and 28 DPI (c). Prior to performing real-time PCR, infected leaves were diagnosed by standard PCR. Subsequently, total DNA extracted from systemically infected leaves at 14 DPI (b) or 28 DPI (c) was used as a template for quantitative PCR using ToYSV DNA-A-specific primers. The fold variation (±SD, n = 3 biological replicates) is shown as copy units of the viral genome. The asterisks indicate significant differences with P < 0.05 according to a Student’s t-test.
Begomovirus infection does not affect resistance-related proteins and defence-related genes in the T474D-overexpressinglines
NIK belongs to the LRRII-RLK subfamily and shares high sequence conservation and a similar receptor configuration with the well-characterized PAMP recognition co-receptor BRI1-associated receptor kinase 1 (BAK1; Sakamoto et al., 2012; Shiu and Bleecker, 2001). To understand whether the pre-activation of the NIK-mediated antiviral response in the T474D-expressing lines would prime typical BAK1-mediated defence responses, such as PTI or SA signalling marker genes, in response to geminivirus infection, we compared the transcriptome changes in infected WT–mock WT vs infected T474D–mock T474D. We detected eight resistance protein-related genes (R genes) that were up-regulated by ToYSV infection in the T474D lines, but not in the WT lines (Table S2). However, significant gene enrichment was not detected for the R gene category, gene silencing category, or abiotic stress response gene category, which includes the immune system category and the response to SA marker genes, using the gene set enrichment analysis (GSEA) method (Table S3). We also compared the ToYSV infection-induced transcriptome of T474D lines with that recently described for the tolerant TYLCV-resistant tomato breeding line CLN2777A (Chen et al., 2013). None of the 40 defence-related genes up-regulated in CLN2777A in response to TYLCV infection were T474D up-regulated genes, although we found some similarities when functional terms were used as the basis for comparison (Table S4). Collectively, these results indicate that typical viral defences, such as activation of SA signalling, gene silencing or the immune system, may not account for the resistant mechanism of the T474D lines. These lines may invoke an alternative mechanism for T474D protection against begomovirus infection.
The enhanced tolerance to begomovirus displayed by the T474D-expressing lines may be associated with the translational control branch of the NIK-mediated antiviral responses
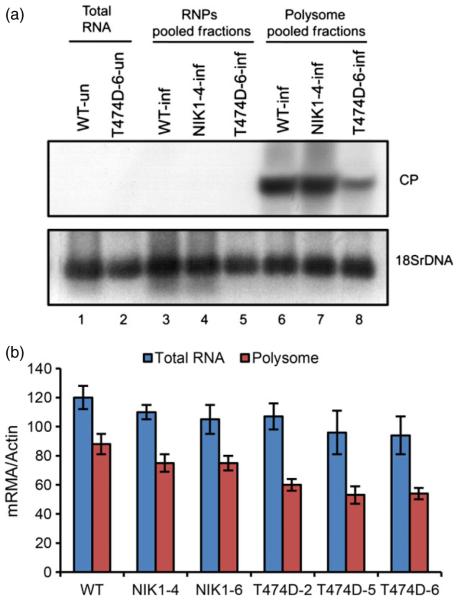
The mock T474D–mock WT combination identified genes affected by T474D expression in the absence of virus infection. These data clearly showed that expression of the T474D mutant in tomato led to a significant down-regulation of translation-related genes and impaired translation, a phenotype associated with the translational control branch of the NIK-mediated antiviral signalling pathway, which has recently been described in Arabidopsis (Zorzatto et al., 2015). Because T474D expression caused a global down-regulation of protein synthesis, we determined whether viral RNA translation was impaired in the T474D-expressing lines. We investigated whether we could detect viral RNA transcripts in actively translating polysome fractions that had been separated from nonpolysomal fractions on sucrose gradients (Figure S6). The polysomal fractions were prepared from infected leaves at 10 DPI, when the accumulation of viral mRNA in transgenic and wild-type leaves was similar (Figure 6b). We observed a significant reduction in the polysome loading of viral mRNA (coat protein mRNA) in systemically infected leaves of the T474D-6-overexpresing line compared to infected wild-type and NIK1-overexpressing leaves (Figure 6a, b). The accumulation of total virus transcripts in all infected lines was confirmed in our RNA sequencing data. These results indicated that the begomovirus was not capable of sustaining high levels of viral mRNA translation in the T474D-6-expressing lines, indicating that suppression of global protein synthesis may effectively protect plant cells against DNA viruses.
Figure 6.
Polysome loading of viral mRNA is reduced in systemically infected leaves in the T47D-6-overexpressing lines. (a) Polysome loading of coat protein (CP) mRNA from ToYSV DNA-A in systemically infected leaves of WT, NIK-overexpressing and T47D-6-overexpressing lines. Polysomesfrom infected WT, NIK1-4-overexpressing and T474D-6-overexpressing lines were isolated from systemic leaves at 10 days postinoculation with tandem copies of DNA-A and DNA-B of ToYSV, as shown in Fig. S6. RNPs refer to a 40S-enriched fraction. Polysome-bound RNA from pooled fractions was extracted with phenol/chloroform/isoamyl alcohol, precipitated with isopropanol, blotted and probed with the coat protein DNA (CP) and 18S rDNA. The identity of the polysome-pooled fraction was confirmed by treatment with 25 mM EDTA prior to the sucrose gradient (data not shown), which releases the mRNA from the polysomes. (b) Quantitation of polysome-associated coat protein viral transcripts in T474D-overexpressing lines by qRT-PCR. Polysomes from infected WT, NIK1-4-overexpressing and T474D-6-overexpressing lines were isolated 10 days postinoculation with infectious ToYSV clones. Polysome-bound RNA from pooled fractions was extracted with phenol/chloroform/isoamyl alcohol, precipitated with isopropanol and quantified by qRT-PCR. Values were normalized to the expression of actin. Error bars represent SD from three measurements.
Discussion
Begomoviruses are one of the largest and most successful groups of plant viruses and cause severe diseases in major crops worldwide, inflicting significant economic losses in many dicotyledonous crops. The tomato-infecting begomoviruses have become an even greater threat to tomato cultivation due to the emergence of new species along with the recent introduction into South America of a new biotype of the whitefly vector Bemisia tabaci, which colonizes tomato plants with high efficiency (Albuquerque et al., 2012; Castillo-Urquiza et al., 2008; Galvão et al., 2003). Current climate changes are expected to further alter the whitefly distribution across the globe, posing a serious threat to agriculture worldwide. Here, we described a novel strategy to control begomovirus infection. By constitutively activating NIK-mediated antiviral signalling, we succeeded in developing a tolerant tomato crop. Tomatoes are usually inflicted by diverse begomoviruses, making engineered tolerant/resistant lines even more difficult to develop. Importantly, the T474D-overexpressing tomato transgenic lines were tolerant to ToYSV and ToSRV, which display highly divergent genomic sequences and hence are phylogenetically separated within the two major groups of begomoviruses found in Brazil (Albuquerque et al., 2012). The NIK-mediated strategy was also effective in conferring tolerance to CaLCuV in the model plant Arabidopsis (Zorzatto et al., 2015). These observations indicate the potential of a sustained NIK-mediated defence pathway to confer broad-spectrum tolerance to begomoviruses in distinct plant species.
The current mechanistic model for the activation of the NIK-mediated antiviral signalling pathway holds that upon an unknown stimulus, the NIK LRR extracellular domain undergoes oligomerization with itself or another receptor, allowing the intracellular kinase domains to transphosphorylate and activate one another. The activation of NIK by phosphorylation on the crucial threonine residue at position 474 leads to the regulated relocation of RPL10 to the nucleus where it interacts to LIMYB to fully down-regulate translation-related genes (Zorzatto et al., 2015). Our current data add a number of relevant insights to the understanding of this layer of defence. First, a comparison between the transcriptomes induced by virus infection in wild-type lines and by ectopic expression of the T474D gain-of-function mutant in transgenic lines indicated that activation of the NIK-mediated signalling pathway triggers a typical response to virus infection. This interpretation is supported by the observation that mock-inoculated T474D-overexpressing lines showed a constitutively infected wild-type transcriptome (Figure 2a). Furthermore, the elimination of the mock T474D DE genes from the raw data of all treatments further indicates that the expression profile induced by the T474D mutant greatly mimics the response to viral infection, as the mock- and infection-induced transcriptomes from each genotype clustered together with high significance (Figure 2b). Second, our data are consistent with the notion that the gain-of-function mutant T474D can sustain an activated NIK-mediated antiviral response in the absence of the virus, further confirming that phosphorylation on Thr-474 is the crucial event that leads to the activation of the kinase. In fact, expression of the T474D mutant potentiated the NIK-mediated response, as it would be expected from expression of a constitutively activated defence receptor NIK. Accordingly, the ectopic expression of the T474D gain-of-function mutant was more effective against begomovirus infection than overexpression of the NIK defence receptor in the tomato transgenic lines.
The experiments presented here shed light on the response underlying NIK-mediated antiviral defences. We showed that constitutive activation of NIK in the T474D lines impaired global translation, and such activation might constitute an excellent strategy for fighting begomovirus infection in host cells. We have also demonstrated recently that constitutive activation of NIK1 and overexpression of LIMYB in Arabidopsis down-regulate translation machinery-related genes, suppress translation and confer tolerance to begomovirus infection (Zorzatto et al., 2015). Because begomoviruses rely completely on the plant translation machinery and cannot circumvent host translational regulation, a global repression of translation is expected to significantly affect virus infection, as observed in the T474D-overexpressing lines. Infact, by directly assessing viral transcripts, we showed that the loading of coat protein mRNA into actively translating polysomes was significantly reduced in systemic infected leaves of T474D-overexpressing plants compared to WT and NIK1-overexpressing lines (Figure 6). This result indicates that the suppression of global protein synthesis may effectively protect plant cells against DNA viruses.
Based on our data, it is clear that the level of translational inhibition mediated by constitutive activation of NIK1 did not impact development in tomato under greenhouse conditions. In the first generation, however, some transgenic lines displayed shorter roots that regained normal biomass and growth in the subsequent generations. As a possible explanation for this phenotype, the T474D-mediated translation inhibition would have maintained the transgenic lines under a constant perception of stress, which, in turn, promoted acclimation to maintain normal growth under greenhouse conditions. We also found that T474D-mediated translation inhibition persisted in different developmental stages of the transgenic lines but to different extents. At later stages of development (28-day-old leaves), we observed a 7% reduction in the level of translation inhibition compared to that of seedlings. This difference in the efficiency of global translation inhibition mediated by ectopic expression of the T474D mutant receptor may reflect an adjustment of transgenic lines towards adaptation at later stages. However, it is very intriguing that ectopic expression of T474D did not impact tomato development despite 19–25% suppression of global translation must be considered in attempts to transfer the T474D-mediated defence strategy to other agronomically relevant crops.
Experimental procedures (a detailed description of experimental procedures is provided as supporting information)
Tomato transformation and RNA sequencing experiment
The clone pK7F-NIK1T474D has been previously described (Santos et al., 2009). It harbours a GFP gene fused in-frame after the last codon of the mutant cDNA T474D under the control of the CaMV 35S promoter. In the mutant cDNA T474D, the threonine residue at position 474 within the activation loop of NIK1 was mutated to an aspartate residue. Leaf discs from in vitro-grown tomato plants (Solanum lycopersicum, cultivar Moneymaker) were transformed with pK7F-NIK1T474D via Agrobacterium-mediated plant transformation (strain LBA4404), as previously described (Carvalho et al., 2008c). The transgenic lines T474D-2, T474D-5 and T474D-6 were selected for further analysis. We also used the previously described NIK1-overex-pressing lines NIK1-4 and NIK1-6 (Carvalho et al., 2008c) for infection assays and RNA-seq analysis. NIK1-4 and NIK1-6 lines harbour the Arabidopsis NIK cDNA fused to a GFP cDNA under the control of the CaMV 35S promoter. The transgenic and wild-type lines were infected at the six-leaf stage with ToYSV-[MG-Bi2], as described below in the infectivity assay. At 10 days postinoculation (dpi), total RNA was isolated using TRIzol for the RNA sequencing experiments. The Illumina RNA sequencing data were obtained using a Genome Analyzer in the Fasteris facilities. A systematic comparison in our tomato data set of five representative normalization methods with and without the correction factors for CG-content and gene length was performed using the Bioconductor packages edgeR (Robinson et al., 2010), DESeq (Yang et al., 2013), and EDASeq (Risso et al., 2011). For differential gene expression (DGE) analysis, we subjected the normalized data provided by the counting table to the most common negative binomial methods present in R/Bioconductor software, such as edgeR (Robinson et al., 2010), DESeq (Anders and Huber, 2010) and baySeq (Hardcastle and Krystyna, 2010). The RNA-seq data were submitted to NCBI-GEO, http://www.ncbi.nlm.nih.gov/geo/info/linking.html, accession number GSM932558.
Infectivity assays
The transgenic and wild-type lines were infected at the six-leaf stage with either ToYSV-[MG-Bi2] or ToSRV infectious clones by biolistic delivery. In each experiment, 20 plants of each line were inoculated with 2 μg of tandemly repeated DNA-A plus DNA-B per plant and grown in a greenhouse under natural light, 70% relative humidity and approximately equal day and night lengths. Viral DNA was quantified by qRT-PCR, and coat protein viral mRNA was quantified by qRT-PCR from polysome-associated RNA prepared from T474D, NIK1 and wild-type infected leaves at 10 dpi.
Supplementary Material
Acknowledgements
This research was financially supported through the following grants from Brazilian Government Agencies: CNPq grants 483659/2012-6, 573600/2008-2, 447578/2014-6 (to E.P.B.F.), FAPEMIG grant CBB-APQ-00070-09 (to E.P.B.F.). O.J.B.B. and D.C were supported by CAPES graduate fellowships; J.P.B.M. was supported by a CNPq graduate fellowship; M.D. and W.A.P. were supported by postdoctoral fellowships from CNPq; A.A.S. was the recipient of a postdoctoral fellowship from CAPES.
Footnotes
Conflict of interest
The authors declare no conflict of interest.
Supporting information
Additional Supporting information may be found in the online version of this article:
Data S1 Supporting Experimental Procedures – detailed description of experimental procedures.
Figure S1 NSP from CaLCuV and ToYSV concentrates in the nucleus when ectopically expressed in N. benthamiana leaves.
Figure S2 Overexpression of T474D mutant receptor in tomato leaves.
Figure S3 Characterization of the T474D-overexpressing lines during the vegetative phase.
Figure S4 Fruit quality and yield of the T474D-overexpressing lines.
Figure S5 Representation of the translational machinery-related genes in the down-regulated changes.
Figure S6 Isolation of polysomal fractions from tomato seedlings.
Table S1 Enriched biological process categories from the GO database using the GSEA method.
Table S2 Resistance protein-related genes differentially expressed in infected T474D (infected T474D-mock T474D).
Table S3 Gene Enrichment Analysis.
Table S4 Functional overlap of DE defence-related genes up-regulated in CLN2777 (resistant) by 3, 5, and 7 dpi and in T474D infected by 10 dpi.
References
- Albuquerque LC, Varsani A, Fernandes FR, Pinheiro B, Martin DP, de Tarso OFP, Lemos TO, Inoue-Nagata AK. Further characterization of tomato-infecting begomoviruses in Brazil. Arch. Virol. 2012;157:747–752. doi: 10.1007/s00705-011-1213-7. [DOI] [PubMed] [Google Scholar]
- Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrade EC, Manhani GG, Alfenas PF, Calegario RF, Fontes EPB, Zerbini FM. Tomato yellow spot virus, a tomato-infecting begomovirus from Brazil with a closer relationship to viruses from Sida sp., forms pseudorecombinants with begomoviruses from tomato but not from Sida. J. Gen. Virol. 2006;87:3687–3696. doi: 10.1099/vir.0.82279-0. [DOI] [PubMed] [Google Scholar]
- Aragão FJ, Faria JC. First transgenic geminivirus-resistant plant in the field. Nat. Biotechnol. 2009;27:1086–1088. doi: 10.1038/nbt1209-1086. [DOI] [PubMed] [Google Scholar]
- Carvalho CM, Machado JPB, Zerbini FM, Fontes EBP. NSP-Interacting GTPase: a cytosolic protein as cofactor for nuclear shuttle proteins. Plant Signal. Behav. 2008a;3:752–754. doi: 10.4161/psb.3.9.6641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho CM, Fontenelle MR, Florentino LH, Santos AA, Zerbini FM, Fontes EPB. A novel nucleocytoplasmic traffic GTPase identified as a functional target of the bipartite geminivirus nuclear shuttle protein. Plant J. 2008b;55:869–880. doi: 10.1111/j.1365-313X.2008.03556.x. [DOI] [PubMed] [Google Scholar]
- Carvalho CM, Santos AA, Pires SR, Rocha CS, Saraiva DI, Machado JP, Mattos EC, Fietto LG, Fontes EPB. Regulated nuclear trafficking of rpL10A mediated by NIK1 represents a defense strategy of plant cells against virus. PLoS Pathog. 2008c;4:e1000247. doi: 10.1371/journal.ppat.1000247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo-Urquiza GP, Beserra JE, Jr, Bruckner FP, Lima AT, Varsani A, Alfenas-Zerbini P, Zerbini FM. Six novel begomoviruses infecting tomato and associated weeds in Southeastern Brazil. Arch. Virol. 2008;153:1985–1989. doi: 10.1007/s00705-008-0172-0. [DOI] [PubMed] [Google Scholar]
- Chen T, Lv Y, Zhao T, Li N, Yang Y, Yu W, He X, Liu T, Zhang B. Comparative transcriptome profiling of a resistant vs. susceptible tomato (Solanum lycopersicum) cultivar in response to infection by tomato yellow leaf curl virus. PLoS ONE. 2013;8:e80816. doi: 10.1371/journal.pone.0080816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day AG, Bejarano ER, Buck KW, Burrell M, Lichtenstein CP. Expression of an antisense viral gene in transgenic tobacco confers resistance to the DNA virus tomato golden mosaic virus. Proc. Natl Acad. Sci. USA. 1991;88:6721–6725. doi: 10.1073/pnas.88.15.6721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florentino LH, Santos AA, Fontenelle MR, Pinheiro GL, Zerbini FM, Baracat-Pereira MC, Fontes EPB. A PERK-like receptor kinase interacts with the geminivirus nuclear shuttle protein and potentiates viral infection. J. Virol. 2006;80:6648–6656. doi: 10.1128/JVI.00173-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontes EPB, Santos AA, Luz DF, Waclawovsky AJ, Chory J. The geminivirus NSP acts as virulence factor to suppress an innate transmembrane receptor kinase-mediated defense signaling. Gene Dev. 2004;18:2545–2556. doi: 10.1101/gad.1245904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gafni Y, Epel BL. The role of host and viral proteins in intra- and intercellular trafficking of geminiviruses. Physiol. Mol. Plant Pathol. 2002;60:231–241. [Google Scholar]
- Galvão RM, Mariano AC, Luz DF, Alfenas PF, Andrade EC, Zerbini FM, Almeida MR, Fontes EPB. A naturally occurring recombinant DNA-A of a typical bipartite begomovirus does not require the cognate DNA-B to infect Nicotiana benthamiana systemically. J. Gen. Virol. 2003;84:715–726. doi: 10.1099/vir.0.18783-0. [DOI] [PubMed] [Google Scholar]
- Hardcastle TJ, Krystyna AK. baySeq: empirical Bayesian methods for identifying differential expression in sequence count data. BMC Bioinformatics. 2010;11:422. doi: 10.1186/1471-2105-11-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashmi JA, Zafar Y, Arshad M, Mansoor S, Asad S. Engineering cotton (Gossypium hirsutum L.) for resistance to cotton leaf curl disease using viral truncated AC1 DNA sequences. Virus Genes. 2011;42:286–296. doi: 10.1007/s11262-011-0569-9. [DOI] [PubMed] [Google Scholar]
- Hong Y, Stanley J. Virus resistance in Nicotiana benthamiana conferred by African cassava mosaic virus replication-associated protein (AC1) transgene. Mol. Plant Microbe Interact. 1996;9:219–225. [Google Scholar]
- Kunik T, Salomon R, Zamir D, Navot N, Zeidan M, Michelson I, Gafni Y, Czosnek H. Transgenic tomato plants expressing the tomato yellow leaf curl virus capsid protein are resistant to the virus. Biotechnology. 1994;12:500–504. doi: 10.1038/nbt0594-500. [DOI] [PubMed] [Google Scholar]
- Lin CY, Tsai WS, Ku HM, Jan FJ. Evaluation of DNA fragments covering the entire genome of a monopartite begomovirus for induction of viral resistance in transgenic plants via gene silencing. Transgenic Res. 2012;21:231–241. doi: 10.1007/s11248-011-9523-9. [DOI] [PubMed] [Google Scholar]
- Lucioli A, Sallustio DE, Barboni D, Berardi A, Papacchioli V, Tavazza R, Tavazza M. A cautionary note on pathogen-derived sequences. Nat. Biotechnol. 2008;26:617–619. doi: 10.1038/nbt0608-617. [DOI] [PubMed] [Google Scholar]
- Mariano AC, Andrade MO, Santos AA, Carolino SMB, Oliveira ML, Baracat-Pereira MC, Brommonshenkel SH, Fontes EPB. Identification of a novel receptor-like protein kinase that interacts with a geminivirus nuclear shuttle protein. Virology. 2004;318:24–31. doi: 10.1016/j.virol.2003.09.038. [DOI] [PubMed] [Google Scholar]
- McGarry RC, Barron YD, Carvalho MF, Hill JE, Gold D, Cheung E, Kraus WL, Lazarowitz SG. A novel Arabidopsis acetyltransferase interacts with the geminivirus movement protein NSP. Plant Cell. 2003;15:1605–1618. doi: 10.1105/tpc.012120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noris E, Accotto GP, Tavazza R, Brunetti A, Crespi S, Tavazza M. Resistance to tomato yellow leaf curl geminivirus in Nicotiana benthamiana plants transformed with a truncated viral C1 gene. Virology. 1996;224:130–138. doi: 10.1006/viro.1996.0514. [DOI] [PubMed] [Google Scholar]
- Reyes MI, Nash TE, Dallas MM, Ascencio-Ibáñez JT, Hanley-Bowdoin L. Peptide aptamers that bind to geminivirus replication proteins confer a resistance phenotype to TYLCV and ToMoV infection in tomato. J. Virol. 2013;87:9691–9706. doi: 10.1128/JVI.01095-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risso D, Schwartz K, Sherlock G, Dudoit S. GC-content normalization for RNA-Seq data. BMC Bioinformatics. 2011;12:480. doi: 10.1186/1471-2105-12-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha CS, Santos AA, Machado JPB, Fontes EPB. The ribosomal protein L10/QM-like protein is a component of the NIK-mediated antiviral signaling. Virology. 2008;380:165–169. doi: 10.1016/j.virol.2008.08.005. [DOI] [PubMed] [Google Scholar]
- Rojas MR, Hagen C, Lucas WJ, Gilbertson RL. Exploiting chinks in the plant’s armor: evolution and emergence of geminiviruses. Annu. Rev. Phytopathol. 2005;43:361–394. doi: 10.1146/annurev.phyto.43.040204.135939. [DOI] [PubMed] [Google Scholar]
- Sakamoto T, Deguchi M, Brustolini OJB, Santos AA, Silva FF, Fontes EPB. The tomato RLK superfamily: phylogeny and functional predictions about the role of the LRRII-RLK subfamily in antiviral defense. BMC Plant Biol. 2012;12:229. doi: 10.1186/1471-2229-12-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderfoot AA, Lazarowitz SG. Cooperation in viral movement: the geminivirus BL1 movement protein interacts with BR1 and redirects it from the nucleus to the cell periphery. Plant Cell. 1995;7:1185–1194. doi: 10.1105/tpc.7.8.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderfoot AA, Lazarowitz SG. Getting it together in plant virus movement: cooperative interactions between bipartite geminivirus movement proteins. Trends Cell Biol. 1996;6:353–358. doi: 10.1016/0962-8924(96)10031-3. [DOI] [PubMed] [Google Scholar]
- Sanderfoot AA, Ingham DJ, Lazarowitz SG. A viral movement protein as a nuclear shuttle: the geminivirus BR1 movement protein contains domains essential for interaction with BL1 and nuclear localization. Plant Physiol. 1996;110:23–33. doi: 10.1104/pp.110.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos AA, Carvalho CM, Florentino LH, Ramos HJ, Fontes EPB. Conserved threonine residues within the A-Loop of the receptor NIK differentially regulate the Kinase function required for antiviral signaling. PLoS ONE. 2009;4:e5781. doi: 10.1371/journal.pone.0005781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos AA, Lopes KVG, Apfta JAC, Fontes EPB. NSP-interacting kinase, NIK: a transducer of plant defence signalling. J. Exp. Bot. 2010;61:3839–3845. doi: 10.1093/jxb/erq219. [DOI] [PubMed] [Google Scholar]
- Shiu SH, Bleecker AB. Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proc. Natl Acad. Sci. USA. 2001;98:10763–10768. doi: 10.1073/pnas.181141598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley J, Frischmuth T, Ellwood S. Defective viral DNA ameliorates symptoms of geminivirus infection in transgenic plants. Proc. Natl Acad. Sci. USA. 1991;87:6291–6295. doi: 10.1073/pnas.87.16.6291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunitha S, Marian D, Hohn B, Veluthambi K. Antibegomoviral activity of the agrobacterial virulence protein VirE2. Virus Genes. 2011;43:445–453. doi: 10.1007/s11262-011-0654-0. [DOI] [PubMed] [Google Scholar]
- Ward BM, Lazarowitz SG. Nuclear export in plants: use of geminivirus movement proteins for a cell-based export assay. Plant Cell. 1999;11:1267–1276. doi: 10.1105/tpc.11.7.1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang E-W, Girke T, Jiang T. Differential gene expression analysis using coexpression and RNA-Seq data. Bioinformatics. 2013;29:2153–2161. doi: 10.1093/bioinformatics/btt363. [DOI] [PubMed] [Google Scholar]
- Zhang SC, Wege C, Jeske H. Movement proteins (BC1 and BV1) of Abutilon mosaic geminivirus are cotransported in and between cells of sink but not of source leaves as detected by green fluorescent protein tagging. Virology. 2001;290:249–260. doi: 10.1006/viro.2001.1185. [DOI] [PubMed] [Google Scholar]
- Zorzatto C, Machado JPB, Lopes KVG, Nascimento KJT, Pereira WA, Brustolini OJB, Reis PAB, Calil IP, Deguchi M, Martins GS, Gouveia BC, Loriato VAP, Silva MAC, Silva FF, Santos AA, Chory J, Fontes EPB. NIK-mediated translation suppression functions as a plant antiviral immunity mechanism. Nature. 2015 doi: 10.1038/nature14171. doi: 10.1038/nature14171. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.