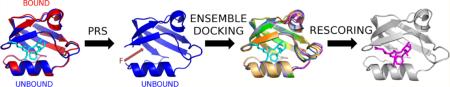
Abstract
Molecular docking serves as an important tool in modeling protein–ligand interactions. However, it is still challenging to incorporate overall receptor flexibility, especially backbone flexibility, in docking due to the large conformational space that needs to be sampled. To overcome this problem, we developed a novel flexible docking approach, BP-Dock (Backbone Perturbation-Dock) that can integrate both backbone and side chain conformational changes induced by ligand binding through a multi-scale approach. In the BP-Dock method, we mimic the nature of binding-induced events as a first-order approximation by perturbing the residues along the protein chain with a small Brownian kick one at a time. The response fluctuation profile of the chain upon these perturbations is computed using the perturbation response scanning method. These response fluctuation profiles are then used to generate binding-induced multiple receptor conformations for ensemble docking. To evaluate the performance of BP-Dock, we applied our approach on a large and diverse data set using unbound structures as receptors. We also compared the BP-Dock results with bound and unbound docking, where overall receptor flexibility was not taken into account. Our results highlight the importance of modeling backbone flexibility in docking for recapitulating the experimental binding affinities, especially when an unbound structure is used. With BP-Dock, we can generate a wide range of binding site conformations realized in nature even in the absence of a ligand that can help us to improve the accuracy of unbound docking. We expect that our fast and efficient flexible docking approach may further aid in our understanding of protein–ligand interactions as well as virtual screening of novel targets for rational drug design.
INTRODUCTION
Molecular docking is an effective tool for predicting the structures of protein–ligand complexes, studying the protein–ligand interactions, and evaluating the binding affinities of such complexes.1 Indeed, it has become the primary component in many drug discovery programs especially for virtual screening.2–6 Although the first docking was pioneered in the early 1980s,4 there are still tremendous research efforts going on to improve the docking algorithms. Particularly, recapitulating the experimentally known binding information is the major challenge in docking, especially when the bound structure is not available.
Most of the earlier docking methods keep the receptor protein as rigid and move the target ligand around the binding site of the protein while performing an energy minimization.5–7 The major problems associated with rigid docking are (i) proteins are not rigid and undergo various types of conformational changes and (ii) simply relying on pure energy minimization is an insufficient approach to predict correct binding affinities.6 Thus, in recent years, docking algorithms have significantly evolved to incorporate full flexibility of the ligand and partial flexibility of the protein.1,5,7–9 However, direct modeling of the protein (i.e., receptor) flexibility still represents a challenging problem due to (i) the high dimensionality of conformational space that must be sampled, which significantly increases the computational time and also results in a higher rate of false-positive solutions, and (ii) complexity of the energy function.7
Some recent flexible docking approaches, such as induced fit docking (IFD), allow the docking simulation to search for a new conformational space to perform direct changes in the binding site conformation.9 However, various IFD methods model flexibility only for a limited number of receptor residues.10–23 Moreover, most of these methods are computationally intensive, making docking difficult for larger systems.9,23 There are also hinge-bent docking algorithms24–27 that allow hinge bending in docking where rigid subdomains are docked separately, and the consistent results are then assembled.1 Like IFD methods, they also have limited ability to handle docking of unbound molecules with significant backbone flexibility.28 In contrast to modeling protein flexibility explicitly, ensemble docking methods account for protein flexibility prior to the actual docking by making use of a limited number of discrete protein conformations such as Rosetta-Backrub,29 MedusaDock,30 AutoDock,31 and IFREDA.32 Interestingly, few of the IFD docking methods also use a pre-existing ensemble of conformations such as FlexX-Ensemble,14 FLIPDock,16,17 FITTED,20–22 and DOCK 4.0.11 The docking time for these approaches scales linearly with the number of structures in the ensemble.33 The integration of multiple receptor conformation (MRC) sampling into the docking algorithm might improve computational speed and help us simplify data management.7 The sources of ensemble generation vary from experimentally determined X-ray or NMR protein structures34–38 to computationally derived protein conformations from molecular dynamics (MD) simulations,6,39 homology models,6 or normal mode analysis.6,40–43 The success of ensemble docking approaches depends on two features of the multiple receptor conformations: (i) a wide range of binding site conformations realized in nature should be sampled in the ensemble of receptors and (ii) the artifact conformations that predict incorrect poses should be excluded. Therefore, it becomes important to mimic nature and sample binding-induced conformations using effective and intelligent sampling strategies while generating ensembles from any of the above-mentioned approaches.6,7
In order to overcome the challenges in generating an ensemble of correct bound-like conformations in a computationally efficient way, we developed a flexible docking scheme called BP-Dock (Backbone Perturbation-Dock) based on perturbation response scanning (PRS).41–44 PRS couples the elastic network model45 (ENM) with linear response theory (LRT).46 With PRS, we simulate the natural course of a binding event by computing fluctuation responses of all the residues in a protein by exerting random external unit force on a single α-carbon atom of the chain, especially those in the binding pocket.41–44 BP-Dock computes a ligand-induced mean-square fluctuation profile for the backbone of a protein by using PRS, which is then followed by all atom energy minimization of the perturbed protein conformation. This two-step multi-scale approach enables us to integrate both backbone and side chain conformational changes of a receptor into docking, and it is computationally efficient to model large-scale backbone movements. Indeed, we have shown that the residue fluctuation responses obtained upon perturbation of a single residue can capture conformational change between unbound and bound conformations.41 Moreover, the ensemble of multiple receptor conformations generated through this approach was successful in capturing the correct binding affinities for the bound (holo) structure of PICK1 (protein interacting with C kinase) protein and its mutants.42
“Bound” docking that reconstructs a complex using the bound structure of the receptor and the ligand is a fairly simple problem in docking. The more challenging one is indeed “unbound” docking where an unbound (apo) form of the structure is used along with the ligand to obtain a complex form. As a matter of fact, the accuracy of the docking methods decreases when the unbound receptor is used.2 The unbound structure can be an experimental structure in the absence of a ligand or a homology model. In the present work, we apply our BP-Dock approach on unbound structures. The two main goals are (i) to check whether the unbound docking with BP-Dock can recapitulate the bound docking results and (ii) to test if the method accurately captures the experimental binding affinities when an unbound receptor structure is used.
We test our flexible docking approach for a data set of protein–peptide as well as protein–small ligand complexes. The data set used for this study comprises five diverse sets of protein–ligand complexes of HIV-1 protease, carbonic anhydrase II, alcohol dehydrogenase, alpha-thrombin, and cytochrome C peroxidase, where we compared the experimental binding affinities of each individual set with the binding energy scores obtained from unbound docking by BP-Dock. In addition to these sets, we also analyze another 20 individual protein complexes with available bound and unbound experimental structures. Overall, the unbound/bound pairs in our data set cover a wide range of root-mean-square distance (RMSD) between bound and unbound conformations ranging from 0.103 to 1.65 Å (Table S1, Supporting Information), which enables us to rigorously test the performance of BP-Dock on unbound structures having a diverse set of RMSDs from the bound structures. Furthermore, 13 proteins ranging from 59 to 537 residue long chains that are in complex with various types of ligands, including peptides with different lengths (from 2 to 10 mers), are chosen to provide an extensive pool of flexible degrees of freedom. We also perform “rigid docking” that does not incorporate the flexibility of the backbone and side chains outside the binding pocket, using both bound and unbound experimental structures. This enables us to compare the performance of unbound docking with BP-Dock with respect to rigid bound and unbound docking. To further determine the accuracy and sensitivity of our docking method, we also perform cross-docking tests for HIV and postsynaptic density-95/Dlg/ZO-1 (PDZ) domain proteins. Overall, our analysis yields that BP-Dock is a computationally efficient approach to incorporate full receptor flexibility to generate MRCs, as also observed in our earlier work.42,43 Ensemble docking using MRC generated from unbound conformation can capture the bound docking results. Moreover, it can improve the binding affinity prediction in several cases. The success of the approach rests on generating a wide range of binding site conformations realized in nature.
METHODS
Benchmark
We analyzed five different diverse sets of protein–ligand complexes (HIV-1 protease (N = 20), carbonic anhydrase II (N = 9), alcohol dehydrogenase (N = 8), alpha-thrombin (N = 13), and cytochrome C peroxidase (N = 18), where N is the number of complexes for each protein set used in the study) and an individual set of another 20 proteins with available bound (holo) and unbound (apo) structures that are retrieved from the Protein Data Bank (PDB).47 The names of the proteins, PDB codes of their corresponding bound and unbound structures, chain length, root mean-square distance between bound and unbound structures, and names of binding ligand/peptide and sequences of peptides are displayed in Table S1 of the Supporting Information. The experimental binding affinities for the five test sets (total 68 test cases) are obtained from LPDB48 and Astex49 databases. The performance of rigid docking versus BP-Dock is also tested in cross-docking studies on the HIV-1 protease set. The HIV-1 protease benchmark set has 20 complexes, and cross-docking tests are performed on 20 × 20 = 400 cases. Furthermore, we also analyze the homology model of the channel-interacting PDZ protein (CIPP). Overall, we have a large and diverse data set of 494 docking cases (including 400 cross-docking cases) to evaluate the performance of BP-Dock.
Ensemble Docking with BP-Dock
To generate binding-induced conformations, we use the perturbation response scanning technique that combines the elastic network model and linear response theory.41,43 In the elastic network model, a protein structure is viewed as a three-dimensional elastic network, and all residue pairs are subjected to a uniform, single-parameter, harmonic potential if they are located within an interaction range or cutoff distance, rc.45,50,51 The overall potential is given by the sum of all harmonic potentials among interacting nodes such that
| (1) |
where γ is the interaction (spring) constant, Rij is the unit vector connecting residue pairs i and j, Aij represents the elements of the adjacency matrix, and is the average distance between residues i and j.45 In this study, however, we weight the interaction strength between all residue pairs by using the inverse of the square distance of their separation rather than using arbitrary cutoff distances.50,52 The expansion of the potential near the equilibrium state can be written in compact notation as
| (2) |
Here, ΔR is the 3N-dimensional vector of fluctuations of all residues, and H is the Hessian, a 3N × 3N matrix composed of second derivatives of potential with respect to the components of position vectors of length N. After obtaining H, a random unit force (F) is applied sequentially to the α-carbon atom of each residue one at a time, and then we record the resulting relative displacement of all residues using LRT. The overall response of residue network is calculated through
| (3) |
where the ΔF vector contains components of externally applied force vectors on each single residue, and H−1 is inverse of the Hessian matrix. The final perturbed coordinates, Rper, for each residue are calculated using
| (4) |
where R0 is a vector containing the initial coordinates of the residues before perturbation, and α is a scaling factor.43,53 In order to present a significant conformational change on the structure after perturbing, we multiply the response fluctuation vector with a scaling factor as PRS is based on LRT. The scaling factor is chosen such that it yields an ensemble of perturbed structures that have a RMSD deviation ranging from 0.25 to 1 Å from the original unbound structure.
Perturbed structures are then clustered using the k-means clustering algorithm54 to discard similar conformations generated from perturbations of different residues in the protein. This step is followed by an all-atom minimization of clustered structures using the AMBER 99SB force field,55 along with a GB solvation model,56 to account for rotameric changes of side chains and also to relieve any strain in the structure. All these steps lead to a set of conformations that constitute multiple binding-induced receptor conformations. Finally, an ensemble docking for all these individual conformations of MRC is performed using RosettaLigand.10,23
Docking with RosettaLigand
The docking simulation for each structure in the ensemble is performed using the RosettaLigand10,23 protocol in the Rosetta program. RosettaLigand incorporates ligand flexibility by changing the torsional angles and backbone of the ligand, while optimizing the side chain of the binding pocket. In this study, we perturb the ligand position and orientation randomly with translation of mean 0.1 Å and rotations of mean 3°, respectively. For each case, coordinates of the ligand are taken from the crystallographic complex of the bound protein. We compute 10,000 trajectories to generate a comprehensive ensemble of conformations of receptor–ligand complexes for each protein, which also produce a well-converged distinct binding funnel in energy score/RMSD plots. Final docked conformations are selected based on the lowest free energy pose in the protein-binding site.10,23 The lowest free energy pose has the lowest Rosetta energy score among all other docked poses. The scoring function of Rosetta is a weighted sum of 12 different energy terms including van der Waals, solvation, hydrogen bonding, torsional, Coulombic, and harmonic restraints.10
Assessing the Scoring Accuracy with X-Score
After selecting the lowest Rosetta energy score pose, we reassess the binding energy score of the complex using X-Score.57 X-Score is an empirical scoring function developed to re-rank the protein–ligand complex obtained from various docking approaches and gives a more accurate estimation of the binding free energies. X-Score was also shown to have the best correlation with the experimental binding affinities as compared to other available scoring functions in a study by Wang et al.58 Likewise, our binding affinities obtained by rescoring the lowest energy pose with X-Score provide a better correlation with experimental affinities.
Modeling Unbound Proteins and Non-Native Peptides
The homology model of CIPP is constructed using MODELER59 with a minimal sequence similarity of 50% to the target. Before introducing flexibility in the homologue structure of CIPP, it is subjected to an energy minimization of 50 steepest descent iterations followed by 1000 conjugate gradient iterations using the AMBER 99SB force field,55 along with a GB solvation model.56 We also model mutated unbound proteins for the HIV-1 protease and cytochrome C peroxidase test sets. The starting unbound structures are obtained from PDB47 (2PC0 for HIV-1 protease and 1CCP for cytochrome C peroxidase). The mutations corresponding to the desired bound protein are introduced in the unbound structure using PyMOL,60 which is followed by an all atom energy minimization on the modeled unbound protein following the same procedure performed on CIPP. This all atom energy minimization helps to accommodate the necessary side-chain rotamer changes around the residue subjected to the point mutation. For a non-native peptide docking to PDZ proteins, we use the original crystal structure of the native peptide and mutate each position in the native peptide to the corresponding amino acid of the desired peptide and perform an all-atom energy minimization on the modeled peptide–protein complex using the AMBER 99SB force field,55 along with a GB solvation model.56
Ensemble Docking with Backrub
We use the backbone sampling method61 from the RosettaBackrub design server29 to generate multiple receptor conformations for ensemble docking. The server utilizes the “Backrub” method for flexible protein backbone modeling that was first described by Davis et al.62 Briefly, this method randomly makes one of three types of moves: (i) a rotamer change (50% of the time), (ii) a local backbone conformational change (Backrub move) consisting of a rigid body rotation of a random peptide segment about the axis connecting the endpoint C-α atoms (25% of the time), or (iii) a composite move with a Backrub change and one or two rotamer changes (25% of the time). After each move, the positions of the C-β and H-α atoms are modified to minimize bond angle strain.61 We dock these ensembles of proteins obtained from Backrub to their respective peptides using RosettaLigand.10
RESULTS AND DISCUSSION
Previously, we have shown that the BP-Dock approach gives better correlation with experimental binding affinities compared to conventional rigid docking for the bound structure of PICK1 protein and its mutants.42 In this study, we extend our approach to the unbound structure in order to test if we can predict the binding affinities of several peptide/ligands when they are docked into an unbound structure.
Docking Results for Five Different Test Sets and Their Correlation with Experimental Binding Affinities
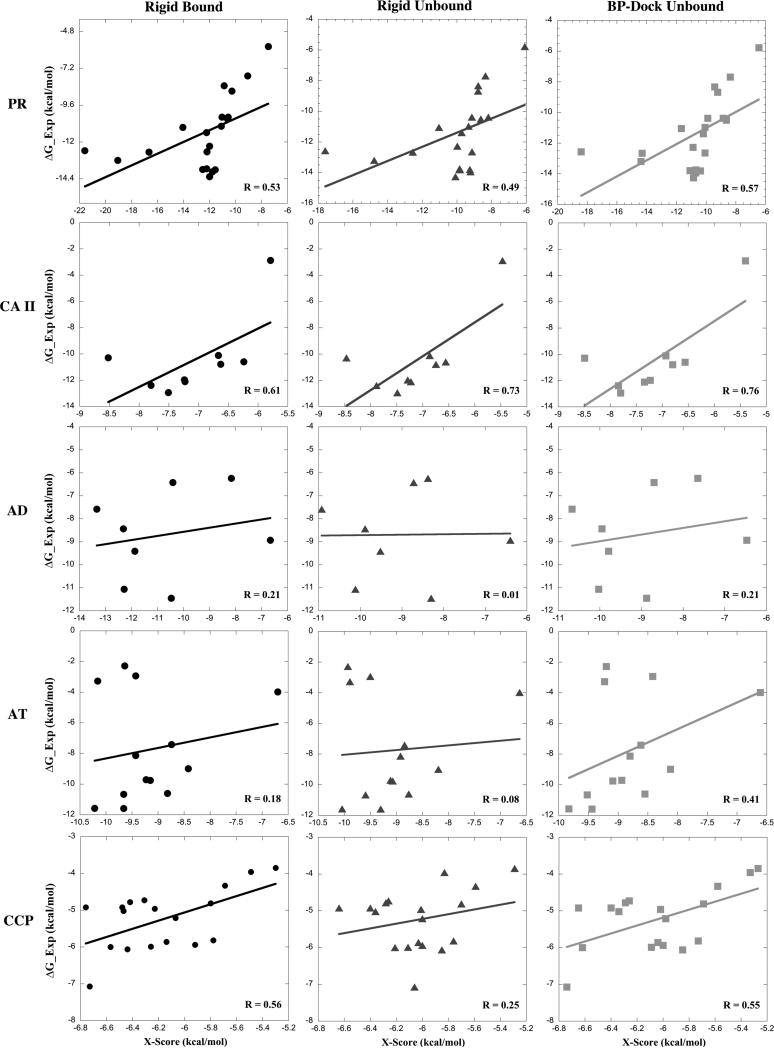
We first compare the performance of our flexible docking with rigid docking for five different sets of protein–ligand complexes: HIV-1 protease (PR), carbonic anhydrase II (CA II), alcohol dehydrogenase (AD), alpha-thrombin (AT), and cytochrome C peroxidase (CCP). For each set, we have different bound structures (i.e., with different ligands) but only one unbound structure. For HIV-1 protease, we perform docking using an apo wide open conformation to test the accuracy of the BP-Dock approach in predicting the binding specificity observed in a closed holo structure using an open unbound form. Thus, we aim to determine whether our BP-Dock approach can capture the different bound conformations with correct binding energies through generating an ensemble of conformations from a single unbound structure in a quick and efficient manner. Rigid docking is also performed on crystal structures for both bound and unbound structures using RosettaLigand.10 For PR and CCP, we use the modeled unbound structure due to point mutations in bound structures (see Methods). The lowest RosettaLigand energy scores and X-Scores for (i) rigid bound and unbound docking and (ii) flexible docking for unbound structures using BP-Dock for all five different data sets are reported in Table I. The available experimental binding affinities for all the test cases are obtained from LPDB48 and Astex49 databases and are reported in Table I. The correlation plots of X-Score energies for (i) rigid bound, (ii) rigid unbound, and (iii) BP-Dock unbound versus the experimental binding free energies for the five test sets are plotted in Figure 1. The X-Score energies of BP-Dock unbound docking have a higher correlation with experimental binding energies compared to rigid unbound docking for all the five test sets. Interestingly, both the BP-Dock unbound X-Score and RosettaLigand energy scores for HIV-1 protease show a much better correlation with experimental binding energies than rigid unbound docking. Indeed, unbound docking by BP-Dock is even better than rigid bound docking for PR and CA II. Moreover, when we compare the RosettaLigand energy scores, we observe that BP-Dock provides better correlation with experimental binding energies than rigid unbound docking for all five test sets (Figure S1, Supporting Information). Strikingly, rigid unbound docking scores are negatively correlated with the experimental binding energies for CA II, AD, and CCP data sets when the complexes are not re-evaluated by X-Score. On the other hand, BP-Dock performs better than rigid unbound docking and is also better compared to rigid bound docking for all the test sets except AD. The overall RMSDs of the ligand from the lowest energy docked poses for each docking case shows a similar trend, indicating that the backbone flexibility introduced by BP-Dock also improves the orientation of the ligand compared to rigid unbound docking (Table S2, Supporting Information).
Table I.
RosettaLigand Energy Scores and X-Scores for Rigid Bound, Rigid Unbound, and Flexible BP-Dock Docking for Unbound Structures with Available Experimental Data
| RosettaLigand score (kcal/mol) |
X-Score (kcal/mol) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| PDB code |
rigid |
BP-Dock |
rigid |
BP-Dock |
||||||
| protein | bound | unbound | chain length | bound | unbound | unbound | bound | unbound | unbound | Exp_ ΔG |
| HIV-1 protease | 1HBV | 2PC0 | 198 | –357.51 | –558.70 | –390.74 | –10.28 | –8.78 | –9.23 | –8.68 |
| 1HEG | 2PC0 | 198 | –64.8 | –563.82 | –402.30 | –10.56 | –8.19 | –8.86 | –10.38 | |
| 1HIH | 2PC0 | 198 | –491.08 | –569.36 | –397.08 | –11.1 | –9.33 | –10.08 | –10.97 | |
| 1HIV | 2PC0 | 198 | –462.25 | –555.32 | –393.39 | –12.2 | –9.13 | –10.08 | –12.64 | |
| 1HPS | 2PC0 | 198 | –394.35 | –586.28 | –416.19 | –16.65 | –12.55 | –14.31 | –12.66 | |
| 1HTE | 2PC0 | 198 | –210.93 | –559.65 | –383.46 | –9.06 | –8.36 | –8.38 | –7.69 | |
| 1HTF | 2PC0 | 198 | –394.32 | –568.62 | –402.59 | –14.05 | –11.04 | –11.68 | –11.04 | |
| 1HTG | 2PC0 | 198 | –528.38 | –582.81 | –413.38 | –19.05 | –14.77 | –14.39 | –13.20 | |
| 1HVI | 2PC0 | 198 | –513.57 | –564.07 | –401.71 | –12.22 | –9.85 | –10.69 | –13.74 | |
| 1HVJ | 2PC0 | 198 | –496.71 | –563.75 | –399.83 | –12 | –10.09 | –10.86 | –14.26 | |
| 1HVK | 2PC0 | 198 | –518.85 | –568.48 | –399.62 | –12.53 | –9.26 | –11.09 | –13.79 | |
| 1HVL | 2PC0 | 198 | –490.64 | –565.61 | –401.29 | –12 | –9.98 | –10.89 | –12.27 | |
| 1HVS | 2PC0 | 198 | –434.77 | –559.28 | –408.65 | –11.57 | –9.84 | –10.39 | –13.81 | |
| 1SBG | 2PC0 | 198 | –368.53 | –568.57 | –405.08 | –11.04 | –9.14 | –9.90 | –10.38 | |
| 4HVP | 2PC0 | 198 | –290.75 | –567.84 | –381.94 | –10.88 | –8.77 | –9.43 | –8.33 | |
| 4PHV | 2PC0 | 198 | –476.11 | –599.01 | –432.82 | –21.58 | –17.60 | –18.41 | –12.56 | |
| 5HVP | 2PC0 | 198 | –418.5 | –563.39 | –398.59 | –10.62 | –8.62 | –8.64 | –10.50 | |
| 9HVP | 2PC0 | 198 | –237.85 | –565.45 | –403.47 | –12.23 | –9.73 | –10.20 | –11.38 | |
| 1A30 | 2PC0 | 198 | –517.82 | –538.30 | –372.12 | –7.48 | –6.09 | –6.45 | –5.77 | |
| 1KZK | 2PC0 | 198 | –572.92 | –545.99 | –394.12 | –11.78 | –9.22 | –10.70 | –13.94 | |
| carbonic anhydrase II | 1OQ5 | 2ILI | 259 | –715.26 | –756.01 | –773.92 | –8.52 | –8.47 | –8.49 | –10.29 |
| 1AVN | 2ILI | 259 | –592.57 | –751.43 | –701.87 | –5.79 | –5.47 | –5.4 | –2.88 | |
| 1CIL | 2ILI | 259 | –721.24 | –752.23 | –767.82 | –7.51 | –7.49 | –7.8 | –12.94 | |
| 1CIM | 2ILI | 259 | –685.56 | –752.41 | –765.94 | –7.23 | –7.23 | –7.34 | –12.1 | |
| 1CIN | 2ILI | 259 | –700.59 | –752.19 | –769.06 | –7.24 | –7.29 | –7.23 | –11.97 | |
| 1CNW | 2ILI | 259 | –623.88 | –758.62 | –773.11 | –6.24 | –6.56 | –6.56 | –10.6 | |
| 1CNX | 2ILI | 259 | –628.09 | –759.78 | –773.72 | –6.67 | –6.87 | –6.93 | –10.11 | |
| 1CNY | 2ILI | 259 | –559.46 | –758.64 | –773.63 | –6.63 | –6.75 | –6.8 | –10.78 | |
| 1OKL | 2ILI | 259 | –755.5 | –756.12 | –772.95 | –7.8 | –7.89 | –7.84 | –12.39 | |
| alcohol dehydrogenase | 1ADB | 8ADH | 374 | –716.52 | –536.87 | –1009 | –10.47 | –8.3 | –8.88 | –11.45 |
| 1ADC | 8ADH | 374 | –635.01 | –539.07 | –997.2 | –10.41 | –8.72 | –8.7 | –6.42 | |
| 1ADF | 8ADH | 374 | –664.17 | –535.33 | –997.55 | –8.16 | –8.38 | –7.65 | –6.24 | |
| 1BTO | 8ADH | 374 | –999 | –454.08 | –962.59 | –6.66 | –6.4 | –6.47 | –8.93 | |
| 1HLD | 8ADH | 374 | –1001.88 | –552.66 | –1009.89 | –13.33 | –10.93 | –10.67 | –7.58 | |
| 1LDE | 8ADH | 374 | –952.57 | –536.52 | –1005.22 | –11.87 | –9.52 | –9.79 | –9.41 | |
| 1LDY | 8ADH | 374 | –930.53 | –540.04 | –1008.14 | –12.28 | –10.12 | –10.03 | –11.06 | |
| 3BTO | 8ADH | 374 | –1088.23 | –546.46 | –1006.28 | –12.31 | –9.89 | –9.95 | –8.43 | |
| alpha-thrombin | 1A4W | 1C5L | 274 | –572.9 | –791.4 | –787.2 | –9.43 | –8.92 | –8.8 | –8.13 |
| 1AE8 | 1C5L | 298 | –758.2 | –790.99 | –784.3 | –8.42 | –8.19 | –8.12 | –8.99 | |
| 1BMM | 1C5L | 295 | –445.95 | –796.63 | –791.14 | –9.15 | –9.07 | –9.09 | –9.75 | |
| 1BMN | 1C5L | 292 | –513.89 | –792.21 | –789.85 | –9.66 | –9.3 | –9.44 | –11.58 | |
| 1D3D | 1C5L | 290 | –626.78 | –794.97 | –789.76 | –10.16 | –9.89 | –9.23 | –3.27 | |
| 1D3P | 1C5L | 290 | –612.8 | –796.24 | –790.58 | –9.43 | –9.5 | –8.42 | –2.93 | |
| 1D4P | 1C5L | 290 | –605.2 | –796.04 | –791.08 | –9.64 | –9.93 | –9.2 | –2.28 | |
| 1DWB | 1C5L | 298 | –798.4 | –783.67 | –777.29 | –6.7 | –6.63 | –6.61 | –3.98 | |
| 1DWC | 1C5L | 298 | –800.78 | –789.23 | –784.77 | –8.82 | –8.76 | –8.55 | –10.6 | |
| 1DWD | 1C5L | 298 | –811.38 | –797.11 | –791.32 | –10.22 | –10.04 | –9.83 | –11.57 | |
| 1HDT | 1C5L | 303 | –217.53 | –795.04 | –789.04 | –9.66 | –9.59 | –9.52 | –10.66 | |
| 1UVS | 1C5L | 268 | –603.35 | –788.95 | –782.21 | –8.74 | –8.84 | –8.62 | –7.41 | |
| 1OYT | 1C5L | 306 | –825.83 | –792.24 | –791.47 | –9.23 | –9.11 | –8.94 | –9.71 | |
| cytochrome C peroxidase | 1AC4 | 1CCP | 291 | –753.91 | –887.07 | –897.53 | –5.3 | –5.29 | –5.27 | –3.85 |
| 1AC8 | 1CCP | 291 | –790.67 | –889.01 | –899.96 | –6.42 | –6.28 | –6.29 | –4.78 | |
| 1AEB | 1CCP | 291 | –790.68 | –889.59 | –900.82 | –5.8 | –5.7 | –5.69 | –4.81 | |
| 1AED | 1CCP | 291 | –788.54 | –886.36 | –897.54 | –6.14 | –6.03 | –6.04 | –5.86 | |
| 1AEE | 1CCP | 291 | –803.13 | –893.8 | –904.99 | –5.49 | –5.83 | –5.33 | –3.96 | |
| 1AEF | 1CCP | 291 | –792.96 | –893.39 | –914.1 | –6.57 | –6.21 | –6.62 | –6 | |
| 1AEG | 1CCP | 291 | –795.32 | –893.45 | –904.04 | –6.26 | –6.11 | –6.09 | –5.99 | |
| 1AEH | 1CCP | 291 | –793.03 | –897.21 | –902.31 | –6.23 | –6.01 | –6.02 | –4.96 | |
| 1AEJ | 1CCP | 291 | –790.57 | –891.97 | –902.97 | –6.07 | –6 | –5.98 | –5.21 | |
| 1AEK | 1CCP | 291 | –793.62 | –891.75 | –902.36 | –6.76 | –6.64 | –6.65 | –4.92 | |
| 1AEM | 1CCP | 291 | –795.62 | –894.37 | –905.32 | –6.48 | –6.4 | –6.4 | –4.92 | |
| 1AEN | 1CCP | 291 | –753.99 | –887.28 | –898.04 | –6.73 | –6.06 | –6.74 | –7.07 | |
| 1AEO | 1CCP | 291 | –764.09 | –894.73 | –904.55 | –6.47 | –6.36 | –6.34 | –5.02 | |
| 1AEQ | 1CCP | 291 | –790.59 | –891.23 | –901.78 | –6.31 | –6.26 | –6.26 | –4.73 | |
| 1AES | 1CCP | 291 | –777.02 | –890.29 | –900.74 | –5.69 | –5.59 | –5.58 | –4.33 | |
| 1AET | 1CCP | 291 | –808.57 | –890.68 | –900.81 | –5.78 | –5.76 | –5.73 | –5.82 | |
| 1AEU | 1CCP | 291 | –756.73 | –891.24 | –901.34 | –5.92 | –6 | –6 | –5.94 | |
| 1AEV | 1CCP | 291 | –783.98 | –891.28 | –902.13 | –6.44 | –5.85 | –5.85 | –6.06 | |
Figure 1.
Correlation plots of binding energy scores evaluated from X-Score vs experimental binding energies for HIV-1 protease (PR), carbonic anhydrase II (CA II), alcohol dehydrogenase (AD), alpha-thrombin (AT), and cytochrome C peroxidase (CCP).
The overall correlation coefficients (R) of X-Score energies with binding experimental energies for all 68 test cases clearly show the success of BP-Dock through incorporation of backbone flexibility (R = 0.65). It is significantly higher than rigid unbound docking (R = 0.56) and also higher than rigid bound docking (R = 0.60). Moreover, when we consider proteins having relatively larger conformational changes upon binding (bound–unbound RMSD > 1 Å), we still observe the same trend, in which the rigid unbound docking cannot capture correct binding conformations for such cases (R = 0.44), whereas BP-Dock provides a better correlation (R = 0.56) in estimating native-like binding affinities, and is even slightly better than rigid bound docking (R = 0.49). This is due to the fact that rigid docking can only optimize side chains lining the binding pocket and cannot sample large backbone movements (or conformational changes) associated with binding unlike BP-Dock. Therefore, incorporating backbone flexibility in an unbound structure becomes even more crucial in proteins with larger RMSD difference. With BP-Dock, we can significantly improve the binding affinity predictions for proteins with larger conformational changes by integrating both backbone and side-chain flexibility through our multi-scale approach.
Cross-Docking Results for the HIV-1 Protease Set
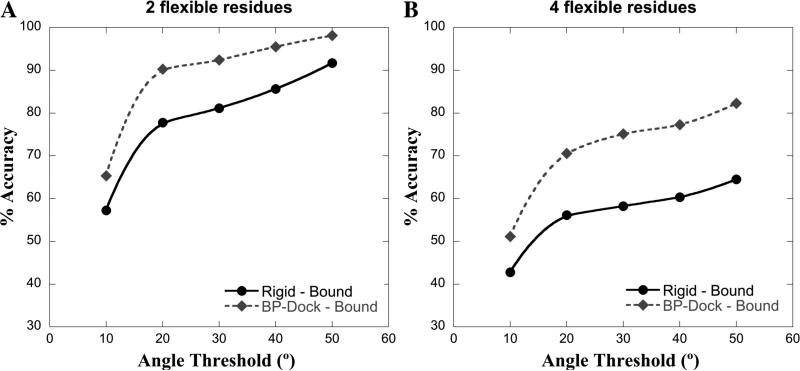
In cross-docking studies, a ligand A (say, from protein A) is docked to a different receptor (say, protein B, bound to a different ligand) to evaluate the performance of a docking method in recapitulating the protein conformational changes associated with ligand binding. Therefore, in this study, we performed cross-docking using the flexible BP-Dock as well as the rigid docking approach on the protein B–ligand A complex for the 20 bound structures from the HIV-1 protease set leading to 20 × 20 = 400 test cases. The lowest RosettaLigand energy docked pose from the protein B–ligand A cross-docking experiment is then compared with the experimental bound structure of protein A–ligand A to check the accuracy of prediction of conformational changes, following the analysis of Osterberg et al.63 and Shin and Seok.64 We investigate the flexibility of two ARG8s and two ILE50s from the two chains of HIV-1 protease. ARG8 and ILE50 are purposefully selected because they have the largest steric clashes caused by swapping ligands.65 Thus, we compare the prediction accuracy of the side-chain χ1 angle of the cross-docked complex (i.e., ligand A from protein A, in complex with receptor from protein B) to χ1 angle of the native complex (i.e., experimental structure of protein A bound to ligand A) for flexible residues ARG8 and ILE50 from the two chains. The predicted χ1 angle is considered accurate if its value is within a range (angle threshold (deg)) of the native χ1 angle. The plots of prediction accuracy of the χ1 angle as a function of the χ1 angle threshold (deg) for the flexible BP-Dock and rigid cross-docking results are shown in Figure 2. The plots for the two flexible residues (ARG8 of the two chains) are shown in Figure 2A and that for four flexible residues (two ARG8s and two ILE50s) are shown in Figure 2B. Clearly, for both the cases, the BP-Dock approach shows better prediction accuracies for χ1 angle compared to the rigid cross-docking, which confirms that even in the case of lower backbone deviation, incorporating backbone flexibility improves proper side-chain orientations during docking.
Figure 2.
Plot of average accuracy of χ1 angle prediction as a function of χ1 angle threshold (deg) for BP-Dock cross-docking and rigid cross-docking on HIV-1 protease bound structures (A) for two flexible residues (two ARG8s) and (B) for four flexible residues (two ARG8s and two ILE50s).
Analysis of Unbound Docking for 20 Individual Bound–Unbound Pairs
Table II shows the RosettaLigand energy scores and X-Scores of 20 individual proteins for (i) rigid bound and unbound docking with RosettaLigand and (ii) flexible docking for unbound structure using BP-Dock. The RMSD (Å) values between the ligand positions of the lowest energy docked poses from rigid bound, rigid unbound, and flexible BP-Dock docking of these 20 individual proteins and that of the bound crystal structure are also shown in Table S2 of the Supporting Information. In most of the cases, rigid docking with a bound structure shows a better affinity prediction as compared to rigid docking of an unbound structure. This is unsurprising because the prediction accuracy of docking calculations decreases with the quality of receptor from bound to unbound protein to modeled structures.2 However, the flexible BP-Dock scheme does a similar or better job in indicating bound-like binding scores for most of the unbound proteins as compared to rigid unbound docking. Overall, these results support the fact that improvement obtained with our flexible docking approach relies on correctly predicting binding relevant motions through perturbation of unbound structures. Moreover, in order to have a better understanding of the advantages and limitations of BP-Dock, we investigate various test cases separately, including the receptors with larger conformational changes upon binding, receptors with larger chains, and those in complex with large peptides.
Table II.
RosettaLigand Energy Scores and X-Scores for Rigid Bound, Rigid Unbound, and Flexible BP-Dock Docking for Unbound Structures
| RosettaLigand score(kcal/mol) |
X-Score (kcal/mol) |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| PDB code |
rigid |
BP-Dock |
rigid |
BP-Dock |
|||||
| protein | peptide/ligand | bound (holo) |
unbound (apo) |
bound (holo) |
unbound (apo) |
unbound (apo) |
bound (holo) |
unbound (apo) |
unbound (apo) |
| PSD-95 | KQTSV | 1BE9 | 1BFE | –230.78 | –166.29 | –273 | –7.27 | –7.38 | –7.32 |
| GRIP | ATVRTYSC | 1N7F | 1N7E | –241.76 | –218.37 | –229.36 | –8.14 | –7.74 | –7.82 |
| syntenin | DSVF | 1OBX | 1NTE | –208.24 | –215.09 | –205.67 | –7.29 | –6.95 | –7.16 |
| syntenin | NEFYA | 1OBY | 1NTE | –225.2 | –221.41 | –211.88 | –7.99 | –7.5 | –7.85 |
| syntenin | FFEEL | NA | 1NTE | –179.21 | –121.47 | –183.71 | –6.83 | –6.24 | –7.02 |
| SH3 domain of GRB2 | RHYRPLPPLP | 1IO6 | 1GFD | –126.24 | –131.36 | –146.09 | –8.12 | –7.4 | –7.48 |
| SH3 domain of GRB2 | VPPPVPPRRR | NA | 1GFD | –119.31 | –99.82 | –127.97 | –7.98 | –7.01 | –7.37 |
| cyclophilin A | AP | 2CYH | 2CPL | –474.43 | –476.3 | –479.82 | –6.65 | –6.21 | –6.52 |
| cyclophilin A | HAGPIA | 1AWQ | 2CPL | –479.63 | –493.49 | –497.5 | –7.86 | –7.1 | –7.16 |
| methyltransferase | NWETF | 1BC5 | 1AF7 | –598.01 | –642.08 | –781.48 | –9.12 | –8.39 | –8.65 |
| aldose reductase | TOL4 | 2FZB | 2ACR | –782.2 | –721.69 | –876.52 | –8.73 | –8.04 | –8.96 |
| aldose reductase | IDD552 | 1T40 | 2ACR | –799.57 | –786.34 | –873.39 | –8.96 | –5.95 | –8.78 |
| carboxypeptidase | HFA | 2CTC | 1M4L | –890.48 | –883.57 | –972.95 | –7.17 | –6.96 | –7.29 |
| carboxypeptidase | FVF | 7CPA | 1M4L | –729.44 | –882.88 | –974.06 | –10.18 | –8.86 | –9.79 |
| TIM | 2PG | 4TIM | 3TIM | –1170.66 | –871.05 | –1454.37 | –6.46 | –6.02 | –6.09 |
| TIM | G3P | 6TIM | 3TIM | –1070.71 | –871.29 | –1457.93 | –6.34 | –5.76 | –6.01 |
| ABP | NLA | 1LRH | 1LR5 | –382.41 | –372.43 | –402.68 | –8.09 | –8.19 | –8.24 |
| acetylcholinesterase | huperzine A | 1GPK | 1EA5 | –1564.29 | –1546.05 | –1651.73 | –8.72 | –8.55 | –8.76 |
| adenosine deaminase | FR233624 | 1UML | 1VFL | –846.1 | –943.34 | –1098.53 | –9.73 | –9.93 | –9.89 |
| quinone reductase 2 | resveratrol | 1SG0 | 1QR2 | –1415.7 | –1310.89 | –1338.84 | –11.55 | –11.22 | –11.67 |
Unbound Docking for Proteins Having Critical Conformational Changes upon Binding
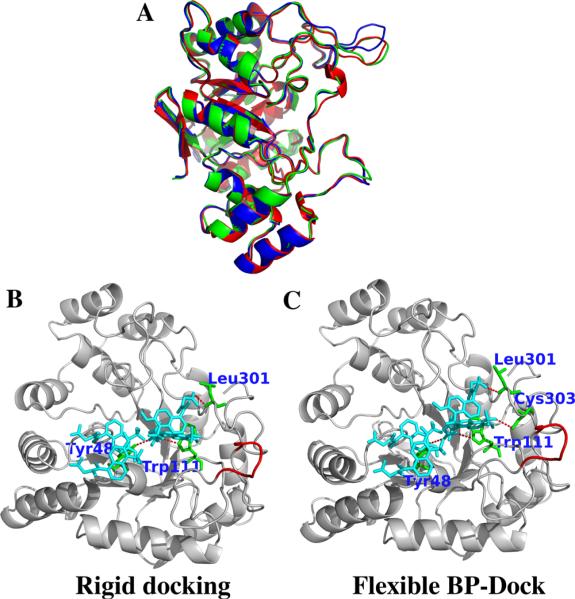
For proteins such as aldose reductase, the bound and unbound conformations do not necessarily have a large RMSD difference; however, loops and regions near the binding pocket may differ significantly. These loops are often related to diverse biological functions that can change their conformation upon ligand binding. For example, the bound (PDB id: 2FZB) and unbound (PDB id: 2ACR) structures of aldose reductase (AR) are quite similar to each other with a RMSD of 0.36 Å; yet there is a significant difference in the loop region near the binding pocket (residues 121–130) (Figure 3A). The all-atom RMSD of the loop between the bound and unbound conformation is ~0.6 Å. Upon applying perturbations (i.e., an external Brownian kick) to the unbound structure of AR and computing response fluctuation profiles of the whole chain, we generate an ensemble of conformations that mimics the complete ligand-binding event. Interestingly, one conformation in the ensemble is very similar to the native bound-like conformation as shown in green in Figure 3A, where the loop perfectly aligns with the bound conformation (shown in red). This indicates the capability of our flexible approach in correctly predicting the binding-induced conformational changes when an unbound form of protein is used, even without the presence of any ligand. Moreover, it also shows that BP-Dock is distinctly different from other multiple receptor docking approaches based on normal mode analysis. Indeed, a recent study has shown that selecting the most relevant mode/modes related to binding is rather difficult in those approaches and makes the method more restricted because some higher frequency modes can be responsible for binding-induced conformational changes.39 However, with the BP-Dock approach, the most relevant modes are automatically induced by perturbing the individual residues of the receptor; therefore, we do not need to search for correct modes that are most related to binding.
Figure 3.
(A) Bound (2FZB) and unbound (2ACR) ribbon diagrams of aldose reductase are shown in red and blue, respectively. The RMSD for the loop region (encircled, Phe121–Val130) between the bound and unbound structures is ~0.6 Å. One of the perturbed conformations using the BP-Dock scheme (shown in green) is similar to the bound structure, especially around the specified loop. Hydrogen bond interactions are shown of the best docked poses from unbound docking of four tolrestat molecules with (B) rigid docking and (C) with flexible BP-Dock. The specified loop region is colored red. The hydrogen bond interactions are studied with Chimera,76 and the images are prepared using PyMOL.60
The flexible BP-Dock results for docking of four tolrestat molecules (TOL4) to unbound AR shows a binding energy prediction of −8.96 kcal/mol (RosettaLigand score, −876.52 kcal/mol), even more favorable than the rigid docking prediction of the bound structure (X-Score, −8.73 kcal/mol; RosettaLigand score, −782.2 kcal/mol). On the other hand, the rigid docking of unbound AR leads to a less favorable binding energy score for TOL4 (X-Score, −8.04 kcal/mol; RosettaLigand score, −721.69 kcal/mol). When we compare the docked poses of unbound conformation from rigid and flexible docking, we observe that the ligand forms only three hydrogen bonds in the case of rigid unbound docking (Figure 3B), whereas TOL4 forms four hydrogen bonds with Tyr 48, Trp111, Leu301, and Cys303 in the case of BP-Dock docking (Figure 3C). The loss of a hydrogen bond could possibly explain a less favorable binding energy score for rigid unbound docking, as these residues have been shown critical for binding.66,67 Previously, Sotriffer et al.66 have shown that the specificity binding region of AR, constituted by the residues Leu300, Trp111, and Thr113, can only be accessible to ligand by the correct orientation of Leu300.66 Interestingly, in the analysis of the BP-Dock pose, we observe that the side chain of Leu300 and Trp111 get shifted to open up a wider space in the binding pocket compared to that of the unbound complex, thus avoiding any clashes with TOL4. This emphasizes that by introducing perturbations and computing the response, our approach may have led to this specific orientation change, which in return made the binding site of aldose reductase much more feasible and approachable to tolrestat, especially near the specificity region.
We also analyze large proteins having different conformational changes upon binding to several different ligands such as carboxypeptidase (CPA) (307 residues) and trypanosomal triosephosphate isomerase (TIM) (250 residues in each chain A and B). For both of these test cases, the previous docking studies have failed in predicting the correct binding poses.10,23 From our docking results, we observe that the RosettaLigand energy scores for rigid bound docking for CPA indicate higher affinity for l-phenyl lactate (or HFA) than for the FVF ligand, in contradiction to experimental results.68,69 On the other hand, RosettaLigand energy scores of flexible BP-Dock for the unbound structure of CPA are in agreement with the experimental observations (Table II). However, rescoring the lowest binding energy poses with X-Score helps in correctly estimating the binding energy preferences for all three types of dockings (rigid bound, rigid unbound, and BP-Dock unbound). Nonetheless, for the SH3 domain of GRB2 and cyclophilin A, we also observe that the RosettaLigand energy scores for rigid docking of the bound structures are less favorable than those for the rigid unbound structures, but rescoring helps again in correcting this anomaly. Furthermore, for the TIM protein, both the RosettaLigand energy scores as well as X-Scores for rigid bound docking fail in correctly predicting the binding affinities for two ligands, 2-phosphoglycerate (2PG) and glycerol-3-phosphate (G3P), whereas BP-Dock succeeds in correctly predicting these differences.70
Analysis of PDZ Domains
We also test whether we can predict the binding selectivities of several PDZ domain proteins (PDZs), where the backbone dynamics are crucial in binding affinity predictions.6,42,71,72 PDZ domains have been categorized into three main classes according to the specificity of the interaction depending on its C-terminal four amino acids of their binding peptides. Class I type PDZs bind to a C-terminal motif with the sequence [X-Ser/Thr-X-U-COOH]. Class II type PDZs prefer the sequence [X-U-X-U-COOH]. Class III type binds to the sequence [X-Glu/Asp-X-U-COOH], where X is any amino acid and U is a hydrophobic amino acid. Although the PDZ binding site is well defined and PDZ motifs are classified based on their sequence type, there is still little information available on the binding affinity and stoichiometry of PDZ binding motifs and blocking peptides.73 We focus on the most common Class I and Class II types for this study.
Among our test PDZ cases, PSD-95 binds to a Class I peptide, whereas GRIP binds to a Class II.6 The docking results for PDZ domain proteins (Table II) show that the rigid unbound docking fails to predict bound-like affinities for both PSD-95 and GRIP. However, BP-Dock unbound docking shows similar or more favorable (for PSD-95) binding energy scores than rigid bound docking. Furthermore, BP-Dock results on the PDZ2 domain of syntenin indicate that it has dual specificity for Class I (IL5R-α) and Class II (syndecan) peptides with a slightly higher affinity toward the Class II peptide. This result is also consistent with experimental observations indicating that it binds slightly better to syndecan (Kd ~ 2.9 μM) than to IL5R-α (Kd ~ 43.8 μM).74 Moreover, we are able to predict that the Merlin (−FFEEL) peptide has the least significant affinity toward the PDZ2 domain of syntenin as is also shown experimentally (Kd ~ 1 mM).74
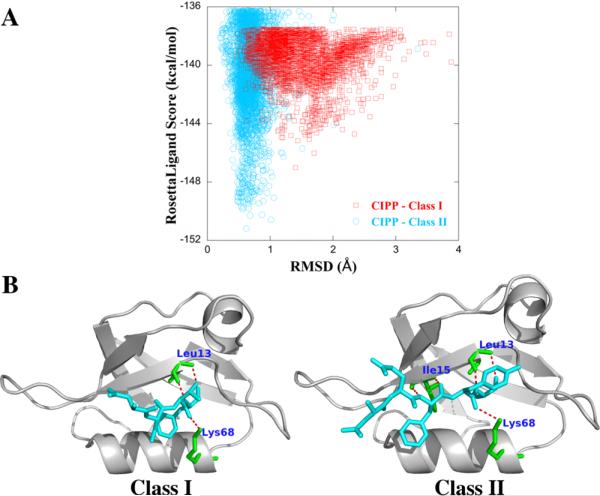
Furthermore, we also look at the binding selectivities of the homologue structure CIPP. One of the most difficult tasks for any docking protocol is to correctly predict the binding affinities for homologue structures. Therefore, in this study, we apply our flexible docking approach to a homology model for CIPP of PDZ, whose binding selectivity has already been verified experimentally.75 Table III shows the lowest RosettaLigand energy scores and X-Scores of the modeled CIPP with Class I (CRIPT and IL5R-α) and Class II (syndecan and Erbin) peptides for rigid and our flexible docking method. The RMSD (Å) values of the lowest energy docked poses from rigid and flexible BP-Dock docking of homologue CIPP are given in Table S3 of the Supporting Information. Figure 4A shows the RosettaLigand energy score vs the RMSD plot for the BP-Dock complex of CIPP with Class I (IL5R-α) and Class II (syndecan) peptides. The formation of a well-converged distinct binding funnel in energy score/RMSD plots indicate successful docking.10 CIPP prefers to bind syndecan with a higher affinity of −6.92 kcal/mol as compared to −6.33 kcal/mol for IL5R-α, which is in agreement with experimental results.75 Further analysis of CIPP complex with both Class I and Class II peptides shows that CIPP residues form hydrogen bonds with both IL5R-α (Class I) and syndecan (Class II) as shown in Figure 4B. The syndecan peptide forms five hydrogen bonds via the interaction of three crucial residues Leu 13, Ile 15, and Lys 68. However, the IL5R-α peptide forms only three hydrogen bonds with Leu 13 and Lys 68. The lesser number of hydrogen bonds formed by the IL5R-α peptide could possibly explain its lower binding affinity with CIPP.
Table III.
Rigid and Flexible Docking Predictions for Homologue Structure of CIPP
| RosettaLigand score (kcal/mol) |
X-Score (kcal/mol) |
|||||
|---|---|---|---|---|---|---|
| homologue protein | peptide | peptide class | rigid unbound | BP-Dock unbound | rigid unbound | BP-Dock unbound |
| CIPP | KQTSV | I | –135.93 | –150.73 | –7.07 | –7.08 |
| DSVF | I | –130.02 | –147 | –6.43 | –6.33 | |
| NEFYA | II | –136.26 | –151.18 | –7.09 | –6.92 | |
| EYLGLDVPV | II | –137.92 | –151.87 | –7.15 | –7.49 | |
Figure 4.
(A) RosettaLigand energy score vs RMSD of the docked complex of CIPP with Class I and Class II peptides of syntenin. (B) Hydrogen bond interactions of CIPP with Class I and Class II peptides are shown as analyzed using the best docked pose obtained from flexible BP-Dock docking. Ribbon diagrams are prepared using PyMOL.60
Flexible Docking Comparison for Ensembles Generated with BP-Dock and RosettaBackrub
Another commonly used method to incorporate backbone movements in Rosetta is the “Backrub” method.61,62 This method randomly perturbs a segment of 2–12 residues through a rigid body rotation by an angle of up to 11–40° to model the conformational changes in a protein.61,62 For comparison, we also generate ensembles of PDZs using the RosettaBackrub server,29 and we use these ensembles for docking with RosettaLigand. Table IV shows the docking results of six PDZ–ligand pairs for the ensembles generated by both Backrub and BP-Dock, and the RMSDs of the ligand from the lowest energy docked poses are reported in Table S4 of the Supporting Information. We observe that by using BP-Dock, we can discriminate the higher binding preferences of syntenin toward syndecan (i.e., the binding energies evaluated from X-Score for the lowest RosettaLigand score complexes are −7.85 kcal/mol and −7.16 kcal/mol for syndecan and IL5R-α peptides, respectively). However, Backrub ensemble docking fails in estimating the binding preferences of CIPP for IL5R-α and syndecan peptides (i.e., the binding energies of the lowest score docked poses are −6.99 kcal/mol for syndecan peptide and −7.18 kcal/mol for IL5R-α peptide).
Table IV.
Comparison of Flexible Docking for Ensembles Generated with BP-Dock vs Backrub
| RosettaLigand score (kcal/mol) |
X-Score (kcal/mol) |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| protein | peptide | peptide classification |
peptide binding specificity |
rigid bound |
BP-Dock unbound |
Backrub unbound |
rigid bound |
BP-Dock unbound |
Backrub unbound |
| PSD-95 | KQTSV | I | I | –230.78 | –273.03 | –221.83 | –7.27 | –7.32 | –6.66 |
| PSD-95 | ATVRTYSC | II | I | NA | –271 | –223.04 | NA | –7.1 | –7.31 |
| GRIP | ATVRTYSC | II | II | –241.76 | –229.36 | –218.37 | –8.14 | –7.82 | –7.74 |
| GRIP | KQTSV | I | II | NA | –223.97 | –215.32 | NA | –7.1 | –7.15 |
| syntenin | DSVF | I | I and II both | –208.24 | –205.67 | –219.09 | –7.29 | –7.16 | –7.18 |
| syntenin | NEFYA | II | 1 and II both | –225.2 | –211.88 | –219.57 | –7.99 | –7.85 | –6.99 |
| CIPP | KQTSV | I | II | NA | –150.73 | –149.6 | NA | –7.08 | –7.23 |
| CIPP | EYLGLDVPV | II | II | NA | –151.87 | –154.6 | NA | –7.49 | –7.29 |
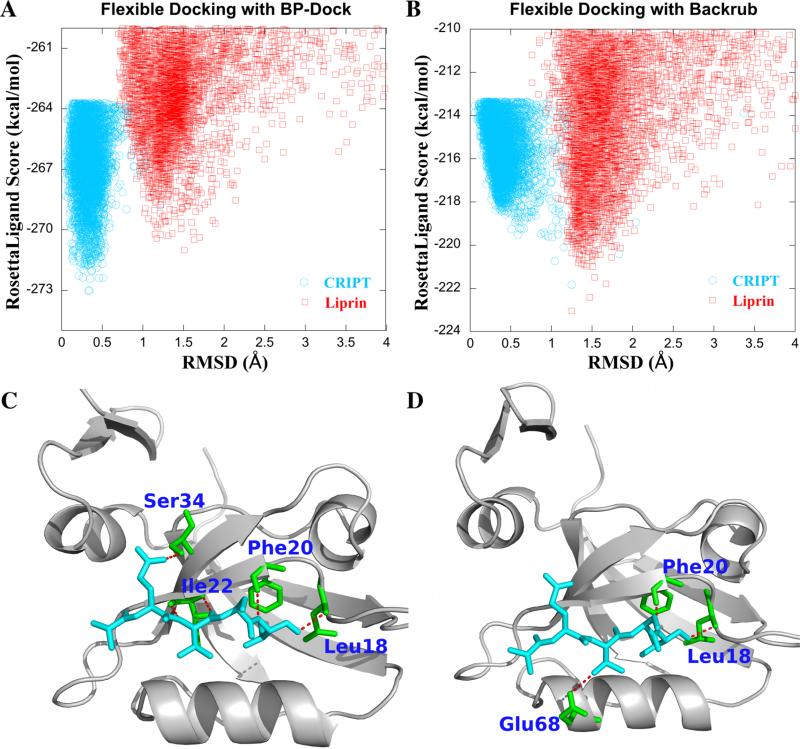
For a more rigorous comparison, we also perform cross-docking on PSD-95 and GRIP. We select liprin, a Class II peptide for docking to PSD-95 and CRIPT, a Class I peptide for docking to GRIP. As shown in Table IV, the binding energy of a BP-Dock pose obtained with X-Score indicates a higher affinity for the Class I peptide compared to the Class II peptide. Similarly, GRIP prefers a Class II peptide in comparison to a Class I peptide. Overall, BP-Dock ensemble docking is successful in predicting the binding affinities of PSD-95 and GRIP. Backrub ensemble docking also correctly predicts the preference of GRIP toward a Class II peptide. However, in the case of PSD-95, the binding energy scores indicate a higher affinity for a Class II peptide (−7.31 kcal/mol) over a Class I peptide (−6.66 kcal/mol), contradictory to the results from previous studies.6 Figure 5A and B shows the self-docking and cross-docking energy score/RMSD plots of PSD-95 when docked using the ensembles of (i) BP-Dock and (ii) Backrub. In order to further investigate the difference in binding energy scores for the two flexible ensemble dockings, we analyze the hydrogen bond pattern of the complexes obtained from these two separate ensemble docking. The lowest energy complex obtained from BP-Dock shows that P-0 and P-2 residues of the Class I peptide form five hydrogen bonds with Leu18, Phe20, Ile22, and Ser34 of PSD-95 (Figure 5C). On the other hand, the docked pose with the lowest energy score obtained from Backrub indicates that the peptide forms only three hydrogen bonds with Leu18, Phe20, and Glu68 (Figure 5D). Moreover, analysis of cross-docking with the Backrub ensemble shows that the number of hydrogen bonds has been increased to five in the case of the Class II peptide docked pose. This may be attributed to an increase in the binding affinity of PSD-95 for the Class II peptide when the Backrub ensemble is used for docking. Overall, this comparison suggests that in unbound ensemble docking, accuracy in predicting binding affinity increases when the ensemble consists of correct binding-induced conformations. Indeed, Backrub and BP-Dock can be merged to increase overall accuracy.
Figure 5.
RosettaLigand energy score vs RMSD of PSD-95 with CRIPT and liprin peptides obtained from the flexible docking with (A) BP-Dock and (B) Backrub ensembles. Hydrogen bond interactions are shown for the CRIPT peptide with PSD-95 as analyzed using the lowest energy docked pose obtained from (C) BP-Dock and (D) Backrub ensembles. Ribbon diagrams are prepared using PyMOL.60
CONCLUSION
Incorporation of backbone flexibility helps in sampling the bound-like conformation, which is crucial for accurate complex geometry and binding affinity predictions especially for docking with unbound structures. This is because small conformational changes in the backbone upon protein binding can lead to significant changes in the side-chain orientations. Our analysis of unbound docking in comparison with bound docking suggests that conformations generated through perturbations should simulate similar changes that occur when a ligand interacts with the receptor during the binding event in order to increase the accuracy of docking. The most intriguing aspect of the BP-Dock approach is that we are able to mimic the induced effects of peptide–ligand binding using the unbound structure, even in the absence of any ligand or peptide. Thus, the BP-Dock approach can be utilized to increase the accuracy of binding scores when unbound structures or unbound models are used.
Supplementary Material
ACKNOWLEDGMENTS
We gratefully acknowledge Brandon Mac Butler for his valuable comments on the manuscript. S.B.O. and A.B. acknowledge funding support from NSF-MCB Grant 1121276. We also thank A2C2 at Arizona State University and Xsede for computer time.
Footnotes
ASSOCIATED CONTENT
Supporting Information
Table S1: PDB codes of unbound and bound structures, chain length, RMSDs between bound and unbound structures, native peptides or ligands, and peptide sequences for the proteins used in this study. Table S2: RMSD (Å) values between the ligand positions of the lowest energy docked poses obtained from rigid bound, rigid unbound, and BP-Dock docking and that of the bound crystal structure. Only heavy atoms of the ligand are taken into account for RMSD calculation. Table S3: RMSD (Å) values between the ligand positions of the lowest energy docked poses obtained from rigid and flexible BP-Dock docking of homologue CIPP and that of the bound crystal structure. Table S4: RMSD (Å) values between the ligand positions of the lowest energy docked poses obtained from rigid bound, BP-Dock unbound, and Backrub unbound docking of PDZ domain proteins and that of the holo crystal structure. Figure S1: Correlation plots of RosettaLigand energy scores vs experimental binding energies for HIV-1 protease (PR), carbonic anhydrase II (CA II), alcohol dehydrogenase (AD), alpha-thrombin (AT), and cytochrome C peroxidase (CCP). This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
REFERENCES
- 1.Andrusier N, Mashiach E, Nussinov R, Wolfson HJ. Principles of flexible protein–protein docking. Proteins. 2008;73:271–289. doi: 10.1002/prot.22170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McGovern SL, Shoichet BK. Information decay in molecular docking screens against holo, apo and modeled conformation of enzymes. J. Med. Chem. 2003;46:2895–2907. doi: 10.1021/jm0300330. [DOI] [PubMed] [Google Scholar]
- 3.Kitchen DB, Decornez H, Furr JR, Bajorath J. Docking and scoring in virtual screening for drug discovery: Methods and applications. Nat. Rev. Drug Discovery. 2004;3:935–949. doi: 10.1038/nrd1549. [DOI] [PubMed] [Google Scholar]
- 4.Kuntz ID, Blaney JM, Oatley SJ, Langridge R, Ferrin TE. A geometric approach to macromolecule–ligand interactions. J. Mol. Biol. 1982;161:269–288. doi: 10.1016/0022-2836(82)90153-x. [DOI] [PubMed] [Google Scholar]
- 5.Zacharias M. Accounting for conformational changes during protein–protein docking. Curr. Opin. Struct. Biol. 2010;20:180–186. doi: 10.1016/j.sbi.2010.02.001. [DOI] [PubMed] [Google Scholar]
- 6.Gerek ZN, Ozkan SB. A flexible docking scheme to explore the binding selectivity of PDZ domains. Protein Sci. 2010;19:914–928. doi: 10.1002/pro.366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Totrov M, Abagyan R. Flexible ligand docking to multiple receptor conformations: A practical alternative. Curr. Opin. Struct. Biol. 2008;18:178–184. doi: 10.1016/j.sbi.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bienstock RJ. Computational drug design targeting protein–protein interactions. Curr. Pharm. Des. 2012;18:1240–1254. doi: 10.2174/138161212799436449. [DOI] [PubMed] [Google Scholar]
- 9.Lexa KW, Carlson HA. Protein flexibility in docking and surface mapping. Q. Rev. Biophys. 2012;45:301–343. doi: 10.1017/S0033583512000066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meiler J, Baker D. RosettaLigand: Protein–small molecule docking with full side-chain flexibility. Proteins. 2006;65:538–548. doi: 10.1002/prot.21086. [DOI] [PubMed] [Google Scholar]
- 11.Huang SY, Zou X. Ensemble docking of multiple protein structures: Considering protein structural variations in molecular docking. Proteins. 2007;66:399–421. doi: 10.1002/prot.21214. [DOI] [PubMed] [Google Scholar]
- 12.Sherman W, Beard HS, Farid R. Use of an induced fit receptor structure in virtual screening. Chem. Biol. Drug Des. 2006;67:83–84. doi: 10.1111/j.1747-0285.2005.00327.x. [DOI] [PubMed] [Google Scholar]
- 13.Sherman W, Day T, Jacobson MP, Friesner RA, Farid R. Novel procedure for modeling ligand/receptor induced fit effects. J. Med. Chem. 2006;49:534–553. doi: 10.1021/jm050540c. [DOI] [PubMed] [Google Scholar]
- 14.Claussen H, Buning C, Rarey M, Lengauer T. FlexE: Efficient molecular docking considering protein structure variations. J. Mol. Biol. 2001;308:377–395. doi: 10.1006/jmbi.2001.4551. [DOI] [PubMed] [Google Scholar]
- 15.Nabuurs SB, Wagener M, de Vlieg J. A flexible approach to induced fit docking. J. Med. Chem. 2007;50:6507–6518. doi: 10.1021/jm070593p. [DOI] [PubMed] [Google Scholar]
- 16.Zhao Y, Sanner MF. FLIPDock: Docking flexible ligands into flexible receptors. Proteins. 2007;68:726–737. doi: 10.1002/prot.21423. [DOI] [PubMed] [Google Scholar]
- 17.Zhao Y, Sanner MF. Protein–ligand docking with multiple flexible side chains. J. Comput.-Aided Mol. Des. 2008;22:673–679. doi: 10.1007/s10822-007-9148-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zacharias M. Rapid protein–ligand docking using soft modes from molecular dynamics simulations to account for protein deformability: Binding of FK506 to FKBP. Proteins. 2004;54:759–767. doi: 10.1002/prot.10637. [DOI] [PubMed] [Google Scholar]
- 19.Zacharias M. Combining elastic network analysis and molecular dynamics simulations by Hamiltonian replica exchange. J. Chem. Theory Comput. 2008;4:477–487. doi: 10.1021/ct7002258. [DOI] [PubMed] [Google Scholar]
- 20.Corbeil CR, Englebienne P, Moitessier N. Docking ligands into flexible and solvated macromolecules. 1. Development and validation of FITTED 1.0. J. Chem. Inf. Model. 2007;47:435–449. doi: 10.1021/ci6002637. [DOI] [PubMed] [Google Scholar]
- 21.Corbeil CR, Englebienne P, Yannopoulos CG, Chan L, Das SK, Bilimoria D, L'heureux L, Moitessier N. Docking ligands into flexible and solvated macromolecules. 2. Development and application of fitted 1.5 to the virtual screening of potential HCV polymerase inhibitors. J. Chem. Inf. Model. 2008;48:902–909. doi: 10.1021/ci700398h. [DOI] [PubMed] [Google Scholar]
- 22.Corbeil CR, Moitessier N. Docking ligands into flexible and solvated macromolecules. 3. Impact of input ligand conformation, protein flexibility, and water molecules on the accuracy of docking programs. J. Chem. Inf. Model. 2009;49:997–1009. doi: 10.1021/ci8004176. [DOI] [PubMed] [Google Scholar]
- 23.Davis IW, Baker D. RosettaLigand docking with full ligand and receptor flexibility. J. Mol. Biol. 2009;385:381–392. doi: 10.1016/j.jmb.2008.11.010. [DOI] [PubMed] [Google Scholar]
- 24.Sandak B, Wolfson HJ, Nussinov R. Flexible docking allowing induced fit in proteins. Proteins. 1998;32:159–174. [PubMed] [Google Scholar]
- 25.Sandak B, Nussinov R, Wolfson HJ. A method for biomolecular structural recognition and docking allowing conformational flexibility. J. Comp. Biol. 1998;5:631–654. doi: 10.1089/cmb.1998.5.631. [DOI] [PubMed] [Google Scholar]
- 26.Schneidman-Duhovny D, Inbar Y, Nussinov R, Wolfson HJ. Geometry based flexible and symmetric protein docking. Proteins. 2005;60:224–231. doi: 10.1002/prot.20562. [DOI] [PubMed] [Google Scholar]
- 27.Schneidman-Duhovny D, Nussinov R, Wolfson HJ. Automatic prediction of protein interactions with large scale motion. Proteins. 2007;69:764–773. doi: 10.1002/prot.21759. [DOI] [PubMed] [Google Scholar]
- 28.Mashiach E, Schneidman-Duhovny D, Peri A, Shavit Y, Nussinov R, Wolfson HJ. An integrated suite of fast docking algorithms. Proteins. 2010;78:3197–3204. doi: 10.1002/prot.22790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lauck F, Smith CA, Friedland GD, Humphris EL, Kortemme T. RosettaBackrub: A web server for flexible backbone protein structure modeling and design. Nucleic Acids Res. 2010;38:W569–W575. doi: 10.1093/nar/gkq369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ding F, Dokholyan NV. Incorporating backbone flexibility in MedusaDock improves ligand-binding pose prediction in the CSAR2011 docking benchmark. J. Chem. Inf. Model. 2013;53:1871–1879. doi: 10.1021/ci300478y. [DOI] [PubMed] [Google Scholar]
- 31.Osterberg F, Morris GM, Sanner MF, Olson AJ, Goodsell DS. Automated docking to multiple target structures: incorporation of protein mobility and structural water heterogeneity in AutoDock. Proteins. 2002;46:34–40. doi: 10.1002/prot.10028. [DOI] [PubMed] [Google Scholar]
- 32.Cavasotto CN, Abagyan RA. Protein flexibility in ligand docking and virtual screening to protein kinases. J. Mol. Biol. 2004;337:209–225. doi: 10.1016/j.jmb.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 33.Korb O, Olsson TS, Bowden SJ, Hall RJ, Verdonk ML, Liebeschuetz JW, Cole JC. Potential and limitations of ensemble docking. J. Chem. Inf. Model. 2012;52:1262–1274. doi: 10.1021/ci2005934. [DOI] [PubMed] [Google Scholar]
- 34.Barril X, Morley SD. Unveiling the full potential of flexible receptor docking using multiple crystallographic structures. J. Med. Chem. 2005;48:4432–4443. doi: 10.1021/jm048972v. [DOI] [PubMed] [Google Scholar]
- 35.Damm KL, Carlson HA. Exploring experimental sources of multiple protein conformations in structure-based drug design. J. Am. Chem. Soc. 2007;129:8225–8235. doi: 10.1021/ja0709728. [DOI] [PubMed] [Google Scholar]
- 36.Philippopoulos M, Lim C. Exploring the dynamic information content of a protein NMR structure: Comparison of a molecular dynamics simulation with the NMR and X-ray structures of Escherichia coli ribonuclease HI. Proteins. 1999;36:87–110. doi: 10.1002/(sici)1097-0134(19990701)36:1<87::aid-prot8>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 37.Carlson HA, McCammon JA. Accommodating protein flexibility in computational drug design. Mol. Pharmacol. 2000;57:213–218. [PubMed] [Google Scholar]
- 38.Kuzu G, Gursoy A, Nussinov R, Keskin O. Exploiting conformational ensembles in modeling protein–protein interactions on the proteome scale. J. Proteome Res. 2013;12:2641–2653. doi: 10.1021/pr400006k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dietzen M, Zotenko E, Hildebrandt A, Lengauer T. On the applicability of elastic network normal modes in small-molecule docking. J. Chem. Inf. Model. 2012;52:844–856. doi: 10.1021/ci2004847. [DOI] [PubMed] [Google Scholar]
- 40.Cavasotto CN, Kovacs JA, Abagyan RA. Representing receptor flexibility in ligand docking through relevant normal modes. J. Am. Chem. Soc. 2005;127:9632–9640. doi: 10.1021/ja042260c. [DOI] [PubMed] [Google Scholar]
- 41.Atilgan C, Gerek ZN, Ozkan SB, Atilgan AR. Manipulation of conformational change in proteins by single-residue perturbations. Biophys. J. 2010;99:933–943. doi: 10.1016/j.bpj.2010.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bolia A, Gerek ZN, Keskin O, Banu Ozkan S, Dev KK. The binding affinities of proteins interacting with the PDZ domain of PICK1. Proteins. 2012;80:1393–1408. doi: 10.1002/prot.24034. [DOI] [PubMed] [Google Scholar]
- 43.Gerek ZN, Ozkan SB. Change in allosteric network affects binding affinities of PDZ domains: Analysis through perturbation response scanning. PLoS Comput. Biol. 2011;7:e1002154. doi: 10.1371/journal.pcbi.1002154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Atilgan C, Atilgan AR. Perturbation-response scanning reveals ligand entry–exit mechanisms of ferric binding protein. PLoS Comput. Biol. 2009;5:e1000544. doi: 10.1371/journal.pcbi.1000544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Atilgan AR, Durell SR, Jernigan RL, Demirel MC, Keskin O, Bahar I. Anisotropy of fluctuation dynamics of proteins with an elastic network model. Biophys. J. 2001;80:505–515. doi: 10.1016/S0006-3495(01)76033-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ikeguchi M, Ueno J, Sato M, Kidera A. Protein structural change upon ligand binding: Linear response theory. Phys. Rev. Lett. 2005;94:078102. doi: 10.1103/PhysRevLett.94.078102. [DOI] [PubMed] [Google Scholar]
- 47.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Roche O, Kiyama R, Brooks CL., 3rd Ligand-protein database: linking protein–ligand complex structures to binding data. J. Med. Chem. 2001;44:3592–3598. doi: 10.1021/jm000467k. [DOI] [PubMed] [Google Scholar]
- 49.Hartshorn MJ, Verdonk ML, Chessari G, Brewerton SC, Mooij WTM, Mortenson PN, Murray CW. Diverse, high-quality test set for the validation of protein–ligand docking performance. J. Med. Chem. 2007;50:726–741. doi: 10.1021/jm061277y. [DOI] [PubMed] [Google Scholar]
- 50.Hinsen K. Analysis of domain motions by approximate normal mode calculations. Proteins. 1998;33:417–429. doi: 10.1002/(sici)1097-0134(19981115)33:3<417::aid-prot10>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 51.Tirion MM. Large Amplitude elastic motions in proteins from a single- parameter, atomic analysis. Phys. Rev. Lett. 1996;77:1905–1908. doi: 10.1103/PhysRevLett.77.1905. [DOI] [PubMed] [Google Scholar]
- 52.Yang L, Song G, Jernigan RL. Protein elastic network models and the ranges of cooperativity. Proc. Natl. Acad. Sci. U.S.A. 2009;106:12347–12352. doi: 10.1073/pnas.0902159106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bahar I, Lezon TR, Yang LW, Eyal E. Global dynamics of proteins: Bridging between structure and function. Annu. Rev. Biophys. 2010;39:23–42. doi: 10.1146/annurev.biophys.093008.131258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.MacQueen JB. Some Methods for Classification and Analysis of Multivariate Observations. Proceedings of 5th Berkeley Symposium on Mathematical Statistics and Probability; Berkeley, CA. 1967; University of California Press; pp. 281–297. [Google Scholar]
- 55.Hornak V, Abel R, Okur A, Strockbine B, Roitberg A, Simmerling C. Comparison of multiple amber force fields and development of improved protein backbone parameters. Proteins. 2006;65:712–725. doi: 10.1002/prot.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tsui V, Case DA. Molecular dynamics simulations of nucleic acids with a generalized Born solvation model. J. Am. Chem. Soc. 2000;122:2489–2498. [Google Scholar]
- 57.Wang R, Lai L, Wang S. Further development and validation of empirical scoring functions for structure-based binding affinity prediction. J. Comput.-Aided Mol. Des. 2002;16:11–26. doi: 10.1023/a:1016357811882. [DOI] [PubMed] [Google Scholar]
- 58.Wang R, Lu Y, Wang S. Comparative evaluation of 11 scoring functions for molecular docking. J. Med. Chem. 2003;46:2287–2303. doi: 10.1021/jm0203783. [DOI] [PubMed] [Google Scholar]
- 59.Sali A, Blundell TL. Comparative protein modeling by satisfaction of spatial restraints. J. Mol. Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 60.The PyMOL Molecular Graphics System, Version 1.3. Schrödinger, LLC; New York: [Google Scholar]
- 61.Friedland GD, Lakomek NA, Griesinger C, Meiler J, Kortemme T. A correspondence between solution-state dynamics of an individual protein and the sequence and conformational diversity of its family. PLoS Comput. Biol. 2009;5:e1000393. doi: 10.1371/journal.pcbi.1000393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Davis IW, Arendall WB, 3rd, Richardson DC, Richardson JS. The backrub motion: How protein backbone shrugs when a sidechain dances. Structure. 2006;14:265–274. doi: 10.1016/j.str.2005.10.007. [DOI] [PubMed] [Google Scholar]
- 63.Österberg F, Morris GM, Sanner MF, Olson AJ, Goodsell DA. Automated docking to multiple target structures: Incorporation of protein mobility and structural water heterogeneity in AutoDock. Proteins. 2002;46:34–40. doi: 10.1002/prot.10028. [DOI] [PubMed] [Google Scholar]
- 64.Shin W, Seok C. GalaxyDock: Protein–ligand docking with flexible protein side-chains. J. Chem. Inf. Model. 2012;52:3225–3232. doi: 10.1021/ci300342z. [DOI] [PubMed] [Google Scholar]
- 65.Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sotriffer CA, Kramer O, Klebe G. Probing flexibility and “induced-fit” phenomena in aldose reductase by comparative crystal structure analysis and molecular dynamics simulations. Proteins. 2004;56:52–66. doi: 10.1002/prot.20021. [DOI] [PubMed] [Google Scholar]
- 67.Steuber H, Zentgraf M, Gerlach C, Sotriffer CA, Heine A, Klebe G. Expect the unexpected or caveat for drug designers: Multiple structure determinations using aldose reductase crystals treated under varying soaking and co-crystallisation conditions. J. Mol. Biol. 2006;363:174–187. doi: 10.1016/j.jmb.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 68.Teplyakov A, Wilson KS, Orioli P, Mangani S. High-resolution structure of the complex between carboxypeptidase A and L-phenyl lactate. Acta Crystallogr., Sect. D: Biol. Crystallogr. 1993;49:534–540. doi: 10.1107/S0907444993007267. [DOI] [PubMed] [Google Scholar]
- 69.Kim H, Lipscomb WN. Comparison of the structures of three carboxypeptidase A-phosphonate complexes determined by X-ray crystallography. Biochemistry. 1991;30:8171–8180. doi: 10.1021/bi00247a012. [DOI] [PubMed] [Google Scholar]
- 70.Noble ME, Wierenga RK, Lambeir AM, Opperdoes FR, Thunnissen AM, Kalk KH, Groendijk H, Hol WG. The adaptability of the active site of trypanosomal triosephosphate isomerase as observed in the crystal structures of three different complexes. Proteins. 1991;10:50–69. doi: 10.1002/prot.340100106. [DOI] [PubMed] [Google Scholar]
- 71).Gerek ZN, Keskin O, Ozkan SB. Identification of specificity and promiscuity of PDZ domain interactions through their dynamic behavior. Proteins. 2009;77:796–811. doi: 10.1002/prot.22492. [DOI] [PubMed] [Google Scholar]
- 72.Smith CA, Kortemme T. Structure-based prediction of the peptide sequence space recognized by natural and synthetic PDZ domains. J. Mol. Biol. 2010;402:460–474. doi: 10.1016/j.jmb.2010.07.032. [DOI] [PubMed] [Google Scholar]
- 73.Songyang Z, Fanning AS, Fu C, Xu J, Marfatia SM, Chishti AH, Crompton A, Chan AC, Anderson JM, Cantley LC. Recognition of unique carboxyl-terminal motifs by distinct PDZ domains. Science. 1997;275:73–77. doi: 10.1126/science.275.5296.73. [DOI] [PubMed] [Google Scholar]
- 74.Kang BS, Cooper DR, Jelen F, Devedjiev Y, Derewenda U, Dauter Z, Otlewski J, Derewenda ZS. PDZ tandem of human syntenin: Crystal structure and functional properties. Structure. 2003;11:459–468. doi: 10.1016/s0969-2126(03)00052-2. [DOI] [PubMed] [Google Scholar]
- 75.Kang BS, Cooper DR, Devedjiev Y, Derewenda U, Derewenda ZS. Molecular roots of degenerate specificity in syntenin's PDZ2 domain: Reassessment of the PDZ recognition paradigm. Structure. 2003;11:845–853. doi: 10.1016/s0969-2126(03)00125-4. [DOI] [PubMed] [Google Scholar]
- 76.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.