Abstract
To provide a useful community resource for orthogonal assessment of NGS analysis software, we present the ICR142 NGS validation series. The dataset includes high-quality exome sequence data from 142 samples together with Sanger sequence data at 730 sites; 409 sites with variants and 321 sites at which variants were called by an NGS analysis tool, but no variant is present in the corresponding Sanger sequence. The dataset includes 286 indel variants and 275 negative indel sites, and thus the ICR142 validation dataset is of particular utility in evaluating indel calling performance. The FASTQ files and Sanger sequence results can be accessed in the European Genome-phenome Archive under the accession number EGAS00001001332.
Keywords: Variant calling, next-generation sequencing, NGS, exome, indel, validation
Introduction
Next-generation sequencing (NGS) approaches have greatly enhanced our ability to detect genetic variation. Over the past decade NGS hardware, software, throughput, data quality and analytical tools have evolved dramatically. Thorough evaluation of each new laboratory and analytical development is challenging but necessary to fully understand how pipeline modification can impact results. To fully assess performance, NGS analysis tools should ideally be run on samples with pre-determined positive and negative sites assessed through orthogonal experimentation such as Sanger sequencing.
Over the past five years, we have generated extensive data on thousands of samples using different NGS instruments, sequencing chemistry, gene panels, exome captures and variant calling tools. Fortuitously, during this process we have generated orthogonal validation data using Sanger sequencing for a core set of 142 samples that were included in the majority of our experiments. We now formally use these samples, which we call the ICR142 NGS validation series, to evaluate NGS variant calling performance after any change to experimental or analytical protocols. This series has proved an extremely useful resource for our assessment of NGS analysis in both the research and clinical settings. We believe that it may also have utility for others, and hence are making it available here.
Materials and methods
We used lymphocyte DNA from 142 unrelated individuals. All individuals were recruited to the BOCS study and have given informed consent for their DNA to be used for genetic research. The study is approved by the London Multicentre Research Ethics Committee (MREC/01/2/18)
Over the last five years we have generated data from the ICR142 validation series using different exome captures which we have analysed with multiple aligner/caller combinations 1– 6. To date we have generated Sanger sequence data for 730 sites amongst the 142 individuals. These sites include variants called by only one aligner and caller combination, increasing the representation of sites which can discriminate performance between methods.
To generate the Sanger sequence data, we performed PCR reactions using the Qiagen Multiplex PCR kit, and bidirectional sequencing of resulting amplicons using the BigDye terminator cycle sequencing kit and an ABI3730 automated sequencer (ABI PerkinElmer). All sequencing traces were analysed with both automated software (Mutation Surveyor version 3.10, SoftGenetics) and visual inspection.
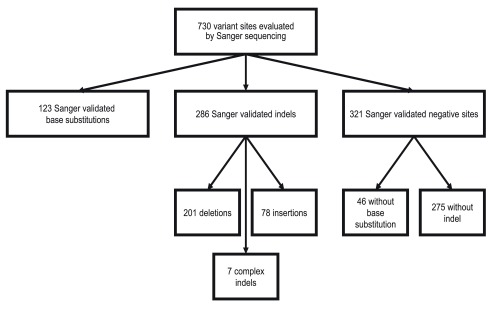
We considered a site negative for a base substitution if the specific base substitution was not present, resulting in 46 negative base substitution sites. We considered a site negative for an indel if no indel, of any kind, was detected in the sequencing trace, resulting in 275 negative indel sites. We annotated confirmed variants with the HGVS-compliant CSN standard using CAVA (version 1.1.0) according to the transcripts designated in Supplementary table 1 7. There were 123 confirmed base substitution variants and 286 confirmed indel variants ( Figure 1, Supplementary table 1).
Figure 1. Description of variant sites evaluated by Sanger sequencing in the ICR142 NGS validation series.
We have also generated high-quality exome sequencing data for the ICR142 NGS validation series. We prepared DNA libraries from 1.5 µg genomic DNA using the Illumina TruSeq sample preparation kit. DNA was fragmented using Covaris technology and the libraries were prepared without gel size selection. We performed target enrichment in pools of six libraries (500 ng each) using the Illumina TruSeq Exome Enrichment kit. The captured DNA libraries were PCR amplified using the supplied paired-end PCR primers. Sequencing was performed with an Illumina HiSeq2000 (SBS Kit v3, one pool per lane) generating 2×101 bp reads. CASAVA v1.8.1 (Illumina) was used to demultiplex and create FASTQ files per sample from the raw base call files.
All of the 730 sites had at least 15× coverage in the exome data, defined as at least 15 reads of good mapping quality (mapping score ≥20). Because these sites are well covered, we can readily assess the variant calling performance of any software tool by applying the pipeline to the exome sequencing data and comparing the variant calls with the Sanger sequencing dataset.
Data availability
The data referenced by this article are under copyright with the following copyright statement: Copyright: © 2016 Ruark E et al.
We have deposited the FASTQ files for all 142 individuals in the European Genome-phenome archive (EGA). The accession number is EGAS00001001332. Details of how to request access to the data are available at: www.icr.ac.uk/icr142.
Researchers and authors that use the ICR142 NGS validation series should reference this paper and should include the following acknowledgement: "This study makes use of the ICR142 NGS validation series data generated by Professor Nazneen Rahman’s team at The Institute of Cancer Research, London”.
Acknowledgements
We are grateful to the Scientific Computing Team at the Institute of Cancer Research for provision of HPC services. We are grateful to Peter Humburg, Andy Rimmer, Manuel Rivas and Peter Donnelly for undertaking some of the aligner/caller comparisons.
Funding Statement
We acknowledge NHS funding to the NIHR Biomedical Research Centre at The Royal Marsden and the ICR. This study was funded by the Institute of Cancer Research, London.
I confirm that the funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
[version 1; referees: 2 approved]
Supplementary material
Supplementary table 1. Sanger sequencing results for 730 sites in the ICR142 NGS validation series. Confirmed variants are annotated according to the designated transcript by CAVA using CSN 7.
The description of the column headings are given below:
Sample – sample name in the ICR142 series
Gene – HGNC symbol
SangerCall – the most 3’ representation annotated with CSN
Type – “bs”, “del”, “ins”, “complex”, or “indel” for base substitutions, simple deletions, simple insertions, complex indels, or negative indel sites, respectively
Transcript – the ENST ID from Ensembl v65 used to annotate the Sanger call
Chr – chromosome
EvaluatedPosition – evaluated hg19 site position, centre of designed amplicon
POS – the left-aligned position in hg19 coordinates for variants called in exome data by Platypus v0.1.5
REF – the reference allele in hg19 for variants called in exome data by Platypus v0.1.5
ALT – the alternative allele in hg19 for variants called in exome data by Platypus v0.1.5
References
- 1. Lunter G, Goodson M: Stampy: a statistical algorithm for sensitive and fast mapping of Illumina sequence reads. Genome Res. 2011;21(6):936–9. 10.1101/gr.111120.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Li H, Durbin R: Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–60. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rivas MA, Beaudoin M, Gardet A, et al. : Deep resequencing of GWAS loci identifies independent rare variants associated with inflammatory bowel disease. Nat Genet. 2011;43(11):1066–73. 10.1038/ng.952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. McKenna A, Hanna M, Banks E, et al. : The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–303. 10.1101/gr.107524.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rimmer A, Mathieson I, Lunter G, et al. : Platypus: An Integrated Variant Caller.2012. Reference Source [Google Scholar]
- 6. SOFTGENETICS: NextgGENe® software for Next Generation (NGS) sequence analysis. Reference Source [Google Scholar]
- 7. Münz M, Ruark E, Renwick A, et al. : CSN and CAVA: variant annotation tools for rapid, robust next-generation sequencing analysis in the clinical setting. Genome Med. 2015;7(1):76. 10.1186/s13073-015-0195-6 [DOI] [PMC free article] [PubMed] [Google Scholar]