Version Changes
Revised. Amendments from Version 1
Figures 2 and 3 in the new version of the article are extended with data which additionally support informational similarity between the E protein from Zika virus and HA1 from the pdmH1N1 influenza virus.
Abstract
The in silico analysis shows that the envelope glycoproteins E of Zika viruses (ZIKV) isolated in Asia, Africa and South and Central America encode highly conserved information determining their interacting profile and immunological properties. Previously it was shown that the same information is encoded in the primary structure of the hemagglutinin subunit 1 (HA1) from pdmH1N1 influenza A virus. This similarity suggests possible repurposing of the seasonal influenza vaccine containing pdmH1N1 component for prevention of the ZIKV infection.
Keywords: Zika virus, vaccine, influenza virus, vaccine repurposing, in silico analysis, hemagglutinin, envelope glycoprotein E, informational spectrum method
Introduction
The recent pandemic of the pdmH1N1 influenza virus and epidemic of the Ebola virus in West Africa showed a lack of preparedness to adequately respond to emerging infectious diseases with potential catastrophic consequences. One of the main obstacles to a fast and efficient response to emerging new infectious disease is insufficient knowledge of the biological, immunological and pathogenic properties of new pathogens and a lack of an appropriate experimental system for testing drugs and vaccines. The new emergence of the ZIKV showed that despite significant progress in molecular biology, biochemistry, immunology, medicine and pharmacology which allows better understanding, prevention and therapy of infectious diseases, the world once again is not prepared for early and decisive action which would prevent hundreds of unnecessary cases of ZIKV infections and potential congenital abnormalities in newborns caused by this virus.
Previously, at the beginning of the HIV epidemic 1, and recently during the swine flu pandemic 2 and Ebola epidemic in West Africa 3, we demonstrated that the bioinformatics tool which is based on the informational spectrum method (ISM) 4 can give some useful information about the host-pathogen interaction and help in selection of drug and vaccine candidates. An essential advantage of ISM over other bioinformatics approaches is in the use of DNA and protein sequences as the only input information which allows analysis of new pathogens. Because data about the sequencing of new pathogens usually are available at the beginning of the outbreaks, the ISM analysis can start immediately and provide some information which could accelerate development of vaccines and drugs.
The ZIKV, native to parts of Africa and Asia, has for the first time been introduced into the Americas. The ZIKV epidemic in Brazil currently is estimated at 440000–1300000 cases, and in February 2016 it has spread to other Latin-American countries, the USA and Europe ( http://www.thelancet.com/pdfs/journals/lancet/PIIS0140-6736(16)00014-3.pdf), threatening to become a pandemic. Recently, the World Health Organization (WHO) declared an international public health emergency ( http://www.who.int/mediacentre/news/statements/2016/emergency-committee-zika-microcephaly/en/). There is no vaccine against the virus or any antiviral treatment.
Here we analyzed ZIKV E proteins using ISM. Results of this in silico analysis revealed that these viral proteins encode the highly conserved information which determines their interacting profile and immunological properties. Previously, we reported that the human interacting profile of HA1 from pdmH1N1 influenza viruses is characterized by the same information 2. This result suggests possible repurposing of the seasonal influenza vaccine containing pdmH1N1 for prevention of ZIKV infection.
Material and methods
Sequences
All sequences of the ZIKV E protein are taken from the NCBI databank ( http://www.ncbi.nlm.nih.gov/nuccore/?term=zika) and are given in Dataset 1 (accessions: KU312314, KU312315, KU312313, KU312312, KJ776791, KJ634273, KF993678, JN860885, EU545988, HQ234499, KF268950, KF268948, KF268949, AY632535, LC002520, DQ859059, HQ234501, HQ234500, FSS13025, AHL43505, AHL43503, AHL43502, AMA12087). Sequences for HA1 from influenza viruses were taken from the GISAID database ( http://platform.gisaid.org) and are given in Dataset 2 (accessions: EPI705910, EPI696955).
Copyright: © 2016 Veljkovic V and Paessler S
Data associated with the article are available under the terms of the Creative Commons Zero "No rights reserved" data waiver (CC0 1.0 Public domain dedication).
Copyright: © 2016 Veljkovic V and Paessler S
Data associated with the article are available under the terms of the Creative Commons Zero "No rights reserved" data waiver (CC0 1.0 Public domain dedication).
Informational spectrum method
The ISM is a virtual spectroscopy technique, developed for the study of protein-protein interactions. The physical and mathematical base of ISM was described in detail elsewhere ( 5 and references therein), and here we will only in brief present this bioinformatics method.
The ISM technique is based on a model of the primary structure of a protein using a sequence of numbers, by assigning to each amino acid the correspondence value of the electron ion interaction potential (EIIP; L 0.0000, I 0.0000, N 0.0036, G 0.0050, V 0.0057, E 0.0058, P 0.0198, H 0.0242, K 0.0371, A 0.0373, Y 0.0516, W 0.0548, Q 0.0761, M 0.0823, S 0.0829, C 0.0829, T 0.0941, F 0.0946, R 0.0959, D 0.1263). The EIIP values are in Rydbergs (Ry).
The obtained numerical sequence is then subjected to a discrete Fourier transformation which is defined as follows:
where x(m) is the m-th member of a given numerical series, N is the total number of points in this series, and X(n) are discrete Fourier transformation coefficients. These coefficients describe the amplitude, phase and frequency of sinusoids, which comprised the original signal. Relevant information encoded in the primary structure is presented in an energy density spectrum which is defined as follows:
In this way, sequences are analyzed as discrete signals. It is assumed that their points are equidistant with the distance d = 1. The maximal frequency in a spectrum defined as above is F = 1/2d = 0.5. The frequency range is independent of the total number of points in the sequence. The total number of points in a sequence influences only the resolution of the spectrum. The resolution of the N-point sequence is 1/n. The n-th point in the spectral function corresponds to a frequency f(n) = nf = n/N. Thus, the initial information defined by the sequence of amino acids can now be presented in the form of the informational spectrum (IS), representing the series of frequencies and their amplitudes.
The IS frequencies correspond to the distribution of structural motifs with defined physicochemical properties determining a biological function of a protein. When comparing proteins, which share the same biological or biochemical function, the ISM technique allows detection of code/frequency pairs which are specific for their common biological properties, or which correlate with their specific interaction. These common informational characteristics of sequences are determined by the cross-spectrum (CS) which is obtained by the following equation:
where Π(i,j) is the j-th element of the i-th power spectrum and C(j) is the j-th element of CS. Peak frequencies in CS represent the common information encoded in the primary structure of analyzed sequences. This information corresponds to the mutual interaction between analyzed proteins or their interaction with the common interactor.
Results and discussion
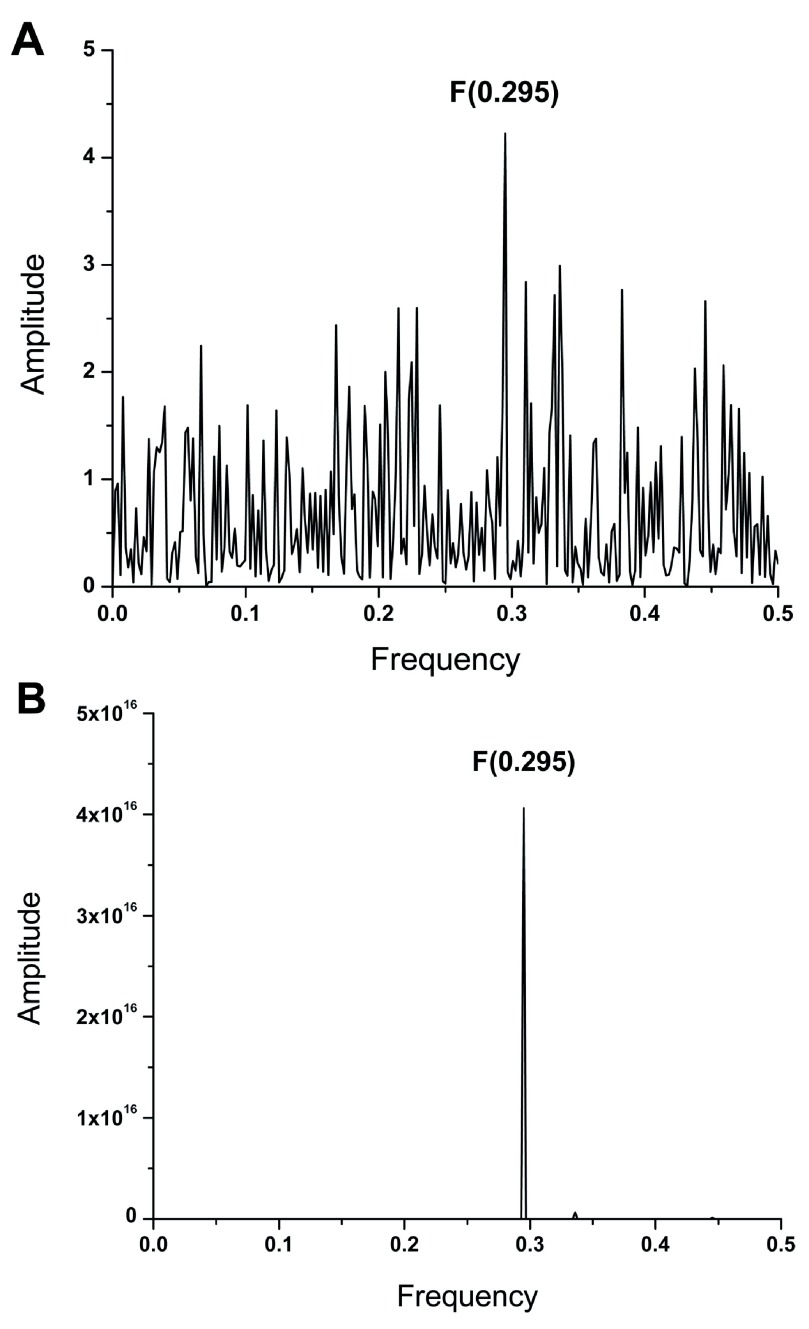
The envelope glycoprotein (protein E) which mediates the virus cell entry is highly conserved and virtually identical in all ZIKV isolated during 2015 in countries of Central and South America. Figure 1A shows the IS of ZIKV E isolated in 2015 in Brazil (AMA12087) which is characterized by a dominant peak at frequency F(0.295). Figure 1B shows the CIS of protein E from ZIKV isolated between 1968 and 2015 in diverse countries of Asia and Africa ( Dataset 1). This CIS contains only one dominant peak at frequency F(0.295). This result suggests that all analyzed ZIKV E encode the same highly conserved information which is represented by the IS frequency F(0.295). According to the ISM concept 6– 9, this information determines the interacting profile of ZIKV E.
Figure 1.
( A) The informational spectrum of ZIKV E protein. ( B) The consensus informational spectrum of protein E from ZIKV isolated in Asia, Africa and South/Central America.
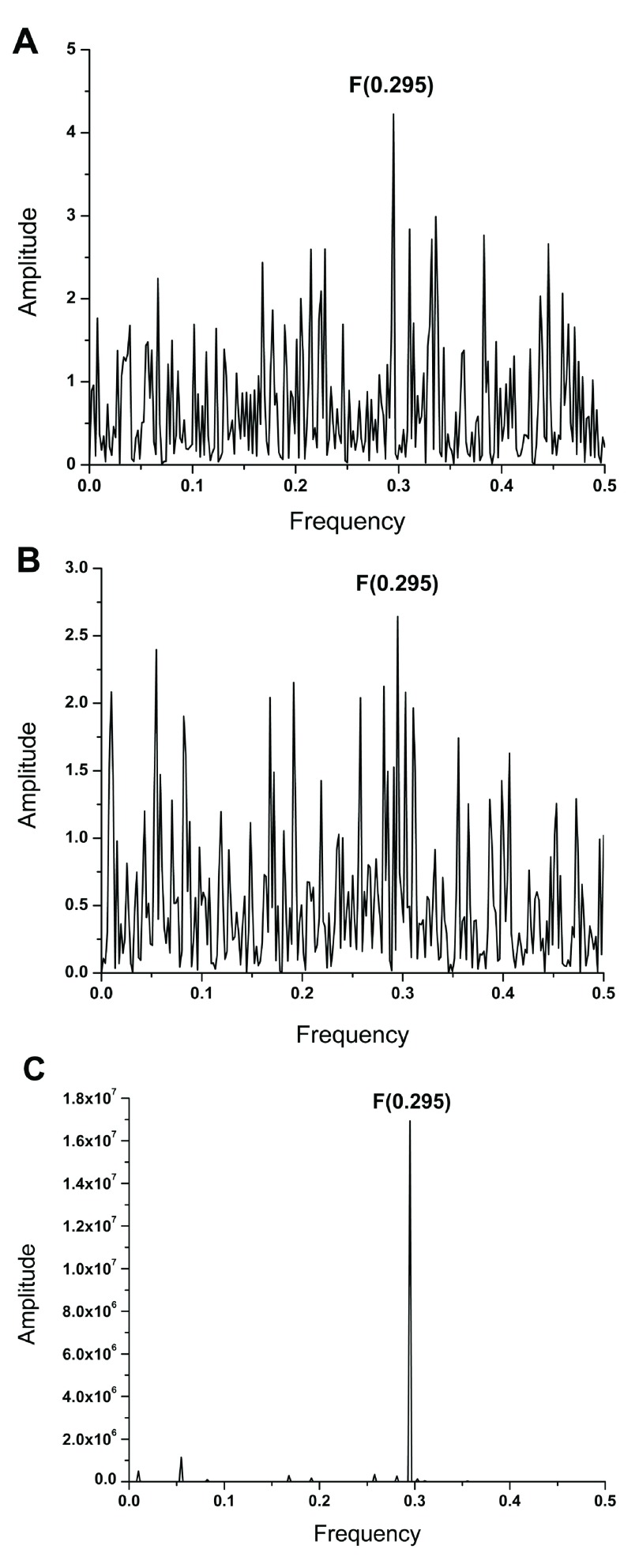
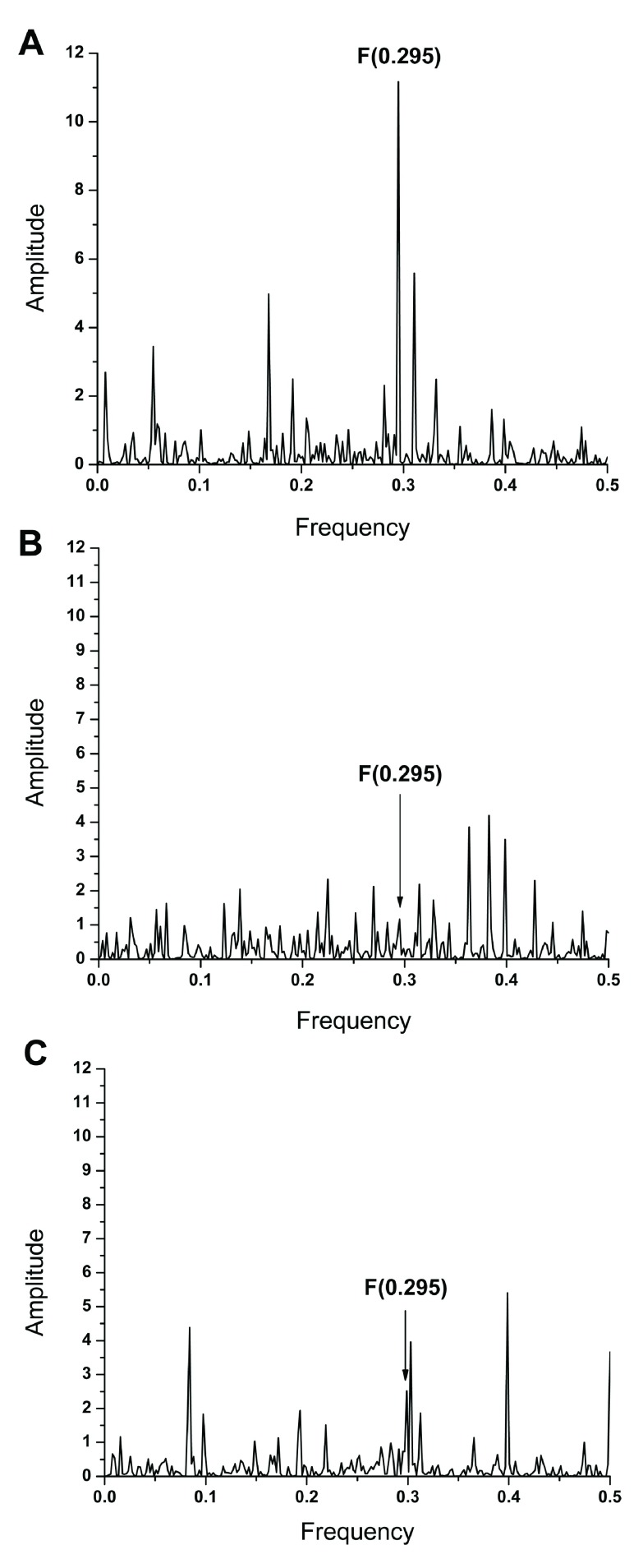
Previously, it has been shown that frequency F(0.295) characterizes the human interacting profile of pdmH1N1 HA1 ( Figure 2C) 2. It has also been shown that antigens which share a common frequency component in their IS are immunologically cross-reactive ( 10 and references therein). Presence of the frequency component F(0.295) in ZIKV E and pdmH1N1 HA indicates that antibodies elicited by this protein of influenza virus could affect interaction between ZIKV E and host proteins. We compared the IS of ZIKV E and HA1 from A/California/07/2009(H1N1) and A/Switzerland/9715293/2013(H3N2) viruses, which are components of the 2015/2016 seasonal influenza vaccine. Results given in Figure 2 and Figure 3A show that ZIKV E and A/California/07/2009(H1N1) encode the common information represented in their IS with the dominant peak at frequency F(0.295). As can be seen in Figure 3B & 3C, this frequency component is not present in the IS of A/Switzerland/9715293/2013(H3N2) HA1. These results indicate that antibodies elicited by the vaccine virus A/California/07/2009(H1N1) could affect interaction between ZIKA E and host proteins mediating virus cell entry.
Figure 2.
( A) The informational spectrum of ZIKV E protein. ( B) The informational spectrum of HA1 from A/California/07/2009(H1N1) influenza virus. ( C) The consensus informational spectrum of HA1 from pdmH1N1 2.
Figure 3.
( A) The cross-spectrum of ZIKV E protein and HA1 from A/California/07/2009(H1N1) influenza virus. ( B) The cross-spectrum of ZIKV E protein and HA1 from A/Switzerland/9715293/2013(H3N2) influenza virus. ( C) The cross-spectrum of HA1 from A/California/07/2009(H1N1) and A/Switzerland/9715293/2013(H3N2) influenza viruses.
Conclusions
The presented results of the in silico analysis of ZIKV E and host factors mediating viral infection suggest that the seasonal influenza vaccines containing pdmH1N1 as a component, could protect to some extent against the ZIKV infection. Because of the lack of prevention and therapy of the ZIKV disease, in a situation when the ZIKV infection is explosively spreading, this possible safe and inexpensive solution is worth being seriously considered.
Data availability
The data referenced by this article are under copyright with the following copyright statement: Copyright: © 2016 Veljkovic V and Paessler S
Data associated with the article are available under the terms of the Creative Commons Zero "No rights reserved" data waiver (CC0 1.0 Public domain dedication). http://creativecommons.org/publicdomain/zero/1.0/
F1000Research: Dataset 1. Sequences of the ZIKV E protein taken from the NCBI databank (accessions: KU312314, KU312315, KU312313, KU312312, KJ776791, KJ634273, KF993678, JN860885, EU545988, HQ234499, KF268950, KF268948, KF268949, AY632535, LC002520, DQ859059, HQ234501, HQ234500, FSS13025, AHL43505, AHL43503, AHL43502, AMA12087)., 10.5256/f1000research.8102.d114325 11
F1000Research: Dataset 2. Sequences of HA of A/California/07/2009(H1N1) and HA of A/Switzerland/9715293/2013(H3N2) taken from the GSAID database (accessions: EPI705910, EPI696955)., 10.5256/f1000research.8102.d114326 12
Funding Statement
The author(s) declared that no grants were involved in supporting this work.
[version 2; referees: 2 approved]
References
- 1. Veljković V, Metlas R: Identification of nanopeptide from HTLV-III, ARV-2 and LAVBRU envelope gp120 determining binding to T4 cell surface protein. Cancer Biochem Biophys. 1988;10(2):91–106. [PubMed] [Google Scholar]
- 2. Veljkovic V, Niman HL, Glisic S, et al. : Identification of hemagglutinin structural domain and polymorphisms which may modulate swine H1N1 interactions with human receptor. BMC Struct Biol. 2009;9:62. 10.1186/1472-6807-9-62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Veljkovic V, Glisic S, Muller CP, et al. : In silico analysis suggests interaction between Ebola virus and the extracellular matrix. Front Microbiol. 2015;6:135. 10.3389/fmicb.2015.00135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Veljković V, Cosić I, Dimitrijević B, et al. : Is it possible to analyze DNA and protein sequences by the methods of digital signal processing? IEEE Trans Biomed Eng. 1985;32(5):337–341. 10.1109/TBME.1985.325549 [DOI] [PubMed] [Google Scholar]
- 5. Veljkovic N, Glisic S, Prljic J, et al. : Discovery of new therapeutic targets by the informational spectrum method. Curr Protein Pept Sci. 2008;9(5):493–506. 10.2174/138920308785915245 [DOI] [PubMed] [Google Scholar]
- 6. Veljkovic V, Veljkovic N, Muller CP, et al. : Characterization of conserved properties of hemagglutinin of H5N1 and human influenza viruses: possible consequences for therapy and infection control. BMC Struct Biol. 2009;9:21. 10.1186/1472-6807-9-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Perovic VR, Muller CP, Niman HL, et al. : Novel phylogenetic algorithm to monitor human tropism in Egyptian H5N1-HPAIV reveals evolution toward efficient human-to-human transmission. PLoS One. 2013;8(4):e61572. 10.1371/journal.pone.0061572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Schmier S, Mostafa A, Haarmann T, et al. : In Silico Prediction and Experimental Confirmation of HA Residues Conferring Enhanced Human Receptor Specificity of H5N1 Influenza A Viruses. Sci Rep. 2015;5: 11434. 10.1038/srep11434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Veljkovic V, Paessler S, Glisic S, et al. : Evolution of 2014/15 H3N2 Influenza Viruses Circulating in US: Consequences for Vaccine Effectiveness and Possible New Pandemic. Front Microbiol. 2015;6:1456. 10.3389/fmicb.2015.01456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Veljkovic V, Glisic S, Veljkovic N, et al. : Influenza vaccine as prevention for cardiovascular diseases: possible molecular mechanism. Vaccine. 2014;32(48):6569–6575. 10.1016/j.vaccine.2014.07.007 [DOI] [PubMed] [Google Scholar]
- 11. Veljkovic V, Paessler S: Dataset 1 in: Possible repurposing of seasonal influenza vaccine for prevention of Zika virus infection. F1000Research. 2016. Data Source [DOI] [PMC free article] [PubMed]
- 12. Veljkovic V, Paessler S: Dataset 2 in: Possible repurposing of seasonal influenza vaccine for prevention of Zika virus infection. F1000Research. 2016. Data Source [DOI] [PMC free article] [PubMed]