Abstract
Protein palmitoylation has been shown to be an important post-translational modification in eukaryotic cells. This modification alters the localization and/or the function of the targeted protein. In the recent years protein palmitoylation has risen in importance in apicomplexan parasites as well. In Toxoplasma gondii, some proteins have been reported to be modified by palmitate. With the development of new techniques that allow the isolation of palmitoylated proteins, this significant post-translational modification has begun to be studied in more detail in T. gondii. Here we describe the palmitoylome of the tachyzoite stage of T. gondii using a combination of the acyl-biotin exchange chemistry method and mass spectrometry analysis. We identified 401 proteins found in multiple cellular compartments, with a wide range of functions that vary from metabolic processes, gliding and host-cell invasion to even regulation of transcription and translation. Besides, we found that more rhoptry proteins than the ones already described for Toxoplasma are palmitoylated, suggesting an important role for this modification in the invasion mechanism of the host-cell. This study documents that protein palmitoylation is a common modification in T. gondii that could have an impact on different cellular processes.
Keywords: Toxoplasma gondii, acyl-biotin exchange, palmitoylome, protein identification, rhoptry, host-cell invasion
1. Introduction
Protein palmitoylation refers to the post-translational addition of palmitoyl-Coenzyme A to cysteine residues of certain proteins [1]. This reversible modification has been shown to play key roles in regulating sub-cellular localization (reviewed in [2]), trafficking [3], enzymatic activity [4,5], protein-protein interaction [6] and gene expression [7,8].
In Toxoplasma gondii, the causative agent of toxoplasmosis, N-terminal palmitoylation has been shown to play a key role in the localization of proteins to the parasite's inner membrane complex (IMC), pellicle and rhoptry membranes. It has been reported that predicted myristoylation and palmitoylation sites of the inner membrane complex sub-compartment protein (ISP) -1,-2, and -3 are important to target these proteins to the IMC since site-directed mutagenesis alters the localization from the IMC to the cytosol [9]. It is important to highlight that these proteins are involved in cell-daughter formation. ISP-4 only contains one predicted palmitoylation site which seems to be sufficient to target this protein to the IMC [10]. Similarly, the myosin light chain 2 (MLC2), a protein that is part of the motility system, has been reported to be anchored to the pellicle likely by N-terminal palmitoylation [11]. More recently, it has been demonstrated that the small heat shock protein 20 (TgHSP20) depends on N-terminal palmitoylation for proper IMC localization [12]. Furthermore, N-terminal myristoylation and palmitoylation are responsible for the attachment of the enzyme cGMP-dependent protein kinase (PKG) to the pellicle [13], and it has been demonstrated that an armadillo repeat only protein (ARO) depends on myristoylation and palmitoylation for its localization on the cytosolic face of rhoptry membranes [14]. Additionally, a recent publication has described that the microneme protein AMA1 is palmitoylated and that its palmitoylation status regulates its own release and the release of other invasion-related proteins [15].
Supporting the importance of protein palmitoylation in the invasion process, it has been shown that inhibition of palmitoylation by 2-bromopalmitate alters invasion as well as the gliding mechanism in T. gondii [16]. Furthermore, inhibition of depalmitoylation enhances those same two processes [17]. This suggests that more proteins than the ones found to date must be targeted by palmitoylation.
Interestingly, it has been reported that T. gondii possesses all the machinery required to add palmitate on a subset of selected proteins since it expresses 18 palmitoyl-acyltransferases (TgPATs) with different localizations, 16 of which are found in the tachyzoite stage and some are unique to apicomplexan organelles important for the invasion of host-cells [18].
Although many important biological aspects of T. gondii are affected by palmitoylation, the identity of the proteins affected by this modification is starting to be uncovered [15]. As such, a T. gondii's palmitoylome provides key information to start unravelling the relevance of protein palmitoylation in this parasite’s biology.
In the present study, isolation of palmitoylated proteins using acyl-biotin exchange strategy (ABE) was coupled to mass spectrometry analysis. This approach led to the identification of 401 proteins displaying a wide array of functions and localizations. Some of them were previously described palmitoylated proteins and most of them were novel palmitoylated proteins. Interestingly, localization of some rhoptry proteins as well as the rhoptries themselves seem to be palmitoylation-dependent. Thus, our work provides further evidence of the importance of protein palmitoylation in the biology of T. gondii.
2. Materials and Methods
2.1 Antibodies and reagents
Specialized and common reagents were from Sigma, unless specified. Dulbecco's Modified Eagle Medium, penicillin and streptomycin were from Life Technologies (CABA, Argentina). N-ethylmaleimide (NEM), Streptavidin-agarose and HPDP-biotin were from Thermo Scientific (IL, USA). Complete protease inhibitor cocktail tablets were from Roche Diagnostics Corporation (IN, USA). ECL Plus was from GE Biosciences (UK). The serum anti-Ty was kindly provided by Dr. Dubremetz (Université de Montpellier, France) as well as anti-ROP4, -ROP5, -ROP7 monoclonal antibodies. Anti-AMA1 antibodies were generously provided by Dr. Ward (University of Vermont, USA).
2.2 Toxoplasma and host-cell cultures
T. gondii tachyzoites of the RH Δhxgprt strain [19] were used throughout the study. Parasites were maintained by serial passage on confluent monolayers of human foreskin fibroblasts (HFFs) in Dulbecco's Modified Eagle Medium supplemented with 10% v/v bovine serum albumin (BSA), 100 i.u. (international units)/ml penicillin and 100 µg/ml streptomycin. Tachyzoites were then physically separated from host cells by passage through a 27G syringe needle and purified from host cell debris using a 3.0 µm filter before use[20].
2.3 Acyl-biotin exchange method on total parasite lysates
ABE of whole parasite lysates was mainly carried out as described by Wan and colleagues [21] with the following modifications. Briefly, parasites were purified by 3.0 µm polycarbonate filter and a total of 1–5 × 109 parasites were used for the assay. Parasites were resuspended in 4 ml of lysis buffer containing 10 mM NEM and sonicated 15" on/off for 10 periods. Then the concentration of NEM was adjusted to 2 mM for overnight treatment. The rest of the procedure was performed as described [21].
2.4 Separation and digestion of proteins
Protein samples were separated by 12% SDS-PAGE. The resultant gel was stained with Coomassie Brilliant Blue R-250. Each lane of the gel was completely cut into individual slices. Each band was then cut into 1 mm3 cubes and further destained with three washes of 50 mM NH4HCO3 in 50% CH3CN with 10 min incubations. Each group of gel cubes was then dehydrated in CH3CN for 10 min and dried in a Speed Vac. Protein samples were reduced by dithiothreitol (DTT) and alkylated by iodoacetamide [22]. A solution of 10 ng/µL trypsin in 50 mM NH4HCO3 was used to re-swell the gel pieces completely at 4°C for 30 min, followed by a 37°C digestion overnight. A small amount of 10% formic acid was then added to stop the digestion. The sample was then centrifuged at 2,800 × g, and the supernatant was collected for LC-MS/MS.
2.5 LC-MS/MS analysis
Five µl of tryptic peptide samples were loaded onto the LC microcapillary column (12 cm × 100 µm inner diameter) packed with C18 reversed-phase resin (5 µm particle size; 20 nm pore size; Magic C18AQ, Michrom Bioresources Inc.), and separated by applying a gradient of 3–60% acetonitrile in 0.1% formic acid for 45 min at a flow rate of 500 nl/min after the flow is split to waste. The flow rate was controlled by a 1000 psi back pressure regulator (IDEX Health & Science LLC, Oak Harbor, WA) which connected flow to waste. The nanospray ESI was fitted onto a linear quadrupole ion trap mass spectrometer (Thermo Electron, San Jose, CA) that was operated in a collision-induced dissociation mode to obtain both MS and tandem MS (MS/MS) spectra. Mass spectrometry data were acquired in a data-dependent acquisition mode, in which a full MS scan from m/z 400–1700 was followed by 10 MS/MS scans of the most abundant ions.
2.6 Protein identification
Obtained MS spectra were searched against the T. gondii ToxoDB (v 26; www.toxodb.org) protein database using Proteome Discovery 1.4 (Thermo Electron, San Jose, CA). The workflow includes Spectrum Files, Spectrum Selector, Sequest search nodes followed by Target Decoy PSM Validator. The search parameters permitted a 2 Da peptide MS tolerance and a 1.0 Da MS/MS tolerance. Oxidation of methionine (M) and carboxymethylation of cysteines (C) were allowed as variable modifications. Up to two missed tryptic peptide cleavages were considered. The proteins, which False Discovery Rate (FDR) is less than 1% at the peptide level. All identified proteins, even the ones with one peptide matched, had been listed in the supplements.
2.7 Data analysis and bioinformatics
Palmitoylated proteins were classified according to all 3 gene ontology (GO) categories (molecular function, biological process and cellular component) taking into account all GOs provided by ToxoDB [23]. Conserved domains in the identified proteins were predicted by Prosite [24] and BLASTp. Predicted presence of signal peptide or transmembrane domains were determined by SignalP 4.1 Server [25] and TMHMM Server v. 2.0 (http://www.cbs.dtu.dk/services/TMHMM/) respectively. For subcellular localization predictions, PSORT II [26] was used, except for those cases where localization to a Toxoplasma specific organelle or compartment was already determined and annotated in ToxoDB. Presence of potentially lipidated residues was predicted by: CSS-Palm 3.0 [27] for palmitoylated cysteines, Myrirstoylator [28] for N-terminal myristoylated glycines and PrePS [29] for prenylated motifs. GO and metabolic pathway enrichment analyses, as well as comparison between T. gondii and P. falciparum proteins were performed using the informatics tools provided by ToxoDB and EuPathDB respectively.
2.8 Treatment with 2-bromopalmitate (2-BP)
For western blot, immunofluorescence and electron-microscopy studies, treatment was performed using 12.5 µM 2-BP which was shown to inhibit palmitoylation while not causing gross cell morphology alterations [16]. For immunofluorescence studies monolayers of HFF cells grown on glass slides were infected with T. gondii tachyzoites in 24-well dishes. After 10 min on ice, parasites were allowed to invade for 1 h at 37 °C. Then, the cells were washed twice with PBS and the media was changed by DMEM containing 1% v/v BSA fatty acid-free and 12.5 µM 2-BP. The infected cells were incubated for 16–18 h at 37°C, then were subjected to indirect immunofluorescence studies. For transmission electron-microscopy (TEM), the treatment was similar except it was performed in T25 culture flasks, and after incubation with 2-BP, cells were fixed and scraped as described in the EM section.
For all 2-BP treatments, a 0 µM (no-treatment) control was included which consisted of DMEM containing 1% v/v BSA fatty acid-free and 2% v/v DMSO.
2.9 Indirect immunofluorescence studies
All steps were carried out at room temperature. Media was discarded and cells were washed twice with PBS, followed by fixation with formaldehyde 4% v/v in PBS for 30 min. After a 1 min wash with PBS, cells were permeabilized with 0.3% v/v Triton X-100/PBS for 20 min and blocked with 3% w/v BSA in PBS for 30 min. Appropriate primary antibodies were added diluted in 3% w/v BSA/PBS for 60 min followed by extensive washes. Alexa conjugated anti-species secondary antibodies (dil 1/4000 in 3% w/v BSA/PBS) were added and incubated for another 60 min followed by extensive washes. Finally samples were mounted with Mowiol 4-88 mounting media (Merck Millipore) and DAPI staining at 5ug/ml.
Primary antibodies used were: monoclonal anti-ROP4 (1/200), monoclonal anti-ROP5 (1/200), monoclonal anti-ROP7 (1/200), mouse anti-Ty (1/300), monoclonalanti-AMA1 (1/200). Parasites were imaged using a Nikon Eclipse E600 microscope and Nikon Digital Sight DS-Qi1Mc camera.
2.10 Transmission electron microscopy
Samples were fixed in a solution containing 2.5 % glutaraldehyde in 0.1 M cacodylate buffer, pH 7.2. Then, cells were washed in buffer and post-fixed for 40 minutes in the dark in a solution containing 1% osmium tetroxide and 1.25% potassium ferrocyanide and 5mM CaCl2 in 0.1 M cacodylate buffer, pH 7.2. The cells were then washed in buffer, dehydrated in acetone and embedded in Polybed© (Polysciences) Epoxy resin. Ultrathin sections were obtained in a Leica EM UC7 ultramicrotome. The sections were stained with uranyl acetate and lead citrate and examined in a Zeiss 900 transmission electron microscope operating at 80 kV.
2.11 Western blot on 2-BP and ABE treated parasites
Intracellular tachyzoites grown in T-175 culture flasks were treated overnight with 12.5 µM 2-BP (together with its corresponding DMSO control) as described in section 2.8. The following day, cells were washed 3 times with PBS and harvested by scraping, then passed through a 27-gauge needle and filtered with a 3 µm polycarbonate syringe filter. Equal amount of parasites were counted and used for both treated and control samples. ABE protocol was scaled down and performed as described in section 2.3, with the exception of the initial treatment with NEM which was performed at 100 mM. An aliquote (20% of total sample) was taken after the biotinylation step to be further used as the input control. The remainder of the sample was pulled down using NeutrAvidin-agarose resin (Thermo Scientific), and then eluted with 5mM DTT. Eluates and inputs were run in parallel in a 10% SDS-PAGE, transferred to a PVDF membrane and western blot was performed using anti-ROP5 monoclonal antibody at a 1:1000 dilution. The bands were quantified using Image J software. Each pull down value was normalized by its respective input, and each sample final value was expressed as a percentage of the hydroxylamine-treated control sample (DMSO, HA+). Three independent experiments were carried out.
3. Results and discussion
3.1 Isolation and identification of palmitoylated proteins
In order to isolate and identify novel palmitoylated proteins from T. gondii, we substituted S-acyl groups for biotin after hydroxylamine cleavage (Figure 1A) based on the method originally described by Drisdel and Green [30] and later modified by Wan [21]. Briefly, free cysteines were blocked with N-ethylmaleimide (NEM), and split in two halves. One half of the sample was treated with neutral hydroxylamine to remove S-acyl groups (HA+ sample, Figure 1B) and exposed cysteines were tagged with biotin. Biotinylated proteins were purified using NeutrAvidin-agarose beads. The other half of the sample was processed without the hydroxylamine treatment (HA− sample, Figure 1B). To increase confidence in the identification of S-acylated proteins, three independent pairs of samples were prepared and analyzed by mass spectrometry. Results from all three replicates led to the identification of 401 non-redundant proteins, from which 310 are known while the rest are hypothetical proteins (see Table S1 for complete list). Several aspects of all the identified proteins were explored using bioinformatics tools, such as prediction of conserved domains, GO terms, presence of transmembrane domains and/or signal peptide, intracellular localization and prediction of protein lipidations (Table S1). In those cases where the predicted palmitoylated cysteine residue was found to be part of the signal peptide (marked in red in Table S1), alternative cysteines were proposed (even if the CSS-Palm 4.0 predictor could not predict them).
Figure 1. Acyl biotin exchange method used in this study.
A) Flowchart depicting the schematics of the ABE technique. B) Representative Coomassie Blue-stained gel of ABE-treated Toxoplasma gondii protein extract. HA+: Hydroxylamine-treated sample. HA-: Tris-treated control (no hydroxylamine). M: molecular mass reference. Both lanes were dissected into small slices and analyzed by MS (mass spectrometry).
Although the ABE approach for identification of palmitoylated proteins has the capacity to detect endogenously biotinylated proteins, this does not represent a major constraint for the application of the technique in T. gondii since the parasite expresses only 2 easy-to-identify biotinylated proteins: ACC (acetyl CoAcarboxylate), a 240kDa apicoplast protein and pyruvate carboxylase, a 130kDa mitochondrial protein [31].
Another difficulty coming from the ABE strategy is the number of false-positive hits that might appear and how to discriminate positive signal from noise. Non-redundant peptides (NRP) were used as a semi-quantitative index to compare relative protein abundance in plus-versus minus-HA samples. The higher the HA +/− NRP ratio for a given protein, the higher the probability for that protein to be indeed palmitoylated. Table S1 shows all non-redundant identified candidate proteins from three independent assays. Normalization of plus/minus-hydroxylamine NRP values was performed as follows: individual NRP values for a given protein were merged (in case that protein was detected more than once in that assay) then divided by the total NRP value for that MS analysis, and multiplied by an approximation of the total NRP per experiment). To calculate the ratio of plus/minus-HA, we divided the normalized plus-HA by the normalized minus-HA NRP value. To avoid division by 0 we used 0.2 as an arbitrary value in those cases where no trace of the protein was detected in the HA- fraction. This list includes proteins already described to be palmitoylated (Table S1 highlighted in blue) such as HSP20 [12], GAP45 and AMA1 [15] and members of the ISP family [9], those predicted to be palmitoylated but without experimental evidence (Table S1 highlighted in grey) such as members of the ISC (IMC structure component), IMC (inner membrane complex) and AC (apical cap) families [32], and those that share homology to known S-acylated proteins from Plasmodium falciparum, a related organism, such as 14-3-3, Hsp70 and proteins of the CDPK family [33] (Table S1 highlighted in green). We also identified TGME49_213550 (TgDHHC 4), TGME49_249380 (TgDHHC13) and TGME49_278850 (TgDHHC2) which are DHHC S-acyl transferases [18]. The first two localize to the plasma membrane while the latter is anchored to the IMC and was shown to be essential for parasite survival [18]. DHHC S-acyl transferases are known to perform auto-S-acylation in order to form a stable catalytic intermediate [34], thus it was expected to find T. gondii DHHC S-acyl transferases in the palmitoylome. It may be possible that due to the limitations of the technique and the relative abundance of these proteins, no other palmitoyl acyl transferases (PATs) could be detected.
3.2 Palmitoylated proteins in T. gondii have a widespread localization and varied functions
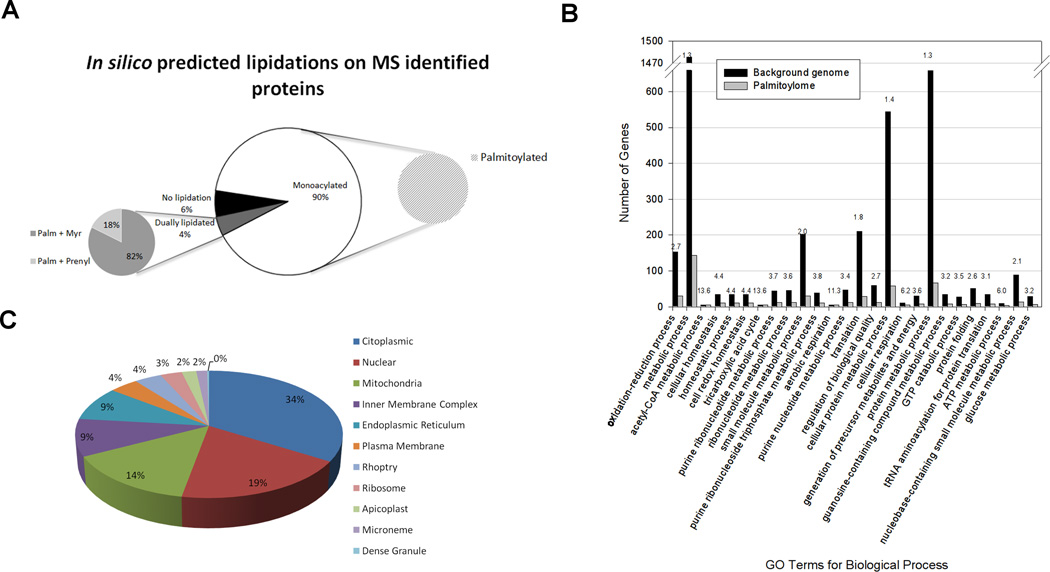
Similarly to what was observed in higher eukaryotes, protein palmitoylation can occur alone or in concert with other lipidic modifications (Figure 2A). Our proteome shows that 90% of the identified proteins are predicted to be monoacylated (all palmitoylated). However, there are some proteins (4% of the total) that would present dual lipidic modifications, either palmitoylation combined with myristoylation (82%) or with prenylation (18%) (Figure 2A).
Figure 2. In silico analyses of experimentally found palmitoylated proteins.
A) Other predicted lipidations. Monoacylated proteins are only palmitate-modified. In dually lipidated proteins, palmitoylation can occur either in combination with myristoylation or with prenylation. B) GO Biological processes enrichment analysis: Bars represent genes associated with a specific GO term both in the background genome and in the palmitoylome. Numbers above the bars indicate fold enrichment of that specific term in the palmitoylome with respect to the background. C) Predicted subcellular localization of the identified palmitoylated proteins.
For a global analysis of our palmitoylome, we performed GO terms enrichment analyses (Table S2). Regarding function and biological processes in which palmitoylated proteins are involved, most of them cluster into groups including metabolic and energy-related processes and protein translation (Figure 2B). However, it is evident that the modification affects the majority of cellular functions since we were able to identify proteins involved in a plethora of roles: from signaling (CDPK3), to gliding motility (myosin A, GAP45 and various filament-like alveolins, members of the recently described IMC, AC and ISC families [32]), translation (translation initiation factor eIF-5A and translation elongation factor 2), among others. Some of these proteins have been previously reported to be palmitoylated: in the case of TgCDPK3, studies have shown that palmitoylation not only occurs but it also affects the localization of the protein and thus its function in the parasite’s ability to egress the host-cell [35]. Likewise, mutagenesis studies on TgGAP45, have demonstrated that both N- and C- terminal acylations are important for proper anchoring of the protein to the parasite’s pellicle and affect vital processes such as gliding, invasion and egress [36].
Considering the variety of functions in which palmitoylation is involved, it was expected that the same diversity was reflected in the localization of the affected proteins. For that, we examined the presence of palmitoylated proteins in different subcellular compartments. GO enrichment analysis of cellular components indicates that the highest values are related to membranous sub-compartments such as mitochondria, pellicle and IMC (Table S2). As for the predicted localization of the proteins we identified, our data shows that an important group of them would localize to the nucleus (19%), and other membranous organelles such as mitochondria (14%), endoplasmic reticulum (9%) and even rhoptries and micronemes which are apicomplexan-specific organelles involved in host-cell invasion (Figure 2C).
Our data show that there are several nuclear proteins that are palmitoylated, including elongation factor 1-alpha (TGME49_294800), eukaryotic initiation factor-4A (TGME49_250770), 14-3-3 (TGME49_263090) (Table S1) and the effect of this modification on them should be further studied. The finding of nuclear palmitoylated proteins is really recent as well as their involvement in the regulation of transcriptional and chromatin-related processes. The first report on this topic came out in 2013 by Park et al. for the nuclear protein Rif1 [37]. In Saccharomyces cerevisiae palmitoylation regulates Rif1 association to the nuclear envelope [8] and thus heterochromatin dynamics. Furthermore, this work mentions that in addition to Rif1, there would be other nuclear palmitoylated proteins. A later report showed that the deacetylase SIRT6 can be activated by free long-chain fatty acids such as myristate and palmitate at physiological concentrations and thus, regulate gene transcription [38]. Finally, in 2014 Chen and co-workers showed that palmitoylation of the acetylase P300 is required for nuclear localization and for acylation of H3 and H4 histones and thus, to control neuronal differentiation [39].
While the finding of nuclear palmitoylated proteins is quite interesting and novel, it was also expected to find palmitoylated proteins in mitochondria. Although there are several reports describing that mitochondrial proteins are palmitoylated in higher eukaryotes [4,5,40], this report suggest that palmitoylation of mitochondrial proteins is found in T. gondii as well.
It is interesting that our global analysis unveiled novel palmitoylated proteins in micronemes and rhoptries as well, since these organelles are a fundamental part of the invasion machinery in T. gondii and some of their secreted proteins are involved in host-cell signaling. However, the effect of palmitate on these proteins remains to be studied. Our global palmitoylome suggests that the microneme proteins AMA1, MIC1, MIC2, MIC4, MIC8 and MIC2-associated protein (M2AP) would be palmitoylated. Regarding rhoptry proteins, ROP4, ROP5, ROP7, ROP2/8, ROP11, ROP13, ROP14, ROP18, ROP39, RON3, RON5 and RON8 would be palmitoylated as well. Many of the rhoptry and microneme proteins found in this report were also identified by a different approach [15] supporting that these proteins are likely palmitoylated.
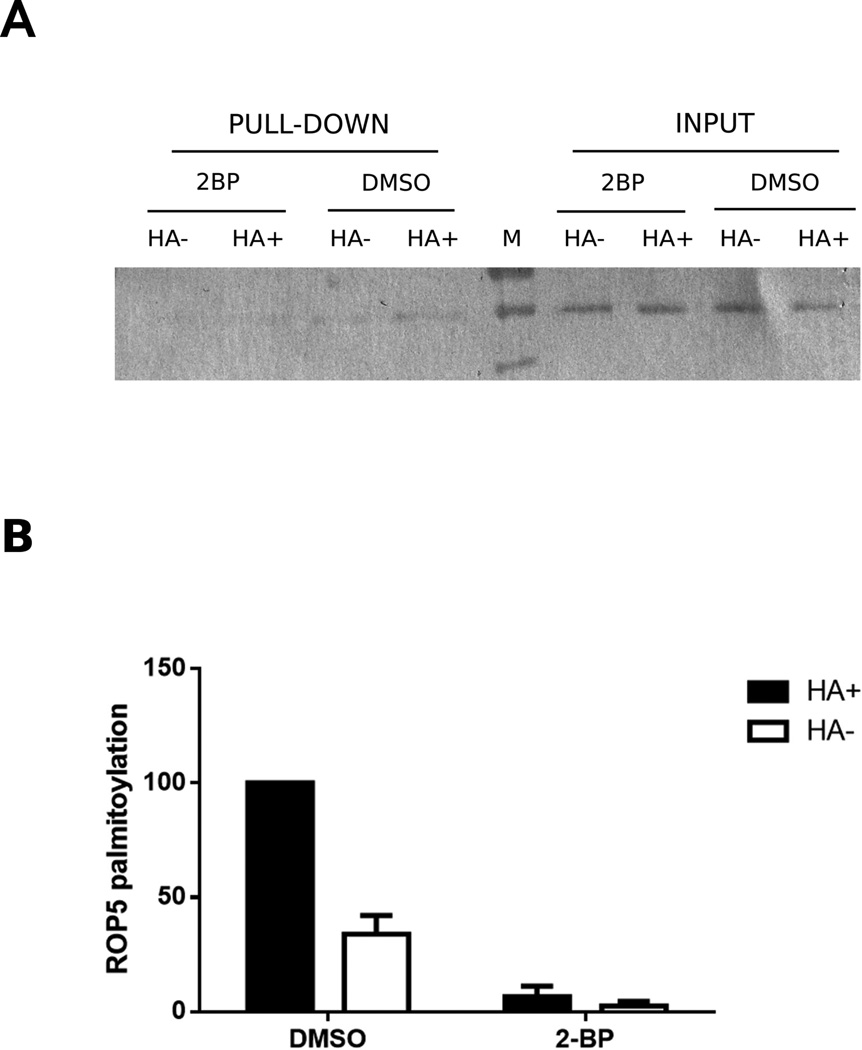
We next decided to validate the palmitoylation status of ROP5, a rhoptry protein not previously described to be palmitoylated and that we found in all the ABE assays performed. In order to confirm the palmitoylation status of ROP5, intracellular parasites were treated with 2-BP to inhibit palmitoylation followed by ABE, eluted proteins were run in a 10% SDS-PAGE gel and western blot using specific anti-ROP5 antibodies was performed. Figure 3A shows that ROP5 is only present in the control (DMSO treated) lanes, while it is almost absent in 2-BP-treated samples, all of which can be clearly observed by the quantification of the intensity of the western blot bands (Figure 3B). This result confirms that ROP5 is palmitoylated. Interestingly, ROP5 is not only involved in invasion, but also in virulence. ROP5 increases ROP18 kinase activity and they act together to enhance parasite’s virulence [41]. Moreover, it forms complexes with pseudo-kinases ROP2/8 and ROP4/7 [41]. Therefore, given the various roles of ROP5 it is possible that changes in its palmitoylation status would regulate its ability to interact with its partners. However, whether palmitoylation of this protein could affect parasite virulence as well as host-cell invasion remains to be studied.
Figure 3. ROP5 is specifically palmitoylated in T. gondii.
A) Representative Western blot anti-ROP5 on 2BP-treated intracellular parasites. During the ABE protocol, both 2-BP treated parasites and control (DMSO) samples were split into two halves: hydroxylamine-treated (HA+) and control (HA−). An aliquot of each sample was taken before the biotinylation step and was used as input, to normalize the quantification of the bands and as a loading control. B) Quantification of the intensity of the bands in WB: each pull-down value was normalized by its respective input, and each sample final value was expressed as a percentage of the hydroxylamine-treated sample (DMSO, HA+). The graph represents the mean and standard deviation of three independent experiments.
Another interesting aspect is that despite the fact that TgDHHC7 -a palmitoyl S-acyl transferase (PAT)- has been found in rhoptry organelles, ROPs 5/8/11/13 and 14 might not be substrates for this PAT. This possibility comes from the fact that ROPs5/8/11 and 13 are secreted proteins that contain a predicted signal peptide. Instead, they might be substrates for other PATs located either in the Golgi apparatus or ER [18]. Although ROP14 does not contain a predicted signal peptide, it has been described to behave as if it were [42]).
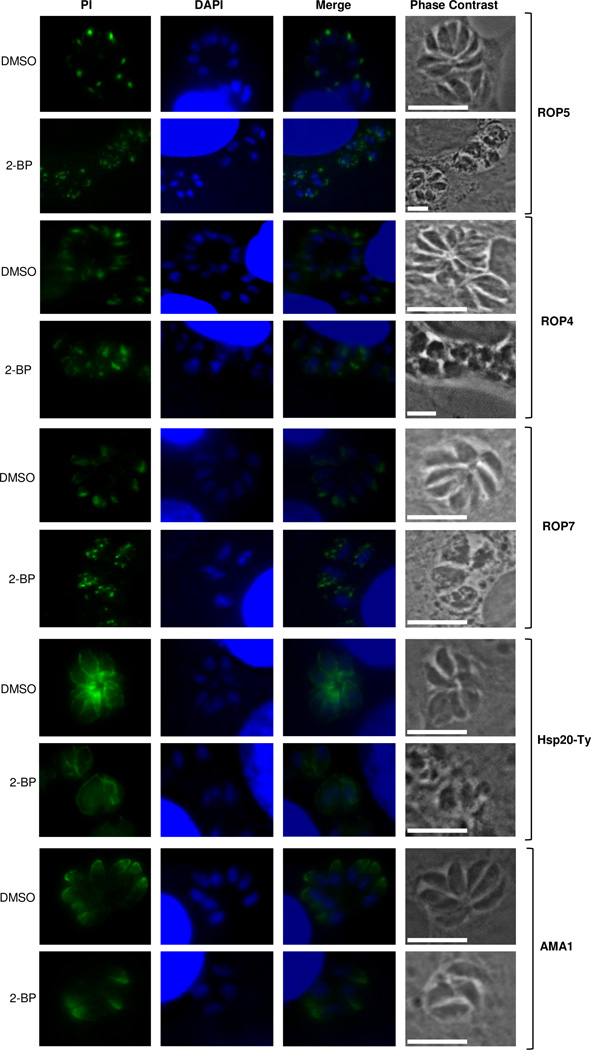
Given our interest in elucidating the influence of palmitoylation on the host-cell invasion machinery, we decided to focus on rhoptry-related proteins. In order to test if palmitoylation of ROP5 was responsible for its localization, we performed immunofluorescence studies on 2-BP treated intracellular parasites using specific monoclonal antibodies against ROP5. Figure 4 shows a clear change in subcellular localization in comparison with the untreated control: while the untreated sample showed a punctuated and apically defined rhoptry localization, 2-BP-treated parasites seem to be unable to direct these proteins to their specific organelle, thus showing a disperse pattern all over the parasite.
Figure 4. Localization of rhoptry proteins in 2-BP-treated and control intracellular tachyzoites.
Columns, from left to right: protein of interest (PI), DAPI, merge, phase contrast. DMSO indicates control (untreated) samples and 2-BP indicates samples treated with 12.5 µM 2-bromopalmitate. Square brackets on the right indicate the corresponding antibodies used: anti-ROP5, -ROP4, -ROP7, -Ty and -AMA1 respectively.
The same phenotypic effect observed for ROP5 was also found in ROP4 and ROP7 (Figure 4). We also included parasites expressing a Ty-tagged version of HSP20 as a control, since it has already been proved that HSP20 localization to the IMC is controlled by palmitoylation [12]. A clear change in HSP20 localization could be observed after 2-BP treatment, with the appearance of a characteristic “membranous” structure. The transmembrane protein AMA1 which according to our study appears to be palmitoylated as well, did not change its subcellular localization after treatment with the palmitoylation inhibitor, which suggests that palmitoylation is not essential for its microneme targeting in the parasite. A similar result was recently found by Foe and coworkers using a click chemistry reaction (copper-catalyzed cycloaddition) [15].
The change in localization of rhoptry proteins observed after 2-BP treatment is in accordance with what was previously reported for TgDHHC7 knock-out parasites, by Frenal et al. [18]: they observed that when TgDHHC7 is not expressed, TgARO -a protein which normally localizes to the periphery of the rhoptries- becomes cytosolic and that the rhoptry organelles are found dispersed through the cytosol and not clustered in the apical pole as expected. Furthermore, Beck and co-workers showed that complementation of TgDHHC7 knock-out parasites with a catalytically inactive TgDHHC7 fails to rescue the apical rhoptry phenotype, suggesting that the tethering of this organelle is dependent on TgDHHC7 activity [43]. As such, palmitoylation of rhoptry proteins is critical for their correct cellular positioning. It has been described that inhibition of palmitoylation prevents the invasion process [16] and that palmitoylation exacerbation enhances the secretion of invasion-associated organelles thus increasing the invasive capacity of tachyzoites [17]. Whether this effect is related to palmitoylation of rhoptry proteins remains to be studied in more detail.
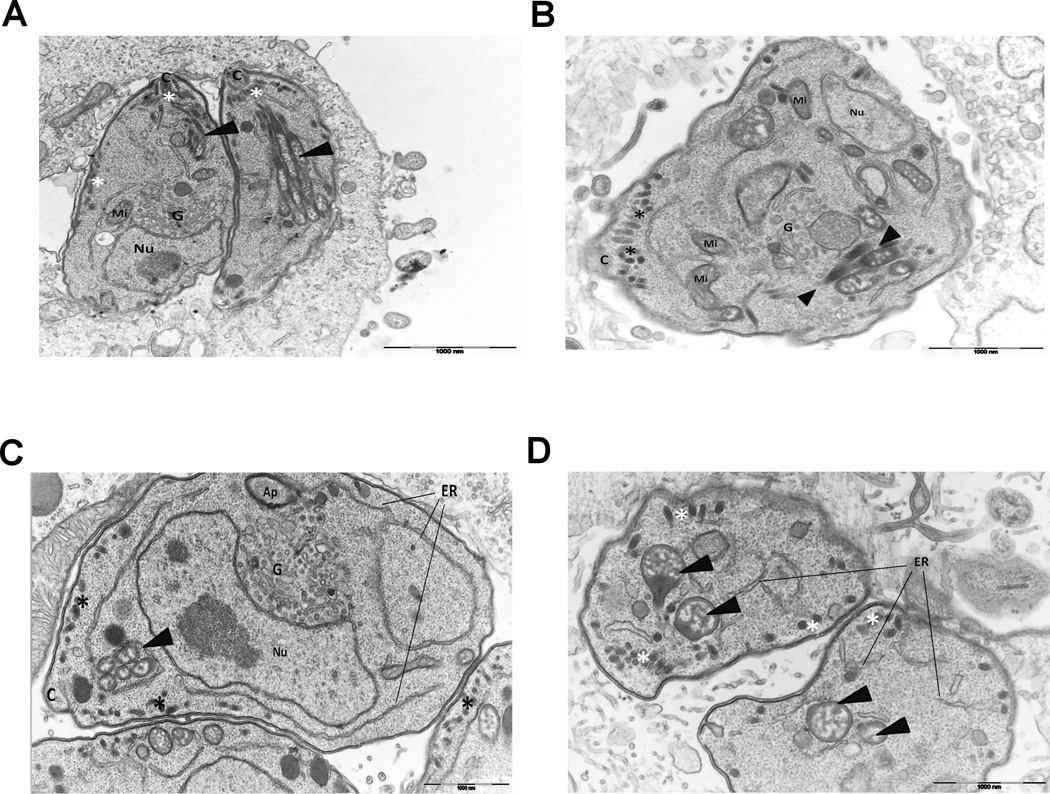
A question that aroused from the observed mislocalization of rhoptry proteins after 2-BP treatment was whether palmitoylation contributes to the general organization and morphology of rhoptries or if just affects the positioning of their protein content. To address this issue, we performed transmission electron microscopy on 2-BP-treated intracellular parasites. Our results (Figure 5) show that in the control - DMSO-treated - samples, Toxoplasma has a very well established internal organization with micronemes and rhoptries all set between the nucleus and the conoid (Figure 5A). Inhibition of palmitoylation by 2-BP, do not consistently alter rhoptry shape or size (Figure 5B). However, abnormal rhoptries, rather spherical than club shaped, were sometimes observed (Figure 5C). Likewise, their positioning inside the parasite seems to be affected: instead of being apically located and converging to the conoid, they appear more disperse in the cytoplasm (Figure 5D). These results are in accordance with was reported for Plasmodium falciparum, a closely related organism, where treatment with 2-BP caused gross mislocalization of rhoptries [44], and also supports our immunofluorescence studies where we observed some ROP proteins scattered in the cytosol after 2-BP treatment. Thus, protein palmitoylation might be affecting the localization and/or the function of some rhoptry proteins, among others, as well as the positioning of the rhoptries but not so much the morphology of the organelles themselves. Golgi apparatus also appears to be affected by the inhibition of protein palmitoylation since it looks enlarged and with more cisternae when compared to the control, and the ER is also more noticeable in 2-BP-treated parasites (Figure 5C). This is not a striking phenomenon, because in the absence of palmitoylation, proper tethering of many proteins to their target locations may fail and they might accumulate in those membranous organelles along the trafficking pathway. Another effect of 2-BP-treatement was that, besides the normal distribution around the base of the conoid, micronemes were observed in large amount scattered in the periphery of the cells (Figure 5, panels C and D). Given that both rhoptries and micronemes are fundamental parts of the host-cell invasion machinery, protein palmitoylation, being a dynamic mechanism, is likely to help control such highly coordinated processes.
Figure 5. Transmission electron microscopy of intracellular parasites.
A) control (DMSO-treated) Longitudinal section of two tachyzoites. Bundles of rhoptries with normal club-shaped morphology can be observed. Micronemes are apically located next to the conoid. B, C and D) 2BP-treated. Some abnormal rhoptries in morphology and localization can be observed, micronemes are scattered in the periphery of the parasite, not just in the apical region and Golgi is enlarged. Arrowheads indicate rhoptries, asterisks indicate micronemes, Nu: host-cell nucleus and, C: conoid, Mi: mitochondria, Ap: apicoplast, G: Golgi apparatus, ER: endoplasmic reticulum.
4. Concluding remarks
T. gondii palmitoylome reveals that this post-translational modification is widespread and a general mechanism of dynamic protein regulation that affects vital processes of this parasite. The information here provided includes proteins already known to be palmitoylated, which validates ABE as an efficient method of choice to identify palmitoylated proteins, but also describes other novel palmitoylated proteins.
In fact, a recent T. gondii palmitoylome was published using a click chemistry approach [15]. Although there are a large number of proteins found by both methods, it is important to note that each method results in the isolation of unique classes of false positives. For this reason is that ABE- and click chemistry-based palmitoylome purification methods are complementary and that all proteins found by any method will require further validation [44].
The fact of having found nuclear palmitoylated proteins is exciting and deserves to be studied in more detail. Furthermore, the identification of palmitoylated proteins in different components of the host-cell invasion machinery (mainly rhoptries but also IMC, micronemes and even dense granules) suggests a key role of this modification in the invasion process. The information here presented will be extremely useful to the whole community since it sets the basis for further studies that could shed light on the role of palmitoylation on particularly selected relevant proteins.
Supplementary Material
Acknowledgments
This work was supported by: ANPCyT grant BID 1728 OC-AR PICT 2010-1494 (MMC), a PIP grant 2010-0190 (MMC) and the proteomic facility is supported by NIH-funded Vermont Genetics Network (P20GM103449). MMC is a researcher from the National Council of Research (CONICET) and UNSAM. MCC and AMA are PhD fellows from CONICET.
Footnotes
Supporting Information
Table S1: List of all non-redundant identified candidate proteins from the three independent ABE assays. The first worksheet tab contains a multiconsensus analysis showing proteins identified in all three assays together with their respective mass spectrometry information and in silico predictions. Highlighted in blue are those already described to palmitoylated, in grey are those predicted to be palmitoylated but without experimental evidence and in green those that have homology to known S-acylated proteins from P. falciparum. Tabs 2, 3 and 4 contain particular information and NRP calculations for each assay, while the last tab is a summary of the calculated NRP HA+/HA- ratios calculated for each protein.
Table S2: Gene Ontology (GO) terms enrichment analyses for cellular component, molecular function and biological processes showing fold enrichment for each term.
References
- 1.Linder ME, Deschenes RJ. New insights into the mechanisms of protein palmitoylation. Biochemistry. 2003;42:4311–4320. doi: 10.1021/bi034159a. [DOI] [PubMed] [Google Scholar]
- 2.Henis YI, Hancock JF, Prior IA. Ras acylation, compartmentalization and signaling nanoclusters (Review) Mol Membr Biol. 2009;26:80–92. doi: 10.1080/09687680802649582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Michaelson D, Ahearn I, Bergo M, Young S, Philips M. Membrane trafficking of heterotrimeric G proteins via the endoplasmic reticulum and Golgi. Mol Biol Cell. 2002;13:3294–3302. doi: 10.1091/mbc.E02-02-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berthiaume L, Deichaite I, Peseckis S, Resh MD. Regulation of enzymatic activity by active site fatty acylation. A new role for long chain fatty acid acylation of proteins. J Biol Chem. 1994;269:6498–6505. [PubMed] [Google Scholar]
- 5.Corvi MM, Soltys CL, Berthiaume LG. Regulation of mitochondrial carbamoyl-phosphate synthetase 1 activity by active site fatty acylation. J Biol Chem. 2001;276:45704–45712. doi: 10.1074/jbc.M102766200. [DOI] [PubMed] [Google Scholar]
- 6.Blaskovic S, Blanc M, van der Goot FG. What does S-palmitoylation do to membrane proteins? FEBS J. 2013;280:2766–2774. doi: 10.1111/febs.12263. [DOI] [PubMed] [Google Scholar]
- 7.Kostiuk MA, Keller BO, Berthiaume LG. Palmitoylation of ketogenic enzyme HMGCS2 enhances its interaction with PPARalpha and transcription at the Hmgcs2 PPRE. FASEB J. 2010;24:1914–1924. doi: 10.1096/fj.09-149765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Park S, Patterson EE, Cobb J, Audhya A, Gartenberg MR, et al. Palmitoylation controls the dynamics of budding-yeast heterochromatin via the telomere-binding protein Rif1. Proc Natl Acad Sci U S A. 2011;108:14572–14577. doi: 10.1073/pnas.1105262108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beck JR, Rodriguez-Fernandez IA, de Leon JC, Huynh MH, Carruthers VB, et al. A novel family of Toxoplasma IMC proteins displays a hierarchical organization and functions in coordinating parasite division. PLoS Pathog. 2010;6:e1001094. doi: 10.1371/journal.ppat.1001094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fung C, Beck JR, Robertson SD, Gubbels MJ, Bradley PJ. Toxoplasma ISP4 is a central IMC sub-compartment protein whose localization depends on palmitoylation but not myristoylation. Mol Biochem Parasitol. 2012;184:99–108. doi: 10.1016/j.molbiopara.2012.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Polonais V, Javier Foth B, Chinthalapudi K, Marq JB, Manstein DJ, et al. Unusual anchor of a motor complex (MyoD-MLC2) to the plasma membrane of Toxoplasma gondii. Traffic. 2011;12:287–300. doi: 10.1111/j.1600-0854.2010.01148.x. [DOI] [PubMed] [Google Scholar]
- 12.De Napoli MG, de Miguel N, Lebrun M, Moreno SN, Angel SO, et al. N-terminal palmitoylation is required for Toxoplasma gondii HSP20 inner membrane complex localization. Biochim Biophys Acta. 2013;1833:1329–1337. doi: 10.1016/j.bbamcr.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Donald RG, Liberator PA. Molecular characterization of a coccidian parasite cGMP dependent protein kinase. Mol Biochem Parasitol. 2002;120:165–175. doi: 10.1016/s0166-6851(01)00451-0. [DOI] [PubMed] [Google Scholar]
- 14.Cabrera A, Herrmann S, Warszta D, Santos JM, John Peter AT, et al. Dissection of minimal sequence requirements for rhoptry membrane targeting in the malaria parasite. Traffic. 2012;13:1335–1350. doi: 10.1111/j.1600-0854.2012.01394.x. [DOI] [PubMed] [Google Scholar]
- 15.Foe IT, Child MA, Majmudar JD, Krishnamurthy S, van der Linden WA, et al. Global Analysis of Palmitoylated Proteins in Toxoplasma gondii. Cell Host Microbe. 2015;18:501–511. doi: 10.1016/j.chom.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alonso AM, Coceres VM, De Napoli MG, Nieto Guil AF, Angel SO, et al. Protein palmitoylation inhibition by 2-bromopalmitate alters gliding, host cell invasion and parasite morphology in Toxoplasma gondii. Mol Biochem Parasitol. 2012;184:39–43. doi: 10.1016/j.molbiopara.2012.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Child MA, Hall CI, Beck JR, Ofori LO, Albrow VE, et al. Small-molecule inhibition of a depalmitoylase enhances Toxoplasma host-cell invasion. Nat Chem Biol. 2013 doi: 10.1038/nchembio.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frenal K, Tay CL, Mueller C, Bushell ES, Jia Y, et al. Global analysis of apicomplexan protein S-acyl transferases reveals an enzyme essential for invasion. Traffic. 2013;14:895–911. doi: 10.1111/tra.12081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Donald RG, Carter D, Ullman B, Roos DS. Insertional tagging, cloning, and expression of the Toxoplasma gondii hypoxanthine-xanthine-guanine phosphoribosyltransferase gene. Use as a selectable marker for stable transformation. J Biol Chem. 1996;271:14010–14019. doi: 10.1074/jbc.271.24.14010. [DOI] [PubMed] [Google Scholar]
- 20.Roos DS, Donald RG, Morrissette NS, Moulton AL. Molecular tools for genetic dissection of the protozoan parasite Toxoplasma gondii. Methods Cell Biol. 1994;45:27–63. doi: 10.1016/s0091-679x(08)61845-2. [DOI] [PubMed] [Google Scholar]
- 21.Wan J, Roth AF, Bailey AO, Davis NG. Palmitoylated proteins: purification and identification. Nat Protoc. 2007;2:1573–1584. doi: 10.1038/nprot.2007.225. [DOI] [PubMed] [Google Scholar]
- 22.Spiess PC, Deng B, Hondal RJ, Matthews DE, van der Vliet A. Proteomic profiling of acrolein adducts in human lung epithelial cells. J Proteomics. 2011;74:2380–2394. doi: 10.1016/j.jprot.2011.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gajria B, Bahl A, Brestelli J, Dommer J, Fischer S, et al. ToxoDB: an integrated Toxoplasma gondii database resource. Nucleic Acids Res. 2008;36:D553–D556. doi: 10.1093/nar/gkm981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sigrist CJ, de Castro E, Cerutti L, Cuche BA, Hulo N, et al. New and continuing developments at PROSITE. Nucleic Acids Res. 2013;41:D344–D347. doi: 10.1093/nar/gks1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 2011;8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 26.Horton P, Nakai K. Better prediction of protein cellular localization sites with the k nearest neighbors classifier. Proc Int Conf Intell Syst Mol Biol. 1997;5:147–152. [PubMed] [Google Scholar]
- 27.Ren J, Wen L, Gao X, Jin C, Xue Y, et al. CSS-Palm 2.0: an updated software for palmitoylation sites prediction. Protein Eng Des Sel. 2008;21:639–644. doi: 10.1093/protein/gzn039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bologna G, Yvon C, Duvaud S, Veuthey AL. N-Terminal myristoylation predictions by ensembles of neural networks. Proteomics. 2004;4:1626–1632. doi: 10.1002/pmic.200300783. [DOI] [PubMed] [Google Scholar]
- 29.Maurer-Stroh S, Eisenhaber F. Refinement and prediction of protein prenylation motifs. Genome Biol. 2005;6:R55. doi: 10.1186/gb-2005-6-6-r55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Drisdel RC, Green WN. Labeling and quantifying sites of protein palmitoylation. Biotechniques. 2004;36:276–285. doi: 10.2144/04362RR02. [DOI] [PubMed] [Google Scholar]
- 31.Jelenska J, Crawford MJ, Harb OS, Zuther E, Haselkorn R, et al. Subcellular localization of acetyl-CoA carboxylase in the apicomplexan parasite Toxoplasma gondii. Proc Natl Acad Sci U S A. 2001;98:2723–2728. doi: 10.1073/pnas.051629998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen AL, Kim EW, Toh JY, Vashisht AA, Rashoff AQ, et al. Novel components of the Toxoplasma inner membrane complex revealed by BioID. MBio. 2015;6:e02357–e02314. doi: 10.1128/mBio.02357-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hodson N, Invergo B, Rayner JC, Choudhary JS. Palmitoylation and palmitoyl-transferases in Plasmodium parasites. Biochem Soc Trans. 2015;43:240–245. doi: 10.1042/BST20140289. [DOI] [PubMed] [Google Scholar]
- 34.Mitchell DA, Mitchell G, Ling Y, Budde C, Deschenes RJ. Mutational analysis of Saccharomyces cerevisiae Erf2 reveals a two-step reaction mechanism for protein palmitoylation by DHHC enzymes. J Biol Chem. 2010;285:38104–38114. doi: 10.1074/jbc.M110.169102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Garrison E, Treeck M, Ehret E, Butz H, Garbuz T, et al. A forward genetic screen reveals that calcium-dependent protein kinase 3 regulates egress in Toxoplasma. PLoS Pathog. 2012;8:e1003049. doi: 10.1371/journal.ppat.1003049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Frenal K, Polonais V, Marq JB, Stratmann R, Limenitakis J, et al. Functional dissection of the apicomplexan glideosome molecular architecture. Cell Host Microbe. 2010;8:343–357. doi: 10.1016/j.chom.2010.09.002. [DOI] [PubMed] [Google Scholar]
- 37.Fox CA, Gartenberg MR. Palmitoylation in the nucleus: a little fat around the edges. Nucleus. 2012;3:251–255. doi: 10.4161/nucl.20391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Feldman JL, Baeza J, Denu JM. Activation of the protein deacetylase SIRT6 by long-chain fatty acids and widespread deacylation by mammalian sirtuins. J Biol Chem. 2013;288:31350–31356. doi: 10.1074/jbc.C113.511261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen X, Du Z, Shi W, Wang C, Yang Y, et al. 2-Bromopalmitate modulates neuronal differentiation through the regulation of histone acetylation. Stem Cell Res. 2014;12:481–491. doi: 10.1016/j.scr.2013.12.010. [DOI] [PubMed] [Google Scholar]
- 40.Kostiuk MA, Corvi MM, Keller BO, Plummer G, Prescher JA, et al. Identification of palmitoylated mitochondrial proteins using a bio-orthogonal azido-palmitate analogue. FASEB J. 2008;22:721–732. doi: 10.1096/fj.07-9199com. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Etheridge RD, Alaganan A, Tang K, Lou HJ, Turk BE, et al. The Toxoplasma pseudokinase ROP5 forms complexes with ROP18 and ROP17 kinases that synergize to control acute virulence in mice. Cell Host Microbe. 2014;15:537–550. doi: 10.1016/j.chom.2014.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bradley PJ, Ward C, Cheng SJ, Alexander DL, Coller S, et al. Proteomic analysis of rhoptry organelles reveals many novel constituents for host-parasite interactions in Toxoplasma gondii. J Biol Chem. 2005;280:34245–34258. doi: 10.1074/jbc.M504158200. [DOI] [PubMed] [Google Scholar]
- 43.Beck JR, Fung C, Straub KW, Coppens I, Vashisht AA, et al. A Toxoplasma palmitoyl acyl transferase and the palmitoylated Armadillo Repeat protein TgARO govern apical rhoptry tethering and reveal a critical role for the rhoptries in host cell invasion but not egress. PLoS Pathog. 2013;9:e1003162. doi: 10.1371/journal.ppat.1003162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jones ML, Collins MO, Goulding D, Choudhary JS, Rayner JC. Analysis of protein palmitoylation reveals a pervasive role in Plasmodium development and pathogenesis. Cell Host Microbe. 2012;12:246–258. doi: 10.1016/j.chom.2012.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.