ABSTRACT
Although molecular biology, genetics, and related special disciplines represent a large amount of empirical data, a practical method for the evaluation and overview of current knowledge is far from being realized. The main concepts and narratives in these fields have remained nearly the same for decades and the more recent empirical data concerning the role of noncoding RNAs and persistent viruses and their defectives do not fit into this scenario. A more innovative approach such as applied biocommunication theory could translate empirical data into a coherent perspective on the functions within and between biological organisms and arguably lead to a sustainable integrative biology.
KEYWORDS: communicative actions, key levels of biocommunication, natural languages, RNA consortia, viruses
Introduction
Current biology is in crisis. Empirical data from various disciplines are so vast that one method designed at coherent overview, interpretation and evaluation cannot be devised.1 With the start of molecular biology, genetics, genetic engineering, microbiology and more recently epigenetics and genome editing the number of special disciplines and related scientific journals has expanded hugely as has empirical information at every level of organization. Additionally, scientists more and more realize that biological organisms, even the simplest, are extraordinarly complex. Theoretical frameworks that could integrate both the complexity and the number of data to orient future research are still lacking.2
Additionally, the philosophy of science's difficulty in constructing correct scientific sentences in theory, observations and experimental set-ups has exacerbated the problem. The reductionist program of a unified method of natural sciences in which all the sentences of a theory can be brought into congruence to sentences with which we describe empirical observations was falsified by Gödel's incompleteness theorem.3 The problems of reductionistic and mechanistic explanatory models of living organisms are increased by the growing evidence that the main narratives of the last decades are false, e.g (1) the central dogma, (2) the belief that non-for-protein-coding DNA is junk, (3) that one gene codes for one protein and (4) that molecular syntax of DNA represents unequivocal meaning/functions.4,5 All these assumptions have dominated research programmes, financial resourcing and curricula for decades as eternally true assumptions. What is needed is a coherent method which integrates all the empirical data into a non-reductionistic perspective on all phenomena of living nature in general and in detail. The biocommunicative approach offers such a road to a sustainable integrative biology.
The social character of language and communication
In contrast to former concepts of information theory, systems theory or similar mathematical theory of language-derived assumptions, empirical data on natural languages and communicative interactions show convincingly that reductionistic approaches cannot identify the core features of these phenomena. Since G.E. Moore we know that every natural language represents a repertoire of signs – whether it be indices, icons or symbols – which are used according to 3 levels of rules, combinatorial (syntactic), contextual (pragmatic) and content-specific (semantics).3 If one level of rules cannot be identified one cannot speak seriously of a natural language.6 Accordingly, natural languages are not context-free or content-free in principle. Only artificial languages can be constructed in such a formalisable way that is free of context.
In natural languages the rules do not represent natural laws and can be changed according to varying circumstances. Most importantly natural languages do not speak themselves but need living agents that are able to use these signs according to the 3 levels of rules. They may acquire this competence genetically—although as in bees it must be triggered by social interactions to be fully developed—or they may be learned and memorised by social interactions in everyday situations. Prior to any scientific language construction one has to be competent in the use of everyday language in which members of a language community learn how to connect utterances with interactional patterns, and meanings with sign sequences (such as words), and how to combine deep grammar meanings (illocutionary acts) with the uttered superficial grammar of sentences.6 This offers an unique capability to transport different meanings with superficially identical sentences e.g., ‘The shooting of the hunters’.7
Empirical data additionally demonstrate that natural language use and competence is a social event, which means it is not possible for one to follow such rules only once. Rule-following is a kind of social interaction.3,8-12 George Herbert Mead demonstrated convincingly that meaning (semantics) is a social event, a social interaction-derived consensus. This contradicts former assumptions of systems theory, information theory and similar derivatives of mathematical theories of language and their core concept of the (coding) sender (and the decoding) receiver narratives which cannot explain how communicating living agents reach a common agreement on the meaning of signs and the goals of cooperation.13
Linguistic competence
In natural languages living agents sharing the ability to use signs according to 3 levels of rules share a limited repertoire of signs and the rules on how they are used are limited. Nevertheless such agents can generate new sequences and new meanings de novo which cannot be predicted by combinations of former sentences.14 This innovation competence can be found in all populations which use natural languages.3 No algorithm is available for innovative sequences out of real-life interactions. This is in stark contrast to former assumptions of systems theory or other mathematical theory of language derivatives. In particular, the transport of meaning in sequences that do not represent this meaning (in extreme cases they may also represent the opposite) leads us to the inherent feature of natural languages and their sign users: that within the superficial grammar of the uttered sequence a deep grammar is transported in parallel which cannot be identified by analysis of the superficial syntax of the sign sequence.
The deep grammar of an uttered sign sequence determines its meaning, i.e., its function for the addressees. This is essential for natural languages and communication processes; the context in which sign sequences such as sentences are used determines the meaning, not the syntax.
Communicative competence
Linguistic competence denotes the ability to build correct sequences of signs such as in sentences. We term communicative competence to install a social interaction.3,14 Former concepts explained communication in most cases simply as information exchange. This is in fact correct but it is a side-result and not the central aspect. With communicative acts individuals in populations act toward one another or as a group with group identity which itself is the result of a consensus found by communicative acts to install conviction, belief, and knowledge-based empirically tested experiences.6 A core tool of communicative actions is a natural language or code. This helps agents to interact on the basis of signs (index, icons, symbols) following 3 levels of rules (syntax, semantics, pragmatics) such as those documented in the variety of constantive, expressive, innovative, imperative, regulative, communicative and operative speech acts. They are used in everyday life to install social bonds and socially coordinated actions, being essential to install and strengthen self-identity and group identity. The whole life of social organisms depends on whether such communicative actions function or do not function. This is evident to anybody who lives in social communities and can be tested in everday life. Social life is dominated by communicative actions and if someone is excommunicated this will have a variety of serious consequences for her/him.9 The coordination and organization of goals in social communities without language and communication are impossible.
Languages and communication in non-human living nature
After the controversy about the language of bees in 1953 Karl von Frisch definitively proved that non-human animals are able to use signs within their language.15 Interestingly, Manfred Eigen 20 years later insisted that the genetic code represents a real natural language with all its attributes and not just a metaphor.16,17 In both cases this was not an anthropomorphic extension of a well-known human ability to non-human animals or the genetic code. It is a serious empirically evident observation that if organisms coordinate their behavior there must be some competence for this based on appropriate tools such as signs. These signs may be chemicals of various forms, tactile, auditive or visible. Also, purely behavioral patterns may express similar messages and therefore can serve as signaling processes. Otherwise we have to adopt old metaphysical explanatory models to explain commonly shard understanding of messages, such as mysterious (“morphogenic”) fields in which organisms are interwoven to experience feelings or thoughts (“meme”) of other members of the swarm that are determined by natural laws of physics and chemistry and are therefore interwoven within the universe of deterministic cause-and-effect reaction models.18,19
Yet natural language and communication experiences are beyond metaphysical explanations. The sole demand is for identification of living organisms that organize behavioral patterns and coordination of behavior within a group of others as described above. From 1987 to 1990 I developed a theory of communicative nature.20 Living nature is structured and organized by language and communication within and among organisms. This means that besides human language and communication every organism within its population is able to use signs with which organisms can differentiate between self and non-self and exchange relevant content concerning common coordinations and organisations of single and group behavior. These sign-mediated interactions we term biocommunication. The biocommunicative approach investigates both communication processes within and among cells, tissues, organs and organisms as sign-mediated interactions and nucleotide sequences as code, i.e., language-like text, which follows in parallel 3 kinds of rules: combinatorial (syntactic), context-sensitive (pragmatic) and content-specific (semantic).
Key levels of biocommunication
At the begining of the biocommunication approach I investigated signaling and sign use within communicative interactions in plants, animals (corals and bees), fungi, bacteria and viruses with respect to 4 levels of biocommunication3 in general (Fig. 1). Having translated empirical data into a coherent method of investigation to constitute a sustainable integrative biology I invited leading experts in their field to publish books on domains of life such as biocommunication in soil microorganisms, biocommunication of plants, biocommunication of fungi, biocommunication of animals, biocommunication of ciliates and viruses.21-26
Figure 1.
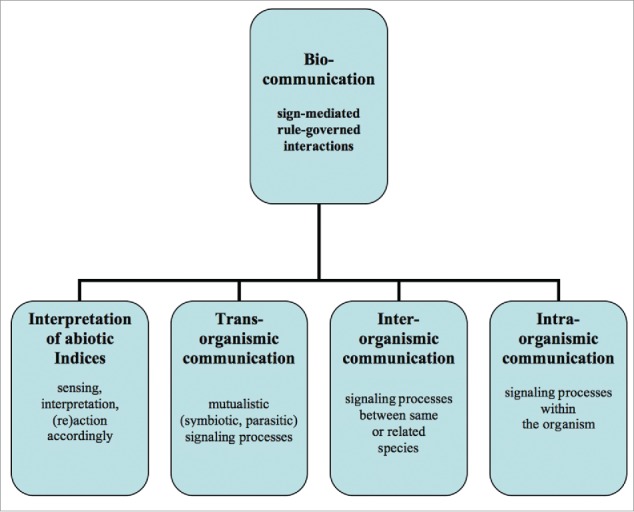
The levels of investigation and categorization according to the biocommunication method.
All this systematized research demonstrated the acceptance and practicability of this method to identify all relevant signaling processes throughout living nature.
Sensing, memory and interpretation of abiotic indices
Sensing, memory and interpretation of recent experiences against the background of memorised experiences according to abiotic indices such as temperature, light, gravity, wind exposure, moisture, etc. All these experiences have to be transported into the organism before an appropriate response behavior can be organized. If something goes wrong within these signaling processes the response behavior may remain rudimentary, deformed or inappropriate for the survival of the organism.
Intraorganismic communication
We can describe all signaling processes within an organism. The organism must differentiate between sensing experiences that come from abiotic indices and receiving biotic information, which means certain behavioral patterns and semiochemicals that are produced not by the organism itself but by those outside.
Interorganismic communication
There is a repertoire of signaling molecules or tactile behavior which derives from species-specific members of a biological community. This means species communities share the ability to generate signals, sense signals, memorise and interpret such signals according to inherited or acquired background knowledge which is stored genetically or by epigenetic markings. As we know from many animals, and even fungi, plants and bacteria, such organism communities can develop ‘dialects’ which are a commonly shared context-dependent ‘culture’ of an ecological niche or specialized environmental circumstances that slightly differ from those of the same species in other ecofields.
Transorganismic communication
Organisms communicate ouside these levels with other organisms that are not members of their species. We can term this trans-organismic. We find such signaling processes in a variety of mutual interactions that extend from all levels of symbiosis until parasitic behavior in that only one participant benefits from these interactions and not the other.
In general no interaction will take place that is coordinated and organized without signaling, including attack or defense behavior. All of these interaction behavioral patterns have been termed as mechanistic in the last decades. However, communicative actions are not mechanistic although they may appear rather conservative and goal oriented. Such actions can change and adapt to a slightly different reaction pattern to derive a completely new behavior out of nothing. This is a key difference from mechanistic and reductionistic explanations.
Context determines meaning: Examples
Competent agents that generate and use signs to coordinate behavior in most cases use semiochemicals (semeion = sign) to transport messages. The repertoire of these semiochemicals which are used as signs in signaling pathways or even in combinations as sign sequences is rather limited in comparison with the variety of behaviors that have to be coordinated. In natural language/code use it is usual to use the same signs and sign sequences to transport different semantic contents (meanings). The context in which interacting living agents are involved decides the meanings of the signs. The following brief examples from plants, fungi, animals and epigenetics will demonstrate this.
Auxin is used in plants' hormonal, morphogenic and transmitter pathways. Because the context of use can be very complex and highly diverse, identifying momentary usage is extremely difficult.27 For synaptic neuronal-like cell-cell communication, plants use neurotransmitter-like auxin28 and presumably also neurotransmitters such as glutamate, glycine, histamine, acetylcholine, and dopamine - all of which they also produce. Auxin is detected as an extracellular signal at the plant synapse in order to react to light and gravity.29 However, it also serves as an extracellular messenger substance sending electrical signals and functions as a synchronisation signal for cell division.30 In intracellular signaling, auxin serves in organogenesis, cell development and differentiation. In the organogenesis of roots, for example, auxin enables cells to determine their position and their identity.31 The cell wall and the organelles it contains help to regulate the signal molecules. Auxin serves also as a growth hormone. Intracellularly, it mediates in cell division and cell elongation. At the intercellular, whole-plant level, it supports cell division in the cambium, and at the tissue level it promotes the maturation of vascular tissue during embryonic development, and organ growth as well as tropic responses and apical dominance.32
In fungi semiochemicals are used to transport certain meanings. Such meanings are subject to change, and rely on different behavioral contexts, which differ under different conditions. In fungi such contexts concern, e.g., cell adhesion, pheromone response, calcium/calmodulin pathways, cell integrity, osmotic growth, cell growth and stress response.33 In this context different modes of behavior can be organized by syntactically identical signaling. For example, cyclic adenosine monophosphate (cAMP) may trigger and inhibit in a variety of fungal species filamentous growth, regulate positive virulence, suppress mating, activate protein kinase or directly or indirectly induce developmental changes.34 Additionally it should be mentioned, that depending on the real-life context, epigenetic regulation can supress or amplify incoming or transmitted secondary metabolites, a rather important signaling resource in fungal organisms. As a result, not for every message, a novel sequence hast to be produced.35
It is well known how bees communicate by using signs, especially moving patterns that serve as signs. Such moving patterns are termed bee dances. Here we will look briefly at the waggle dance, which describes the direction of the destination in terms of the respective position of the sun and defines the distance. As the ability to dance is genetically fixed the whole ability to dance a waggle dance that transports the meaning depends on social interactions in the developmental stage of young bees, which means it relies also on the culture of the real lifeworld of the bees. Karl von Frisch demonstrated the existence of bee dialects in that the same dance behavior can indicate rather different distances depending on the cultural background of the bees and their customs and traditions. The waggle dance as part of the foundation of a new colony informs bees about appropriate new hives that scouts have found. The same dance behavior is used when the new hive is colonised and workers fly out to gather food for the hive.15 In different situational contexts such as (1) searching for a new hive and (2) food gathering the same sign sequence is used to motivate other scouts to see if the hive is suitable and motivates worker bees to gather food.
Interestingly, at the genetic level evolution has also found a technique to trigger certain environmental experiences of organisms which can be stored as a memory tool in repeated experiences for better, faster response behavior which aids better adaptation. Especially with the rise of epigenetics it has become obvious that it is not the syntax of the genetic text which serves as a primary information pool and as a blueprint for the development of an organism but the epigenetic markings.36-41 Such epigenetic markings arise from methylation patterns on the genome or histone modifications that mark special pathways for gene expression and transcription in which long non-coding RNAs are determined that in late transcription are split up into short non-coding RNAs and microRNAs relevant to all cellular processes such as transcription, RNA editing, splicing, translation, immunity and repair.
For example, under extreme stress or dramatic environmental change, epigenetic markings can change.38 It has been reported that such stressful situations can reactivate the genomic sequences of grandparents and great-grandparents of plants if the genetic features of the parents are not sufficient to react appropriately to the stressful situation.42,43 As we know, retroposons are stress-inducible elements that are not solely active in plants but also in animals. During mammalian maternal stress which occurs early in fetal life such retroposons activation can induce non-Mendelian-inherited epigenetic traits.44,45
Cells, tissues, organs, organisms
Interestingly in this systematised biocommunication approach on all levels of all domains of life it becomes increasingly clear that such signaling processes occur not solely within cells but also between cells, cellular communities such as tissues and organs. All these biocommunicative processes must function so that the whole organism can perform its interactions within its species-specific community. All these parts of an organism must communicate constantly to coordinate organ functions, the cells of each specific tissue must be replicated correctly in time and space and if such organismic communication processes are deformed or damaged for various reasons this may lead to disease and death. The cells of tissues and organs share a special kind of evolutionary history and actually a group identity which is epigenetically imprinted. This ensures the constant reproduction of tissue-specific cells at the right place and the right time and if forerunners die they must be substituted to be fully functional for the whole organism.46,47 In this respect we can define a disease always as a result of disturbed, incomplete or damaged communication pathways. Major diseases such as cancer are represented by a communication breakdown between cells and within cells which damages their regulation and the communication within a tissue or organ.
Interacting RNA consortia, persistent viruses and their defectives
The biocommunicative approach investigates communication processes both within and among cells, tissues, organs and organisms as sign-mediated interactions, and nucleotide sequences as code, i.e., language-like text, which follows in parallel 3 kinds of rules: combinatorial (syntactic), context-sensitive (pragmatic) and content-specific (semantic).
Natural genome editing from a biocommunicative perspective is competent agent-driven generation and integration of meaningful nucleotide sequences into pre-existing genomic content arrangements and the ability to (re-)combine and (re-)regulate them according to context-dependent (i.e., adaptational) purposes of the host organism. This means the driving force of diversity, genomic change and genetic innovation are not chance mutations (error replication) and their biological selection but competent agents that edit host genomes and can produce de novo genetic sequences in abundance inherently.14,48,49 What are the agents with which we can replace the error replication narrative?
The success story of RNA biology of the last decade corrected the prevailing assumption that non-coding RNAs represent junk, evolutionary senseless remnants of former coding sequences. As we know now, non-coding RNAs are key to all relevant processes in replication, transcription, translation, immunity and repair in all steps and substeps.50 Without non-coding RNAs no cellular life would function. Interestingly, there are strong indicators that suggest that RNAs pre-existed cellular life.51-55 Experiments tested how single-stranded short RNAs fold back to themselves and form stem loops with base-pairing parts and non-base-pairing loops. Especially the single stranded RNA loops are those locations which bind to both internal and external (‘non-self’) loops. These are core features for forming stem loop groups. More interesting was the finding that single RNA stem loops do not fit into biological selection profiles because their sole reaction type is a physico-chemical one.56,57 If there is a bigger group of such RNA stem loops the biological selection procedure , which is absent in the single stem loop mode, kicks off. From the biocommunication perspective the group behavior of RNA stem loops is at the beginning of life, prior to the innovation of cellular life, which all depend on the regulatory actions of non-coding RNAs, with each molecule forming part of a stem or a loop.56 This led me to suggest RNA sociology as a coherent narrative to explain group building and group identities of RNA stem loops.46,57
In parallel the comeback of virology took place. In contrast to the long-held assumption that viruses are parasites which escape from cells phylogenomic analyses were able to demonstrate that the variety of genetic sequences of viruses that are not part of any cellular genome predate the existence of viruses in cells.58-64 The virus-first perspective states that viruses and virus like agents such as mobile genetic elements are the most abundant biological entities on this planet, outnumbering cellular genomes 10-fold. It was empirically tested in parallel whether the predominant viral lifestyle is a lytic cell death and/or disease causing mechanism found in infection epidemics or a con-evolutionary persistent lifestyle in which genetic sequences of host organisms as well as cytoplasm are the preferred habitat of these infectious agents.65 In this respect it must be added that RNA viruses and retroviruses could predate the evolution of DNA viruses. All known RNA viruses share RNA genomes which are formed of the above-mentioned stem loop structures which have incredible replication and innovation rates, previously termed error replication. If genetic sequences represented by RNA viruses are solely molecular entities which strictly underlie physico- chemical laws then replication in this narrative could only mean reproduction of a 1:1 clone. If this were not the case and slight changes followed such reproduction then this was termed error because the ideal copy was not the result. This was a main assumption of Manfred Eigen.16,17 If we adopt the biocommunication perspective such innovation and generation of genetic diversity is not error but the pure productivity of competent agents.66,67
If such viruses or virus-like infectious agents such as mobile genetic elements invade host cellular organisms not lytically but persistently they follow the unique behavioral pattern detected by virologist Luis Villarreal: The addiction module.68,69 Looking at the abundance of RNA consortia that seek to invade cellular host genomes we must conclude that host genomes are a rather limited resource. So there is intense competition to reach their goal. There is never a sole agent that invades but an abundance of ever-present invading RNA (quasi)species.70 If one population invades host genomes more than one competing invading population is present. So the fight is not solely an invading RNA population versus a host immune system, but additionally competing invading RNA populations. If persistent invasion is successful in all cases the competing RNA species are involved as is the immune system of the host. They counterbalance each other and the immune system takes care of both or all involved.71,72 We can find such persistent infection events in most counterbalanced “addiction” modules such as toxin/antitoxin, restriction/modification or other insertion/deletion modules.73 Also, the strict regulatory modules in the eukaryotic replication modus of excision of introns forming a coherent line of exonic sequences that build a protein coding sequence for translation follows such motifs, involving an abundance of co-regulatory RNA consortia such as ribosomal units, editosome, spliceosome, tRNAs and microRNAs.74,75
Interestingly, most persistent viruses lose their infectious lifestyle and remnants of such viruses remain as exapted and coopted (‘domesticated’) regulatory tools for cellular needs that are very important for adaptation and regulatory processes in the host cell (e.g., SINEs, LINEs, ALUs). Even the spliced out and edited RNAs and the degraded RNAs in several cut-and-degrade processes remain useful tools for reuse in newly built RNA consortia, either for immune functions or other RNA group identities relevant in key regulatory processes of cellular host genomes.76
In this respect from the biocommunicative perspective natural genome editing has been performed from the beginning of biological evolution until today by competent agents that are counterregulated in multiple ways. We may term them natural genetic identity producers.70 It therefore can be predicted that in every regulation of cellular genomes we will find some non-coding RNAs essentially involved that are co-apted tools of former infection events: Frantisek Baluska noted ‘Without infection, no evolution.’77
Conclusion
The biocommunication approach complements former molecular biology, genetics and evolutionary theory and depends on the empirical results that are documented in all these disciplines and subdisciplines. In contrast to these mainstream paradigms of the last decades the biocommunication approach can offer an integrative method of interpretation based on the most recent available empirical knowledge about natural languages and codes and the results of natural communication communities. This offers researchers an entry to a sustainable integrative biology which can overview each detail within the realm of a coherent explanatory model, something which was missing until today. Additionally the biocommunication approach may lead and orientate a variety of research goals in this direction for better predictions, a better chance of more efficient investigations and a better output of results relevant to the development of pharmaceutical drugs and therapies.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
References
- [1].Brenner S. Turing centenary: Life's code script. Nature 2012; 482:461; PMID:22358811; http://dx.doi.org/ 10.1038/482461a [DOI] [PubMed] [Google Scholar]
- [2].Baluška F, Witzany G. At the dawn of a new revolution in life sciences. World J Biol Chem 2013; 4:13-15; PMID:23710294; http://dx.doi.org/ 10.4331/wjbc.v4.i2.13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Witzany G. Biocommunication and natural genome editing, Dordrecht: Springer; 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Shapiro JA. Revisting the central Dogma in the 21st century. Ann NY Acad Sci 2009; 1178:6-28. [DOI] [PubMed] [Google Scholar]
- [5].Mattick JS. Deconstructing the dogma: a new view of the evolution and genetic programming of complex organisms. Ann N Y Acad Sci 2009; 1178:29-46; PMID:19845626; http://dx.doi.org/ 10.1111/j.1749-6632.2009.04991.x [DOI] [PubMed] [Google Scholar]
- [6].Witzany G. Life: The communicative structure, Norderstedt, Libri BoD, 2000. [Google Scholar]
- [7].Witzany G, Baluska F. Life's code script does not code itself. The machine metaphor for living organisms is outdated. EMBO Rep 2012; 13:1054-1056; PMID:23146891; http://embor.embopress.org/content/13/12/1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].McCarthy T. Translator's introduction In: Habermas J. The theory of communicative action, 1 Boston, MA: Beacon Press, ix; 1984. [Google Scholar]
- [9].Habermas J. Actions, speech acts, linguistically mediated interactions and the lifeworld. Phil Probl Tod 1994; 1:45-74. [Google Scholar]
- [10].Austin J. How to do things with words. Oxford: Clarendon Press; 1962. [Google Scholar]
- [11].Searle J. Speech acts: an essay in the philosophy of language. London: Cambridge University Press; 1969. [Google Scholar]
- [12].Tomasello M. Origins of human communication. Cambridge, MA: MIT Press; 2008. [Google Scholar]
- [13].Witzany G. Can mathematics explain the evolution of human language? Comm Int Biol 2012; 4:1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Witzany G. Language and communication as universal requirements for life In Kolb V, editor. Astrobiology: an evolutionary approach. Boca Raton: CrC Press; 2014. 349-369 p. [Google Scholar]
- [15].Frisch K von. Bees: their vision, chemical senses and language. Ithaca: Cornell University Press; 1971. [Google Scholar]
- [16].Eigen M. Selforganization of matter and the evolution of biological macromolecules. Naturwissenschaften 1971; 58:465-523; PMID:4942363. [DOI] [PubMed] [Google Scholar]
- [17].Eigen M, Winkler R. The laws of the game: How the principles of nature govern chance Princeton: Princeton University Press; 1981. [Google Scholar]
- [18].Sheldrake R. A. new science of life: the hypothesis of formative causation. Toronto: Saunders of Toronto Ltd; 1981. [Google Scholar]
- [19].Dawkins R. The selfish gene. Oxford: Oxford University Press; 1976. [Google Scholar]
- [20].Witzany G. Natur der sprache - sprache der natur. sprachpragmatisache philosophie der biologie. Würzburg: Königshausen & Neumann; 1993. [Google Scholar]
- [21].Witzany G. (ed). Biocommunication in soil microorganisms. Dordrecht: Springer; 2011. [Google Scholar]
- [22].Witzany G, Baluska F (eds). Biocommunication of plants. Heidelberg: Springer; 2012. [Google Scholar]
- [23].Witzany G. (ed). Biocommunication of fungi. Dordrecht: Springer; 2012. [Google Scholar]
- [24].Witzany G. (ed). Viruses: essential agents of life. Dordrecht: Springer; 2012. [Google Scholar]
- [25].Witzany G. (ed). Biocommunication of animals. Dordrecht: Springer; 2014. [Google Scholar]
- [26].Witzany G. (ed). Biocommunication of ciliates. Dordrecht: Springer; 2016. [Google Scholar]
- [27].Baluska F, Volkmann D, Menzel D. Plant synapses: actin based domains for cell to cell communication. Trends Plant Sci 2005; 10:106-11; PMID:15749467; http://dx.doi.org/ 10.1016/j.tplants.2005.01.002 [DOI] [PubMed] [Google Scholar]
- [28].Schlicht M, Strnad M, Scanlon MJ, Mancuso S, Hochholdinger F, Palme K, Volkmann D, Menzel D, Baluska F. Auxin immunolocalization implicates vesicular neurotransmitter like mode of polar auxin transport in root apices. Plant Sign Beh 2006; 1:122-130; PMID:19521492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Baluska F, Mancuso S, Volkmann D, Barlow P. Root apices as plant command centres: the unique‚ ‘brain like’ status of the root apex transition zone. Biologia (Bratisl) 2004; 59:7-19. [Google Scholar]
- [30].Campanoni P, Blasius B, Nick P. Auxin transport synchronizes the pattern of cell division in a tobacco cell line. Plant Physiol 2003; 133:1251-1260; PMID:14612587; http://dx.doi.org/ 10.1104/pp.103.027953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Casson SA, Lindsey K. Genes and signalling in root development. New Phytol 2003; 158:11-38; http://dx.doi.org/ 10.1046/j.1469-8137.2003.00705.x [DOI] [Google Scholar]
- [32].Friml J, Wisniewska J. Auxin as an intercellular signal. In: Fleming AJ, editor. Intercellular Communication in Plants. Ann Plant Rev 2005; 16:1-26. [Google Scholar]
- [33].Leeder AC, Palma-Guerrero J, Glass NL. The social network: deciphering fungal language. Nat Rev Microbiol 2011; 9:4440-4451; PMID:21572459; http://dx.doi.org/ 10.1038/nrmicro2580 [DOI] [PubMed] [Google Scholar]
- [34].Lengeler KB, Davidson RC, D'Souza C, Harashima T, Shen WC, Wang P, Pan X, Waugh M, Heitman J. Signal transduction cascades regulating fungal developmewnt and virulence. Microbiol Mol Biol Rev 2000; 64:746-785; PMID:11104818; http://mmbr.asm.org/content/64/4/746.long [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Cichewicz R. Epigenetic regulation of secondary metabolite biosynthetic genes in fungi, In: Witzany G. (ed). Biocommunication of fungi. Dordrecht: Springer; 2012. 57-69 p. [Google Scholar]
- [36].Van Speybroeck L, Van de Vijver G, de Waele D (eds). From epigenesis to epigenetics: the genome in context. Ann NY Acad Sci 2003. 981 p. [Google Scholar]
- [37].Jablonka E, Raz G. Transgenerational epigenetic inheritance: prevalence, mechanisms, and implications for the study of heredity and evolution. Q Rev Biol 2009; 84:131-176; PMID:19606595. [DOI] [PubMed] [Google Scholar]
- [38].Slotkin RK, Martienssen R. Transposable elements and the epigenetic regulation of the genome. Nat Rev Genet 2007; 8:272-285; PMID:17363976; http://www.nature.com/nrg/journal/v8/n4/full/nrg2072.html [DOI] [PubMed] [Google Scholar]
- [39].Mattick JS, Amaral PP, Dinger ME, Mercer TR, Mehler MF. RNA regulation of epigenetic processes. Bioessays 2009; 31:51-59; PMID:19154003; http://www.ncbi.nlm.nih.gov/pubmed/19154003 [DOI] [PubMed] [Google Scholar]
- [40].Mattick JS. RNA as a substrate for epigenome-environment interactions:RNA guidance of epigenetic processes and the expansion of RNA editing in animals underpins development, phenotypic plasticity, learning and cognition. Bioessays 2010; 32:548-552; PMID:20544741; http://dx.doi.org/ 10.1002/bies.201000028 [DOI] [PubMed] [Google Scholar]
- [41].Mercer TR, Mattick JS. Structure and function of long non-coding RNAs in epigenetic regulation. Nat Struct Mol Biol 2013; 20:300-307; PMID:23463315; http://dx.doi.org/ 10.1038/nsmb.2480 [DOI] [PubMed] [Google Scholar]
- [42].Lolle SJ, Victor JL, Young JM, Pruitt RE. Genome-wide non-mendelian inheritance of extragenomic information in Arabidopsis. Nature 2002; 434:505-509; PMID:15785770; http://dx.doi.org/ 10.1038/nature05251 [DOI] [PubMed] [Google Scholar]
- [43].Pearson H. Cress overturns Textbook genetics. Nature 2005; 434:351-360; http://dx.doi.org/ 10.1038/news050321-8 [DOI] [Google Scholar]
- [44].Huda A, Jordan IK. Epigenetic regulation of mammalian genomes by transposable elements. Ann NY Acad Sci 2009; 1178:276-284; PMID:19845643; http://dx.doi.org/ 10.1111/j.1749-6632.2009.05007.x [DOI] [PubMed] [Google Scholar]
- [45].Conley AB, Jordan IK. Endogenous Retroviruses and the Epigenome In: Witzany G. (ed). Viruses: essential agents of life. Dordrecht: Springer; 2012. 309-323 p. [Google Scholar]
- [46].Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet 2003; 33:245-254; PMID:12610534; http://dx.doi.org/ 10.1038/ng1089 [DOI] [PubMed] [Google Scholar]
- [47].Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature 2007; 447:425-432; PMID:17522676; http://dx.doi.org/ 10.1038/nature05918 [DOI] [PubMed] [Google Scholar]
- [48].Villarreal LP, Witzany G. The DNA habitat and its RNA inhabitants: at the dawn of RNA sociology. Genom Ins 2013; 6:1-12; PMID:26217106; http://dx.doi.org/ 10.4137/GEI.S11490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Witzany G. Crucial steps to life: From chemical reactions to code using agents. BioSystems 2016; 140:49-57; PMID:26723230; http://dx.doi.org/ 10.1016/j.biosystems.2015.12.007 [DOI] [PubMed] [Google Scholar]
- [50].Witzany G. Noncoding RNAs: persistent viral agents as modular tools forcellular needs. Ann NY Acad Sci 2009; 1178:244-267; PMID:19845641; http://dx.doi.org/ 10.1111/j.1749-6632.2009.04989.x [DOI] [PubMed] [Google Scholar]
- [51].Hayden EJ, Lehman N. Self-assembly of a group I intron from inactive oligonucleotide fragments. Chem Biol 2006; 13:909-918; PMID:16931340; http://dx.doi.org/ 10.1016/j.chembiol.2006.06.014 [DOI] [PubMed] [Google Scholar]
- [52].Smit S, Yarus M, Knight R. Natural selection is not required to explain universal compositional patterns in rRNA secondary structure categories. RNA 2006; 12:1-14; PMID:16373489; http://dx.doi.org/ 10.1261/rna.2183806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Vaidya N, Manapat ML, Chen IA, Xulvi-Brunet R, Hayden EJ, Lehman N. Spontaneous network formation among cooperative RNA replicators. Nature 2012; 491:72-77; PMID:23075853; http://dx.doi.org/ 10.1038/nature11549 [DOI] [PubMed] [Google Scholar]
- [54].Larson BC, Jensen RP, Lehman N. The chemical origin of behavior is rooted in abiogenesis. Life 2012; 2:313-322; PMID:25371268; http://dx.doi.org/ 10.3390/life2040313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Vaidya N, Walker SI, Lehman N. Recycling of informational units leads to selection of replicators in a prebiotic soup. Chem Biol 2013; 20:241-252; PMID:23438753; http://dx.doi.org/ 10.1016/j.chembiol.2013.01.007 [DOI] [PubMed] [Google Scholar]
- [56].Higgs PG, Lehman N. The RNA world: molecular cooperation at the origins of life. Nat Rev Genet 2015; 16:7-17; PMID:25385129; http://dx.doi.org/ 10.1038/nrg3841 [DOI] [PubMed] [Google Scholar]
- [57].Draper WE, Hayden EJ, Lehman N. Mechanisms of covalent self-assembly of the Azoarcus ribozyme from four fragment oligonucleotides. Nuc Acids Res 2008; 26:520-531; PMID:18048415; http://dx.doi.org/ 10.1093/nar/gkm1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Villarreal LP. Viruses and the evolution of life. Washington: ASM Press; 2005. [Google Scholar]
- [59].Ryan F. Virolution. London: Harper Collins; 2009. [Google Scholar]
- [60].Forterre P, Prangishvili D. The great billion-year war between ribosome-and capsid-encoding organisms (cells and viruses) as the major source ofevolutionary novelties In: Witzany G, editor. Natural genetic engineering and natural genome editing Hoboken: John Wiley & Sons; 2010. 65-77 p. [Google Scholar]
- [61].Koonin EV. On the origin of cells and viruses: primordial virus world scenario In: Witzany G. (ed). Natural genetic engineering and natural genome editing. Hoboken, John Wiley & Sons; 2010. 47-64 p. [Google Scholar]
- [62].Villarreal LP, Witzany G. Viruses are essential agents within the stem and roots of the tree of life. J Theor Biol 2010; 262:698-710; PMID:19833132; http://dx.doi.org/ 10.1016/j.jtbi.2009.10.014 [DOI] [PubMed] [Google Scholar]
- [63].Rohwer F, Youle M, Maughan H, Hisakawa N. Life in our phage world. San Diego: Wholon; 2014. [Google Scholar]
- [64].Seligmann H, Raoult D. Unifying view of stem-loop hairpin RNA as origin of current and ancient parasitic and non-parasitic RNAs, including in giant viruses. Curr Opin Microbiol 2016; 31:1-8; PMID:26716728; http://dx.doi.org/ 10.1016/j.mib.2015.11.004 [DOI] [PubMed] [Google Scholar]
- [65].Villarreal LP. Origin of group identity: viruses, addiction and cooperation. New York, NY: Springer; 2009. [Google Scholar]
- [66].Witzany G. From molecular entities to competent agents: viral infection derived consortia act as natural genetic engineers In: Witzany G, editor. Viruses: essential agents of life. Dordrecht: Springer; 2012. 407-419 p. [Google Scholar]
- [67].Witzany G. Pragmatic turn in biology: from biological molecules to genetic content operators. World J Biol Chem 2014; 5:279-285; PMID:25225596; http://dx.doi.org/ 10.4331/wjbc.v5.i3.279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Villarreal LP. The addiction module as a social force In: Witzany Gn. (ed). Viruses: essential agents of life. Dordrecht: Springer; 2012. 107-145 p. [Google Scholar]
- [69].Villarreal LP. Force for ancient and recent life: viral and stem-loop RNA consortia promote life. Ann NY Acad Sci 2014; 1341:25-34; PMID:25376951; http://dx.doi.org/ 10.1111/nyas.12565 [DOI] [PubMed] [Google Scholar]
- [70].Villarreal LP, Witzany G. Rethinking quasispecies theory: from fittest type to cooperative consortia. World J Biol Chem 2013; 4:79-90; PMID:24340131; http://dx.doi.org/ 10.4331/wjbc.v4.i4.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Villarreal LP. Viruses and host evolution: virus-mediated self identity In: Lopez-Larrea C. (ed). Self and non-self. Austin: Landes Bioscience and Springer Science Business Media; 2011. 185-217 p. [Google Scholar]
- [72].Villarreal LP. Viral ancestors of antiviral systems. Viruses 2011; 3:1933-1958; PMID:22069523; http://dx.doi.org/ 10.3390/v3101933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Mruk I, Kobayashi I. To be or not to be: regulation of restriction-modification systems and other toxin-antitoxin systems. Nucl Acids Res 2014; 42:70-86; PMID:23945938; http://dx.doi.org/ 10.1093/nar/gkt711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Witzany G. Noncoding RNAs: persistent viral agents as modular tools forcellular needs. Ann NY Acad Sci 2009; 1178:244-267; PMID:19845641; http://dx.doi.org/ 10.1111/j.1749-6632.2009.04989.x [DOI] [PubMed] [Google Scholar]
- [75].Witzany G. The agents of natural genome editing. J Mol Cell Biol 2011; 3:181-189; PMID:21459884; http://dx.doi.org/ 10.1093/jmcb/mjr005 [DOI] [PubMed] [Google Scholar]
- [76].Villarreal LP, Witzany G. When competing viruses unify: evolution, conservation, and plasticity of genetic identities. J Mol Evol 2015; 80:305-318; PMID:26014757; http://dx.doi.org/ 10.1007/s00239-015-9683-y [DOI] [PubMed] [Google Scholar]
- [77].Baluška F. Cell-Cell channels, viruses, and evolution: via infection, parasitism, and symbiosis toward higher levels of biological complexity. Ann NY Acad Sci 2009; 178:106-19; PMID:19845631; http://dx.doi.org/ 10.1111/j.1749-6632.2009.04995.x [DOI] [PubMed] [Google Scholar]


