Abstract
RNA motifs can be defined broadly as recurrent structural elements containing multiple intramolecular RNA–RNA interactions, as observed in atomic-resolution RNA structures. They constitute the modular building blocks of RNA architecture, which is organized hierarchically. Recent work has focused on analyzing RNA backbone conformations to identify, define and search for new instances of recurrent motifs in X-ray structures. One current view asserts that recurrent RNA strand segments with characteristic backbone configurations qualify as independent motifs. Other considerations indicate that, to characterize modular motifs, one must take into account the larger structural context of such strand segments. This follows the biologically relevant motivation, which is to identify RNA structural characteristics that are subject to sequence constraints and that thus relate RNA architectures to sequences.
Introduction
What is an RNA motif?
No single definition exists for RNA motifs, as they can be proposed and analyzed at different levels of RNA structure. As discussed in a previous review, RNA motifs can be broadly defined as recurrent structural elements, subject to constraints [1]. This review is complementary to a recent review of new high-resolution RNA structures that exhaustively catalogued new and recurrent motifs [2••]. Therefore, we do not attempt to comprehensively discuss each newly reported motif, but rather aim to critically review evolving notions of recurrent RNA motifs in the context of RNA function and evolution, and how to identify, find and classify them.
Types of RNA motifs
We can distinguish two main classes of motifs — those that operate at the level of RNA sequence and those that entail a specific three-dimensional (3D) structure, characterized by a set of 3D coordinates. An example of a sequence motif is the Shine–Dalgarno sequence of bacterial mRNAs or the Sm-binding sites of some eukaryotic non-coding RNAs [3]. At an intermediate level of analysis, the secondary structure (2D) of an RNA is prominent because it can be calculated quite accurately from sequence information, usually through a combination of thermodynamic and comparative sequence analyses [4]. At the level of secondary structure, the RNA double helix is the fundamental motif. Once helices are specified, other motifs become apparent, which, at the level of secondary structure, are classified as hairpin (or terminal) loops, internal loops (including bulges), and multihelix or junction loops. However, this description of RNA structure is incomplete, as it takes no account of non- Watson–Crick base pairing and most tertiary interactions that stabilize the native architecture.
Secondary structure motifs
How much information can we retrieve from analysis of secondary (2D) structures? Zorn et al. [5] calculate and compare frequency distributions of Watson–Crick (WC) paired nucleotides in helical stems and of nominally unpaired nucleotides in hairpin, internal and junction loops, as they appear in the secondary structures of the 16S and 23S rRNAs. Thus, they treat all bases in ‘loops’ as unpaired, even though a large fraction of them form non- WC base pairs, as is evident from high-resolution 3D structures, which have been available since 2000 [6–8]. Definitions of RNA motifs restricted to secondary structure are not necessarily connected to RNA function or evolution. A further point is that, in such an analysis, all junctions, regardless of the number of helices, are grouped together as the same motif. In fact, it is well known that stable four-way junctions can be constructed without ‘unpaired’ bases, whereas stable geometrically defined three-way junctions require non-helical nucleotides, forming stabilizing non-WC base pairs [9,10].
Motifs considered at the level of secondary structure are also the focus of a recent study that employs RNAMotif, a widely used secondary structure definition and search algorithm [11], to search for RNA aptamers in genomic sequences [12•]. The authors have chosen aptamers for which the corresponding X-ray crystal structures have been solved to test their search algorithm. However, a large part of the 3D information in these structures is ignored by the search models employed, so that the search is conducted essentially at the level of 2D motifs. The results of this analysis have not yet been subjected to experimental verification.
The ability to calculate the probabilities of functional motifs occurring in libraries of random-sequence RNA molecules as a function of library size, sequence length and base composition is useful for planning in vitro selection (SELEX) experiments and for theoretical considerations regarding the role of RNA molecules in the origin of life. Knight and co-workers [13••,14,15] have approached these issues computationally. The outcome of such calculations depends critically on how motifs are defined. Although in their most recent contribution, the presence of supporting double helices is taken into account, the actual substrate-binding or catalytic motifs are treated as single-stranded motifs, subject to independent sequence constraints. The presence in such motifs of non-WC base pairs is not taken into account and it is difficult at this time to assess the effects of such approximations on the statistical outcome.
Graph theory has been used to represent RNA secondary structure in various ways for some time [16,17]. In a series of recent articles, Schlick and co-workers [18–21] have promoted the use of two types of graphs, tree graphs and dual graphs, to represent RNA 2D structure. In tree graphs, edges represent helices and vertices represent hairpin, internal and junction loops. In dual graphs, this is reversed. Dual graphs can also represent pseudoknots, which tree graphs cannot. For each known RNA, both representations are available on the RNA-As-Graphs web site ([19]; http://monod.biomath.nyu.edu/rna/rna.php), which catalogues these graphs according to the number of vertices (V) and the topological complexity (identified with the second-smallest eigenvalue of the Laplacian matrix of the graph). In addition, graph theory is used to enumerate possible graphs with the same value of V, to systematically catalog possible RNA secondary structures. However, only graphs with the same V-value can be directly compared for topological complexity using the second eigenvalue. The applicability of this approach to homologous RNA molecules, especially large functional RNAs such as group I introns and rRNAs, which vary widely in the number of stems and loops and therefore in their V-values, may therefore be limited.
Karklin et al. [22•] introduced a labeled dual graph representation of RNA secondary structure, and developed a similarity measure to compare and distinguish RNA molecules belonging to different families of homologs. In such graphs, helices are represented as nodes labeled with the number of WC base pairs, whereas edges are the nominally single-stranded regions that connect helices to each other (hairpin, internal and junction loops), labeled with the number of nucleotides they comprise. As the authors point out, the accuracy of this approach depends on the accuracy of the secondary structures. However, a further implicit assumption is made in this approach, namely that homologous RNA molecules are conserved fundamentally at the level of secondary structure. In fact, 2D structure is less conserved than 3D structure, and it is the 3D structure of an RNA molecule that is subject to natural selection. A dramatic example of this was recently revealed with the publication of X-ray crystal structures of the specificity (S) domain of A and B type RNase P molecules [23–25]. The A and B architectures present similar features, but the two secondary structures display significant differences [26]. Furthermore, detailed structural analyses of internal and hairpin loop motifs show that motifs with different numbers of nucleotides can adopt similar 3D structures, except for variations in the number of looped-out bases. Examples include T-loops [27], and simple internal loops consisting of a single trans Hoogsteen/sugar edge (sheared) base pair and one to three unpaired, looped-out bases [28].
Representations of RNA three-dimensional structure
Different representations can be used to describe molecular structure information [29]. The most basic representation, used by the 3D structure databases, is the Cartesian coordinates of individual atoms, from which other representations can be derived. However, the large number of variables makes the Cartesian representation awkward when comparing structures or searching for recurrent motifs. Internal coordinates (torsion angles) significantly reduce the number of variables and remove the need to align structures to a common coordinate system. Therefore, several such approaches have been developed (see below). A further simplification is the use of pseudo-torsion angles [30]. An approach using distance matrices has also been applied [29]. Finally, symbolic representations involving higher levels of abstraction have been described [31,32]. These approaches seek to capture the biologically most relevant structural information at the appropriate granularity, by averaging over the minor variations of structure typical of non-covalent interactions, such as hydrogen bonding, to identify features that connect 3D structure explicitly to sequence data [33••]. Regardless of representation, the aim of motif analysis is to cluster motifs that share structural features into geometrically similar classes. As pointed out by Reijmers et al. [29], the outcome of clustering experiments depends largely on the way the data are represented.
Huang et al. [34•] used the 3D coordinates of fifteen atoms per RNA residue, including three base atoms and all the backbone and sugar atoms, to calculate the RMSD distance between two RNA fragments of the same length after they are superposed in 3D. They applied the method to compare all hairpin loops of fixed size in a set of RNA 3D structures, including the large rRNAs. The RMSD distances between all pairs of sequence segments were used to cluster the motifs. UPGMA was applied to produce dendograms of the hairpin loop structures. The algorithm is limited in its ability to find motifs involving different strand segments (composite motifs) or insertions. Therefore, the authors also clustered subsets of nucleotides within longer hairpin loops, and recovered GNRA or UNCG tetraloops with inserted nucleotides, that is, ‘pentaloops’ or ‘hexaloops’ that are GNRA or UNCG hairpins with insertions in characteristic positions.
Harrison et al. [35] describe a reduced vectorial representation of RNA 3D structure designed to convert the problem of searching for recurrent 3D motifs to the subgraph isomorphism problem, for which algorithms are known from graph theory. These methods were first developed for searching substructures in libraries of structures of small molecules, and then applied to proteins and carbohydrates, and recently to RNA [36]. For RNA structure searching, two pairs of pseudo-atoms forming two vectors represent each base. These vector pairs compose the nodes of labeled graphs, one node per base. The relative positions of the bases in the 3D structure are captured by edges connecting the nodes, and labeled with the distance between the start and end points of the vectors composing each node. The problem of searching for a 3D motif is thus reduced to the problem of finding subgraph isomorphisms of graphs representing query motifs in graphs representing structures in the RNA database. Harrison et al. [35] use their approach to search for non-WC base pairs and other small motifs. The Ullman algorithm they use scales with n factorial (n!), where n is the number of nodes in the query motif (subgraph); it is not clear how practical this approach is for searching structures to find larger motifs representing entire hairpin or internal loops.
ARTS (alignment of RNA tertiary structures) is a new computational method that compares and aligns pairs of 3D nucleic acid structures (RNA or DNA) to identify common substructures [37•]. Each nucleotide is represented by the position of its phosphate group. The program seeks the rigid transformation of one structure onto another that superimposes the largest number of phosphate groups of one structure onto the phosphate groups of the second structure, within a specified distance error. ARTS can also be used to discover new motifs.
Classifying backbone conformations
Local motifs (i.e. hairpin and internal loops) result in distinct and reproducible backbone conformations. Therefore, several groups have focused on analyzing and classifying RNA backbone conformations to identify new motifs and search for recurrent motifs in complex high-resolution 3D RNA structures.
Schneider et al. [38••] analyzed and classified the backbone conformations of the 5S and 23S rRNAs of the 50S ribosomal subunit from Haloarcula marismortui using Fourier averaging of the six 3D distributions of torsion angles. They identified 18 non-A-type conformations and 14 A-RNA-related conformations, and determined their corresponding torsion angles. Hershkovitz et al. [39] binned the continuous torsional information into a limited number of discrete values and used pattern recognition methods to find structural recurrences. They found they could represent backbone conformations using a small alphabet, consistent with the fact that four torsion angles contain the bulk of the structure information. Recently, Hershkovitz et al. [40•] applied classical statistical signal processing techniques (‘vector quantization’ or k-means clustering) to more rigorously classify RNA nucleotide conformations. Torsion angle information is lost in scalar quantization or binning. This problem is addressed in vector quantization (VQ). With 4D VQ, applied to the four ‘identifier angles’ (α, γ, δ and ζ), about 60 4D clusters were found, indicating about 60 fundamentally distinct nucleotide conformational states within globular RNAs. This compares with 38 configurational classes identified using binning of torsion angles taken one angle at a time (‘visual binning’). 7D VQ was also carried out and combined with a merging stage to merge conformations based on cluster centroid proximity and on structural constraints. Thus, all A-form helical clusters were merged into a single cluster. This reduces the number of clusters to 26, which further simplifies structure classification compared to the 38 bins identified manually. Richardson and co-workers [41•,42] applied quality filtering techniques to reduce noise levels in the backbone torsion angle distributions from an 8636-residue RNA database. The signal that emerged for half-residue torsion angle distributions for α-β-γ and δ-ε-ζ was plotted and contoured in 3D. About a dozen distinct peaks were observed in the distributions and combined in pairs to define complete RNA backbone conformers. The RNA backbone conformations were reparsed into base-to-base ‘suites’ comprising seven variables, with sugar pucker specified at both ends. Their analysis produced a small library of 42 RNA backbone conformers. Thus, all three of these independent methods of analysis show that the torsion angles of the RNA backbone are quite constrained as to the number of distinct conformations that can result without steric clashes, a concept that was already apparent in the early days of nucleic acid stereochemistry [43,44].
The pseudo torsion angles and , which, in older notation, were designated ωv and ωv, can be used to generate a reduced representation of an RNA backbone configuration — the ‘RNA worm’ — a 3D trajectory described using η, θ and the position of each nucleotide in the sequence as the coordinates [30,45]. 2D η–θ plots correspond formally to the Ramachandran plots of φ – ψ torsion angles used to analyze protein conformation. The program Primos was written to search 3D RNA structures for recurrent RNA worms [45]. Szép et al. [46] used Primos to identify additional occurrences of an RNA strand segment with a sharp turn that they observed in an oligonucleotide X-ray structure and that they called the ‘hook-turn’. All hook-turns identified in large RNA structures occur where the strands of a duplex separate so that they can interact with other RNA regions. One strand doubles back to interact with itself in the 5′-helical region. A careful analysis of hook-turns reveals other characteristic structural elements involving base–base or base–sugar interactions.
The computer program COMPADRES (Comparative Algorithm to Discover Recurring Elements of Structure) implements a novel algorithm, based on the RNA worm representation of the backbone, to identify new recurrent backbone conformations of RNA molecules in the structure database without prior knowledge [47••]. The algorithm compares all short RNA worms in the structure database against each other to discover recurrences within user-supplied tolerances. Applying this algorithm, Wadley and Pyle identified four new recurrent backbone conformations comprising five or more nucleotides, which they named for their shapes: π-turns (type 1 and type 2), Ω-turns, α-loops and C2′-endo-mediated flipped adenosine motifs. The authors note that Ω-turns exhibit some common base-pairing features, but lack a clear basepairing pattern. They discuss the bases forming WC base pairs, but overlook non-WC pairs that also occur in Ω-turns. When non-WC pairs are annotated in the secondary structure, a common base-pairing pattern emerges, as shown in Figure 1, which displays the structural annotation of each Ω-turn reported by Wadley and Pyle [47••] in the context of its interactions with other RNA strand segments. Figure 1 shows that Ω-turns, viewed in context, also form characteristic ordered arrays of non-WC base pairs, even though they may be embedded in various kinds of motifs, including a K-turn, an internal loop or a three-way junction. At this stage, there is no agreement in the field as to where to draw the line between a motif and a submotif, with some workers maintaining that any substructure that occurs more than once in the structure database, and is “large enough to be interesting”, qualifies as an RNA motif in its own right [47••]. The present analysis indicates that Ω-turns, considered in the larger structural context in which they are embedded, can be viewed effectively as submotifs. This example supports the notion that criteria of independence and modularity should be applied to distinguish motifs from submotifs [1] and motifs from the molecules they compose.
Figure 1.
Five examples of the new Ω-turn motif, as proposed by Wadley and Pyle [47••], in their structural contexts: (a) 23S rRNA from H. marismortui (PDB code 1JJ2), (b) 16S rRNA from Thermus thermophilus (PDB code 1N32), (c) RNA aptamer (PDB code 1NTB), (d) 23S rRNA from H. marismortui (PDB code 1JJ2) and (e) 23S rRNA from H. marismortui (PDB code 1JJ2). The upper panel shows 3D representations highlighting the conservation of the backbone conformation of the five nucleotides composing each Ω-turn (shown in red). The lower panel shows schematic representations of each Ω-turn in its structural context, annotated with symbols for base-pairing [31] and base-stacking [48] interactions. The nucleotides composing each Ω-turn are shown in red and nucleotides in the syn conformation are indicated in bold. To prepare Figure 1, each Ω-turn was visually inspected in its structural context, and annotated for base-pairing [31] and base-stacking [48] interactions. This analysis clearly shows that motifs comprising Ω-turns share other common structural characteristics and are subject to sequence constraints. Thus, the first base of each Ω-turn forms a WC base pair. In three out of the five cases reported, the second base forms a trans Hoogsteen/sugar edge (sheared) pair [47••]. In the third case, the second base is a uridine, which cannot make a trans Hoogsteen/sugar edge pair, but rather interacts with C18 to form a cis Hoogsteen/sugar edge pair. However, the position of this second base is the same as in the other Ω-turns. In the fifth case, the corresponding base (G1417 in H. marismortui 23S rRNA) is in the syn glycosidic configuration and, were it to rotate back to the more common anti configuration, it would form the same type of base pair with A1678. In each Ω-turn, the fourth base is in the syn glycosidic configuration and is extruded from the helix formed by the preceding nucleotides, so as to form a cis WC base pair with the base belonging to the other strand that pairs with the second base of the Ω-turn strand. The third base also base pairs, in a variable fashion, but always forming a trans base pair.
Motifs defined by global features
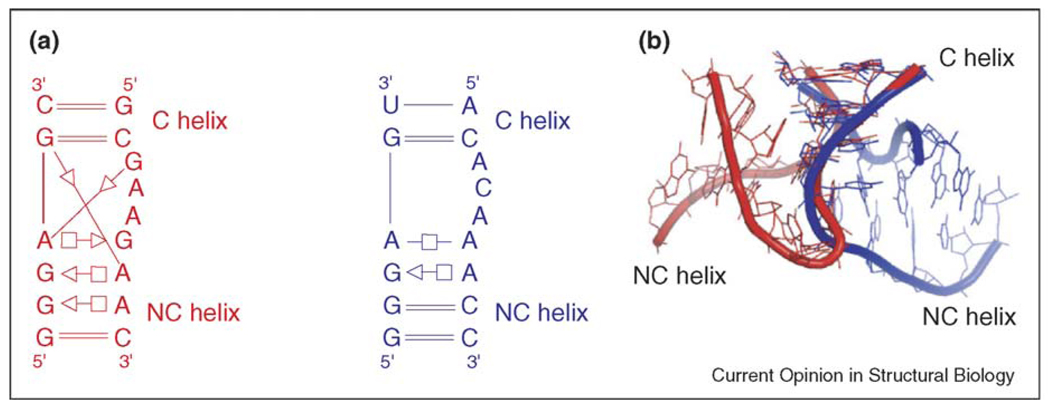
In the striking crystal structure of the Azoarcus group I intron [48,49], Strobel and co-workers noticed a sharp bend between two helical segments, which they named the ‘reverse kink-turn’ [50•]. Such a name implies a close relationship with the previously named kink-turn [51]. In fact, the only common feature of these two motifs is that they produce a sharp bend or kink between two double-stranded elements. However, the kink-turn bends toward the minor/shallow groove and is stabilized by base–base A-minor motifs, whereas the reverse kink-turn bends toward the major/deep groove and is not stabilized by base–base interactions. Annotated drawings comparing the structures of representative kink-turns and reverse kink-turns are shown in Figure 2. This shows that these motifs are fundamentally different. Each motif is characterized by a different set of non-WC base pairs and therefore the motifs do not share sequence signatures (consensus sequences).
Figure 2.
Kink-turn and reverse kink-turn. (a) Schematic structures of the helix 7 kink-turn in 23S rRNA from H. marismortui (shown in red) and the reverse kink-turn in the P9/P9.0 junction of the Azoarcus intron (shown in blue). The structures are annotated for base-pairing interactions using the geometric nomenclature of Leontis and Westhof [31]. (b) Superposition of the canonical helices (C helix) from the 3D structures of the kink-turn (red) and the reverse kink-turn (blue). In the kink-turn structure, the non-canonical helix (NC helix) is oriented in the minor/shallow groove, whereas in the reverse kink-turn structure, the NC helix is in the major/deep groove.
The SCOR classification
The SCOR database aims to comprehensively classify local RNA motifs that appear in 3D structures, focusing on internal and hairpin loops [52,53••]. Although SCOR features 3D data, it is in fact organized using categories defined by secondary structure motifs (http://scor.lbl.gov/scor.html). Thus, only local versions of motifs are provided, omitting structurally similar composite motifs that share the same core of base-pairing and stacking interactions. 3D information is not yet fully exploited in SCOR annotations or classifications; thus, a single dashed line is used to represent all non-WC base pairs in schematic diagrams regardless of the geometric type of each non-WC base pair. For example, internal loops containing n non-WC base pairs, where n = 1, 2, 3, …, are all classified together for a given value of n, regardless of the nature or the order of the component non-WC base pairs. The result is that quite heterogeneous motifs are grouped together while bona fide similarities are overlooked.
Base-pairing patterns and RNA motifs
Keeping in mind that sequences are the more fundamental biological data, other workers have focused on base-pairing patterns and their symbolic representation, and have pointed out that defined backbone configurations are necessary to form ordered arrays of non-WC base pairs [1]. In 2001, a systematic geometry-driven nomenclature was proposed for non-WC RNA base pairs, along with easy to remember annotations for drawing schematic diagrams [31]. Using the nomenclature, all observed and chemically allowed base pairs can be classified into geometric families and isosteric subfamilies that identify those base combinations that can substitute during evolution while preserving 3D structure [54]. Moreover, this approach makes it possible to write computer programs to identify and classify base pairs in 3D structures [55–57]. Alternative classifications use names that are not related directly to the pairing geometry, do not provide ways to annotate 2D diagrams or to automate base-pair identification, and neglect hydrogen bonds involving the 2′-hydroxyl group [58,59].
It is now apparent from crystal structures that RNA architecture is dominated by the continuous stacking of bases. In addition, some arrays of non-WC base pairs prevail because of favorable stacking patterns coupled with standard and stereochemically satisfying sugar– backbone conformations [60]. Significant efforts are still necessary to reconcile these alternative approaches so as to define and classify the major types of recurrent backbone conformations, and associate them with specific occurrences of non-WC base pairs.
The 3D structure of RNA double helices is very regular compared to that of DNA helices. Sequence-dependent differences are more subtle for RNA than for DNA, and are due primarily to near- or non-isosteric base-pair substitutions, including wobble pairs (G/U or A/C), and homopurine (A/G or A/A) and homopyrimidine (C/U, U/U and C/C) pairs, which significantly distort the backbone conformation. Just as RNA helices require the stacking of two or more adjacent WC base pairs, RNA motifs result from combinations of two or more, usually stacked, non-WC base pairs. In this view, individual non-WC base pairs constitute the building blocks of RNA motifs, but do not themselves form integral motifs.
At the level of 3D motifs, more attention is therefore directed to the single-stranded regions of RNA molecules, hairpin or internal loops and multihelix junction loops, than to the helical regions. We now have sufficient numbers of high-resolution structures to conclude that most of the bases in these ‘loop’ regions of structured RNA molecules are paired in non-WC geometries and stacked to form specific structures—3D motifs. Because such a large proportion of bases in ‘loops’ are base paired, a useful definition of RNA motifs at this level is “an ordered array of non-WC base pairs under constraints”. It is worth noting here that, depending on the crystallo-graphic resolution, the presence or absence of individual hydrogen bonds may be difficult to ascertain. It may therefore be difficult to distinguish and classify motifs on the sole basis of hydrogen bonds. As 3D motifs may be local or composite, both types should be included in searches and classifications. Local motifs involve exclusively nucleotides that are close to each other in the secondary structure, that is, they belong to the same hairpin or internal loop. Composite motifs are formed when three or more strands converge to form an ordered array of non-WC base pairs.
Consensus sequences and motif signatures
Consensus sequences are used frequently to describe protein and RNA motifs. Known motifs from different sources are aligned, and the frequency of each residue type is calculated for each column of the alignment and displayed as a sequence logo [61]. This is only appropriate for RNA motifs that are strictly single stranded. To better describe RNA motifs that include WC base pairs, Gorodkin et al. [62] introduced an RNA structure logo that includes mutual information for paired positions [62]. The non-WC base pairs that compose RNA 3D motifs are also subject to pairwise sequence constraints. A more complete description of a recurrent RNA 3D motif, the sequence signature, includes information about the base pairs that can substitute at paired positions and the positions at which insertions and deletions occur. A recent study of two recurrent 3D motifs, the kink-turn and the C-loop, analyzes the sequence variations of all occurrences of these motifs known from crystal structures and derives sequence signatures of this type for each motif [33••]. This paper demonstrates the usefulness of isostericity matrices [54] for analyzing RNA motifs comprising non-WC base pairs, and outlines the steps for productively iterating motif analysis and sequence alignment. The flow chart shown in Figure 3 illustrates the role of isostericity matrices in 3D structural analysis, 3D motif identification and classification, sequence analysis to produce accurate structure-based sequence alignments and 3D modeling, all with the goal of increasing understanding of RNA function and evolution.
Figure 3.
Flow chart illustrating the use of isostericity matrices to integrate 3D structural and sequence information to produce accurate alignments and model 3D structures based on sequence. Isostericity matrices for non-WC base pairs organized in geometric families were proposed based on analysis of high-resolution atomic structures [54], as indicated in path 1. Sequence signatures of RNA motifs identified in 3D structures are deduced by analyzing homologous RNA molecules that have the same motif (path 2). Isostericity matrices are employed to productively iterate between sequence alignment and sequence signature to arrive at accurate, structure-based alignments (path 2). Sequence signatures for recurrent motifs identified in different crystal structures are defined with reference to isostericity matrices (path 3). For families of homologous RNA molecules for which no 3D structure exists (path 4), WC covariations and energy minimization (path 5) can be used to determine common 2D structures, which in turn define hairpin, internal and junction loops in which 3D motifs may occur. Sequence signatures of known motifs are used to propose motifs for loops and to refine alignments of loop regions in an iterative manner (paths 4 and 5). Motif substitutions at corresponding positions in the alignments can also be identified (path 4).
Conclusions
Biological data are fundamentally sequence data. Given the ease of obtaining sequence data and the difficulty of determining 3D structures at high resolution, there will always be more sequence data than structural data. The key challenge for RNA structural and computational biologists and bioinformaticians is to fully integrate these two types of data with a common ontology [63]. The fact that structured RNA molecules are mosaics of recurrent modular motifs means that high-resolution 3D information about one molecule may be useful in analyzing the sequences of another molecule, whether or not the molecules are homologous. For each modular 3D motif identified, we need to define the sequence constraints, as these allow one to identify the motif in other sequences. In this respect, a classification solely based on an analysis of secondary structures is of limited use. For structural biologists, the ability to describe, compare and superimpose using mathematically elegant and powerful tools is the next step after structure determination. Several such tools have been described above, with their advantages and limitations.
However, in the future, there is the hope that the accumulated knowledge of 3D structures, when properly integrated, could be applied to the fundamental problems of searching for non-coding RNA genes in genomic sequences and constructing 3D models of RNA molecules based on known sequences. Finally, we would like to conclude by noting that the ultimate purpose of classification should be borne in mind in order to avoid unnecessary proliferation of confusing jargon. A Tower of Babel of structural acronyms would be erected if every recurrent element of structure at every possible degree of granularity is given a distinct name, without regard to the ways in which these elements combine to create integral structural and functional units or modules.
Acknowledgements
NBL acknowledges grant support from the National Institutes of Health (2 R15 GM055898-03) and the American Chemical Society (PRF# 42357 -AC 4).
References and recommended reading
Papers of particular interest, published within the annual period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Leontis NB, Westhof E. Analysis of RNA motifs. Curr Opin Struct Biol. 2003;13:300–308. doi: 10.1016/s0959-440x(03)00076-9. [DOI] [PubMed] [Google Scholar]
- 2. Holbrook SR. RNA structure: the long and the short of it. Curr Opin Struct Biol. 2005;15:302–308. doi: 10.1016/j.sbi.2005.04.005. Recently determined crystallographic structures are reviewed. All new and recurrent RNA motifs in recent structures are identified and described.
- 3.Khusial P, Plaag R, Zieve GW. LSm proteins form heptameric rings that bind to RNA via repeating motifs. Trends Biochem Sci. 2005;30:522–528. doi: 10.1016/j.tibs.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 4.Mathews DM, Zuker M. Predictive methods using RNA sequences. In: Baxevanis AD, Ouellette BFF, editors. Bioinformatics: A Practical Guide to the Analysis of Genes and Proteins. John Wiley & Sons; 2005. pp. 144–171. [Google Scholar]
- 5.Zorn J, Gan HH, Shiffeldrim N, Schlick T. Structural motifs in ribosomal RNAs: implications for RNA design and genomics. Biopolymers. 2004;73:340–347. doi: 10.1002/bip.10525. [DOI] [PubMed] [Google Scholar]
- 6.Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science. 2000;289:905–920. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- 7.Schuwirth BS, Borovinskaya MA, Hau CW, Zhang W, Vila-Sanjurjo A, Holton JM, Cate JH. Structures of the bacterial ribosome at 3.5 Å resolution. Science. 2005;310:827–834. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- 8.Wimberly BT, Brodersen DE, Clemons WM, Jr, Morgan-Warren RJ, Carter AP, Vonrhein C, Hartsch T, Ramakrishnan V. Structure of the 30S ribosomal subunit. Nature. 2000;407:327–339. doi: 10.1038/35030006. [DOI] [PubMed] [Google Scholar]
- 9.Lilley DM. Structures of helical junctions in nucleic acids. Q Rev Biophys. 2000;33:109–159. doi: 10.1017/s0033583500003590. [DOI] [PubMed] [Google Scholar]
- 10.Lescoute A, Westhof E. Topology of three-way junctions in folded RNAs. RNA. 2006;12:83–93. doi: 10.1261/rna.2208106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Macke TJ, Ecker DJ, Gutell RR, Gautheret D, Case DA, Sampath R. RNAMotif, an RNA secondary structure definition and search algorithm. Nucleic Acids Res. 2001;29:4724–4735. doi: 10.1093/nar/29.22.4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Laserson U, Gan HH, Schlick T. Predicting candidate genomic sequences that correspond to synthetic functional RNA motifs. Nucleic Acids Res. 2005;33:6057–6069. doi: 10.1093/nar/gki911. Structural features of RNA aptamers were used to search genomic sequences using RNAMotif.
- 13. Knight R, De Sterck H, Markel R, Smit S, Oshmyansky A, Yarus M. Abundance of correctly folded RNA motifs in sequence space, calculated on computational grids. Nucleic Acids Res. 2005;33:5924–5935. doi: 10.1093/nar/gki886. The probabilities of functional motifs occurring in libraries of randomsequence RNA molecules are calculated at the level of secondary structure as a function of library size, sequence length and base composition.
- 14.Legiewicz M, Lozupone C, Knight R, Yarus M. Size, constant sequences, and optimal selection. RNA. 2005;11:1701–1709. doi: 10.1261/rna.2161305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Knight R, Yarus M. Finding specific RNA motifs: function in a zeptomole world? RNA. 2003;9:218–230. doi: 10.1261/rna.2138803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Benedetti G, Morosetti S. A graph-topological approach to recognition of pattern and similarity in RNA secondary structures. Biophys Chem. 1996;59:179–184. doi: 10.1016/0301-4622(95)00119-0. [DOI] [PubMed] [Google Scholar]
- 17.Le SY, Nussinov R, Maizel JV. Tree graphs of RNA secondary structures and their comparisons. Comput Biomed Res. 1989;22:461–473. doi: 10.1016/0010-4809(89)90039-6. [DOI] [PubMed] [Google Scholar]
- 18.Kim N, Shiffeldrim N, Gan HH, Schlick T. Candidates for novel RNA topologies. J Mol Biol. 2004;341:1129–1144. doi: 10.1016/j.jmb.2004.06.054. [DOI] [PubMed] [Google Scholar]
- 19.Fera D, Kim N, Shiffeldrim N, Zorn J, Laserson U, Gan HH, Schlick T. RAG: RNA-as-graphs web resource. BMC Bioinformatics. 2004;5:88. doi: 10.1186/1471-2105-5-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gan HH, Fera D, Zorn J, Shiffeldrim N, Tang M, Laserson U, Kim N, Schlick T. RAG: RNA-as-graphs database–concepts, analysis, and features. Bioinformatics. 2004;20:1285–1291. doi: 10.1093/bioinformatics/bth084. [DOI] [PubMed] [Google Scholar]
- 21.Gan HH, Pasquali S, Schlick T. Exploring the repertoire of RNA secondary motifs using graph theory; implications for RNA design. Nucleic Acids Res. 2003;31:2926–2943. doi: 10.1093/nar/gkg365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Karklin Y, Meraz RF, Holbrook SR. Classification of non-coding RNA using graph representations of secondary structure. Pac Symp Biocomput. 2005:4–15. Labeled dual graphs are introduced to compare 2D RNA structures and distinguish RNA molecules that belong to different families of homologues.
- 23.Krasilnikov AS, Xiao Y, Pan T, Mondragon A. Basis for structural diversity in homologous RNAs. Science. 2004;306:104–107. doi: 10.1126/science.1101489. [DOI] [PubMed] [Google Scholar]
- 24.Krasilnikov AS, Yang X, Pan T, Mondragon A. Crystal structure of the specificity domain of ribonuclease P. Nature. 2003;421:760–764. doi: 10.1038/nature01386. [DOI] [PubMed] [Google Scholar]
- 25.Torres-Larios A, Swinger KK, Krasilnikov AS, Pan T, Mondragon A. Crystal structure of the RNA component of bacterial ribonuclease P. Nature. 2005;437:584–587. doi: 10.1038/nature04074. [DOI] [PubMed] [Google Scholar]
- 26.Westhof E, Massire C. Structural biology. Evolution of RNA architecture. Science. 2004;306:62–63. doi: 10.1126/science.1104482. [DOI] [PubMed] [Google Scholar]
- 27.Nagaswamy U, Fox GE. Frequent occurrence of the T-loop RNA folding motif in ribosomal RNAs. RNA. 2002;8:1112–1119. doi: 10.1017/s135583820202006x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leontis NB, Stombaugh J, Westhof E. Motif prediction in ribosomal RNAs lessons and prospects for automated motif prediction in homologous RNA molecules. Biochimie. 2002;84:961–973. doi: 10.1016/s0300-9084(02)01463-3. [DOI] [PubMed] [Google Scholar]
- 29.Reijmers TH, Wehrens R, Buydens LM. The influence of different structure representations on the clustering of an RNA nucleotides data set. J Chem Inf Comput Sci. 2001;41:1388–1394. doi: 10.1021/ci0103626. [DOI] [PubMed] [Google Scholar]
- 30.Olson WK. Configurational statistics of polynucleotide chains. Macromolecules. 1980;13:721–728. doi: 10.1021/ma60045a006. [DOI] [PubMed] [Google Scholar]
- 31.Leontis NB, Westhof E. Geometric nomenclature and classification of RNA base pairs. RNA. 2001;7:499–512. doi: 10.1017/s1355838201002515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gendron P, Lemieux S, Major F. Quantitative analysis of nucleic acid three-dimensional structures. J Mol Biol. 2001;308:919–936. doi: 10.1006/jmbi.2001.4626. [DOI] [PubMed] [Google Scholar]
- 33. Lescoute A, Leontis NB, Massire C, Westhof E. Recurrent structural RNA motifs, isostericity matrices and sequence alignments. Nucleic Acids Res. 2005;33:2395–2409. doi: 10.1093/nar/gki535. Isostericity matrices for non-WC base pairs were used to analyze the structures and sequence alignments of two recurrent RNA motifs, kink-turns and C-loops. Sequence signatures were derived for each motif that might be used to identify and align motifs in other RNA sequences.
- 34. Huang HC, Nagaswamy U, Fox GE. The application of cluster analysis in the intercomparison of loop structures in RNA. RNA. 2005;11:412–423. doi: 10.1261/rna.7104605. Hairpin tetraloops in 3D RNA structures are clustered according to their geometric similarity using an RMSD measure calculated using 15 backbone and base atoms per nucleotide. Major clusters included the GNRA- and UNCG-type hairpin loops.
- 35.Harrison AM, South DR, Willett P, Artymiuk PJ. Representation, searching and discovery of patterns of bases in complex RNA structures. J Comput Aided Mol Des. 2003;17:537–549. doi: 10.1023/b:jcam.0000004603.15856.32. [DOI] [PubMed] [Google Scholar]
- 36.Artymiuk PJ, Spriggs RV, Willett P. Graph theoretic methods for the analysis of structural relationships in biological macromolecules. J Am Soc Inf Sci Tech. 2005;56:518–528. [Google Scholar]
- 37. Dror O, Nussinov R, Wolfson H. ARTS: alignment of RNA tertiary structures. Bioinformatics. 2005;21:II47–II53. doi: 10.1093/bioinformatics/bti1108. The authors present a new computational method to compare two nucleic acid structures (RNAs or DNAs) that detects a priori unknown common substructures.
- 38. Schneider B, Moravek Z, Berman HM. RNA conformational classes. Nucleic Acids Res. 2004;32:1666–1677. doi: 10.1093/nar/gkh333. Fourier averaging of the six 3D distributions of torsion angles, followed by clustering, identified 14 A-type (helical) and 18 non-A-type RNA conformations and their torsion angles.
- 39.Hershkovitz E, Tannenbaum E, Howerton SB, Sheth A, Tannenbaum A, Williams LD. Automated identification of RNA conformational motifs: theory and application to the HM LSU 23S rRNA. Nucleic Acids Res. 2003;31:6249–6257. doi: 10.1093/nar/gkg835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hershkovitz E, Sapiro G, Tannenbaum A, Williams LD. Statistical analysis of RNA backbone. IEEE/ACM Transactions on Computational Biology and Bioinformatics. 2006;3:33–46. doi: 10.1109/TCBB.2006.13. The authors applied tools from statistical signal processing to search for clusters in RNA conformational space. VQ allows several torsion angles to be clustered simultaneously.
- 41. Murray LJ, Richardson JS, Arendall WB, Richardson DC. RNA backbone rotamers–finding your way in seven dimensions. Biochem Soc Trans. 2005;33:485–487. doi: 10.1042/BST0330485. Quality filtering techniques are applied to RNA backbone dihedral angle distributions within sugar-to-sugar ‘suites’. A small library of RNA backbone rotamers is identified that describes almost all RNA backbones in experimental structures.
- 42.Murray LJ, Arendall WB, III, Richardson DC, Richardson JS. RNA backbone is rotameric. Proc Natl Acad Sci USA. 2003;100:13904–13909. doi: 10.1073/pnas.1835769100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sundaralingam M. Stereochemistry of nucleic acids and their constituents. Biopolymers. 1969;7:821–860. [Google Scholar]
- 44.Sundaralingam M, Mizuno H, Stout CD, Rao ST, Liedman M, Yathindra N. Mechanisms of chain folding in nucleic acids. The (omega, omega) plot and its correlation to the nucleotide geometry in yeast tRNAPhe1. Nucleic Acids Res. 1976;3:2471–2484. doi: 10.1093/nar/3.10.2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Duarte CM, Wadley LM, Pyle AM. RNA structure comparison, motif search and discovery using a reduced representation of RNA conformational space. Nucleic Acids Res. 2003;31:4755–4761. doi: 10.1093/nar/gkg682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Szep S, Wang J, Moore PB. The crystal structure of a 26-nucleotide RNA containing a hook-turn. RNA. 2003;9:44–51. doi: 10.1261/rna.2107303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wadley LM, Pyle AM. The identification of novel RNA structural motifs using COMPADRES: an automated approach to structural discovery. Nucleic Acids Res. 2004;32:6650–6659. doi: 10.1093/nar/gkh1002. New software is described that identifies new recurrent backbone conformations in RNA structures without prior knowledge. Four new conformations comprising five or more nucleotides are defined.
- 48.Adams PL, Stahley MR, Gill ML, Kosek AB, Wang J, Strobel SA. Crystal structure of a group I intron splicing intermediate. RNA. 2004;10:1867–1887. doi: 10.1261/rna.7140504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Adams PL, Stahley MR, Kosek AB, Wang J, Strobel SA. Crystal structure of a self-splicing group I intron with both exons. Nature. 2004;430:45–50. doi: 10.1038/nature02642. [DOI] [PubMed] [Google Scholar]
- 50. Strobel SA, Adams PL, Stahley MR, Wang J. RNA kink turns to the left and to the right. RNA. 2004;10:1852–1854. doi: 10.1261/rna.7141504. A new motif featuring a sharp bend toward the major (deep) groove of the RNA helix is described.
- 51.Klein DJ, Schmeing TM, Moore PB, Steitz TA. The kink-turn: a new RNA secondary structure motif. EMBO J. 2001;20:4214–4221. doi: 10.1093/emboj/20.15.4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tamura M, Hendrix DK, Klosterman PS, Schimmelman NR, Brenner SE, Holbrook SR. SCOR: structural classification of RNA, version 2.0. Nucleic Acids Res. 2004;32:D182–D184. doi: 10.1093/nar/gkh080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Klosterman PS, Hendrix DK, Tamura M, Holbrook SR, Brenner SE. Three-dimensional motifs from the SCOR, structural classification of RNA database: extruded strands, base triples, tetraloops and U-turns. Nucleic Acids Res. 2004;32:2342–2352. doi: 10.1093/nar/gkh537. The SCOR database is a compilation and classification of RNA hairpin and internal loop motifs. Several new motifs are described in this article.
- 54.Leontis NB, Stombaugh J, Westhof E. The non-Watson-Crick base pairs and their associated isostericity matrices. Nucleic Acids Res. 2002;30:3497–3531. doi: 10.1093/nar/gkf481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lemieux S, Major F. RNA canonical and non-canonical base pairing types: a recognition method and complete repertoire. Nucleic Acids Res. 2002;30:4250–4263. doi: 10.1093/nar/gkf540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang H, Jossinet F, Leontis N, Chen L, Westbrook J, Berman H, Westhof E. Tools for the automatic identification and classification of RNA base pairs. Nucleic Acids Res. 2003;31:3450–3460. doi: 10.1093/nar/gkg529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jossinet F, Westhof E. Sequence to structure (S2S): display, manipulate and interconnect RNA data from sequence to structure. Bioinformatics. 2005;21:3320–3321. doi: 10.1093/bioinformatics/bti504. [DOI] [PubMed] [Google Scholar]
- 58.Lee JC, Gutell RR. Diversity of base-pair conformations and their occurrence in rRNA structure and RNA structural motifs. J Mol Biol. 2004;344:1225–1249. doi: 10.1016/j.jmb.2004.09.072. [DOI] [PubMed] [Google Scholar]
- 59.Nagaswamy U, Larios-Sanz M, Hury J, Collins S, Zhang Z, Zhao Q, Fox GE. NCIR: a database of non-canonical interactions in known RNA structures. Nucleic Acids Res. 2002;30:395–397. doi: 10.1093/nar/30.1.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Correll CC, Freeborn B, Moore PB, Steitz TA. Metals, motifs, and recognition in the crystal structure of a 5S rRNA domain. Cell. 1997;91:705–712. doi: 10.1016/s0092-8674(00)80457-2. [DOI] [PubMed] [Google Scholar]
- 61.Schneider TD, Stephens RM. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 1990;18:6097–6100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gorodkin J, Heyer LJ, Brunak S, Stormo GD. Displaying the information contents of structural RNA alignments: the structure logos. Comput Appl Biosci. 1997;13:583–586. doi: 10.1093/bioinformatics/13.6.583. [DOI] [PubMed] [Google Scholar]
- 63.Leontis NB, Altman RB, Berman HM, Brenner SE, Brown JW, Engelke DR, Harvey SC, Holbrook SR, Jossinet F, Lewis SE, et al. The RNA ontology consortium: an open invitation to the RNA community. RNA. 2006;12:533–541. doi: 10.1261/rna.2343206. [DOI] [PMC free article] [PubMed] [Google Scholar]