Abstract
Background
Feline diabetes mellitus (DM) shares many pathophysiologic features with human type 2 DM. Human genome‐wide association studies have identified genes associated with obesity and DM, including melanocortin 4 receptor (MC4R), which plays an important role in energy balance and appetite regulation.
Hypothesis/Objectives
To identify single nucleotide polymorphisms (SNPs) in the feline MC4R gene and to determine whether any SNPs are associated with DM or overweight body condition in cats.
Animals
Two‐hundred forty domestic shorthaired (DSH) cats were recruited for the study. Of these, 120 diabetics were selected (60 overweight, 60 lean), along with 120 nondiabetic controls (60 overweight and 60 lean). Males and females were equally represented.
Methods
A prospective case‐control study was performed. Genomic DNA was extracted from blood samples and used as template for PCR amplification of the feline MC4R gene. The coding region of the gene was sequenced in 10 cats to identify polymorphisms. Subsequently, genotyping by restriction fragment length polymorphism (RFLP) analysis assessed MC4R:c.92C > T allele and genotype frequencies in each group of cats.
Results
No significant differences in MC4R:c.92C>T allele or genotype frequencies were identified between nondiabetic overweight and lean cats. In the overweight diabetic group, 55% were homozygous for the MC4R:c.92C allele, compared to 33% of the lean diabetics and 30% of the nondiabetics. The differences between the overweight diabetic and the nondiabetics were significant (P < .01).
Conclusions and Clinical Importance
We identified a polymorphism in the coding sequence of feline MC4R that is associated with DM in overweight DSH cats, similar to the situation in humans.
Keywords: Endocrinology, Feline, Genetic markers, Genetics, Molecular biology, Pancreas
Abbreviations
- AGRP
agouti‐related protein
- BCS
body condition score
- DM
diabetes mellitus
- dNTP
deoxyribonucleotide triphosphate
- DSH
domestic shorthaired
- gDNA
genomic DNA
- GWAS
genome‐wide association studies
- IGF‐1
insulin growth factor‐1
- MC4R
melanocortin 4 receptor
- OR
odds ratio
- PCR
polymerase chain reaction
- RFLP
restriction fragment length polymorphism
- RVC
Royal Veterinary College
- SNP
single nucleotide polymorphisms
- UTR
untranslated region
- α‐MSH
alpha melanocyte stimulating hormone
Diabetes mellitus (DM) is one of the most common feline endocrinopathies.1 In the majority of diabetic cats, the disease is considered to result from similar pathophysiologic mechanisms as human type 2 DM.1 Although the majority of cats suffer from type 2‐like DM, other types of DM do occur, resulting from a variety of underlying disorders that impact pancreatic beta cell function (eg, pancreatitis)2 or that cause marked insulin resistance (eg, growth hormone‐secreting pituitary tumor in acromegaly).3, 4 DM is most commonly seen in domestic shorthaired (DSH) cats. Burmese cats are overrepresented in the diabetic population, whereas other pure breeds seem underrepresented.5, 6 These differences in breed susceptibility suggest that genetic factors are involved in feline DM.
Two important factors may contribute to development of type 2 DM in humans.7 First, the presence of insulin resistance, which is considered to be primarily a consequence of environmental factors such as excessive caloric intake and decreased activity, plays a role. Insulin resistance has been shown to be inherited in several studies in Caucasians and other ethnic groups, suggesting a genetic component.8, 9, 10 The second important aspect is the presence of pancreatic β‐cell dysfunction, in which the β‐cells are less able to compensate for an increase in functional demand and which, as a consequence, undergo apoptosis more readily. β‐cell dysfunction has been linked to several genetic susceptibility factors.11, 12, 13 In studies of humans, some individuals seem to have decreased insulin sensitivity and DM even in the absence of environmental factors such as obesity, whereas others have substantial insulin resistance, but do not seem to progress to becoming overtly diabetic. Thus, type 2 DM is a complex multifactorial interaction, in which the combination of environment and genetic background plays a key role in the development of the disease. Advances in molecular genetic techniques, availability of the complete human genome sequence, and single nucleotide polymorphism (SNP) chips to allow performance of genome‐wide association studies (GWAS) have led to identification of genes associated with type 2 DM in humans.14, 15, 16, 17, 18, 19, 20, 21, 22, 23 To date, there are no studies that have identified specific genes that predispose cats to DM, although given a similar pathophysiology and environmental risk factors, it seems likely that feline DM could share a similar genetic basis with the human disease.
The melanocortin 4 receptor (MC4R) gene, encoding a transmembrane G‐protein–coupled receptor, has been implicated as a susceptibility gene in human DM and obesity.24, 25, 26, 27 This receptor is mainly expressed in the hypothalamus and plays an important role in the regulation of energy balance and appetite.28 In a situation of positive energy balance, MC4R is stimulated by alpha ‐melanocyte stimulating hormone (α‐MSH), leading to a feeling of satiety. During periods of starvation, the activity of MC4R is inhibited by its inverse antagonist, agouti‐related protein (AGRP), leading to a feeling of hunger.28 Mutations in this gene are the most common single genetic cause of human obesity, accounting for up to 6% of cases. A correlation between MC4R mutations and human type 2 DM has been shown in several studies,26, 29, 30 including a recent meta‐analysis, which showed an odds ratio (OR) of 1.10 (95% CI 1.07–1.13).31 Given the influence this gene has on susceptibility to obesity and DM in human beings and given the fact that obesity also poses an important risk factor for DM1, 32 in cats, MC4R was considered to be a logical candidate gene in the search for genetic factors predisposing to DM in cats. The aim of this study was to confirm the sequence of feline MC4R, to identify SNPs in its coding region, and to perform a case‐control study to detect a possible gene association with feline DM and obesity.
Materials and Methods
Animals
Blood samples (EDTA or non‐anticoagulated) from diabetic cats were recruited via the UK Companion Animal Diabetes Register, based at the Royal Veterinary College (RVC), from first opinion veterinary practices in the UK, with samples submitted with a standardized sample submission form (http://www.rvc.ac.uk/cic/documents/GHsubmissionformV1328Jan10DL.pdf), that included details of concurrent diseases and clinical signs, body weight and body condition score (BCS) at and before diagnosis, diet and insulin type and dose. The BCS assessment was carried out according to the routinely used 9‐ and 5‐point BCS systems, information about how to assess the BCS of cats according to these systems was included in the submission form. Blood samples from nondiabetic cats were obtained from the RVC Genetic Archive, consisting of retrospective residual samples (after completion of diagnostic testing) from referral cases seen at RVC hospitals. Ethical approval for sample collection had been obtained previously from the local ethical committee. For the SNP discovery phase, 10 cats (5 Burmese: 2 diabetic, 3 nondiabetic; 5 DSH: 3 diabetic, 2 nondiabetic) were assessed. For the genotyping phase, a total of 240 DSH cats were analyzed, consisting of 4 groups (n = 60 each; with males and females equally represented) of overweight diabetic, lean diabetic, overweight nondiabetic, and lean nondiabetic cats.
The diabetic cats were selected on the basis of the presence of increased serum fructosamine concentration, persistent hyperglycemia, requiring insulin therapy, and having been diagnosed as diabetic by the attending veterinarian for longer than 4 weeks. Insulin growth factor‐1 (IGF‐1) was measured in all samples, those cats with IGF‐1 concentration<800 ng/mL were allocated either to the overweight diabetic group (BCS > 4/5 or 5/9) or to lean diabetic group (BCS ≤ 4/5 or 5/9). Those cats that were overweight before diagnosis and had lost weight before the diagnosis of DM were included in the overweight group, because weight loss is considered one of the landmark clinical signs of DM. Those cats with known concurrent diseases that could have an impact on body weight were excluded from the study. Based on evaluation of medical records and available diagnostic information, nondiabetic cats >9 years of age that had no clinical or biochemical evidence of DM, specifically hyperglycemia or glycosuria, were selected. Cats with clinical or biochemical evidence of other diseases that could lead to changes in body weight (eg, hyperthyroidism, renal disease, gastrointestinal disease, hepatic disease) were excluded from the study. However, because these samples were obtained from an archive of retrospective clinical material, additional prospective diagnostic testing was not performed. All of the nondiabetic cats were seen in a clinic staffed by board‐certified veterinary specialists. There were no known familial relationships among any of the cats included in this study.
Feline MC4R SNP Discovery
The initial phase focused on resequencing the coding region of the feline MC4R gene to identify SNPs. Genomic DNA (gDNA) was extracted from EDTA blood samples or blood clots using a blood genomic DNA extraction kit,1 according to the manufacturer's instructions. Feline MC4R‐specific primers located in the 5′ untranslated region (UTR; Sense: 5′‐CTCAGAACTTTCGGGCAGAC‐3′; starting at position ‐103) and in the 3′ UTR (Antisense: 5′‐ACCCATGCCTTACACAGAGG‐3′; starting at position 1078) were designed according to the available feline MC4R sequence (http://www.ensembl.org/Felis_catus/Info/Index; ENSFCAG00000006540. Assembly ASM18133v1, Genbank Assembly ID: GCA_000181335.1). Polymerase chain reaction was performed in 25 μL volumes using 0.1 μL of DNA polymerase (Immolase DNA polymerase2 ) with 1 μL of gDNA as a template. Each reaction also contained 13 μL of water, 5 μL of 5× PCR‐enhancing additive (Hi‐Spec additive2), 2.5 μL of ammonium sulfate buffer (NH4 buffer2), 1.25 μL MgCl2 2 (2.5 mM final concentration), 0.25 μL 250 μM dNTP2, and 2 μL of primer mix (each at 20 pmol/μL). Using a PCR cycler,3 reactions were heated to 95°C for 10 minutes, followed by 30 cycles consisting of 94°C for 40 seconds, 65°C for 30 seconds, and 72°C for 2 minutes, with a final extension step at 72°C for 10 minutes. PCR products were separated by horizontal gel electrophoresis using a 2% agarose gel and analyzed under 590 nm UV light.4 Subsequently, DNA was extracted using a gel extraction Kit (Genelute gel extraction Kit5 ) according to the manufacturer's instructions. Amplicons then were submitted for sequencing6 and compared to each other and with the reference feline genome sequence using sequence analysis software .7
MC4R Genotyping by RFLP Analysis
Genomic DNA was extracted from EDTA blood or blood clots and PCR performed as described in the SNP discovery section. Amplified DNA then was purified using a PCR clean‐up kit (Genelute PCR clean‐up Kit5), subjected to digestion using a restriction enzyme (BstOI enzyme8), which recognizes a restriction site (CC'(A/T)GG) incorporating the MC4R:c.92C allele, absent in the MC4R:c.92T allele. The digestion reaction consisted of 0.5 μL of BstOI restriction enzyme, 0.5 μL of BSA, 0.5 μL of Buffer (Buffer C8), and 3.5 μL of purified PCR product. Reactions were incubated for 1 hour at 60°C in a water bath and analyzed by agarose gel electrophoresis as described above. In samples in which gel interpretation was potentially ambiguous, genotyping by RFLP was repeated using a higher DNA concentration. If an unambiguous result still was not obtained, manual sequencing was performed.
Statistical Analysis
Statistical analysis was performed using SPSS software package.9 The Mann–Whitney U‐test was used to analyze non parametric variables. The Pearson's two‐tailed chi‐squared test was used to compare MC4R:c.92C>T SNP genotype and allele frequencies among groups, and the OR with 95% confidence interval (CI) was calculated. Bonferroni correction was applied for multiple group comparisons. Significance was considered at P < .01.
Results
Animals
The median age of the overweight diabetic cats was 12 years (range, 6–18 years); the median weight was 6.6 kg (range, 5.1–10.0 kg). The median age of the lean diabetic cats was 13 years (range, 5–18 years); the median weight was 4.15 kg (range, 2.6–6.0 kg).
For the overweight nondiabetic group (30 male, 30 female), the mean age was 12 years (range, 9.1–20 years) and the median weight 5.7 kg (range, 3.4–8.3 kg). For the lean nondiabetic group (30 male, 30 female), the median age was 13.9 years (range, 9–19 years) and the mean weight 3.9 kg (range, 2.5–5.7 kg).
The age difference between diabetics and nondiabetics was not statistically significant (P = .074), whereas the obese cats were significantly heavier than the lean cats (P < .001).
Polymorphisms in Feline MC4R
Initial sequencing results showed the presence of 3 polymorphisms. A non synonymous SNP in position 92 of the coding sequence (MC4R:c.92C>T) was found in the DSH cats only and led to an amino acid change from leucine to proline in the protein sequence. Figure 1 shows the sequencing results from the initial SNP discovery phase (10 cats) and represents the fragment of the coding sequence of MC4R where the non synonymous SNP was detected. Two additional synonymous SNPs (MC4R:c297C>T and MC4R:c303C>T) also were present in both the DSH and the Burmese cats.
Figure 1.
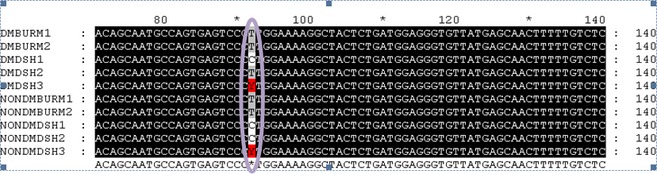
Initial single nucleotide polymorphism (SNP) discovery phase, fragment of the coding sequence of feline MC4R in which the non synonymous SNP was detected (purple circle). This SNP (MC4R:c.92 C>T) was detected exclusively in domestic shorthaired (DSH) cats and leads to leucine being replaced by proline in the translated protein. DMBURM1, diabetic Burmese cat1; DMBURM2, diabetic Burmese cat 2; DMDSH1, diabetic DSH cat 1; DMDSH2, diabetic DSH cat 2; DMDSH3, diabetic DSH cat 3; NONDMBURM1, nondiabetic Burmese cat 1; NONDMBURM2, nondiabetic Burmese cat 2; NONDMDSH1, nondiabetic DSH cat 1; NONDMDSH2, nondiabetic DSH cat 2; NONDMDSH3, nondiabetic DSH cat 3; A, adenine; C, cytosine; G, guanine; T, thymine; Y, heterozygous cats (C and T alleles).
MC4R:c.92C Homozygous Genotype in Obese Diabetic DSH Cats
A case‐control study was performed, focusing on the MC4R:c.92C>T SNP, to determine whether there was any association with increased body weight or DM. The RFLP assay was found to be a rapid and simple test to identify MC4R genotypes in cats (Fig 2). In the 2 nondiabetic groups, there was no significant difference in allele or genotype frequencies comparing the overweight and lean cats (Table 1), and data from these cats subsequently were combined into a single nondiabetic control group. There were no differences in allele frequencies, comparing the lean and overweight diabetic groups (n = 60 each group) and the nondiabetic control group (n = 120) (Table 2). However, in the overweight diabetic group (n = 60), the majority of cats (55%) were homozygous for the MC4R:c.92C allele (Table 2) compared with the nondiabetic control group (n = 120), where 30% were homozygous C, a difference that was statistically significant (P = .002). In contrast, there was no difference in genotype distribution when comparing the lean diabetic cats (n = 60) with the nondiabetic cats (n = 120) (P = .7) and when comparing lean diabetics (n = 60) with the obese diabetic cats (n = 60) (P = .02). Overweight cats that were homozygous for the mutation had a statistically significantly higher OR for DM of 3.67 (95% CI, 1.68–7.95, P = .0007) when compared to nonhomozygous obese cats.
Figure 2.
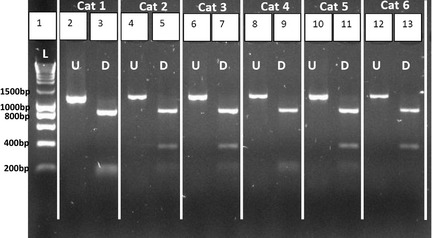
Representative BstOI restriction fragment length polymorphism (RFLP) results. The lane to the left of each digested polymerase chain reaction (PCR) product corresponds to aliquots of the PCR product before digestion (see lanes 2, 4, 6, 8, 10, 12). Lane 1: DNA ladder I (0.2–10 kb). Homozygous C cats have restriction sites in positions 151 and 315 of the PCR product, resulting in 1 fragment of 885 bp and 2 fragments of smaller size: 150 and 165 bp (eg, lanes 3 and 9). Homozygous T cats have 1 restriction site at position 315 of the PCR product, resulting in 1 fragment of 885 bp and 1 fragment of 315 bp (eg, lane 13); Heterozygous cats showed 1 fragment of 885 bp, 1 fragment of 315 bp, and 2 smaller fragments of 150 and 165 bp (eg, lanes 5, 7, and 11). L, ladder; U, undigested; D, digested.
Table 1.
Summary of MC4R:c92T>C RFLP results for control cats
| Genotype Frequency | Genotype Frequency | Allele Frequency | Allele Frequency | |||
|---|---|---|---|---|---|---|
| CC | CT/TT | C | T | |||
| Lean control | 0.35 (n = 21) | 0.65 (n = 39) | P = .19 | 0.61 | 0.39 | P = .31 |
| Overweight control | 0.25 (n = 15) | 0.75 (n = 45) | 0.53 | 0.47 | ||
MC4R, melanocortin 4 receptor; RFLP, restriction fragment length polymorphism; C, cytosine; T, thymine.
Statistical analysis performed using two‐tailed Pearson's chi‐square test. Significance established at a P value <.01.
Table 2.
Association of MC4R:c.92 CC homozygous genotype with diabetes in obese DSH cats
| CC | CT/TT | Total | |
|---|---|---|---|
| Nondiabetica , b | 30% (n = 36) | 70% (n = 84) | 120 |
| Overweight diabetica , c | 55% (n = 33) | 45% (n = 27) | 60 |
| Lean diabeticb , c | 33% (n = 20) | 66% (n = 40) | 60 |
MC4R, melanocortin 4 receptor; DSH, domestic shorthaired; C, cytosine; T, thymine.
Pearson's chi‐square comparison of CC genotype and CT/TT genotypes between nondiabetic DSH cats and overweight diabetic DSH cats, P = .002.
Pearson's chi‐square comparison of CC genotype and CT/TT genotypes between nondiabetic DSH cats and lean diabetic DSH cats, P = .7.
Pearson's chi‐square comparison of CC genotype and CT/TT genotypes between overweight diabetic DSH cats and lean diabetic DSH cats, P = .02.
Discussion
The present study demonstrates that a genetic factor (homozygosity for the MC4R:c.92C allele) may be associated with DM in overweight DSH cats. Three SNPs were identified in the feline MC4R coding sequence, with 2 of these (MC4R:c297C>T and MC4R:c303C>T) representing synonymous changes and therefore less likely to impact protein structure or function than the non synonymous SNP (MC4R:c.92C>T). Additional polymorphisms may occur upstream (ie, in the gene regulatory regions) or downstream of the coding region, but these areas were not investigated in the current study. Because only 10 cats were evaluated in the SNP discovery phase, it is possible that other less common SNPs are present in feline MC4R. However, because only SNPS with a minor allele frequency >0.1 are normally used in case‐control association studies, relatively rare SNPs would not be deemed suitable and therefore were excluded from subsequent genetic analysis.
Previous studies in humans have shown that mutations in the MC4R gene are associated with obesity, which increases the risk of those individuals developing type 2 DM.26, 29, 30, 31 The current study failed to demonstrate any association between the MC4R:c.92C>T polymorphism and overweight body condition in nondiabetic DSH cats, but the sample size was relatively small. Rather than having a direct effect on regulating satiety and food intake and consequently influencing body weight, these results suggest that this polymorphism may be associated with progression to overt DM in overweight cats. Alternatively, this polymorphism might be in linkage disequilibrium with another causative polymorphism or mutation elsewhere in the genome.
Under physiologic conditions, MC4R is constitutively active, leading to an appetite suppression signal until negative energy balance develops. At this time, a decrease in leptin concentration and a subsequent increase in the inverse agonist AGRP occur. AGRP binding inactivates the MC4R receptor, leading to an increase in appetite.28 Functional studies performed in humans have demonstrated that mutations that influence the amino acid sequence in the N‐terminal region of the protein generate receptors that are unable to maintain their constitutive activity, leading to decreased appetite suppression and increased food intake.33 In addition, several studies have evaluated the role of MC4R in energy metabolism, but conflicting results have been reported.34, 35, 36, 37 Notwithstanding discrepancies in the published literature, studies in mice have indicated that mutations of the MC4R receptor can lead to a reduction in metabolic rate, indicating that this receptor might play additional roles in energy metabolism and there may be interspecies differences in receptor function. The non synonymous MC4R:c92C>T SNP identified in DSH cats causes an amino acid change from leucine to proline at amino acid position 30.
The GWAS of human type 2 DM, which identified MC4R as a susceptibility gene, involved relatively large numbers of cases and controls.25, 38 Unfortunately, such large‐scale genetic studies are difficult to replicate in companion animals, with the result that many studies are relatively underpowered. Regardless, GWAS of several diseases of dogs have yielded results, even with relatively low numbers, when performed within breeds.39, 40, 41 This is likely to be similar for genetic studies of cats, if performed within relatively inbred pedigree populations, but in the DSH cat population, which is more outbred, larger sample sizes are likely to be required for case‐control genotyping studies. Nevertheless, a significant difference was found in the current study, even with relatively small numbers of DSH cats, suggesting that this gene might play a major role in susceptibility to DM in overweight cats.
The current study has a number of limitations. The diabetic cases and controls were breed‐ and sex‐matched, but not age‐matched. Older cats were proactively recruited as controls to decrease the chances of including cats that might become diabetic later in life. Previous studies have demonstrated that the risk of developing DM in cats increases with age,42 and therefore, even though the nondiabetic (overweight and lean) populations were older than the diabetic populations, this is likely to be beneficial rather than detrimental. BCS information was obtained from all cats, including information about BCS before diagnosis when available. Because one of the landmark clinical signs of DM is weight loss, some cats that were overweight before developing the disease may have been included in the lean group. Although an attempt was made to exclude cats that had concurrent diseases that could alter body weight, this was not always possible. This could have changed the results of the study. The overall prevalence of the SNP in the diabetic group (lean and obese), however, still is significantly higher than in the nondiabetic group. Given the effect that MC4R mutations have on appetite regulation in humans, it would have been valuable to have obtained information about each cat in terms of appetite and food intake. Although some of this information was available for some of the cats, it proved difficult to incorporate these data into the analysis or selection criteria because of the subjectivity of evaluating food intake and appetite, particularly in multi‐cat households and when different diets and feeding regimes (eg, ad libitum or not) were being used.
The current findings are of interest not only in advancing our knowledge of the pathogenesis of feline DM but also in terms of clinical relevance. Thus, MC4R genotyping of overweight cats might be useful in estimating the relative risk of the cat progressing to overt DM, given an OR of 3.67 comparing the overweight diabetic group with the overweight nondiabetic group. Because feline DM is likely to represent a complex genetic disorder, identification of other susceptibility genes is warranted and, if confirmed to be present, a test based on genetic profiling of multiple genes potentially could be used to establish an overall genetic risk for development of DM. This might then be useful for identifying cats with a greater risk of developing the disease, so that preventative measures (eg, dietary management) could be implemented to decrease other risk factors (eg, development of obesity) that might precipitate disease in a genetically susceptible individual.
In conclusion, we have demonstrated that a genetic factor may influence susceptibility to DM in cats. The feline MC4R:c.92C>T polymorphism may be involved in progression to overt DM in overweight cats, but it remains to be established whether or not a causal relationship is present, or if it is simply an association. If the polymorphism does have functional consequences, this does not seem to be directly related to increased susceptibility to overweight body condition and might, instead, play a role in glycemic control in the presence of insulin resistance or influence pancreatic beta cell function in response to chronic hyperglycemia. Functional studies to investigate the physiologic relevance of this polymorphism and GWAS to identify other susceptibility genes for feline DM are warranted.
Acknowledgments
Conflict of Interest Declaration: The authors disclose no conflict of interest.
Footnotes
GenElute blood genomic DNA extraction kit; Sigma, Poole, UK
Bioline, London, UK
Eppendorf Master Cycler; Eppendorf AG, Hamburg, Germany
ImageMaster VDS; Pharmacia Biotech/GE Healthcare, Buckinghamshire, UK
Sigma‐Aldrich, Dorset, UK
GATC Biotech Ltd The London BioScience Innovation Centre, London, UK
Genedoc software (Nicholas et al, 1997), http://www.nrbsc.org/gfx/genedoc/index.html
Promega, Southampton, UK
SPSS software, IBM, Portsmouth, UK
References
- 1. Rand JS, Fleeman LM, Farrow HA, et al. Canine and feline diabetes mellitus: Nature or nurture? J Nutr 2004;134(8 Suppl):2072S–2080S. [DOI] [PubMed] [Google Scholar]
- 2. Forcada Y, German AJ, Noble PJ, et al. Determination of serum fPLI concentrations in cats with diabetes mellitus. J Feline Med Surg 2008;10:480–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Niessen SJ. Feline acromegaly: An essential differential diagnosis for the difficult diabetic. J Feline Med Surg 2010;12:15–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Niessen SJ, Petrie G, Gaudiano F, et al. Feline acromegaly: An underdiagnosed endocrinopathy? J Vet Intern Med 2007;21:899–905. [DOI] [PubMed] [Google Scholar]
- 5. Rand JS, Bobbermien LM, Hendrikz JK, Copland M. Over representation of Burmese cats with diabetes mellitus. Aust Vet J 1997;75:402–405. [DOI] [PubMed] [Google Scholar]
- 6. McCann TM, Simpson KE, Shaw DJ, et al. Feline diabetes mellitus in the UK: The prevalence within an insured cat population and a questionnaire‐based putative risk factor analysis. J Feline Med Surg 2007;9:289–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bell GI, Polonsky KS. Diabetes mellitus and genetically programmed defects in beta‐cell function. Nature 2001;414:788–791. [DOI] [PubMed] [Google Scholar]
- 8. Malecki MT. Genetics of type 2 diabetes mellitus. Diabetes Res Clin Pract 2005;68(Suppl 1):S10–S21. [DOI] [PubMed] [Google Scholar]
- 9. Martin BC, Warram JH, Rosner B, et al. Familial clustering of insulin sensitivity. Diabetes 1992;41:850–854. [DOI] [PubMed] [Google Scholar]
- 10. Lillioja S, Mott DM, Howard BV, et al. Impaired glucose tolerance as a disorder of insulin action. Longitudinal and cross‐sectional studies in Pima Indians. N Engl J Med 1988;318:1217–1225. [DOI] [PubMed] [Google Scholar]
- 11. Grant SF, Thorleifsson G, Reynisdottir I, et al. Variant of transcription factor 7‐like 2 (TCF7L2) gene confers risk of type 2 diabetes. Nat Genet 2006;38:320–323. [DOI] [PubMed] [Google Scholar]
- 12. Waters KM, Stram DO, Hassanein MT, et al. Consistent association of type 2 diabetes risk variants found in europeans in diverse racial and ethnic groups. PLoS Genet 2010;6:e1001078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mathews AE, Mathews CE. Inherited beta‐cell dysfunction in lean individuals with type 2 diabetes. Diabetes 2012;61:1659–1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Weedon MN, Clark VJ, Qian Y, et al. A common haplotype of the glucokinase gene alters fasting glucose and birth weight: Association in six studies and population‐genetics analyses. Am J Hum Genet 2006;79:991–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lyssenko V, Lupi R, Marchetti P, et al. Mechanisms by which common variants in the TCF7L2 gene increase risk of type 2 diabetes. J Clin Invest 2007;117:2155–2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Frayling TM, Timpson NJ, Weedon MN, et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science 2007;316:889–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zeggini E, Weedon MN, Lindgren CM, et al. Replication of genome‐wide association signals in UK samples reveals risk loci for type 2 diabetes. Science 2007;316:1336–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Frayling TM. Genome‐wide association studies provide new insights into type 2 diabetes aetiology. Nat Rev Genet 2007;8:657–662. [DOI] [PubMed] [Google Scholar]
- 19. Winckler W, Weedon MN, Graham RR, et al. Evaluation of common variants in the six known maturity‐onset diabetes of the young (MODY) genes for association with type 2 diabetes. Diabetes 2007;56:685–693. [DOI] [PubMed] [Google Scholar]
- 20. Perry JR, Frayling TM. New gene variants alter type 2 diabetes risk predominantly through reduced beta‐cell function. Curr Opin Clin Nutr Metab Care 2008;11:371–377. [DOI] [PubMed] [Google Scholar]
- 21. Hennig BJ, Fulford AJ, Sirugo G, et al. FTO gene variation and measures of body mass in an African population. BMC Med Genet 2009;10:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yajnik CS, Janipalli CS, Bhaskar S, et al. FTO gene variants are strongly associated with type 2 diabetes in South Asian Indians. Diabetologia 2009;52:247–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Florez JC, Jablonski KA, Bayley N, et al. TCF7L2 polymorphisms and progression to diabetes in the Diabetes Prevention Program. N Engl J Med 2006;355:241–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Qi L, Kraft P, Hunter DJ, Hu FB. The common obesity variant near MC4R gene is associated with higher intakes of total energy and dietary fat, weight change and diabetes risk in women. Hum Mol Genet 2008;17:3502–3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Loos RJ, Lindgren CM, Li S, et al. Common variants near MC4R are associated with fat mass, weight and risk of obesity. Nat Genet 2008;40:768–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Xi B, Takeuchi F, Chandak GR, et al. Common polymorphism near the MC4R gene is associated with type 2 diabetes: Data from a meta‐analysis of 123,373 individuals. Diabetologia 2012;55:2660–2666. [DOI] [PubMed] [Google Scholar]
- 27. Wen J, Ronn T, Olsson A, et al. Investigation of type 2 diabetes risk alleles support CDKN2A/B, CDKAL1, and TCF7L2 as susceptibility genes in a Han Chinese cohort. PLoS One 2010;5:e9153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cone RD. Anatomy and regulation of the central melanocortin system. Nat Neurosci 2005;8:571–578. [DOI] [PubMed] [Google Scholar]
- 29. Cauchi S, Ezzidi I, El Achhab Y, et al. European genetic variants associated with type 2 diabetes in North African Arabs. Diabetes Metab 2012;38:316–323. [DOI] [PubMed] [Google Scholar]
- 30. Thearle MS, Muller YL, Hanson RL, et al. Greater impact of melanocortin‐4 receptor deficiency on rates of growth and risk of type 2 diabetes during childhood compared with adulthood in Pima Indians. Diabetes 2012;61:250–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cauchi S, Stutzmann F, Cavalcanti‐Proenca C, et al. Combined effects of MC4R and FTO common genetic variants on obesity in European general populations. J Mol Med 2009;87:537–546. [DOI] [PubMed] [Google Scholar]
- 32. Slingerland LI, Fazilova VV, Plantinga EA, et al. Indoor confinement and physical inactivity rather than the proportion of dry food are risk factors in the development of feline type 2 diabetes mellitus. Vet J 2009;179:247–253. [DOI] [PubMed] [Google Scholar]
- 33. Srinivasan S, Lubrano‐Berthelier C, Govaerts C, et al. Constitutive activity of the melanocortin‐4 receptor is maintained by its N‐terminal domain and plays a role in energy homeostasis in humans. J Clin Invest 2004;114:1158–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tao YX. The melanocortin‐4 receptor: Physiology, pharmacology, and pathophysiology. Endocr Rev 2010;31:506–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chen AS, Metzger JM, Trumbauer ME, et al. Role of the melanocortin‐4 receptor in metabolic rate and food intake in mice. Transgenic Res 2000;9:145–154. [DOI] [PubMed] [Google Scholar]
- 36. Kring SI, Holst C, Toubro S, et al. Common variants near MC4R in relation to body fat, body fat distribution, metabolic traits and energy expenditure. Int J Obes (Lond) 2010;34:182–189. [DOI] [PubMed] [Google Scholar]
- 37. Cole SA, Butte NF, Voruganti VS, et al. Evidence that multiple genetic variants of MC4R play a functional role in the regulation of energy expenditure and appetite in Hispanic children. Am J Clin Nutr 2010;91:191–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chambers JC, Elliott P, Zabaneh D, et al. Common genetic variation near MC4R is associated with waist circumference and insulin resistance. Nat Genet 2008;40:716–718. [DOI] [PubMed] [Google Scholar]
- 39. Seppala EH, Koskinen LL, Gullov CH, et al. Identification of a novel idiopathic epilepsy locus in Belgian Shepherd Dogs. PLoS One 2012;7:e33549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tsai KL, Noorai RE, Starr‐Moss AN, et al. Genome‐wide association studies for multiple diseases of the German Shepherd Dog. Mamm Genome 2012;23:203–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mogensen MS, Karlskov‐Mortensen P, Proschowsky HF, et al. Genome‐wide association study in Dachshund: Identification of a major locus affecting intervertebral disc calcification. J Hered 2011;102(Suppl 1):S81–S86. [DOI] [PubMed] [Google Scholar]
- 42. Panciera DL, Thomas CB, Eicker SW, Atkins CE. Epizootiologic patterns of diabetes mellitus in cats: 333 cases (1980–1986). J Am Vet Med Assoc 1990;197:1504–1508. [PubMed] [Google Scholar]


