Abstract
There are still no highly sensitive and unique biomarkers for measurement of puberty onset. Circulating miRNAs have been shown to be promising biomarkers for diagnosis of various diseases. To identify circulating miRNAs that could be served as biomarkers for measuring chicken (Gallus gallus) puberty onset, the Solexa deep sequencing was performed to analyze the miRNA expression profiles in serum and plasma of hens from two different pubertal stages, before puberty onset (BO) and after puberty onset (AO). 197 conserved and 19 novel miRNAs (reads > 10) were identified as serum/plasma-expressed miRNAs in the chicken. The common miRNA amounts and their expression changes from BO to AO between serum and plasma were very similar, indicating the different treatments to generate serum and plasma had quite small influence on the miRNAs. 130 conserved serum-miRNAs were showed to be differentially expressed (reads > 10, P < 0.05) from BO to AO, with 68 up-regulated and 62 down-regulated. 4829 putative genes were predicted as the targets of the 40 most differentially expressed miRNAs (|log2(fold-change)|>1.0, P < 0.01). Functional analysis revealed several pathways that were associated with puberty onset. Further quantitative real-time PCR (RT-qPCR) test found that a seven-miRNA panel, including miR-29c, miR-375, miR-215, miR-217, miR-19b, miR-133a and let-7a, had great potentials to serve as novel biomarkers for measuring puberty onset in chicken. Due to highly conserved nature of miRNAs, the findings could provide cues for measurement of puberty onset in other animals as well as humans.
Introduction
Precocious puberty is a pathologic status for humans, and could result in serious impact on health and development, including increase obesity, diabetes and cancer risks [1]. Puberty onset is a complicated process that comprises many genes and regulatory factors. Recently, much progress has been made in identifying components that regulate puberty onset [2]. Notably, a set of genes including KISS1/GPR54 [3], TAC3/TACR3 [4] and LIN28 [5] are found to play important roles in initiating puberty. However, association analysis of variants in genes with puberty onset are often not consistent among different groups [6,7]. Furthermore, the conclusion suggesting that timing of puberty for girls initiated earlier than in the past has long been a controversial topic [8].One of the main reasons is due to lack of unanimous standards to measure puberty. This, in turn, limits the discovery of new factors related to puberty onset.
To accurately estimate puberty onset is challenging [9]. In the last decade, there was no new measures of puberty emerged [8]. Physical alterations, thelarche, pubarche as well as age at menarch described by Tanner and vaginal opening [10] are the primary indicators used to measure female puberty onset in humans and mammals respectively. These are not accurate and efficient markers. Some blood reproductive hormones have also been widely applied to measure puberty, but due to the pulsatile secretion manner, their levels fluctuate easily and usually could not match to a specific pubertal stage [11,12]. So, to find new type biomarkers with higher sensitivity and specificity for puberty onset measurement is necessary.
microRNAs are small non-coding RNA molecules that suppress gene expression post-transcriptionally, and function important roles in diverse biological processes [13]. In addition to endogenous presence in cells, miRNAs can also be actively released into extracellular fluids through exosomes or microvesicles [14,15]. These circulating miRNAs can hold stably in body fluids, not only connect a new cell to cell communication mechanism but also their expression pattern changes are linked to body physiological and pathologic status. A number of circulating miRNAs have been identified as accurate biomarkers for disease diagnosis [16,17,18,19,20] and even for milk quality control [21,22]. This sheds light on measurement of puberty onset.
As an important model organism, chicken’s sexual development is very distinctive when compared to mammals and humans [23]. In our previous study, we had roughly determined the timing of transition from juvenility to puberty onset in Wenchang chicken, a Chinese indigenous chicken breed, through measuring inner gonad development and visible crown growth [24]. In order to find circulating miRNA biomarkers that could be served as biomarkers for measuring puberty onset in Wenchang chicken, in this study, we investigated the expression profile of miRNAs in the serum and plasma of chicken during puberty onset. The results demonstrate that chicken serum and plasma contains large amounts of miRNAs. A seven-miRNAs panel is identified as potential biomarkers for measuring chicken puberty onset. Considering the characteristics of miRNA functional conservation, the results will contribute to measurement of puberty onset in other animals as well as humans.
Materials and Methods
Ethics statement
The Wenchang chicken breed used in the present study is not endangered. The animals were allowed access to feed and water freely. Before sacrifice, the chickens were anaesthetized by giving them water mixed with diazepam and ethanol. When they became unconscious, the electric shocks were carried to minimize suffering. All animal experiments were approved by the Poultry institute, Chinese Academy of Agricultural Science, Yangzhou, China and Institutional Animal Care and Use Committees in College of Animal Science & Technology, Nanjing Agricultural University, Nanjing, China.
Collection of serum and plasma samples
To generate serum, 4.0~5.0ml venous blood was collected into tubes free of anticoagulant. The whole blood was allowed to stand for 1 hour at 4°C before being centrifuged at 300×g for 5 minutes, and the supernatant by a final centrifugation at 800×g for 10 minutes.
To generate plasma, 4.0~5.0ml blood was collected into tubes with EDTA, directly centrifuged at 300×g for 5 minutes to spin down the blood cells, then followed by a second centrifugation at 800×g for 10 minutes to completely remove cellular components. The resultant serum and plasma were aliquoted into eppendorf tubes and stored at -80°C. All the centrifugation steps were performed at 4°C. Blood sample was processed and serum or plasma was frozen within 2 hours after blood was derived.
Small RNA library preparation and sequencing
Collected serum and plasma from 6 hens at the age of 13 weeks were used to construct two small RNA libraries in this study. The 6 animals, including 3 BO (before puberty onset) ones and 3 AO (after puberty onset) ones, were from a mating family as described in detail previously [24]. Each library contained one full-sib and two half-sibs.
Total RNA was extracted from the serum and plasma using TruSeq Small RNA Sample Pre Kits (Illumine, San Diego, USA) according to the manufacturer’s instructions. Total RNA quality was checked with a Bioanalyzer 2100 (Agilent Technologies, USA). The RIN was > 8.0 and A260/A280 was > 2.1 for all samples. Total sera or plasma RNA of chickens from the same status were mixed with equal amounts to construct two pooled libraries. The overall flow of the sequencing procedure is as follows: small RNAs ranging from 18 to 35nt in length was purified from 15% polyacrylamide gels, then ligated to 5, and 3, adapters. Reverse transcription was performed, and followed by PCR amplification. The purified PCR products (~140bp) were used directly for cluster generation and sequencing analysis using the Illumina’s Solexa Sequencer according to the manufacturer’s instructions (LC-Bioscience, Hangzhou, China).
Sequence data analysis
Sequence data analysis was done using AGGT101-miR tool. After deleting poor quality reads, adaptor pollution reads and reads less than 18nt, the clean reads were obtained.
The clean reads of small RNAs were aligned to the reference chicken (G.gallus) genome to identify known miRNAs. The sequences that matched perfectly to known miRNAs (miRBase V21.0) were determined as conserved miRNAs. Other small RNAs (rRNA, tRNA, snRNA and snoRNA) were annotated by blasting against the Rfram, Repbase and ncRNA databases.
The unannotated small RNA sequences were aligned to the reference chicken (G.gallus) genome to find potential precursor sequences for novel miRNAs. Novel miRNAs were predicted by RNA-fold tools following the criteria: (1) number of nucleotides in one bulge in stem (< = 12); (2) number of base pairs in the stem region of the predicted hairpin (> = 16); (3) cutoff of free energy (kcal/mol< = -15); (4) length of hairpin (up and down stems + terminal loop> = 50); (5) length of hairpin loop (< = 20); (6) number of nucleotides in one bulge in mature region (< = 8); (7) number of biased errors in one bulge in mature region (< = 4); (8) number of biased bulges in mature region (< = 2); (9) number of errors in mature region (< = 7); (10)number of base pairs in the mature region of the predicted hairpin (> = 12); (11)percent of mature in stem (> = 80). Furthermore, the raw reads > = 10 at least one time point.
To identify differentially expressed miRNAs, the number of conserved miRNAs was normalized to the total number of reads in each sample that matched the chicken (G.gallus) genome. P-values for differentially expressed miRNAs (Serum:BO vs AO; Plasma:BO vs AO) were calculated by Fisher,s exact-test and Chi square (2×2) test.
Quantitative real-time PCR analysis
The candidate miRNAs for further quantitative real-time PCR (RT-qPCR) analysis were selected as the following criteria: (1) in the list of differentially expressed serum-miRNAs; (2) with more than middle expression levels (raw reads> = 40); (3) association with reproduction events, including gonad development, muscle development, glucose metabolism, fat metabolism, insulin metabolism, sex hormone synthesis and secretion.
Total RNAs of sampled sera and plasma were reverse-transcribed by PrimeScript® RT reagent Kit (TAKARA, DRR037A). The primers were designed by Primer 5.0 (ABI). 5ul RT reaction system included: denatured RNA and RT primer (2 uM) 3.0ul, 5×PrimeScript®Buffer 1.0ul, RNase Free dH2O 0.6ul, PrimeScript® RT Enzyme Mix I 0.4ul. The RT reactions were performed as follows: 42°C for 15 minutes, 85°C for 5 seconds and hold at 4°C. 20ul real-time PCR reaction system included: 2×SYBR Green Mix With ROX 10.0ul, ddH2O 8.2ul, Primer mix (10 uM) 0.8ul, RT product 1ul. The PCR reactions were performed as follows: 50°C for 2 minutes, 95°C for 2 minutes, then 40 cycles with 94°C for 15 seconds and 60°C for 30 seconds.
All experiments were performed on ABI 7900 HT sequence detection system. Each reaction was carried out with 3 replicates. snRNA U6 was used as the control for RT-qPCR. The relative expression level of each miRNA to U6 snRNA was normalized as ΔCq = Cq miRNA-Cq U6RNA [25]. Comparison of relative expression level in different stages was determined using the 2-ΔΔCq method [26]. Statistical significance analysis of the expression change was performed by one-way ANOVA in SPSS 20.0.
MiRNA target prediction and functional analysis
Target genes of differentially expressed miRNAs were predicted by TargetScan and miRanada. To acquire a higher prediction accuracy, only common target genes were considered. Gene Ontology (GO) annotation and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis were retrieved using DAVID (http://david.abcc.ncifcrf.gov/).
The global work flow above was showed as Fig 1.
Fig 1. Global work chart of identification of potential circulating miRNAs for measuring chicken puberty onset.
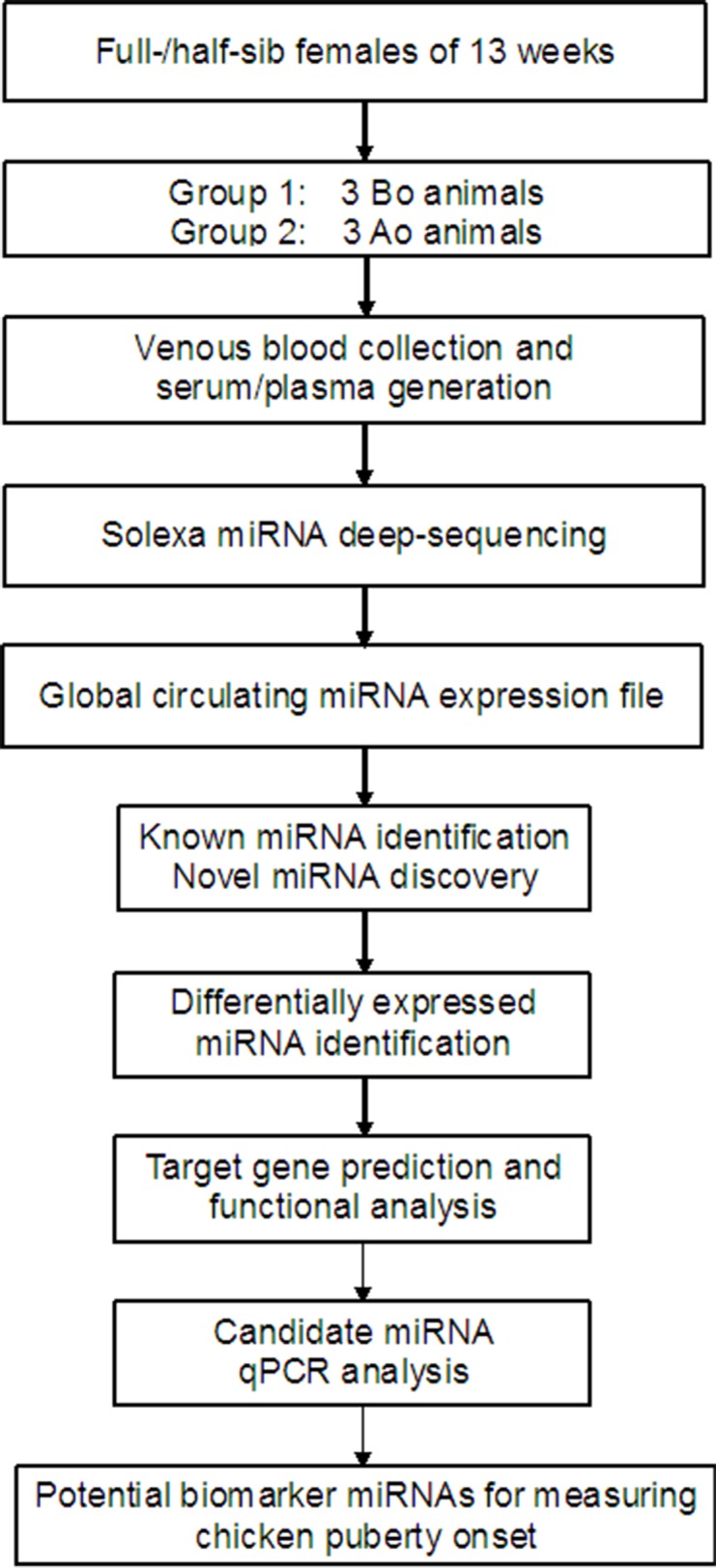
BO, before puberty onset; AO, after puberty onset.
Results
Small RNA library construction and sequencing
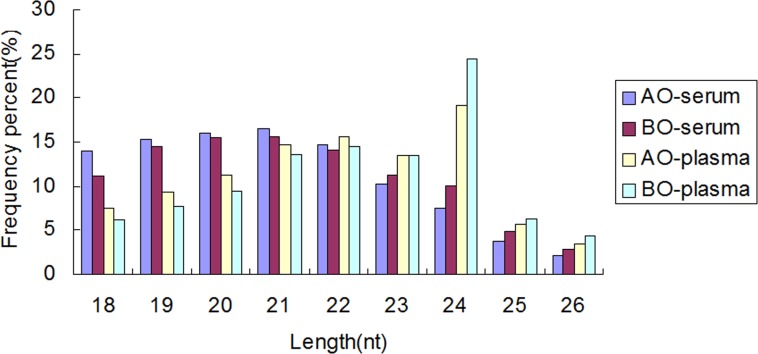
To investigate the miRNA expression profile in chicken serum and plasma, two small RNA libraries representing before puberty onset (BO) and after puberty onset (AO) status were constructed from full-/ half-sib animals. About 1.5~2.0 ml serum/plasma for each animal was used for RNA extraction, the RNA concentration was 30~40ng/ml. For each pool, RNAs were mixed at equal concentration (150ng of total RNA). High-throughput Solexa sequencing yielded 13,566,503 (BO) and 8,736,644 (AO) raw reads for serum small RNA libraries, and 9,522,247 (BO) and 10,082,482 (AO) raw reads for plasma small RNA libraries. After filtered low quality sequences, 7,552,415 (BO) and 4,268,347 (AO) clean reads for serum, 5,960,124 (BO) and 6,694,759(AO) clean reads for plasma were obtained respectively. The histograms of the reads length distribution showed majority were 20nt ~ 24nt (Fig 2). Of these, 205,107 (BO) and 218,200 (AO) unique small RNAs for serum, 214,226 (BO) and 221,870 (AO) unique small RNAs for plasma were identified.
Fig 2. Frequency distribution of the sequence lengths in serum and plasma.
The X axis shows sequence size. The Y axis shows the percents of sequence lengths. The majority of sequences for both libraries were 20nt ~ 24nt. BO, before puberty onset; AO, after puberty onset.
Identification of conserved miRNAs
To identify conserved miRNAs in chicken serum and plasma, the small RNAs were aligned to current miRBase (Release V21.0). Sequences with perfect matching to known chicken (G.gallus) miRNAs were considered as conserved miRNAs. In total, 398 conserved sequences were annotated as chicken miRNAs. To obtain higher reliable results, only the miRNAs with raw reads >10 at least one time point were considered. Then 192 were sorted as common miRNAs, with only 1 serum-specific miRNAs and 4 plasma-specific miRNAs. All 197 conserved miRNAs detected by sequencing were listed in S1 Table.
Discovery of novel miRNAs
To predict novel miRNAs, the unannotated reads were analyzed using RNAfold tools. A total of 100 novel miRNAs expressed in serum or plasma met the predictive criteria. The abundance of these novel miRNAs was relatively lower, only 19 ones with raw reads >10 at least one time point (S2 Table). So, this might be the reason they were not found previously.
Differential expression profiles of conserved miRNAs between BO and AO
To compare the differential expression of miRNAs in the serum/plasma of BO versus AO chickens, the number of miRNAs in each sample was normalized to the total number of reads. It was of note that the list of differential expression miRNAs in plasma was the same with that in serum. The expression of 4 plasma-specific miRNAs was not significant in AO chickens compared to BO counterparts. So only the differential expression miRNAs in serum were showed in S3 Table. In total, 130 miRNAs were considered to be differentially expressed (P < 0.05), with 68 up-regulated and 62 down-regulated. 40 miRNAs had more than two fold expression changes (|log2(fold-change)|> = 1.0) from BO to AO (Table 1).
Table 1. The differentially expressed serum-miRNAs with more than two fold-changes between BO and AO.
| miRNAs | Fold-change | Log2(fold-change) | p-value |
|---|---|---|---|
| gga-miR-3536 | 16.48 | 4.04 | < 0.001 |
| gga-miR-499-3p | 13.73 | 3.78 | < 0.001 |
| gga-miR-24-5p | 12.09 | 3.60 | < 0.001 |
| gga-miR-27b-5p | 6.04 | 2.60 | 0.005 |
| gga-miR-2188-5p | 5.83 | 2.54 | < 0.001 |
| gga-miR-193b-5p | 5.60 | 2.49 | < 0.001 |
| gga-miR-3528 | 5.37 | 2.42 | < 0.001 |
| gga-miR-451 | 4.25 | 2.09 | < 0.001 |
| gga-miR-1736-3p | 4.21 | 2.07 | < 0.001 |
| gga-miR-365-5p | 3.85 | 1.94 | < 0.001 |
| gga-miR-144-3p | 3.83 | 1.94 | < 0.001 |
| gga-miR-1552-3p | 3.57 | 1.84 | 0.007 |
| gga-miR-92-5p | 3.30 | 1.72 | < 0.001 |
| gga-miR-365-3p | 3.14 | 1.65 | < 0.001 |
| gga-miR-193b-3p | 2.95 | 1.56 | < 0.001 |
| gga-miR-221-5p | 2.75 | 1.46 | 0.011 |
| gga-miR-142-3p | 2.58 | 1.37 | < 0.001 |
| gga-miR-183 | 2.38 | 1.25 | < 0.001 |
| gga-miR-29b-3p | 2.32 | 1.22 | < 0.001 |
| gga-miR-101-5p | 2.30 | 1.20 | < 0.001 |
| gga-miR-1451-5p | 2.16 | 1.11 | < 0.001 |
| gga-miR-142-5p | 2.15 | 1.11 | < 0.001 |
| gga-miR-19a-3p | 2.00 | 1.00 | < 0.001 |
| gga-miR-216b | 0.50 | -1.01 | < 0.001 |
| gga-miR-216a | 0.49 | -1.02 | 0.024 |
| gga-miR-1664-3p | 0.46 | -1.13 | < 0.001 |
| gga-miR-375 | 0.45 | -1.16 | < 0.001 |
| gga-miR-193a-3p | 0.41 | -1.30 | 0.003 |
| gga-miR-193a-5p | 0.38 | -1.40 | < 0.001 |
| gga-miR-449c-5p | 0.34 | -1.54 | < 0.001 |
| gga-miR-31-5p | 0.33 | -1.60 | 0.002 |
| gga-miR-217-5p | 0.24 | -2.04 | < 0.001 |
| gga-miR-20a-3p | 0.24 | -2.09 | < 0.001 |
| gga-miR-133a-3p | 0.24 | -2.05 | < 0.001 |
| gga-miR-155 | 0.22 | -2.15 | < 0.001 |
| gga-miR-460b-5p | 0.17 | -2.53 | < 0.001 |
| gga-miR-29c-3p | 0.15 | -2.70 | < 0.001 |
| gga-miR-9-5p | 0.15 | -2.70 | < 0.001 |
| gga-miR-19b-3p | 0.15 | -2.80 | < 0.001 |
| gga-miR-460b-3p | 0.09 | -3.45 | < 0.001 |
BO indicated before puberty onset, AO indicated after puberty onset.
Target prediction and functional analysis of differential expression miRNAs
To further explore the roles of differentially expressed miRNAs, putative target genes of the most differentially expressed 40 miRNAs (|Log2 (fold-change)|> = 1.0) were predicted by integrating TargetScan and miRanda. In total, 4829 common target genes were found (data is not shown).
GO annotation (S4 Table) showed the putative target genes were significantly enriched (counts > 30, P < 0.05) in protein transport and protein catabolism biological processes. The KEGG analysis (S5 Table) suggested that MAPK signaling pathway, focal adhesion, regulation of actin cytoskeleton, endocytosis, ubiquitin mediated proteolysis and calcium signaling pathway were the most enriched pathways (counts > 50, P < 0.01).
RT-qPCR validation of candidate miRNAs
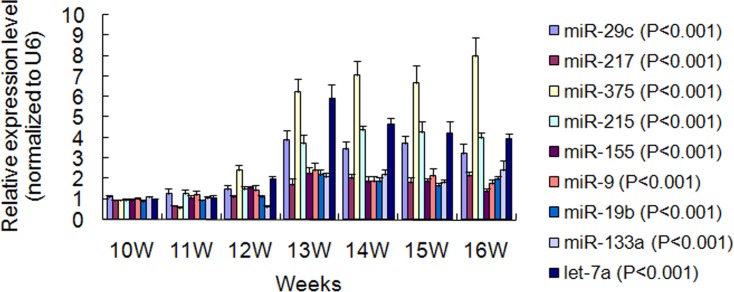
To identify miRNAs that can be served as potential biomarkers for measuring puberty onset in chicken. RT-qPCR validation of 9 candidate miRNAs was performed in serum from 10 to 16 weeks. The primers were listed in S6 Table. The results demonstrated that expression of control U6 was very stable, with quantification cycle (Cq) difference between groups less than 0.6. The Cq of the 9 miRNAs also had smaller variation between samples in one group (S7 Table). The single peak in dissociation curve indicated higher specificity of PCR products. The melting temperature (Tm) was 80.0~90.0°C. As illustrated in Fig 3, 7 miRNAs including miR-29c, miR-217, miR-375, miR-215, miR-19b, miR-133a and let-7a had relatively low and stable expression levels (P < 0.05) in early period, and increased significantly (P < 0.01) from 12 to 13 weeks when the gonads entered into rapid development. More importantly, the increased higher expression levels for these 7 ones could keep or show further increment until age at the first egg. Although the expression levels of miR-155 and miR-9 also increased significantly (P < 0.01) from 12 to 13 weeks, they then dropped significantly (P < 0.05, P < 0.01) and recovered to lower levels as early period.
Fig 3. Relative expression changes of 9 candidate serum-miRNAs in different stages.
Bars show standard deviations for replicates (n = 6).
Discussion
It has been suggested that circulating miRNAs are derived from multiple tissues. Specific miRNAs are enriched in exosomes in a cell-type-dependent manner [14]. miRNAs are found to express widely in gonad tissues and function roles in reproductive events [27,28,29,30]. Especially, the observations in model organisms and mammals have shown a potential link between miRNAs and puberty onset [31,32,33,34]. Our previous study also reveals miRNAs as novel partners involved in chicken puberty onset [24].This supports the possibility of developing circulating miRNA biomarkers to measure puberty onset.
The Solexa deep sequencing was performed to analyze the miRNA expression profiles in serum and plasma of hens from two different pubertal stages, before puberty onset (BO) and after puberty onset (AO). In total, 197 conserved miRNAs were identified in chicken serum and plasma. The co-expressed miRNA amounts (192/197) and their expression trends from BO to AO between serum and plasma were very similar, indicating that the different treatments to generate serum and plasma had nearly no influence on miRNAs. Many of our detected miRNAs had been found to express in various tissues, which further confirmed the wide origins of circulating miRNAs. Interestingly, some hypothalamic miRNAs involved in timing chicken puberty revealed by our previous study [24] were also abundantly expressed in serum and plasma. A recent report has confirmed small non-coding RNAs can transfer through mammalian placenta and directly regulate fetal gene expression [35]. Rom et al (2015) [36] found miR-98 and let-7g were protectors of the blood-brain barrier under neuroinflammatory conditions. Yet, whether these miRNAs are derived from the hypothalamus needs further investigation.
40 differentially expressed miRNAs were found during transition from BO to AO. To knowledge the global networks regulated by these differential miRNAs, target genes prediction and functional analysis were performed. The results suggest these differentially expressed miRNAs are involved in various biological pathways. Prominent among the pathways, MAPK signaling pathway has been suggested to play roles in female sexual maturation [37]. Wnt signaling pathway is believed to be a significant contributor to the regulation of ovarian follicle maturation and steroidogenesis [38]. mTOR signaling pathway is found to couple the nutritional status to regulate puberty onset in rats [39]. ErbB and GnRH signaling pathway is required for the timely initiation of puberty [40].
To identify circulating miRNAs can be served as biomarkers for measuring puberty onset in chicken is our main focus. The complex events involved in initiation of puberty require functional changes not only in neuroendocrine regulation genes and gonadal development genes, but also in energy metabolism-related genes. Therefore, there should be multiple miRNAs targeting those genes involved in puberty onset. Circulating miRNAs can derive from the gonad themselves or from metabolic tissues. According to previous reports and our sequencing results, 9 miRNAs, including miR-29c-3p, miR-375, miR-215-5p, miR-9-5p, miR-19b-3p, miR-133a-3p, let-7a, miR-217-5p and miR-155 were determined as candidates. Firstly, these miRNAs are differentially expressed from BO to AO with more than middle expression levels. Those ideal BO-specific or AO-specific miRNAs are not chosen just because of their lower expression, which will be a limitation in practical test. Of course, this criterion (raw reads> = 40) had disadvantage. It may not be suitable for other species. Secondly, they are associated with reproduction. Of the 9 members, miR-217 and miR-375 are regulators of chicken ovary maturity [29]. The developmental change of let-7a in hypothalamus is suggested to result in puberty onset in rats [33]. miR-375 and let-7a also play roles in regulation of insulin sensitivity and glucose metabolism [41,42,43]. miR-29c is linked to function in superior ovulation rate and kidding rate [44], and is a signature under high glucose conditions and a biomarker for anabolic steroids treatment [45]. miR-155 is found to be a molecular switch that regulates fat metabolism [46]. Circulating miR-19b is a marker of fatty liver [47]. Furthermore, miRNA-217, miRNA-155, miR-19b and miR-9 have target genes that are associated with puberty onset, such as FSHR, LEPR and circadian clock genes. The combination of the 9 serum miRNAs should be more reliable and unique than a single miRNA for puberty onset measurement.
To further validate the applicability of the nine miRNAs, RT-qPCR assays were performed in a set of individual serum samples and at multiple developmental stages. Seven of the nine miRNAs, except for miR-155 and miR-9, had a relatively consistent expression in earlier developmental phases, increased significantly when puberty initiated and hold stably or showed further increment until laying the first egg. Accordingly, a panel of seven miRNAs was identified as potential biomarkers to measure puberty onset in chicken. One of the weakness in our study is that the effectiveness of the seven-miRNA panel has not been confirmed in other chicken breeds. In addition, use of the seven-miRNAs panel can not differentiate the developmental progress after the initiation of puberty.
Conclusions
The present study is the first to examine the miRNA expression profile in the serum and plasma of chickens during puberty onset. The results show that numerous miRNAs are present in chicken serum and plasma. A seven serum-miRNA panel is identified as potential biomarkers for measuring chicken puberty onset. Due to highly conserved nature of miRNAs, the findings from our study will provide cues for other animals as well as human precocious puberty research.
Supporting Information
(XLS)
(XLS)
The miRNAs with |log2(fold-change)|> 1.0 were indicated in bold.
(XLS)
Go terms with counts of genes > 30 were indicated in bold.
(XLS)
KEGG pathway with counts of genes > 50 were indicated in bold.
(XLS)
(XLS)
(XLS)
Acknowledgments
We thank Shuangjie Zhang and Jinhong Lu for providing helps in animal sampling.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This research was supported by the National Natural Science Foundation of China (31201799) received by WH, Agricultural breed, technology and model innovation of Jiangsu Province SXGC(2015)300 received by WH and Agricultural science and technology innovation project of Jiangsu Province CX(15)1009 received by JMZ. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Lakshman R FN, Luben R, Bingham S, Khaw K, Wareham N, Ong KK. (2008) Association between age at menarche and risk of diabetes in adults: results from the EPIC-Norfolk cohort study. Diabetologia 51: 781–786. 10.1007/s00125-008-0948-5 [DOI] [PubMed] [Google Scholar]
- 2.Lomniczi A, Wright H, Castellano JM, Sonmez K, Ojeda SR (2013) A system biology approach to identify regulatory pathways underlying the neuroendocrine control of female puberty in rats and nonhuman primates. Horm Behav 64: 175–186. 10.1016/j.yhbeh.2012.09.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Seminara SB, Messager S, Chatzidaki EE, Thresher RR, Acierno JSJ, et al. (2003) The GPR54 gene as a regulator of puberty. N Engl J Med 349: 1614–1627. [DOI] [PubMed] [Google Scholar]
- 4.Topaloglu AK, Reimann F, Guclu M, Yalin AS, Kotan LD, et al. (2009) TAC3 and TACR3 mutations in familial hypogonadotropic hypogonadism reveal a key role for Neurokinin B in the central control of reproduction. Nat Genet 41: 354–358. 10.1038/ng.306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ong KK, Elks CE, Li S, Zhao JH, Luan J, et al. (2009) Genetic variation in LIN28B is associated with the timing of puberty. Nat Genet 41: 729–733. 10.1038/ng.382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gajdos ZK, Butler JL, Henderson KD, He C, Supelak PJ, et al. (2008) Association studies of common variants in 10 hypogonadotropic hypogonadism genes with age at menarche. J Clin Endocrinol Metab 93: 4290–4298. 10.1210/jc.2008-0981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elks CE PJ, Sulem P, Chasman DI, Franceschini N, He C (2010) Thirty new loci for age at menarche identified by a meta-analysis of genome-wide association studies. Nat Genet 42: 1077–1085. 10.1038/ng.714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lorah DD, Frank MB (2011) Puberty and its measurement: a decade in review. Journal of research on adolescence 21: 180–195. [Google Scholar]
- 9.Parent AS, Teilmann G, Juul A, Skakkebaek NE, Toppari J, et al. (2003) The timing of normal puberty and the age limits of sexual precocity: variations around the world, secular trends, and changes after migration. Endocr Rev 24: 668–693. [DOI] [PubMed] [Google Scholar]
- 10.Yingling VR, Khaneja A (2006) Short-term delay of puberty causes a transient reduction in bone strength in growing female rats. Bone 38: 67–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dorn LD, Dahl RE, Woodward HR, Biro F (2006) Defining the Boundaries of Early Adolescence: A User's Guide to Assessing Pubertal Status and Pubertal Timing in Research With Adolescents. Applied Developmental Science 10: 30–56. [Google Scholar]
- 12.Shirtcliff EA, Dahl RE, Pollak SD (2009) Pubertal development: Correspondence between hormonal and physical development. Child Development 80: 327–337. 10.1111/j.1467-8624.2009.01263.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bartel DP (2009) MicroRNAs: target recognition and regulatory functions. Cell 136: 215–233. 10.1016/j.cell.2009.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Squadrito ML, Baer C, Burdet F, Maderna C, Gilfillan GD, et al. (2014) Endogenous RNAs modulate microRNA sorting to exosomes and transfer to acceptor cells. Cell Rep 8: 1432–1446. 10.1016/j.celrep.2014.07.035 [DOI] [PubMed] [Google Scholar]
- 15.Villarroya-Beltri C G- VC, Sánchez-Cabo F, Pérez-Hernández D, Vázquez J, Martin-Cofreces N, Martinez-Herrera DJ, et al. (2013) Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat Commun 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qin H, Chen GX, Liang MY, Rong J, Yao JP, et al. (2013) The altered expression profile of microRNAs in cardiopulmonary bypass canine models and the effects of mir-499 on myocardial ischemic reperfusion injury. J Transl Med 11: 154 10.1186/1479-5876-11-154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Botta-Orfila T, Morato X, Compta Y, Lozano JJ, Falgas N, et al. (2014) Identification of blood serum micro-RNAs associated with idiopathic and LRRK2 Parkinson's disease. J Neurosci Res 92: 1071–1077. 10.1002/jnr.23377 [DOI] [PubMed] [Google Scholar]
- 18.Akat K M MMD, Morozov P (2014) Comparative RNA-sequencing analysis of myocardial and circulating small RNAs in human heart failure and their utility as biomarkers. PNAS 111(30): 11151–11156. 10.1073/pnas.1401724111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang C, Wang C, Chen X, Chen SD, Zhang YN, Zhi F, et al. (2013) Identification of seven serum microRNAs from a genome-wide serum microRNA expression profile as potential noninvasive biomarkers for malignant astrocytomas. Int J Cancer 132(1): 116–127. 10.1002/ijc.27657 [DOI] [PubMed] [Google Scholar]
- 20.Liu H, Zhu L, Liu B, Yang L, Meng X, et al. (2012) Genome-wide microRNA profiles identify miR-378 as a serum biomarker for early detection of gastric cancer. Cancer Lett 316(2): 196–203. 10.1016/j.canlet.2011.10.034 [DOI] [PubMed] [Google Scholar]
- 21.Chen X, Gao C, Li H, Huang L, Sun Q, et al. (2010) Identification and characterization of microRNAs in raw milk during different periods of lactation, commercial fluid, and powdered milk products. Cell Res 20: 1128–1137. 10.1038/cr.2010.80 [DOI] [PubMed] [Google Scholar]
- 22.Zheng Y, Chen KL, Zheng XM, Li HX, Wang GL (2014) Identification and bioinformatics analysis of microRNAs associated with stress and immune response in serum of heat-stressed and normal Holstein cows. Cell Stress Chaperones 19: 973–981. 10.1007/s12192-014-0521-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paczoska-Eliasiewicz HE, Proszkowiec-Weglarz M, Proudman J, Jacek T, Mika M, et al. (2006) Exogenous leptin advances puberty in domestic hen. Domest Anim Endocrinol 31: 211–226. [DOI] [PubMed] [Google Scholar]
- 24.Han W Z, Wang K, Su Y, Zhu Y, Song C, Li G, et al. (2015) High-throughput sequencing reveals hypothalamic microRNAs as novel partners involved in timing the rapid development of chicken (Gallus gallus) gonads. PLoS One 10(6):e0129738 10.1371/journal.pone.0129738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mestdagh P, Van Vlierberghe P, De Weer A, Muth D, Westermann F, et al. (2009) A novel and universal method for microRNA RT-qPCR data normalization. Genome Biol 10(6): 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- 27.Carletti MZ, Christenson LK (2009) MicroRNA in the ovary and female reproductive tract. J Anim Sci 87: E29–38. 10.2527/jas.2008-1331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luo L, Ye L, Liu G, Shao G, Zheng R, et al. (2010) Microarray-based approach identifies differentially expressed microRNAs in porcine sexually immature and mature testes. PLoS One 5: e11744 10.1371/journal.pone.0011744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kang L, Cui X, Zhang Y, Yang C, Jiang Y (2013) Identification of miRNAs associated with sexual maturity in chicken ovary by Illumina small RNA deep sequencing. BMC Genomics 14: 352 10.1186/1471-2164-14-352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu J, Zhu H, Song W, Li M, Liu C, et al. (2014) Identification of conservative microRNAs in Saanen dairy goat testis through deep sequencing. Reprod Domest Anim 49: 32–40. 10.1111/rda.12217 [DOI] [PubMed] [Google Scholar]
- 31.Zhu H, Shah S, Shyh-Chang N, Shinoda G, Einhorn WS, et al. (2010) Lin28a transgenic mice manifest size and puberty phenotypes identified in human genetic association studies. Nat Genet 42: 626–630. 10.1038/ng.593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu G, Park MY, Conway SR, Wang JW, Weigel D, et al. (2009) The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell 138: 750–759. 10.1016/j.cell.2009.06.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sangiao-Alvarellos S, Manfredi-Lozano M, Ruiz-Pino F, Navarro VM, Sanchez-Garrido MA, et al. (2013) Changes in hypothalamic expression of the Lin28/let-7 system and related microRNAs during postnatal maturation and after experimental manipulations of puberty. Endocrinology 154: 942–955. 10.1210/en.2012-2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Boulan L, Martin D, Milan M (2013) bantam miRNA promotes systemic growth by connecting insulin signaling and ecdysone production. Curr Biol 23: 473–478. 10.1016/j.cub.2013.01.072 [DOI] [PubMed] [Google Scholar]
- 35.Li J ZY, Li D, Liu Y, Chu D, Jiang X, Hou D, et al. (2015) Small non-coding RNAs transfer through mammalian placenta and directly regulate fetal gene expression. Protein Cell 6(6): 391–396. 10.1007/s13238-015-0156-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rom S DH, Zuluaga-Ramirez V, Reichenbach N L, Persidsky Y (2015) miR-98 and let-7g* protect the blood-brain barrier under neuroinflammatory conditions. J Cereb Blood Flow Metab 10.1038/jcbfm.2015.154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kolesarova A, Sirotkin AV, Mellen M, Roychoudhury S (2015) Possible intracellular regulators of female sexual maturation. Physiol Res 64: 379–386. [DOI] [PubMed] [Google Scholar]
- 38.Hernandez Gifford JA (2011) The role of WNT signaling in adult ovarian folliculogenesis. Reproduction 150: R137–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roa J, Tena-Sempere M (2012) Energy balance and puberty onset: emerging role of central mTOR signaling. Trends Endocrinol Metab 21: 519–528. [DOI] [PubMed] [Google Scholar]
- 40.Fortes MR, Reverter A, Nagaraj SH, Zhang Y, Jonsson NN, et al. (2011) A single nucleotide polymorphism-derived regulatory gene network underlying puberty in 2 tropical breeds of beef cattle. J Anim Sci 89: 1669–1683. 10.2527/jas.2010-3681 [DOI] [PubMed] [Google Scholar]
- 41.Frost RJ, Olson EN (2009) Control of glucose homeostasis and insulin sensitivity by the Let-7 family of microRNAs. Proc Natl Acad Sci U S A 108: 21075–21080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhu H, Shyh-Chang N, Segre AV, Shinoda G, Shah SP, et al. (2011) The Lin28/let-7 axis regulates glucose metabolism. Cell 147: 81–94. 10.1016/j.cell.2011.08.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lovis P, Gattesco S, Regazzi R (2008) Regulation of the expression of components of the exocytotic machinery of insulin-secreting cells by microRNAs. Biol Chem 389: 305–312. 10.1515/BC.2008.026 [DOI] [PubMed] [Google Scholar]
- 44.Ling YH, Ren CH, Guo XF, Xu LN, Huang YF, et al. (2014) Identification and characterization of microRNAs in the ovaries of multiple and uniparous goats (Capra hircus) during follicular phase. BMC Genomics 15: 339 10.1186/1471-2164-15-339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Becker C, Riedmaier I, Reiter M, Tichopad A, Pfaffl MW, et al. (2011) Changes in the miRNA profile under the influence of anabolic steroids in bovine liver. Analyst 136: 1204–1209. 10.1039/c0an00703j [DOI] [PubMed] [Google Scholar]
- 46.Chen Y, Siegel F, Kipschull S, Haas B, Frohlich H, et al. (2013) miR-155 regulates differentiation of brown and beige adipocytes via a bistable circuit. Nat Commun 4: 1769 10.1038/ncomms2742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pirola CJ, Fernandez Gianotti T, Castano GO, Mallardi P, San Martino J, et al. (2014) Circulating microRNA signature in non-alcoholic fatty liver disease: from serum non-coding RNAs to liver histology and disease pathogenesis. Gut 64: 800–812. 10.1136/gutjnl-2014-306996 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLS)
(XLS)
The miRNAs with |log2(fold-change)|> 1.0 were indicated in bold.
(XLS)
Go terms with counts of genes > 30 were indicated in bold.
(XLS)
KEGG pathway with counts of genes > 50 were indicated in bold.
(XLS)
(XLS)
(XLS)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.