Abstract
Comparative genomic analysis of the nuclear receptor family suggests that the testicular receptor 2, Nr2c1, undergoes positive selection in the human-chimpanzee clade based upon a significant increase in nonsynonymous compared to synonymous substitutions. Previous in situ analyses of Nr2c1 lacked the temporal range and spatial resolution necessary to characterize cellular expression of this gene from early to mid gestation, when many nuclear receptors are key regulators of tissue specific stem or progenitor cells. Thus, we asked whether Nr2c1 protein is associated with stem cell populations in the mid-gestation mouse embryo. Nr2c1 is robustly expressed in the developing olfactory epithelium. Its expression in the olfactory epithelium shifts from multiple progenitor classes at early stages to primarily transit amplifying cells later in olfactory epithelium development. In the early developing central nervous system, Nr2c1 is limited to the anterior telencephalon/olfactory bulb anlagen, coincident with Nestin-positive neuroepithelial stem cells. Nr2c1 is also seen in additional cranial sensory specializations including cells surrounding the mystacial vibrissae, the retinal pigment epithelium and Scarpa’s ganglion. Nr2c1 was also detected in a subset of mesenchymal cells in developing teeth and cranial bones. The timing and distribution of embryonic expression suggests that Nr2c1 is primarily associated with the early genesis of mammalian cranial sensory neurons and craniofacial skeletal structures. Thus, Nr2c1 may be a candidate for mediating parallel adaptive changes in cranial neural sensory specializations such as the olfactory epithelium, retina and mystacial vibrissae and in non-neural craniofacial features including teeth.
Keywords: Nr2c1, TR2, Olfactory epithelium, Pax7, Ncam, Ascl1
1. Introduction
Nuclear receptors (NRs), which evolved approximately 400 million years ago (Owen and Zelent, 2000; Thornton, 2001), function as intracellular transcription factors that regulate gene expression, usually in response to lipophilic ligands. In a number of developing and mature tissues, including the skin, lungs, gut, gonads and nervous system, these nuclear signaling proteins are associated with stem cell regulation (Jeong and Mangelsdorf, 2009). There is evidence of strong purifying selection acting to stabilize the majority of NR genes throughout primate evolution, consistent with necessary functional constraints across this essential family of transcriptional regulators. A recent analysis of NR sequence evolution in primates, however, shows that the testicular orphan receptor 2, Nr2c1 (previously known as TR2), an orphan receptor, for which the endogenous ligand has yet to be identified (Enmark and Gustafsson, 1996; Benoit et al., 2006), underwent a shift in selection pressure specifically on the human–chimpanzee clade (Baker et al., 2015, in review). This divergence in selection indicates that Nr2c1 may have diverse roles in mammalian development. Thus, we asked whether the cellular expression of Nr2c1 in a mammalian model species, the mouse, provides insight into its likely function.
Orphan receptors are quite likely the most ancient of the NRs, and they are thought to act during early embryonic development as well as in adult tissues (Enmark and Gustafsson, 1996; Laudet, 1997; Boukhtouche et al., 2006; Jolly et al., 2011). Accordingly, it seems likely that Nr2c1 influences developmental as well as homeostatic processes. Nevertheless, there is little insight into likely functions of Nr2c1 during development in any vertebrate species. To address this lack of foundational insight, we completed a thorough cellular characterization of Nr2c1 expression in the mouse embryo during mid and late gestation. Previous low-resolution in situ hybridization analysis in mice suggests that Nr2c1 may have a functional role in regulating stem cells during early development, perhaps via interactions with estrogen receptors or other NRs (Young et al., 1998; Hu et al., 2002; Lee et al., 2002). Nevertheless, the lack of precise cellular localization makes further interpretation difficult. Thus, we localized Nr2c1 protein in parallel with molecular markers of stem and transit amplifying cells, as well as early post-mitotic neurons in the developing mouse embryo at mid- and late gestation.
We found that Nr2c1 mRNA is substantially expressed in the developing peripheral nervous system, and minimally in the central nervous system during mid-gestation in the mouse. Nr2c1 protein, based upon its immuno-cytochemical cellular localization, is a potential regulator of progenitor cells in cranial sensory specializations, including the olfactory epithelium, the mystacial vibrissae, the retina, and cranial sensory ganglia. Nr2c1 is also expressed in developing craniofacial skeletal structures including apparently neural crest-derived bones and teeth. Accordingly, Nr2c1 may selectively regulate stem cells in cranial structures, including sensory specializations in the nose, eyes, and ears.
2. Results
2.1. Nr2c1 is robustly expressed in the developing olfactory epithelium
We used qPCR to measure Nr2c1 transcript levels in whole embryos at E8.5 and E9.5 (Fig. 1A, left) to survey early expression of Nr2c1 at the start of midgestation. Nr2c1 was readily detectable, and its expression level remained relatively consistent between E8.5 and E9.5. To further refine this assessment, we microdissected E8.5–E10.5 embryos to identify whether Nr2c1 expression is concentrated in specific compartments. Nr2c1 was not observed at higher levels in dissected E8.5 neural plate. We dissected E9.5 embryos by transecting the embryo at levels corresponding to key segmental boundaries (separating the telencephalic, mesencephalic, and rhombencephalic segments) to assess whether Nr2c1 message is concentrated in any of those neural domains or their associated structures (e.g. pharyngeal arches). Expression in all but the rhombencephalic segment was at levels approximating that observed in whole E9.5 embryos. To evaluate whether Nr2c1 was significantly expressed in the developing olfactory placode/pit we examined Nr2c1 expression in isolated olfactory epithelium (LaMantia et al., 2000) from E9.5 embryos. Nr2c1 is modestly increased in the olfactory epithelium, relative to the forebrain, in the E9.5 embryo. We then asked whether Nr2c1 expression levels were maintained, diminished or increased in the olfactory epithelium at E10.5 when progenitor specification has occurred, and neurogenesis has begun. We found a substantial increase in Nr2c1 expression at E10.5 (Fig. 1B). Thus, the nascent olfactory epithelium appears to be a site of localized and increasing expression of Nr2c1 in the mid-gestation mouse embryo.
Fig. 1.
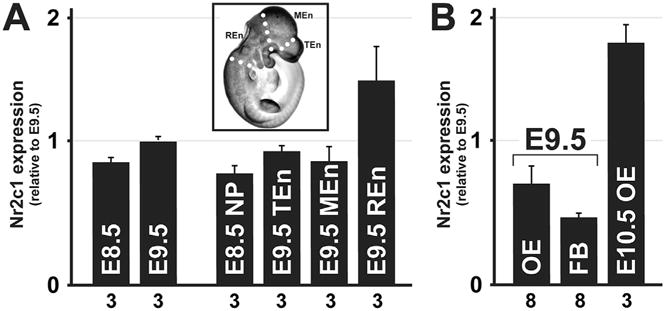
qPCR of microdissected embryos confirms Nr2c1 is robustly expressed in olfactory epithelium at E10.5. A. Relative expression of Nr2c1 was assessed for whole embryos (E8.5 and E9.5) and microdissected embryos (E8.5 anterior neural plate/folds, and E9.5 embryos dissected at boundaries of the telencephalon, mesencephalon, and rhombencephalon). Inset illustrates dissection boundaries on sample E10.5 embryo. Key: NP: neural plate; TEn: telencephalon; MEn: mesencephalon; REn: rhombencephalon. B. qPCR of microdissected and enzymatically separated tissue compartments of the frontonasal process (the olfactory epithelium and forebrain neurepithelium of an E9.5 embryo, and isolated olfactory epithelium from an E10.5 embryo). Key: OE: olfactory epithelium; FB: forebrain. Error bars represent S.E.M; n for each measurement is noted along the axis.
2.2. Nr2c1 is expressed in a subset of olfactory stem cells
The enhanced expression of Nr2c1 in the olfactory epithelium during initial stem cell specification and neurogenesis suggested that this region of the peripheral nervous system might be ideal for assessing the association of Nr2c1 with tissue specific stem, progenitor, or differentiated cell classes. Our recent studies identified specific markers for olfactory stem and transit amplifying cells (Tucker et al., 2010). Thus we used dual immunofluorescent localization of Nr2c1 with these additional markers to determine whether there was specific expression of Nr2c1 in distinct proliferative or post-mitotic cell classes in the mid-gestation embryo.
Cellular localization of Nr2c1 in the E10.5 olfactory epithelium shows that it is primarily a nuclear protein at this stage, and confocal microscopic assessment shows that Nr2c1 is concentrated in puncta within the nucleus (Fig. 2A–D). To determine whether Nr2c1 is selectively expressed in distinct cell classes in the olfactory epithelium we evaluated co-expression with three additional markers: Pax7, which identifies the slowly dividing presumed olfactory stem cell population (Murdoch et al., 2010), Ascl1, which identifies a rapidly dividing transit-amplifying precursor population (Tucker et al., 2010), and Ncam, which identifies post-mitotic olfactory neurons (Calof and Chikaraishi, 1989). At E10.5, we found that a small fraction of cells that robustly express Nr2c1 also express Pax7 (Fig. 2B, C, and E). Substantially more cells co-express Nr2c1 and Ascl1 (Fig. 2G, H, and J). In contrast, Ncam-expressing, presumably recently post-mitotic neurons in the olfactory epithelium do not express Nr2c1 (Fig. 2K–N); however, there is modest nuclear expression of Nr2c1 in Ncam-labeled cells of the migratory mass (Fig. 2O–Q), which is primarily composed of migrating neurons (Miller et al., 2010). Some of these cells may be olfactory ensheathing cell precursors (Barnett and Riddell, 2004) and thus retain a progenitor versus post-mitotic identity, despite the expression of the neuronal marker Ncam. Thus, at E10.5, Nr2c1 is associated primarily with proliferative stem and transit amplifying cells in the differentiating olfactory epithelium.
Fig. 2.
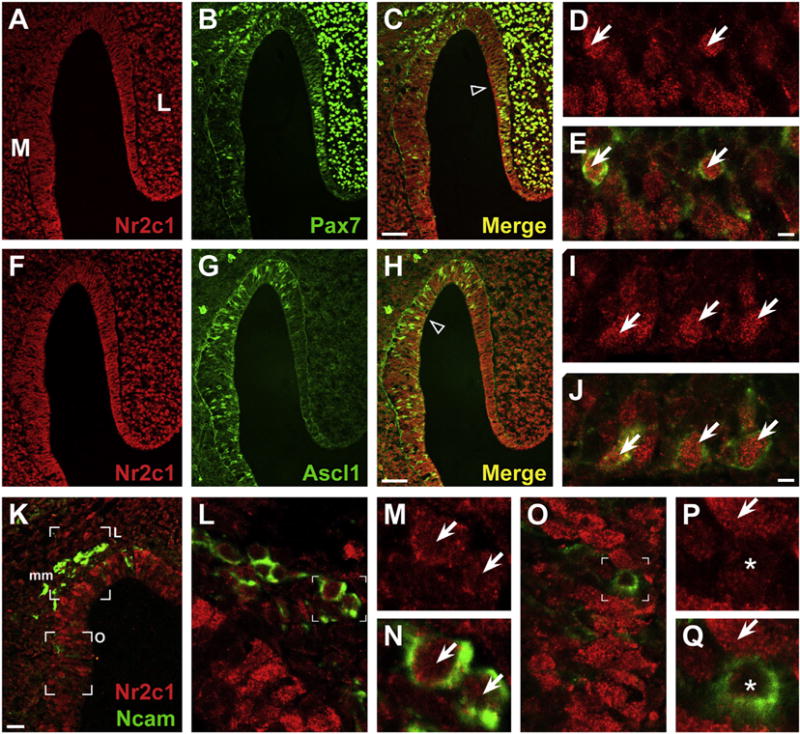
Fluorescent immunolocalization of Nr2c1 and markers of stem cell populations in mouse olfactory epithelium at E10.5 shows that a subset of Nr2c1-expressing cells are olfactory stem cells. A–E. Co-expression of Nr2c1 (red) and Pax7 (green), a marker for pluripotent stem cells in the OE. A subset of Nr2c1-expressing cells coexpress Pax7 (arrows, D–E). F–J. Co-expression of Nr2c1 (red) and Ascl1, a marker for a rapidly dividing transit-amplifying precursor population in the OE. Numerous Nr2c1-expressing cells co-express Ascl1 (arrows, I,J). K–P. Co-expression of Nr2c1 (red) and Ncam (green), a marker for post-mitotic neurons, both in the OE as well as in the adjacent migratory mass (mm). Some Ncam-positive neurons in the migratory mass show Nr2c1 expression (L–N, arrows), although this expression is less robust than the expression in the adjacent OE (L). O–Q. Within the OE, cells that robustly express Nr2c1 are not co-labeled with Ncam (P–Q, arrows), and Ncam-labeled cells express Nr2c1 at low levels (P–Q, asterisks). All panels imaged by confocal microscope (Left: 20×, Right: 63×). Scale bars for C, H, K are 25 μm; E, J: 5 μm.
At E11.5, Nr2c1 is still expressed throughout the olfactory epithelium, although its expression appears more robust in the medial versus lateral region (Fig. 3). Moreover, at E11.5, Nr2c1 appears to have shifted from primarily nuclear to primarily cytoplasmic expression. In the E11.5 olfactory epithelium, the Pax7 population is clearly separated from the Ascl1 population, with the former clearly on the lateral aspect (Fig. 3B), and the latter spatially separated along the medial aspect (Fig. 3G). Nr2c1 is no longer robustly associated with Pax7 labeled olfactory stem cells (Fig. 3D and E, asterisks), although the two proteins are occasionally recognized in the same cell (Fig. 3D and E, arrow). In contrast, cells in the medial olfactory epithelium frequently co-express Nr2c1 and Ascl1 (Fig. 3F–J). At E11.5, Ascl1 expression is mostly nuclear, while Nr2c1 is now shifted to primarily cytoplasmic expression (Fig. 3I and J). By E11.5, the expression of Ncam in newly generated neurons in the medial olfactory epithelium is far more substantial; however, most Ncam positive cells do not express Nr2c1 (e.g. Fig. 3K–O). A small number of Ncam labeled cells co-express Nr2c1 at E11.5 (Fig. 3N, O, arrow); however, these cells may be terminally dividing progenitors that also express Ascl1 or other related basic helix-loop-helix (bHLH) genes (Cau et al., 1997; Beites et al., 2009) as they cease division and begin to acquire neuronal identity.
Fig. 3.
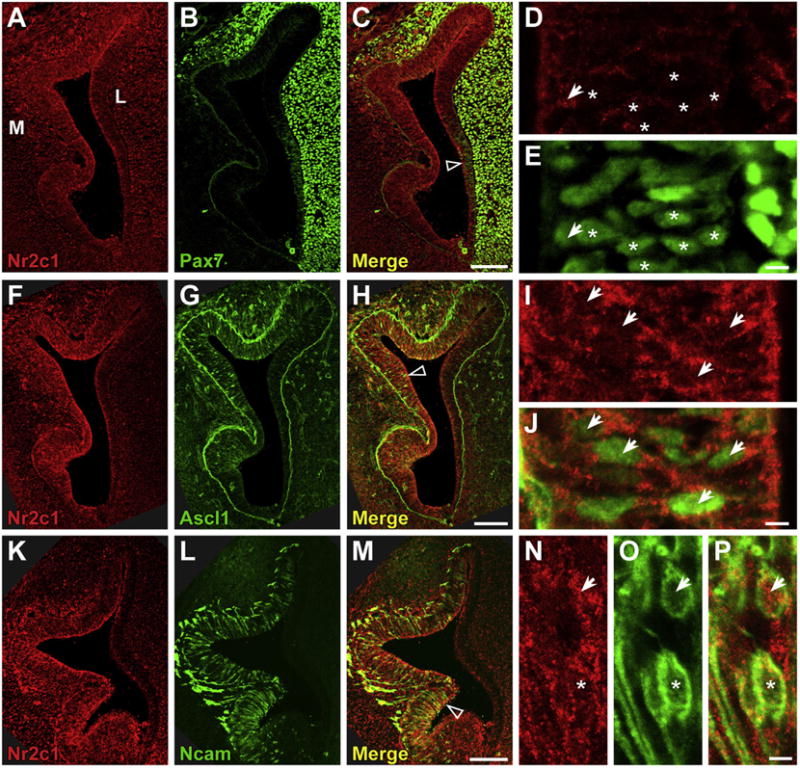
Fluorescent immunolocalization of Nr2c1 and markers of stem cell populations in mouse olfactory epithelium at E11.5 shows significant co-localization with Ascl1. A–E. Coexpression of Nr2c1 and Pax7 in the E11.5 OE. Nr2c1 expression is most robust in the medial aspect, while Pax7 is primarily in the lateral OE and lateral mesenchyme. Previous experiments have shown that the lateral OE cells that express modest levels of Pax7 are stem cells. Most Pax7-expressing cells in the OE do not express Nr2c1 (asterisks, D–E), although there are a small number of double-labeled cells with modest Nr2c1 expression (arrows, D–E). F–J. Co-expression of Nr2c1 and Ascl1 shows that most Ascl1 expressing cells co-express cytoplasmically-localized Nr2c1 (arrows, I, J). K–P. Co-expression of Nr2c1 and Ncam shows that there is some co-expression in the OE. Some Ncam-positive neurons in the OE have modest levels of cytoplasmic Nr2c1 expression (N–O, arrow), although Nr2c1 is absent from others (N–P, asterisks). All panels imaged by confocal microscope (Left: 20×, Right: 63×). Scale bars for C, H, M are 100 μm; E, J, O: 5 μm.
2.3. Nr2c1 expression in the maturing olfactory epithelium and the brain
At later developmental stages, although Nr2c1 expression is still present throughout the olfactory epithelium, it is also more widespread. Robustly labeled cells can be found throughout the entire olfactory epithelium at E17.5 (Fig. 4A). Expression is observed in both the thicker, presumptive neural epithelium, as well as in the thinner, presumptive respiratory epithelium (Fig. 4A and B). The expression of Nr2c1 at these later stages also appears to be primarily cytoplasmic, as observed in multiple regions of three individuals, unlike the expression observed at earlier stages. As the olfactory epithelium retains an active stem cell pool that is capable of regenerating both neural and respiratory epithelium into adulthood, we investigated whether Nr2c1 is still co-expressed in subsets of presumed progenitors. We found that Nr2c1 is robustly expressed in small subsets of Ascl1-positive cells that are found in discrete patches within the E16.5 olfactory epithelium (Fig. 4C), suggesting that Nr2c1 is still coincident with this key marker for rapidly dividing transit-amplifying precursor populations. We also assessed whether these patterns of Nr2c1 expression were also observed in the vomeronasal organ, the specialized domain of the olfactory epithelium that contains pheromone receptors. We found that Nr2c1 is expressed in the vomeronasal organ at E14.5 (Fig. 4D), and E17.5 (Fig. 4E), although vomeronasal expression at both stages is less robust than it is in the adjacent olfactory epithelium.
Fig. 4.
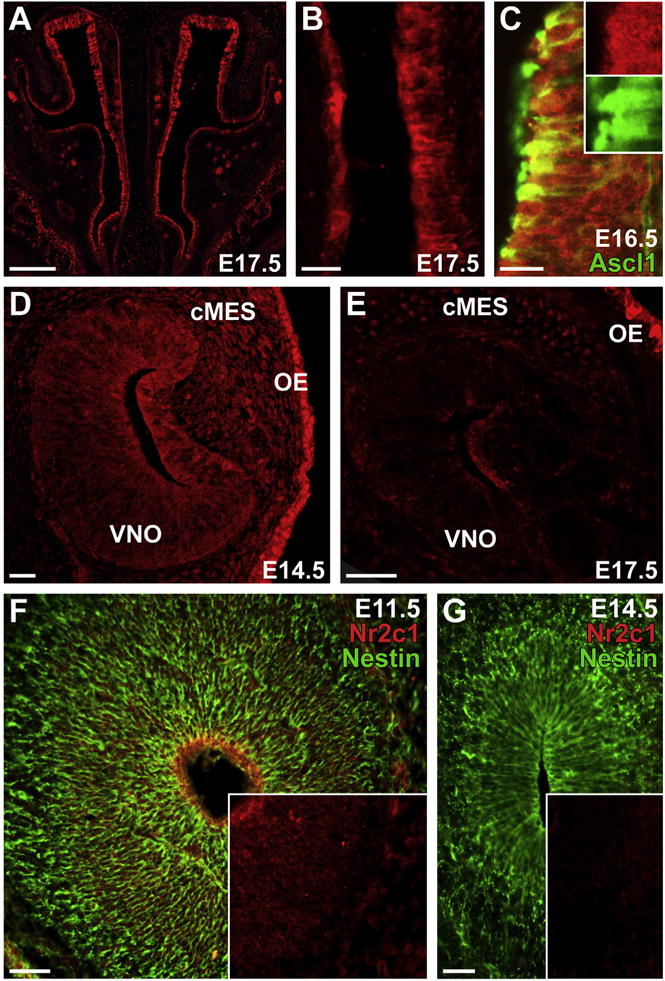
Nr2c1 is expressed in the OE, the vomeronasal organ (VNO), and craniofacial mesenchyme (cMES) at E14.5–17.5. A. Nr2c1 is robustly expressed throughout the entire OE at E17.5. B. Higher-magnification image of E17.5 OE shows clear expression in both the thicker (presumptive neural) and thinner (presumptive respiratory) epithelium. C. Confocal image (20×) showing Nr2c1 and Ascl1 co-localized in small patches of the OE. Inset: 1.5× magnification and channel separation. D. Confocal image (20×) showing expression of Nr2c1 in the vomeronasal organ (VNO) at E14.5; expression of Nr2c1 is present, but at lower levels in the VNO and adjacent cMES relative to the adjacent OE. E. Expression of Nr2c1 is diminished in the VNO at E17.5, relative to the adjacent cMES and OE. F. Nr2c1 is robustly expressed at the ventricular surface of the olfactory bulb at E11.5, coincident with the apical endfeet of the Nestin-positive neuroepithelial stem cells (radial glia). G. Expression of Nr2c1 is absent from the olfactory bulb at E14.5, although numerous Nestin-positive radial glia are present. Insets for F, G: Nr2c1 channel only. Scale bars for A, are 200 μm; B: 25 μm; C: 20 μm; D: 25 μm; E–F; 50 μm.
We found limited expression of Nr2c1 in the central nervous system. At E11.5 there is Nr2c1 expression in the ventricular zone of the anterior forebrain, the presumed olfactory bulb rudiment (Fig. 4F). Expression diminishes outside of this region, and only low levels are observed in the anterior dorsal telencephalon. By E14.5 Nr2c1 expression in these regions is greatly reduced and expression is not seen in anterior regions of the cortex or the now distinct olfactory bulb (Fig. 4B). Limited, diffuse staining, hardly above background, was seen in more caudal regions of the brain, including the cortex, ventral forebrain structures, and thalamus between E10.5 and E14.5.
2.4. Nr2c1 is expressed in supporting cells around cranial sensory specializations
In addition to the robust expression in the developing olfactory epithelium we also localized Nr2c1 in several other embryonic sensory structures. Nr2c1 is seen in the developing otic vesicle at E10.5 (Fig. 5A and B), as well as in the adjacent vestibuloacoustic ganglia (cranial nerve VIII). Nr2c1 is heavily expressed in the supporting cells of the mystacial vibrissae (Fig. 5C and D); however, it does not coincide with the NCAM-positive neural processes that innervate the vibrissae (Fig. 5D). At E14.5, the developing eye shows distinctive Nr2c1 expression at the margin of the retina, coincident with the presumptive retinal pigment epithelium (Fig. 5E and inset). Expression in the neural retina is largely absent, although there is some low-level diffuse signal in the ciliary marginal zone (arrow, Fig. 5E). Thus, Nr2c1 is expressed in cranial sensory specializations beyond the olfactory epithelium and vomeronasal organ, although not exclusively in their neural components.
Fig. 5.
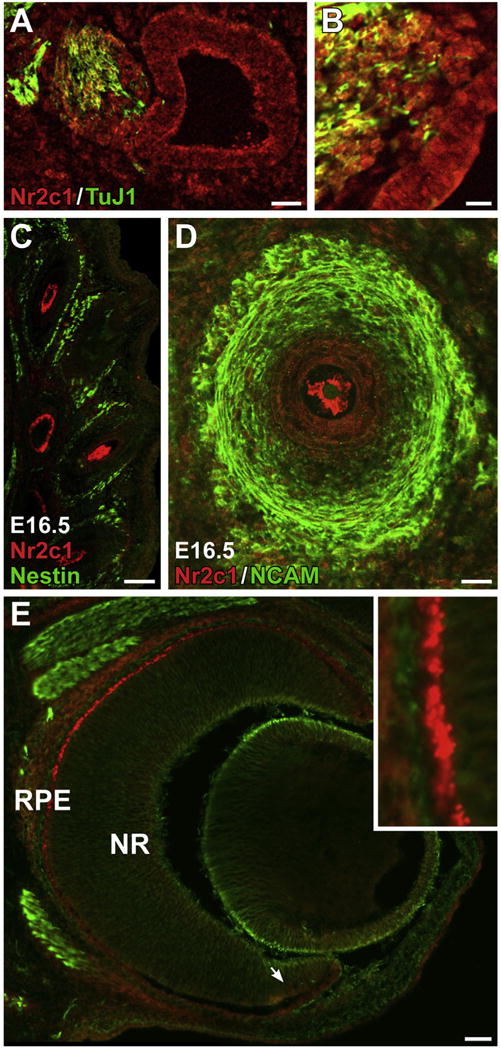
A–B. Nr2c1 is expressed in the placodally-derived neurepithelium of the otic vesicle at E10.5, as well as in the adjacent otic ganglion, as viewed in a saggital section. B. Higher magnification view of otic ganglia/otic vesicle from adjacent section. C–D. Expression of Nr2c1 in support cells of the facial vibrissae, as observed in a coronal section of an E16.5 embryo. C. Low-magnification image across multiple vibrissae shows that Nr2c1 labeled cells have distinctive morphologies, depending on the depth of the cross-section of the vibrissae. D. Confocal optical section (20×) illustrating support cells. E. Nr2c1 is robustly expressed in the presumptive retinal pigment epithelia of the eye at E14.5. Arrow: ciliary marginal zone. Inset: 5× magnification.
2.5. Nr2c1 expression in craniofacial primordia
We finally assessed the apparent expression of Nr2c1 in subsets of non-neural developing craniofacial structures. Nr2c1 is expressed in the developing teeth (Fig. 6A). There is robust labeling in the dental pulp (Fig. 6B), and Nr2c1 expression is co-localized with a subset of Nestin-positive presumptive dental mesenchymal stem cells (arrows). Nr2c1 expression is also observed in a subset of presumptive odontoblasts at the margin of the dental pulp (Fig. 6C) as well as in the mesenchymal stem cells and presumptive osteoblasts in the developing alveolar bone (Fig. 6D). At E17.5, modest expression is also apparent in the craniofacial mesenchyme, including the developing septal bone adjacent to the olfactory epithelium and vomeronasal organ at E14.5 and E17.5 (Fig. 6E and F). This expression is particularly robust in a subset of ventral facial bones, including the septal and frontal bones (Fig. 6E and F). Although expression is more robust at later stages, labeled mesenchymal cells are apparent from the earliest stages we examined, and at E10.5 cells expressing Nr2c1 were found within in the craniofacial mesenchyme, most robustly in the lateral nasal process (see Fig. 2A).
Fig. 6.
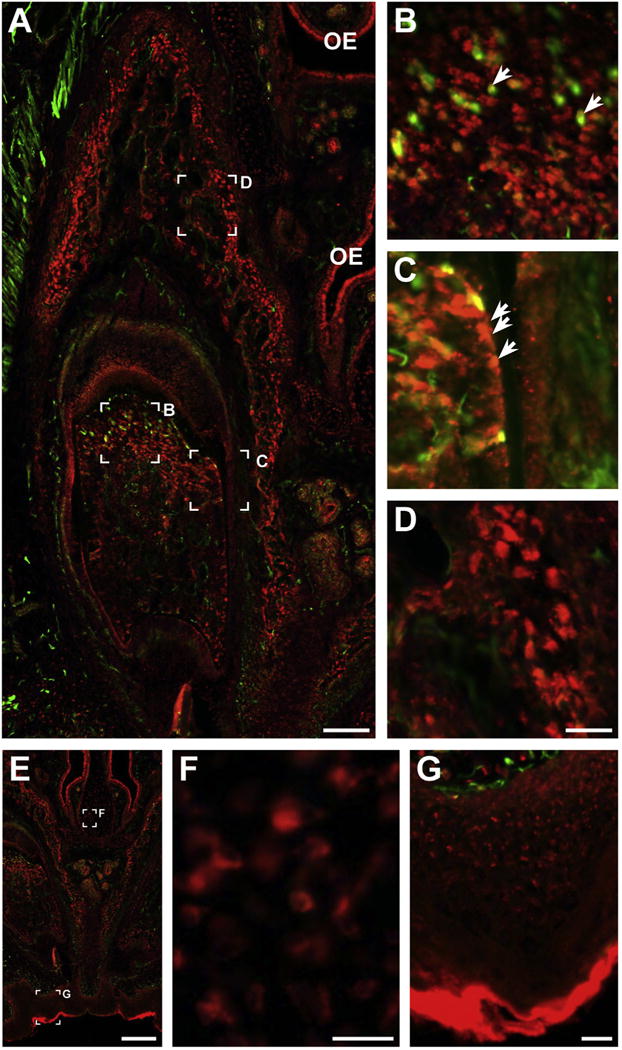
Nr2c1 expression in craniofacial primordia. A. Section of E17.5 upper incisor, stained for Nr2c1 (red) and Nestin (green). B–D. 4× magnification of specific regions of A, illustrating specific features. B. Nr2c1 shows robust labeling in the dental pulp. Nr2c1 expression is found in the vicinity of, and co-localized with a subset of Nestin-positive presumptive dental mesenchymal stem cells (arrows). C. Expression in a subset of presumptive odontoblasts (arrows) at the margin of the dental pulp. D. Expression in presumptive osteoblasts and mesenchymal stem cells in the developing alveolar bone. E–G. Expression of Nr2c1 in craniofacial structures, presumptive septal bone (F) and presumptive bone of the palatine process (F) at E17.5. Note: E–G are alternate views of image shown in Fig. 4A.
3. Discussion
We have found that Nr2c1 is associated with a wider array of neural and non-neural craniofacial cells, particularly those in the olfactory epithelium. These observations expand on previous observations that associated Nr2c1 more generally with embryonic stem (ES) cells and the developing nervous system (Hu, 2002; Lee et al., 2002; Shyr et al., 2009). Nr2c1 is expressed in the olfactory epithelium at all of the embryonic ages we studied, although its expression changes as the olfactory epithelium differentiates. Initially it is expressed robustly in Pax7-and Ascl1-positive olfactory stem and transit amplifying cell populations (Murdoch et al., 2010; Tucker et al., 2010). As development proceeds its expression becomes primarily cytoplasmic and limited to transit amplifying and potentially some recently post-mitotic neurons. The association between Nr2c1 expression and pluripotentiality becomes less evident as development proceeds: while our co-localization analysis clearly shows that Nr2c1 identifies a subpopulation of pluripotent stem cells at E10.5, its expression in the olfactory epithelium becomes more widespread by E17.5. Nr2c1 is also expressed at other cranial epithelial sites, in locations that include significant stem or progenitor cell populations associated with sensory specializations and epithelial appendages. For example, a subset of the Nr2c1-expressing cells in the dental papilla co-express Nestin, which is characteristic of a stem cell population at that site (Sonoyama et al., 2008). In other cases, the close identification with stem or progenitor cell populations is transient, or less clear Expression in the anterior forebrain/presumed olfactory bulb rudiment at E10.5 does not persist. There is also persistent expression of Nr2c1 in the vibrissae, but this expression does not appear to be coincident with the significant population of Nestin-expressing stem cells that remain pluripotent well into adulthood (Oshima et al., 2001; Amoh et al., 2012).
It is not clear whether the variability in subcellular localization from nucleus to cytoplasm we have observed indicates a change in Nr2c1 function. At the earliest stages of olfactory epithelium development, we observed clear nuclear localization of Nr2c1; however, at later stages, and in most other tissues, we observed cytoplasmic localization of the protein. It is possible that this shift may indicate a relative change in transcriptional activity. Alternatively, the change in subcellular localization may indicate either the presence or absence of an unidentified cofactor or ligand that regulates subcellular localization.
We have also established the expression of Nr2c1 in the craniofacial mesenchyme, including in developing teeth and in a subset of craniofacial bones. As a significant subset of the mesenchymal population involved in dental development is neural crest-derived (Sharpe, 2001), it is possible that robust Nr2c1 expression reflects the significant numbers of neuroectodermally-derived stem cells in these regions. Similarly, the cranial neural crest contributes significantly to the mesenchymal population that forms craniofacial bones and other mesenchymally-derived structures (including vascular smooth muscle and pericytes) particularly in the anterior and ventral regions of the head.
Contrary to our initial expectations, however, we did not find significant expression of Nr2c1 in the developing CNS. While we saw clear Nr2c1 expression in the anterior forebrain/nascent olfactory bulb, this expression was not maintained as development proceeded. We did not observe significant levels of Nr2c1 expression in more caudal regions of the brain at any gestational age. Thus, our observations are consistent with Nr2c1 playing a role in the development of cranial placodal neurepithelium and neural crest-derived skeletal/dental structures, but not in the central nervous system.
Our localization of Nr2c1 during mid-gestation was performed in mice. Accordingly, relating these results to the function and evolutionary changes in human NR2C1 remains a challenge. Our results suggest the possibility that Nr2c1 in the mouse may be involved in regulating the progenitor or early differentiation state of a wide array of cells in cranial sensory specializations and in neural crest-derived non-neural craniofacial structures. Evolutionary changes in this gene in the human-chimpanzee clade suggest that Nr2c1 may be a candidate for mediating adaptive changes in olfaction and other cranial sensory mechanisms, as well as in neural crest-derived craniofacial structures, such as the face and dentition that distinguish modern humans and chimpanzees.
4. Materials and methods
4.1. Animals
CD1 mice were used for these analyses. Embryo stages were determined based upon timed pregnancies, with the vaginal plug date identified as day 0.5 of gestation (thus E0.5). We collected embryos at E8.5, E9.5, E10.5, E14.5 and E16.5. All animal procedures were performed according to protocols approved by the Institutional Animal Care and Use Committee (IACUC) at The George Washington University.
4.2. RNA extraction, cDNA synthesis, and quantitative RT-PCR (qPCR)
Tissues were homogenized in Trizol (Invitrogen), RNA extracted from whole embryos or micro-dissected samples, and cDNA prepared as described previously (Maynard et al., 2013). RNA was quantified, and genomic DNA removed using DNA-free Turbo DNAse treatment (Ambion). First-strand cDNA pools were generated from RNA using ImPrompII reverse transcriptase (Promega) with random hexamer primers. GAPDH served as a control to calibrate cDNA templates used in qPCR assays. Assays were prepared using an epMotion liquid handling system. Quantitative analysis was performed using a CFX384 qPCR platform (Bio-Rad). For additional rigor, all analyses were performed with two independent primers for Nr2c1, but because the two primers gave equivalent results in all cases, we only report results from the first set here. Primer sequences are shown in Table 1.
Table 1.
Sequences of oligonucleotide primers used for qPCR analysis (listed as synthesized, in 5′–3′ orientation).
| Transcript | Forward primer | Reverse primer | Amplicon size (bp) |
|---|---|---|---|
| Gapdh | CTGACGTGCCGCCTGGAGAAA | GTTGGGGGCCGAGTTGGGATAGG | 346 |
| Nr2c1 (A) | CTGCGGTGGGGATATGCCTGTG | GCTACGTGCGACTCGCTCAGCAG | 252 |
| Nr2c1 (B) | GACAAAGCATCAGGGCGTCATTAC | GGCGTTGCGGCTAGAGGGCTTC | 333 |
4.3. Immunohistochemical analysis
Mouse embryos were dissected, fixed with 4% paraformaldehyde for 1 h, cryoprotected by infiltration of 30% sucrose in PBS, and embedded in OCT embedding compound (Sakura). Specimens were cryo-sectioned at 12 μm. All sections were preincubated in 2% bovine serum albumin. Nr2c1 rabbit anti-human polyclonal (LSBio, 1:500) primary antibody as well as mouse anti-Pax7 (Developmental Studies Hybridoma Bank, deposited by Atsushi Kawakami, 1:100), Rat anti-Ncam (EMD/Chemicon MAB310, 1:400), mouse anti-Ascl1 (BD Pharmingen, 24B72D11.1, 1:500), mouse anti-β3 Tubulin (Covance TuJ1, 1:3000), or chicken anti-Nestin (Novus, 1:2000) antibodies. Depending on the primary antibody combinations, the secondary antibodies used were either AlexaFluor 546 goat-anti-rabbit IgG (Molecular Probes), and Alex-aFluor 488 goat anti-mouse or rat IgG or anti-chicken IgY. (Molecular Probes). Assays were performed with multiple combinations of secondary antibodies to verify results. To ensure that our antibody reagents were robust and specific, we performed negative controls (immunostaining without either primary or secondary antibody) on representative sections at all ages and saw no evidence of non-specific staining, and confirmed that immunolabeling was the same using an independent anti-Nr2c1 antibody from a different vendor (TR2 rabbit polyclonal IgG; Santa Cruz, 1:500) that was prepared against a different fragment of the protein (data not shown). Images were captured either using a Leica DM6000 epifluorescence macroscope with digital capture, using a motorized stage to facilitate tiling and image stitching, or captured on a Zeiss LM-510 confocal microscope, where noted. Qualitative assessments of cell frequency and staining are based on examinations of at least four sections including at least two independent embryos.
Supplementary Material
Acknowledgments
We thank Elizabeth Paronett for assistance in histology and imaging.
This work was funded by a National Science Foundation Doctoral Dissertation Research Improvement Grant (BSC-1455625), a Wenner-Gren Foundation Dissertation Fieldwork Grant (#8735), a NSF IGERT grant (DGE-0801634), support from GW OVPR and The GW School of Medicine for the GW Institute of Neuroscience Biomarker Core Facility, and NIH DC011534 (ASL).
Appendix A. Supplementary data
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.gep.2015.12.002.
References
- Amoh Y, Mii S, Aki R, Hamada Y, Kawahara K, Hoffman RM, Katsuoka K. Multipotent nestin-expressing stem cells capable of forming neurons are located in the upper, middle and lower part of the vibrissa hair follicle. Cell Cycle. 2012;11(18):3513–3517. doi: 10.4161/cc.21803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker JL, Dunn K, Mingrone J, Wood BA, Karpinski BA, Sherwood CC, Wildman DE, Maynard TM, Bielawski JP. Functional Divergence of the Nuclear Receptor NR2C1 as a Modulator of Pluripotentiality During Hominid Evolution. Genetics. 2016 Apr 13; doi: 10.1534/genetics.115.183889. [epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnett SC, Riddell JS. Olfactory ensheathing cells (OECs) and the treatment of CNS injury: advantages and possible caveats. J Anat. 2004;204:57–67. doi: 10.1111/j.1469-7580.2004.00257.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beites CL, Kawauchi S, Calof AL. Olfactory neuron patterning and specification. Dev Neurobiol. 2009;7:145–156. doi: 10.1016/b978-008045046-9.01046-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benoit G, Cooney A, Giguere V, Ingraham H, Lazar M, Muscat G, Perlmann T, Renaud JP, Schwabe J, Sladek F, et al. International Union of Pharmacology. LXVI. Orphan nuclear receptors. Pharmacol Rev. 2006;58:798–836. doi: 10.1124/pr.58.4.10. [DOI] [PubMed] [Google Scholar]
- Boukhtouche F, Vodjdani G, Jarvis CI, Bakouche J, Staels B, Mallet J, Mariani J, Lemaigre-Dubreuil Y, Brugg B. Human retinoic acid receptor-related orphan receptor alpha1 overexpression protects neurones against oxidative stress-induced apoptosis. J Neurochem. 2006;96:1778–1789. doi: 10.1111/j.1471-4159.2006.03708.x. [DOI] [PubMed] [Google Scholar]
- Calof AL, Chikaraishi DM. Analysis of neurogenesis in a mammalian neuroepithelium: proliferation and differentiation of an olfactory neuron precursor in vitro. Neuron. 1989;3(1):115–127. doi: 10.1016/0896-6273(89)90120-7. [DOI] [PubMed] [Google Scholar]
- Cau E, Gradwohl G, Fode C, Guillemot F. Mash1 activates a cascade of bHLH regulators in olfactory neuron progenitors. Development. 1997;124:1611–1621. doi: 10.1242/dev.124.8.1611. [DOI] [PubMed] [Google Scholar]
- Enmark E, Gustafsson JA. Orphan nuclear receptors–the first eight years. Mol Endocrinol. 1996;10:1293–1307. doi: 10.1210/mend.10.11.8923456. [DOI] [PubMed] [Google Scholar]
- Hu YC, Shyr CR, Che W, Mu XM, Kim E, Chang C. Suppression of estrogen receptor-mediated transcription and cell growth by interaction with TR2 orphan receptor. J Biol Chem. 2002;277:33571–33579. doi: 10.1074/jbc.M203531200. [DOI] [PubMed] [Google Scholar]
- Hu YC. Suppression of estrogen receptor-mediated transcription and cell growth by interaction with TR2 orphan receptor. J Biol Chem. 2002;277:33571–33579. doi: 10.1074/jbc.M203531200. [DOI] [PubMed] [Google Scholar]
- Jeong Y, Mangelsdorf DJ. Nuclear receptor regulation of stemness and stem cell differentiation. Exp Mol Med. 2009;41:525–537. doi: 10.3858/emm.2009.41.8.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolly S, Journiac N, Naudet F, Gautheron V, Mariani J, Vernet-der Garabedian B. Cell-autonomous and non-cell-autonomous neuroprotective functions of RORα in neurons and astrocytes during hypoxia. J Neurosci. 2011;31:14314–14323. doi: 10.1523/JNEUROSCI.1443-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaMantia AS, Bhasin N, Rhodes K, Heemskerk J. Mesenchymal/epithelial induction mediates olfactory pathway formation. Neuron. 2000;28:411–425. doi: 10.1016/s0896-6273(00)00121-5. [DOI] [PubMed] [Google Scholar]
- Laudet V. Evolution of the nuclear receptor superfamily: early diversification from an ancestral orphan receptor. J Mol Endocrinol. 1997;19:207–226. doi: 10.1677/jme.0.0190207. [DOI] [PubMed] [Google Scholar]
- Lee YF, Lee HJ, Chang C. Recent advances in the TR2 and TR4 orphan receptors of the nuclear receptor superfamily. J Steroid Biochem Mol Biol. 2002;81:291–308. doi: 10.1016/s0960-0760(02)00118-8. [DOI] [PubMed] [Google Scholar]
- Maynard T, Gopalakrishna D, Meechan D, Paronett E, Newbern J, LaMantia A. 22q11 gene dosage establishes an adaptive range for sonic hedgehog and retinoic acid signaling during early development. Hum Mol Genet. 2013;22(2):300–312. doi: 10.1093/hmg/dds429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murdoch B, DelConte C, García-Castro MI. Embryonic Pax7-expressing progenitors contribute multiple cell types to the postnatal olfactory epithelium. J Neurosci. 2010;30(28):9523–9532. doi: 10.1523/JNEUROSCI.0867-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller AM, Treloar HB, Greer CA. Composition of the migratory mass during development of the olfactory nerve. J Comp Neurol. 2010;24:4825–4841. doi: 10.1002/cne.22497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshima H, Rochat A, Kedzia C, Kobayashi K, Barrandon Y. Morphogenesis and renewal of hair follicles from adult multipotent stem cells. Cell. 2001;104:233–245. doi: 10.1016/s0092-8674(01)00208-2. [DOI] [PubMed] [Google Scholar]
- Owen GI, Zelent A. Origins and evolutionary diversification of the nuclear receptor superfamily. Cell Mol Life Sci. 2000;57:809–827. doi: 10.1007/s000180050043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpe PT. Neural crest and tooth morphogenesis. Adv Dent Res. 2001;15:4–7. doi: 10.1177/08959374010150011001. [DOI] [PubMed] [Google Scholar]
- Shyr CR, Kang HY, Tsai MY, Liu NC, Ku PY, Huang KE, Chang C. Roles of testicular orphan nuclear receptors 2 and 4 in early embryonic development and embryonic stem cells. Endocrinology. 2009;150:2454–2462. doi: 10.1210/en.2008-1165. [DOI] [PubMed] [Google Scholar]
- Sonoyama W, Liu Y, Yamaza T, Tuan RS, Wang S, Shi S, Huang GT. Characterization of the apical papilla and its residing stem cells from human immature permanent teeth: a pilot study. J Endod. 2008;34(2):166–171. doi: 10.1016/j.joen.2007.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton JW. Evolution of vertebrate steroid receptors from an ancestral estrogen receptor by ligand exploitation and serial genome expansions. Proc Natl Acad Sci U S A. 2001;98:5671–5676. doi: 10.1073/pnas.091553298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker ES, Lehtinen MK, Maynard T, Zirlinger M, Dulac C, Rawson N, Pevny L, LaMantia AS. Proliferative and transcriptional identity of distinct classes of neural precursors in the mammalian olfactory epithelium. Development. 2010;137:2471–2481. doi: 10.1242/dev.049718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young WJ, Lee YF, Smith SM, Chang C. A bidirectional regulation between the TR2/TR4 orphan receptors (TR2/TR4) and the Ciliary Neurotrophic Factor (CNTF) signaling pathway. J Biol Chem. 1998;273:20877–20885. doi: 10.1074/jbc.273.33.20877. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


