Abstract
IMPORTANCE
Evolutionary medicine may provide insights into human physiology and pathophysiology, including tumor biology.
OBJECTIVE
To identify mechanisms for cancer resistance in elephants and compare cellular response to DNA damage among elephants, healthy human controls, and cancer-prone patients with Li-Fraumeni syndrome (LFS).
DESIGN, SETTING, AND PARTICIPANTS
A comprehensive survey of necropsy data was performed across 36 mammalian species to validate cancer resistance in large and long-lived organisms, including elephants (n = 644). The African and Asian elephant genomes were analyzed for potential mechanisms of cancer resistance. Peripheral blood lymphocytes from elephants, healthy human controls, and patients with LFS were tested in vitro in the laboratory for DNA damage response. The study included African and Asian elephants (n = 8), patients with LFS (n = 10), and age-matched human controls (n = 11). Human samples were collected at the University of Utah between June 2014 and July 2015.
EXPOSURES
Ionizing radiation and doxorubicin.
MAIN OUTCOMES AND MEASURES
Cancer mortality across species was calculated and compared by body size and life span. The elephant genome was investigated for alterations in cancer-related genes. DNA repair and apoptosis were compared in elephant vs human peripheral blood lymphocytes.
RESULTS
Across mammals, cancer mortality did not increase with body size and/or maximum life span (eg, for rock hyrax, 1% [95%CI, 0%–5%]; African wild dog, 8%[95%CI, 0%–16%]; lion, 2%[95%CI, 0% –7%]). Despite their large body size and long life span, elephants remain cancer resistant, with an estimated cancer mortality of 4.81% (95%CI, 3.14%–6.49%), compared with humans, who have 11% to 25%cancer mortality. While humans have 1 copy (2 alleles) of TP53, African elephants have at least 20 copies (40 alleles), including 19 retrogenes (38 alleles) with evidence of transcriptional activity measured by reverse transcription polymerase chain reaction. In response to DNA damage, elephant lymphocytes underwent p53-mediated apoptosis at higher rates than human lymphocytes proportional to TP53 status (ionizing radiation exposure: patients with LFS, 2.71% [95%CI, 1.93%–3.48%] vs human controls, 7.17%[95%CI, 5.91%–8.44%] vs elephants, 14.64%[95%CI, 10.91%–18.37%]; P < .001; doxorubicin exposure: human controls, 8.10% [95%CI, 6.55%–9.66%] vs elephants, 24.77%[95%CI, 23.0%–26.53%]; P < .001).
CONCLUSIONS AND RELEVANCE
Compared with other mammalian species, elephants appeared to have a lower-than-expected rate of cancer, potentially related to multiple copies of TP53. Compared with human cells, elephant cells demonstrated increased apoptotic response following DNA damage. These findings, if replicated, could represent an evolutionary-based approach for understanding mechanisms related to cancer suppression.
The mechanisms that prevent accumulation of genetic damage and subsequent uncontrolled proliferation of somatic cells in multicellular organisms remain poorly understood. A greater number of cells and cell divisions increases the chance of accumulating mutations resulting in malignant transformation.1 If all mammalian cells are equally susceptible to oncogenic mutations, then cancer risk should increase with body size (number of cells) and species life span (number of cell divisions). The Peto paradox describes the observation that cancer incidence across animals does not appear to increase as theoretically expected for larger body size and life span.2,3 To our knowledge, the cellular mechanism for this phenomenon of cancer resistance has never been demonstrated experimentally in organisms other than rodents.4–6
TP53 (encoding the protein p53 [RefSeq NM_000546]) is a crucial tumor suppressor gene, mutated in the majority of human cancers.7 Referred to as the “guardian of the genome,” inactivation of p53 leads to 3 cancer cell characteristics including suppression of apoptosis, increased proliferation, and genomic instability.8,9 Humans contain 1 copy (2 alleles) of TP53, and both functioning alleles are crucial to prevent cancer development. Absence of 1 functional allele leads to Li-Fraumeni syndrome (LFS), a cancer predisposition with more than a 90% lifetime risk for cancer, multiple primary tumors, and early childhood cancers.10,11 Understanding the cellular mechanism of cancer suppression in animals could benefit humans at high risk of cancer, such as patients with LFS, and even the healthy, aging population.
This study investigated the cancer rate in different mammals (including elephants), identified potential molecular mechanisms of cancer resistance, and compared response to DNA damage in elephants with that in healthy human controls and individuals with LFS.
Methods
Ethical and scientific institutional review board approval was obtained from each participating research organization for all elephant and human participation, including written informed consent from human participants. Experiments were performed on peripheral blood lymphocytes (PBLs) from African and Asian elephants, from a representative clinical cohort of patients with LFS enrolled in a separate study (the Cancer Genetics Study, University of Utah), and from age-matched human controls without a significant family history of cancer also enrolled in the Cancer Genetics Study. Patients with LFS were selected for inclusion as a representative sample based on TP53 mutation status, varied cancer history, and availability for blood draw. Human subject materials were collected at the University of Utah from June 2014 to July 2015. Laboratory experiments were also performed on African elephant fibroblasts, human fibroblasts, and HEK293 cells to confirm these findings.
Necropsy data were examined from zoo animals to determine if empirical evidence supports that cancer incidence does not increase with body size or life span. Fourteen years of necropsy data collected by the San Diego Zoo12 was compiled and tumor incidence was calculated for 36 mammalian species, spanning up to 6 orders of magnitude in size and life span.13 Data from the Elephant Encyclopedia14 were analyzed on the cause of death in captive African (Loxodonta africana) and Asian (Elephas maximus) elephants to estimate age incidence and overall lifetime cancer risk. Using the cancer transformation model from Calabrese and Shibata,15 the percentage decrease in cellular mutation rate was calculated to account for a 100× increase in cell mass (the difference between elephants and humans) without cancer development.
Genomic sequence analysis was next performed on the publicly available scaffolds of the African elephant genome in the Ensembl database (release 72; http://www.enssemble.org/) and the NCBI Gene database (http://www.ncbi.nlm.nih.gov/gene), with examination of cancer-related genes including oncogenes and tumor suppressors. TP53 sequence alignments were explored in related species, and African and Asian elephant TP53 retrogenes were cloned and resequenced. Capillary sequencing was performed on single elephants to avoid issues of single-nucleotide polymorphisms between elephants. Whole genome sequencing (Illumina HiSeq 2500) was performed on freshly extracted DNA from an African elephant at 40× average sequence coverage, with more than 100× coverage within areas of TP53.
Functional molecular analysis of TP53 and its retrogenes was performed on peripheral blood mononuclear cells from African and Asian elephants and fibroblasts from an African elephant. To determine if TP53 retrogenes are expressed in the elephant, reverse transcription–polymerase chain reaction was performed on RNA collected from African elephant peripheral blood mononuclear cells and African elephant fibroblasts. Polymerase chain reaction primers were designed to distinguish the TP53 retrogenes from the ancestral sequence and splice variants. Human vs elephant DNA repair efficiency (measured by double-strand breaks indicated by number of phospho-histone H2AX [pH2AX] foci), apoptosis (annexin V [AV] and propidium iodide [PI] by flow cytometry and Apotox-Glo, Promega), and cell cycle arrest (Apotox-Glo, Promega) were compared at different time points (1, 5, 10, 18, 24, and 72 hours) after DNA damage (doxorubicin, 0.005–30 µM; and ionizing irradiation, 0.5, 2, 5, 6, 10, and 20 Gy). Late apoptosis was defined as AV+PI+ and early apoptosis was defined as AV+PI−. Experiments were performed in either triplicate or quadruplicate. p53 plays a critical role in p21 and mouse double minute 2 homolog (Mdm2 or E3 ubiquitin-protein ligase Mdm2) protein induction following DNA damage,16,17 so p21 immunoblots were performed to validate a p53-dependent DNA damage response in elephant cells. p53 retrogene 9 (GenBank KF715863) was cloned into an expression vector to produce a protein fused to an epitope from the Myc protein. HEK293 cells were transfected with this Myc-tagged p53 retrogene 9 expression vector and p53 retrogene protein expression was measured by immunoblot using an antibody to the Myc tag. Retrogene protein product was co-immunoprecipitated from HEK293 cell lysates with Myc antibody, followed by immunoblots for phospho-p53 (serine-15) and Mdm2. The HEK293 cell line was chosen for these experiments because it is a human cell line (human embryonic kidney) that is easy to transfect and measure protein expression.
Cross-species lifetime cancer incidence was estimated by the number of animals in each species that reportedly died of cancer. A logistic regression model was fit to determine if body mass and maximum life span are variables associated with cancer incidence (R software, version 3.2.1). Additionally, all combinations of mass, life span, and mass-specific basal metabolic rate were examined for evidence of cancer association. An inverse cancer association was specifically tested in the largest existing terrestrial mammal, the elephant. For the DNA damage analysis, a χ2 test was used to compare pH2AX foci, and an unpaired 2-sided t test with α = .05 was used for apoptosis and cell cycle arrest (R software, version 3.2.1, and Graph-Pad Prism, version 6.0e). Both a linear regression and a Jonckheere-Terpstra test were used to assess if apoptotic response decreased with age.
Details of the experimental methods are further described in the eAppendix in the Supplement.
Results
Zoo Necropsies and Cancer Mortality
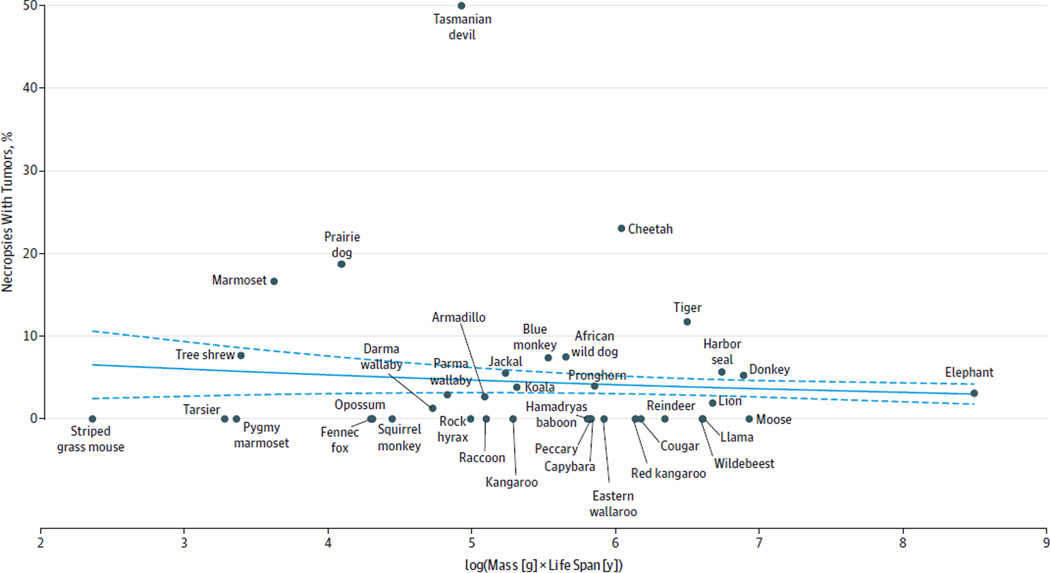
The 36 mammalian species analyzed spanned from the striped grass mouse (weight, 51 g, with a maximum life span of 4.5 years) to the elephant (weight, 4800 kg, with a maximum life span of 65 years). Cancer risk did not increase with mammalian body size and maximum life span among 36 species analyzed (eg, for rock hyrax, 1% [95% CI, 0%–5%]; African wild dog, 8% [95% CI, 0%–16%]; lion, 2% [95% CI, 0%–7%]) (Figure 1). No significant relationship was found with any combinations of mass, life span, and basal metabolic rate and cancer incidence (eFigure 1 and eTable 1 in the Supplement). Among 644 annotated elephant deaths from the Elephant Encyclopedia database, the lifetime cancer incidence was 3.11% (95% CI, 1.74%–4.47%) (Table 1). To obtain a more conservative estimate, an inferred cancer incidence was calculated for cases that lacked adequate details for the cause of death, leading to an estimated elephant cancer mortality rate of 4.81% (95% CI, 3.14%–6.49%). Based on an algebraic model of carcinogenesis,15 a 2.17-fold decrease in mutation rate was calculated as sufficient to protect elephants from cancer development given their 100× increased cellular mass compared with humans.
Figure 1. Cancer Incidence Across Species by Body Size and Life Span.
The mammalian species studied span the striped grass mouse to the elephant. Cancer incidence is not associated with mass and life span, as shown by the logistic regression (model fit shown as blue line; 95%CIs shown as dashed lines). Each data point in the graph is supported by a minimum of 10 necropsies for the included mammals (San Diego Zoo) and 644 annotated deaths for elephants (Elephant Encyclopedia database). The risk of cancer depends on both the number of cells in the body and the number of years over which those cells can accumulate mutations; therefore, cancer incidence is plotted as a function of mass × life span. All data with 95%CIs are presented in eTable 1 in the Supplement.
Table 1.
Cause of Death in 644 Elephantsa
| Age Range, y |
Total Necropsies |
No. of Elephants | Observed % With Cancer (95% CI) |
Inferred % With Cancer (95% CI) |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Euthanized, Noncancer |
Noncancer Disease |
Exogenous Mortality |
Euthanized, Unspecified |
Disease, Unspecified |
Euthanized, Cancer |
Cancer | ||||
| 0–5 | 125 | 15 | 77 | 28 | 1 | 2 | 0 | 2 | 1.60 (0.00–4.24) | 2.40 (0.00–5.44) |
| 6–15 | 83 | 20 | 36 | 19 | 4 | 1 | 1 | 2 | 3.61 (0.00–8.02) | 6.02 (0.58–11.47) |
| 16–25 | 121 | 35 | 48 | 25 | 7 | 2 | 2 | 2 | 3.31 (0.00–6.69) | 4.96 (0.86–9.05) |
| 26–35 | 108 | 27 | 51 | 15 | 8 | 4 | 3 | 0 | 2.78 (0.00–6.11) | 3.70 (0.00–7.60) |
| 36–45 | 94 | 32 | 27 | 13 | 12 | 5 | 0 | 5 | 5.32 (0.47–10.16) | 6.38 (1.18–11.58) |
| 46–55 | 70 | 14 | 23 | 7 | 7 | 17 | 1 | 1 | 2.86 (0.00–7.37) | 5.71 (0.00–11.59) |
| ≥56 | 43 | 3 | 7 | 6 | 7 | 19 | 1 | 0 | 2.33 (0.00–8.16) | 6.98 (0.00–15.29) |
| Lifetime, 0–≥56 |
644 | 146 | 269 | 113 | 46 | 50 | 8 | 12 | 3.11 (1.74–4.47) | 4.81 (3.14–6.49) |
Observed cancers are reported as the percentage of deaths annotated as being caused by cancer or by euthanasia due to cancer. Inferred cancer risk assumes that cancer occurs at the same fraction of deaths in cases with unspecified causes as those with specified causes. Exogenous causes of mortality include accidents (eg, falling in the enclosure) and animal fights that cause fatal injury.
African Elephant Genome Analysis
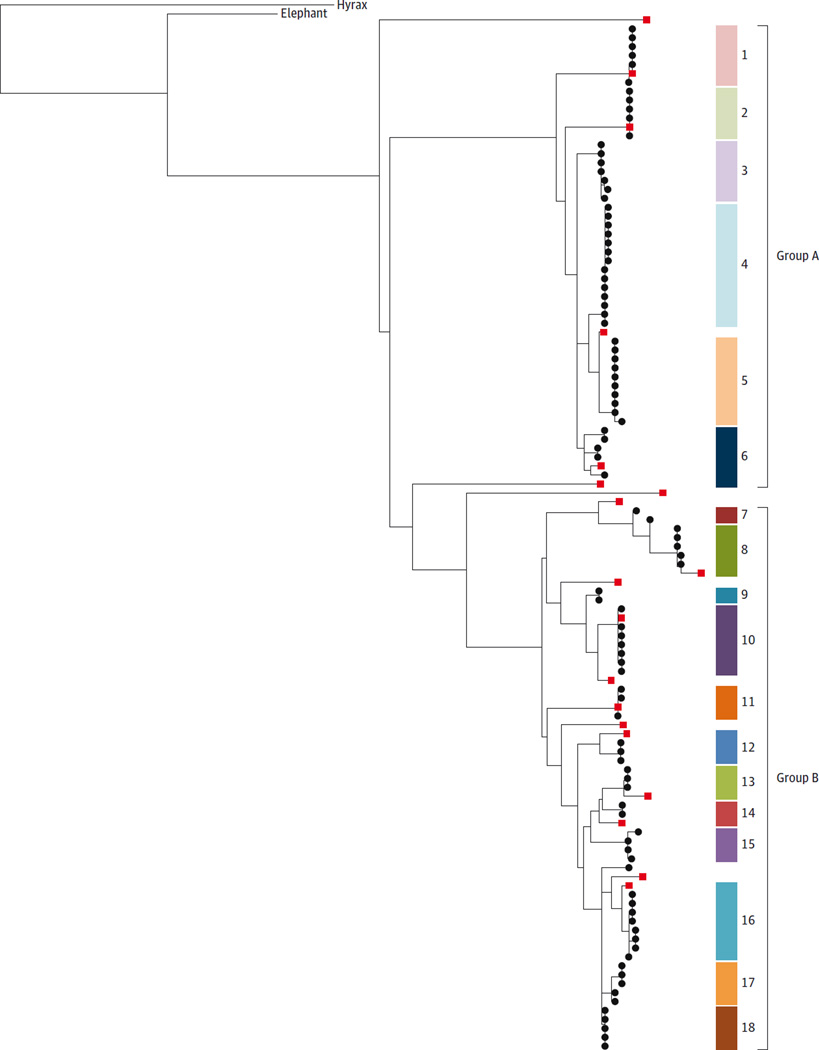
The African elephant (L africana) draft genome LoxAfr3 contains 19 copies of TP53. The human haploid genome contains 1 copy of TP53, while Ensembl and GenBank annotate a large number of TP53 paralogs in the African elephant genome (12 and 20 haploid copies, respectively; eTable 2 in the Supplement). Elephant sequence alignments revealed 1 TP53 copy with a comparable gene structure to TP53 found in other mammalian species (ancestral copy). The other 19 copies lack true introns, suggesting that they originated from retrotransposition (retrogenes). Cloning and resequencing confirmed at least 18 distinct retrogene copies in the African elephant in a maximum likelihood phylogeny, supported by multiple clones clustered into 2 main subtrees (groups A and B; Figure 2). Whole-genome sequencing with deep coverage confirmed 1 ancestral copy and 19 total retrogene copies, similar to the TP53 20 total copies annotated in GenBank. Eleven of the 18 retrogenes from the capillary sequencing were similar but not identical to previous Gen-Bank annotations and local whole genome sequencing data (eTable 2 in the Supplement). High variance in coverage across reference TP53 copies may indicate additional TP53 elephant copies not yet successfully assembled. There was no evidence for 8 of the published retrogene copies, possibly because of undersampling of clones, misassembly in the published genome, or differences between individual elephants. An additional 7 cloned sequences had support from multiple clones but were not found in either database. Further TP53 copies in the genome may also have been undetected by the polymerase chain reaction primers. The Asian elephant DNA was also found to contain 15 to 20 copies of group A and B TP53 retrogenes (eFigure 2 in the Supplement).
Figure 2. Group A and Group B TP53 Retrogenes in the African Elephant.
A maximum likelihood phylogeny was used to cluster the sequenced TP53 retrogene clones and to confirm the number of unique genes uncovered in the African elephant genome. The phylogeny allows for visualization of TP53 retrogene similarity to one another as well as their relationship to the ancestral TP53 sequence in the elephant and hyrax. The capillary sequenced clones from this study are shown as black circles and published sequences from GenBank are shown as red squares. Gene identifiers and genomic coordinates are given in eTable 2 in the Supplement. Phylogenic analysis reveals at least 18 distinct clusters of processed TP53 copies (shown as colored blocks numbered 1 to 18). These clusters fall into 2 groups, labeled group A and group B. The branch labeled “elephant” is the coding sequence of the ancestral TP53, and “hyrax” represents the coding sequences from the hyrax TP53. The hyrax, on the upper left, is used as the outgroup to show that the hyrax and elephant ancestral TP53 sequences are more similar to each other than to the retrogenes, and also that the retrogenes evolved after the split between hyrax and elephant. The distances between the retrogene sequences display their relationship based on sequence similarity but do not represent precise evolutionary time estimates. These data were generated with DNA from 1 elephant to control for polymorphic bases between individual elephants.
TP53 Retrogene Transcription and Translation
Reverse transcription–polymerase chain reaction on RNA from African elephant peripheral blood mononuclear cells and fibroblasts exposed to 2 Gy of radiation demonstrated TP53 retrogene expression. Products of the expected sizes were observed, separating the 2 groups of retrogenes (eFigure 3 in the Supplement). Sanger sequencing confirmed their identities as retrogenes from group A and/or group B (eFigure 4 in the Supplement). Transfected HEK293 cells showed p53 retrogene 9 protein expression by immunoblotting that increased with DNA damage similar to p53 in human fibroblasts exposed to DNA damage (eFigure 5, A–B, in the Supplement). Co-immunoprecipitation of lysates from the transfected HEK293 cells exposed to 6 Gy of ionizing radiation displayed phosphorylation of the Myc-tagged p53 elephant retrogene at serine-15 along with 90 kDa Mdm2, indicating Mdm2 binding (eFigure 5C in the Supplement).
Elephant Cell Response to DNA Damage
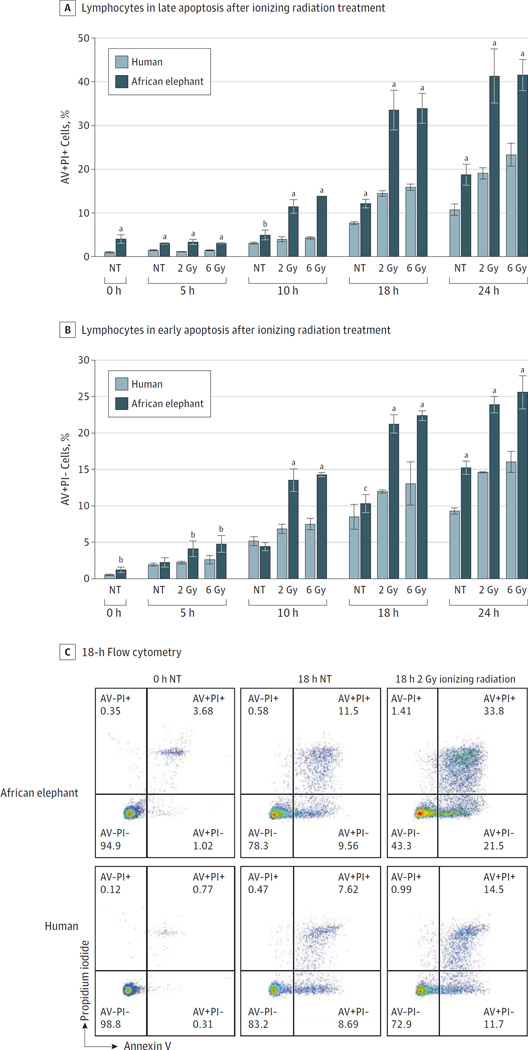
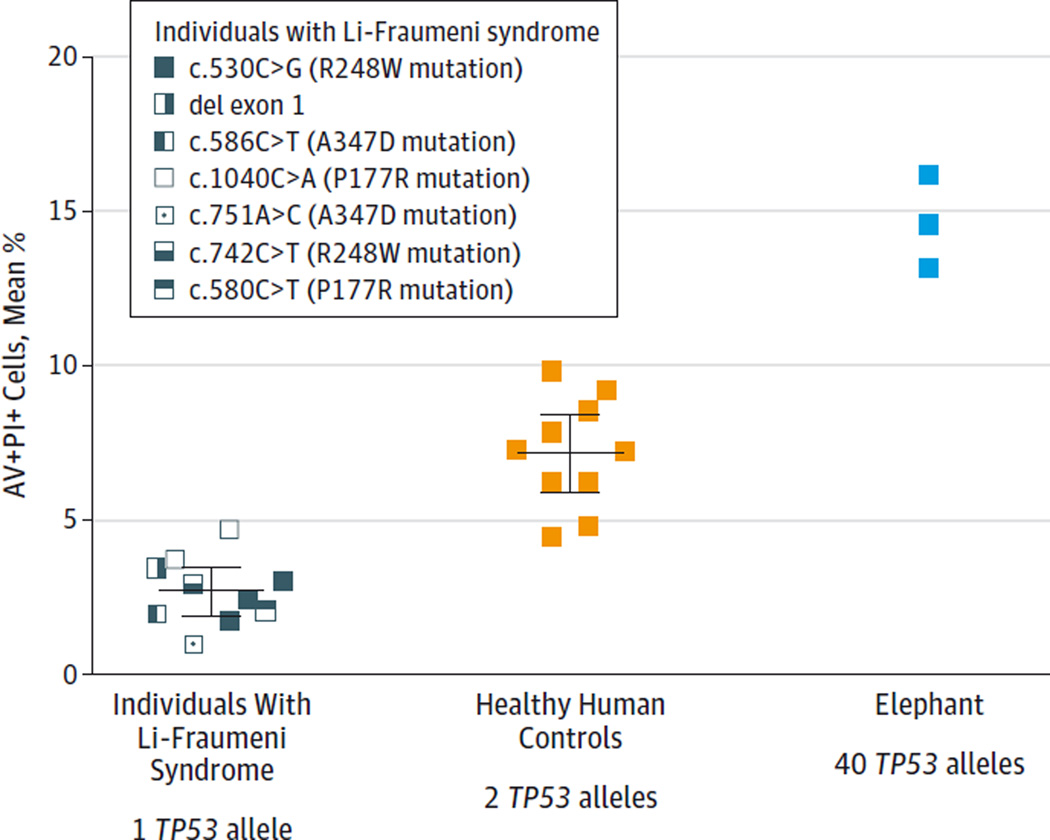
Lymphocytes undergo p53-dependent apoptosis in response to DNA damage,18,19 while fibroblasts undergo both p53-dependent apoptosis and cell cycle arrest,20–22 and both elephant cell types were tested accordingly. African elephant PBLs demonstrated apoptosis at significantly elevated rates compared with human PBLs after 18 hours when exposed to 2 Gy of ionizing radiation (late apoptosis: 33.20% [95% CI, 28.31%–38.09%] vs 14.07% [95% CI, 13.13%–15.01%]; P < .001; early apoptosis: 21.07% [95% CI, 19.61%–22.52%] vs 11.73% [95% CI, 11.35%–12.11%]; P < .001) (Figure 3, A–C) and when exposed to 5 µM of doxorubicin (24.77% [95% CI, 23.0%–26.53%] vs 8.10% [95% CI, 6.55%–9.66%]; P < .001) (eFigure 6 in the Supplement). Peripheral blood lymphocytes from individuals with LFS (n = 10), healthy controls (n = 10), and 1 African elephant (tested in 3 independent experiments) treated with 2 Gy of ionizing radiation revealed different levels of apoptosis (apoptosis calculated at 18 hours by subtracting the percentage of AV+PI+ cells treated with 2 Gy of ionizing radiation from the percentage of AV+PI+ cells cultured without treatment). Cells of patients with LFS underwent significantly less apoptosis (2.71%; 95% CI, 1.93%–3.48%) compared with healthy human PBLs (7.17%; 95% CI, 5.91%–8.44%; P < .001) and elephant PBLs (14.64%; 95% CI, 10.91%–18.37%; P < .001) (Figure 4 and eTable 3 in the Supplement).
Figure 3. African Elephant and Human Peripheral Blood Lymphocytes and Sensitivity to Ionizing Radiation.
A, The percentage of late apoptosis (annexin V positive [AV+] and propidium iodide positive [PI+]) and B, early apoptosis (AV+PI−) in elephant peripheral blood lymphocytes compared with human peripheral blood lymphocytes in response to 2 Gy and 6 Gy of ionizing radiation are graphed. Significant differences computed with a 2-sided t test between human and elephant at 0, 5, 10, 18, and 24 hours are indicated. Error bars represent 95% CIs. C, Representative scatter plots from flow cytometry are shown from the 0- and 18-hour time points. NT indicates no treatment.
aP <. 001.
bPanel A: NT at 10 hours, P = .008. Panel B: NT at 0 hours, P = .002; 2 Gy at 5 hours, P = .003; 6 Gy at 5 hours, P = .004.
cP = .03.
Figure 4. Apoptosis Response Relative to Number of Copies of TP53.
Percentage of apoptosis is shown for peripheral blood lymphocytes treated with 2 Gy of ionizing radiation from 10 individuals with Li-Fraumeni syndrome (with 1 functioning TP53 allele), 10 healthy controls (with 2 TP53 alleles), and 1 African elephant tested in 3 independent experiments (with 40 TP53 alleles). Ionizing radiation–induced apoptosis increased proportionally with additional copies of TP53 and inversely correlated with cancer risk. Experiments performed in quadruplicate for each individual and each colored box represents the mean percentage of cells in late apoptosis as measured by flow cytometry (percentage of annexin V–positive [AV+] and propidium iodide–positive [PI+] treated cells minus AV+PI+ untreated cells). The healthy control lymphocytes underwent more apoptosis than those from LFS patients (P < .001), and elephant lymphocytes underwent more apoptosis than those from healthy controls (P < .001 by 2-sided t test). Horizontal lines indicate the combined mean for all data points in each group with error bars indicating 95%CIs.
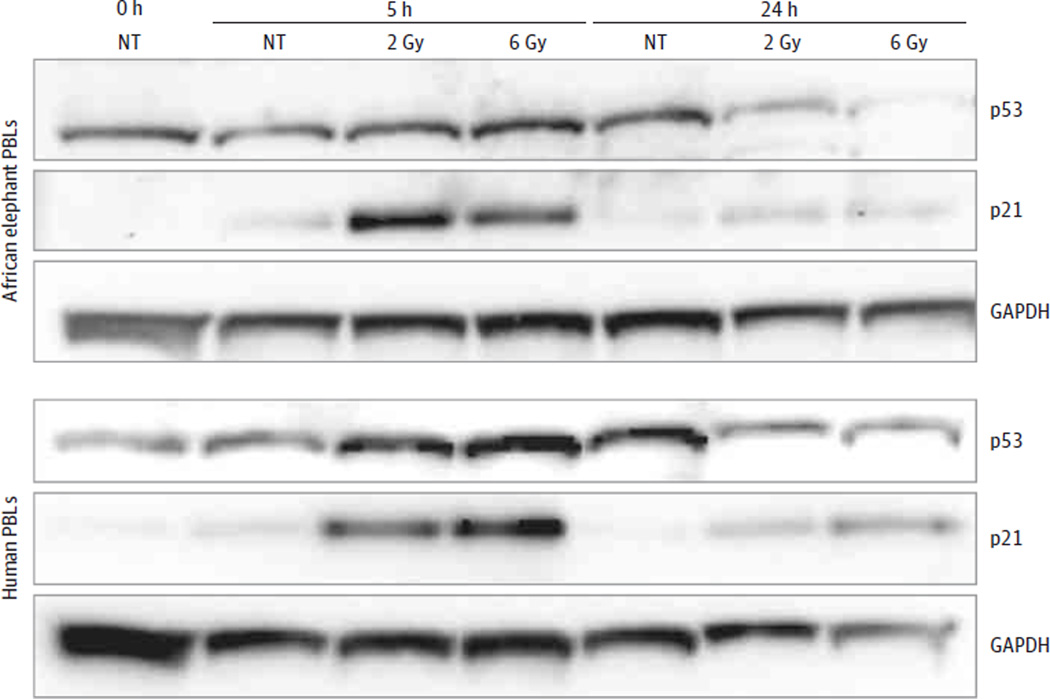
No significant difference was detected in pH2AX foci following ionizing radiation between human and elephant PBLs, indicating that the increased apoptosis in elephants cannot be attributed to more DNA damage (Table 2, Figure 5, and eFigure 7 in the Supplement). This increased apoptosis was observed in different lymphocyte wash conditions (eFigure 8 in the Supplement). Unlike increasing TP53 mRNA levels seen in human PBLs after ionizing radiation, gene expression of ancestral and retrogene TP53 did not increase in elephant PBLs (eFigure 9 in the Supplement). Both elephant and human PBLs showed p53 and p21 protein expression following ionizing radiation exposure (Figure 6). More p21 protein expression was observed at 5 hours in elephant PBLs treated with 0.5 Gy of ionizing radiation (20.1-fold increase; 95% CI, 8.72- to 31.5-fold) compared with human PBLs (3.5-fold increase; 95% CI, 1.7- to 5.31-fold; P = .004) (eFigure 10, A–B, in the Supplement). Elephant fibroblasts also showed increased p21 protein expression following 2 Gy of ionizing radiation at 5 hours (1.9-fold increase) compared with no increase in human fibroblasts (eFigure 10C in the Supplement). Similar to lymphocytes, elephant vs human fibroblasts showed evidence of increased apoptosis after 10 µM of doxorubicin as measured by increased caspase activity relative to dimethyl sulfoxide–treated fibroblasts (elephant: 9.1-fold increase [95% CI, 7.93- to 10.25-fold] vs human: 2.24-fold increase [95% CI, 1.5- to 2.98-fold]; P < .001) and additionally showed reduced viability consistent with cell cycle arrest after 0.5 Gy of ionizing radiation (elephant: 80.81% [95% CI, 68.86%–92.75%] vs human: 95.87% [95% CI, 90.73%–101.0%]; P = .01) (eFigure 11 in the Supplement; some of the elephant fibroblast experiments do not have P values because they were designed to demonstrate p21 protein expression and not powered for statistical comparison).
Table 2.
pH2AX Foci in Human and African Elephant Cells After 2 Gy of Ionizing Radiationa
| Treatment and Time Frame | Viable Cells With Indicated No. of pH2AX Foci, % | |||||||
|---|---|---|---|---|---|---|---|---|
| 0–5 Foci | 6–10 Foci | 11–15 Foci | 16–≥20 Foci | |||||
| Human | Elephant | Human | Elephant | Human | Elephant | Human | Elephant | |
| No treatment, 1 h | 97.3 | 98.7 | 2.7 | 1.3 | 0.0 | 0.0 | 0.0 | 0.0 |
| No treatment, 5 h | 97.7 | 98.0 | 2.3 | 1.3 | 0.0 | 0.7 | 0.0 | 0.0 |
| No treatment, 24 h | 99.7 | 99.7 | 0.3 | 0.3 | 0.0 | 0.0 | 0.0 | 0.0 |
| 2 Gy, 1 h | 23.0 | 26.3 | 25.3 | 33.7 | 19.0 | 17.0 | 32.7 | 23.0 |
| 2 Gy, 5 h | 46.7 | 51.0 | 32.7 | 39.0 | 14.3 | 9.0 | 6.3 | 1.0 |
| 2 Gy, 24 h | 94.3 | 92.3 | 5.3 | 7.3 | 0.3 | 0.0 | 0.0 | 0.0 |
Foci indicate remaining DNA double-stranded breaks. Cells are binned by the number of phospho-histone H2AX (pH2AX) foci and demonstrate no significant difference in the rate of DNA damage repair between human and elephant (P > .05 by χ2 test).
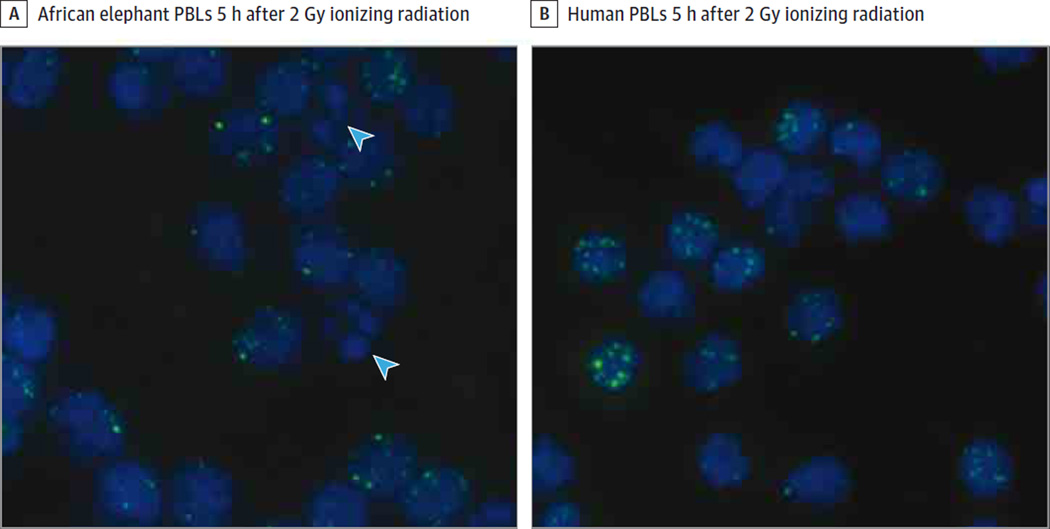
Figure 5. Visualization of Apoptosis and DNA Damage in Human and Elephant Cells After Ionizing Radiation.
DAPI, a nuclear stain that binds to DNA (blue), and phospho-histone H2AX foci (green) labeled peripheral blood lymphocytes (PBLs) 5 hours after 2 Gy of ionizing radiation show similar amounts of DNA damage. Apoptosis, rarely observed in the human cells, is visualized in the elephant cells (blue arrowheads indicate apoptotic cells with DNA fragmentation, identified by nuclear blebbing). Images displayed at 40× magnification.
Figure 6. p21 and p53 Protein Expression After Ionizing Radiation.
Western blot at the indicated time points after ionizing radiation shows p21 and p53 protein expression in elephant and human lymphocytes. The p53 antibody detects only nonphosphorylated protein. GAPDH indicates glyceraldehyde 3-phosphate dehydrogenase, a protein-loading control; PBL, peripheral blood lymphocyte; NT, no treatment.
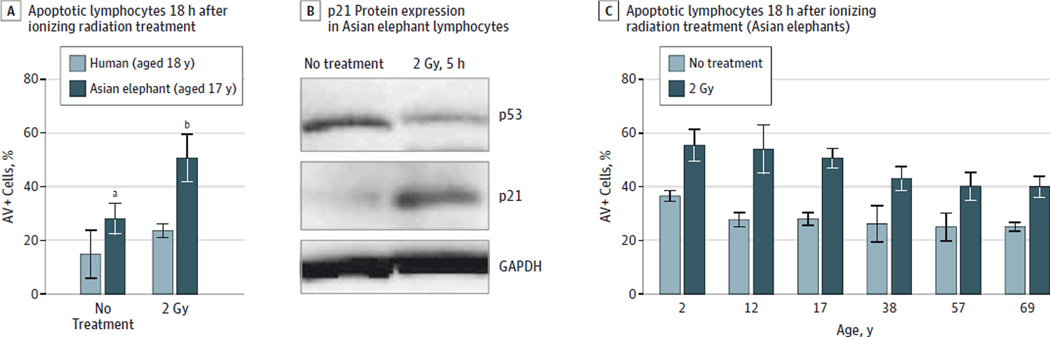
As a post hoc analysis, the same experiments were repeated in PBLs from multiple Asian elephants (n = 6) of different ages (2, 12, 17, 38, 57, and 69 years old). Asian elephant lymphocytes also demonstrated an increased rate of apoptosis (50.63%; 95% CI, 41.71%–59.53%) relative to human cells (23.67%; 95%CI, 21.18%–26.15%; P < .001) when exposed to 2 Gy of ionizing radiation (18-hour culture) and an increase in p21 expression (Figure 7, A–B). Additionally, the apoptotic response in PBLs decreased with the age of Asian elephants when tested with both a linear regression and a Jonckheere-Terpstra test, which allows for nonlinear relationships (Figure 7C) (2-year-oldelephantwith2Gy radiation at 18hours, 52.53% [95% CI, 35.86%–69.2%] and 69-year-old elephant, 40.03% [95% CI, 30.64%–49.43%]; P = .002 by linear regression; P < .001 by Jonckheere-Terpstra test). These age-related results should be interpreted as exploratory and hypothesis generating.
Figure 7. Asian Elephant Cells and DNA Damage Response.
A, An example is shown of percentage of annexin V–positive (AV+) (apoptotic) lymphocytes from a 17-year-old Asian elephant compared with AV+ lymphocytes from an 18-year-old human 18 hours after ionizing radiation exposure. Error bars represent 95%CIs and significant differences computed with a 2-sided t test are indicated. B, Evidence of p21 protein expression is seen 5 hours after 2 Gy of ionizing radiation in Asian elephant lymphocytes. C, The apoptotic response in Asian elephant lymphocytes is shown to decrease with age (P = .002 by linear regression and P < .001 by Jonckheere-Terpstra tests). A single elephant of each indicated age was tested in triplicate. GAPDH indicates glyceraldehyde 3-phosphate dehydrogenase, a protein-loading control.
aP = .006.
bP < .001.
Discussion
Peto first made the observation more than 35 years ago that larger and longer-lived mammals develop less cancer than expected,3,23,24 but the evolutionary and functional mechanisms for this phenomenon have been studied only in rodents.4–6 To our knowledge, this study offers the first supporting evidence based on empirical data that larger animals with longer life spans may develop less cancer, especially elephants. The cancer mortality rate for elephants was found to be less than 5% compared with a cancer mortality rate for humans of 11% to 25%.25 Additionally, TP53 amplification was identified in elephants, and the effect TP53 amplification may have on apoptotic response to DNA damage was explored. These findings support the concept of an evolutionary-based approach for cancer suppression.
TP53 plays a central role in cancer suppression and response to DNA damage through apoptosis and cell cycle arrest.8,26,27 Patients with LFS inherit only 1 functioning TP53 allele and may have a lifetime risk of cancer approaching 100%.10,11 Conversely, inserting additional copies of constitutively active TP53 in mice confers cancer resistance with accelerated aging,28 while redundant TP53 alleles under the endogenous promoter generate cancer-resistant laboratory mice that age normally.29 The evolution of the elephant would have involved a strong selective pressure to naturally suppress cancer in a long-lived animal 100 000 times the size of a mouse. Female elephants reproduce and raise offspring throughout their entire life span of 50 to 80 years, older males have higher status and more reproductive opportunities,30 and herds with older matriarchs may have higher fitness.31 The enormous mass, extended life span, and reproductive advantage of older elephants would have selected for an efficient and fail-safe method for cancer suppression. The multiple copies of TP53 and the enhanced p53-mediated apoptosis observed in elephants may have evolved to offer such cancer protection.
The data suggest a lower threshold for DNA damage before triggering p53-dependent apoptosis in elephants than in humans, a possible evolutionary strategy to avoid cancer by efficiently removing mutant cells. Consistent with previous evidence that increasing TP53 gene dosage increases transcriptional regulation of p53 target genes,17,27 apoptotic rates in lymphocytes increased proportionally among patients with LFS (1 TP53 functioning allele), human controls (2 TP53 alleles), and elephants (40 TP53 alleles). Elephant cells exposed to DNA damage showed increased p21 expression, a downstream target of p53 activation. Also, p53 retrogenes were up-regulated and translated when transfected into human cells treated with ionizing radiation and doxorubicin. These combined observations suggest that the increased cell death in elephants may be mediated by p53 and enhanced by the additional TP53 retrogenes.
Retrotransposed genes, often called pseudogenes, can play functional roles in biology.32,33 Based on the study results, the TP53 retrogenes may functionally increase elephant cell response to DNA damage by triggering p53-dependent apoptosis rather than increasing DNA repair. Apoptosis can prevent mutations from propagating to future cell generations through removal of mutated clones. The elephant cells appeared twice as sensitive to DNA damage–induced apoptosis as human cells. Increasing apoptosis effectively lowers the ongoing mutation rate for the entire cell population and, as calculated, this 2-fold decrease in the somatic mutation rate (doubling of apoptosis) in elephants could explain the 100× increase in cell mass without cancer transformation.15,34 The Asian elephant genome contained 15 to 20 TP53 retrogene copies, suggesting that TP53 retrogene amplification predated the split of the African and Asian elephant species approximately 6.6 million to 8.8 million years ago.35 The hyrax (Procavia capensis) is the closest elephant relative with an available genome assembly (proCap1) and contains only 1 copy (2 alleles) of TP53. The hyrax and elephant lineages diverged 54 million to 65 million years ago,36 making this time frame the upper bound of when these TP53 retrogenes evolved.
A consistent age-related decrease in apoptosis was found in Asian elephants. Age-related decline in apoptotic response has been observed in murine T cells,37 human PBLs,38 and human sperm.39 Young elephants rapidly grow in less than 10 years from a birth weight of 100 kg to more than 3000 kg at reproductive age, a 30-fold increase in cellular mass with more than 1 kg of weight gain per day. Such a high rate of cell division and expansion in the growing elephant requires an especially efficient system of cancer prevention.
The study of cancer and apoptosis across species has several limitations. Cancer mortality rates in humans are often reported as deaths per 100 000 per year,25 and sufficient sample sizes of animals are difficult to find for comparison. The cross-species mortality rates in this study included estimates based on small numbers of captive animals with wide confidence intervals. More data need to be collected to confidently demonstrate the absence of correlation of mass and life span with cancer mortality. Environmental factors also play a role in cancer development, and it is unclear how captivity influences cancer rates through diet, stress, physical activity, and reproduction. The expected life span of captive African and Asian elephants is decreased,40 and this analysis may not have fully captured the elderly elephant population most expected to develop cancer. Adding to the complexity, humans are treated with modern medicine and may have an artificially extended life span, which, along with carcinogenic exposures like smoking, increases the lifetime risk of cancer death. Neither the African nor Asian elephant genome has been formally assembled and, consequently, elephant-specific molecular agents such as phosphorylated p53 elephant antibodies to measure elephant p53 activation are challenging to obtain. Studying the p53 pathway requires certain assumptions, such as that p21 and Mdm2 protein levels truly reflect p53 activity, as they do in humans. Although the data are suggestive, it is still unknown if elephant TP53 retrogenes produce functional protein. These retrogenes may serve as either functional or nonfunctional protein decoys for degradation (eFigure 12 in the Supplement), explaining the co-immunoprecipitation of Mdm2 with TP53 elephant retrogenes. With further assembly of the elephant genome, future experiments with genomic technologies like RNA sequencing will prove helpful in understanding the functional differences reflected in the increased apoptosis found in elephants.
Conclusions
Compared with other mammalian species, elephants appeared to have a lower-than-expected rate of cancer, potentially related to multiple copies of TP53. Compared with human cells, elephant cells demonstrated increased apoptotic response following DNA damage. These findings, if replicated, could represent an evolutionary-based approach for understanding mechanisms related to cancer suppression.
Supplementary Material
Acknowledgments
Drs Abegglen and Schiffman report a patent pending on use of elephant TP53 to prevent or treat cancer. Dr Waddell reports receipt of grants from the National Institutes of Health.
Funding/Support: This work was supported in part by US Department of Energy grant DE-FG02-97ER25308 and National Institutes of Health grants P01 CA91955, R01 CA149566, R01 CA170595, R01 CA185138, and R01 CA140657, R01 LM008626, and Congressionally Directed Medical Research Programs’ Breast Cancer Research Program award BC132057. The Huntsman Cancer Institute (HCI) Nuclear Control Program provided pilot support through the Cancer Center Support Grant, and National Cancer Institute grant CA24014 supported HCI core facilities. This research was made possible through funding from Soccer for Hope, Ringling Bros and Barnum & Bailey Circus (Feld Entertainment), and the Primary Children’s Hospital (PCH) Pediatric Cancer Research Program, supported by the Intermountain Healthcare Foundation and the PCH Foundation.
Role of the Funder/Sponsor: The funders had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; or decision to submit the manuscript for publication.
Additional Contributions: We acknowledge the collaboration of the Hogle Zoo (Salt Lake City, Utah), the Oakland Zoo (Oakland, California), and the Ringing Bros Center for Elephant Conservation (Polk City, Florida) and recognize the assistance of the following in obtaining samples from elephants: Eric Peterson, AD (Hogle Zoo), Lauren LeCoque, BS (Hogle Zoo), and Jeff Kinzley, BS (Oakland Zoo). We thank the following at the University of Utah for technical and analytical assistance: Bella Johnson, BA, Marcus Vranes, BS, Kiera Jorgensen, Sharanya Raghunath, PhD, Clint Mason, PhD, and Jamie Gardiner, BS. We recognize the following for helpful experimental discussions: Douglas Brash, PhD (Yale University) and Bradley Cairns, PhD (University of Utah). We acknowledge Kenneth Jahan for help with compiling cancer incidence data from the San Diego Zoo, and we thank George Church, PhD (Harvard University), for providing the African elephant fibroblasts. We acknowledge the Genome Analysis Core at the Helen Diller Family Comprehensive Cancer Center, the Genomics Core Facility at the University of California, San Francisco, and the University of Utah’s Flow Cytometry, DNA Sequencing, and High-Throughput Genomics Core Facilities. None of these individuals were compensated for their contributions except for Core Facilities.
Glossary
- AV
annexin V
- LFS
Li-Fraumeni syndrome
- Mdm2
mouse doubleminute 2 homolog
- PBL
peripheral blood lymphocyte
- PI
propidium iodide
Footnotes
Author Contributions: Dr Schiffman had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis. Drs Abegglen and Caulin contributed equally to this work. Drs Maley and Schiffman jointly directed this work.
Study concept and design: Abegglen, Waddell, Bhaskara, Maley, Schiffman.
Acquisition, analysis, or interpretation of data: Abegglen, Caulin, Chan, Lee, Robinson, Campbell, Kiso, Schmitt, Waddell, Jensen, Schiffman.
Drafting of the manuscript: Abegglen, Caulin, Chan, Maley, Schiffman.
Critical revision of the manuscript for important intellectual content: Abegglen, Lee, Robinson, Campbell, Kiso, Schmitt, Waddell, Bhaskara, Jensen, Maley, Schiffman.
Statistical analysis: Abegglen, Caulin, Chan, Waddell, Jensen
Obtained funding: Schmitt, Waddell, Maley, Schiffman.
Administrative, technical, or material support: Campbell, Kiso, Schmitt, Waddell, Bhaskara, Maley, Schiffman.
Study supervision: Maley, Schiffman.
Conflict of Interest Disclosures: All authors have completed and submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. No other disclosures were reported.
Disclaimer: The findings, opinions, and recommendations expressed herein are those of the authors and not necessarily those of the universities where the research was performed or the National Institutes of Health.
REFERENCES
- 1.Tomasetti C, Vogelstein B. Cancer etiology: variation in cancer risk among tissues can be explained by the number of stem cell divisions. Science. 2015;347(6217):78–81. doi: 10.1126/science.1260825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Caulin AF, Maley CC. Peto’s Paradox: evolution’s prescription for cancer prevention. Trends Ecol Evol. 2011;26(4):175–182. doi: 10.1016/j.tree.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peto R, Roe FJC, Lee PN, Levy L, Clack J. Cancer and ageing in mice and men. Br J Cancer. 1975;32(4):411–426. doi: 10.1038/bjc.1975.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tian X, Azpurua J, Hine C, et al. High-molecular-mass hyaluronan mediates the cancer resistance of the naked mole rat. Nature. 2013;499(7458):346–349. doi: 10.1038/nature12234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gorbunova V, Hine C, Tian X, et al. Cancer resistance in the blind mole rat is mediated by concerted necrotic cell death mechanism. Proc Natl Acad Sci U S A. 2012;109(47):19392–19396. doi: 10.1073/pnas.1217211109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Seluanov A, Hine C, Bozzella M, et al. Distinct tumor suppressor mechanisms evolve in rodent species that differ in size and lifespan. Aging Cell. 2008;7(6):813–823. doi: 10.1111/j.1474-9726.2008.00431.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hollstein M, Sidransky D, Vogelstein B, Harris CC. p53 mutations in human cancers. Science. 1991;253(5015):49–53. doi: 10.1126/science.1905840. [DOI] [PubMed] [Google Scholar]
- 8.Lane DP. p53, Guardian of the genome. Nature. 1992;358(6381):15–16. doi: 10.1038/358015a0. [DOI] [PubMed] [Google Scholar]
- 9.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 10.McBride KA, Ballinger ML, Killick E, et al. Li-Fraumeni syndrome: cancer risk assessment and clinical management. Nat Rev Clin Oncol. 2014;11(5):260–271. doi: 10.1038/nrclinonc.2014.41. [DOI] [PubMed] [Google Scholar]
- 11.Testa JR, Malkin D, Schiffman JD. Connecting molecular pathways to hereditary cancer risk syndromes. Am Soc Clin Oncol Educ Book. 2013:81–90. doi: 10.1200/EdBook_AM.2013.33.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Griner LA. Pathology of Zoo Animals. San Diego, CA: Zoological Society of San Diego; 1983. [Google Scholar]
- 13.deMagalhães JP, Costa J. A database of vertebrate longevity records and their relation to other life-history traits. J Evol Biol. 2009;22(8):1770–1774. doi: 10.1111/j.1420-9101.2009.01783.x. [DOI] [PubMed] [Google Scholar]
- 14.Elephant Encyclopedia. [Accessed November 2012];1995–2012 http://www.elephant.se. [Google Scholar]
- 15.Calabrese P, Shibata D. A simple algebraic cancer equation: calculating how cancers may arise with normal mutation rates. BMC Cancer. 2010;10:3. doi: 10.1186/1471-2407-10-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Macleod KF, Sherry N, Hannon G, et al. p53-dependent and independent expression of p21 during cell growth, differentiation, and DNA damage. Genes Dev. 1995;9(8):935–944. doi: 10.1101/gad.9.8.935. [DOI] [PubMed] [Google Scholar]
- 17.Yoon H, Liyanarachchi S, Wright FA, et al. Gene expression profiling of isogenic cells with different TP53 gene dosage reveals numerous genes that are affected by TP53 dosage and identifies CSPG2 as a direct target of p53. Proc Natl Acad Sci U S A. 2002;99(24):15632–15637. doi: 10.1073/pnas.242597299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heinrichs S, Deppert W. Apoptosis or growth arrest: modulation of the cellular response to p53 by proliferative signals. Oncogene. 2003;22(4):555–571. doi: 10.1038/sj.onc.1206138. [DOI] [PubMed] [Google Scholar]
- 19.Lowe SW, Ruley HE, Jacks T, Housman DE. p53-dependent apoptosis modulates the cytotoxicity of anticancer agents. Cell. 1993;74(6):957–967. doi: 10.1016/0092-8674(93)90719-7. [DOI] [PubMed] [Google Scholar]
- 20.Antoccia A, Sgura A, Berardinelli F, et al. Cell cycle perturbations and genotoxic effects in human primary fibroblasts induced by low-energy protons and x/gamma-rays. J Radiat Res. 2009;50(5):457–468. doi: 10.1269/jrr.09008. [DOI] [PubMed] [Google Scholar]
- 21.Kastan MB, Zhan Q, el-Deiry WS, et al. A mammalian cell cycle checkpoint pathway utilizing p53 and GADD45 is defective in ataxia-telangiectasia. Cell. 1992;71(4):587–597. doi: 10.1016/0092-8674(92)90593-2. [DOI] [PubMed] [Google Scholar]
- 22.Attardi LD, de Vries A, Jacks T. Activation of the p53-dependent G1 checkpoint response in mouse embryo fibroblasts depends on the specific DNA damage inducer. Oncogene. 2004;23(4):973–980. doi: 10.1038/sj.onc.1207026. [DOI] [PubMed] [Google Scholar]
- 23.Peto R. Epidemiology, multistage models, and short-term mutagenicity tests. Paper presented at: The Origins of Human Cancer, Cold Spring Harbor Conferences on Cell Proliferation; Cold Spring Harbor, NY. 1977. [Google Scholar]
- 24.Nunney L. Lineage selection and the evolution of multistage carcinogenesis. Proc Biol Sci. 1999;266(1418):493–498. doi: 10.1098/rspb.1999.0664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136(5):E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 26.Bates S, Vousden KH. p53 in signaling checkpoint arrest or apoptosis. Curr Opin Genet Dev. 1996;6(1):12–18. doi: 10.1016/s0959-437x(96)90004-0. [DOI] [PubMed] [Google Scholar]
- 27.Lakin ND, Jackson SP. Regulation of p53 in response to DNA damage. Oncogene. 1999;18(53):7644–7655. doi: 10.1038/sj.onc.1203015. [DOI] [PubMed] [Google Scholar]
- 28.Tyner SD, Venkatachalam S, Choi J, et al. p53 mutant mice that display early ageing-associated phenotypes. Nature. 2002;415(6867):45–53. doi: 10.1038/415045a. [DOI] [PubMed] [Google Scholar]
- 29.García-Cao I, García-Cao M, Martín-Caballero J, et al. “Super p53” mice exhibit enhanced DNA damage response, are tumor resistant and age normally. EMBO J. 2002;21(22):6225–6235. doi: 10.1093/emboj/cdf595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Holliseter-Smith JA, Poole JH, Archie EA, et al. Age, musth and paternity success in wild male African elephants, Loxodonta africana. Anim Behav. 2007;74:287–296. [Google Scholar]
- 31.McComb K, Shannon G, Durant SM, et al. Leadership in elephants: the adaptive value of age. Proc Biol Sci. 2011;278(1722):3270–3276. doi: 10.1098/rspb.2011.0168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pei B, Sisu C, Frankish A, et al. The GENCODE pseudogene resource. Genome Biol. 2012;13(9):R51. doi: 10.1186/gb-2012-13-9-r51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pink RC, Wicks K, Caley DP, Punch EK, Jacobs L, Carter DR. Pseudogenes: pseudo-functional or key regulators in health and disease? RNA. 2011;17(5):792–798. doi: 10.1261/rna.2658311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Caulin AF, Graham TA, Wang LS, Maley CC. Solutions to Peto’s paradox revealed by mathematical modelling and cross-species cancer gene analysis. Philos Trans R Soc Lond B Biol Sci. 2015;370(1673):370. doi: 10.1098/rstb.2014.0222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rohland N, Malaspinas AS, Pollack JL, Slatkin M, Matheus P, Hofreiter M. Proboscidean mitogenomics: chronology and mode of elephant evolution using mastodon as outgroup. PLoS Biol. 2007;5(8):e207. doi: 10.1371/journal.pbio.0050207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Eizirik E, Murphy WJ, O’Brien SJ. Molecular dating and biogeography of the early placental mammal radiation. J Hered. 2001;92(2):212–219. doi: 10.1093/jhered/92.2.212. [DOI] [PubMed] [Google Scholar]
- 37.Spaulding CC, Walford RL, Effros RB. The accumulation of non-replicative, non-functional, senescent T cells with age is avoided in calorically restricted mice by an enhancement of T cell apoptosis. Mech Ageing Dev. 1997;93(1–3):25–33. doi: 10.1016/s0047-6374(96)01808-8. [DOI] [PubMed] [Google Scholar]
- 38.Camplejohn RS, Gilchrist R, Easton D, et al. Apoptosis, ageing and cancer susceptibility. Br J Cancer. 2003;88(4):487–490. doi: 10.1038/sj.bjc.6600767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Singh NP, Muller CH, Berger RE. Effects of age on DNA double-strand breaks and apoptosis in human sperm. Fertil Steril. 2003;80(6):1420–1430. doi: 10.1016/j.fertnstert.2003.04.002. [DOI] [PubMed] [Google Scholar]
- 40.Wiese R, Willis K. Calculation of longevity and life expectancy in captive elephants. Zoo Biol. 2004;23:365–373. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.