Abstract
The structural remodeling of collagens is important in several biological processes including wound healing, tendon repair and adaptation, fibrosis and morphogenesis. Multiphoton microscopy is efficient in the induction of highly specific second harmonic generation (SHG) signal from non-centrosymmetric macromolecules such as fibrillar collagens. Although the detectors in the reflection geometry have been normally employed for capturing the backward scattered SHG considering the wide range of engineered thick tissue applications, there are still questions about the generated 3D collagen structures because of the directional pattern of SHG signals. The present study dealt with an in vitro collagen-fibroblast raft or bioartificial tendon model where the stimulation of fibroblast cells induced lateral orientation of collagen Type I fibers. The SHG signals originating from 3D collagen matrix were captured simultaneously in both forward and backward scattering directions. Our structural analysis indicates that collagen fibers formed in such in vitro model systems are predominantly of uniform sizes and are aligned preferentially in the lateral direction. The criss-cross arrangements of laterally oriented fibers are evident in the initial stages of contraction but eventually those laterally oriented collagen fibers are found to be aligned in parallel to each other as well as to the fibroblasts after an extended period of contraction. Our comprehensive quantitative assessment of simultaneously captured forward and backward 3D SHG image datasets, which includes the SHG signal decay, fiber diameter, cell dimensions, colocalization profiles, the 3D voxel volumes and Fourier analysis, indicates strong correlation of structural features identified in forward and backward directions.
1. Introduction
Interactions of cells with the collagen matrix are the basis of many fundamental biological processes including the formation and organization of tissue structures (morphogenesis and wound repair), as well as the remodeling of the pre-existing tissue matrix as a result of both pathological and adaptive processes. Therefore, a detailed analysis of the collagen matrix in relevant in vitro models is of significance in gaining relevant structural information. When examining structural remodeling as well as the accumulation and organization of collagen macromolecules, a minimally invasive analytical method which provides specificity, sensitivity and spatial resolution is particularly useful. Such imaging methodologies can be used for several biomedical applications including quantifying physiological and biochemical processes, elucidating relevant biological mechanisms, as well as evaluating the effects of drugs on these processes. From the biotechnology point of view, such structural remodeling information can provide vital information in tissue engineering applications (Griffith and Swartz, 2006).
Multiphoton microscopy, which uses ultra-short femto-second laser pulses as an excitation source, produces multiphoton excitation fluorescence (MPEF) from exogenous fluorescent labels tagged to various cellular macromolecular objects and induces a highly specific second harmonic generation (SHG) signal from non-centrosymmetric macromolecules such as fibrillar collagens (Zipfel et al., 2003a and Zipfel et al., 2003b). While MPEF arises from an absorptive process which produces excitation fluorescence, SHG does not arise from an absorptive process but produces highly specific signals. Specifically, SHG is a frequency doubling optical process, in which photons interacting with anisotropic materials are effectively combined to form new photons with twice the energy. SHG is a second order nonlinear optical process that can only arise from materials lacking a center of symmetry (i.e. materials lacking generalized mirror symmetry) such as fibrillar collagens (Campagnola and Loew, 2003). When an intense light interacts with such materials, the vibrating electric field of the incident beam results in polarization of the material, and in general the optical response of the materials can be described by expressing the induced polarization (P) as a power series of the optical field or the strength of electric field vector (E ) of the incident light, P=χ1E1+χ2E2+χ3E3+…, here χ1 is the linear susceptibility related to the refractive index Re{χ1, and the absorption cross section, Im{χ1, and this first term as such describes linear absorption; the second term (χ2 describes SHG, and the third term (χ3encompasses MPEF and third harmonic generation. The MPEF involves real energy transition of electrons whereas the SHG involves only virtual energy transition. As a result, using the ultra-short (femto-second) pulsed lasers, one can generate SHG signal with a similar response time which is several orders of magnitude faster than the nanosecond response time of the MPEF signal. This allows very fast and highly sensitive SHG detection. Additionally, because the photons from SHG process have a shorter wavelength than MPEF photons, SHG signals can be separated from MPEF and can be captured simultaneously to provide spatially resolved, quantitative imaging of complex biological structures in 3D with spectral specificities. SHG and MPEF signals have been successfully used to probe the cell-mediated structural remodeling of extracellular collagen matrix as well as the related cellular morphologies in 3D space for an in vitro collagen-fibroblast raft model system which has wide applications in various in vitro wound healing and diseases model systems (Abraham et al., 2010, Pena et al., 2010, Wolf et al., 2009 and Zoumi et al., 2002). The detectors in the reflection geometry have been normally employed for capturing the backward scattered SHG as well as the MPEF signals. Despite these developments, there are still questions about such backward generated 3D collagen structures because of the directional pattern of SHG signals ( Han et al., 2005, Nadiarnykh et al., 2007 and Zipfel et al., 2003b). From the structural point of view, the collagen molecules assemble progressively into higher ordered structures, beginning with the nanometer level (collagen molecules-triple helix, 1.3 nm) that then become organized into fibrils/fibers (~50–500 nm), which assemble into fascicles (~50–300 μm) (Puxkandl et al., 2002). Previous theoretical studies as well as experimental work performed on tendons have demonstrated that fibrils with the axial size DA in the order of the SHG wavelength (λSHG), i.e. DA ~ λSHG, exhibit predominantly forward directed SHG, while fibrils with an axial size far less than the SHG wavelength, i.e. DA ≪ λSHG, produce nearly equal backward and forward SHG signals (Zipfel et al., 2003b). However, these findings could not account for similar experiments performed on opaque sclera and transparent cornea which are known to made up of thick and relatively thin fibrils respectively (Han et al., 2005). Indeed, the opaque sclera which is made of relatively thick collagen fibrils (i.e.DA ~ λSHG) produces equally strong SHG signals in both forward and backward directions while cornea which is made out of relatively thin fibrils (DA ≪ λSHG) produces very weak backward SHG signal. The same directional pattern questions also arise in the fibrillar collagen raft models which is increasingly used in various in vitro biological model systems. The primary objective of this study is to visualize and quantify the spatial remodeling of fibrillar collagen Type 1 in cellular collagen rafts or bio-artificial tendons (BATs) over extended period of contractions. These rafts are generally made from pepsin digested fibrillar collagens ( Triantafillopoulos et al., 2004), and to our knowledge 3D SHG image data or fine structural data of such raft systems have never been reported before. In order to have comprehensive structural information and to see if any structural differences because of above said SHG directional pattern questions, we have captured SHG signals originating from 3D collagen matrix simultaneously in both forward and backward scattering directions. We have quantified and compared both forward and backward 3D image data sets by examining SHG signal decay as a function of depth, fiber diameter, colocalization profiles, co-localization coefficients, 3D voxel volume and by performing Fourier analysis. Our detailed analysis of this opaque cellular collagen raft system indicates that the backward SHG detection, which is practical for engineered thick tissue sections, provides the same relevant structural and quantitative details as the forward scattered SHG.
2. Materials and methods
2.1. Cells and collagen matrix
Collagen rafts, also known as bioartificial tendons (BATs), were prepared according to the procedure presented elsewhere (Triantafillopoulos et al., 2004). Briefly, subconfluent fibroblast cells (C3H10T1/2 line) were detached in 1 ml of trypsin for 10 min, diluted with DMEM containing 10% FBS, passed through a 70 μm filter to exlude cell clumps, and counted with a hemocytometer. Approximately 5 × 106 cells were centrifuged for 7 min and re-suspended in 5 ml of Type I collagen solution consisting of 3.5 mg/ml collagen (Purecol, Advanced Biometrix), 1 ml of DMEM, and 0.5 ml FBS, brought to 7.4 pH with 0.1 N NaOH immediately prior to use. 150 μl of cell-collagen mixture was pipetted into each well of an untreated 6-well tissue train plate (FlexCell International) according to the manufacturer’s instructions. The gel was allowed to set for 2 h and become anchored to the fabric tethers for 2 h, and the resulting collagen rafts (BATs) were covered with 2 ml of DMEM containing either 1% or 10% FBS and 0.25 mM ascorbic acid (Sigma) and allowed to contract for 2 and 7 days in a humidified incubator at 37 °C with 5% CO2.
2.2. Cell fixation and staining
After 2 days or 7 days of incubation, cells were fixed using 3.7% formaldehyde for 30 min, permeabilized with 0.1% triton X-100 for 30 min, and blocked in 1% BSA. Cells were then incubated in Alexa Fluor 488 phalloidin (Molecular Probes, Eugene, OR, USA) for 30 min.
2.3. MPEF and SHG microscopy methods
The laser used for SHG as well as the MPEF was a mode-locked femto-second Ti:Sapphire Tsunami (Spectra-Physics, Mountain View, CA) synchronously pumped by a Millenia Xs J (Spectra-Physics) diode-pumped solid-state laser capable of delivering up to 10 W pumping power at 532 nm. The Tsunami laser, tunable between 700 and 1000 nm, capable of producing pulse repetition rate of 80-MHz, consistently produced pulses of 100-fs width between 790 and 950 nm. The average power at different wavelengths was recorded by a Spectra-Physics 407a power meter. Wavelength identification and selection was made possible with the use of an IST laser spectrum analyzer (IST, Horseheads, NY) coupled to a TDS 210 oscilloscope (Tektronix, Beaverton, OR). The laser output was attenuated using neutral density filters (New Focus, San Jose, CA) and the average power was consistently maintained below the damage threshold of the samples. The power attenuated laser was directed to a Leica AOBS RS scan head (4000 Hz) coupled with Leica upright microscope system (Heidelberg, Germany). The laser beam was focused on the specimen through a Lecia water immersion objective, 63 X/1.2 NA HCX PL APO. The lateral resolution (Rxy) and axial resolution (Rz) values of this objective were computed as 0.257 and 0.935 μm respectively. Upon entering the microscope, the laser beam was directed to the scanning mirrors, then through a 670 nm long pass dichroic mirror (DCXRU 690, Chroma Technology, USA) and subsequently focused on the specimens through the water dipping objective lens. The backscattered emission from the sample was collected through the same objective lens (Fig. 1). Leica Confocal Software TCS SP2 was used for the image acquisition. Non-descanned detectors both in the reflection as well as in the transmission geometries and spectral scanning mode in the reflection geometry were used for capturing the 3D images as well as for the spectral signal measurements respectively. In the reflection mode non-descanned PMT detectors (R6357, Hamamatsu, Shizuoka, Japan), a 700 nm short pass filter (E700SP, Chroma Technology, USA) was used to prevent the scattered IR laser radiation from reaching the detector and a 455 long pass dichroic beam splitter (455 DCXRU, Chroma Technology, USA) was used to separate SHG signal from the MPEF signal. SHG signal in forward direction was captured using a non-descanned detector (R6357, Hamamatsu, Shizuoka, Japan) in the transmission geometry equipped with similar IR block filter and a 445/60 nm band pass filter (MP 445/60, Chroma Technology) and 1.4 NA condenser.
Figure 1.
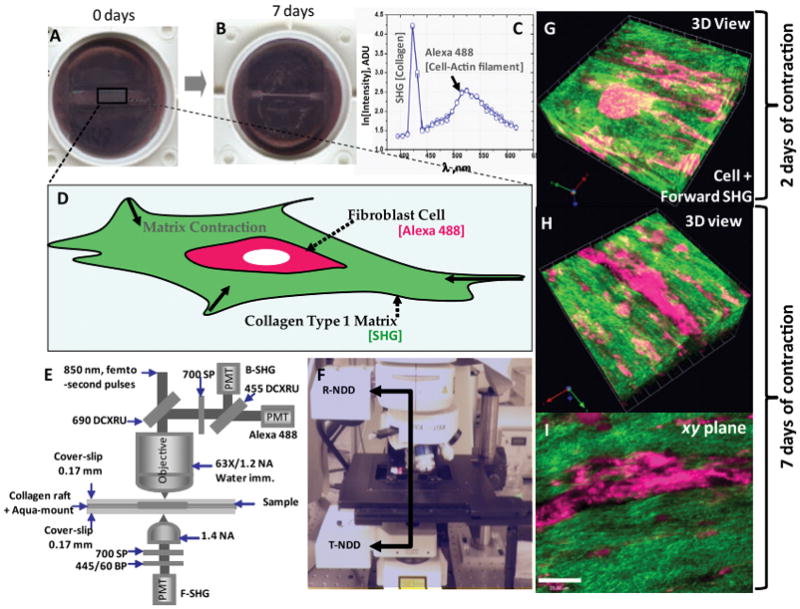
(A and B) Photographs of the fibroblast–collagen gel systems in culture wells at 0 days and after 7 days of contraction. (C) The SHG and MPEF spectral scan data measured from the fibroblast-collagen gel systems. The detected signals show SHG originating from the fibrillar collagen Type 1 matrix which is manifested by a narrow peak arising only at the expected wavelength 425 nm which is half of the excitation wavelength 850 nm and Alexa 488 signal originating from actin filaments of fibroblasts. (D) Schematics of the in vitro fibroblast-mediated collagen gel contraction model, where fibroblasts are seeded into a 3-dimensional collagen matrix. (E and F) Multiphoton microscopy setup: Femto-second IR laser pulses tuned to 850 nm is directed through a 690 nm DCXRU dichroic mirror before being focused on the specimen through a high resolution 63X/1.2 NA HCX PL APO water immersion objective. The backscattered emissions from the sample is collected through the objective lens and directed to the non-de-scanned PMT detectors in the reflection geometry (R-NDD) using the same 690 nm DCXRU dichroic mirror for generating images. A 700 nm short pass filter is used to prevent the scattered IR laser radiation from reaching the non-de-scanned PMT detectors and a 455 nm DCXRU dichroic mirror is used to separate SHG signal from the MPEF signal originating from the Alexa 488 signals. SHG signal in forward direction is captured using a non-descanned detector in the transmission geometry (T-NDD) equipped with similar IR block filter (700 SP) and a 445/60 nm band pass filter and 1.4 NA oil condenser. (G) Representative 3D rendered forward SHG image originating from the collagen Type 1 matrix (green color) overlaid with the MPEF image of stimulated fibroblasts (red color) after 2 days of contraction. The stimulated collagen raft system reveals that the fibroblasts induce lateral orientation of collagen fibers. The fibrillar collagen appears to have criss-cross oriented fiber morphology. (H and I) Similar 3D rendered image and an xy-plane image after 7 days of contraction. The lateral orientation of fibrillar collagen and the formation of long laterally oriented collagen fibers are particularly evident after 7 days of contraction (Scale bar: 25 μm).
All SHG and MPEF spectral measurements were performed using the de-scanned PMT detector (R6357, Hamamatsu) located inside the scan head where the emission signals were delivered through the Leica AOBS detection system with the maximum confocal pinhole setting at 600-μm via the spectral dispersion prism. The width of the slits in front of each PMT could be software adjusted such that each PMT could detect spectral regions spanning from a ~5 nm bandwidth up to the overall spectral capacity of the system (400–800 nm). With this instrument configuration, a series of individual images were collected using a narrow detection window with a width of ~10 nm, and each image detected at this specific emission wavelength band (i.e. ~10 nm) provided a data point in the spectral graph.
For 3D image data set acquisition, the multiphoton excitation beam was first focused at the maximum signal intensity focal position within the tissue sample and the appropriate PMT levels (both the gain and offset levels) were then selected to obtain the pixel intensities within range of 0–255 (8-bit images) using a color gradient function. Later on, the beginning and end of the 3D stack (i.e. the top and the bottom optical sections) were set based on the signal level degradation. A series of 2D Images for a selected 3D stack volume were then acquired at slow scan speed i.e. 10 s per 512 × 512 pixels. The 3D stack images with optical section thickness (z-axis) of approximately 0.25 μm were captured from collagen raft volumes. For each tissue volume reported here, z-section images were compiled and finally the 3-dimensional image restoration was performed using Volocity software (Improvisions, UK). We collected at least four 3D image datasets representing ~60 μm, ~240 optical sections with optical section thickness ~0.25 μm from four different collagen rafts after 2 and 7 days of contraction.
2.4. Quantification
2.4.1. Voxels and voxel intensity
Volocity software was used to quantify voxels and voxel intensities. We applied a noise removal filter with a kernel size of 3 × 3 to all 3D image data. For the analysis, the lower threshold level in the histogram was first set to positive first standard deviation (+1σ) of the mean voxel intensity value to exclude all possible background voxel values. The sum of all voxels and voxel intensities above this threshold value is determined and reported.
2.4.2. Fiber diameter
MATLAB software (Mathworks, Natick, MA) was used to determine the overall fiber diameter. In this analysis, the lower threshold level in the histogram was first set to the positive first standard deviation (+1σ) of the mean voxel intensity value to exclude all possible background voxel values. A grid containing five vertical and horizontal lines was placed over the image (Fig. 2A) and wherever the lines cross one of the fibers, the program recorded the pixels that the fiber covers. Fig. 2A1 shows plot of the intensity profile along the middle vertical line in the image (3rd line from left). In this plot, the x-axis is the pixel location starting at the top of the image (0) and ending at the bottom of the image (512) and the y-axis is the pixel intensities. Each peak represents a collagen fiber and the full width half maximum (FWHM) of these fibers were determined by fitting peaks to Gaussian distribution. The program then averages FWHM values for all five horizontal and vertical line profiles and the values are then multiplied by the pixel size value to obtain the fiber diameter in microns. Note that the fiber sizes are only measured for optical sections in between 10 and 40 μm collected in both forward and backward directions and their average values with standard deviations are reported.
Figure 2.
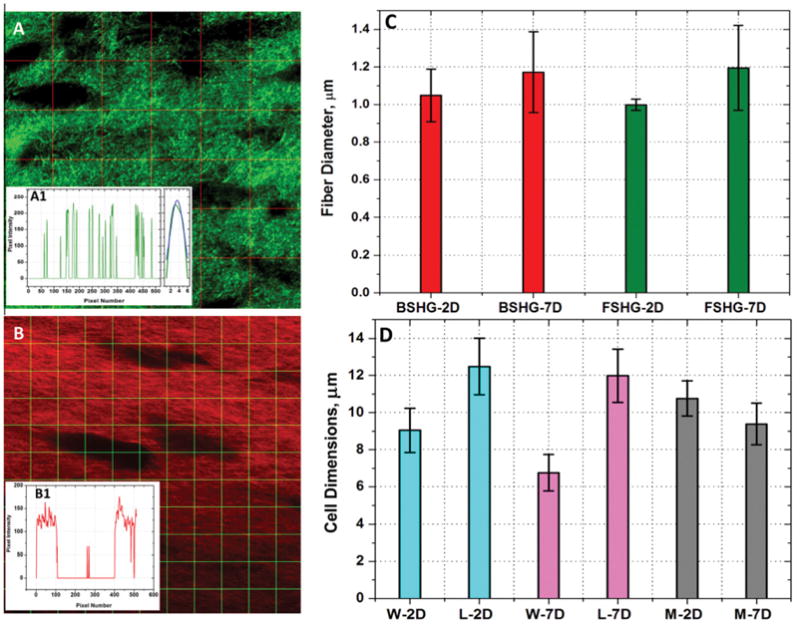
Representative image for fiber diameter measurement (A) and the insert (A1, Left) shows plot of the intensity profile along the middle vertical line in the image (3rd line from left). In the plot the x-axis is the pixel number starting at the top of the image (0) and ending at the bottom of the image (512) and the y-axis is the respective pixel intensity values. Each peak represents a collagen fiber and the full width half maximum (FWHM) of these fibers were determined by fitting peaks to Gaussian distribution (A1, Right). Representative image for cell size measurement (B) and the insert (B1) shows the plot of the intensity profile along the middle horizontal line (5th from top). In our cell size determination, any values greater than 15 pixels (or 3.7 μm), was considered as a cell. From the graph, we can see that along this profile there are two cells large enough to meet the criteria. (C) Collagen fiber diameter measured from both forward and backward SHG images after 2 and 7 days of contraction. BSHG, FSHG and D denote backward SHG, forward SHG and days of contraction respectively. Overall there is only marginal difference in fiber size determined from the forward and backward SHG images regardless of the extent of contraction. (D) Cell dimensions measured from backward SHG images after 2 and 7 days of contraction. W, L, M and D denote cell width, cell length, overall cell size (average of width plus length) and days of contraction respectively. Width wise, there is a significant reduction in cell size over the extent of contraction (p-value <0.05).
2.4.3. Cell dimensions
MATLAB software was used to determine the overall pore size. First, the image has been noise filtered with a 3 × 3 kernel, then thresholded to exclude low intensity values. A grid containing 10 vertical and horizontal lines was placed over the image (Fig. 2B) and wherever the lines cross one of the cells, the program recorded the number of pixels that cell covers. Fig. 2B1 shows the plot of the intensity profile along the middle horizontal line (5th from left). The x-axis is the pixel number location starting at the top of the image (0) and ending at the bottom of the image (512). The y-axis is the pixel intensity at that location. In the plot (Fig. 2B1), the cells are represented by the areas on the graph where there are several consecutive dark pixels. In our cell size determination, any values greater than 15 pixels (or 3.7 μm), was considered as a cell. From the plot (Fig. 2B1), we can see that along this profile there are two pores that large enough to meet the criteria. The program performs this calculation for each line in the image and records the averages as the horizontal (length) and vertical values (width) for that image. Note that the cell dimensions are only measured for optical sections in between 10 and 40 μm collected only in the backward direction (since there is no directional pattern associated with the cell dimensions) and their average values with standard deviations are reported.
2.4.4. Colocalization analysis
Volocity software was used to perform colocalization analysis. The most commonly used quantitative estimate of colocalization of voxel intensities is the Pearsons correlation coefficient (Rp) ( Manders et al., 1993 and Manders et al., 1992). This is applied to measure colocalization between forward and backward 3D SHG image dataset. The purpose of this colocalization coefficient is to characterize the degree of overlap between SHG signals collected in the forward and backward directions simultaneously. By definition Rp is a measure of the covariance between two signals which depends on the amount of colocalized signals in both channels, and is given by, Rp = Σi(Fi − FA) × (Bi − BA)/√[Σi(Fi − FA)2 × Σi(Bi − BA)2], where Fi and Bi are the voxel intensities of the forward and backward SHG emissions, respectively. FA and BA are the average intensities of forward and backward SHG emissions, respectively. By definition, Rp is a robust colocalization quantity and is unaffected by the detector (PMT) settings such as gain and offset levels of individual channels. A positive number for the Rp reflects the partial overlap of signals with 1.0 representing a complete overlap, while a value of zero indicates the random overlap or placement. For a particular SHG detection, the ratio of the colocalized voxel intensities (Fcolocalized or Bcolocalized) to the total voxel intensities, define the overlapping fraction for that particular SHG signal, MF= Σi Fcolocalized/ΣiFi,MB=ΣiBcolocalized/ΣiBi. By definition, MF and MB are unaffected by the PMT gain level, but are affected by the PMT offset level.
2.4.5. Fourier transform image analysis
MATLAB software was used to perform Fourier analysis. The 2D Fourier transform, which converts complex spatial patterns (represented by changes in pixel intensity values) into directionally dependent frequency components, has been used to determine the fiber orientation and anisotropy of the collagen fibers (Petroll, 2006). Fast Fourier transform (FFT) was performed on each image of dimensions 512 × 512 pixels, converting it from spatial (x, y) to frequency domain (u, v) using MATLAB according to where (x, y) represents the spatial coordinates of the image and (u, v) indicates the spatial frequency corresponding to (x, y) in the Fourier domain. The transform coefficient, F (u, v), which has both real and imaginary components, is expressed in its absolute form, P (u, v), which is the magnitude of the transform equation, based on P(u, v) = |F(u, v)|. Plotting P (u, v), which shows frequency distribution of the 2D original image, results in a power spectrum. The FT power spectrum thus generated was rotated 90° to align the frequency distribution data with the original 2D image. Using polar coordinates (r, θ), the magnitude values from the center to the periphery of the rotated FT power spectrum were calculated along the radial direction. Pixels close to the center were excluded from the mean pixel intensity calculations since they represent low-frequency information. An orientation index (OI) was then used to quantify the degree of orientation of the collagen fibrils along a given angle (θm ), using the equation, . Thus, an OI of 100% represents an image with all fibers aligned along a particular direction, while an OI of 0% is representative of random alignment.
3. Results and analysis
3.1. Collagen matrix remodeling in the presence of cell contraction
Samples were imaged using SHG signals as detailed in Section 2. Representative forward SHG image originating from the collagen matrix (A) overlaid with the MPEF image (B) of contracted fibroblast cells are shown in Fig. 1. A broad range of infrared laser wavelength of excitation wavelengths (800–900 nm) with a scan interval of 10 nm was employed to detect the SHG signals from the collagen Type I matrix. We found that SHG signal peaked at an excitation wavelength of 850 nm. The emission spectrum obtained from the wavelength scan (λ -scan) revealed strong SHG signal manifested by a narrow peak at half of the excitation wavelength. This SHG signal is spectrally clean, arising only at the expected wavelength. The same wavelength was used to generate MPEF signal from the Alexa 488 tagged to actin filaments. The forward SHG signal from the collagen Type I matrix as well as the MPEF signal originating from the filamentous actin (Alexa Fluor 488 phalloidin) were captured simultaneously using the non-de-scanned external detectors in the transmission and reflection geometries respectively. A 455 DCXRU dichroic beam splitter was used in the reflection geometry to separate backscattered SHG signal from the MPEF signal and a 445/60 nm band pass filter was used in the transmission geometry to capture only the forward scattered SHG. The images of this stimulated collagen raft system generally revealed that the fibroblast cells are highly elongated and these cells induce lateral orientation of collagen fibers. The fibrillar collagen appears to have crisscross oriented fiber morphology after two days of contraction (Fig. 1G). The lateral orientation of fibrillar collagen and the formation of long laterally oriented collagen fibers, however, are particularly evident after 7 days of contraction (Fig. 1H and I).
3.2. Collagen matrix remodeling in presence of cell contraction: forward vs. backward SHG
3.2.1. Voxels and voxel Intensity
The SHG signals originating from the 3D collagen matrix were captured simultaneously in both forward and backward scattering directions as described in Section 2. Representative forward and backward SHG 3D images originating from the collagen matrix after 2 and 7 days are shown in Fig. 3. Similar filters as described in the above section were employed for capturing these 3D SHG image datasets. The mean backward SHG intensity per layer, i.e. total layer intensity/total layer area, was found to be relatively stronger than the respective forward SHG intensity particularly in the top optical sections of the rafts (Fig. 3E and F). However, the ratio of total illuminated voxels, VF/VB, where VF and VB are the total illuminated voxels in the forward and backward directions respectively, was found to be close to unity (0.99 ± 0.06). A possible reason for this higher mean SHG signal in the backward direction would be that the SHG detected in the backward direction can arise from direct coherent or phase matched emission from collagen fibers, as well as the reflection of forward directed coherent or phase matched SHG signal which undergoes multiple scattering events and result in an incoherent backward component ( Han et al., 2005 and Nadiarnykh et al., 2007). This incoherent component can be very significant in our collagen raft case because of its opaque nature similar to sclera as reported elsewhere (Han et al., 2005). For the collagen raft thickness investigated, both the forward and backward scattered SHG signal intensities, layer by layer, were found to be decayed as well as we go deeper into the collagen raft (Fig. 3E and F). The decay is relatively strong in the case of backward directed SHG signals particularly after 7 days of contraction. Such a sharp increase in decay constant can be attributed to increase in collagen fiber density in the collagen rafts after 7 days of contraction and thereby increase in degree of scattering of backward directed SHG as we go deep into the collagen raft. As noted earlier, the backward detected SHG includes both the direct coherent backward directed SHG emission plus reflections of forward directed coherent SHG signals which undergo multiple scattering events resulting in an incoherent backward component. As we scan deeper into the raft, the scattering of backward directed incoherent and coherent SHG signals becomes significant eventually leading to weaker or no detectable signals in the backward direction. There still can be detectable SHG signal in the forward direction due to the coherent or phase matched condition of the forward detected SHG signals.
Figure 3.
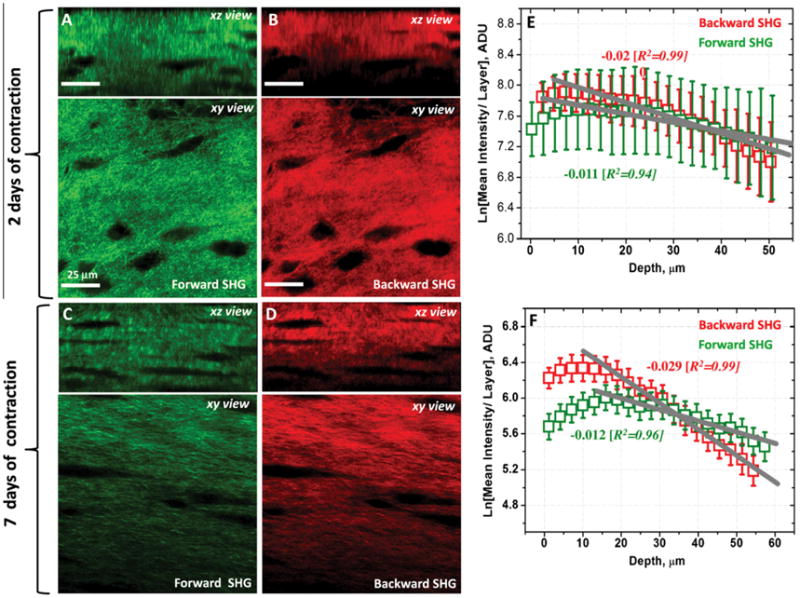
(A, B, C and D) Representative forward detected SHG and backward SHG images originating from the contracted collagen matrix in the stimulated cellular environment after 2 and 7 days of contraction. Both xy- and xz- are shown to provide sufficient spatial distribution details of the collagen matrix in the 3D space. [See also Supplementary file: 3D Slice Overlay-7 days of contraction.wmv]. (E and F) The natural logarithm of mean intensity per layer (i.e. total voxel intensity/total layer area) of forward and backward SHG images as a function of collagen raft depth after 2 and 7 days of contraction. Both forward and backward scattered SHG signal intensities decay as a function of the depth. The decay is relatively strong in the case of backward directed SHG signals particularly after 7 days of contraction.
3.2.2. Fiber diameter and cell dimensions
Fig. 2C shows overall collagen fiber diameter measured from both forward and backward SHG images. The FWHM values of forward and backward Gaussian SHG peaks (i.e. fiber diameters) were found to be 0.999 ± 0.030 and 1.049 ± 0.140, respectively after 2 days of contraction and 1.195 ± 0.226 and 1.172 ± 0.215 respectively after 7 days of contraction. Overall there is only marginal difference in fiber diameter determined from the forward and backward SHG images regardless of the extent of contraction. Width wise (dimension measured from the vertical lines in Fig. 2B), there is a significant reduction in cell size over the extent of contraction from 9.042 ± 1.191 (2 days) to 6.760 ± 0.981 (7 days) [p-value <0.05] although the lengthwise (dimension measured from the horizontal lines in Fig. 2B) the differences are marginal, 12.482 ± 1.526 (2 days) versus 11.986 ± 1.440 (7 days). The overall dimension of cells, which is an average of width plus length, is found to be 10.762 ± 0.948 and 9.384 ± 1.119 after 2 and 7 days of contractions respectively. In summary, the overall fiber diameter remains the same while cells become narrower over the extent of contraction.
3.2.3. Colocalization analysis
Representative overlay image of forward and backward SHG signals after 2 days of contraction is shown in Fig. 4. The scatter plot where forward SHG voxel intensities are plotted as a function of backward SHG voxel intensities showed linear relationship between forward and backward SHG signals (Fig. 4B). The computed overlapping fractions MF and MB, was found to be close to unity (0.99 ± 0.01, 0.99 ± 0.01, respectively) again suggesting voxel to voxel overlap of forward and backward scattered signals. The Pearson coefficient Rp, which is a robust measurements and unaffected by the detector (PMT) settings such as gain and offset levels of individual channels, was found to be about 0.81 ± 0.06. This again indicates strong colocalization and high degree of positive correlation of both the forward and backward SHG signals. The assessment of lateral (lines in Fig. 4C and D) and axial (lines in Fig. 4E and F) line profiles, where also the positively and negatively correlated voxel intensity values of (Fi-FA)×(Bi-BA)were plotted as a function of the line distance, indicate higher degree of correlation of both the forward and backward SHG signals (Fig. 4G and 4H). Note that in the correlation coefficient, i.e. (Fi-FA)×(Bi-BA), Fi and FA are the individual and mean intensities of the voxels for forward detected SHG signals, respectively, and Bi and BA are the individual and mean intensities of the voxels for backward detected SHG signals respectively. Voxels for which both Fi and Bi are above the mean intensity show positive correlation ( Fig. 4D and E and blue line in Fig. 4G and H). Voxels for which either channel is below the mean intensity while the other channel is above the mean value show negative correlation or non colocalization (gray line in Fig. 4G and H). In our current data set such negative correlation or non-colocalization does not exist between the forward and the backward SHG signals, and as a result those voxel intensity values were found to be close to zero (so does the gray lines in Fig. 4G and H). The colocalization analysis performed on 3D images after 7 days of contraction provided similar results with regard to the colocalization quantities and profiles (data not shown).
Figure 4.
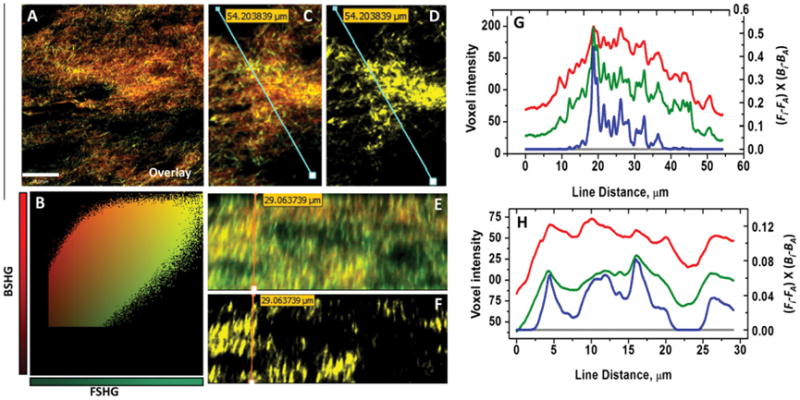
(A and B) Representative overlaid xy-plane image of both forward and backward SHG images and a scatter plot, in which forward SHG voxel intensities are plotted as a function of backward SHG voxel intensities, shows linear relationship between forward and backward SHG signals. (C) Overlaid xy-plane image of forward and backward SHG images which shows the position of line profiles correspond to forward SHG signal (green line in G) and backward SHG (red line in plot G). (D) The positively correlated xy-plane image which shows areas of positive correlation and the position of line profile corresponds to positively correlated voxel intensities (blue line in plot G). (E) Overlaid xz-plane image of forward and backward SHG images which shows the position of line profile correspond to forward SHG signals (green line in plot H) and backward SHG signals (red line in plot H). (F) The positively correlated xz-plane image which shows areas of positive correlation and the position of line profile corresponds to positively correlated voxel intensities (blue line in plot H). (G) Lateral (xy-plane) line profiles of forward and backward SHG signals where voxel intensities (y -axis, left) are plotted as a function of line distance. Also shown colocalization profile (blue line) and non-colocalization profile (gray line) where positively and negatively correlated voxel intensities of (Fi-FA)×(Bi-BA) (y-axis, right) are plotted as a function of the line distance. Note that the negatively correlated or non-colocalized voxels are non-existent in xy-plane. (H) Axial (xz-plane) line profiles of forward and backward SHG signals where voxel intensities (y -axis, left) are plotted as a function of line distance. Also shown colocalization profile (blue line) and non-colocalization profile (gray line) where positively and negatively correlated voxel intensities of (Fi-FA)×(Bi-BA) (y-axis, right) are plotted as a function of the line distance. Note that the negatively correlated or non-colocalized pixels are non-existent in xz-plane as well. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.2.4. Fourier analysis
The results of Fourier analysis of both forward and backward SHG are summarized in Fig. 5. The plot of power spectral density (Fig. 5E) shows that the high frequency components of both forward and backward SHG signals are condensed at multiple directions, indicating criss-cross arrangements of collagen fibers after 2 days of contraction. It is interesting to note the high frequency components of both forward and backward SHG signals overlap completely each other. The computed orientation index, which quantifies the preferred orientations of the collagen fibers, varies based on the depth of the collagen raft (Fig. 5G). As evidenced from the Fig. 5G, the computed orientation index values of both forward and backward SHG images again overlap strongly each other regardless of the depth of the collagen rafts. Since the fibers are preferentially oriented in criss-cross pattern, the mean orientation index values of collagen fibers for both forward and backward SHG signals are found to be low and their mean numerical values are found to be 15.53 ± 6.79 and 14.20 ± 2.91 respectively in the case of 2 days of contraction. In sharp contrast, as evidenced from the plot of the power spectral density shown in Fig. 4F, the collagen fibers are preferentially oriented in one direction rather than criss-cross after 7 days of contraction. As expected, the computed layer-by-layer orientation index values were found to be significantly higher (Fig. 5H) and their mean numerical values found to be 53.71 ± 9.01, 53.24 ± 7.12 for the forward and backward SHG images respectively. Thus, the high frequency component distributions as well as the fiber orientation properties of both forward and backward SHG images are found to be nearly identical regardless of the degree of contraction and the length of the contraction experiments. In summary, the Fourier transform analysis, which is a powerful tool to quantify the frequency distribution as well as the fiber orientation properties, indicates that the SHG signal captured in the backward direction from the collagen raft provides the same quantitative information as that of the forward captured SHG signal.
Figure 5.
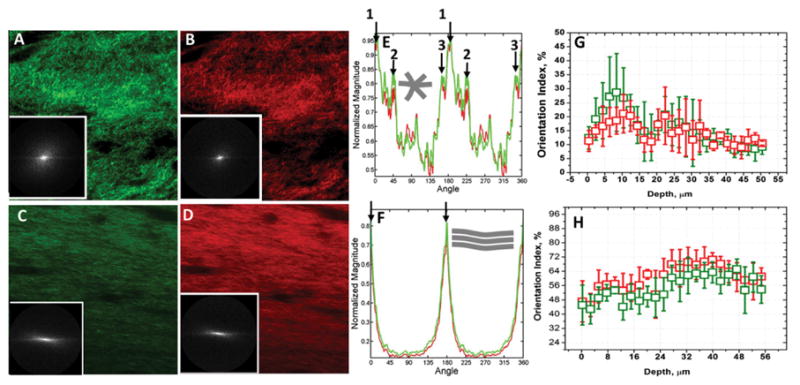
(A, B, C and D) Representative forward and backward SHG images originating from the contracted collagen matrix after 2 and 7 days of contraction are shown. Fig. inserts show their respective power density spectrum (PSD). (E) PSD plot of collagen matrix after two days of contraction shows the high frequency components of both forward and backward SHG signals are condensed at multiple angles and directions, indicating criss-cross arrangements collagen fibers. (F) PSD plot after 7 days of contraction shows that the collagen fibers are preferentially oriented in one direction rather than criss-cross arrangement. (G) Computed orientation index of collagen raft after 2 days of contraction as a function of depth. Since the fibers are preferentially oriented in criss-cross pattern in this case, the mean orientation index values of collagen fibers for both forward and backward SHG signals are found to be low. Inserted schematic shows collagen fibers oriented in multiple directions. (H) Computed orientation index of collagen raft after 7 days of contraction as a function of depth. Computed layer-by-layer orientation index values are significantly higher. Inserted schematic shows collagen fibers preferentially oriented in one direction.
4. Discussion and conclusions
In recent years, there has been a considerable interest in the area of tissue engineered in vitro model systems because such a model approach can be very useful in understanding the human organ physiology and pathophysiology in vitro in addition to their obvious potential uses as artificial organs ( Griffith and Swartz, 2006). It is highly likely that engineered tissue in vitro models will find widespread use in the future as a more efficient means of conducting pre-clinical drug testing ( DiMasi et al., 2003). These engineered 3D in vitro tissue models, particularly using human cells, will allow us to conveniently test pharmacological and physiological responses, thus bridging the gap between conventional 2D in vitro models and in vivo physiological studies. Using this system, for instance, we have recently identified that genes related to collagen synthesis are mechanically regulated in a cycle-, strain-, and frequency-dependent manner (Scott et al., 2011a and Scott et al., 2011b).
These 3D in vitro models often involve collagen Type I matrix since this genetic type of fibrillar collagen is the most abundant connective tissue element in human tissues (Choe et al., 2006). The imaging modalities so far used to understand the structural reorganization of in vitro models involving collagen matrices are mainly confocal/differential interference contrast methods (Petroll, 2006) and electron microscopy based techniques (Choe et al., 2003). The former involves staining of collagen macromolecules for visualization, and the latter involves heavy chemical processing including dehydration which could consequently affect the collagen organization, volume fraction as well as the cell structural features. Both of these methods provide insufficient 3-dimensional spatial resolution, depth discrimination as well as specificity due to the limitations imposed by these methods. To this end, the multiphoton microscopy which can be operated in multiple modalities including SHG and MPEF where SHG can provide detailed 3D structural information about fibrillar collagens without exogenous staining while MPEF can potentially identify other endogenously fluorescent extracellular components or exogenously stained cells (Abraham et al., 2010, Pena et al., 2010 and Zoumi et al., 2002). Because of the larger thickness of these 3D in vitro models in practical applications, the SHG signals from the remodeled fibrillar collagen matrix have been normally collected in the backward direction in such experiments. However, using well designed experiments, several investigators (Cox et al., 2003 and Zipfel et al., 2003b) have demonstrated that second harmonic signal has a unique propagation pattern in which the fine collagen structures scatter equally in all directions, while thick collagen structures scatter predominantly in the forward direction. Considering the collagen fiber size distribution in real in vitro model systems and applications, it is very important that we apply the forward and backward SHG detection schemes and investigate further the collagen structural differences (backward SHG vs. forward SHG) to generate a more comprehensive understanding of collagen matrix remodeling in such in vitro model systems.
The cellular collagen rafts described here are based on pepsin digested fibrillar collagen Type 1 where the fiber size scales with SHG wavelength. Consequently only predominantly forward scattered SHG signals are expected since these thick fibers are in the regime of DA ~ λ SHG (Zipfel et al., 2003b). However, the backward scattered SHG signals are found to be equally strong from this opaque cellular collagen raft system, similar to the observations in opaque sclera tissue (Han et al., 2005), thus providing all the quantitative information relevant to such collagen rafts. Our quantitative assessment including the Fourier analysis of simultaneously captured forward and backward 3D SHG image datasets from a laterally oriented collagen Type I contraction model indicates strong correlation between structural features identified in the forward and backward directions. For the collagen raft thickness investigated, layer-by-layer for approximately 60 μm opaque collagen rafts, we have shown here that both forward and backward scattered SHG signals decayed as a function of depth despite the fact that the mean SHG intensity of forward SHG images decayed more or less the same regardless of the extent of contraction. The measured overall fiber diameter, colocalization profiles, the colocalization coefficients, the 3D voxel volumes and the Fourier analysis collectively indicated a high degree of correlation for both the forward and backward SHG signals.
From the structural point of view, our quantitative analysis indicates that collagen fibers and cell dimensions formed in such collagen Type I raft in vitro model systems are predominantly of uniform sizes and those fibers are aligned preferentially in lateral directions. Although the overall hydrated collagen fiber diameter is found to nearly identical after 2 and 7 days of contraction, there is a considerable change in overall cell dimensions. The cells appeared to contract laterally while cells nearly maintain their longitudinal dimension. The criss-cross arrangements of those laterally oriented fibers are evident in the initial stages of contraction but eventually those laterally oriented collagen fibers are aligned in parallel to the fibroblast after more extended periods of contraction. The collagen raft model presented here thus produces lateral arrangements of collagen Type I fibers running parallel to each other as well as with reference to the fibroblasts after extended period of contraction (7 days). This evolved microstructural features is consistent with the structural features identified in tendon tissues (Fung et al., 2010, Scott et al., 2011a and Scott et al., 2011b). As an extension to this work, it would be interesting to construct such cellular collagen raft models from combinations of genetic types of fibrillar collagens such as collagen Type I and Type III. The collagen Type I, for instance, is known to form thick fibers while collagen Type III preferentially forms relatively thin fibers (Lapiere et al., 1977). Constructing such a fibrillar collagen raft model and collecting SHG signals simultaneously in the forward and the backward directions could provide valuable insights and key answers to the questions regarding the directional pattern of the SHG signals as well as the structural features of such composite tissue constructs.
Supplementary Material
Acknowledgments
Images were generated and processed at the Cellular Imaging and Biophysics Core Facility at the James Hogg Research Centre (JHRC), Institute for Heart + Lung Health at St. Paul’s Hospital, an imaging facility created from Canada Foundation for Innovation funds. TA acknowledges salary support from the JHRC and the St. Paul’s Hospital during the course of this work. Cells were cultured using facilities provided by the Centre for Hip Health and Mobility, a research centre established through the Canada Foundation for Innovation and the BC Knowledge Development fund. This work was partially supported by an NSERC Discovery Grant and Michael Smith Scholar award (AS). AS thanks Dr. Michael Underhill and Arthur Sampaio for their valuable ideas and advice, and Ashwariya Sharma and Gloria Fong for excellent technical assistance.
References
- Abraham T, Carthy J, McManus B. Collagen matrix remodeling in 3-dimensional cellular space resolved using second harmonic generation and multiphoton excitation fluorescence. J Struct Biol. 2010;169:36–44. doi: 10.1016/j.jsb.2009.07.023. [DOI] [PubMed] [Google Scholar]
- Campagnola PJ, Loew LM. Second-harmonic imaging microscopy for visualizing biomolecular arrays in cells, tissues and organisms. Nat Biotechnol. 2003;21:1356–1360. doi: 10.1038/nbt894. [DOI] [PubMed] [Google Scholar]
- Choe MM, Sporn PH, Swartz MA. An in vitro airway wall model of remodeling. Am J Physiol Lung Cell Mol Physiol. 2003;285:L427–L433. doi: 10.1152/ajplung.00005.2003. [DOI] [PubMed] [Google Scholar]
- Choe MM, Tomei AA, Swartz MA. Physiological 3D tissue model of the airway wall and mucosa. Nat Protoc. 2006;1:357–362. doi: 10.1038/nprot.2006.54. [DOI] [PubMed] [Google Scholar]
- Cox G, Kable E, Jones A, Fraser I, Manconi F, et al. 3-dimensional imaging of collagen using second harmonic generation. J Struct Biol. 2003;141:53–62. doi: 10.1016/s1047-8477(02)00576-2. [DOI] [PubMed] [Google Scholar]
- DiMasi JA, Hansen RW, Grabowski HG. The price of innovation: new estimates of drug development costs. J Health Econ. 2003;22:151–185. doi: 10.1016/S0167-6296(02)00126-1. [DOI] [PubMed] [Google Scholar]
- Fung DT, Sereysky JB, Basta-Pljakic J, Laudier DM, Huq R, et al. Second harmonic generation imaging and Fourier transform spectral analysis reveal damage in fatigue-loaded tendons. Ann Biomed Eng. 2010;38:1741–1751. doi: 10.1007/s10439-010-9976-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith LG, Swartz MA. Capturing complex 3D tissue physiology in vitro. Nat Rev Mol Cell Biol. 2006;7:211–224. doi: 10.1038/nrm1858. [DOI] [PubMed] [Google Scholar]
- Han M, Giese G, Bille J. Second harmonic generation imaging of collagen fibrils in cornea and sclera. Opt Express. 2005;13:5791–5797. doi: 10.1364/opex.13.005791. [DOI] [PubMed] [Google Scholar]
- Lapiere CM, Nusgens B, Pierard GE. Interaction between collagen type I and type III in conditioning bundles organization. Connect Tissue Res. 1977;5:21–29. doi: 10.3109/03008207709152608. [DOI] [PubMed] [Google Scholar]
- Manders EMM, Verbeek FJ, Aten JA. Measurement of colocalization of objects in dual-color confocal Images. J Microsc-Oxford. 1993;169:375–382. doi: 10.1111/j.1365-2818.1993.tb03313.x. [DOI] [PubMed] [Google Scholar]
- Manders EMM, Stap J, Brakenhoff GJ, Vandriel R, Aten JA. Dynamics of 3-dimensional replication patterns during the S-Phase, analyzed by double labeling of DNA and confocal microscopy. J Cell Sci. 1992;103:857–862. doi: 10.1242/jcs.103.3.857. [DOI] [PubMed] [Google Scholar]
- Nadiarnykh O, Lacomb RB, Campagnola PJ, Mohler WA. Coherent and incoherent SHG in fibrillar cellulose matrices. Opt Express. 2007;15:3348–3360. doi: 10.1364/oe.15.003348. [DOI] [PubMed] [Google Scholar]
- Pena AM, Fagot D, Olive C, Michelet JF, Galey JB, et al. Multiphoton microscopy of engineered dermal substitutes: assessment of 3-D collagen matrix remodeling induced by fibroblast contraction. J Biomed Opt. 2010;15:056018. doi: 10.1117/1.3503411. [DOI] [PubMed] [Google Scholar]
- Petroll WM. Differential interference contrast and confocal reflectance imaging of collagen organization in three-dimensional matrices. Scanning. 2006;28:305–310. doi: 10.1002/sca.4950280602. [DOI] [PubMed] [Google Scholar]
- Puxkandl R, Zizak I, Paris O, Keckes J, Tesch W, et al. Viscoelastic properties of collagen: synchrotron radiation investigations and structural model. Philos Trans R Soc Lond B Biol Sci. 2002;357:191–197. doi: 10.1098/rstb.2001.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott A, Danielson P, Abraham T, Fong G, Sampaio AV, et al. Mechanical force modulates scleraxis expression in bioartificial tendons. J Musculoskelet Neuronal Interact. 2011;11:124–132. [PubMed] [Google Scholar]
- Scott A, Sampaio A, Abraham T, Duronio C, Underhill TM. Scleraxis expression is coordinately regulated in a murine model of patellar tendon injury. J Orthop Res. 2011;29:289–296. doi: 10.1002/jor.21220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triantafillopoulos IK, Banes AJ, Bowman KF, Jr, Maloney M, Garrett WE, Jr, et al. Nandrolone decanoate and load increase remodeling and strength in human supraspinatus bioartificial tendons. Am J Sports Med. 2004;32:934–943. doi: 10.1177/0363546503261700. [DOI] [PubMed] [Google Scholar]
- Wolf K, Alexander S, Schacht V, Coussens LM, von Andrian UH, et al. Collagen-based cell migration models in vitro and in vivo. Semin Cell Dev Biol. 2009;20:931–941. doi: 10.1016/j.semcdb.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel WR, Williams RM, Webb WW. Nonlinear magic: multiphoton microscopy in the biosciences. Nat Biotechnol. 2003;21:1369–1377. doi: 10.1038/nbt899. [DOI] [PubMed] [Google Scholar]
- Zipfel WR, Williams RM, Christie R, Nikitin AY, Hyman BT, et al. Live tissue intrinsic emission microscopy using multiphoton-excited native fluorescence and second harmonic generation. Proc Natl Acad Sci USA. 2003;100:7075–7080. doi: 10.1073/pnas.0832308100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoumi A, Yeh A, Tromberg BJ. Imaging cells and extracellular matrix in vivo by using second-harmonic generation and two-photon excited fluorescence. Proc Natl Acad Sci USA. 2002;99:11014–11019. doi: 10.1073/pnas.172368799. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


