Abstract
The pluripotent state of embryonic stem (ES) cells provides a unique perspective on regulatory programs that govern self-renewal and differentiation, and somatic cell reprogramming. Here we review the highly connected protein and transcriptional networks that maintain pluripotency, and how they are intertwined with factors that affect chromatin structure and function. The complex interrelationships between pluripotency and chromatin factors are illustrated by X-chromosome inactivation, regulatory control by non-coding RNAs, and environmental influences on cell states. Recent findings suggest a model in which environmental cues and growth conditions may direct the fate of cells transitioning a “plastic state” induced during reprogramming.
Introduction
Embryonic stem (ES) cells have attracted special attention on account of their unique properties and extraordinary potential in regenerative medicine. ES cells are distinguished by unlimited self-renewal and the capacity to differentiate into any cell type, the hallmarks of pluripotency. The remarkable ease with which somatic cells are converted to an “ES-like” state (or induced pluripotent, iPS cells) by expression of 4 transcription factors (Oct4, Sox2, KLF4, c-Myc), or other combinations (Stadtfeld and Hochedlinger, 2010; Takahashi and Yamanaka, 2006), has focused interest on the regulatory mechanisms by which pluripotency is established and maintained. In this review we aim to integrate recent findings regarding the connections of a core ES cell transcriptional network, chromatin remodeling and modification, and somatic cell reprogramming.
In the first part of this review, we discuss how transcription factors, in concert with chromatin regulators, establish interconnected networks that maintain pluripotency. We further elucidate the mechanisms by which opposing chromatin regulators keep ESCs in a self-renewing pluripotent state that is poised for rapid initiation of differentiation into any cell type.
Unique chromatin structure of pluripotent cells
Chromatin -- chromosomal DNA as packaged with histones -- provides the cellular context for gene expression and cell fate determination. Changes in chromatin structure are brought about through chemical modification of histones (e.g. acetylation, methylation, demethylation, ubiquitination), and DNA methylation, as well as the action of DNA-binding proteins and chromatin remodeling enzyme complexes. The chromatin of ES cells is “open” (see Gaspar-Maia et al., 2011)(Figure 1). At the histological level stainable, transcriptionally silent constitutive heterochromatin is dispersed and less evident than in other cell types. The exchange of both histone and non-histone proteins, including heterochromatin protein 1 (HP1), linker histone H10, and core histones H2B, and H3, in chromatin is hyperdynamic. As differentiation of ES cells proceeds, heterochromatin appears heterogeneous and clustered in distinct blocks, and hyperdynamic proteins become immobilized on chromatin. The open nature of ES cell chromatin is also reflected in global transcriptional hyperactivity. Interestingly, expression of several chromatin remodeling factors is enhanced, heralding a role for such factors in maintaining chromatin plasticity in ES cells. Recent findings demonstrating that cells of the day 3.5 mouse blastocyst exhibit a similar open chromatin conformation is reassuring in relating the chromatin state of ES cells to an in vivo context.
Figure 1. Properties of open and closed chromatin.
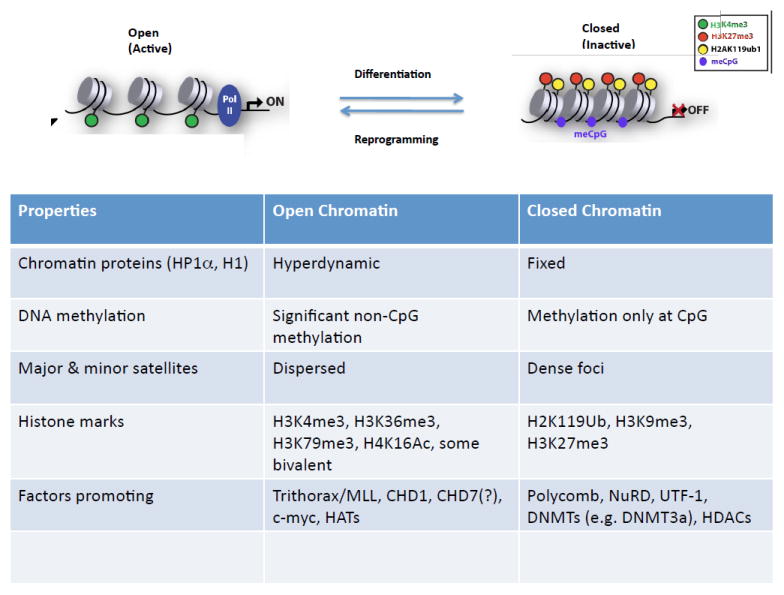
For details, (see Gaspar-Maia et al., 2011).
Networks for pluripotency
A core protein interaction network
Study of gene expression of preimplantation mouse embryos and transcriptional profiling of ES cells led to candidate transcription factors involved in early developmental fate decisions and pluripotency (see Young, 2011). Oct4, first identified as a PU-domain factor specific for preimplantation embryos, was later shown to be essential for pluripotent cell formation and cooperate with Sox2 on composite DNA bindings sites. The third member of the basic pluripotency core, Nanog, was discovered as an ES-cell associated transcript (ecat) and as a factor promoting leukemia inhibitory factor (Lif)-independent growth of ESCs. While Oct4, Sox2, and Nanog constitute bona fide core factors, additional ecat, such as Dax-1, Rex1, Sall4, and Tcl1, function within a larger regulatory network supporting pluripotency.
To determine potential relationships between the core factors and discover additional critical factors, several groups performed proteomic studies based on affinity purification of Oct4, Nanog, and Sox2, coupled with iterative purification of associated proteins and microsequencing (Liang et al., 2008; Pardo et al., 2010; Wang et al., 2006). As opposed to highly stable protein complexes, those containing Oct4, Sox2, and Nanog appear less stable, heterodisperse, and non-stoichiometric. As precise conditions influence the repertoire of associated proteins determined in proteomic studies, a consensus view of the interactome surrounding Oct4, Nanog, and Sox2 is presented (Figure 2).
Figure 2. Protein interaction network supporting pluripotency and connections to chromatin complexes.
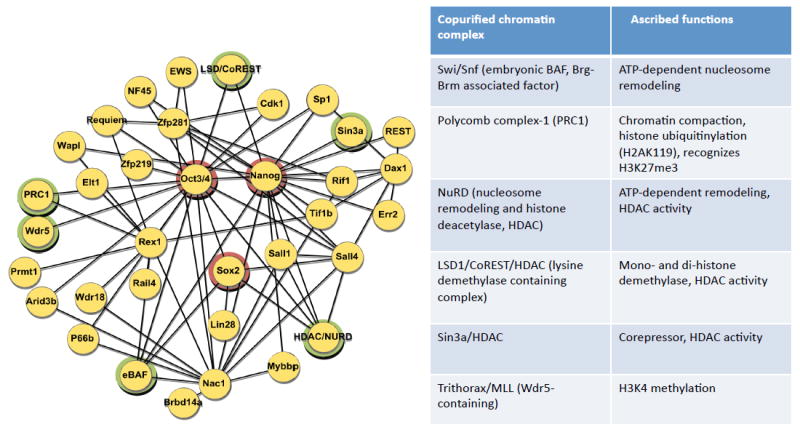
Protein-protein interactions derived from microsequencing of protein complexes purified from ESCs are shown on the left. The triad of pluripotency factors, Oct4, Nanog, and Sox2, are circled in red. Components of chromatin remodeling or modifying complexes are highlighted in green circles. On the right, several of the protein complexes associated with the pluripotency protein network are listed with their associated functional activities.
Remarkably, virtually all factors critical for the maintenance of pluripotency can now be placed within the protein interactome of Oct4, Nanog, and Sox2 (Wang et al., 2006). The majority of proteins within the interactome are essential for early development based on knockout or knockdown studies. Although these pluripotency components are connected to one another based on proteomics, it remains uncertain which interactions are direct, or alternatively indirect through cell-specific or more widely expressed proteins. The dynamic aspects of the protein complexes may provide a means by which variations in protein concentration influence the stability and stoichiometry of specific complexes and contribute to maintenance of the pluripotent state.
Target gene network supporting pluripotency
Comprehensive studies of chromatin occupancy by Oct4, Sox2, and Nanog, complemented by analyses of numerous other transcription factors and chromatin marks, have revealed an extraordinary degree of regulatory connections among proteins of the network (Boyer et al., 2005; Chen et al., 2008; Kim et al., 2008). Initial studies of Oct4 and Nanog demonstrated potential autoregulation, feed-forward, and cross-regulation (based on promoter occupancy). Subsequent work demonstrated additional complexity and remarkable combinatorial binding. The genes encoding numerous proteins within the expanded pluripotency network are targets of Oct4 and Nanog, and the respective proteins bind to regulatory elements of Oct4 and Nanog. The pattern of target gene occupancy conforms to a network with multiple, prominent “hubs” (Kim et al., 2008.). At the level of transcription factor-target interaction, pluripotency factors occupy regulatory regions of chromatin-related components. Some of the target gene relationships of the pluripotency network are summarized in Figure 3.
Figure 3. Transcriptional regulatory interactions in ESCs.
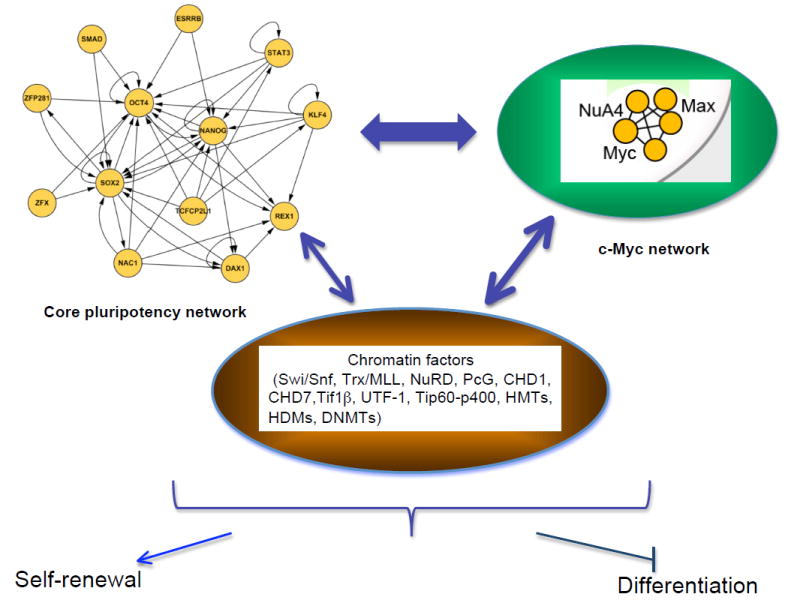
The core network depicted on the upper right illustrates target gene relationships. The c-myc network to the upper left represents the sub-network defined by common target genes (see Kim et al, 2010). The factors in the core and c-myc regulatory networks cross-regulate each other, and regulate, and are regulated by, chromatin factor components illustrated in the center. The output of these complex regulatory interactions is maintenance of self-renewal and blocking of lineage-specific differentiation.
The target genes bound by the major pluripotency factors segregate into two classes: those expressed and not expressed in ESCs. Among the targets of individual factors, such as Oct4 and Nanog, both expressed and non-expressed genes are nearly equally represented. The expressed genes generally include those anticipated to be required for maintenance of self-renewal or pluripotency. Numerous genes involved in lineage-specific differentiation are found among non-expressed targets (Boyer et al., 2005). This overall view is consistent with a dual role for the pluripotency factors: positive action promoting self-renewal and the ES cell state and negative regulation of genes promoting differentiation (Figure 3). Comprehensive chromatin occupancy studies reveal a consistent theme: target loci whose promoters and/or enhancers are bound by multiple proteins within the pluripotency network tend to be expressed, whereas those bound by one or a few of the proteins are non-expressed (Chen et al., 2008; Kim et al., 2008). The behavior of the “common” transcription factor gene targets may reflect enhanced chromatin accessibility afforded by associated chromatin factors, or the need for the cumulative action of multiple weak transcriptional activators. The set of common targets includes numerous other transcription factors of unknown function, as well as several chromatin-related proteins. The common target genes are typically expressed in ESCs and then turned off upon ES cell differentiation. These genes tend to be highly enriched for the active chromatin mark H3K4me3 in ESCs, and lose this mark and acquire a repressive mark, such as H3K27me3, upon differentiation.
Among the target gene or enhancer regions bound by multiple pluripotency factors, the predicted consensus binding motif conforms to an Oct4 or composite Oct4-Sox2 consensus site. This striking finding underscores the centrality of Oct4 and suggests that Oct4 binding recruits other factors to critical regions and promotes assembly of multiprotein factor complexes. Oct4-dependence of chromatin structure surrounding the Nanog locus in ESCs is consistent with this view (Levasseur et al., 2008).
c-Myc, pluripotency, and transcriptional plasticity
c-Myc is a pervasive transcription factor associated with active transcription and open chromatin. c-Myc serves as a major regulator of cell proliferation and stimulates iPS cell reprogramming (Nakagawa et al., 2010; Wernig et al., 2008). Lif/STAT3 control of the ES cell state is Myc-dependent (Cartwright et al., 2005). Given overlapping expression and function of c-Myc and N-Myc, it has proven challenging to determine contributions of c-Myc to maintenance of pluripotency. Recent findings, however, point to important roles in forestalling lineage-specific differentiation, in part through direct repression of GATA6 expression (Smith et al., 2010; Varlakhanova et al., 2010). c-Myc also contributes importantly to the control of proliferation through regulation of cell cycle genes, as well as miRNAs (Lin et al., 2009a).
c-Myc protein recruits multiple activities implicated in chromatin modification or structure, including histone acetyltransferases (GCN5, p300), chromatin remodeling complexes, histone deacetylasess (HDACs), and histone demethylases (Lin et al., 2009b). Consistent with these interactions, induction of c-Myc expression increases histone acetylation and histone methylation, including H3K4me3 deposition. c-Myc null embryos are highly deficient in global H3K4me3 (Lin et al., 2009b). In ESCs, ~3000 promoters are bound by c-Myc (Kim et al., 2008; Kim et al., 2010a; Lin et al., 2009a). c-Myc would be anticipated to bind some pluripotency and chromatin modifier gene targets just based on the overall frequency of sites. For example, Sox2 is bound by c-Myc, constituting one potential link to the pluripotency protein network. Generally, though, genes bound by c-Myc tend to segregate apart from common targets of the core network, exhibit a higher frequency of the active H3K4me3 mark, and tend to be expressed rather than inactive or repressed Nonetheless, perhaps 10% of c-Myc gene targets are repressed by c-Myc. By applying the “common” gene target approach to c-Myc and its interacting proteins (Kim et al., 2010a), a distinct “Myc network“ that is readily separable from the core network has been defined. During iPS cell generation, the c-Myc module targets are activated prior to engagement of the endogenous core module (Stadfeld and Hochedlinger, 2010). These findings implicate targets of the Myc network in the early phase of reprogramming, possibly through facilitating chromatin accessibility. The c-Myc module is highly represented in ES cell-associated transcriptional signatures that have been widely used in assessing the relatedness of cancer and embryonic cells (Kim et al., 2010a).
In addition to the above activities, c-Myc plays a critical role in transcription pause release at a substantial fraction of transcribed genes (Rahl et al., 2010), and therefore may broadly affect pathways essential to factor-induced reprogramming through that route. L-Myc, a c-Myc relative, is active in cellular reprogramming but not oncogenic for iPS cells. Therefore, the activities of c-Myc for reprogramming may be distinct from those promoting cellular transformation (Nakagawa et al., 2010).
With regard to the potential roles of c-Myc in influencing chromatin structure, the identification of the Tip60-p400 complex (also known as NuA4 HAT) as a c-Myc associated complex (Kim et al., 2010a) is of particular note. Tip60-p400 was also identified in a focused screen of chromatin factors as essential for maintaining the ES cell state (Fazzio et al., 2008). The multisubunit Tip60-p400 complex has two chromatin regulatory activities. Tip60 serves as a protein acetyltransferase. p400, a member of the Swi2/Snf2 family, functions in exchange of histones H2AZ-H2B within nucleosomes. RNAi inhibition of components of the Tip60-p400 complex leads to differentiation of ESCs (Fazzio et al., 2008). H2AZ, which is often found at sites bound by Polycomb repressive complex-2 (PRC-2, see below), is required for proper ES cell differentiation (Creyghton et al., 2008). Induction of c-Myc expression promotes incorporation of H2AZ in target promoters. Based on gene expression profiling, it has been suggested that Tip60-p400 and Nanog lie within a common pathway as Nanog depletion leads to reduced p400 binding at targets (Fazzio et al., 2008). Alternatively, common gene target analysis places Tip60-p400 targets closer to a c-Myc regulated network, consistent with proteomic findings (Kim et al., 2010a).
Although overexpression of c-Myc has been described as dispensable for somatic cell reprogramming (Nakagawa et al., 2010; Wernig et al., 2008), it is likely that endogenous c-Myc (or its relatives) participates, given its direct connections to chromatin activities and to the pluripotency network.
Links between core pluripotency and chromatin factor complexes
Multiple mechanisms and levels of control ensure globally “open” chromatin in ESCs, while also permitting repression of differentiation-related genes that are activated upon exit from the pluripotent state. This balance reflects interplay between the critical regulatory factors essential for pluripotency and chromatin remodeling and modification complexes. It is notable that critical factors within the pluripotency interactome are linked directly through protein interactions to a variety of chromatin modifying or remodeling complexes, including the ATP-dependent SWItch/Sucrose NonFermentable (Swi/Snf) remodeling, the nucleosome remodeling and histone deacetylation (NuRD) chromatin remodeling, Polycomb, Trithorax/MLL/Wdr5 (Ang) and histone deacetylase (HDAC)/Sin3a complexes (Figure 2).
Oct4-,Nanog-, and Sox2- associated proteins include components of the Swi/Snf (or BAF, Brg/Brahma associated factors) complex, a molecular machine that moves nucleosomes along DNA. Swi/Snf complexes are found in all cells, but the precise composition varies based on inclusion of alternative subunits (Lessard and Crabtree, 2010). The combinatorial assembly of Swi/Snf complexes underlies developmental-stage specific epigenetic control. Within ESCs the complex is characterized by the presence of the core subunit Brg1, BAF155, and BAF60A. Deletion of Brg1, BAF155(Smarcc1), BAF47(Smarcb1), and BAF250 leads to pre-implantation lethality and disruption of ES cell pluripotency. Overexpression of Swi/Snf components has been reported to enhance reprogramming by Oct4, Sox2, and KLF4 (Singhal et al., 2010).
The NuRD complex associates with Oct4 and Nanog, as well as other critical pluripotency factors, such as Sall4. Moreover, HDACs associate with core factors as part of HDAC/Sin3a or HDAC/CoREST/LSD1 complexes. Loss of MBD3, a core component of NuRD, undermines pluripotency of ESCs in part through a failure to block trophectoderm differentiation (Kaji et al., 2007; Zhu et al., 2009).
Transcription intermediary factor-1b (TIF1b, or TRIM28 and KAP1), a scaffold protein that recruits chromatin complexes and functions in transcriptional repression, interacts with several proteins within the pluripotency network, including Oct4 and Nanog (Seki et al., 2010; Wang et al., 2006). Previously, TIF1b was linked to silencing and formation of heterochromatin through interaction with HP1, the histone methyltransferase SETDB1, and NuRD. Nonetheless, TIF1b was identified through a genome-wide siRNA screen for factors required to sustain Oct4-driven GFP expression (Hu et al., 2009). Recently, Seki and colleagues (Seki et al., 2010) reported that a phosphorylated form of TIF1b interacts with the ESC-specific form of the Swi/Snf complex, localizes to euchromatin, influences some pluripotency and chromatin remodeling genes, and modulates iPS cell generation (Seki et al., 2010). In part, this may involve recruitment of Oct4 to phosphorylated TIF1bat target genes, such as Nanog.
It is poorly understood how the various interactions between core pluripotency factors and chromatin factors are mediated, and to what extent the associations are direct or indirect. In principle, the interactions may provide a means for recruiting chromatin factors to target genes bound by the transcription factors. Alternatively, prebound chromatin complexes may establish a suitable chromatin “milieu” and facilitate the assembly of transcription factors at their sites of action. Of note, Sall4, which lies within the pluripotency factor interactome (Wang et al., 2006; Wu et al., 2006) and is critical for pluripotency of ESCs as well as extraembryonic endoderm stem cells (Lim et al., 2008) contains a highly conserved N-terminal NuRD-binding sequence (Kidder et al., 2009; Lauberth and Rauchman, 2006). Structural studies reveal that this motif docks with RbAp48, a histone-binding NuRD core subunit shared with HDAC/sin3a and PRC2 (Lejon et al., 2011). It is provocative that loss of Lin-53, a RbAp48 homologue in C. elegans, removes the barrier to direct reprogramming of germ cells into neurons by the transcription factor Che-1. The effect of Lin-53 loss is mimicked by HDAC inhibition (Tursun et al., 2011).
Evidence also suggests that binding of chromatin factors to gene regulatory elements of the pluripotency factors provides a means for crosstalk. For example, Swi/Snf complexes occupy the Oct4, Sox2, Nanog, Sall4, and c-Myc genes among many others (Lessard and Crabtree, 2010). Indeed, ~60-70% of the target genes of Oct4, Nanog, or Sox2 are bound by Brg1.The consequences of Brg1 binding to target genes in ESCs appear to be complex. It has been proposed that ESC-specific genes, such as Nanog and Oct4, are “tonically” repressed by Brg1 in order to maintain expression within optimal limits. In addition, it has been suggested that Swi/Snf is important for repression of pluripotency genes on differentiation, as well as for facilitating chromatin compaction (Schaniel et al., 2009). As SwiSnf complexes can promote or repress gene expression, further work is needed to clarify how their diverse, presumably cell-context-dependent, actions contribute to pluripotency and the exit to differentiation.
The pluripotency regulatory network is also directly linked to the control of histone modifying proteins/complexes, as illustrated by the Jmj-family H3K9 demethylases Jmjd1a and Jmjd2c, which reverse H3K9me2 and H3K9me3, respectively (Loh et al., 2007). Both Jmjd1a and Jmjd2c lie downstream of Oct4 and are regulated positively through its action. Nonetheless, depletion of either factor in ESCs leads to differentiation, though with differing phenotypes. Loh et al (Loh et al., 2007) propose that Jmjd1a and Jmjd2c act on the Tcl1 and Nanog genes, respectively, as downstream targets. Thus, Oct4 directly controls epigenetic regulators that further act on target genes that encode proteins whose functions are critical to the pluripotency network. Given the multiplicity of Jmj-family proteins, we may anticipate additional examples of this regulatory circuitry.
Regulation of ES cell chromatin structure by opposing systems
The interplay between self-renewal and differentiation in ESCs is reflected in large part by the levels of the active mark H3K4me3 and the repressive mark H3K27me3 at target genes and more globally. The complexity of pathways operating to modulate these histone modifications and global chromatin architecture is only now becoming apparent. While ESCs favor a transcriptionally “permissive” state, potent repressor pathways are critical for keeping expression of differentiation-promoting genes off and for sequencing exit from pluripotency.
Polycomb as a repressive system
As a major repressive system in development, Polycomb Group (PcG) proteins have received attention as silencers of differentiation pathways in pluripotent cells. PcG proteins act in two different multiprotein complexes, known as PRC1 and PRC2 (see Margueron and Reinberg, 2011). Four core proteins -- EED, Suz12, Ezh2, and RbAp46/48 -- comprise PRC2. PRC1 is considerably more diverse, as it is composed of core subunits Ring1A and 1B with a variety of other proteins. Through the aegis of Ezh2 or the related protein Ezh1 (Margueron et al., 2008; Shen et al., 2008), PRC2 catalyzes di- and tri-methylation of histone H3 lysine 27. H3K27me3 also binds to EED and stimulates activity of the complex (Xu et al., 2010). This repressive mark further serves as a docking site for PRC1, which catalyzes monoubiquitination of histone H2A at lysine 119. Domains marked by H3K27me3 may be quite large (>100kb) or on the scale of a few kilobases.
Initial chromatin occupancy studies revealed that PRC2 and PRC1 components bind numerous differentiation-related genes that are silent but “poised” for expression in ESCs (Boyer et al., 2006). These targets display a “bivalent” chromatin mark, defined by the presence of active H3K4me3 and repressive H3K27me3 marks (Bernstein et al., 2006). Upon ES cell differentiation, PcG-bound targets are expressed in concert with loss of H3K27me3. In the simplest interpretation, polycomb-mediated repression is essential for maintenance of pluripotency. This conclusion is inconsistent with the capacity of EED-null ESCs to give rise to all three germ layers (Chamberlain et al., 2008). Subsequent studies reveal added complexity, particularly with respect to PRC2, and provide a more nuanced view of the role of PcG in pluripotency (Shen et al., 2009; Shen et al., 2008).
An unanticipated finding in proteomic studies was identification of Jarid2 (or Jmj), the founding member of the Jmj (JumonjiC) family of proteins, as a tightly associated component of PRC2 purified from ESCs and required for proper ES cell differentiation (Landeira and Fisher, 2011). Members of the Jmj family are typically lysine demethylases, such as aforementioned Oct4-regulated Jmjd1a and Jmjd2c. In mice Jarid2 is essential for development of the neural tube and the heart, though precise mutant phenotypes are highly sensitive to genetic background. In genome-wide chromatin occupancy studies in ESCs, Jarid2 binding extensively (>90%) overlaps that of other PRC2 components and H3K27me3. Jarid2 appears to facilitate recruitment of the PRC2 complex to chromatin, possibly through its affinity for GC-rich DNA (Li et al., 2010). Paradoxically, Jarid2 is enzymatically inactive, as it lacks conserved residues for cofactor binding, and H3K27me3 is not as affected upon its loss as predicted by its role in recruitment to chromatin. A possible role of Jarid2 in recruiting PRC1 and poised RNA polymerase II to PcG targets has been suggested (Landeira et al., 2010). Jarid2 is a common target of multiple pluripotency factors and rapidly downregulated on differentiation. Thus, the structure of PRC2 during differentiation must be dynamic.
A homologue of Drosophila Polycomb-like (Nekrasov et al., 2007; Walker et al., 2010), PLC2 (or MTF2, metal response element binding transcription factor 2), stimulates PRC2 activity, and its loss leads to altered properties of ESCs and impaired differentiation (Ahmed et al., 2010; Shen et al., 2009). PLC2 is recruited to a subset of PRC2 targets. Recent evidence suggests that under some circumstances PLC2-containing PRC2 may strongly influence the subsequent repressive action of PRC1 without an evident change in H3K27me3.
Available findings indicate that PcG function is critical to the balance of ES cell self-renewal and differentiation, particularly in sequencing transcriptional events necessary to exit the pluripotent state and culminate in successful lineage-specification (Shen et al., 2009; Shen et al., 2008). Additionally, potential regulatory interactions may exist between Swi/Snf complexes and PcG. Chromatin occupancy of genes encoding various PcG components has been interpreted as consistent with opposition of Swi/Snf and polycomb function (Lessard and Crabtree, 2010). Recent genetic findings also point to an antagonistic relationship between PcG and Swi/Snf function in control of specific genes (e.g. Ink4a/ARF in mouse embryo fibroblasts) and in oncogenesis (Wilson et al., 2010).
How the composition and modification of PcG complexes change during differentiation is likely to provide new insights into cell fate transitions (see Margueron and Reinberg, 2011). For example, Ezh2 is a substrate for various kinases, including Akt, CDK1 and CDK2. Phosphorylation of Ezh2 has different reported consequences depending on the specific modified residue. Effects on recruitment to chromatin, H3K27 methylation activity, binding to the long non-coding RNA HOTAIR, and differentiation have been described.
Although it is generally presumed that the histone modifying activities of PRC2 and PRC1 are synonymous with repressive function, the situation is not so straightforward. PRC1 compacts chromatin structure and represses Hox gene gene expression independent of histone ubiquitination (Eskeland et al., 2010; Francis et al., 2004). In the absence of the core Ring1B subunit of PRC1, Hox genes are modestly derepressed and chromatin decompaction occurs. Moreover, PRC1 may prevent expression at bivalent genes in part through impaired transcription elongation, perhaps countering the actions of c-Myc in promoting expression. To add to this complexity, in some circumstances PcG complexes act redundantly in repression, independent of H3K27me3 (Leeb et al., 2010). Finally, PcG proteins have recently been implicated in long-range contacts in 3D-nuclear space that lead to corepression of Hox genes in Drosophila (Bantignies et al., 2011). Hence, much remains to be explored regarding the mechanisms by which PcG complexes repress gene expression and alter chromatin structure.
Trithorax as an agent of active gene expression and self-renewal
Classical studies in Drosophila revealed functional antagonism between polycomb and Trithorax (Trx) with respect to Hox gene expression. Whereas polycomb is associated with the repressive H3K27me3 mark, Trx complexes write the active H3K4me3 mark. Mammalian Trx, a homologue of yeast COMPASS, contains a histone methyltransferase (Set1a/b, MLL1-4) and a subunit that recognizes H3K4me3 (Wdr5), as well as other components (Ash2, RBbp5, Dpy-30, etc.). Recently, Trx has been linked to the pluripotency network through study of Dpy-30 (Jiang et al., 2011) and Wdr5 in ESCs (Ang et al., 2011). Protein complexes containing Oct4 interact, either directly or indirectly, with Wdr5 and recruit it to target genes, many of which are also bound by Nanog and Sox2. Furthermore, Wdr5 is required to maintain local and global H3K4me3 and to sustain self-renewal. Depletion of Dpy-30 fails to affect self-renewal but impairs neural differentiation. Interestingly, while substantial target gene overlap is seen between Oct4 and Wdr5, an equivalent, or higher degree, is seen with c-Myc targets. This finding suggests that Wdr5 (and hence, Trx) may link the core and c-Myc regulatory submodules. In this context, it is of interest that c-Myc may interact with MLL complexes and therefore either recruit MLL complexes to specific targets or conversely stabilize MLL complexes at their targets.
Contribution of CpG-binding proteins to local chromatin structure
CpG islands (CGIs) are prominent in mammalian genomes. Commonly, promoters are embedded within CGIs that lack DNA methylation, and are marked by H3K4me3. Recent studies suggest that CGIs influence local chromatin structure through the recruitment of CpG-binding proteins, such as Cfp1 (Thomson et al., 2010). Histone modification is directed by the presence of CGIs in the absence of a promoter, and hence, influenced by the genetic “environment”. These findings are consistent with the association of Cfp1 with the Setd1 histone H3K4 methyltransferase/COMPASS complex (Lee and Skalnik, 2005). Cfp1-null ESCs exhibit various defects, including a decrease in global cytosine DNA methylation, reduced levels of heterochromatin, and reduced H3K4me3 mark at CGIs (Tate et al., 2009), and are unable to differentiate in vitro, a phenotype reminiscent of loss of the MBD3 subunit of NuRD. CGIs may further sculpt local chromatin structure through recruitment of histone demethylases. For example, CGIs recruit the H3K36-specific demethylases KDM2A (jhdm1a, FbxL), leading to depletion of H3K36me2 (Blackledge et al., 2010). Recruitment is blocked by CpG DNA methylation. Although H3K39 methylation is enriched on active genes and appears antagonistic to PRC2 (Yuan et al., 2011), experiments have been unsuccessful to date in defining the consequences of KDM2A loss in ESCs (Blackledge et al., 2010).
Global chromatin regulators
Through a focused RNAi screen in ESCs, Gaspar-Maia and colleagues (Gaspar-Maia et al., 2009) identified the chromatin–remodeling enzyme, Chd1, as essential for pluripotency and sustained Oct4 expression. Chd1 is a member of the ATPase SNF2-helicase family, recognizes HeK4me2/3 through its chromodomains, and localizes to active genes in euchromatin. Chd1 has been associated with transcriptional activation in numerous settings. The Chd1 locus appears to be a target of the pluripotency network. Depletion of Chd1 in ESCs is associated with an increase in foci of heterochromatin marks, such as H3K9me3 and HP1g, as well as reduced exchange of linker histone H1. Chd1-depleted ESCs retained features of pluripotent cells but also tended to differentiation along the neuronal lineage. Despite the association of Chd1 with ~30% of genes marked by H3K4me3, gene expression changes in its absence were paradoxically limited. Chd1 is just one of presumably numerous factors that contribute to proper maintenance of open chromatin structure in pluripotent cells.
Another chromodomain protein, Chd7, is highly associated with p300 at enhancers characterized by binding of pluripotency factors, open chromatin regions, and gene activity. Despite these relationships, loss of Chd7 fails to perturb ES cell self-renewal, pluripotency, or reprogramming (Schnetz et al., 2010).
Recent findings suggest that undifferentiated embryonic cell transcription factor 1 (UTF1) may counterbalance effects of proteins such as Chd1 on overall chromatin structure. UTF-1 is a tightly chromatin-associated protein that occupies >1700 target genes, including many that overlap with pluripotency factors and c-Myc (Kooistra et al., 2010). UTF1-depleted ESCs continue to self-renew but are defective in differentiation. Upon UTF1-depletion, expression of numerous genes is altered, but notably ~90% are upregulated, a finding consistent with the prior assignment of UTF1 as a repressor. UTF1-depletion is also associated with increased release of nucleosomes from chromatin on micrococcal nuclease treatment. These observations implicate UTF1 in preventing chromatin decondensation and possibly limiting transcriptional promiscuity in the setting of the open chromatin state of ESCs.
Developmental potential and chromatin structure
In the second part of this review, we discuss cellular model systems that link changes in developmental potential with alterations in chromatin structure. First, we discuss the process of X inactivation, which dynamically changes with the differentiation state of cells and documents intriguing connections between pluripotency transcription factors and chromatin structure. We then summarize emerging data that suggest important roles for short and long non-coding RNAs in regulating chromatin structure in pluripotent cells. Finally, we elucidate the influence environmental cues have on chromatin structure and cellular state in the context of normal differentiation and cellular reprogramming, and speculate on the exciting possibility to generate any specialized cell type by combining transcription factor expression with specific growth conditions.
X inactivation as a model to study coupling of pluripotency factors and chromatin structure
X chromosome inactivation in females
The intricate relationship of pluripotency and epigenetic programs is highlighted by X chromosome inactivation (XCI), the mechanism in placental mammals that ensures proper gene dosage of X-linked genes in females compared with males by randomly inactivating one of the two X chromosomes in female cells (Lee, 2009). The active and inactive X chromosomes provide models for open and closed chromatin, respectively. The paternal X chromosome is inactive in cleavage stage embryos and remains inactive in extraembryonic cells that form the placenta. However, XCI is reversed in the pluripotent inner cell mass (ICM) of the preimplantation embryo, resulting in two active (paternal and maternal) X chromosomes. Following implantation, random XCI ensues in differentiating cells and is stably maintained in all somatic daughter cells throughout life with the exception of female primordial germ cells (PGCs) that undergo another round of XCI reversion. Remarkably, the reprogramming of female somatic cells by nuclear transfer, cell fusion or transcription factor expression also reverses XCI, indicating that these approaches can mimic developmental XCI reprogramming (Navarro and Avner, 2009). Thus, XCI provides a unique platform for studying heterochromatin formation and its reversal during normal development.
XCI is a multistep process involving (i) choice of the future active X (Xa), (ii) initiation of silencing of the future inactive X (Xi), (iii) maintenance of the Xi silencing throughout development, and (iv) reversal of XCI in ICM cells and PGCs. Here, we briefly review how these events are regulated by non-coding (nc) RNAs, pluripotency factors, and selected chromatin regulators.
Roles of non-coding RNAs and chromatin factors during XCI
The X inactivation center (XIC) harbors several long non-coding (lnc)RNAs that are involved in the choice and initiation of Xi silencing (Lee, 2009)(Figure 4A). These ncRNAs include Xist, expressed from the Xi, and its antisense transcript Tsix, expressed from the Xa. The antagonism between Xist and Tsix transcripts determines which X remains active and which becomes inactivated. Xist is a 17 kb lncRNA that initiates XCI by associating with the Ezh2 subunit of PRC2, leading to H3K27me3 and consequent spreading of silencing over the entire X (Lee, 2009). Female mouse ESCs serve as an in vitro model to recapitulate XCI; ESCs carry two Xa and express Tsix from both chromosomes to repress Xist transcription. Upon differentiation, Xist becomes strongly upregulated on the future Xi while Tsix expression is downregulated and absent in differentiated cells (Figure 4B). The maintenance of an Xi is dependent on multiple epigenetic marks, such as DNA and histone methylation, late replication and hypoacetylation of histone H4.
Figure 4. Pluripotency factors and X chromosome inactivation.
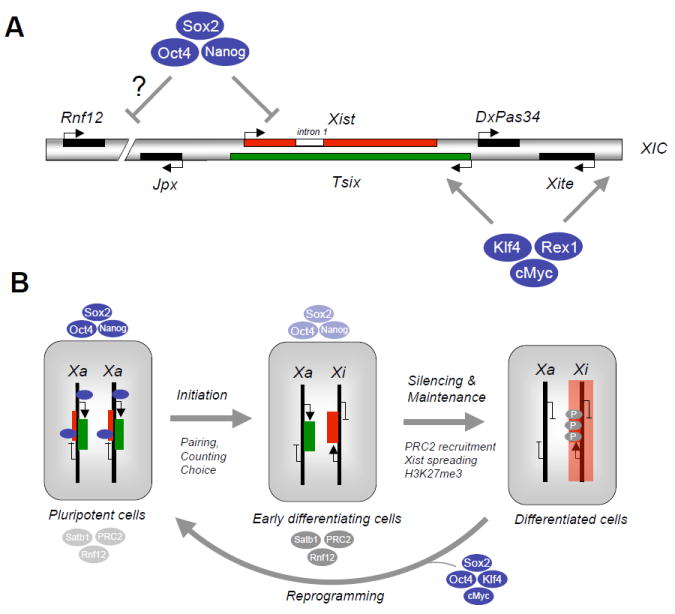
(A) Schematic depiction of X Inactivation Center (XIC) on the X chromosome with positions of non-coding RNAs Xist, Tsix, DxPas34, Xite, Jpx and the dose-dependent Xist activator Rnf12 as indicated. In undifferentiated female embryonic stem cells (ESCs), Xist (intron 1) and possibly Rnf12 are occupied and transcriptionally suppressed by Oct4, Sox2 and Nanog while Xite and Tsix are bound and transcriptionally activated by Klf4, Rex1 and cMyc. (B) In female ESCs, Xist is silenced while Tsix is activated by the pluripotency factors shown in (A). Upon differentiation, X chromosome inactivation ensues through a multistep process that involves initiation (pairing and counting of X chromosomes to choose the future inactive and active X, Xi and Xa), silencing and maintenance of the silenced X. The initiation and onset of silencing are tightly linked with the downregulation of pluripotency factors and the concomitant upregulation of chromatin regulators that mediate XCI, such as PRC2 (recruited by Xist RNA to mediate spreading of silencing along the entire X), and Satb1 (organization of active chromatin into loops). Silencing of the inactive X further results in H3K27 trimethylation by PRC2. Introduction of Oct4, Sox2, Klf4 and cMyc into differentiated cells gives rise to induced pluripotent stem cells, which is accompanied by X chromosome reactivation in mouse.
Ectopic Xist expression from an autosome induces XCI in undifferentiated ESCs but remains without consequence in in vitro differentiated cells, presumably due to the lack of cofactors essential for XCI (Wutz and Jaenisch, 2000). In contrast, widespread ectopic Xist expression in male transgenic mice results in severe anemia and death due to inappropriate XCI in hematopoietic progenitors (Savarese et al., 2006). Subsequent work has identified Satb1 and Satb2 as the elusive “competence factors” present in hematopoietic precursors (Agrelo et al., 2009). These proteins are critical for Xist’s potential to silence the X during regular XCI in differentiating embryonic cells and upon ectopic Xist expression in adult mice. Satb1 was previously shown to organize transcriptionally active chromatin into loops. Thus, Xist may pull inactive genes of the X into repressive chromosome territories with Satb1, possibly serving as an anchor in this chromosomal reorganization (Agrelo et al., 2009).
While much has been learned regarding the earliest steps of XCI during differentiation, the mechanism by which the paternally silenced X becomes reactivated specifically in the ICM and later in PGCs remains elusive. Recent evidence documents a direct role for several pluripotency factors during the reversion of XCI in the ICM (Navarro and Avner, 2009).
Direct control of XCI reversal by pluripotency factors
Oct4, Sox2 and Nanog bind directly to intron 1 of Xist in undifferentiated female ESCs to suppress its expression (Navarro et al., 2008), while Rex1, c-Myc and Klf4 associate with the Tsix promoter to stimulate its expression (Navarro et al.)(Figure 4A). Consistent with this finding, cells that reactivate XCI in the ICM strictly correlates with Nanog expresison, and Nanog-deficient blastocysts fail to undergo XCI reprogramming (Silva et al., 2009). Unexpectedly, deletion of intron 1 of Xist, encompassing all known pluripotency binding sites, is insufficient to activate Xist expression in ESCs, suggesting that other targets of Oct4, Nanog and Sox2 may exist that suppress Xist transcription. One such target may be the X-linked ubiquitin ligase Rnf12 (Barakat et al., 2011; Jonkers et al., 2009), which functions as a dose-dependent activator of Xist transcription. Indeed, the Rnf12 gene promoter is occupied by pluripotency factors (Kim et al., 2008; Marson et al., 2008).
Pluripotency factors have been suggested to play additional roles during XCI by influencing the processes of X chromosome “pairing” and “counting” (Donohoe et al., 2009). “Pairing” denotes the physical association of both Xs to establish asymmetries between the future Xa and Xi and provides the basis for “counting” to ensure that only cells with two Xs undergo XCI (Lee, 2009). Specifically, Oct4 protein associates with Ctcf, a chromatin insulator protein involved in X chromosome pairing. Accordingly, pairing and counting are abrogated in Oct4-deficient cells, resulting in two Xi.
Given that exit from pluripotency correlates with the onset of XCI, transcriptional repressors of pluripotency genes could also be involved in XCI. Indeed, the XCI competence factor Satb1 physically associates with and inhibits the Nanog and Klf4 promoters in differentiating ESCs (Savarese et al., 2009). Together, these results demonstrate a direct role for pluripotent transcription factors and their repressors at multiple steps of XCI. While Oct4, Sox2 and Nanog are also expressed in PGCs (Durcova-Hills et al., 2008) that undergo X reactivation, a direct role for these factors in germ cell XCI reprogramming remains to be experimentally tested.
Cellular reprogramming and X chromosome reactivation
The reactivation of the somatically silenced X upon overexpression of Oct4, Sox2, c-Myc and Klf4 in generation of iPSC from female fibroblasts lends support for direct involvement of pluripotency factors in XCI reversal and chromatin remodeling (Maherali et al., 2007). Differentiation of these iPSCs results in random XCI, indicating that introduction of reprogramming factors is sufficient to erase the epigenetic imprint of the previously inactive X chromosome. However, since XCI reversal occurs late during reprogramming (Stadtfeld et al., 2008), additional molecules that act downstream of Oct4, Sox2, and Klf4 must be involved. Of note, pluripotent epiblast stem cells (EpiSCs), which are derived from postimplantation embryos (Brons et al., 2007; Tesar et al., 2007), express Oct4 and Sox2 at comparable levels as ESCs but nevertheless exhibit XCI (Guo et al., 2009), suggesting that these factors are insufficient to reprogram the silenced X. However, overexpression of Klf4 or Nanog, which are downregulated in EpiSCs relative to ESCs, facilitates the conversion of EpiSCs into ESC-like cells as well as XCI reactivation (Guo et al., 2009; Silva et al., 2009).
It remains unclear if the same coupling of XCI and pluripotency factors applies to human ESCs. Human ESCs resemble mouse EpiSCs more than mouse ESCs and invariably exhibit signs of XCI (Hoffman et al., 2005; Silva et al., 2008). Consistent with this, human iPSCs retain the inactive X chromosome of their somatic donor cell (Tchieu et al., 2010). However, ectopic expression of OCT4, KLF2 and KLF4 endows human ESCs with a mouse-like state that shows two Xa, providing evidence that the function of pluripotency transcription factors in XCI reprogramming is conserved (Hanna et al., 2010).
Evidence for a direct involvement of pluripotency factors in telomere extension, which is, like XCI reversion, another molecular process tightly associated with the acquisition of pluripotency in iPSCs, stems from the recent observation that OCT4 and NANOG bind directly to the TERC and DKC1 genes, which are critical for telomerase activity (Agarwal et al., 2010).
Role of non-coding RNAs in regulating chromatin state and pluripotency
Accumulating evidence suggests that the principles of RNA-mediated gene control during XCI may apply to several other loci and cellular processes. The ncRNA machinery in the cell provides a complex repertoire of regulatory molecules that can be subdivided into (i) Argonaute associated small RNAs ranging from 20-30nt including microRNAs, endogenous small interfering RNAs (siRNAs) and PIWI-interacting RNAs (piRNAs), (ii) short RNAs (20-200nt long) associated with promoter and enhancer regions, and (iii) lnc RNAs (several kbs in length) including large intergenic ncRNA (lincRNAs) (Pauli et al., 2011). Here, we will focus only on miRNAs and lncRNAs since there is some evidence for their involvement in pluripotency, chromatin structure and reprogramming (Figure 5).
Figure 5. Non-coding RNAs modulate ESC self renewal and differentition as well as cellular reprogramming.
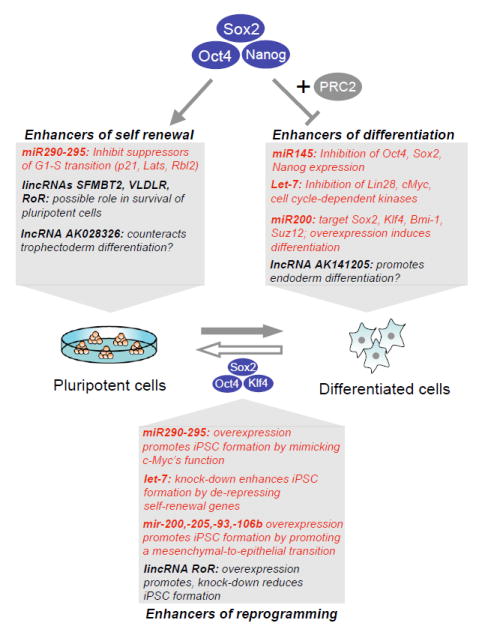
Shown are examples of microRNAs (in red) and long non-coding (lnc)RNAs (in black) that are occupied and either activated by Oct4, Sox2 and Nanog, or silenced by the same factors in combination with the PRC2 repressor complex in pluripotent cells, and their roles in self renewal and differentiation. Manipulation of several non-coding RNAs in the context of induced pluripotent stem cell (iPSC) formation has been shown to enhance cellular reprogramming. Note that some miRNAs, such as members of the miR-200 family, seem to directly target PRC1 and PRC2 components, such as Bmi-1 and Suz12, respectively.
Role of miRNAs in pluripotency and reprogramming
miRNAs are short ncRNAs that inhibit gene expression mostly by destabilizing and repressing target RNAs (Pauli et al., 2011), although they may in certain cellular contexts activate translation (Vasudevan et al., 2007). Their biogenesis depends on the RNA processing enzymes Dicer, Drosha and its essential cofactor Dgcr8. Mice mutant for Dicer, Drosha or Dgcr8 die early during gestation (Pauli et al., 2011), documenting that miRNAs are essential for normal embryogenesis. Moreover, deletion of either enzyme in ESCs results in severe growth and differentiation defects, indicating roles in self-renewal and pluripotency (Pauli et al., 2011). In an elegant complementation approach, Blelloch and colleagues found that miR-290-295 microRNA family members rescue the observed proliferation defect of Dgcr8 knockout ESCs by inhibiting suppressors of G1-S progression such as Lats2, p21 and Rbl2 (Wang et al., 2008). In contrast, introduction of mature let-7 miRNA, which is absent from pluripotent cells and expressed in many differentiated cells, into Dgcr8-deficient ESCs suppresses their self-renewal (Melton et al., 2010). Let-7 targets include regulators of self-renewal, such as Lin28, c-Myc and cyclin-dependent kinases. Of note, the precursor for let-7 is present in both undifferentiated ESCs and mature cells. However, its biogenesis in undifferentiated ESCs is suppressed by the action of Lin28. Thus, the antagonism between Lin28 and let-7 establishes a bistable switch typical of many miRNAs that controls the transition between the undifferentiated (Lin28 on, let-7 off) and differentiated (Lin28 off, let-7 on) cell state.
How are miRNAs themselves regulated at the transcriptional level? Genome-wide binding studies suggest that pluripotency transcription factors including Oct4, Sox2, Nanog and Tcf3 occupy the promoters of ESC-specific miRNA genes (Marson et al., 2008). In addition, many differentiation-associated miRNA gene loci, such as let-7, are bound by the same pluripotency factors in combination with components of the repressive PRC2 complex in ESCs, resulting in their transcriptional suppression. The dual role pluripotency factors play in binding to active and repressed miRNA genes in ESCs is akin to that seen for protein-coding genes involved in self-renewal and differentiation (Boyer et al., 2006; Lee et al., 2006). Intriguingly, another differentiation-associated miRNA, miR-145, which targets Oct4, Sox2 and Klf4 transcripts for degradation, is itself transcriptionally repressed by Oct4 in undifferentiated ESCs, establishing a negative feedback loop that ensures rapid suppression of the self-renewal program upon initiation of differentiation (Xu et al., 2009).
Consistent with their role in regulating ESC self-renewal, the modulation of miRNAs affects the reprogramming of somatic cells into iPSCs. For example, ectopic expression of ESC-specific miRNAs from the miR-290-295 cluster in fibroblasts enhances the formation of iPSCs in the absence, but not in the presence, of c-Myc (Judson et al., 2009), suggesting that the miRNAs are downstream effectors of c-Myc during cellular reprogramming. In agreement with let-7’s inhibitory effect on ESC self-renewal, its suppression promotes the derivation of iPSCs (Melton et al., 2010). The finding that ectopic expression of Lin28, which is critical for the biogenesis of let-7, is sufficient to reprogram somatic cells in combination with Oct4, Sox2 and Nanog (Yu et al., 2007), is in further accordance with this result. Some of the earliest events fibroblasts undergo upon overexpression of reprogramming factors are the downregulation of mesenchymal genes and the subsequent upregulation of epithelial genes, features resembling a mesenchymal-to-epithelial transition (MET). Indeed, overexpression of miR-200 or miR-205, which have previously been shown to promote MET by suppressing mesenchymal genes such as Snail and Slug, indeed enhances the formation of iPSCs from mouse fibroblasts (Polo and Hochedlinger, 2010).
Role of lncRNAs in chromatin structure, pluripotency and reprogramming
In addition to the association of lncRNAs with XCI and genomic imprinting (Pauli et al., 2011), recent studies in mouse and human ESCs have identified hundreds of so-called large intergenic non-coding RNAs (lincRNAs), which are implicated in the control of ESC self-renewal and pluripotency (Guttman et al., 2009; Khalil et al., 2009). By interrogating genome-wide intergenic H3K4me3/H3K36me3 chromatin territories, indicative of PolII transcripts, in combination with a parallel cDNA sequencing approach (Guttman et al., 2010), over 900 ESC-specific lincRNAs were discovered in total. Of these, a third appear to be bound by Oct4 and Nanog in their promoter regions, directly linking their transcription with the core pluripotency network.
What is the mechanism of lncRNA-mediated gene regulation? While different lncRNAs may utilize different modes of gene regulation (Pauli et al., 2011), increasing evidence suggests that lncRNAs involved in XCI, genomic imprinting and Hox gene regulation associate with components of histone modifying complexes, such as PRC2, G9a, LSD1, CoREST and SMCX, as well as repressor proteins, such as hnRNP-K (Khalil and Rinn, 2011), leading to epigenetic silencing of target genes. Although most reported lncRNAs have been associated with the silencing of target genes, a recent study suggests that lncRNAs may also activate transcription of nearby genes, thus functioning like enhancers (Orom et al., 2010). Whether this entails the recruitment of activating histone modifying enzymes or alternative mechanisms awaits further experimentation. The majority of described lncRNAs appear to act in cis by influencing local chromatin structure. However, some lncRNAs such as Jpx (Tian et al., 2010), HOTAIR (Rinn et al., 2007) and lincRNA-p21 (Huarte et al., 2010) are proposed to function in trans, resulting in the targeting of hundreds of genomic loci throughout the genome (Gupta et al., 2010; Huarte et al., 2010).
Because no enzymatic activity is known to process lincRNAs, as is the case with Drosha/Dcgr8 and miRNAs, it has been impossible to study the functional consequences of ablating all lincRNAs in ESCs. However, gain and loss of function experiments have been performed on individual lincRNAs and suggest an involvement in cellular differentiation, proliferation and reprogramming. Rinn, Daley and colleagues have recently identified ten lincRNAs that are upregulated in human iPSCs compared with ESCs, indicating a possible role in cellular reprogramming (Loewer et al., 2010). Indeed, knock-down and overexpression of one of these lincRNAs, lincRNA-RoR, abrogated and slightly enhanced, respectively, the formation of iPSCs from human fibroblasts, providing the first evidence for the involvement of a lincRNA in cellular reprogramming. Another study reported two lncRNAs whose regulatory sequences are bound by Oct4 and Nanog, respectively, and which appear to control the self-renewal capacity of mouse ESCs (Sheik Mohamed et al., 2010).
The lncRNA Gtl2, which is part of the ~1Mb long Dlk1-Dio3 imprinted locus, becomes aberrantly silenced by DNA hypermethylation and histone hypoacetylation during cellular reprogramming into iPSCs (Stadtfeld et al., 2010). The silenced status of this cluster in 90% of iPSCs tighly correlates with the developmental failure of these iPSCs to contribute efficiently to tissues in mice as well as to produce entirely iPSC-derived animals. Gtl2 is is a maternally expressed gene that is thought to negatively regulate the paternally expressed Dlk1 gene, involved in fetal growth, within the same gene cluster. The paternal allele is normally silenced by DNA methylation and histone deacetylation. We speculate that enforced expression of reprogramming factors, either directly or indirectly, recruits DNA and histone modifying enzymes, such as de novo DNA methyltransferases and histone deacetylases to this locus, which result in stable chromatin silencing. Consistent with this finding is the observation that binding sites for Oct4 and Nanog have been identified in the upstream region of the Gtl2 locus (Navarro et al., 2010). Moreover, proteomic studies on Oct4 and Nanog interacting proteins have identified prime suspects for chromatin silencing, such as Dnmt3a and components of the NURD complex (Pardo et al., 2010; van den Berg et al., 2010) (Liang et al., 2008; Pardo et al.; van den Berg et al.; Wang et al., 2006). Since many pluripotency factors are also expressed in PGCs, which undergo global erasure of genomic imprints during normal development, we hypothesize that the reprogramming of imprints, akin to the reactivation of XCI in germ cells, might be regulated by some of these factors. It remains unclear, however, why this results in biallelic silencing in iPSCs rather than in biallelic expression as seen in PGCs. It is further puzzling that no other imprinted loci are affected during iPSC generation, suggesting that Dlk1-Dio3 imprinting may be regulated differently from most other imprinted genes.
It should be noted that other ncRNAs have recently been identified in ESCs based on their interaction with PRC2. These include a new class of small RNAs (50-200nt) that are produced around the transcriptional start sites of repressed Polycomb targets and might assist recruitment of PRC2 to their promoters (Kanhere et al., 2010). A different study has used RIP-seq to isolate over 9,000 ncPRC2-associated non-coding RNAs of different lengths that include antisense, intergenic and promoter-associated transcripts, which are produced in proximity to imprinted loci, cancer-associated genes and stem cell-related bivalent domains (Zhao et al., 2010). Of note, small bidirectional transcripts have also been detected at enhancer elements in neurons and ESCs (Creyghton et al., 2010; Kim et al., 2010b). The function of these ncRNAs in regulating chromatin structure and gene expression in pluripotent cells remains unclear.
Environmental influences on chromatin structure and cellular state
Here we discuss recent findings interrogating the effects of environmental factors on the chromatin and developmental states of pluripotent cells (Figure 6). We further propose that cell fates may be diverted to desired lineages by a combination of pluripotency factor expression and culture conditions.
Figure 6. Examples of culture-induced epigenetic and developmental changes in pluripotent mouse cells.
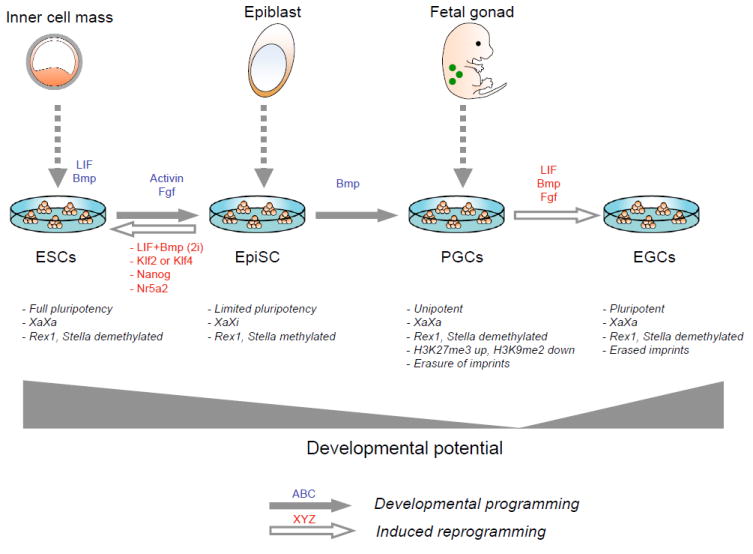
Embryonic stem cells (ESCs), derived from the inner cell mass of blastocysts, are maintained in an undifferentiated state in the presence of LIF and Bmp. Exchange of LIF and Bmp with Fgf and activin induces their differentiation into epiblast stem cells (EpiSCs), which are normally derived from the epiblast of postimplantation embryos and have limited differentiation potential. The ESC-to-EpiSC transition is accompanied by characteristic epigenetic changes, such as X inactivation and methylation silencing of Rex1 and Stella genes, which can be reversed by replating cells in LIF/Bmp or 2i, or upon overexpression of Klf2, Klf4, Nanog, or Nr5a2. When exposed to Bmp, EpiSCs continuously give rise to unipotent primordial germ cells (PGCs) that undergo genome-wide epigenetic remodeling, X reactivation and erasure of genomic imprinting. In the presence of LIF, Bmp and Fgf, these PGCs undergo dedifferentiation into pluripotent embryonic germ cells (EGCs).
Mouse ESCs can be maintained in a self-renewing pluripotent state in the presence of LIF/Stat3 and Bmp/Smad/Id signaling (Wray et al., 2010). The effects of LIF and Bmp are mimicked by growth of ESCs in the presence of two chemical inhibitors, dubbed “2i”, of the MAP kinases Erk1 and Erk2 and gycogen synthase kinase 3 (Gsk3) (Ying et al., 2008). Genomic targets of the core pluripotency triad, Oct4, Sox2 and Nanog, are frequently co-occupied by the downstream effectors of LIF and Bmp signaling, Stat3 and Smad1, as well as by the histone acetyltransferase p300, thus providing a link between growth factor signaling, the core pluripotency network and chromatin regulation (Chen et al., 2008).
Reprogramming of germ cells into pluripotent cells
Two typical examples for culture-induced changes of epigenetic and developmental state are the conversion of PGCs and derivative spermatogonial stem cells (SSCs) into pluripotent stem cells (Hochedlinger and Jaenisch, 2006). When explanted in culture, PGCs give rise to embryonic germ cells (EGCs) in the presence of Fgf2, Lif and Scf (Matsui et al., 1992; Resnick et al., 1992) or, alternatively, 2i and Lif (Leitch et al., 2010) (Figure 6). Importantly, PGCs are unipotent and hence can only produce sperm or oocytes in vivo, while derivative EGCs are pluripotent and contribute to all tissues in mice, including germ cells.
During early stages of PGC reprogramming in Fgf2/Lif/Scf, the germ cell specification factor Blimp1/Prdm1 becomes downregulated while its repressed targets c-Myc and Klf4 are upregulated (Durcova-Hills et al., 2008). Given that Oct4 and Sox2 are already expressed in PGCs, this result suggests that PGCs have an inherent potential to become pluripotent, which is normally blocked by the transcriptional repressor protein Blimp1. Of note, Blimp1 cooperates with the arginine methyltransferase Prmt5 during PGC specification (Ancelin et al., 2006). Recent data suggest that translocation of Prtm5 from the nucleus to the cytosol during PGC-to-EGC conversion, as well as during ESC derivation from ICM cells, mediates histone H2A methylation, which in turn leads to the suppression of differentiation-associated genes (Tee et al., 2010). Prmt5 expression, in combination with Oct4 and Klf4, is also sufficient to induce pluripotency from murine fibroblasts (Nagamatsu et al., 2011). Furthermore, Prmt5 physically interacts with Stat3, which is critical for EpiSC-to-ESC conversion and cellular reprogramming into iPSCs (Yang et al., 2010), thus providing an interesting link between the Lif/Stat3 signaling pathway, chromatin structure and the establishment of a pluripotent ground state.
Other signaling pathways that have been previously shown to enhance the conversion of PGCs into EGCs include activation of AKT (Kimura et al., 2008), and inhibition of PTEN (Kimura et al., 2003) or p53 (Kimura et al., 2008), which seem to confer survival and self-renewal potentials to PGCs. While the relevant downstream effectors of these pathways at the chromatin level remain unclear, it is interesting to note that H3K27me3 methylation is suppressed through phosphorylation of Ezh2 by activated AKT (Cha et al., 2005). Thus, Ezh2-suppressed targets critical for the conversion of PGCs into EGCs may become derepressed by activation of AKT signaling.
Similar to PGCs, SSCs give rise at extremely low frequency (0.01%) to pluripotent ESC-like germline stem cells (called mGSCs or gPSCs) when grown in the presence of Lif and serum (Kanatsu-Shinohara et al., 2004; Ko et al., 2009). Interestingly, the gene expression and methylation patterns between SSCs and gPSCs are very similar, with many pluripotency loci being demethylated and transcribed in both cell types (e.g., Oct4, ERas, Sox2, Rex1) while few pluripotency promoters are methylated and silenced (e.g., Nanog, Fbx15, Fgf4) (Imamura et al., 2006). At the protein level, however, Sox2 is undetectable and Oct4 levels are substantially reduced compared with gPSCs, suggesting that posttranscriptional mechanisms operate in SSCs to prevent their inappropriate dedifferentiation into pluripotent cells. Thus, PGCs and SSCs utilize various mechanisms to efficiently suppress the full pluripotency program in vivo whereas explantation in culture seems to remove these constraints and facilitates spontaneous conversion into pluripotent cells.
Effects of Fgf, Jak/Stat, and Bmp signaling on the epigenetic and differentiation state of cells
Another example of an environment-induced change of epigenetic and developmental states of cells is the interconversion of ESCs and EpiSCs (Figure 6). The differentiation of ESCs into EpiSCs, which mimics the normal progression of preimplantation ICM cells into postimplantation epiblast, is achieved by the replacement of Lif and Bmp in established ESCs with Fgf and activin (Guo et al., 2009). Resultant EpiSCs resemble EpiSCs derived directly from embryos in their epigenetic profile (e.g, XCI, methylation of Stella and Rex1 promoter regions) and limited differentiation potential (e.g., capacity to form teratomas but inability to contribute to chimeras).
Replating of EpiSCs in Bmp/Lif or 2i/Lif gives rise, at extremely low frequencies, to reverted ESC-like cells that show reactivation of the silenced X chromosome and demethylation of Stella and Rex1 promoters (Bao et al., 2009). A recent report has linked this reversion to the inhibition of Fgf signaling by 2i, which appears to relieve Fgf’s suppressive effect on Klf2 expression (Greber et al.). In a related study, Smith and coworkers identified an additional role for Jak/Stat3 signaling in EpiSC-to-ESC conversion (Yang et al., 2010). Notably, activation of Jak signaling as well as overexpression of Nanog (Silva et al., 2009) or Klf2/Klf4 (Guo et al., 2009; Hall et al., 2009), all of which facilitate an EpiSC-into-ESC conversion, also promoted the progression of partially reprogrammed iPSCs into fully reprogrammed iPSCs, indicating commonalities among these different types of reprogramming. Lastly, exposure of EpiSCs to Bmp4 promotes the delineation of PGCs and subsequently the derivation of EGCs in culture, which showed epigenetic changes typical for germ cell maturation, including reactivation of XCI and erasure of imprinted gene methylation (Hayashi and Surani, 2009). Taken together, these result show that Fgf, Jak/Stat and Bmp signaling dynamically regulate the interconversion of ESC, EpiSCs and PGCs/EGCs, hence linking extracellular signaling pathways to changes in the epigenetic configuration and differentiation state of pluripotent cells.
JAK signaling also appears to contribute to the self renewal of ESCs in a Lif-independent fashion by interfering with the binding of the heterochromatin factor HP1a at key pluripotency genes, including Nanog (Griffiths et al., 2011). Specifically, constitutive active JAK signaling in ESCs results in the phosphorylation of histone H3 tyrosine 41 (H3Y41), thereby displacing HP1a from many targets involved in the self renewal of ESCs. These ESCs grow in the absence of 2i or Lif and do not activate Stat3. Consistent with this, inhibition of JAK signaling with a small molecule results in inappropriate differentiation of such cells, which can be rescued by Nanog overepression. This result uncovers a previously unrecognized role for JAK signaling in directly communicating with the pluripotency network by controlling chromatin accessibility at crucial self-renewal genes.
The derivation of human ESCs in low oxygen provides another example for the influence of environmental factors on the epigenetic state of pluripotent cells (Lengner et al., 2010). Specifically, the generation of hESCs in physiological oxygen preserves cells in a pre X-inactivation state that is reminiscent of mouse ESCs, which carry two Xa. Interestingly, low oxygen levels also prevent the spontaneous differentiation of human ESCs (Ezashi et al., 2005; Lengner et al., 2010) and enhance the derivation of iPSCs from fibroblasts (Utikal et al., 2009; Yoshida et al., 2009), suggesting that hypoxic culture conditions in general are beneficial for the establishment and maintenance of very primitive pluripotent cells. While the mechanisms underlying these observations remain unclear, it is possible that elevated levels of hypoxia-induced Hif-2a which positively regulates Oct4 at the transcriptional level, contributes to these effects (Covello et al., 2006).
Environmental factors and alternative cellular states
The observation that changes of environmental factors and/or forced expression of transcription factors generate alternate pluripotent cell states (EpiSCs, ESCs, and EGCs) (Figure 6) raises the intriguing possibility that non-iPSC fates might be produced directly from somatic cells upon overexpression of Oct4, Sox2, Klf4 and c-Myc when exposed to appropriate culture conditions. Indeed, fibroblasts expressing these four factors and cultivated in the presence of Fgf and activin appear to give rise directly to EpiSCs, although differentiation from a rare iPSC intermediate cannot be excluded (Han et al., 2011).
Remarkably, when reprogramming factors were expressed in fibroblasts for a brief time insufficient to produce iPSCs and followed by a change of culture conditions conducive for cardiomyocyte growth, cardiomyocytes that activated a cardiac reporter, exhibited action potentials and spontaneously twitched were produced (Efe et al., 2011). These results suggest that early during the reprogramming process, the chromatin may become sufficiently plastic to assume different cellular states, which are selected for by the extracellular signals provided. That is, in the presence of ESC growth factors, conversion to pluripotency is favored while the presence of growth factors specific for other cell lineages may facilitate conversion into alternative differentiation states (Figure 7).
Figure 7. Proposed synergism between pluripotency gene expression and growth factors in changing cellular identity.
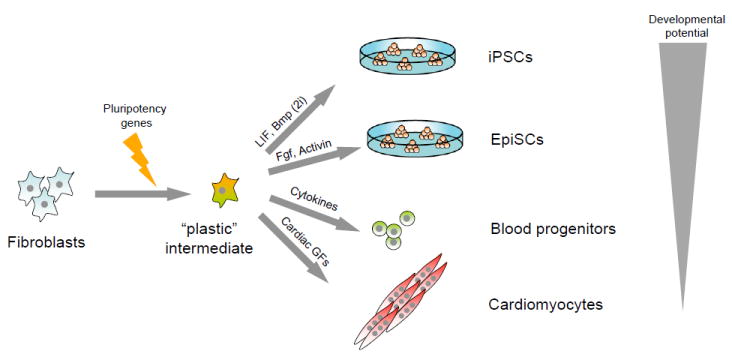
Introduction of individual or combinations of pluripotency genes into fibroblasts may generate a hypothetical “plastic” intermediate that is amenable to further reprogramming into induced pluripotent stem cells (iPSCs) or epiblast stem cells (EpiSCs) when exposed to LIF/Bmp (2i) or Fgf/activin, respectively. Alternatively, such intermediate cells may be converted directly into blood progenitors or cardiomyocytes when exposed to hematopoeitic cytokines or cardiac growth factors, respectively. Note that the developmental potency of resultant cells appears to depend on the provided growth conditions.
Another provocative study, which supports the notion that pluripotency gene expression in differentiated cells may give rise to alternative cell fates, reported that expression of Oct4 is sufficient to convert human dermal fibroblasts into CD45+ hematopoietic progenitor-like cells in vitro (Szabo et al., 2010). Upon exposure of fibroblast-derived CD45+ progenitors to different hematopoietic cytokines, cells with myeloid, erythroid and megakaryocytic phenotypes were observed that could, to a limited extent, engraft in irradiated mice. An alternative explanation for these results, which cannot be ruled out, is that rare pre-existing CD45+ progenitors present in the heterogeneous fibroblast population were expanded rather than generated de novo by ectopic Oct4 expression. Consistent with this interpretation, ectopic Oct4 expression in mice expands adult progenitor cells rather than induces dedifferentiation of mature cells (Hochedlinger et al., 2005).
Non-physiological binding to lineage-specific target genes is one mechanism by which forced pluripotency factor expression might induce alternative differentiated cell fates. Support for this hypothesis derives from the observation that partially reprogrammed iPSCs exhibit aberrant expression and pluripotency factor binding of differentiation-specific genes (Mikkelsen et al., 2008; Sridharan et al., 2009), and fibroblasts overexpressing Oct4 show abnormal binding to hematopoietic targets that are normally occupied by Oct1 and Oct2 in the differentiated state (Szabo et al., 2010). Thus, overexpression of individual ESC-specific transcription factors in somatic cells might endow them with the fate of any other cell type whose generation depends on a homologous transcription factor recognizing the same genomic target site. In analogy, several Pre-B cell-specific enhancers that are normally bound and activated by Sox4 in mature cells, are occupied and silenced by Sox2 and FoxD3 in undifferentiated ESCs (Liber et al., 2010). Sox2 binding seems to mediate the deposition of activating histone H3K4 methylation marks at these repressed genes in ESCs, thus endowing them with a poised state that is readily activated upon differentiation by exchange with the lymphoid-specific Sox4 transcription factor. These observations are further reminiscent of the recently reported pattern of poised enhancer elements identified in mouse and human ESCs based on depletion for histone H3 lysine 27 acetylation (H3K27ac) (Creyghton et al., 2010; Rada-Iglesias et al., 2011). Such poised enhancers are frequently cooccupied by Oct4 and Sox2, which may be involved in the recruitment of suppressive chromatin factors.
Concluding thoughts
Genomic and proteomic studies document that pluripotency transcription factors form multiprotein complexes that associate with diverse chromatin regulators in ESCs to control gene expression. Remarkably, by engaging with either activating or inhibiting chromatin complexes, the same factors either activate genes important for self-renewal or silence genes required for differentiation. Non-coding RNAs have emerged as possible mediators to assemble and deposit these chromatin complexes to their targets. Moreover, accumulating evidence demonstrates that changes in environmental cues have profound effects on the chromatin state and cell fate. A better understanding of how diverse growth factor pathways signal to chromatin will yield critical insights into mechanisms of normal development and provide a framework for attempts to change the identity of one cell type into that of any other cell type by manipulating defined proteins. If a “plastic state” is achieved during factor-induced reprogramming prior to activation of the endogenous ESC program, as hinted at by recent studies, new strategies for the generation of cells representative of specific lineages may be greatly enhanced.
Contributor Information
Stuart H. Orkin, Email: Stuart_Orkin@DFCI.harvard.edu.
Konrad Hochedlinger, Email: Khochedlinger@helix.mgh.harvard.edu.
References
- Agarwal S, Loh YH, McLoughlin EM, Huang J, Park IH, Miller JD, Huo H, Okuka M, Dos Reis RM, Loewer S, et al. Telomere elongation in induced pluripotent stem cells from dyskeratosis congenita patients. Nature. 2010;464:292–296. doi: 10.1038/nature08792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrelo R, Souabni A, Novatchkova M, Haslinger C, Leeb M, Komnenovic V, Kishimoto H, Gresh L, Kohwi-Shigematsu T, Kenner L, et al. SATB1 defines the developmental context for gene silencing by Xist in lymphoma and embryonic cells. Dev Cell. 2009;16:507–516. doi: 10.1016/j.devcel.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ancelin K, Lange UC, Hajkova P, Schneider R, Bannister AJ, Kouzarides T, Surani MA. Blimp1 associates with Prmt5 and directs histone arginine methylation in mouse germ cells. Nat Cell Biol. 2006;8:623–630. doi: 10.1038/ncb1413. [DOI] [PubMed] [Google Scholar]
- Ang Y-S, Tsai S-Y, Lee D-F, Monk J, Su J, Ratnakumar K, Ding J, Ge Y, Darr H, Chang B, et al. Wdr5 mediates self-renewal and reprogramming via the embryonic stem cell core transcriptional network. Cell. 2011 doi: 10.1016/j.cell.2011.03.003. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bantignies F, Roure V, Comet I, Leblanc B, Schuettengruber B, Bonnet J, Tixler V, Mas A, Cavalli G. Polycomb-dependent regulatoryu contacts between distant Hox loci in Drosophila. Cell. 2011;144:214–226. doi: 10.1016/j.cell.2010.12.026. [DOI] [PubMed] [Google Scholar]
- Cartwright P, McLena C, Sheppard A, Rivett D, Jones K, Dalton S. LIF/STAT3 controls ES cell self-renewal and pluripotency by a myc-dependent mechanism. Development. 2005;132:885–896. doi: 10.1242/dev.01670. [DOI] [PubMed] [Google Scholar]
- Chamberlain SJ, Yee D, Magnuson T. Polycomb repressive complex 2 is dispensable for maintenance of embryonic stem cell pluripotency. Stem Cells. 2008;26:1496–1505. doi: 10.1634/stemcells.2008-0102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creyghton MP, Markoulaki S, Levine SS, Hanna J, Lodato MA, Sha K, Young RA, Jaenisch R, Boyer LA. H2AZ is enriched at polycomb complex target genes in ES cells and is necessary for lineage commitment. Cell. 2008;135:649–661. doi: 10.1016/j.cell.2008.09.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donohoe ME, Silva SS, Pinter SF, Xu N, Lee JT. The pluripotency factor Oct4 interacts with Ctcf and also controls X-chromosome pairing and counting. Nature. 2009;460:128–132. doi: 10.1038/nature08098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durcova-Hills G, Tang F, Doody G, Tooze R, Surani MA. Reprogramming primordial germ cells into pluripotent stem cells. PLoS One. 2008;3:e3531. doi: 10.1371/journal.pone.0003531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazzio TG, Huff JT, Panning B. An RNAi screen of chromatin proteins identifies Tip60-p400 as a regulator of embryonic stem cell identity. Cell. 2008;134:162–174. doi: 10.1016/j.cell.2008.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo G, Yang J, Nichols J, Hall JS, Eyres I, Mansfield W, Smith A. Klf4 reverts developmentally programmed restriction of ground state pluripotency. Development. 2009;136:1063–1069. doi: 10.1242/dev.030957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi K, Surani MA. Self-renewing epiblast stem cells exhibit continual delineation of germ cells with epigenetic reprogramming in vitro. Development. 2009;136:3549–3556. doi: 10.1242/dev.037747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochedlinger K, Jaenisch R. Nuclear reprogramming and pluripotency. Nature. 2006;441:1061–1067. doi: 10.1038/nature04955. [DOI] [PubMed] [Google Scholar]
- Jiang H, Shukla A, Wang X, Chen W-y, Bernstein B, Roeder R. Role for Dpy-30 in ES cell-fate specification by regulation of H3K4 methylation within bivalent domains. Cell. 2011;144:513–525. doi: 10.1016/j.cell.2011.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judson RL, Babiarz JE, Venere M, Blelloch R. Embryonic stem cell-specific microRNAs promote induced pluripotency. Nat Biotechnol. 2009;27:459–461. doi: 10.1038/nbt.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil A, Rinn J. RNA-protein interactions in human health and disease. Semin Cell Dev Biol Feb. 2011 doi: 10.1016/j.semcdb.2011.02.016. 17 epub (ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Chu J, Shen X, Wang J, Orkin SH. An extended transcriptional network for pluripotency of embryonic stem cells. Cell. 2008;132:1049–1061. doi: 10.1016/j.cell.2008.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura T, Tomooka M, Yamano N, Murayama K, Matoba S, Umehara H, Kanai Y, Nakano T. AKT signaling promotes derivation of embryonic germ cells from primordial germ cells. Development. 2008;135:869–879. doi: 10.1242/dev.013474. [DOI] [PubMed] [Google Scholar]
- Kooistra S, van den Boom V, Thummer R, Johannes F, Wardenaar R, Tesson B, Veehoff L, Fusetti F, O’Neill L, Turner B, et al. Undifferentiated embryonic cell transcription factor 1 regulates ESC chromatin organization and gene expression. Stem Cells. 2010;28:1703–1714. doi: 10.1002/stem.497. [DOI] [PubMed] [Google Scholar]
- Landeira D, Fisher A. Inactive yet indispensable: the tale of Jarid2. Trends in Cell Biology. 2011;21:74–79. doi: 10.1016/j.tcb.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JT. Lessons from X-chromosome inactivation: long ncRNA as guides and tethers to the epigenome. Genes Dev. 2009;23:1831–1842. doi: 10.1101/gad.1811209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessard JA, Crabtree GR. Chromatin regulatory mechanisms in pluripotency. Annu Rev Cell Dev Biol. 2010;26:503–532. doi: 10.1146/annurev-cellbio-051809-102012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CH, Jackson AL, Guo J, Linsley PS, Eisenman RN. Myc-regulated microRNAs attenuate embryonic stem cell differentiation. Embo J. 2009a;28:3157–3170. doi: 10.1038/emboj.2009.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CH, Lin C, Tanaka H, Fero ML, Eisenman RN. Gene regulation and epigenetic remodeling in murine embryonic stem cells by c-Myc. PLoS One. 2009b;4:e7839. doi: 10.1371/journal.pone.0007839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maherali N, Sridharan R, Xie W, Utikal J, Eminli S, Arnold K, Stadtfeld M, Yachechko R, Tchieu J, Jaenisch R, et al. Directly reprogrammed fibroblasts show global epigenetic remodeling and widespread tissue contribution. Cell Stem Cell. 2007;1:55–70. doi: 10.1016/j.stem.2007.05.014. [DOI] [PubMed] [Google Scholar]
- Margueron R, Reinberg D. The polycomb complex PRC2 and its mark in life. Nature. 2011;469:343–349. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marson A, Levine SS, Cole MF, Frampton GM, Brambrink T, Johnstone S, Guenther MG, Johnston WK, Wernig M, Newman J, et al. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell. 2008;134:521–533. doi: 10.1016/j.cell.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa M, Takizawa N, Narita M, Ichisaka T, Yamanaka S. Promotion of direct reprogramming by transformation-deficient Myc. Proc Natl Acad Sci USA. 2010;107:14152–14157. doi: 10.1073/pnas.1009374107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro P, Avner P. When X-inactivation meets pluripotency: an intimate rendezvous. FEBS Lett. 2009;583:1721–1727. doi: 10.1016/j.febslet.2009.03.043. [DOI] [PubMed] [Google Scholar]
- Polo JM, Hochedlinger K. When fibroblasts MET iPSCs. Cell Stem Cell. 2010;7:5–6. doi: 10.1016/j.stem.2010.05.018. [DOI] [PubMed] [Google Scholar]
- Savarese F, Davila A, Nechanitzky R, De La Rosa-Velazquez I, Pereira CF, Engelke R, Takahashi K, Jenuwein T, Kohwi-Shigematsu T, Fisher AG, et al. Satb1 and Satb2 regulate embryonic stem cell differentiation and Nanog expression. Genes Dev. 2009;23:2625–2638. doi: 10.1101/gad.1815709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savarese F, Flahndorfer K, Jaenisch R, Busslinger M, Wutz A. Hematopoietic precursor cells transiently reestablish permissiveness for X inactivation. Mol Cell Biol. 2006;26:7167–7177. doi: 10.1128/MCB.00810-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaniel C, Ang YS, Ratnakumar K, Cormier C, James T, Bernstein E, Lemischka I, Paddison P. Smarcc1/Baf155 couples self-renewal gene repression with changes in chromatin structure in mouse embryonic stem cells. Stem Cells. 2009;27:2929–2991. doi: 10.1002/stem.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki Y, Kurisaki A, Watanabe-Susaki K, Nakajima Y, Nakanishi M, Arai Y, Shiota K, Sugino H, Asashima M. TIF1beta regulates the pluripotency of embryonic stem cells in a phosphorylation-dependent manner. Proc Natl Acad Sci USA. 2010;107:10926–10931. doi: 10.1073/pnas.0907601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva J, Nichols J, Theunissen TW, Guo G, van Oosten AL, Barrandon O, Wray J, Yamanaka S, Chambers I, Smith A. Nanog is the gateway to the pluripotent ground state. Cell. 2009;138:722–737. doi: 10.1016/j.cell.2009.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadfeld M, Hochedlinger K. Induced pluripotency: history, mechanisms, and applications. Genes and Development. 2010;24:2239–2263. doi: 10.1101/gad.1963910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtfeld M, Maherali N, Breault D, Hochedlinger K. Defining molecular cornerstones during fibroblast to iPS cell reprogramming in mouse. Cell Stem Cell. 2008;2:230–240. doi: 10.1016/j.stem.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo E, Rampalli S, Risueno R, Schnerch A, Mitchell R, Fiebig-Comyn A, Levadoux-Martin M, Bhatia M. Direct conversion of human fibroblasts to multilineage blood progenitors. Nature. 2010;468:521–526. doi: 10.1038/nature09591. [DOI] [PubMed] [Google Scholar]
- Wang J, Rao S, Chu J, Shen X, Levasseur DN, Theunissen TW, Orkin SH. A protein interaction network for pluripotency of embryonic stem cells. Nature. 2006;444:364–368. doi: 10.1038/nature05284. [DOI] [PubMed] [Google Scholar]
- Wang Y, Baskerville S, Shenoy A, Babiarz JE, Baehner L, Blelloch R. Embryonic stem cell-specific microRNAs regulate the G1-S transition and promote rapid proliferation. Nat Genet. 2008;40:1478–1483. doi: 10.1038/ng.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wutz A, Jaenisch R. A shift from reversible to irreversible X inactivation is triggered during ES cell differentiation. Mol Cell. 2000;5:695–705. doi: 10.1016/s1097-2765(00)80248-8. [DOI] [PubMed] [Google Scholar]
- Xu C, Bian C, Yang W, Galka M, Ouyang H, Chen C, Qiu W, Liu H, Jones A, MacKenzie F, et al. Binding of different histone marks differentially regulates the activity and specificty of polycomb repressive complex 2 (PRC2) Proc Natl Acad Sci USA. 2010;107:19266–19271. doi: 10.1073/pnas.1008937107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu N, Papagiannakopoulos T, Pan G, Thomson JA, Kosik KS. MicroRNA-145 regulates OCT4, SOX2, and KLF4 and represses pluripotency in human embryonic stem cells. Cell. 2009;137:647–658. doi: 10.1016/j.cell.2009.02.038. [DOI] [PubMed] [Google Scholar]
- Young R. Control of embryonic stem cell state. Cell. 2011;144:940–954. doi: 10.1016/j.cell.2011.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan W, Xu M, Huang C, Liu N, Chen S, Zhu B. H3K36 methylation antagonizes PRC2 mediated H3K27 methylation. J Biol Chem. 2011;286:7983–7989. doi: 10.1074/jbc.M110.194027. [DOI] [PMC free article] [PubMed] [Google Scholar]


