Abstract
The high prevalence of cartilage diseases and limited treatment options create a significant biomedical burden. Due to the inability of cartilage to regenerate itself, introducing chondrocyte progenitor cells to the affected site is of significant interest in cartilage regenerative therapies. Tissue engineering approaches using human mesenchymal stem cells (MSCs) are promising due to their chondrogenic potential, but a comprehensive understanding of the mechanisms governing the fate of MSCs is required for precise therapeutic applications in cartilage regeneration. TGF-β is known to induce chondrogenesis by activating SMAD signaling pathway and upregulating chondrogenic genes such as SOX9; however, the epigenetic regulation of TGF-β-mediated chondrogenesis is not understood. In this report, we found that TGF-β dramatically induced the expression of KDM4B in MSCs. When KDM4B was overexpressed, chondrogenic differentiation was significantly enhanced while KDM4B depletion by shRNA led to a significant reduction in chondrogenic potential. Mechanistically, upon TGF-β stimulation, KDM4B was recruited to the SOX9 promoter, removed the silencing H3K9me3 marks, and activated the transcription of SOX9. Furthermore, KDM4B depletion reduced the occupancy of SMAD3 in the SOX9 promoter, suggesting that KDM4B is required for SMAD-dependent coactivation of SOX9. Our results demonstrate the critical role of KDM4B in the epigenetic regulation of TGF-β-mediated chondrogenic differentiation of MSCs. Since histone demethylases are chemically modifiable, KDM4B may be a novel therapeutic target in cartilage regenerative therapy.
Keywords: Transforming growth factor-β, KDM4B, Chondrogenesis, SOX9, Mesenchymal stem cells, Differentiation
Introduction
Human mesenchymal stem cells (MSCs) are progenitor cells with self-renewal capacity and differentiation potential into distinct cell lineages, including osteoblasts, chondrocytes, and adipocytes [1–3]. These unique characteristics of MSCs make them a promising tool in regenerative medicine [4]. MSC-mediated cartilage regeneration is of particular interest because: (a) articular cartilage is an avascular tissue with limited regenerative stem cells; (b) it cannot repair injuries to itself spontaneously; and (c) harvesting valuable cartilaginous tissue from unaffected areas may cause additional injury to an otherwise healthy site [5, 6]. Occasional success in cartilage regeneration was reported in which autologous MSCs were expanded ex vivo and reimplanted via collagen gel into areas of articular cartilage defects in osteoarthritis patients [7]. However, the use of MSCs in cartilage repair remains limited in clinical settings and requires further molecular insight into MSC lineage commitment to chondrogenesis.
Although MSCs used in cell therapies are mostly isolated from adult bone marrow, harvesting MSCs from bone marrow is an invasive procedure with potential side effects for the donor [8]. Moreover, bone marrow stem cells (BMSCs) have a restricted proliferative capacity in culture [8]. Due to these shortcomings, alternative sources of MSCs have been extensively researched including MSCs from umbilical cord, fat tissue, and craniofacial tissues [1]. However, such MSCs from adult tissues are heterogeneous and variable among donors, and isolation of homogenous populations of MSCs has been challenging [9]. To overcome these issues, human embryonic stem cell-derived MSCs (ES-MSCs) are being used as a possible alternative source for MSCs [2, 10]. ES-MSCs display similar biological and functional phenotypes as BMSCs but with higher osteogenic potential, comparable chondrogenic potential, and higher proliferation rate with less immunogenicity [10]. As such, ES-MSCs may provide a promising alternative source for MSCs to be used in bone and cartilage engineering.
Transforming growth factor β (TGF-β) is a potent regulator of chondrocyte proliferation and differentiation. The underlying mechanisms of TGF-β-mediated chondrogenesis are the subject of extensive research [11, 12]. TGF-β signaling, mediated by specific sets of SMAD transducers, has important functions in the regulation of cell proliferation, differentiation, apoptosis, and migration, as well as the synthesis and degradation of extracellular matrix [13]. Generally, upon binding of TGF-β, type II receptor, TGF-β-RII, recruits and phosphorylates TGF-β-RI, leading to phosphorylation and activation of SMAD2 or SMAD3 (SMAD2/3) [14]. Phosphorylated SMAD2/3 combines with co-SMAD (SMAD4), and the SMAD heteromeric complex translocates into the nucleus where it regulates the transcription of target genes by directly binding to the SMAD-binding element (SBE) on DNA sequence [14–16]. One of these target genes is SOX9, which is the master chondrogenic transcription factor expressed in all chondrocyte progenitors and chondrocytes [17]. SOX9, an HMG-domain transcription factor, regulates other chondrocyte-specific marker genes such as Collagen2a1 (COL2A1), Collagen1a2 (COL1A2), and Aggrecan (ACAN) [18]. SOX9, SOX5, and SOX6, collectively referred as the SOX trio, cooperatively upregulate the expression of type II, IX, and XI collagens and Aggrecan [19]. This increased expression promotes early stages of chondrocyte differentiation and enhances the transactivation ability of SOX9 as a homodimer [20, 21]. While the molecular regulation mechanisms of chondrogenesis at the transcriptional level are well-understood, the epigenetic regulation of these chondrogenic genes remains elusive.
Epigenetic mechanisms regulate gene expression by modifying the chromatic architecture and the accessibility of genes while leaving the primary nucleotide sequence untouched [22–24]. Epigenetic mechanisms work in conjunction with genetic mechanisms to determine the fate of MSCs [25–27]. Histone demethylases, one of many epigenetic machineries, have demonstrated their significant roles in MSC fate determination by removing gene silencing epigenetic marks on lineage-specific genes to facilitate differentiation toward a particular lineage [28–30]. We previously reported that KDM4B, a Jumonji-C domain containing histone demethylase, is a major epigenetic regulator in MSC osteogenic differentiation [31]. KDM4B selectively demethylates H3K9me3, an epigenetic mark associated with gene repression, to H3K9me1 while leaving H3K9me2 untouched [29, 32–34]. By removing H3K9me3 at pericentric heterochromatin and activating osteogenic gene transcription, KDM4B plays a critical role in directing osteogenic differentiation in MSCs [32]. Less is known however, about the role of KDM4B in chondrogenic differentiation. Therefore, we examined whether KDM4B also plays a role in TGF-β-mediated chondrogenesis.
Experimental Procedures
Cell Isolation and Culture
Primary human BMSCs were purchased from Texas A&M Health Science Center College of Medicine Institute for Regenerative Medicine at Scott & White Hospital. Culturing of ES-related work was approved by UCLA Embryonic stem cells research oversight (ESCRO) committee (IRB: 10-001711-CR-00001). H1 Human ESCs (WA-01, XY) were obtained from UCLA Broad Stem Cell Research Center. Human ESCs (passages 35–45) were cultured as cell aggregates on a mitotically inactivated mouse embryonic feeder layer, as previously described [35]. To induce differentiation of human ESCs, ESC aggregates were detached by type IV collagenase (1 mg/ml, Invitrogen, Carlsbad, CA, http://www.invitrogen.com) and were cultured in suspension for 7 days to generate embryoid bodies (EBs). Then the EBs were plated into tissue culture dishes, and grown in Dulbecco’s modified Eagle’s medium (DMEM), containing 15% fetal bovine serum (FBS), 1% l-glutamine, 1% nonessential amino acid, and 1% Penicillin-Streptomycin (MSC medium, all from Invitrogen). After three to five passages, these cells were sorted for MSC surface markers, CD73, CD90, and CD146 using flow cytometry [2, 36] (Fig. 1). Cell gating was based on comparison with isotype negative controls. Antibodies used include allophycocyanin-conjugated CD73, fluorescein isothiocyanate-conjugated CD90, and phycoerythrin-conjugated CD146 antibody (all from Biolegend, San Diego, CA, http://www.biolegend.com). The chondrogenic inducing medium consisted of high glucose DMEM supplemented with 10% FBS, 100nM sodium pyruvate, 40 µg/ml proline, 100 nM dexamethasone, 200 µM ascorbic acid (Sigma-Aldrich, St. Louis, MO, http://www.sigmaaldrich.com), and 10 ng/ml TGFβ3 (R&D, Minneapolis, MN, http://www.rndsystems.com). Media was changed every 2 days.
Figure 1.
Immunophenotype of embryonic stem cell-derived mesenchymal stem cell (ES-MSCs) by fluorescence activated cell sorting. (A–C): Histogram representing the immunophenotype of the ES-MSCs using the following positive markers: CD73 (A), CD90 (B), CD146 (C). The cells were stained with isotype controls or allophycocyanin-conjugated CD73 antibody, fluorescein isothiocyanate-conjugated CD90 antibody, and phycoerythrin-conjugated CD146 antibody, respectively. (D): Representative scatter plots showing CD73+/CD90+/CD146 + MSCs which make up 10.3% of the unsorted population. Abbreviations: SSC, side scatter.
Virus Preparation and Infection
Viral packaging was prepared as described previously [37, 38]. For viral infection, MSCs were plated overnight and then infected with lentiviruses or retroviruses in the presence of polybrene (6 µg/ml; Sigma-Aldrich) for 6 hours. The cells were then selected with puromycin for 3 days. Resistant clones were pooled and knockdown/overexpression was confirmed via real-time reverse transcriptase-polymerase chain reaction (real-time RT-PCR) or Western blot analysis. The target sequences for short hairpin RNA (shRNA) were: KDM4Bsh1, 5′-CCTGCCTCTAGGTTCATAA-3′; and KDM4Bsh3, 5′-GCTACGAAGTGAACTTCGA-3′.
Alcian Blue Staining
To induce MSC differentiation, MSCs were grown in chondrogenic-inducing media. Media was changed every 2 days. After 3 weeks of differentiation in vitro, Alcian blue staining (Sigma-Aldrich) was performed as previously described. For quantification, stained Alcian blue was eluted with 6M guanidine HCl for 6 hours at room temperature. The optical absorbance was measured at 650 nm using a microplate reader.
Real-time RT-PCR
Total RNA was isolated from MSCs using Trizol reagents (Invitrogen). Two microgram aliquots of RNA were synthesized using random hexamers and reverse transcriptase according to the manufacturer’s protocol (Invitrogen). Real-time RT-PCR reactions were performed using the QuantiTect SYBR Green PCR kit (Qiagen, Chatsworth, CA, http://www1.qiagen.com) and the Icycler iQ Multi-color Real-time PCR Detection System. KDM4B mRNA levels were assessed after 8 hours of induction with either chondrogenic media or TGF-β. Chondrogenic marker gene expression levels were evaluated after 2 weeks of chondrogenic induction. The primers for COL2A1 were: forward, 5′-CTCCTGGAGCATCTGGAGAC-3′; reverse: 5′-ACCACGATCACCCTTGACTC-3′. The primers for SOX9 were: forward, 5′-TTCTGGGAATGTTTCAGCAG; reverse: 5′-GTTCTGAGAGGCACAGGTGA-3′. The primers for ACAN were: forward, 5′-TCGAGGACAGCGAGGCC-3′; reverse, 5′-TCGAGGGTGTAGCGTGTAGAGA-3′. The primers for KDM4B were: forward, 5′-CGGGTTCTATCTTTGTTTCTCTCACCCG-3′; reverse, 5′-AAGGAAGCCTCTGGAACACCTG-3′.
Chromatin Immunoprecipitation Assays
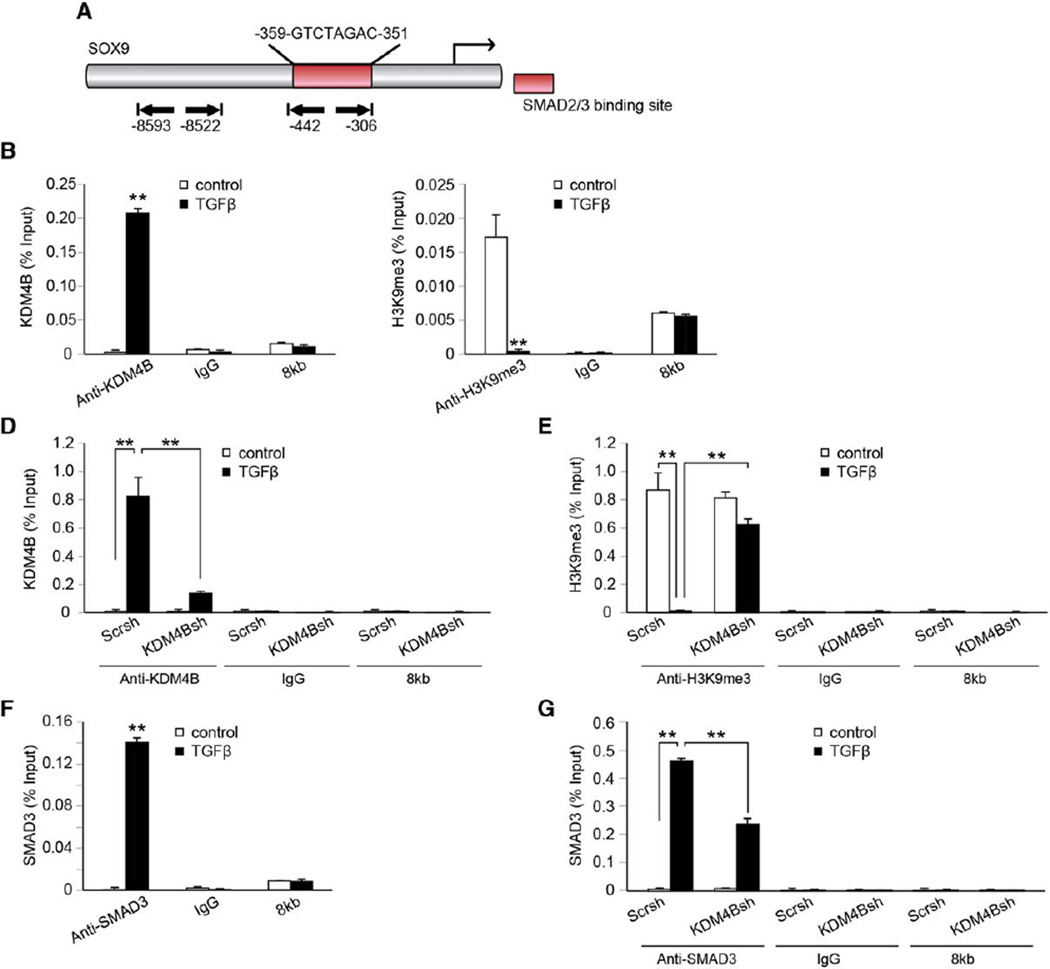
The chromatin immunoprecipitation (ChIP) assay was performed using a ChIP DNA extraction kit (Upstate, Charlottesville, VA, http://www.upstate.com) according to the manufacturer’s protocol as described previously [37]. Cells were incubated with a dimethyl 3,3′ dithiobispropionimidate-HCl solution (5 mmol; Pierce, Rockford, IL, http://www.piercenet.com) for 10 minutes at room temperature, followed by formaldehyde treatment for 15 minutes in a 37 °C water bath. For each ChIP reaction, 2 × 106 cells were used. All resulting precipitated DNA samples were quantified with real-time PCR. Data were expressed as a percentage of input DNA. Antibodies for ChIP assays were purchased from the following commercial sources: monoclonal anti-SMAD3 (Cell Signaling, Danvers, MA, http://www.cellsignal.com); polyclonal anti-KDM4B (Millipore, Billerica, MA, http://www.millipore.com); polyclonal anti-H3K9me3 (Abcam, Cambridge, U.K., http://www.abcam.com). The promoter analysis at the SOX9 promoter region revealed putative SMAD2/3 binding sites from −359 to −351 (Fig. 6A). Based on this information, we designed the ChIP primer sequence to evaluate the binding of KDM4B to the SOX9 promoter region. The primers for SOX9 were: forward, 5′-CGAATACTGCAAACTCCAGCT-3′; reverse, 5′-CGAATCTTGTGTGTGTGTGTGTG-3′. The primers for 8kb were: forward, 5′-GCATAAGGAGCCATCTGCCA-3′; reverse, 5′-ACAGTGGGACATCTTGATCACT-3′.
Figure 6.
KDM4B is required for SOX9 expression in mesenchymal stem cells (MSCs) by removal of H3K9me3 marks. (A): Schematics of SOX9 promoter denoting chromatin immunoprecipitation-polymerase chain reaction amplified region (−442 bp to −306 bp) encompassing the SMAD binding element and the control region 8 kb upstream of the transcription start site (−8,593 bp to −8,522 bp). (B): KDM4B occupancy at SOX9 promoter, as determined by ChIP, following TGF-β treatment. (C): Occupancy of H3K9me3 at the SOX9 promoter following TGF-β treatment. (D): Knockdown of KDM4B inhibited the TGF-β-induced recruitment of KDM4B. (E): Knockdown of KDM4B suppressed the removal of H3K9me3 at the SOX9 promoter following TGF-β treatment. (F): SMAD3 occupancy at the SOX9 promoter after TGF-β treatment. (G): TGF-β promoted SMAD3 recruitment to SOX9 promoter, but this recruitment was inhibited by KDM4B depletion. For all panels, cells were treated with 10 ng/ml TGF-β for 8 hours. **, p < .01. Abbreviation: TGF-β, transforming growth factor-β.
Statistical Analysis
All of the quantitative data was represented as the mean±SD. Each experiment was performed with a sample number of 3 to 4 and repeated at least twice. The results from the representative experiment were presented. Statistical significance was analyzed by Student’s t test (α50.05). A p value * less than .05 or p value ** less than .01 were considered statistically significant.
Results
Induction of KDM4B by TGF-β in MSCs
TGF-β, a potent regulator of chondrocyte proliferation and differentiation, induces MSCs to undergo chondrogenesis in vitro [2, 11, 12]. We investigated the potential role of the histone demethylase, KDM4B, in TGF-β-induced chondrogenic differentiation of MSCs. To ensure the purity of our MSCs, we used immune-phenotyping to detect cell surface markers and isolated MSCs based on the expression of CD73, CD90, and CD146 by fluorescence-activated cell sorting (FACS) analysis (Fig. 1A–1C) [2, 36]. FACS revealed that 10.5% of ES-MSCs were CD73+/CD90+/CD146+ (Fig. 1D). Upon treatment with chondrogenic inducing media containing TGF-β, ES-MSCs underwent chondrogenic differentiation as evidenced by Alcian blue staining after prolonged treatment for 21 days (Fig. 2A). Additionally, the expression of chondrogenic marker genes, COL2A1, ACAN, and SOX9, was evaluated at 0, 3, 7, 14 days of chondrogenic induction, and the expression was upregulated in a time-dependent manner with the greatest increase at 14 days of chondrogenic differentiation (Fig. 2B). Consistently, protein levels of SOX9 and ACAN were significantly elevated after 14 days of chondrogenic differentiation (Fig. 2C).
Figure 2.
TGF-β increased KDM4B expression in ES-MSCs and BMSCs. (A, F): Alcian blue staining of ES-MSCs (A) and BMSCs (F) after 3-week treatment of chondrogenic media. (B, G): real-time RT-PCR of chondrogenic marker genes at 0, 3, 7, 14 days of chondrogenic media induction in ES-MSCs. (B) and 14 days of chondrogenic media induction in BMSCs (G). (C): Western blot analysis of chondrogenic genes at 14 days of chondrogenic differentiation. (D): KDM4B mRNA level following media induction for up to 24 hours. (E, H): KDM4B mRNA levels in ES-MSCs (E) and BMSCs (H) following TGF-β treatment (10 ng/ml). **, p < .01. Abbreviations: BMSC, bone marrow stem cells; ES-MSC, embryonic stem cell-derived mesenchymal stem cell; TGF-β, transforming growth factor-β.
Previously, we found that KDM4B played an important role in MSC cell fate determination toward osteogenic differentiation [31]. Here, we examined if KDM4B could play a role in chondrogenesis. Indeed, KDM4B mRNA expression was significantly upregulated in a time-dependent manner during chondrogenic induction (Fig. 2D), with the most significant upregulation by 90-fold occurring after 8 hours of induction (Fig. 2D). In addition, to determine whether KDM4B was involved specifically in TGF-β-mediated chondrogenesis, we examined the expression of KDM4B in ES-MSCs upon TGF-β treatment. Our results showed that TGF-β alone induced KDM4B expression to a similar degree as chondrogenic induction media, showing up to a 75-fold increase at 8 hours after treatment (Fig. 2E). Consistently, similar upregulation of KDM4B during chondrogenesis was observed in BMSCs (Fig. 2F–2H), suggesting that KDM4B likely plays an important role in TGF-β mediated chondrogenesis in human MSCs.
KDM4B is Required for Chondrogenic Differentiation of MSCs
To further investigate the direct role of KDM4B in chondrogenesis of MSCs, we overexpressed Flag-tagged KDM4B using a retroviral construct. Overexpression of KDM4B significantly enhanced ES-MSCs’ chondrogenic potential as demonstrated by Alcian blue staining (Fig. 3B). In addition, the overexpression of KDM4B significantly enhanced gene expression of COL2A1, ACAN, and SOX9 (Fig. 3C). These results suggest that KDM4B promotes chondrogenic differentiation by upregulating the expression of multiple chondrogenic marker genes.
Figure 3.
Overexpressed KDM4B enhances chondrogenic differentiation of ES-MSCs. (A): KDM4B mRNA level in ES-MSCs ectopically expressing FLAG-tagged KDM4B (ES-MSC/flag-KDM4B) compared to empty vector control (ES-MSC/V). (B): Alcian blue staining of ES-MSC/flag-KDM4B and control ES-MSC/V cells after 3 weeks of chondrogenic medium induction. (C): Real-time reverse transcriptase polymerase chain reaction of chondrogenic marker genes at 14 days of chondrogenic media induction in ESMSC. **, p < .01. Abbreviation: ES-MSC, embryonic stem cell-derived mesenchymal stem cell.
Conversely, to examine whether KDM4B is required for the chondrogenic differentiation of MSCs, we knocked down KDM4B in ES-MSCs using a lentivirus expressing KDM4B shRNAs that targets the 3′ untranslated region (UTR) of KDM4B mRNA. To rule out off-target effects of shRNA, we used two different shRNA sequences (KDM4Bsh1 and KDM4Bsh3), both of which resulted in efficient depletion of KDM4B in MSCs as confirmed by real-time RT-PCR (Fig. 4A). When ES-MSCs were induced to undergo chondrogenic differentiation, ES-MSC/KDM4Bsh1 and ES-MSC/ KDM4Bsh3 cells lost their capacity to differentiate into chondrocytes, as demonstrated by Alcian blue staining (Fig. 4B). These cells also exhibited significant suppression of COL2A1, ACAN, and SOX9 gene expression upon induction of chondrogenic differentiation (Fig. 4C).Western blot analysis showed that the induction of SOX9 and ACAN is significantly reduced in KDM4B depleted cells (Fig. 4D). Consistent results were obtained using BMSCs (Fig. 4E–4G), suggesting that KDM4B is required for chondrogenic differentiation.
Figure 4.
The knockdown KDM4B inhibits chondrogenic differentiation of ES-MSCs and BMSCs. (A, E): Depletion of KDM4B in ES-MSCs (A) and BMSCs (E) via two different shRNA (-KDM4Bsh1 and -KDM4Bsh3) significantly inhibited the KDM4B induction after 8 hours of TGF-β treatment, as determined by real-time RT-PCR. (B, F): Alcian blue staining of KDM4B-depleted ES-MSCs (B) and BMSCs (F) following 3 weeks of chondrogenic medium induction. (C, G): Knockdown of KDM4B inhibited chondrogenic marker gene expression at basal levels in ES-MSCs (C) and BMSCs (G). (D): Western blot analysis of chondrogenic genes at 14 days of chondrogenic differentiation. *, p < .05; **, p < .01. Abbreviations: BMSC, bone marrow stem cells; ES-MSC, embryonic stem cell-derived mesenchymal stem cell; TGF-β, transforming growth factor-β.
To confirm the specific effects of KDM4B in chondrogenic differentiation of MSCs, we strategically restored KDM4B expression in MSCs that already express KDM4B shRNA. We introduced KDM4B in MSCs that harbor KDM4B shRNA targeting the 3′ UTR region of KDM4B mRNA. By doing so, shRNA targets only the endogenous KDM4B but not ectopically introduced KDM4B. Flag-tagged KDM4B in ES-MSC/KDM4Bsh/Flag- KDM4B cells restored KDM4B expression, as evidenced by real-time RT-PCR (Fig. 5A). When ES-MSC/KDM4Bsh/Flag-KDM4B cells were induced to undergo chondrogenic differentiation, the inhibited chondrogenic potential due to KDM4B depletion was restored as demonstrated by Alcian blue staining (Fig. 5B). Similarly, the suppressed expression of COL2A1, ACAN, and SOX9 as a result of KDM4B depletion was rescued when KDM4B was reintroduced, indicating that KDM4B specifically regulates chondrogenic differentiation of human MSCs (Fig. 5C).
Figure 5.
Rescue of KDM4B expression in KDM4B-depleted ES-MSCs restored chondrogenic potential. (A): KDM4B depletion in ESMSCs was rescued by ectopic expression of flag-KDM4B (ES-MSC/KDM4Bsh/flag-KDM4B). KDM4B mRNA expression was compared among control (ES-MSC/V), knockdown (ES-MSC/KDM4Bsh/V), and rescue (ES-MSC/KDM4Bsh/flag-KDM4B) groups, after 8 hours of TGFβ treatment. (B): Alcian blue staining of control, KDM4B-knockdown and rescue ES-MSCs. (C): Real-time reverse transcriptase polymerase chain reaction of chondrogenic marker genes at 14 days of chondrogenic media induction. **, p < .01. Abbreviations: ES-MSC, embryonic stem cell-derived mesenchymal stem cell; TGF-β, transforming growth factor-β.
KDM4B Epigenetically Regulates SOX9 Gene Expression in MSCs by Removing the H3K9me3 Mark
KDM4B is a histone demethylase that activates the target gene expressions by removing the repressive epigenetic mark, H3K9me3 [32–34, 39]. SOX9 is known to be a master regulator of chondrogenesis, and we found that SOX9 mRNA expression was regulated by KDM4B [17]. In order to examine whether KDM4B regulates MSC chondrogenic differentiation by epigenetically modulating SOX9 expression, we performed ChIP assay to assess the physical occupancy of KDM4B and determine the changes in the histone methylation status at the SOX9 promoter region (Fig. 6A).
Indeed, upon TGF-β treatment, KDM4B bound to the SOX9 promoter region (Fig. 6B) and the amount of H3K9me3 decreased (Fig. 6C), suggesting that KDM4B was recruited to the SOX9 promoter region and removed the silencing epigenetic mark, H3K9me3, thereby activating SOX9 gene expression. In ES-MSCs depleted of KDM4B, TGF-β-induced recruitment of KDM4B at the SOX9 promoter region was significantly inhibited (Fig. 6D) along with increased levels of the substrate, H3K9me3 (Fig. 6E). As a control, we could not detect KDM6B occupancy 8 kb upstream of the transcription start site, and the knockdown of KDM4B did not alter the level of H3K9me3 silencing epigenetic marks on that region (Fig. 6B–6E). Taken together, our results suggest that KDM4B mediates TGF-induced SOX9 activation through the removal of H3K9me3 from the SOX9 promoter.
TGF-β mediates chondrogenic induction via SMAD2/3 binding to the promoter of SOX9 for its gene activation [8]. We designed our ChIP primers to encompass the SBE of the SOX9 promoter (Fig. 6A). Since TGF-β induced KDM4B binding to proximity of the SBE, it is possible that KDM4B may interplay with SMAD2/3 to directly affect TGF-β-mediated SOX9 activation. Indeed, when ES-MSCs were treated with TGF-β, the occupancy of SMAD3 was increased at the SBE in the SOX9 promoter region (Fig. 6F). Conversely, when MSCs depleted of KDM4B were induced with TGF-β, along with a decrease in KDM4B binding to the SMAD-binding site, the occupancy of SMAD3 was also significantly decreased in the SOX9 promoter region (Fig. 6G). These findings indicate that KDM4B is required for recruitment of SMAD3 during TGF-β mediated SOX9 activation.
Discussion
While epigenetic machineries governing MSCs into osteogenesis and adipogenesis are becoming increasingly elucidated, the understanding of such regulation in chondrogenic differentiation remains largely elusive. In this study, we identified the critical role of a histone demethylase in chondrogenic differentiation; KDM4B removes repressive epigenetic mark, H3K9me3, from the SOX9 promoter region and activates the transcription of SOX9, a master regulator of chondrogenesis. Furthermore, we found that KDM4B is required for SMAD3 binding at the SOX9 promoter during its gene activation. Our results provide the first demonstration that the histone demethylase, KDM4B, epigenetically mediates SOX9 activation to promote TGF-β-mediated chondrogenesis, suggesting its potential value for regenerative therapy in cartilage diseases.
Few studies have shown the role of epigenetic regulators in cartilage formation, particularly in the context of modulating the expression of SOX9 and other chondrogenic genes. Histone acetyltransferases, P300 and Tip60, were found to promote chondrogenesis, while histone deacetylation by HDAC1 was shown to inhibit cartilage differentiation [40–43]. A previous study using fibroblastic cell lines showed that Arid5b recruits a histone demethylase, Phf2, to remove H3K9me2 silencing marks from the promoter of COL2A1 during chondrogenesis [44]. In osteoarthritic cartilage, the epigenetic status of the SOX9 promoter is altered with increased silencing histone methylation marks, H3K9me3 and H3K27me3, and decreased histone acetylation [45]. These findings suggest that histone acetylation and methylation appear to be two major epigenetic mechanisms by which chondrogenesis is regulated. Recently, Herlofsen et al. performed genome-wide quantitative epigenetic analysis of BMSCs undergoing chondrogenic differentiation and reported that increased expression of associated genes are most closely predicted by a high increase in H3K4me3 and H3K36me3, concluding that histone methylation, rather than DNA methylation, provides the primary epigenetic regulation of MSC chondrogenic differentiation [46]. Here in our study, we were able to illuminate the important role of a histone demethylase in the epigenetic control of the master chondrogenic gene SOX9 and provide novel insights to the dynamic control of genetic program in MSC chondrogenic differentiation.
KDM4B has been characterized to selectively remove H3K9me3, a repressive epigenetic mark, leading to target gene activation [29, 32–34]. This gene activating function of KDM4B is observed clinically in numerous cancers with overexpressed KDM4B [47–50].We previously reported that KDM4B is upregulated by bone morphogenetic protein (BMP)4/7, a potent inducer of osteogenic differentiation [31]. This BMP-induced KDM4B played a critical role in MSC osteogenic commitment by removing H3K9me3 silencing marks at the DLX5 promoter [31]. BMPs mainly induce gene expression through SMAD1/4 signaling and SMAD depletion inhibited KDM4B expression, suggesting that this BMP-mediated KDM4B induction was dependent on SMAD signaling [31]. Interestingly, we found that KDM4B is also significantly induced by TGF-β, which signals through SMAD2/3, thereby leading to chondrogenic commitment by epigenetic modulation of a different target gene, SOX9. These results suggest the complexity of the mechanistic control of KDM4B activation. Both members of TGF superfamily, BMP4/7 and TGF-β, can induce KDM4B levels signaling through different types of SMAD proteins, suggesting the central role of KDM4B in both osteogenic and chondrogenic lineage commitment. Given that KDM4B is also known to inhibit adipogenic differentiation, the multiple roles played by KDM4B in MSC cell fate determination highlight its potential in regenerative therapy [31].
Furthermore, our study showed that SMAD3 binding onto the SOX9 promoter upon TGF-β treatment is promoted by KDM4B, as KDM4B depletion resulted in decreased occupancy of SMAD3 at the SOX9 promoter. Therefore, the physical presence of SMAD3 in the nucleus is not sufficient to activate SOX9 expression, but additional regulatory mechanisms involving KDM4B are required. Removing H3K9me3 on the SOX9 promoter by KDM4B might facilitate the recruitment of SMAD2/3 and other transcriptional coactivators to the SOX9 promoter. Additionally, KDM4B might be in direct interaction with SMAD proteins and mutually enhances their binding to the SOX9 promoter. The induction of KDM4B by TGF-β might form a positive feedback loop to activate SOX9 transcription. These results emphasize the importance of an epigenetic component in the programming of cell fate, as determined by the delicate orchestration of gene activation and repression at specific time points, all directed toward a terminally differentiated phenotype of MSCs.
In this study, we provide insights into TGF-β-stimulated MSC chondrogenic differentiation. Our findings support the following model: SMAD2/3 is activated with TGF-β treatment and forms a heteromeric complex with SMAD4, which then translocates into the nucleus (Fig. 7). Concurrently, TGFβ- induced KDM4B also binds to the SBE and removes H3K9me3 from the SOX9 promoter, thereby promoting SMAD2/3 binding to the SOX9 promoter. Based on the established association between p300 and SMAD2/3 on the SOX9 promoter, KDM4B might coordinate with p300 to modulate both the histone methylation and acetylation status of the SOX9 promoter [40, 42]. Future studies will aim to elucidate in greater depth of the complex interaction between KDM4B and other epigenetic regulators in the chondrogenesis of MSCs.
Figure 7.
KDM4B may be a crucial regulator in TGF-β-directed chondrogenesis. Model illustrating KDM4B induction by TGF-β and its recruitment to SOX9 promoter. TGF-β stimulation also leads to phosphorylation and activation of SMAD 2/3, which in turn bind with SMAD4 and translocate into the nucleus. TGF-β-induced KDM4B also binds to the SBE and removes H3K9me3 from the SOX9 promoter to activate the transcription of SOX9. KDM4B likely facilitates recruitment of SMAD2/3 via complex formation and with removal of H3K9me3. Abbreviation: TGF-β, transforming growth factor.
Cartilage engineering with MSCs is highly relevant in clinical settings due to the limited ability of avascular cartilage tissue to regenerate and repair itself. The therapeutic function of MSCs relies on the intricate mechanisms that control the fate and delineation of these multipotent cells [51–54]. Our findings revealed that KDM4B stimulated chondrogenic differentiation by epigenetically regulating the SOX9 expression of the TGF-β pathway. Further understanding of the pathways that MSCs undergo when differentiating into chondrocytes can help guide MSCs to make lineage-specific decisions for therapeutic applications. In addition, while epigenetic drugs are currently limited to DNA methyltransferases and histone deacetylases, our novel finding that KDM4B is an important epigenetic link in directing MSC fate into chondrogenesis and modulating the SOX9 gene may have innovative clinical implications for the treatment of human cartilage and other diseases.
Conclusion
Our results demonstrate for the first time that the histone demethylase, KDM4B, coordinates with SMAD2/3 to epigenetically activate SOX9 transcription during TGF-β-mediated chondrogenesis. These findings provide an important epigenetic link in directing MSC fate toward chondrogenesis, thereby creating a foundation for future studies and revealing KDM4B as a potential new target in cartilage regenerative therapy.
Significance Statement.
Given the high prevalence of cartilage diseases and the challenges with cartilage regeneration, it is critical to identify molecular pathways and regulators to improve therapeutic options for cartilage regeneration. This study identifies a key epigenetic regulator, histone demethylase KDM4B, in promoting chondrogenic differentiation of mesenchymal stem cells. Mechanistically, KDM4B removes a repressive epigenetic mark, H3K9me3, from the SOX9 promoter and induces the transcription of SOX9, a master regulator of chondrogenesis. These findings are significant in stem cell biology as KDM4B may serve as a novel therapeutic agent in MSC-mediated cartilage regenerative medicine.
Acknowledgments
This work was supported by NIH/NIDCR grants K08DE024603-01 (C.H.), R01DE16513, and R01AR063089 (C.Y.W.).
Footnotes
Author Contributions
C.-Y.W. and C.H.: conception and design, financial support, and administrative support; B.Y. and P.D.: provision of study material or patients; H-L.L., B.Y., P.D., and C.H.: collection and/or assembly of data; H-L.L., C-Y.W., and C.H.: data analysis and interpretation; H-L.L.., B.Y., P.D., C.-Y.W., and C.H.: manuscript writing; C-Y.W.: final approval of manuscript.
Disclosure Of Potential Conflicts Of Interest
The authors indicate no potential conflicts of interest.
References
- 1.Salem HK, Thiemermann C. Mesenchymal stromal cells: Current understanding and clinical status. Stem Cells. 2010;28:585–596. doi: 10.1002/stem.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arpornmaeklong P, Brown SE, Wang Z, et al. Phenotypic characterization, osteoblastic differentiation, and bone regeneration capacity of human embryonic stem cell-derived mesenchymal stem cells. Stem Cells Dev. 2009;18:955–968. doi: 10.1089/scd.2008.0310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kolf CM, Cho E, Tuan RS. Mesenchymal stromal cells. Biology of adult mesenchymal stem cells: regulation of niche, self-renewal and differentiation. Arthritis Res Ther. 2007;9:204. doi: 10.1186/ar2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Quarto R, Mastrogiacomo M, Cancedda R, et al. Repair of large bone defects with the use of autologous bone marrow stromal cells. N Engl J Med. 2001;344:385–386. doi: 10.1056/NEJM200102013440516. [DOI] [PubMed] [Google Scholar]
- 5.Buckwalter JA. Articular cartilage: Injuries and potential for healing. J Orthop Sports Phys Ther. 1998;28:192–202. doi: 10.2519/jospt.1998.28.4.192. [DOI] [PubMed] [Google Scholar]
- 6.Risbud MV, Sittinger M. Tissue engineering: Advances in in vitro cartilage generation. Trends Biotechnol. 2002;20:351–356. doi: 10.1016/s0167-7799(02)02016-4. [DOI] [PubMed] [Google Scholar]
- 7.Wakitani S, Imoto K, Yamamoto T, et al. Human autologous culture expanded bone marrow mesenchymal cell transplantation for repair of cartilage defects in osteoarthritic knees. Osteoarthritis Cartilage. 2002;10:199–206. doi: 10.1053/joca.2001.0504. [DOI] [PubMed] [Google Scholar]
- 8.Charbord P. Bone marrow mesenchymal stem cells: Historical overview and concepts. Hum Gene Ther. 2010;21:1045–1056. doi: 10.1089/hum.2010.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Indumathi S, Harikrishnan R, Rajkumar JS, et al. Immunophenotypic comparison of heterogenous non-sorted versus sorted mononuclear cells from human umbilical cord blood: A novel cell enrichment approach. Cytotechnology. 2015;67:107–114. doi: 10.1007/s10616-013-9663-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li O, Tormin A, Sundberg B, et al. Human embryonic stem cell-derived mesenchymal stroma cells (hES-MSCs) engraft in vivo and support hematopoiesis without suppressing immune function: implications for off-the shelf ES-MSC therapies. PLoS One. 2013;8:e55319. doi: 10.1371/journal.pone.0055319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Im GI, Jung NH, Tae SK. Chondrogenic differentiation of mesenchymal stem cells isolated from patients in late adulthood: the optimal conditions of growth factors. Tissue Eng. 2006;12:527–536. doi: 10.1089/ten.2006.12.527. [DOI] [PubMed] [Google Scholar]
- 12.Lorda-Diez CI, Montero JA, Martinez-Cue C, et al. Transforming growth factors beta coordinate cartilage and tendon differentiation in the developing limb mesenchyme. J Biol Chem. 2009;284:29988–29996. doi: 10.1074/jbc.M109.014811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tayalia P, Mooney DJ. Controlled growth factor delivery for tissue engineering. Adv Mater. 2009;21:3269–3285. doi: 10.1002/adma.200900241. [DOI] [PubMed] [Google Scholar]
- 14.Heldin CH, Miyazono K, ten Dijke P. TGF-beta signalling from cell membrane to nucleus through SMAD proteins. Nature. 1997;390:465–471. doi: 10.1038/37284. [DOI] [PubMed] [Google Scholar]
- 15.Derynck R, Zhang YE. Smad-dependent and Smad-independent pathways in TGF-beta family signalling. Nature. 2003;425:577–584. doi: 10.1038/nature02006. [DOI] [PubMed] [Google Scholar]
- 16.Sporn MB, Roberts AB. The transforming growth factor-betas: Past, present, and future. Ann N Y Acad Sci. 1990;593:1–6. doi: 10.1111/j.1749-6632.1990.tb16095.x. [DOI] [PubMed] [Google Scholar]
- 17.Ng LJ, Wheatley S, Muscat GE, et al. SOX9 binds DNA, activates transcription, and coexpresses with type II collagen during chondrogenesis in the mouse. Dev Biol. 1997;183:108–121. doi: 10.1006/dbio.1996.8487. [DOI] [PubMed] [Google Scholar]
- 18.Bi W, Deng JM, Zhang Z, et al. Sox9 is required for cartilage formation. Nat Genet. 1999;22:85–89. doi: 10.1038/8792. [DOI] [PubMed] [Google Scholar]
- 19.Lefebvre V, Behringer RR, de Crombrugghe B. L-Sox5, Sox6 and Sox9 control essential steps of the chondrocyte differentiation pathway. Osteoarthritis Cartilage. 2001;9(Suppl A):S69–S75. doi: 10.1053/joca.2001.0447. [DOI] [PubMed] [Google Scholar]
- 20.Ikeda T, Kamekura S, Mabuchi A, et al. The combination of SOX5, SOX6, and SOX9 (the SOX trio) provides signals sufficient for induction of permanent cartilage. Arthritis Rheum. 2004;50:3561–3573. doi: 10.1002/art.20611. [DOI] [PubMed] [Google Scholar]
- 21.Han Y, Lefebvre V. L-Sox5 and Sox6 drive expression of the aggrecan gene in cartilage by securing binding of Sox9 to a far-upstream enhancer. Mol Cell Biol. 2008;28:4999–5013. doi: 10.1128/MCB.00695-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dwivedi RS, Herman JG, McCaffrey TA, et al. Beyond genetics: Epigenetic code in chronic kidney disease. Kidney Int. 2011;79:23–32. doi: 10.1038/ki.2010.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Galm O, Esteller M. Beyond genetics— The emerging role of epigenetic changes in hematopoietic malignancies. Int J Hematol. 2004;80:120–127. doi: 10.1532/ijh97.04075. [DOI] [PubMed] [Google Scholar]
- 24.Li YY. Genetic and epigenetic variants influencing the development of nonalcoholic fatty liver disease. World J Gastroenterol. 2012;18:6546–6551. doi: 10.3748/wjg.v18.i45.6546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aranda P, Agirre X, Ballestar E, et al. Epigenetic signatures associated with different levels of differentiation potential in human stem cells. PLoS One. 2009;4:e7809. doi: 10.1371/journal.pone.0007809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hermann A, Maisel M, Storch A. Epigenetic conversion of human adult bone mesodermal stromal cells into neuroectodermal cell types for replacement therapy of neurodegenerative disorders. Expert Opin Biol Ther. 2006;6:653–670. doi: 10.1517/14712598.6.7.653. [DOI] [PubMed] [Google Scholar]
- 27.Teven CM, Liu X, Hu N, et al. Epigenetic regulation of mesenchymal stem cells: A focus on osteogenic and adipogenic differentiation. Stem Cells Int. 2011;2011:201371. doi: 10.4061/2011/201371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Agger K, Christensen J, Cloos PA, et al. The emerging functions of histone demethylases. Curr Opin Genet Dev. 2008;18:159–168. doi: 10.1016/j.gde.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 29.Klose RJ, Kallin EM, Zhang Y. JmjC-domain-containing proteins and histone demethylation. Nat Rev Genet. 2006;7:715–727. doi: 10.1038/nrg1945. [DOI] [PubMed] [Google Scholar]
- 30.Shi Y. Histone lysine demethylases: Emerging roles in development, physiology and disease. Nat Rev Genet. 2007;8:829–833. doi: 10.1038/nrg2218. [DOI] [PubMed] [Google Scholar]
- 31.Ye L, Fan Z, Yu B, et al. Histone demethylases KDM4B and KDM6B promotes osteogenic differentiation of human MSCs. Cell Stem Cell. 2012;11:50–61. doi: 10.1016/j.stem.2012.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fodor BD, Kubicek S, Yonezawa M, et al. Jmjd2b antagonizes H3K9 trimethylation at pericentric heterochromatin in mammalian cells. Genes Dev. 2006;20:1557–1562. doi: 10.1101/gad.388206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cloos PA, Christensen J, Agger K, et al. The putative oncogene GASC1 demethylates tri- and dimethylated lysine 9 on histone H3. Nature. 2006;442:307–311. doi: 10.1038/nature04837. [DOI] [PubMed] [Google Scholar]
- 34.Whetstine JR, Nottke A, Lan F, et al. Reversal of histone lysine trimethylation by the JMJD2 family of histone demethylases. Cell. 2006;125:467–481. doi: 10.1016/j.cell.2006.03.028. [DOI] [PubMed] [Google Scholar]
- 35.Choo A, Lim SK. Derivation of mesenchymal stem cells from human embryonic stem cells. Methods Mol Biol. 2011;690:175–182. doi: 10.1007/978-1-60761-962-8_12. [DOI] [PubMed] [Google Scholar]
- 36.Russell KC, Phinney DG, Lacey MR, et al. In vitro high-capacity assay to quantify the clonal heterogeneity in trilineage potential of mesenchymal stem cells reveals a complex hierarchy of lineage commitment. Stem Cells. 2010;28:788–798. doi: 10.1002/stem.312. [DOI] [PubMed] [Google Scholar]
- 37.Fan Z, Yamaza T, Lee JS, et al. BCOR regulates mesenchymal stem cell function by epigenetic mechanisms. Nat Cell Biol. 2009;11:1002–1009. doi: 10.1038/ncb1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Park BK, Zhang H, Zeng Q, et al. NF-kappaB in breast cancer cells promotes osteolytic bone metastasis by inducing osteoclastogenesis via GM-CSF. Nat Med. 2007;13:62–69. doi: 10.1038/nm1519. [DOI] [PubMed] [Google Scholar]
- 39.Klose RJ, Bird AP. Genomic DNA methylation: The mark and its mediators. Trends Biochem Sci. 2006;31:89–97. doi: 10.1016/j.tibs.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 40.Furumatsu T, Tsuda M, Yoshida K, et al. Sox9 and p300 cooperatively regulate chromatin-mediated transcription. J Biol Chem. 2005;280:35203–35208. doi: 10.1074/jbc.M502409200. [DOI] [PubMed] [Google Scholar]
- 41.Hattori T, Coustry F, Stephens S, et al. Transcriptional regulation of chondrogenesis by coactivator Tip60 via chromatin association with Sox9 and Sox5. Nucleic Acids Res. 2008;36:3011–3024. doi: 10.1093/nar/gkn150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tsuda M, Takahashi S, Takahashi Y, et al. Transcriptional co-activators CREB-binding protein and p300 regulate chondrocyte-specific gene expression via association with Sox9. J Biol Chem. 2003;278:27224–27229. doi: 10.1074/jbc.M303471200. [DOI] [PubMed] [Google Scholar]
- 43.Liu CJ, Zhang Y, Xu K, et al. Transcriptional activation of cartilage oligomeric matrix protein by Sox9, Sox5, and Sox6 transcription factors and CBP/p300 coactivators. Front Biosci. 2007;12:3899–3910. doi: 10.2741/2359. [DOI] [PubMed] [Google Scholar]
- 44.Hata K, Takashima R, Amano K, et al. Arid5b facilitates chondrogenesis by recruiting the histone demethylase Phf2 to Sox9-regulated genes. Nat Commun. 2013;4:2850. doi: 10.1038/ncomms3850. [DOI] [PubMed] [Google Scholar]
- 45.Kim K, K I, Park YS, Im GI. Changes in the epigenetic status of the SOX-9 promoter in human osteoarthritic cartilage. J Bone Miner Res. 2013;28:1050–1060. doi: 10.1002/jbmr.1843. [DOI] [PubMed] [Google Scholar]
- 46.Herlofsen SR, Bryne JC, Hoiby T, et al. Genome-wide map of quantified epigenetic changes during in vitro chondrogenic differentiation of primary human mesenchymal stem cells. BMC Genomics. 2013;14:105. doi: 10.1186/1471-2164-14-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Northcott PA, Nakahara Y, Wu X, et al. Multiple recurrent genetic events converge on control of histone lysine methylation in medulloblastoma. Nat Genet. 2009;41:465–472. doi: 10.1038/ng.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li W, Zhao L, Zang W, et al. Histone demethylase JMJD2B is required for tumor cell proliferation and survival and is overexpressed in gastric cancer. Biochem Biophys Res Commun. 2011;416:372–378. doi: 10.1016/j.bbrc.2011.11.045. [DOI] [PubMed] [Google Scholar]
- 49.Toyokawa G, Cho HS, Iwai Y, et al. The histone demethylase JMJD2B plays an essential role in human carcinogenesis through positive regulation of cyclin-dependent kinase 6. Cancer Prev Res. 2011;4:2051–2061. doi: 10.1158/1940-6207.CAPR-11-0290. [DOI] [PubMed] [Google Scholar]
- 50.Fu L, Chen L, Yang J, et al. HIF-1alpha-induced histone demethylase JMJD2B contributes to the malignant phenotype of colorectal cancer cells via an epigenetic mechanism. Carcinogenesis. 2012;33:1664–1673. doi: 10.1093/carcin/bgs217. [DOI] [PubMed] [Google Scholar]
- 51.Tang Y, Wu X, Lei W, et al. TGF-beta1-induced migration of bone mesenchymal stem cells couples bone resorption with formation. Nat Med. 2009;15:757–765. doi: 10.1038/nm.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Medici D, Shore EM, Lounev VY, et al. Conversion of vascular endothelial cells into multipotent stem-like cells. Nat Med. 2010;16:1400–1406. doi: 10.1038/nm.2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kronenberg HM. Gs signaling in osteoblasts and hematopoietic stem cells. Ann N Y Acad Sci. 2010;1192:327–329. doi: 10.1111/j.1749-6632.2009.05251.x. [DOI] [PubMed] [Google Scholar]
- 54.Huebsch N, Arany PR, Mao AS, et al. Harnessing traction-mediated manipulation of the cell/matrix interface to control stemcell fate. Nat Mater. 2010;9:518–526. doi: 10.1038/nmat2732. [DOI] [PMC free article] [PubMed] [Google Scholar]