Abstract
Purpose of review
Autophagy is an evolutionarily-conserved cellular program for the turnover of organelles, proteins, and other macromolecules, involving the lysosomal degradation pathway. Emerging evidence suggests that autophagy can play a central role in human metabolism as well as impact diverse cellular processes including organelle homeostasis, cell death and proliferation, lipid and glycogen metabolism, and the regulation of inflammation and immune responses. The purpose is this review is to examine recent evidence for the role of autophagy in cellular metabolism, and its relevance to select human diseases that involve disorders of metabolism.
Recent findings
Recent studies suggest that autophagy may play multiple roles in metabolic diseases, including diabetes and its complications, metabolic syndrome and obesity, myopathies and other inborn errors of metabolism, as well as other diseases that may involve altered mitochondrial function.
Summary
Strategies aimed at modulating autophagy may lead to therapies for diseases in which altered cellular and tissue metabolism play a key role.
Keywords: Autophagy, Diabetes, Metabolism, Metabolic Disease, Mitophagy, Myopathy, Obesity
Introduction
Macro-autophagy (abbreviated as ‘autophagy’) is an inducible cellular process that facilitates the degradation of cellular organelles (e.g., mitochondria, peroxisomes, ribosomes), proteins, and other macromolecular substrates (e.g., lipids, RNA, glycogen), and invading microorganisms)[1-2]. Autophagy may be non-selective, in reference to bulk degradation of cytosol, or highly selective, involving the identification (through ubiquitination), and targeting (by cargo adaptor proteins such as p62SQSTM1), of specific substrates for degradation [3]. This process depends on membrane components potentially originating from subcellular membranes (e.g., endoplasmic reticulum (ER), plasma membrane), which are recruited to the phagophore, and then elongated to form double-membrane compartments called autophagosomes [4]. Autophagy cargos are assimilated into autophagosomes, which subsequently fuse to lysosomes, the subcellular site where the cargos are degraded by lysosomal enzymes [5](Figure 1).
Figure 1. Autophagic Pathway.
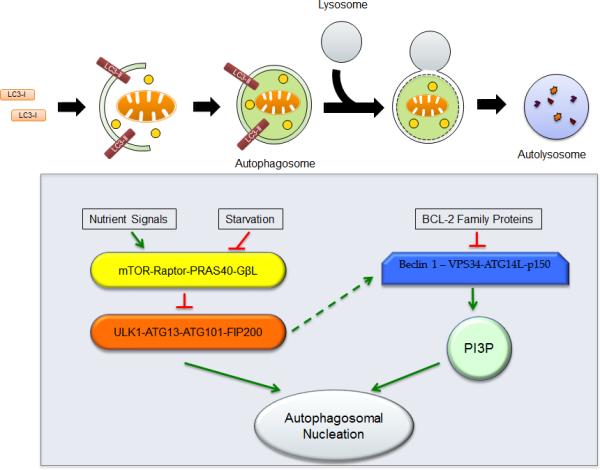
The autophagic pathway proceeds through several steps, including the initiation of autophagosome formation, the elongation and maturation of the autophagosome and the fusion of the lysosome with the autophagosome (Upper panel). Following fusion, the hydrolytic enzymes from the lysosomes break down the autophagosomal contents, and the remaining debris is then released for metabolic recycling.
The autophagy pathway responds to various stress cues and signals from both inside and outside of the cell (Lower Panel). While nutrient signals induce activity of the mammalian target of rapamycin (mTOR) signaling complex 1 (mTORC1), starvation cues directly block the activity of mTORC1. Inhibition of mTORC1 results in activation of UNC-51-like kinase 1 (ULK1), which leads to the initiating step of autophagy, while activation of mTORC1 inhibits the activity of ULK1, thereby preventing the onset of autophagy. The Beclin 1 – interactive complex, comprised of Beclin 1, class III phosphatidylinositol-3-kinase (PIK3C3, or VPS34), and ATG14L, also regulates the autophagic pathway. This complex, when stimulated, induces the activity of phosphatidylinositol-3-phosphate (PI3P), leading to autophagosomal nucleation. ULK1 may also induce the activity of the Beclin 1 – interactive complex, thereby enhancing the production of PI3P, which triggers the autophagosomal nucleation.
Autophagy can exert many diverse functions in cellular regulation, including roles in organelle homeostasis, protein degradation, programmed cell death, regulation of inflammation, as well as innate and adaptive immunity (e.g., efferocytosis, antigen presentation, antibody production, and bacterial clearance)[6-7]. Increasing evidence suggests that autophagy may influence the pathogenesis of human diseases, including cancer, neurodegenerative diseases, and diseases of the lung, liver, kidney, and cardiovascular system [2, 8-15]. Several inherited myopathies are associated with aberrant autophagy [16-17]. Recent investigations have explored the functions of autophagy in the pathogenesis of metabolic disorders such as diabetes, insulin resistance, and obesity [18-21], and the progression of aging [22-23]. This review will focus on the central role of autophagy as a partner with cellular metabolism, with emphasis on select human diseases where disturbance of ordinary metabolism plays a key role.
Molecular Regulation of Autophagy
Mammalian autophagy is regulated by a complex network of autophagy-related proteins (ATGs) that include over 30 molecular species originally identified in yeast. The pathway is highly conserved with many corresponding mammalian homologues identified [24-25]. Autophagy falls under strict regulation by nutrient and growth factor-sensitive signaling pathways, which include the mammalian target of rapamycin (mTOR), and the 5'-adenosine monophosphate (AMP)-activated protein kinase (AMPK) signaling pathways [26-27](Figure 1). Nutrient deprivation induces autophagy through inhibition of mTOR, which resides in a multi-protein complex, mTORC1. In response to stimulation by nutrients or growth factors, mTORC1 negatively regulates the ULK1/2 complex, which results in autophagy suppression [28]. Depletion of cellular energy charge, as reflected by accumulation of adenosine monophosphate (AMP), stimulates autophagy through activation of AMPK, which regulates mTORC1 and ULK1/2 [29].
Autophagy is also regulated by the Beclin 1 (Atg6) interactome, which associates with VPS34, a class III phosphatidylinositol-3-kinase (PI3KCIII) and other proteins (e.g., ATG14L or UVRAG; Ambra1, Rubicon). The Beclin 1 complex is negatively regulated by the Class I PI3K/Akt pathway, as well by anti-apoptotic Bcl-2 family proteins. The production of phosphatidylinositol-3-phosphate (PI3P) by this complex regulates autophagosome formation [30]. Recent studies have elucidated functional links between ULK1/2 and the Beclin1 complex. Phosphorylation of Ambra1 by ULK1 releases the Beclin1 complex from cytoskeletal sequestration by dyneins, permitting the initiation of autophagy [31]. Furthermore, Ser14 phosphorylation of Beclin 1 by ULK1 enhances PI3KCIII activity [32]. Autophagosome elongation is catalyzed by two ubiquitin-like conjugation systems: the Atg5-Atg12 conjugation system, and the Atg8 conjugation system. In mammals, microtubule-associated protein-1 light chain 3B (LC3B) and its related Atg8 homologues are conjugated to phosphatidylethanolamine. Subsequently, the lipidconjugated form of LC3B (i.e., LC3B-II) is incorporated in autophagosomal membranes [4, 24].
Role of Autophagy in Cellular Metabolism
By virtue of its role as a degradative process for cellular macromolecules, autophagy may play a role in cellular survival, by renewing the bioavailability of metabolic precursors during nutrient deficiency states [33]. Autophagy, which releases amino acids, lipids, and other metabolic precursors, as the result of lysosomal breakdown of complex molecules, may have a profound impact on tissue metabolism [33]. Conversely, autophagy may be upregulated by metabolic derangements that result in the accumulation of damaged or disordered macromolecules [34]. The latter may include misfolded or aggregated proteins that may accumulate in cells as a consequence of chemical or physical stress (e.g., ER stress), or as the result of mutation [35]. Furthermore, autophagy may respond to conditions that cause disruption of mitochondrial integrity and function [36].
Selective Autophagy Pathways
The selective targeting of distinct substrates to the autophagy pathway (termed selective autophagy) has been described for several distinct cargoes, including mitochondria (mitophagy), peroxisomes (pexophagy), ribosomes (ribophagy), and others [4, 24]. Mitophagy is regulated by the phosphatase and tensin homolog deleted in chromosome 10 (PTEN)-induced putative kinase 1 (Pink1) and Parkinson protein-2 (Park2; Parkin)[37]. Mutations in the PINK1 and PARK2 genes are associated with recessive familial Parkinson's [38]. PINK1 is stabilized on dysfunctional mitochondria, where it recruits cytosolic Parkin, an E3 ubiquitin protein ligase. Parkin catalyzes the polyubiquitination of mitochondrial outer membrane proteins, which identify depolarized mitochondria for degradation [39]. In addition to dysfunctional organelles, autophagy can degrade ubiquitinated protein aggregates, in a selective process termed “aggrephagy”. Ubiquitinated substrates including mitochondria and protein aggregates are subsequently recognized and targeted to autophagosomes by p62SQSTM1 and other autophagy cargo adaptor proteins [3]. The p62SQSTM1 recognizes ubiquitinated substrates through its ubiquitin-associated domain, and recognizes LC3 through its LC3-interacting region [40].
Autophagy in Diabetes
Type II Diabetes is characterized by insulin resistance and pancreatic islet β cell dysfunction, while Type I diabetes is characterized by the autoimmune destruction of pancreatic B-cells, resulting in insulin deficiency. Recent studies indicate that autophagy can play important roles in the pathogenesis of Type I and Type II diabetes, and associated complications [18-20].
Autophagy can act as mediator of pancreatic β-cell homeostasis [41]. β-cell specific deletion of Atg7 resulted in mice that were hyperglycemic and glucose intolerant, and displayed reduced β-cell mass, and decline in insulin production [42-43]. Furthermore, Atg7 deficient β-cells displayed accumulations of ubiquitinated proteins, mitochondrial dysfunction and reduced insulin production [42-43]. Stimulation of autophagy improved β-cell function and reduced ER stress in a model of diabetes caused by an insulin misfolding mutation [44].
Recent studies indicate potential role for autophagy in diabetic nephropathy, a condition associated with decline in kidney function, and proteinuria [45]. The accumulation of cellular organelle and protein damage caused by hyperglycemia is associated with glomerular epithelial cell (podocyte) dysfunction and apoptosis. Autophagy was inhibited in podocytes by hyperglycemia in vitro, and in diabetic mice, leading to loss of epithelial barrier function [46]. Treatment with chemical chaperones restored podocyte autophagy in vivo. In contrast, hyperglycemia was observed to induce static markers of autophagy in podocytes, dependent on reactive oxygen species production [47]. Additional experiments to monitor autophagic flux under diabetic conditions may resolve these discrepancies. Kidney-specific Atg7 deletion sensitized mice to acute kidney injury caused by ischemia/reperfusion or nephrotoxic agents [48]. Thus, inducible autophagy may act as a protective response against xenobiotic stress in renal proximal tubules. Autophagy may be impaired in kidney tubules during obesity, sensitizing the kidney to injury in diabetic states [49].
Diabetic cardiomyopathy is associated with ventricular dysfunction, cardiomyocyte apoptosis, and with autophagy suppression in cardiomyocytes, which may involve downregulation of the AMPK pathway, upregulation of the mTOR pathway, and inhibition of Beclin 1 through interaction with Bcl-2 [50-52]. Conversely, autophagy activation by stimulation of the AMPK pathway prevented apoptosis in glucose-stimulated cardiomyocytes [53]. Chronic metformin administration conferred cardioprotection in mouse models of Type I diabetes, in an AMPK-dependent manner [51-52]. Overexpression of the stress protein heme oxygenase-1 (HO-1) conferred cardioprotection in the streptozotocin-induced diabetes model in mice through downregulation of cardiomyocyte apoptosis and upregulation of autophagy [54]. In contrast, knockout of Beclin 1 or Atg7 resulted in cardioprotection in Type I diabetes, though activation of compensatory autophagy pathways may have contributed to the observed phenotype [55].
Pathogenic accumulation of glycogen may be important in several metabolic diseases including diabetic cardiomyopathy. A selective autophagy-dependent process termed “glycophagy” may function in the amelioration of glycogen accumulation. Recent studies indicate that glycogen accumulation in fasting may be more prevalent in female heart tissue. The resulting induction of glycophagy was associated with cardioprotection [56].
Finally, aberrant inflammatory responses, including inflammasome-associated cytokines production (e.g., IL-1β) may play a pathogenic role in promoting insulin-resistance and the progression of Type II diabetes [57]. Our recent studies suggest that autophagy can play a key role in dampening inflammasome-associated cytokines production through stabilizing mitochondria [58]. Thus, candidate therapeutics which dampen inflammasome responses)(e.g., IL-1β receptor anagonists), in combination with therapies that activate autophagy (e.g., the AMPK activator metformin), may have potential for the treatment of diabetes and its complications [59].
Autophagy in Obesity
Genetic deletion of autophagy proteins promoted the storage of triglycerides into lipid droplets (LD) in the liver, suggesting that autophagy acts as a regulator of lipid metabolism and storage [60]. The selective autophagic degradation of LD, termed “lipophagy” may serve to regenerate free fatty acids during nutrient deficiency states [61]. Defects in this pathway may be associated with hepatic diseases, vascular diseases (atherosclerosis), obesity and metabolic syndrome, and organ dysfunction in aging [61]. Hepatocyte-specific deletion of Atg7 in mice resulted in fatty liver [60].
Autophagy was previously suggested to be downregulated in the liver in mouse models of obesity and insulin resistance. Genetic interference of Atg7 resulted in insulin resistance and promoted hepatic ER stress [62]. In contrast to these findings, the liver-specific knockout of Atg7 protected mice from diet-induced obesity and insulin resistance [63]. Furthermore, skeletal muscle-specifc deletion of Atg7 resulted in lean mice that were protected against diet-induced obesity and insulin resistance. This phenotype was accompanied by increased fatty acid oxidation and browning of white adipose tissue. These changes were associated with mitochondrial dysfunction and production of fibroblast growth factor 21 (Fgf21) [63].
Autophagy may play a role in adipocyte homeostasis and differentiation. Static markers of autophagy were found elevated in adipose tissue in a rat model of insulin resistance [64]. White adipose tissue-specific deletion of autophagy proteins (eg., Atg7) resulted in lean mice with reduced white adipose tissue mass, and increased brown adipose tissue [65]. Furthermore, Atg7 deficiency in adipocytes promoted insulin sensitivity in these mice [66]. Atg7 deletion specific to Myf5+ progenitor cells impaired brown adipose differentiation, and resulted in brown adipose tissue dysfunction, as well as reduced muscle mass and glucose intolerance in mice [67].
Exercise was shown to increase autophagy in cardiac and skeletal muscle, adipose tissue, and pancreatic β-cells. The induction of autophagy in mice by exercise resulted in protection against glucose-intolerance caused by high fat diet [68]. Mice bearing a phospho-deficient Bcl-2 mutation were unable to induce autophagy by exercise, and lost the protective phenotype with respect to glucose tolerance [69].
Recent studies indicate that stimulation of autophagy in the brain can alter the regulation of food intake. Starvation-induced autophagy led to increases in hypothalmic production of Agouti-related peptide hormone, a regulator of food intake. Neuron-specific knockout of Atg7 resulted in a lean phenotype, and reduction of food intake in response to fasting [70].
Autophagy in Glycogen storage disease
Glycogen storage disease type II (GSDII; also called Pompe disease) is an autosomal recessive disease caused by genetic deficiencies in acid α-1,4-glucosidase (GAA). Genetic deficiencies of GAA result in partial or complete inhibition of lysosomal degradation of glycogen, and consequently accumulation of glycogen-rich lysosomes, and autophagosomes [71]. The disease affects primarily skeletal and respiratory muscles, and may target vascular smooth muscle. Accumulations of autophagosomes in this disease may interfere with the function of muscle fibers [71]. Skeletal muscle biopsies from GSDII patients display accumulations of membrane-bound glycogen occurring in association with increased autophagic vacuoles [72]. Autophagic “flux”, or lysosomal degradative activity, was impaired in both infantile and late onset GSDII patients. Accumulations of p62SQSTM1 containing aggregates in skeletal muscle biopsies, indicative of impaired autophagy flux, correlated with disease progression and atrophy in late onset GSDII patients [73-74]. Furthermore, autophagic flux was required for GAA maturation, and for processing of recombinant GAA [74]. Enzyme replacement therapy (ERT) for this disease is more effective at reversing the associated cardiac pathology relative to skeletal muscle pathology [75]. The efficacy of ERT in late onset GSDII patients was associated with restoration of autophagic flux in muscle and downregulation of atrophy related gene expression [74]. In a murine model of Pompe disease (GAA−/− mice), suppression of autophagy by muscle-specific knockout of Atg5 aggravated the muscle pathology, whereas muscle-specific knockout of Atg7 improved the efficiency of ERT in this model [76]. Transcription factor E3, and EB (TFE3, TFEB), mediators of lysosomal biogenesis and exocytosis, may represent novel therapeutic targets for this disease [77-78]. Upregulation of TFEB promoted increased exocytosis of glycogen containing autophagolysosomes in cell culture models of Pompe disease [78].
Autophagy in inherited myopathies
Danon's disease is an X-linked dominant cardiomyopathy caused by deficiency of lysosomal-associated membrane protein 2 (LAMP2). The disease is characterized by accumulation of autophagosomes and glycogen in cardiac and skeletal muscle cells [16]. LAMP2 functions in autophagosomal-lysosomal fusion. Additionally, the isoform LAMP2a functions as a receptor for chaperone-mediated autophagy, a subtype of autophagy in which proteins bearing specific recognition motifs are directly targeted to lysosomes [79]. Evidence of autophagic vacuolar myopathy was also recently observed in mice deficient in vacuolar sorting protein-15 (Vps15), a regulatory partner of Vps34/PI3KCIII [80]. Interestingly, Vps15 overexpression reversed glycogen accumulation in myocytes from Danon's disease patients [80].
X-linked myopathy with excessive autophagy (XMEA) is a rare myopathy caused by mutations in the VMA21 gene, an essential assembly chaperone of the vacuolar-ATPase, the principle mammalian proton pump [17]. The disease is characterized by atrophy and accumulation of autophagic vacuoles in skeletal muscle [81]. VMA21 deficiency results in incomplete lysosomal acidification, which impairs autophagic processing of substrates, and pathological accumulation of autophagosomes [82].
Autophagy in other inherited disorders
Valosin containing protein (p97/VCP) mutations cause a group of disorders that include hereditary inclusion body myopathy, Paget's disease of bone, fronto-temporal dementia (IBMPFD); and can also contribute to the pathogenesis of familial amyotrophic lateral sclerosis (ALS). Homozygous knock-in VCP mutant mice (VCPR155H/R155H) carrying the common R155H mutations develop muscle, heart, brain and bone pathology, as well as evidence of autophagy and ubiquitin processing defects [83].
Paget's disease is an inherited disorder of bone metabolism [84]. Mutations of the autophagy cargo adaptor p62SQSTM1 gene, especially in the ubiquitin-binding region, have been linked to Paget's disease. p62SQSTM1 is a multifunctional protein that regulates selective autophagy processes, but may also influence cellular signaling pathways including the NF-κB and Nrf2/Keap1-dependent signaling pathways [85-86].
Inherited α1-anti-trypsin (α1-AT) deficiency causes liver dysfunction resulting from accumulation of mutant α1-AT protein. Autophagy may represent a compensatory mechanism for clearance of the mutant α1-AT protein. Upregulation of TFEB promoted autophagy and improved hepatic dysfunction in a mouse model of α1-AT deficiency [87]. Elevated markers of autophagy were recently reported in lung tissues from α1-AT deficiency patients with chronic lung disease [88].
Conclusion
Autophagy plays a complex role in tissue homeostasis by acting as a mediator of mitochondrial homeostasis, a degradative pathway for cellular debris, and as a pathway for the recycling of metabolic precursors. Further studies are needed to understand how metabolic changes incurred by autophagy stimulation impact the progression of human diseases. Autophagy dysfunction may be an important contributor to metabolic diseases such as inherited myopathies, diabetes, obesity and metabolic syndrome (Figure 2). Recent studies indicate protective functions of inducible or basal autophagy in diabetic disorders. Autophagy stimulating drugs such as metformin, which activates the AMPK pathway, have been proposed as potential therapies in diabetic cardiomyopathy and nephropathy. Autophagy is also proposed to play a role in lipid metabolism, adipocyte differentiation and homeostasis, regulation of food intake, and physiological responses to exercise, and thus has a complex role in obesity. Finally, the studies described in this review underscore a critical role for autophagy in the pathogenesis and therapeutic potential approaches to inherited metabolic diseases. The multifunctional nature of autophagy, as well as contrasting studies on the relative function of autophagy in these conditions warrants additional investigation before therapeutic targeting of autophagy in human disease.
Figure 2. Role of autophagy in various metabolic diseases.
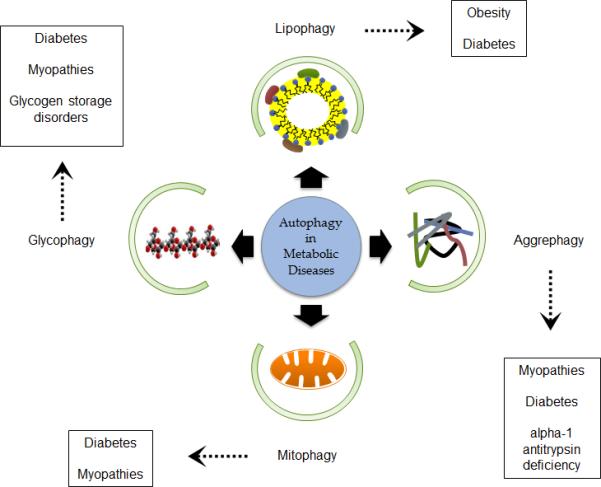
Autophagy can be separated into different functional categories where the targeted molecules and compounds can be linked with various metabolic diseases. Breakdown of lipid droplets (Lipophagy), protein aggregates (Aggrephagy), mitochondria (Mitophagy) and glycogen (Glycophagy) are all thought to be involved in metabolic diseases, such as obesity, myopathies and diabetes. Defective or impaired autophagy in targeting these molecules can be an agent in inducing metabolic diseases, as improper autophagy would result in failure to break down and recycle cellular molecules. For example, obesity may be due to defective lipophagy, diabetes and myopathies due to ineffective glycophagy and /or aggrephagy/mitophagy. Therapeutic strategies for ameliorating these metabolic disorders may include inducing autophagy via disabling the sequestration of vital autophagic proteins by inhibiting their complex formation, by gene therapy approaches, or by manipulating input signaling with pharmaceuticals.
Key Points.
Autophagy plays a central role in cellular metabolism by facilitating the turnover of organelles and proteins, clearing subcellular debris, and recycling metabolic precursors from the breakdown of complex molecules.
Impaired or dysfunctional autophagy may play a pathogenic role in metabolic disorders such as diabetes, glycogen storage disorders, and myopathies.
Autophagy has a complex relationship with insulin resistance and obesity, with functional roles in adipocyte differentiation, skeletal muscle differentiation, regulation of food intake, breakdown and storage of lipid droplets, and physiological responses to exercise.
Stimulation or restoration of autophagy (e.g., with AMPK activators or gene therapy approaches), coupled with strategies to reduce inflammatory responses, may show promise in metabolic diseases such as Type II diabetes.
Acknowledgements
This work was supported by NIH grants P01 HL108801, RO1 HL060234 and R01 HL079904.
Footnotes
Conflict of Interest:
There are no conflicts of interest.
References and recommended reading
Papers of particular interest, published within the annual period of review, have been highlighted as:
• of special interest
•• of outstanding interest.
- 1.Mizushima N, Komatsu M. Autophagy: renovation of cells and tissues. Cell. 2011;147:728–741. doi: 10.1016/j.cell.2011.10.026. [DOI] [PubMed] [Google Scholar]
- 2.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3•.Rogov V, Dötsch V, Johansen T, Kirkin V. Interactions between autophagy receptors and ubiquitin-like proteins form the molecular basis for selective autophagy. Mol Cell. 2014;53:167–178. doi: 10.1016/j.molcel.2013.12.014. [A recent review on the mechanisms of selective autophagy, with an emphasis on cargo adaptor and ubiquitin-like modifier proteins.] [DOI] [PubMed] [Google Scholar]
- 4•.Klionsky DJ, Codogno P. The mechanism and physiological function of macroautophagy. J Innate Immun. 2013;5:427–433. doi: 10.1159/000351979. [A recent review on the physiological significance of autophagy.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kelekar A. Autophagy. Ann N Y Acad Sci. 2005;1066:259–271. doi: 10.1196/annals.1363.015. [DOI] [PubMed] [Google Scholar]
- 6•.Boya P, Reggiori F, Codogno P. Emerging regulation and functions of autophagy. Nat Cell Biol. 2013;15:713–720. doi: 10.1038/ncb2788. [An overview on the functional significance of autophagy with emphasis on physiological significance and novel aspects.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7•.Deretic V, Saitoh T, Akira S. Autophagy in infection, inflammation and immunity. Nat Rev Immunol. 2013;13:722–737. doi: 10.1038/nri3532. [This paper provides a comprehensive summary of the role of autophagy in innate and adaptive immune system function.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8•.Choi AM, Ryter SW, Levine B. Autophagy in human health and disease. N Engl J Med. 2013;368:651–662. doi: 10.1056/NEJMra1205406. [This article provides a concise current overview of the role of autophagy in major human diseases.] [DOI] [PubMed] [Google Scholar]
- 9•.Rubinsztein DC, Codogno P, Levine B. Autophagy modulation as a potential therapeutic target for diverse diseases. Nat Rev Drug Discov. 2012;11:709–730. doi: 10.1038/nrd3802. [This article summarizes the role of autophagy in human diseases with an emphasis on therapeutic targeting.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10••.Helgason GV, Holyoake TL, Ryan KM. Role of autophagy in cancer prevention, development and therapy. Essays Biochem. 2013;55:133–151. doi: 10.1042/bse0550133. [This review discusses the role of autophagy in cancer and carcinogenesis with an emphasis on therapy.] [DOI] [PubMed] [Google Scholar]
- 11•.Nixon RA. The role of autophagy in neurodegenerative disease. Nat Med. 2013;19:983–997. doi: 10.1038/nm.3232. [This article summarizes the role of autophagy in the pathogenesis of neurodegenerative disorders including Alzheimer’s disease and Parkinson's disease.] [DOI] [PubMed] [Google Scholar]
- 12.Ryter SW, Nakahira K, Haspel JA, Choi AM. Autophagy in pulmonary diseases. Annu Rev Physiol. 2012;74:377–401. doi: 10.1146/annurev-physiol-020911-153348. [DOI] [PubMed] [Google Scholar]
- 13•.Schneider JL, Cuervo AM. Liver autophagy: much more than just taking out the trash. Nat Rev Gastroenterol Hepatol. 2013;11:187–200. doi: 10.1038/nrgastro.2013.211. [Recent review article on the role of autophagy in liver homeostasis and disease.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14•.Wang Z, Choi ME. Autophagy in kidney health and disease. Antioxid Redox Signal. 2014;20:519–537. doi: 10.1089/ars.2013.5363. [Recent review article on the role of autophagy in liver homeostasis and disease.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15•.Lavandero S, Troncoso R, Rothermel BA, et al. Cardiovascular autophagy: Concepts, controversies and perspectives. Autophagy. 2013;9(10) doi: 10.4161/auto.25969. [Epub ahead of print] [Summary overview on the possible functions of autophagy in cardiac homeostasis and disease.] [DOI] [PubMed] [Google Scholar]
- 16.Nishino I, Fu J, Tanji K, et al. Primary LAMP-2 deficiency causes X-linked vacuolar cardiomyopathy and myopathy (Danon disease). Nature. 2000;406:906–910. doi: 10.1038/35022604. [DOI] [PubMed] [Google Scholar]
- 17.Kalimo H, Savontaus ML, Lang H, et al. X-linked myopathy with excessive autophagy: a new hereditary muscle disease. Ann Neurol;1988;23:258–265. doi: 10.1002/ana.410230308. [DOI] [PubMed] [Google Scholar]
- 18•.Wilson CM, Magnaudeix A, Yardin C, Terro F. Autophagy dysfunction and its link to Alzheimer's Disease and Type II Diabetes Mellitus. CNS Neurol Disord Drug Targets. 2013 Sep 18; doi: 10.2174/18715273113126660146. [Epub ahead of print] [This review explores autophagy dysfunction as a common link in the pathogenesis of diabetes and Alzheimer's Disease.] [DOI] [PubMed] [Google Scholar]
- 19•.Wang Y, Li YB, Yin JJ, et al. Autophagy regulates inflammation following oxidative injury in diabetes. Autophagy. 2013;9:272–277. doi: 10.4161/auto.23628. [This review explores the concept that autophagy can act as a regulator of inflammation, a primary determinant in the pathogenesis of diabetes.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20•.Ouyang C, You J, Xie Z. The interplay between autophagy and apoptosis in the diabetic heart. J Mol Cell Cardiol. 2013 doi: 10.1016/j.yjmcc.2013.10.014. [Epub ahead of print] [This paper examines the relationship of apoptosis and autophagy in the diabetic heart.] [DOI] [PubMed] [Google Scholar]
- 21•.Christian P, Sacco J, Adeli K. Autophagy: Emerging roles in lipid homeostasis and metabolic control. Biochim Biophys Acta. 2013;1831:819–824. doi: 10.1016/j.bbalip.2012.12.009. [This paper represents an overview of the complex role of autophagy in lipid metabolism.] [DOI] [PubMed] [Google Scholar]
- 22•.Tan CC, Yu JT, Tan MS, Jiang T, Zhu XC, Tan L. Autophagy in aging and neurodegenerative diseases: implications for pathogenesis and therapy. Neurobiol Aging. 2014;35:941–957. doi: 10.1016/j.neurobiolaging.2013.11.019. [Recent overview of the function of autophagy in ageing, with emphasis on organelle and protein turnover.] [DOI] [PubMed] [Google Scholar]
- 23.Rubinsztein DC, Mariño G, Kroemer G. Autophagy and aging. Cell. 2011;146:682–695. doi: 10.1016/j.cell.2011.07.030. [DOI] [PubMed] [Google Scholar]
- 24••.Feng Y, He D, Yao Z, Klionsky DJ. The machinery of macroautophagy. Cell Res. 2014;24:24–41. doi: 10.1038/cr.2013.168. [This review represents the most updated summary of the molecular mechanisms for autophagy regulation.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parzych KR, Klionsky DJ. An overview of autophagy: morphology, mechanism, and regulation. Antioxid Redox Signal. 2014;20:460–473. doi: 10.1089/ars.2013.5371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Inoki K, Kim J, Guan KL. AMPK and mTOR in cellular energy homeostasis and drug targets. Annu Rev Pharmacol Toxicol. 2012;52:381–400. doi: 10.1146/annurev-pharmtox-010611-134537. [DOI] [PubMed] [Google Scholar]
- 27.Alers S, Löffler AS, Wesselborg S, Stork B. Role of AMPK-mTOR-Ulk1/2 in the regulation of autophagy: cross talk, shortcuts, and feedbacks. Mol Cell Biol. 2012;32:2–11. doi: 10.1128/MCB.06159-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28•.Wong PM, Puente C, Ganley IG, Jiang X. The ULK1 complex: sensing nutrient signals for autophagy activation. Autophagy. 2013;9:124–137. doi: 10.4161/auto.23323. [An updated summary of the role of the ULK1 complex in autophagy regulation.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29•.Egan DF, Shackelford DB, Mihaylova MM, et al. Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science. 2011;331:456–461. doi: 10.1126/science.1196371. [This article provides a link between autophagy regulation through ULK1 and the AMPK-dependent energy sensing pathway.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.He C, Levine B. The Beclin 1 interactome. Curr Opin Cell Biol. 2010;22:140–149. doi: 10.1016/j.ceb.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Di Bartolomeo S, Corazzari M, Nazio F, et al. The dynamic interaction of AMBRA1 with the dynein motor complex regulates mammalian autophagy. J Cell Biol. 2010;191:155–168. doi: 10.1083/jcb.201002100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32••.Russell RC, Tian Y, Yuan H, et al. ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nat Cell Biol. 2013;15:741–750. doi: 10.1038/ncb2757. [This paper describes a mechanism by which the ULK1/2 regulatory complex directly regulates the Beclin 1 complex, thereby providing a missing link in autophagy regulation.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rabinowitz JD, White E. Autophagy and metabolism. Science. 2010;330:1344–1348. doi: 10.1126/science.1193497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kroemer G, Mariño G, Levine B. Autophagy and the integrated stress response. Mol Cell. 2010;40:280–293. doi: 10.1016/j.molcel.2010.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yamamoto A, Simonsen A. The elimination of accumulated and aggregated proteins: a role for aggrephagy in neurodegeneration. Neurobiol Dis. 2011;43:17–28. doi: 10.1016/j.nbd.2010.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36•.May AI, Devenish RJ, Prescott M. The many faces of mitochondrial autophagy: making sense of contrasting observations in recent research. Int J Cell Biol. 2012;2012:431684. doi: 10.1155/2012/431684. [Recent review on the regulation and functional significance of mitophagy.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Youle RJ, Narendra DP. Mechanisms of mitophagy. Nat Rev Mol Cell Biol. 2011;12:9–14. doi: 10.1038/nrm3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morris HR. Genetics of Parkinson's disease. Ann Med. 2005;37:86–96. doi: 10.1080/07853890510007269. [DOI] [PubMed] [Google Scholar]
- 39••.Jin SM, Youle RJ. The accumulation of misfolded proteins in the mitochondrial matrix is sensed by PINK1 to induce PARK2/Parkin-mediated mitophagy of polarized mitochondria. Autophagy. 2013;9:1750–1757. doi: 10.4161/auto.26122. [This paper demonstrates that the signal for mitophagy may involve mitochondrial protein aggregation.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40•.Birgisdottir ÅB, Lamark T, Johansen T. The LIR motif-crucial for selective autophagy. J Cell Sci. 2013;126:3237–3247. doi: 10.1242/jcs.126128. [A recent review on selective autophagy with emphasis on functional domains of cargo adaptor proteins.] [DOI] [PubMed] [Google Scholar]
- 41•.Mazza S, Maffucci T. Autophagy and pancreatic β-cells. Vitam Horm. 2014;95:145–164. doi: 10.1016/B978-0-12-800174-5.00006-5. [This paper describes the functional role of autophagy in pancreatic β-cell homeostasis.] [DOI] [PubMed] [Google Scholar]
- 42.Quan W, Lim YM, Lee MS. Role of autophagy in diabetes and endoplasmic reticulum stress of pancreatic β-cells. Exp Mol Med. 2012;44:81–88. doi: 10.3858/emm.2012.44.2.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jung HS, Chung KW, Won Kim J, et al. Loss of autophagy diminishes pancreatic beta cell mass and function with resultant hyperglycemia. Cell Metab. 2008;8:318–324. doi: 10.1016/j.cmet.2008.08.013. [DOI] [PubMed] [Google Scholar]
- 44••.Bachar-Wikstrom E, Wikstrom JD, Ariav Y, et al. Stimulation of autophagy improves endoplasmic reticulum stress-induced diabetes. Diabetes. 2013;62:1227–1237. doi: 10.2337/db12-1474. [This paper shows that stimulation of autophagy can reduce ER stress and improve protein clearance in a diabetic model caused by insulin misfolding.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45•.Yamahara K, Yasuda M, Kume S, et al. The role of autophagy in the pathogenesis of diabetic nephropathy. J Diabetes Res. 2013;2013:193757. doi: 10.1155/2013/193757. [This paper describes the role of autophagy dysfunction in the pathogenesis of kidney disease during diabetes.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46•.Fang L, Zhou Y, Cao H, et al. Autophagy attenuates diabetic glomerular damage through protection of hyperglycemia-induced podocyte injury. PLoS One. 2013;8:e60546. doi: 10.1371/journal.pone.0060546. [The paper demonstrates high basal autophagy in cultured kidney epithelial cells, its impairment by hyperglycemia, and restoration by chemical chaperones.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47•.Ma T, Zhu J, Chen X, et al. High glucose induces autophagy in podocytes. Exp Cell Res. 2013;319:779–789. doi: 10.1016/j.yexcr.2013.01.018. [The paper demonstrates induction of autophagic markers in cultured kidney epithelial cells subjected to hyperglycemia.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48•.Jiang M, Wei Q, Dong G, et al. Autophagy in proximal tubules protects against acute kidney injury. Kidney Int. 2012;82:1271–1283. doi: 10.1038/ki.2012.261. [Renal proximal tubule-specific knockout of Atg7 sensitized mice to renal injury induced by nephrotoxins.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49••.Yamahara K, Kume S, Koya D, et al. Obesity-mediated autophagy insufficiency exacerbates proteinuria-induced tubulointerstitial lesions. J Am Soc Nephrol. 2013 doi: 10.1681/ASN.2012111080. In press. [This paper suggests that downregulation of autophagy in obesity may render the kidney susceptible to injury in diabetic states.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50•.Zou MH, Xie Z. Regulation of interplay between autophagy and apoptosis in the diabetic heart: new role of AMPK. Autophagy. 2013;9:624–625. doi: 10.4161/auto.23577. [This paper discusses the potential for stimulation of autophagy through the AMPK pathway as a therapeutic strategy in diabetic cardiomyopathy.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xie Z, He C, Zou MH. AMP-activated protein kinase modulates cardiac autophagy in diabetic cardiomyopathy. Autophagy. 2011;7:1254–1255. doi: 10.4161/auto.7.10.16740. [This paper demonstrates that chronic administration of metformin stimulates autophagy in the heart through the AMPK pathway.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52•.Xie Z, Lau K, Eby B, et al. Improvement of cardiac functions by chronic metformin treatment is associated with enhanced cardiac autophagy in diabetic OVE26 mice. Diabetes. 60:1770–1778. doi: 10.2337/db10-0351. 201. [This paper demonstrates that chronic administration of metformin stimulates autophagy through the AMPK pathway and preserves cardiac function in diabetic cardiomyopathy.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53•.He C, Zhu H, Li H, et al. Dissociation of Bcl-2-Beclin1 complex by activated AMPK enhances cardiac autophagy and protects against cardiomyocyte apoptosis in diabetes. Diabetes. 2013;62:1270–1281. doi: 10.2337/db12-0533. [This paper suggests that AMPK activation in the heart activates autophagy through disruption of the Bcl-2-Beclin1 interaction.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54•.Zhao Y, Zhang L, Qiao Y, et al. Heme oxygenase-1 prevents cardiac dysfunction in streptozotocin-diabetic mice by reducing inflammation, oxidative stress, apoptosis and enhancing autophagy. PLoS One. 2013;8:e75927. doi: 10.1371/journal.pone.0075927. [Heme oxygenase-1 conferred cardioprotection in a Type I diabetes model associated with activation of autophagy.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55•.Xu X, Kobayashi S, Chen K, et al. Diminished autophagy limits cardiac injury in mouse models of type 1 diabetes. J Biol Chem. 2013;288:18077–18092. doi: 10.1074/jbc.M113.474650. [This paper suggest that downregulation of cardiac autophagy can be cardioprotective in Type 1 diabetes.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56••.Reichelt ME, Mellor KM, Curl CL, et al. Myocardial glycophagy - a specific glycogen handling response to metabolic stress is accentuated in the female heart. J Mol Cell Cardiol. 2013;65:67–75. doi: 10.1016/j.yjmcc.2013.09.014. [This paper shows that autophagy plays a selective role in cardiac glycogen metabolism in females.] [DOI] [PubMed] [Google Scholar]
- 57.Wen H, Gris D, Lei Y, Jha S, Zhang L, Huang MT, Brickey WJ, Ting JP. Fatty acid- induced NLRP3-ASC inflammasome activation interferes with insulin signaling. Nat Immunol. 2011;12:408–415. doi: 10.1038/ni.2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nakahira K, Haspel JA, Rathinam VA, et al. Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nat Immunol. 2011;12:222–230. doi: 10.1038/ni.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Choi AM, Nakahira K. Dampening insulin signaling by an NLRP3 'meta-flammasome'. Nat Immunol. 2011;12:379–380. doi: 10.1038/ni.2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Singh R, Kaushik S, Wang Y, et al. Autophagy regulates lipid metabolism. Nature. 2009;458:1131–1135. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61•.Singh R, Cuervo AM. Lipophagy: connecting autophagy and lipid metabolism. Int J Cell Biol. 2012;2012:282041. doi: 10.1155/2012/282041. [A recent review on the role of autophagy in lipid metabolism and associated metabolic disorders.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yang L, Li P, Fu S, et al. Defective hepatic autophagy in obesity promotes ER stress and causes insulin resistance. Cell Metab. 2010;11:467–478. doi: 10.1016/j.cmet.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63••.Kim KH, Jeong YT, Oh H, et al. Autophagy deficiency leads to protection from obesity and insulin resistance by inducing Fgf21 as a mitokine. Nat Med. 2013;19:83–92. doi: 10.1038/nm.3014. [This paper suggests that hepatocyte or skeletal muscle specific deletion of Atg7 promotes insulin sensitivity through enhanced production of Fgf21.] [DOI] [PubMed] [Google Scholar]
- 64.Kosacka J, Koch K, Gericke M, et al. The polygenetically inherited metabolic syndrome of male WOKW rats is associated with enhanced autophagy in adipose tissue. Diabetol Metab Syndr. 2013;5:23. doi: 10.1186/1758-5996-5-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang Y, Goldman S, Baerga R, et al. Adipose-specific deletion of autophagy-related gene 7 (atg7) in mice reveals a role in adipogenesis. Proc Natl Acad Sci U S A. 2009;106:19860–19865. doi: 10.1073/pnas.0906048106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66••.Singh R, Xiang Y, Wang Y, et al. Autophagy regulates adipose mass and differentiation in mice. J Clin Invest. 2009;119:3329–3339. doi: 10.1172/JCI39228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67••.Martinez-Lopez N, Athonvarangkul D, Sahu S, et al. Autophagy in Myf5+ progenitors regulates energy and glucose homeostasis through control of brown fat and skeletal muscle development. EMBO Rep. 2013;14:795–803. doi: 10.1038/embor.2013.111. [This report shows that deletion of Atg7 in myogenic Myf5+ progenitors impairs the brown adipose tissue and skeletal muscle differentiation, and results in glucose intolerance.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68•.He C, Sumpter R, Jr, Levine B. Exercise induces autophagy in peripheral tissues and in the brain. Autophagy. 2012;8:1548–1551. doi: 10.4161/auto.21327. [Exercise induced autophagy confers proection against high fat diet induced glucose intolerance.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69•.He C, Bassik MC, Moresi V, et al. Exercise-induced BCL2-regulated autophagy is required for muscle glucose homeostasis. Nature. 2012;481:511–515. doi: 10.1038/nature10758. [Exercise induced autophagy through dissociation of Bcl2-Beclin 1 complexes.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kaushik S, Rodriguez-Navarro JA, Arias E, et al. Autophagy in hypothalamic AgRP neurons regulates food intake and energy balance. Cell Metab. 2011;14:173–183. doi: 10.1016/j.cmet.2011.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Raben N, Wong A, Ralston E, Myerowitz R. Autophagy and mitochondria in Pompe disease: nothing is so new as what has long been forgotten. Am J Med Genet C Semin Med Genet. 2012;160C:13–21. doi: 10.1002/ajmg.c.31317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72•.Katona I, Weis J, Hanisch F. Glycogenosome accumulation in the arrector pili muscle in Pompe disease. Orphanet J Rare Dis. 2014;9:17. doi: 10.1186/1750-1172-9-17. doi:10.1186/1750-1172-9-17. [The paper represents a histological assessment of glycogen autophagy in human skeletal muscle biopsies from Pompe patients.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73••.Nascimbeni AC, Fanin M, Masiero E, et al. Impaired autophagy contributes to muscle atrophy in glycogen storage disease type II patients. Autophagy. 2012;8:1697–1700. doi: 10.4161/auto.21691. [Impaired autophagy flux was shown here to correlate with disease progression and atrophy in late onset GSDII patients.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nascimbeni AC, Fanin M, Masiero E, et al. The role of autophagy in the pathogenesis of glycogen storage disease type II (GSDII). Cell Death Differ. 2012;19:1698–1708. doi: 10.1038/cdd.2012.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75•.Prater SN, Patel TT, Buckley AF, et al. Skeletal muscle pathology of infantile Pompe disease during long-term enzyme replacement therapy. Orphanet J Rare Dis. 2013;8:90. doi: 10.1186/1750-1172-8-90. [This paper examines the clinical efficacy of α-glucosidase replacement therapy in Pompe disease patients.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Raben N, Schreiner C, Baum R, et al. Suppression of autophagy permits successful enzyme replacement therapy in a lysosomal storage disorder-murine Pompe disease. Autophagy. 2010;6:1078–1089. doi: 10.4161/auto.6.8.13378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77•.Martina JA, Diab HI, Lishu L, et al. The nutrient-responsive transcription factor TFE3 promotes autophagy, lysosomal biogenesis, and clearance of cellular debris. Sci Signal. 2014;7:ra9. doi: 10.1126/scisignal.2004754. [This paper suggests TFE3 can be used in gene therapy for Pompe disease, to activate autophagy and promote lysosomal exocytosis.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78•.Spampanato C, Feeney E, Li L, et al. Transcription factor EB (TFEB) is a new therapeutic target for Pompe disease. EMBO Mol Med. 2013;5:691–706. doi: 10.1002/emmm.201202176. [This paper suggests TFEB can be used in gene therapy for Pompe disease, to activate autophagy and promote lysosomal exocytosis.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79•.Cuervo AM, Wong E. Chaperone-mediated autophagy: roles in disease and aging. Cell Res. 2014;24:92–104. doi: 10.1038/cr.2013.153. [A current review on the mechanisms and functions of chaperone-mediated autophagy.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80••.Nemazanyy I, Blaauw B, Paolini C, et al. Defects of Vps15 in skeletal muscles lead to autophagic vacuolar myopathy and lysosomal disease. EMBO Mol Med. 2013;5:870–890. doi: 10.1002/emmm.201202057. [This paper shows that Vps15 mutations can cause disease associated with dysfunctional autophagy.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Crockett CD, Ruggieri A, Gujrati M, et al. Late-adult onset of X-linked myopathy with excessive autophagy (XMEA). Muscle Nerve. 2014 Feb 1; doi: 10.1002/mus.24197. doi: 10.1002/mus.24197. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82•.Ramachandran N, Munteanu I, Wang P, et al. VMA21 deficiency prevents vacuolar ATPase assembly and causes autophagic vacuolar myopathy. Acta Neuropathol. 2013;125:439–457. doi: 10.1007/s00401-012-1073-6. [This paper shows that VMA21 mutations can cause disease associated with dysfunctional autophagy.] [DOI] [PubMed] [Google Scholar]
- 83••.Nalbandian A, Llewellyn KJ, Kitazawa M, et al. The homozygote VCP(R¹ H/R¹ H) mouse model exhibits accelerated human VCP-associated disease pathology. PLoS One. 2012;7:e46308. doi: 10.1371/journal.pone.0046308. [Disorders involving valosin containing protein mutations were shown to involve aberrant autophagy and protein ubiquitination.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chung PY, Van Hul W. Paget's disease of bone: evidence for complex pathogenetic interactions. Semin Arthritis Rheum. 2012;41:619–641. doi: 10.1016/j.semarthrit.2011.07.005. [DOI] [PubMed] [Google Scholar]
- 85•.Rea SL, Walsh JP, Layfield R, et al. New insights into the role of sequestosome 1/p62 mutant proteins in the pathogenesis of Paget's disease of bone. Endocr Rev. 2013;34:501–524. doi: 10.1210/er.2012-1034. [The paper reviews the possible functional significance of p62 mutations in the pathogenesis of Paget’s disease.] [DOI] [PubMed] [Google Scholar]
- 86•.Ichimura Y, Waguri S, Sou YS, et al. Phosphorylation of p62 activates the Keap1-Nrf2 pathway during selective autophagy. Mol Cell. 2013;51:618–631. doi: 10.1016/j.molcel.2013.08.003. [This article demonstrates a relationship between the selective autophagy cargo adaptor protein p62 and the regulation of the antioxidant stress response.] [DOI] [PubMed] [Google Scholar]
- 87•.Pastore N, Blomenkamp K, Annunziata F, et al. Gene transfer of master autophagy regulator TFEB results in clearance of toxic protein and correction of hepatic disease in alpha-1-anti-trypsin deficiency. EMBO Mol Med. 2013;5:397–412. doi: 10.1002/emmm.201202046. [The paper shows that gene therapy targeting autophagy can improve protein clearance in α-1 anti-trypsin deficiency.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chen ZH, Kim HP, Sciurba FC, et al. Egr-1 regulates autophagy in cigarette smoke- induced chronic obstructive pulmonary disease. PLoS One. 2008;3:e3316. doi: 10.1371/journal.pone.0003316. [DOI] [PMC free article] [PubMed] [Google Scholar]


