Abstract
For both plant (e.g., potato) and animal (e.g., salmon) species, unveiling the genetic architecture of complex traits is key to the genetic improvement of polyploids in agriculture. F1 progenies of a biparental cross are often used for quantitative trait loci (QTL) mapping in outcrossing polyploids, where haplotype reconstruction by identifying the parental origins of marker alleles is necessary. In this paper, we build a novel and integrated statistical framework for multilocus haplotype reconstruction in a full-sib tetraploid family from biallelic marker dosage data collected from single-nucleotide polymorphism (SNP) arrays or next-generation sequencing technology given a genetic linkage map. Compared to diploids, in tetraploids, additional complexity needs to be addressed, including double reduction and possible preferential pairing of chromosomes. We divide haplotype reconstruction into two stages: parental linkage phasing for reconstructing the most probable parental haplotypes and ancestral inference for probabilistically reconstructing the offspring haplotypes conditional on the reconstructed parental haplotypes. The simulation studies and the application to real data from potato show that the parental linkage phasing is robust to, and that the subsequent ancestral inference is accurate for, complex chromosome pairing behaviors during meiosis, various marker segregation types, erroneous genetic maps except for long-range disturbances of marker ordering, various amounts of offspring dosage errors (up to ∼20%), and various fractions of missing data in parents and offspring dosages.
Keywords: polyploidy, outbred population, double reduction, preferential pairing, ancestral inference
POLYPLOIDY occurs in some animals such as salmon but is pervasive in plants, including many important crop species such as potato (Solanum tuberosum) and alfalfa (Medicago sativa). Understanding the genetic architecture of complex traits in polyploids plays a fundamental role in their genetic improvement. Numerous statistical methods have been developed for quantitative trait locus mapping in humans, animal, and plant species with diploid genomes. In contrast, corresponding studies in polyploids are very few, although an analogous linear model framework was introduced for tetraploid mapping populations at least 15 years ago (Xie and Xu 2000; Hackett et al. 2001).
In the linear (mixed) models for quantitative trait locus mapping in diploid and polyploid species, the genetic component of a quantitative trait requires the calculation of genetic predictors (covariates), often expressed as the probabilities that the alleles at putative quantitative trait loci (QTL) are derived from particular parental chromosomes conditional on the observed genotypic data of mapping individuals and their parents. The haplotype reconstruction for calculating genetic predictors in diploids has been well developed (Mott et al. 2000; Broman et al. 2003; Liu et al. 2010; Zheng et al. 2015). The aim of this work is haplotype reconstruction in a full-sib tetraploid family.
Compared to diploids, there are several challenges for haplotype reconstruction in polyploids. First, traditional marker systems such as dominant amplified fragment length polymorphism (AFLP) and codominant simple sequence repea (SSR) do not provide full information for a straightforward estimation of the number of copies of each allele (dosage). This is so because each score (gel band pattern) may correspond to multiple allelic dosages, and some alleles may not be revealed in a gel band pattern (the null alleles). However, new genotyping technologies, e.g., the Illumina Infinium platform and sequencing-based methods such as genotyping-by-sequencing, allow accurate estimation of the dosage of high-density single-nucleotide polymorphisms (SNPs) throughout the genome (Voorrips et al. 2011; Garcia et al. 2013; Hackett et al. 2013; Li et al. 2014). We focus on analyzing the increasingly available SNP dosage data.
Second, the pairing and segregation of chromosomes during meiosis are more complex in polyploids than in diploids. Polyploids are traditionally classified into allopolyploids and autopolyploids. Allopolyploids are derived from hybridization of two different species and subsequent chromosome doubling, and thus homologous chromosomes are more likely to pair together than homeologous chromosomes (Sybenga 1994). However, in autopolyploids, all the homologous chromosomes can pair during meiosis and form either random (nonpreferential) bivalents or multivalents, leading to polysomic inheritance. To maximize flexibility, we do not make a strict distinction between allopolyploids and autopolyploids, so both preferential bivalent pairing and quadrivalent pairing are possible a priori in tetraploids. Quadrivalent pairing can lead to a phenomenon called double reduction; i.e., corresponding parts of two sister chromatids of a chromosome sort into the same gamete (Mather 1936).
Last but not least, the parents of a mapping population are often outbred (not homozygous), which requires the crucial reconstruction of parental linkage phases across all SNP markers within each linkage group. For traditional marker systems such as AFLP and SSR in a full-sib tetraploid family, Luo et al. (2001) developed a general expectation-maximization algorithm to obtain the most likely phases for all pairs of markers and then reconstructed the phase of the complete linkage group using a heuristic algorithm. This procedure was extended to analyze SNP dosage data (Hackett et al. 2013). However, this still requires manual assignment of some SNP marker phases, and its application to large data sets may be slow.
Our approach to haplotype reconstruction in outcrossing tetraploids consists of multilocus parental linkage phasing and subsequent ancestral inference from SNP dosage data. It builds on an integrated network modeling of tetraploid inheritance, accounting for preferential bivalent and quadrivalent pairing. The multilocus linkage phasing is an automatic marker analysis involving no manual manipulation of intermediate results. Conditional on the estimated parental linkage phases, ancestral inference is performed to calculate posterior genotype probabilities at all marker locations based on a hidden Markov model (HMM) derived from the network model. Hackett et al. (2013) developed a similar HMM, except that their model assumes bivalent pairing and does not account for quadrivalent pairing. Leach et al. (2010) developed multilocus autotetrasomic linkage analysis using a HMM for traditional AFLP and SSR marker data conditional on marker ordering and parental linkage phases; their method accounts for a mixture of random bivalent and quadrivalent pairing, and it also may be used for ancestral inference in autotetraploids.
We assume a given genetic linkage map, i.e., the ordering of SNP markers and the intermarker genetic distances, that may be constructed from two-locus linkage analysis (Luo et al. 2001, 2006; Hackett et al. 2013). Specifically, for each linkage group, the two-locus analysis produces the recombination fraction and the LOD score for all pairs of markers, which can be used to construct the map using, e.g., the least-squares procedure implemented in JoinMap software (Stam 1993).
To evaluate the robustness of our new haplotype reconstruction method called TetraOrigin, we simulate many scenarios, including various chromosome pairing behaviors, various marker segregation types, and erroneous genetic linkage maps. We also study the impact of missing dosage data among parents and offspring and the effects of errors in offspring dosage data. Then we apply TetraOrigin to real potato data (Hackett et al. 2013). For both simulation studies and application to real data, we compare TetraOrigin with the methodology described in Hackett et al. (2013), henceforth abbreviated to H2013.
Methods
The model overview
Consider a full-sib tetraploid family with two parents denoted P1 and P2. We model the genetic inheritance independently across linkage groups and thus consider only one group. We label the four homologous or homeologous chromosomes of parent P1 1, 2, 3, and 4, assuming that chromosomes I and II are homologous and that so are chromosomes III and IV. The four chromosomes of parent P2 are similarly labeled 5, 6, 7, and 8. The ordering of the two parents and the ordering of the four chromosomes within a parent are otherwise arbitrary. Denote by the dosage of offspring at locus and the dosage of parent p = P1, P2 at locus t. The ordering and genetic locations of markers are assumed to be known, and all the markers are biallelic.
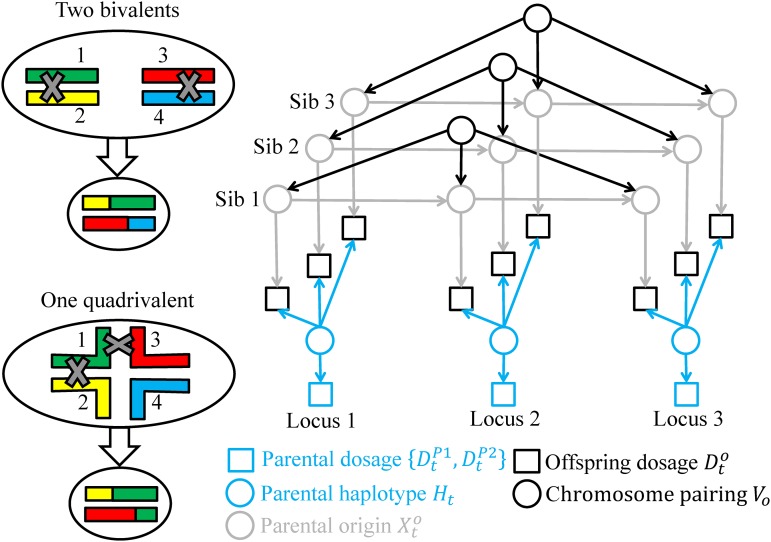
An overview of the model for analyzing dosage data and is shown in Figure 1. The offspring dosage data are not independent across offspring, which provides information on estimating the parental haplotype . Given the parental haplotype and offspring o, the dosage data are not independent across markers, which provides information on estimating the chromosome pairing when producing offspring o. Denote this by .
Figure 1.
The network model. Left panels refer to the possible gametes produced by either bivalent pairing or quadrivalent pairing in parent P1, where the four homologous/homeologous chromosomes are labeled by different colors. The right panel refers to the directed acyclic graph of the model for No = 3 offspring at Nt = 3 marker loci, where the rectangles denote known dosage data, the circles random variables, and the solid arrows probabilistic relationships described in the Methods section. Conditional on the chromosome pairings , the network model becomes a HMM along the chromosomes with the latent variables being the parental haplotypes and the parental origins .
Conditional on the parental haplotype H and the chromosome pairing Vo, we model parental origins along the four chromosomes of offspring o by a discrete-time Markov chain. The Markov chain can be described by an initial distribution specifying the probability that the parental origin at the first locus is at the jth state for and an transition probability matrix specifying how the parental origin state changes from locus t into the next. The state space of the Markov chain depends on the chromosome pairing Vo, which will be described in the following gamete and zygote models. For convenience, we specify , Ht, and Vo by either their discrete values or integer labels starting from 1.
The data likelihood
We first consider that there is no error and no missing data in the parental dosages. At a given locus t, the posterior probability follows a discrete uniform distribution over all possible combinations of haplotypes compatible with parental dosages. For example, the dosages for parents P1 and P2 are 1 and 2, respectively. Then there are four possible haplotypes for P1, 1000, 0100, 0010, and 0001, and six possible haplotypes for P2, 1100, 1010, 1001, 0110, 0101, and 0011, where we denote by 0 and 1 the two alleles at the locus, and the dosage refers to the number of copies of allele 1. Thus, at this locus is one of the possible combinations, e.g., , at equal probability.
If a parent dosage is missing, we treat all five dosages as equally probable with probability 1/5. When modeling errors are seen in the parent dosages, we assume that each observed dosage is the true dosage with probability and that all the other four possible dosages are equally probable with probability . We denote after accounting for missing data and dosage error (the dependence on parental dosages is not shown).
Let ε be the error probability for the offspring dosage data and the likelihood . For missing dosage data, we set the likelihood conditional on the pattern of missing data. We assume that the observed dosage takes one of the other four possible dosages with probability 1/4, given that an error occurs. Thus, it holds that if the observed dosage is the same as the dosage value that is derived from and and otherwise.
The gamete model
Bivalent chromosome pairing:
A gamete is produced from a pair of bivalents in the tetraploid parent. For example, we consider the bivalent formation in parent P1. Let the bracket () denote the bivalent formed between chromosomes . After crossover between chromosomes of the bivalent , the resulting chromosome consists of mosaic blocks with the parental origins and . We model the parental origins along the resulting chromosome as a discrete-time Markov chain. The parental origin at the first locus can be equally or . The transition probability matrix is given by
where r is the known recombination frequency between two loci, or it can be calculated from the genetic distance (e.g., Haldane mapping function).
The crossover events are assumed to occur independently for the two bivalents when producing a diploid gamete. Thus, the discrete-time Markov chain for the parental origins along the two chromosomes can be derived easily. Let denote the two bivalent pairs, and it may take one of three possible combinations, , , or . The parental origin state at the first locus can be , , , or , each with equal probability . The gamete transition matrix is given by , a Kronecker product between the transition matrices for the two bivalents.
Quadrivalent chromosome pairing:
A gamete is produced from a multivalent in the tetraploid parent, e.g., P1. The biological meiosis process of quadrivalent pairing is very complicated, and the resulting gamete genotypes at two linked loci depend on many factors, such as the configuration of the four chromosomes and the locations of the two loci relative to the centromere (e.g., Stift et al. 2010). We build a simple discrete-time Markov chain for the parental origins along the two chromosomes, aiming to account for the phenomenon of double reduction resulting from quadrivalent pairing.
The parental origin process is assumed to be independent along each of the two chromosomes within the gamete produced. Along one chromosome, the parental origin state at the first locus can be 1, 2, 3, or 4, each with equal probability 1/4; the transition matrix is given by
so the parental origin changes into one of the other three possible values with equal probability 1/3 given that a transition occurs between the two loci with probability r. Along the two chromosomes of the diploid gamete, the parental origin state at the first locus can be one of 16 phased genotypes with equal probability, and the transition matrix is given by .
Among the 16 phased genotypes, there are four with double reduction: 11, 22, 33, and 44. The prior probability of double reduction is thus 1/4, which is also the highest value given for the maximum possible double-reduction rate (Luo et al. 2006); values of 1/6, 1/7, and 1/8 are also mentioned (Mather 1935; Sybenga 1972; Voorrips and Maliepaard 2012). However, our prior probability will hardly affect ancestral inference when marker data are substantial.
The zygote model
The two gametes constituting a zygote are assumed to be produced independently during meiosis. Let , where denotes the two bivalents or the quadrivalent formed in parent when producing offspring o. Let be the probability that a gamete is produced from quadrivalent formation and be the extra probability of pairing between homologous chromosomes with respect to that between homeologous chromosomes. Consider, e.g., a P1 gamete that is produced via bivalent formation with probability , and the resulting pair of bivalents is , , and with probabilities , , and , respectively, assuming that is the preferred homologous pairing. For the bvModel, we set a priori and , so or can be equally one of the three possible bivalent pairings; for the fullModel, we set a priori and , so or can be equally one of the four possible chromosome pairings.
Consider, e.g., that offspring o is produced by bivalent formations in both parents, with and . Along the four chromosomes of offspring o, the parental origin at the first locus can be equally one of possible combinations (e.g., ). The transition matrix is given by , a Kronecker product between the gamete transition matrices.
If the P1 gamete is produced from a quadrivalent formation, . The zygote transition matrix and thus the parental origin can be equally one of possible states. Similarly for the P2 gamete resulting from a multivalent formation, where is the same as except the state labels. If both gametes are produced from a quadrivalent formation, there are equally possible states, and .
Parental linkage phasing
Phasing algorithm:
We estimate the parental haplotype H by maximizing the marginal likelihood , where is the marginal likelihood for offspring o with dosage data . In addition, we denote by the posterior probability of for offspring o, which will be used in the phasing algorithm. The calculation of and is described in the next section on ancestral inference. The maximization is an adaptation of the Metropolis algorithm (e.g., Gelman et al. 2004), where the acceptance always increases the target function . The algorithm proceeds as follows:
A0. Sample a starting parental haplotype H. To set an overdispersed starting point, we randomly sample to be one of possible values compatible with the parental dosages and , for .
A1. Sample independently for offspring . Conditional on H, set that maximizes the posterior probability over all possible chromosome pairing values .
A2. Sample a proposal and from their posterior distribution conditional on , which will be described in algorithms B and C.
A3. Calculate . If , set and return to A1; if , stop the algorithm, and if , reject the proposal and return to A1.
In addition, we stop a single phasing run of algorithm A if it rejects in consecutive iterations or it reaches the prefixed maximum number of iterations . By default, we set and based on our simulation studies.
To find the global maximization, we perform multiple runs of algorithm A and select the one with the largest target function value. We repeat algorithm A until the largest among the so-far phasing runs has been obtained times including the current run or the number of runs reaches the prefixed threshold . By default, we set and based on our simulation studies.
To increase computational efficiency, we exclude quadrivalent formations, so can take only one of the nine possible values. As shown in the simulated studies, the phasing algorithm is very robust to the quadrivalent inheritance.
Proposal sampling:
To sample the parental haplotypes, we first extend the forward algorithm (Rabiner 1989) to calculate the marginal posterior probabilities of latent and at locus t, integrating out the previous parental haplotypes and parental origins . Denote by the dosage data up to locus t. For offspring o, we denote by and , where according to the Bayesian theorem (Gelman et al. 2004). We denote by and , where according to the Bayesian theorem (Gelman et al. 2004). The algorithm proceeds as follows:
B0. Initialize , for , , and . Then calculate . Remember that is the initial distribution for the discrete-time Markov chain of offspring o and that is used as the prior haplotype probability after accounting for the missing data and errors in parental dosages. We obtain and by normalizing and , respectively.
- B1. For , compute
We obtain and by normalizing and , respectively.
Based on the probabilities and calculated forwardly by algorithm B, we sample the parental haplotypes and the parental origins backwardly:
C0. Sample from ; sample from independently for , conditional on the haplotype .
-
C1. For ,
Conditional on , compute and .
Sample from the unnormalized probabilities .
Conditional on , sample from the unnormalized probabilities independently for .
Ancestral inference
Let H be the parental haplotype estimated by the phasing algorithm A or any given parental haplotype. Denote by the dosage data for offspring o. For each of the 9 (bvModel) or 16 (fullModel) possible values of , we calculate the marginal likelihood and the posterior probability by integrating out the latent parental origins using the forward-backward algorithm (Rabiner 1989) independently for .
We divide offspring into four possible types, 22, 24, 42, and 44, where the first digit denotes the bivalent (digit 2) or quadrivalent (digit 4) formation in parent P1, and the second digit, for parent P2. We set each offspring to the type with the largest probability. For example, the posterior probability of offspring o being type 22 can be obtained by summing the posterior probability over 9 of the 16 possible values for bivalent pairing. The posterior probability of the chromosome pairing is given by
where according to the law of total probability. Here the prior probability is assumed to be independent of parental haplotype H, and it is determined by the preferential probability and the quadrivalent probability . We have for the bvModel and for the fullModel.
After assigning the chromosome pairing type for each offspring, we obtain the final marginal posterior probability
where the summation is taken over all the possible chromosome pairing values that are compatible with the assigned type. Note that the possible ancestral origin states depend on during the weighted summation.
Data availability
The TetraOrigin package has been implemented in Mathematica 9.0 (Wolfram Research 2012) and is freely available under the GNU General Public License from the website https://github.com/chaozhi/TetraOrigin.git. The full data sets for the 14 simulation scenarios and the real potato data extracted from Hackett et al. (2013) are included in the package.
Results
Simulation experiments
We evaluate the performance and robustness of our method on haplotype reconstruction by intensive simulation studies in full-sib tetraploid families. We simulate 14 scenarios using PedigreeSim v2.0 (Voorrips and Maliepaard 2012), differing with respect to preferential and/or quadrivalent pairing, marker segregation type, and accuracy of genetic maps (Table 1). We simulate only one linkage group. The full data set of each scenario consists of 200 offspring and 1200 SNPs randomly distributed along four homologous/homeologous chromosomes of 120 cM in length; the centromere is located at 40 cM.
Table 1. The 14 simulated scenarios classified into the three types.
| Type | Data set | Marker segregation type | Preferential pairing | Quadrivalent pairing |
|---|---|---|---|---|
| A | DStd-M | Mixed | 0 | 0 |
| DPref-M | Mixed | 1/2 | 0 | |
| DQuad-M | Mixed | 0 | 2/3 | |
| DPrefQuad-M | Mixed | 1/2 | 1/2 | |
| B | DStd-01 | Nulliplex-simplex | 0 | 0 |
| DStd-02 | Nulliplex-duplex | 0 | 0 | |
| DStd-11 | Simplex-simplex | 0 | 0 | |
| DStd-12 | Simplex-duplex | 0 | 0 | |
| DStd-13 | Simplex-triplex | 0 | 0 | |
| DStd-22 | Duplex -duplex | 0 | 0 | |
| C | DStd-M-V0.25 | Mixed | 0 | 0 |
| DStd-M-V1 | Mixed | 0 | 0 | |
| DStd-M-Local | Mixed | 0 | 0 | |
| DStd-M-Long | Mixed | 0 | 0 |
For the mixed-segregation type, each parental allele at a SNP site is a dosage allele with probability ½, so the expected relative proportions are 8, 6, 8, 24, 8, and 9 for the types nulliplex-simplex, nulliplex-duplex, simplex-simplex, simplex-duplex, simple-triplex, and duplex-duplex, respectively. The nulliplex-simplex refers to the unordered parental dosages 01, 03, 14, and 34 and so on for the other types. Note that the type C data sets are derived from DStd-M by disturbing the genetic map.
The 14 scenarios can be divided into three types. Type A has different combinations of preferential and quadrivalent pairings. We denote by DStd-M the standard data set without preferential pairing and without quadrivalent pairing, where -M refers to the mixed segregation types owing to mixed parental dosages. Type B has six scenarios with the data sets denoted by DStd-st, where the segregation type st = 01, 02, 11, 12, 13, or 22 refers to the unordered dosages of two parents at each SNP locus; the segregations with the parental dosages 03, 14, 23, 24, 33, and 34 are equivalent to one of the preceding segregation types by switching dosage alleles into nondosage alleles for parents and offspring (see Table 1). Type C has four scenarios for studying sensitivity to erroneous genetic maps, and they are derived from DStd-M by disturbing the intermarker distances or the marker ordering while keeping the dosages unchanged. We denote by -V0.25 (-V1) the disturbance of intermarker distances by a gamma distribution with mean being the original distance and variance being 0.25 (1), by -Local the disturbance of marker ordering by partitioning chromosomes into segments of 10 consecutive markers and swapping two markers within each segment, and by -Long for each of the 10 pairs of markers being chosen randomly within the same linkage group (not necessarily within the same segment). We consider only one linkage group for each simulation scenario and perform the disturbances independently for each sub–data set.
We carry out multilocus haplotype reconstruction for each of the 14 scenarios using the network model illustrated in Figure 1 and described in detail in the Methods section. The haplotype reconstruction is divided into two stages: parental linkage phasing and ancestral inference. For each stage, two models can be used: bvModel, where only preferential or nonpreferential bivalent pairings are modeled, and fullModel, where both bivalent and quadrivalent pairings are modeled. We evaluate separately the results obtained from each stage. In addition, we study the impact of missing data by analyzing DStd-M with a given fraction of dosages regarded as missing and the effects of dosage errors based on data sets derived from DStd-M by changing the correct dosage to another random dosage in a specified fraction of data points.
Evaluation of parental linkage phasing
To test the effect of sample size, for each full data set, we extract six nested subsets with the number of offspring No = 10, 20, and 100 and the number of markers Nt = 75 and 300 obtained by sampling every sixteenth and fourth of the 1200 markers, respectively. For each subset, we perform phasing analysis using the bvModel because the fullModel is more computationally intensive. Because the permutation of the four haplotypes within each parent is arbitrary and nonidentifiable from dosage data, we label the ordering of the most probable parental haplotypes obtained from the simulated data sets so that the number of mismatches with their true (simulated) values is minimized and keep the same labeling for real data where the true values are not known. Table 2 shows the comparisons of estimated parental haplotypes with their true values.
Table 2. Estimates of parental linkage phases.
| Data set | No = 10 | No = 20 | No = 100 | |||
|---|---|---|---|---|---|---|
| Nt = 75 | Nt = 300 | Nt = 75 | Nt = 300 | Nt = 75 | Nt = 300 | |
| DStd-M | 6 (−9.9)a | 8 (0.0003) | 0 | 0 | 0 | 0 |
| DPref-M | 56 (−36.7) | 4 (0) | 0 | 0 | 0 | 0 |
| DQuad-M | 40 (−4.6) | 0 | 0 | 0 | 0 | 0 |
| DPrefQuad-M | 8 (−14.9) | 130 (55.5) | 0 | 0 | 0 | 0 |
| DStd-01 | 0 | 0 | 0 | 0 | 0 | 0 |
| DStd-02 | 56 (0) | 160 (0) | 24 (0) | 184 (0) | 60 (0) | 144 (0) |
| DStd-11 | 70 (−104.0) | 0 | 0 | 0 | 0 | 0 |
| DStd-12 | 4 (0.002) | 16 (0.0001) | 0 | 0 | 0 | 0 |
| DStd-13 | 0 | 860 (0) | 0 | 0 | 196 (0) | 860 (0) |
| DStd-22 | 12 (0.001) | 360 (0) | 68 (0) | 0 | 68 (0) | 332 (0) |
| DStd-M-V0.25 | 0 | 8 (−0.0003) | 0 | 0 | 0 | 0 |
| DStd-M-V1 | 54 (−70.4) | 4 (0) | 0 | 0 | 0 | 0 |
| DStd-M-Local | 128 (−75.3) | 4 (0) | 0 | 0 | 0 | 0 |
| DStd-M-Long | 106 (−34.7) | 98 (483.6) | 30 (17.9) | 56 (361.7) | 6 (100.3) | 36 (114.0) |
Each cell gives the number of mismatched alleles between estimated and true parental haplotypes, where the value in parentheses is the log likelihood given the estimated haplotypes minus that given the true haplotypes. For each of the 14 simulation scenarios (Table 1), six sub–data sets are specified by the number No = 10, 20, and 100 offspring and the number Nt = 75 and 300 markers.
For example, 6 is the number of mismatched alleles out of the total 8Nt = 600 alleles, and −9.9 is given by logl(estimate H) − logl(true H) (see the phasing algorithm).
We first examine the phasing results for the sample sizes No = 20 and 100. For the type A scenarios, the estimated parental haplotypes match the true values, despite the presence of preferential and/or quadrivalent pairings. For the type B scenarios, most of the estimations match the true values, except for some segregation types such as st = 02, 13, and 22 (Table 2). We may obtain different mismatches if we repeat the phasing analysis with random starting values, but these estimations are always equivalent to their true values in terms of marginal likelihood (Table 2). The mismatches are shown in Supplemental Material, Figure S1, where the dosage and nondosage alleles are switched at some markers for segregation type st = 02, but there is no clear pattern for st = 13 or 22. For each of the type B scenarios, we repeated the phasing analysis at least twice, and the segregation types st = 01, 11, and 12 did not show such equivalent mismatches. For the type C scenarios, the phasing analyses are robust to the noise of intermarker distances and the local disturbance of marker ordering but not to long-range disturbances. In the latter case, the marginal likelihoods for the estimated parental haplotypes are larger than those for the true values, indicating that the phasing analysis cannot be improved without jointly re-estimating the marker ordering (Table 2).
For the smallest sample size No = 10, the parental phasing varies with repeated analysis and thus is unreliable. This inconsistency of results may occur because many similar haplotypes are indistinguishable based on the small data sets; the phasing results are improved with higher marker density, as shown by the larger likelihood values. However, there is no visible effect of marker density for the larger sample sizes No = 20 and 100.
Furthermore, we evaluate the effect of missing data on parental linkage phasing by using DStd-M with No = 200 and Nt = 300. Dosages are assumed to be missing at random. We perform phasing analysis for the missing fraction among two parents being 0, 0.1, 0.2, 0.5, 0.75, and 1 and the missing fraction among offspring being 0, 0.1, 0.2, 0.5, and 0.75. For all 30 combinations of missing fractions, we successfully recover the true parental haplotypes.
Evaluation of ancestral inference
For each simulated data set, we perform ancestral inference (i.e., for each progeny individual we obtain its genetic composition in terms of parental homolog segments) and compare this to their true compositions. To study the effects of marker density, for each of the 14 full data sets we extract sub–data sets by sampling every ith marker for , 2, 4, 8, and 16 such that the marker subsets are nested and recursively reduced by half; the average intermarker distance of 0.1 cM for the full data set is doubled repeatedly until we reach 1.6 cM for the smallest sub–data set.
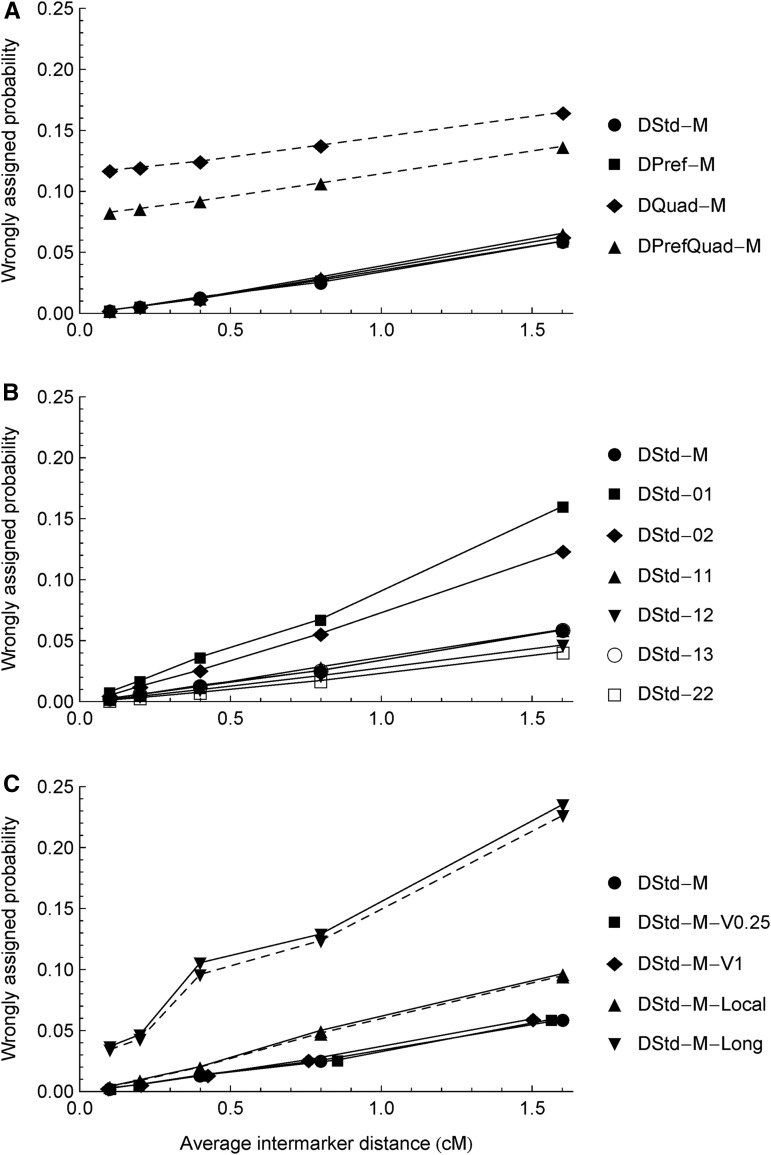
Figure 2 shows the effects of marker density on the wrongly assigned probability for each of the 14 full data sets. The wrongly assigned probability is given by one minus the posterior probability of being the true ancestral state, averaged over all markers and offspring. As expected, the wrongly assigned probabilities increase roughly linearly with the average intermarker distances. Figure 2A shows that the effects of density from the fullModel (solid lines) depend little on the chromosome pairings; the wrongly assigned probabilities from DQuad-M and DPrefQuad-M using the bvModel (dashed lines) are larger than those using the fullModel, and the excess amounts approximate the average double-reduction probabilities (Figure 3). Figure 2B shows that segregation types 01 and 02 are less informative for ancestral inference than other types.
Figure 2.
Density dependencies of ancestral inference for the three types of the 14 simulation scenarios (Table 1). The y-axis is the wrongly assigned probability, one minus the posterior probability of being the true ancestral state, averaged over the No = 200 offspring and all markers. The increasing intermarker distances correspond to the numbers of markers NT = 1200, 600, 300, 150, and 75 on a 120-cM chromosome, respectively. The symbols connected by the solid lines denote the results obtained from the fullModel and the dashed lines from the bvModel. Except for DQuad-M, DPrefQuad-M, DStd-M-Local, and DStd-M-Long, the results from the bvModel and the fullModel are the same because all the offspring are identified as being produced only by bivalent pairing in the fullModel. In B, the results for DStd-11 and DStd-13 largely overlap with those for DStd-M.
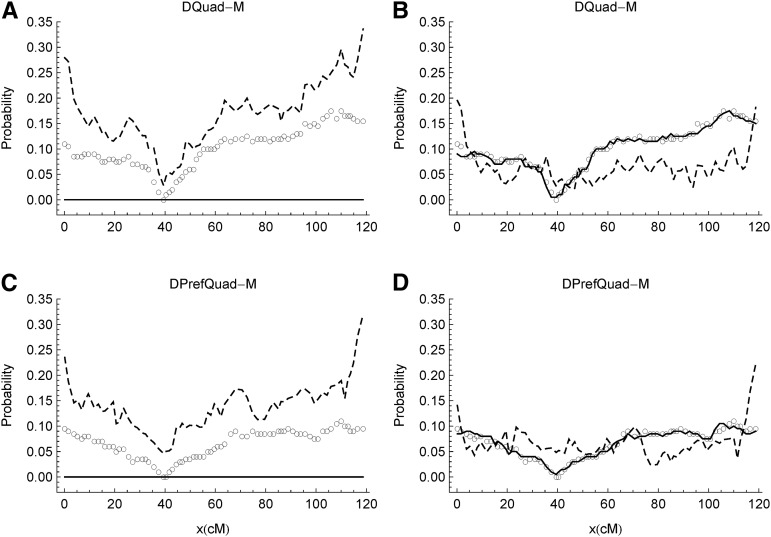
Figure 3.
Estimation of double reduction along the chromosomes obtained from DQuad-M (top panels) and DPrefQuad-M (bottom panels). Each data set has NO = 200 offspring and Nt = 75 markers. The left panels are from the bvModel, and the right panels are from the fullModel. The solid lines denote the estimations, the posterior probabilities of double-reduction states averaged over the offspring at a given marker; the empty circles denote the true values, the fractions of offspring being in double-reduction states. The dashed lines denote the wrongly assigned probabilities along the chromosomes, one minus the posterior probability of being the true ancestral state, averaged over all offspring but not markers.
Figure 2C shows the sensitivities of ancestral inference to an erroneous genetic linkage map. The noises of intermarker distances have little effect on the wrongly assigned probabilities. The local disturbances of marker ordering almost double the wrongly assigned probabilities, though they do not affect the parental linkage phasing (Table 2). The long-distance disturbances have the greatest deleterious effects on ancestral inference because they also deteriorate the phasing accuracy (Table 2). For both the local and long-distance disturbances of marker ordering, the wrongly assigned probabilities obtained from the fullModel are slightly larger than those from the bvModel, probably because for some offspring quadrivalents are wrongly assigned (Figure S2).
Figure 3 shows the estimations of double reduction along chromosomes obtained from DQuad-M and DPrefQuad-M using both the bvModel and the fullModel. For both data sets, the true double-reduction probabilities are zero at the centromere and increase toward the telomeres. Because the bvModel does not allow double reduction, the wrongly assigned probabilities along chromosomes are positively correlated with the true double-reduction fractions (Figure 3, A and B). In contrast, the fullModel estimates the double reduction very well, and thus the wrongly assigned probabilities are small and uncorrelated with the true double-reduction fractions along chromosomes (Figure 3, C and D); the wrongly assigned probabilities are relatively large at the chromosomal ends because the marker information is available only from one side.
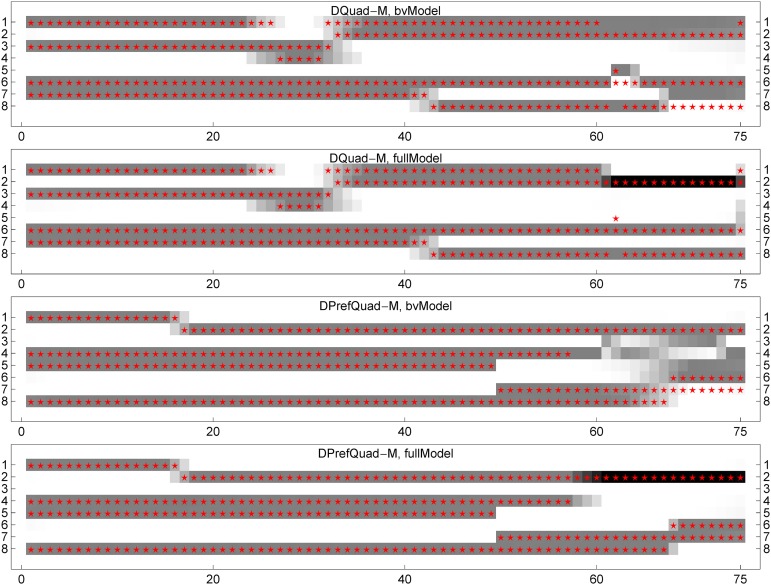
Figure 4 shows the posterior haplotype probabilities along the chromosomes for two offspring, one from each of DQuad-M and DPrefQuad-M. The haplotype probability refers to the probability of each allele at a locus being one of the eight parental origins of the two parents. The gradual changes of gray levels around the recombination breakpoints indicate the large uncertainties of identifying the true ancestral states there. Both the bvModel and the fullModel estimate the parental origins very well in the chromosomal region without double reduction. The fullModel successfully identifies the true double-reduction states on the right end of chromosomes (Figure 4), while the bvModel identifies parental origins correctly for only two or three of four alleles at a locus. Figure S2 shows similar results from the posterior genotype probabilities along the chromosomes.
Figure 4.
The posterior haplotype probabilities for two typical offspring. The top two panels show the results obtained from DQuad-M using the bvModel and the fullModel, respectively; the bottom two panels, for the offspring from DPrefQuad-M. The x-axis refers to the Nt = 75 SNP markers, and the y-axis refers to the eight parental chromosomal origins of two parents. The red stars denote the true ancestral origins, and the gray levels denote the haplotype probabilities, with white = 0 and black =0.5; the haplotype probabilities at each marker sum to 1. The black bands indicate the double reduction where two copies of alleles have the same parental origin.
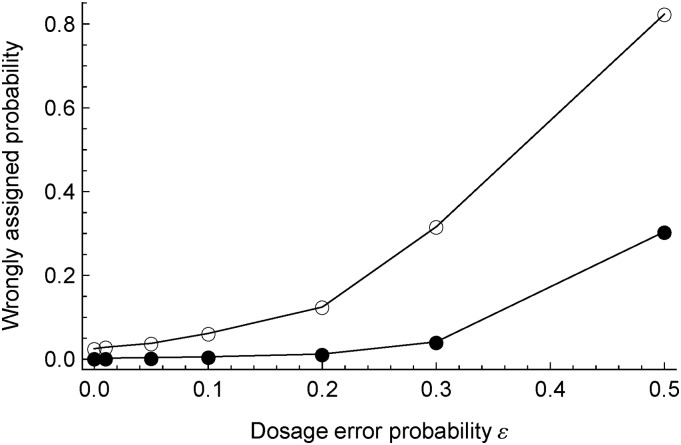
Effects of dosage errors
To evaluate the effects of dosage errors, we analyze the data sets derived from DStd-M by combining two marker densities Nt = 150 and 1200 and seven dosage error probabilities ε = 0, 0.01, 0.05, 0.1, 0.2, 0.3, and 0.5. All the No = 200 offspring are included. Each derived data set is obtained by applying errors to offspring dosages using the dosage error model described in the Methods section. We do not apply errors on parental dosages partly because of the intensive computational load but mainly because these errors might be detectable in real data based on offspring dosages. We use the true dosage error probabilities and the bvModel in the following phasing analysis and ancestral inference.
Table S1 shows the results of the estimated parental linkage phases. The true parental haplotypes are successfully recovered from the sparse marker data sets with ε ≤ 0.2 and from the dense marker data sets with ε ≤ 0.3. For the data sets with large dosage error probability, the phasing algorithm failed to find the global maximization when the number of phasing runs reached the default threshold .
Figure 5 shows the effects of offspring dosage errors on ancestral inference conditional on true parental haplotypes. The wrongly assigned probability increases nonlinearly with the offspring dosage error probability: the dosage errors have very little effect on ancestral inference for the data sets with dosage error probability less than ∼0.2.
Figure 5.
Effect of offspring dosage error on ancestral inference, conditional on true parental linkage phases. The filled (open) circles refer to the derived DStd-M with the number No = 200 offspring and the number Nt = 1200 (Nt = 50) SNP markers.
Comparisons using simulated data
We compare the performance of TetraOrigin with H2013 (Hackett et al. 2013) using the simulated data sets. For H2013, the parental linkage phasing involves heuristic algorithms for placing nonsimplex markers with respect to the phased framework of simplex markers (segregation type 01) and subsequent manual assignment for the remaining markers. Because only a few simplex markers are available for small simulated data sets and extensive manual work is required for large data sets, we perform the comparisons on ancestral inference but not on parental linkage phasing.
Table 3 shows the comparisons of ancestral inferences conditional on the true parental haplotypes, where H2013 cannot be applied to DQuad-M and DPrefQuad-M. Except for DStd-M-Long, the wrongly assigned probabilities for the sparse marker data (Nt = 75) using H2013 are around two times larger than those using TetraOrigin and 1.5 times larger than the dense marker data (Nt = 300). As shown in Table S2, the differences become smaller when the estimations are compared in terms of the wrongly called probability. Here the wrongly called probability is given by one minus the fraction of the calls being the true states, where the calls are determined by the ancestral states with the maximum posterior probabilities. Consistently, Figure S3 shows that the posterior genotype probabilities along the chromosomes obtained from H2013 are noisier than those from TetraOrigin.
Table 3. Comparisons of ancestral inference between TetraOrigin and H2013 for the 14 simulation scenarios (Table 1).
| Data set | No = 100, Nt = 75 | No = 100, Nt = 300 | ||||
|---|---|---|---|---|---|---|
| TetraOrigin (bvModel) | TetraOrigin (fullModel) | H2013 | TetraOrigin (bvModel) | TetraOrigin (fullModel) | H2013 | |
| DStd-M | 0.061 | 0.061 | 0.144 | 0.015 | 0.015 | 0.023 |
| DPref-M | 0.060 | 0.060 | 0.136 | 0.014 | 0.014 | 0.022 |
| DQuad-M | 0.164 | 0.062 | N/A | 0.122 | 0.012 | N/A |
| DPrefQuad-M | 0.146 | 0.065 | N/A | 0.101 | 0.013 | N/A |
| DStd-01 | 0.157 | 0.157 | 0.329 | 0.038 | 0.038 | 0.053 |
| DStd-02 | 0.126 | 0.126 | 0.222 | 0.027 | 0.027 | 0.036 |
| DStd-11 | 0.059 | 0.059 | 0.149 | 0.013 | 0.013 | 0.022 |
| DStd-12 | 0.043 | 0.043 | 0.114 | 0.011 | 0.011 | 0.018 |
| DStd-13 | 0.060 | 0.060 | 0.158 | 0.014 | 0.014 | 0.023 |
| DStd-22 | 0.042 | 0.043 | 0.098 | 0.008 | 0.008 | 0.016 |
| DStd-M-V0.25 | 0.064 | 0.064 | 0.148 | 0.014 | 0.014 | 0.021 |
| DStd-M-V1 | 0.067 | 0.067 | 0.155 | 0.015 | 0.015 | 0.025 |
| DStd-M-Local | 0.100 | 0.101 | 0.171 | 0.020 | 0.020 | 0.026 |
| DStd-M-Long | 0.222 | 0.229 | 0.287 | 0.097 | 0.107 | 0.066 |
For each scenario, two sub–data sets are specified by the number No = 100 offspring and the number Nt = 75 or 300 markers. Each cell gives the wrongly assigned probability conditional on the true parental haplotypes. The DQuad-M and DPrefQuad-M data sets could not be analyzed with H2013.
Table 3 and Table S2 indicate that TetraOrigin extracts more information from marker data than H2013. As a result, for DStd-M-Long, TetraOrigin performs generally a bit worse than H2013, indicating that TetraOrigin is more sensitive to the long-range disturbances of marker ordering.
Comparisons using real potato data
We evaluate TetraOrigin by using real SNP dosage data from potato (Hackett et al. 2013) for a comparison of results with H2013. For the potato mapping population of parents and 190 offspring, 1093 of the 5378 polymorphic SNPs are assigned to the constructed genetic map of 12 chromosomes (Hackett et al. 2013). We set the dosage error probability to be 0.01 for the dosage data, assuming no dosage error for the two parents. We analyze each chromosome (linkage group) independently.
Table 4 shows the comparisons for each of the 12 chromosomes between TetraOrigin (bvModel) and H2013. The parental haplotypes estimated by the two methods are the same for seven chromosomes, and for the other chromosomes, there are a few mismatches mainly around the telomeres (Figure S4). The haplotypes estimated by TetraOrigin are strongly supported in terms of marginal likelihood (Table 4).
Table 4. Comparisons between TetraOrigin (bvModel) and H2013 obtained from the real potato data.
| Chromosome: | I | II | III | IV | V | VI | VII | VIII | IX | X | XI | XII |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. of SNPs: | 142 | 120 | 74 | 152 | 119 | 122 | 89 | 85 | 91 | 104 | 85 | 118 |
| No. of mismatchesa: | 0 | 0 | 6 | 0 | 0 | 0 | 0 | 0 | 6 | 12 | 6 | 4 |
| Δloglb: | 0 | 0 | 164.4 | 0 | 0 | 0 | 0 | 0 | 21.4 | 1248.4 | 227.7 | 309.3 |
| Consistencyc: | 0.91 | 0.91 | 0.84 | 0.88 | 0.93 | 0.88 | 0.91 | 0.90 | 0.72 | 0.86 | 0.70 | 0.92 |
The number of mismatches between the parental haplotypes estimated from TetraOrigin and those from H2013.
The log likelihood given the TetraOrigin estimated parental haplotypes minus that given the H2013 estimated parental haplotypes.
The fraction of ancestral states called by their maximum posterior probabilities that are consistent between two methods.
We perform ancestral inference by TetraOrigin (bvModel) and H2013, conditional on their estimated parental haplotypes. We call the ancestral states at all the markers for each offspring by their maximum posterior probabilities and calculate the fraction of consistent calls between the two methods. As shown in Table 4, on average, of the calls are consistent, while, of course, the true ancestral states are unknown.
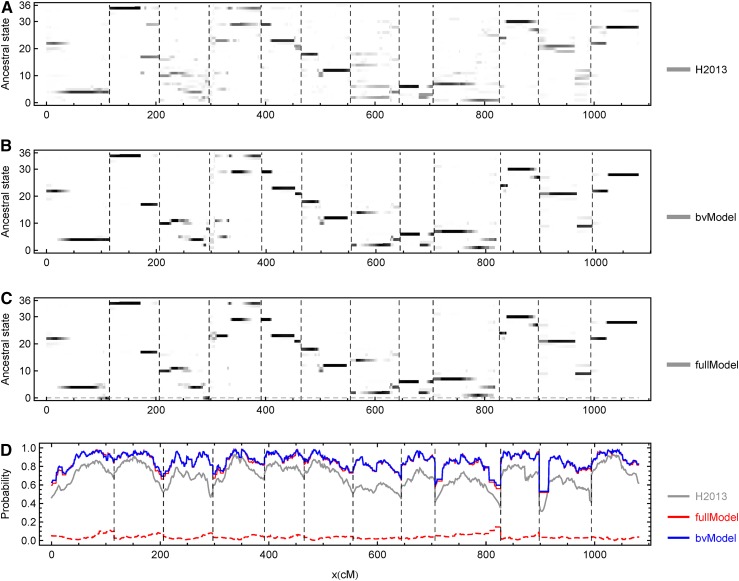
Figure 6 (A–C) shows the genome-wide posterior genotype probabilities for a typical offspring obtained by H2013 and TetraOrigin (bvModel and fullModel). Consistent with the simulation studies (Figure S3), the results from H2013 are noisier than those from TetraOrigin. For example, the ancestral states around 600 cM (chromosome VII) and 980 cM (chromosome XI) are more likely to be identified unambiguously in TetraOrigin. The results from TetraOrigin (fullModel) are different from those from TetraOrigin (bvModel) only for chromosomes I, III, and IV (Figure 6C). The results from TetraOrigin (fullModel) indicate double-reduction segments on the right ends of chromosomes I and III, while the tiny double-reduction segment around the middle of chromosome IV (∼330 cM) is likely to be artifactual.
Figure 6.
Evaluation of ancestral inference along each chromosome obtained from the real potato data set. The dashed vertical lines indicate the chromosomal boundaries. (A–C) The posterior genotype probabilities for an offspring obtained from H2013, TetraOrigin (bvModel), and TetraOrigin (fullModel), respectively. The probabilities are represented by the gray levels, with white = 0 and black = 1. The dashed horizontal line in C denotes the collapsed genotypes with double reduction in the fullModel. (D) The maximum posterior probabilities averaged over the 190 offspring. The gray, blue, and red lines refer to the results obtained from H2013, TetraOrigin (bvModel), and TetraOrigin (fullModel), respectively; the results from the bvModel and fullModel overlap. The red dashed line denotes the estimated double-reduction probabilities by the fullModel.
Figure 6D shows the maximum posterior genotype probabilities along the chromosomes averaged over the 190 offspring. The probabilities obtained from TetraOrigin are much larger than those from H2013. TetraOrigin (fullModel) indicates that the average double-reduction probability is around 0.04, larger than the estimate based only on a subset of markers of a different potato mapping population (Bourke et al. 2015). As expected, double reduction occurs more frequently at the telomeres than near the centromere (Figure 6D).
Discussion
We have developed a novel statistical framework for multilocus haplotype reconstruction in outcrossing tetraploids from SNP dosage data, where the two-stage implementation of parental linkage phasing and ancestral inference is built on an integrated network model of tetraploid inheritance in TetraOrigin. Simulation studies demonstrate that the new haplotype reconstruction is robust to preferential bivalent and quadrivalent pairing during meiosis, to the six marker segregation types, to erroneous genetic maps (except in the case of long-range disturbances in marker ordering), and to the various fractions of missing data in parents and offspring dosages.
We have compared the performance of TetraOrigin and the methodology described in Hackett et al. (2013), H2013. For ancestral inference, H2013 cannot be applied to data sets with quadrivalent pairings, and otherwise, the results are noisier than those from TetraOrigin. For parental linkage phasing, H2013 uses a heuristic multilocus algorithm and requires manual manipulation of intermediate results, and thus, it is less accurate and more time-consuming than TetraOrigin, which uses a fully probabilistic multilocus algorithm (Table 4). In addition, the algorithm for phase reconstruction by H2013 starts from a framework of simplex SNPs and thus has potential difficulties in analyzing data sets with a limited number of simplex SNPs, although this has not been a problem in practice (C. Hackett, personal communication).
Modeling quadrivalent pairing has been a theoretically challenging topic in quantitative genetics, and most studies have been built on the pioneering work of Fisher (1947) and Mather (1936). Fisher (1947) proposed a conceptual two-locus tetrasomic model where all 136 gamete genotypes are classified into 11 modes of gamete formation according to the occurrence of double-reduction and recombination events between two loci. Luo et al. (2004) established a deterministic relationship between the coefficient of double reduction at two linked loci and the recombination fraction between them, which subsequently has been applied to two- and multilocus linkage analysis (Luo et al. 2006; Leach et al. 2010).
In contrast to Leach et al. (2010), we have built a simpler model of quadrivalent pairing, assuming that the ancestral origins along a chromosome follow a time-homogeneous Markov chain independently between two homologous (homeologous) chromosomes of a diploid gamete. Remarkably, our nonmechanistic model produces very good estimations of double reduction (Figure 3), indicating that the marker data (200 offspring and 75 markers within the linkage group) provide enough information on double reduction. Also, the accurate estimation may be due to the equivalence between the transition probability matrix of gamete genotypes in our model and that derived by Leach et al. (2010) based on the 136 two-locus gamete genotypes, although the transition in our model refers to the 16 phased (ordered) genotypes from one locus to the next, instead of the 10 unphased genotypes in Leach et al. (2010).
The simulation studies show that the haplotype reconstruction is not sensitive to intermarker distances, indicating that the assumption of no genetic interference is not critical and that improving the accuracy of the intermarker distances by multilocus linkage analysis may be of marginal value to haplotype reconstruction and thus to QTL mapping. We have also assumed implicitly that there is no selection and thus no segregation distortion. This assumption is used as a prior in TetraOrigin, and the calculated posterior probabilities of parental origins may contain segregation distortion information passed from marker data when parent and offspring dosages are known in distortion regions. However, if parental dosage information is missing in regions with distorted segregation, that might result in incorrect estimation of parental origin and QTL mapping.
TetraOrigin is very capable of handling missing data and dosage errors in parents and offspring. However, accounting for possible dosage errors in parents results in a more than 10-fold reduction in the computational speed of parental linkage phasing because the total number of phases over all the possible dosages per locus increases dramatically. For example, for DStd-M with 200 offspring and 300 markers, the running times on a standard desktop are around 10 and 144 min for the missing fractions of parental dosages being 0 and 1, respectively, assuming no parental dosage error. When accounting for parental dosage error, the running time for the missing fraction 0 of parental dosages would be similar to that for the missing fraction 1. Thus, it is advantageous to perform quality control of parental dosages based on offspring dosages. Accounting for dosage error in the offspring imposes no such computational cost, and error rates of up to in sparse data sets and in dense data sets are handled well (Figure 5). This flexibility may be pertinent to genotyping-by-sequencing data; in particular, relatively low genome coverage typically results in higher error frequency.
The nulliplex-simplex and nulliplex-duplex segregation types are the least informative for ancestral inference, yet these markers, along with simplex-simplex markers, have been used most commonly in mapping studies. If this is the case here, a lack of informative markers on the genetic map may limit the accuracy of estimation by our proposed method. However, if a genome sequence is available, the unmapped markers still may be roughly positioned. More important, current approaches in tetraploid species use most or all marker types (Hackett et al. 2013).
The network model can, in principle, be applied to higher, even-ploidy levels, but we would need to improve the phasing algorithm for computational efficiency. For example, the number of possible bivalent pairings in a tetraploid parent is 3, which increases to 15 for hexaploids and to 105 for octoploids. Thus, the number of combinations of bivalent pairings in two parents increases from for tetraploids, to for hexaploids, and to for octoploids. As a result, we cannot simply perform the maximization step with respect to all the possible combinations of bivalent pairings, at least for octoploid or higher polyploid species. At the tetraploid level, the rigorous framework of TetraOrigin yields accurate posterior genotype probabilities that are needed for downstream QTL analysis in outcrossing tetraploids, even in the absence of parental dosage information.
Acknowledgments
We acknowledge Peter Bourke and Fred A. van Eeuwijk for their helpful comments. R.E.V. and J.J. acknowledge financial support from the TKI polyploids project, ”A genetic analysis pipeline for polyploid crops” (project number BO-26.03-002-001). Work by C.A.H. was supported by the Scottish government’s Rural and Environment Science and Analytical Services Division (RESAS).
Author contributions: J.H. and M.C.A.M.B. initiated and conceived the study. C.Z. and M.C.A.M.B. designed the experiments. C.Z. developed the models and the algorithms, implemented the TetraOrigin software, and performed the analyses using TetraOrigin. R.E.V., J.J., and M.C.A.M.B. contributed to model development. R.E.V. generated the simulated data sets using PedigreeSim software. C.A.H. performed the ranalyses using the H2013 method. C.Z. wrote the first draft of the manuscript, and R.E.V., C.A.H., J.H., and M.C.A.M.B. contributed substantially to revisions. The authors declare no conflicts of interest.
Footnotes
Communicating editor: G. A. Churchill
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.115.185579/-/DC1.
Literature Cited
- Bourke P. M., Voorrips R. E., Visser R. G. F., Maliepaard C., 2015. The double reduction landscape in tetraploid potato as revealed by a high-density linkage map. Genetics 201: 853–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broman K. W., Wu H., Sen S., Churchill G. A., 2003. R/qtl: Qtl mapping in experimental crosses. Bioinformatics 19: 889–890. [DOI] [PubMed] [Google Scholar]
- Fisher R. A., 1947. The theory of linkage in polysomic inheritance. Philos. Trans. R. Soc. Lond. B Biol. Sci. 233: 55–87. [Google Scholar]
- Garcia A. A. F., Mollinari M., Marconi T. G., Serang O. R., Silva R. R., et al. , 2013. Snp genotyping allows an in-depth characterisation of the genome of sugarcane and other complex autopolyploids. Sci. Rep. 3: 3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelman A., Carlin J., Stern H., Rubin D., 2004. Bayesian Data Analysis. Chapman & Hall/CRC, Boca Raton, FL. [Google Scholar]
- Hackett C. A., Bradshaw J. E., McNicol J. W., 2001. Interval mapping of quantitative trait loci in autotetraploid species. Genetics 159: 1819–1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackett C. A., McLean K., Bryan G. J., 2013. Linkage analysis and qtl mapping using snp dosage data in a tetraploid potato mapping population. PLoS One 8: e63939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leach L. J., Wang L., Kearsey M. J., Luo Z. W., 2010. Multilocus tetrasomic linkage analysis using hidden Markov chain model. Proc. Natl. Acad. Sci. USA 107: 4270–4274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Wei Y., Acharya A., Jiang Q., Kang J., et al. , 2014. A saturated genetic linkage map of autotetraploid alfalfa (Medicago sativa L.) developed using genotyping-by-sequencing is highly syntenous with the Medicago truncatula genome. G3 4: 1971–1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu E. Y., Zhang Q., McMillan L., Pardo-Manuel de Villena F., Wang W., 2010. Efficient genome ancestry inference in complex pedigrees with inbreeding. Bioinformatics 26: i199–i207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Z. W., Hackett C. A., Bradshaw J. E., McNicol J. W., Milbourne D., 2001. Construction of a genetic linkage map in tetraploid species using molecular markers. Genetics 157: 1369–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Z. W., Zhang R. M., Kearsey M. J., 2004. Theoretical basis for genetic linkage analysis in autotetraploid species. Proc. Natl. Acad. Sci. USA 101: 7040–7045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Z. W., Zhang Z., Leach L., Zhang R. M., Bradshaw J. E., et al. , 2006. Constructing genetic linkage maps under a tetrasomic model. Genetics 172: 2635–2645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mather K., 1935. Reductional and equational separation of the chromosomes in bivalents and multivalents. J. Genet. 30: 53–78. [Google Scholar]
- Mather K., 1936. Segregation and linkage in autotetraploids. J. Genet. 32: 287–314. [Google Scholar]
- Mott R., Talbot C. J., Turri M. G., Collins A. C., Flint J., 2000. A method for fine mapping quantitative trait loci in outbred animal stocks. Proc. Natl. Acad. Sci. USA 97: 12649–12654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabiner L., 1989. A tutorial on hidden Markov models and selected applications in speech recognition. Proc. IEEE 77: 257–286. [Google Scholar]
- Stam P., 1993. Construction of integrated genetic-linkage maps by means of a new computer package—joinmap. Plant J. 3: 739–744. [Google Scholar]
- Stift M., Reeve R., van Tienderen P. H., 2010. Inheritance in tetraploid yeast revisited: segregation patterns and statistical power under different inheritance models. J. Evol. Biol. 23: 1570–1578. [DOI] [PubMed] [Google Scholar]
- Sybenga J., 1972. General cytogenetics. North-Holland, Amsterdam. [Google Scholar]
- Sybenga J., 1994. Preferential pairing estimates from multivalent frequencies in tetraploids. Genome 37: 1045–1055. [DOI] [PubMed] [Google Scholar]
- Voorrips R. E., Maliepaard C. A., 2012. The simulation of meiosis in diploid and tetraploid organisms using various genetic models. BMC Bioinformatics 13: 248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voorrips R. E., Gort G., Vosman B., 2011. Genotype calling in tetraploid species from bi-allelic marker data using mixture models. BMC Bioinformatics 12: 172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfram Research , 2012. Mathematica, v9.0 edn Wolfram Research, Inc., Champaign, IL. [Google Scholar]
- Xie C. G., Xu S. H., 2000. Mapping quantitative trait loci in tetraploid populations. Genet. Res. 76: 105–115. [DOI] [PubMed] [Google Scholar]
- Zheng C. Z., Boer M. P., van Eeuwijk F. A., 2015. Reconstruction of genome ancestry blocks in multiparental populations. Genetics 200: 1073–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The TetraOrigin package has been implemented in Mathematica 9.0 (Wolfram Research 2012) and is freely available under the GNU General Public License from the website https://github.com/chaozhi/TetraOrigin.git. The full data sets for the 14 simulation scenarios and the real potato data extracted from Hackett et al. (2013) are included in the package.