Abstract
A century of genetic analysis has revealed that multiple mechanisms control the distribution of meiotic crossover events. In Drosophila melanogaster, two significant positional controls are interference and the strongly polar centromere effect. Here, we assess the factors controlling the distribution of crossovers (COs) and noncrossover gene conversions (NCOs) along all five major chromosome arms in 196 single meiotic divisions to generate a more detailed understanding of these controls on a genome-wide scale. Analyzing the outcomes of single meiotic events allows us to distinguish among different classes of meiotic recombination. In so doing, we identified 291 NCOs spread uniformly among the five major chromosome arms and 541 COs (including 52 double crossovers and one triple crossover). We find that unlike COs, NCOs are insensitive to the centromere effect and do not demonstrate interference. Although the positions of COs appear to be determined predominately by the long-range influences of interference and the centromere effect, each chromosome may display a different pattern of sensitivity to interference, suggesting that interference may not be a uniform global property. In addition, unbiased sequencing of a large number of individuals allows us to describe the formation of de novo copy number variants, the majority of which appear to be mediated by unequal crossing over between transposable elements. This work has multiple implications for our understanding of how meiotic recombination is regulated to ensure proper chromosome segregation and maintain genome stability.
Keywords: meiosis, whole-genome sequencing, crossing over, noncrossover gene conversion, interference
The proper segregation of homologous chromosomes at the first meiotic division is essential for the production of viable haploid gametes. In most instances, proper homolog segregation is assured by the formation of crossovers (COs), reciprocal recombination events that link homologous chromosomes together. COs arise at a subset of programmed double-strand breaks (DSBs) that are induced during early prophase by Spo11. DSBs not repaired as COs must be repaired by another mechanism, such as noncrossover gene conversion (NCO) events, sister chromatid exchange events, or by nonhomologous end joining (Do et al. 2013). In many organisms, COs and NCOs occur more frequently at specific regions of the genome, termed hotspots (Lichten and Goldman 1995; Hey 2004). The protein PRDM9 directs the formation of DSBs to these regions in some organisms (Baudat et al. 2010). Other organisms, such as Saccharomyces cerevisiae, lack a PRDM9-like protein but still have hotspots of recombination, and still other organisms lack PRDM9 and display a more even distribution of COs and NCOs (Auton et al. 2013; Singhal et al. 2015), suggesting that PRDM9-independent mechanisms may influence DSB formation. No equivalent of PRDM9 has been identified in D. melanogaster or other species within the Drosophila genus (Heil and Noor 2012; Manzano-Winkler et al. 2013).
Drosophila oocytes experience ∼11–17 DSBs per meiosis that are restricted to the euchromatin (Jang et al. 2003; Mehrotra and McKim 2006; Lake et al. 2013). How the position of these DSBs is determined and their fate (whether they become COs or NCOs) is poorly understood. Based on previous studies, the overall distribution of COs in D. melanogaster oocytes appears to be controlled by multiple mechanisms, most notably crossover interference and the centromere effect (Dobzhansky 1930; Beadle 1932; Offermann and Muller 1932; Lindsley and Sandler 1977). The identification of these mechanisms began with the finding that the genetic distance between phenotypic markers examined was not consistent with the physical location of the genes on polytene maps for any of the five major chromosome arms (Dobzhansky 1930). This suggested that the frequency of crossing over was not proportional to physical distance. Indeed, as noted by Lindsley and Sandler (1977), the frequency of exchange is lowest in both the centromere-proximal euchromatin and telomeric regions and highest in the medial region of the chromosomes (Supplemental Material, Figure S1). Later studies showed that the reduced level of exchange in the proximal euchromatin reflects the activity of the centromere effect, which strongly reduces crossing over in a polar fashion in centromere-proximal regions of the genome (Beadle 1932; Offermann and Muller 1932; Sturtevant and Beadle 1936; Yamamoto and Miklos 1977). Recent work in S. cerevisiae has shown that the Ctf19 inner kinetochore subcomplex suppresses centromere-proximal COs by suppressing pericentric DSBs, the first demonstration of a specific protein or complex contributing to the centromere effect (Vincenten et al. 2015). Other studies suggest that the telomeres may also suppress exchange in a polar fashion, although the effect is substantially weaker than near the centromeres (reviewed in Hawley 1980).
The distribution of COs is also influenced by crossover interference, which can act over long distances. First described in Drosophila by Sturtevant and Muller (Sturtevant 1913, 1915; Muller 1916), interference prevents a second CO from forming near an existing CO, typically ensuring the wide spacing of double crossover (DCO) events. Although interference in other organisms appears to be mediated by modification of the synaptonemal complex (SC) in response to COs (Sym and Roeder 1994; Libuda et al. 2013; Zhang et al. 2014), it remains unclear whether the SC also plays a role in mediating interference in D. melanogaster (Page and Hawley 2001). Finally, there is little information in Drosophila as to what degree, if any, interstitial sites or domains play in controlling the frequency of crossing over in specific euchromatic regions.
Several groups have employed whole-genome sequencing (WGS) to search for regions of increased crossing over, or recombination hotspots, in D. melanogaster. One method is to identify COs in pedigrees generated through controlled crossing schemes. Using this design, two recent studies failed to find strong evidence of hotspots in D. melanogaster, but did identify evidence for intervals of higher or lower rates of crossing over either within the region studied or at the whole-genome level (Comeron et al. 2012; Singh et al. 2013). These observations suggest that traditional recombination hotspots may not exist in Drosophila (Manzano-Winkler et al. 2013). A second approach, which infers recombination rates from population genetic data, also indicated that Drosophila likely does not have hotspots (Chan et al. 2012).
While it is known that interference and the centromere effect control crossover distribution, very little is known about the factors that control the distribution of NCOs in Drosophila. Although early genetic studies suggested that NCOs do not exert interference on COs or respond to interference from COs (Hilliker and Chovnick 1981), these studies looked at only a small number of loci, and only one of them (the rosy locus) in great detail. A recent study using WGS to analyze progeny that were allowed to freely recombine for 1, 2, 5, or 10 generations has shown that, unlike COs, NCO sites appear to be evenly spaced throughout the genome (Comeron et al. 2012). However, this study did not specifically investigate the joint distribution of COs and NCOs and their relationship to each other after a single round of meiosis. Thus, the effect, if any, of interference on NCOs has yet to be investigated on a genome-wide scale after a single round of meiosis in wild-type individuals.
In the present study, we determined the precise position of CO and NCO events on all five major D. melanogaster chromosome arms in 196 single meioses. We found a paucity of COs in the centromere-proximal one-half of most chromosome arms, consistent with the influence of the centromere effect on crossing over. Furthermore, our data suggest that the degree to which interference controls CO positioning may vary across the genome. However, proximity to the centromere does not seem to reduce the frequency of NCOs in this region, suggesting that NCOs are not sensitive to the centromere effect. We also observed NCOs near sites of crossing over and near other NCO events, supporting the hypothesis that NCOs are not sensitive to interference.
Unbiased sequencing of a large number of closely related individuals allows for the recovery of unexpected meiotic events. For example, we recovered several DCOs much smaller than any previously observed in Drosophila. In addition, we observed three NCO events that appear to be the result of discontinuous repair, demonstrating the value of studying a large number of individual meiotic outcomes to elucidate novel or rare repair outcomes. Finally, analysis of all 196 individual male genomes revealed eight large copy number variants (CNVs) ranging in size from 17 kb to 855 kb, most of which appear to have been the result of unequal crossing over between transposable elements (TEs). This leads to a revision of the standard model of TE copy number control through ectopic recombination and suggests that as in humans, TE-mediated copy number variation plays an important role in creating genetic heterogeneity in D. melanogaster.
Methods
Fly stocks and husbandry
Lab strains of w1118 and Canton-S were isogenized as described in Miller et al. (2012). w1118 was isogenic for all four chromosomes, while Canton-S was heterozygous for the fourth chromosome as well as for 15,718 SNPs along 3.9 Mb of 2R, from 2R:21,413,827 to the telomere. All flies were kept on standard cornmeal–molasses and maintained at 25°.
DNA preparation, sequencing, alignment, and SNP calling
DNA for individual flies was prepared from single adult males using the Qiagen DNeasy Blood and Tissue Kit. DNA from parental lines was prepared from males and females. All flies were starved for 4 hr before freezing at −80° for at least 1 hr. One microgram of DNA from each was fragmented to 250-bp fragments using a Covaris S220 sonicator by adjusting the treatment time to 85 sec. Libraries were prepared using a Nextera DNA Sample Prep Kit and Bioo Scientific NEXTflex DNA Barcodes. The resulting libraries were purified using the Agencourt AMPure XP system (Beckman Coulter) then quantified using a bioanalyzer (Agilent Technologies) and a Qubit fluorometer (Life Technologies). For the first batch of 98 individual male flies, libraries were pooled into four groups and were run in four lanes each of an Illumina HiSequation 2500 instrument on either a 150-bp or 100-bp paired-end flowcell (Table S1). For the second batch of 98 individual male flies, libraries were pooled into four groups and run in two lanes each of an Illumina HiSequation 2500 instrument on a 125-bp paired-end flowcell (Table S1). For all runs, HiSeq Control software 2.0.12.0 and Real-Time Analysis version 1.17.21.3 were used. Secondary Analysis version CASAVA-1.8.2 was run to demultiplex reads and generate FASTQ files. Alignment to the D. melanogaster reference genome (dm6, University of California, Santa Cruz) was performed using bwa version 0.7.7-r441 (Li and Durbin 2009). After alignment, Picard and GATK were used to mark duplicate reads and perform local realignment around InDels (McKenna et al. 2010). SNPs were identified using SAMtools version 0.1.19-44428cd and BCFtools version 0.1.19 (Li et al. 2009).
Males were numbered based on whether their father was homozygous w1118 or Canton-S and the number of their heterozygous mother. For example, male cs12.3 had a Canton-S father, its mother was female number 12, and it was the third male selected for DNA extraction. Sibling numbers may not be continuous, as males with low DNA concentrations after DNA extraction were not selected for sequencing.
Identification of sites of crossing over and gene conversion
Parental SNPs with quality scores ≥220 and a read depth of at least 20 were used to identify CO and NCO events in offspring. Only locations with a SNP present in one parent and a reference allele in the other parent were considered in subsequent analysis of the offspring. For each offspring, SNPs with a quality score <200 and read depth <10 were omitted from analysis. For the hemizygous X chromosome, instances where the parent of origin switched from one stock to another were flagged as sites of a potential meiotic event. For the autosomes, the same strategy was used except candidate events were flagged when the parent of origin switched from either a single parent to both parents or from both parents to a single parent of origin. Each putative CO and NCO event was then visually validated using the Integrative Genomics Viewer (Thorvaldsdottir et al. 2013). No CO events were excluded based on visual observation. Events flagged as potential NCOs that were due to local misalignment were excluded. While performing data analysis, we found that lower quality thresholds for SNP calling in either parents or offspring resulted in a high number of false positive NCO events, with the overwhelming majority due to nearby InDel polymorphisms or low-complexity sequence.
Validation of NCO events by PCR
To verify the accuracy of NCO identification, 47 of 291 NCOs (16%) were validated by PCR and Sanger sequencing (Table S4); Phusion polymerase (New England Biolabs) was used according to the manufacturer’s instructions. All 47 NCOs validated as real.
Calculation of NCO tract length and conversion rate
To jointly estimate the rate of NCO events and NCO tract length, we used the maximum likelihood (ML) approach modified from Miller et al. (2012). This method accounts for variable spacing between SNPs by taking into account the likelihood that a DSB fated to become an NCO gene conversion occurred within the span of neighboring SNPs of arbitrary distance, but the conversion tract failed to extend far enough to allow conversion to be seen. Using the entire distribution of distances between unconverted SNPs and the positioning of converted SNPs, we jointly estimate the per-base rate of NCO-fated DSB formation and the tract length parameter, modeled as a geometric process. This allows us to estimate the genome-wide rate of DSB formation and tract length considering the fact that some NCO conversion events will be missed. Since estimation of NCO tract length is difficult when spans between SNPs are large, we first jointly estimated the ML NCO rate and tract length parameters using 225 of 291 conversion tracts in which the distance between the converted and unconverted SNPs on both the left and right side of the NCO tract was <1 kb. We then fixed the tract length parameter and determined the ML NCO rate parameter using 286 of the 291 NCO events defined by SNPs closer than 10 kb apart or that were not part of discontinuous repair tracts.
Motif searching with MEME
To test for the presence of a motif enriched in or around sites of COs, we used MEME version 3.9.0 (Bailey and Elkan 1994; Bailey et al. 2006) to search the sequence surrounding 201 single crossovers (SCOs) defined by polymorphisms ≤500 bp apart (Table S2, Figure S5C). To account for factors acting outside of the apparent CO interval, the search window was expanded to include 1 kb upstream and downstream of each CO interval. We searched for motifs 5–12 bp long. To create a background distribution of motifs, we performed 100 trials where 201 COs were randomly placed along the five major chromosome arms with CO lengths randomly determined to be between 11 and 500 nucleotides long. Four motifs, [AT]GC[TA]GC[TA]GC[AT]GC[TA], ATAT[AG]TA[TC]ATAT, [TGC][TGC]TGGCCA[ACG][ACG], and AA[TA]T[GT][CA]A[AT]TT (Figure S6) were found to be significantly enriched in the observed CO spans but were also found in ≥21 of our randomly sampled CO intervals, suggesting that they are unlikely to be real.
Statistical methods and modeling
The probability of recovering the observed number of SCO, DCO, and triple crossover (TCO) or greater events was calculated by randomly distributing 541 CO events among 980 chromosome arms. Observed and expected values based on 100,000 trials can be found in Table S7.
An expected distribution of distance between DCO events was created by conservatively assuming equal numbers of COs across the five chromosome arms of 196 individual flies and distributing 541 CO events randomly among 980 chromosome arms in 100,000 trials. The average distance between randomly placed DCO events was calculated to be 8.1 Mb.
An expected distribution of the distance between randomly distributed COs and NCOs was created by placing 541 COs and 291 NCO events randomly along the five major chromosome arms. The distance between events occurring on the same arm was then calculated. If one NCO and two COs occurred, then the distance from the NCO to both COs was calculated. If two NCOs and one CO occurred, then the distance from each NCO to the CO was calculated. The observed average distance between a CO and NCO was 8.4 Mb, the expected average distance was 8.8 Mb based on 100,000 trials assuming uniform distribution of both NCO and CO events randomly placed on 980 chromosome arms. Similarly, the expected number of two or more NCO events per chromosome arm and the distance between multiple NCO events per chromosome arm was created by 100,000 trials of randomly placing 291 NCO events along the five major chromosome arms.
Similarly, to determine if NCOs are shifted with respect to the centromere, 291 NCO events were randomly distributed among the five major chromosome arms and the number of NCOs in the proximal one-third of each chromosome arm was calculated. The observed percentage of NCO events in the proximal one-third of each chromosome arm was: X: 26%, 95% CI: 22–46%; 2L: 27%, 95% CI: 22–47%; 2R: 18%, 95% CI: 21–45%; 3L: 27%, 95% CI: 22–45%; and 3R: 29%, 95% CI: 21–45%. A total of 100,000 trials were performed to calculate the confidence interval for each arm.
Data availability
Strains are available on request. Sequencing data from this project has been deposited at the National Center for Biotechnology Information (NCBI) under the following project numbers: PRJNA285112 (isogenized w1118 and Canton-S parental stocks) and PRJNA307070 (196 individual males). Scripts used to align genome data, to call SNPs, to identify CO and NCO sites, and to estimate NCO rate and tract length can be found at https://github.com/danrdanny/2016_CO_NCO_Paper.
Results
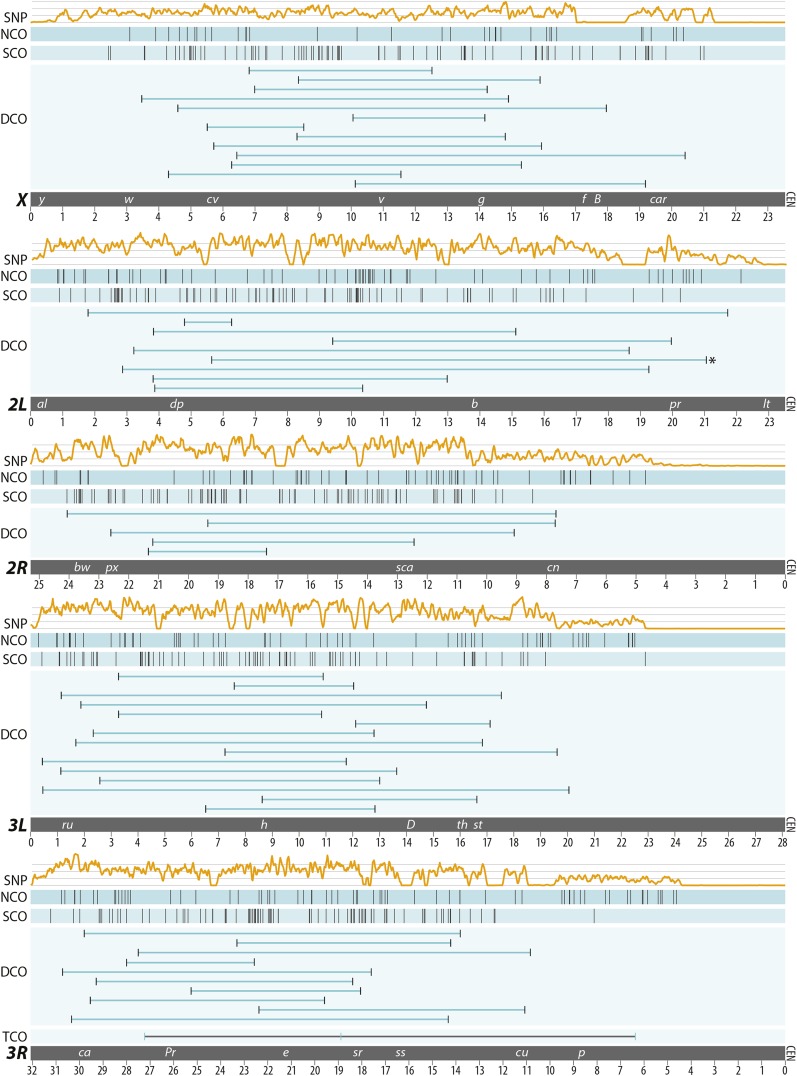
We directly assessed the number and position of COs and NCOs in D. melanogaster using females obtained by crossing two divergent, isogenic stocks—w1118 and Canton-S—which are wild-type laboratory lines commonly used in meiotic segregation assays (Page et al. 2008; Miller et al. 2012; Collins et al. 2014). Isogenized parental lines were found to be different at 486,549 SNPs by WGS. Specifically, we identified on average 1 SNP every 379 base pairs on the X, 1/192 bp on 2L, 1/295 bp on 2R, 1/241 bp on 3L, and 1/302 bp on 3R. Heterozygous F1 females were crossed to either homozygous w1118 males or homozygous Canton-S males and 196 of the resulting F2 male offspring were individually whole-genome sequenced (Figure S2). The F2 males were sequenced to an average X-chromosome depth of 24× (minimum average: 8×, maximum average: 45×) and an average autosomal depth of 45× (minimum average: 14×, maximum average: 87×) (Table S1). By analyzing the euchromatic portion of the genome, 541 sites of crossing over and 291 NCOs were identified by changes in the haplotype origin along the maternally transmitted chromosomes (see Methods) (Figure 1, Figure S3, Figure S4, Table S2, and Table S3). We observed no significant difference in the number or distribution of recombination events recovered from heterozygous females that were the progeny of reciprocal crosses between w1118 and Canton-S (Figure S2).
Figure 1.
Distribution of 541 COs and 291 NCOs recovered in this study. Each panel represents one of the five major D. melanogaster chromosome arms (the 1.4-Mb 4th chromosome was not examined in this study). The centromere (CEN) resides on the right side of each panel. The top track in each panel shows the SNP density observed when comparing the Canton-S and w1118 stocks. Note that SNP density drops to zero in the centromere-proximal regions of most chromosome arms, reflecting the recent addition of previously unmapped sequence to the latest D. melanogaster genome release. The NCO and SCO tracks show the locations of all NCOs and single COs recovered, respectively; the DCO tracks show the locations and spans of all double COs. One DCO on 2L (denoted by *) was partly the result of unequal crossing over between two transposable elements. One TCO was recovered on 3R. The centromere effect shifts crossovers distally on the autosomal arms; note that close to 80% of the SCOs on each autosomal arm occur in the distal one-half of the chromosome, but that frequency is only 60% in the distal one-half of the X chromosome. Commonly used visual markers are shown in the bottom track of each panel; descriptions of each can be found at FlyBase (http://www.flybase.org). Chromosome coordinates are in megabases along the x-axis.
To assess the number of false-positive NCOs recovered, we randomly selected 28 of 79 NCO events defined by a single SNP and 19 of 41 NCO events defined by two SNPs for sequence verification, because we considered those the most likely to be false-positive NCO events. We validated all of the 47 selected NCO events by PCR and Sanger sequencing, giving us high confidence that the remaining NCO events are, in fact, real (Table S4).
Distribution of single COs
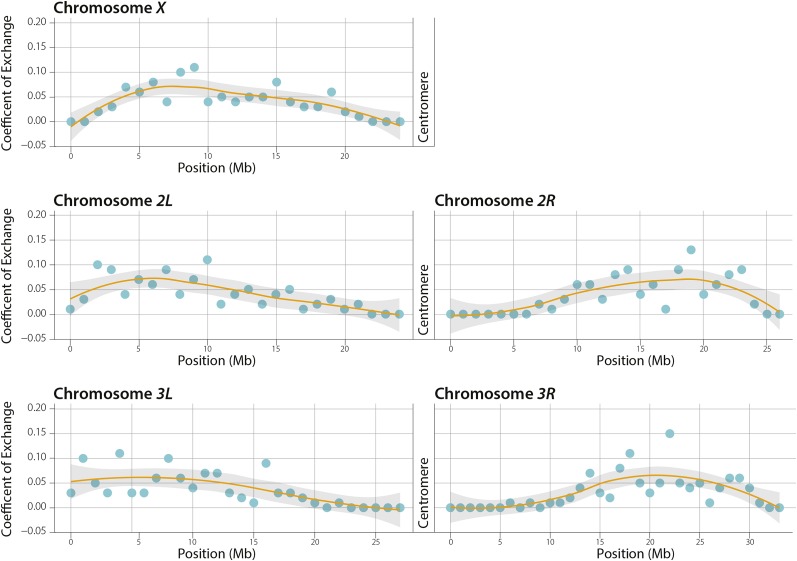
The observed pattern of COs followed a distribution expected by traditional phenotypic marker analysis, with the four autosomal arms displaying a paucity of COs in the centromere-proximal euchromatic sequence due to the centromere effect, and a less pronounced telomere effect shifting COs away from the telomeric regions on all five major chromosome arms (Figure 2). For example, for the four autosomal arms, 72–83% of the SCOs were in the distal one-half of the chromosome arm (Table S2), demonstrating the ability of the centromere effect to alter the proximal distribution of SCO events.
Figure 2.
Coefficient of exchange. COs are plotted in 1-Mb intervals for the five major chromosome arms. The orange line is a best fit of the data and the gray shaded area indicates the 95% confidence interval of the best-fit line. The centromere effect is apparent along the four autosomal arms and the less pronounced telomere effect is apparent along all five arms.
Although the four autosomal arms displayed a relative paucity of centromere-proximal COs, a diminished centromere effect was seen on the X chromosome, with 59% of the SCOs occurring in the distal one-half of the chromosome arm (Figure 1, Table S2). This result parallels observations made in previous genetic studies (Baker and Hall 1972; Lindsley and Sandler 1977; Page et al. 2007) and was not unexpected because the X chromosome has a large block of heterochromatin residing between the centromere and the euchromatin that buffers the distance over which the centromere effect may act (Yamamoto and Miklos 1977; Lindsley and Zimm 1992). Indeed, studies have shown that the frequency of crossing over in the centromere-proximal euchromatin of the X chromosome can be greatly reduced simply by deleting large blocks of proximal heterochromatin (Yamamoto and Miklos 1978).
Double crossovers and crossover interference
In D. melanogaster, researchers have traditionally used a limited number of variably spaced visible markers to measure recombination on each chromosome. (Commonly used markers and their approximate locations are shown in Figure 1.) Using this method of analysis, a CO event occurring distal to the most distally located visual marker on a chromosome would not be evident. Importantly, the distal CO may be part of a DCO that would thus be scored as an SCO instead. Similarly, a small DCO occurring between two adjacent visual markers would also be concealed using standard recombination assays.
As anticipated from studies dating back to Weinstein (1918), we recovered far more SCOs, fewer chromatids that did not experience a crossover event (or parental chromatids), fewer DCOs, and fewer triple crossovers (TCOs) per chromosome arm than expected by chance (Table S5, chi-square P < 0.0001, based on 100,000 trials of randomly distributing 541 COs among 980 chromosome arms). Using WGS after a single round of meiosis further allowed us to precisely measure the distance between each crossover of a DCO and to identify closely spaced DCOs between visual markers, which could be missed by traditional recombination analysis. For example, the small 1.5-Mb DCO recovered between dp and b on 2L (Figure 1) may not have been apparent using visual markers alone. Similarly, several of the SCO events, such as the distalmost SCO on 2R (Figure 1), may also have been missed.
We recovered 52 chromatids with DCOs (Figure 1, Figure S3, Figure S4, and Table S6). Specifically, we identified 13 DCOs on the X chromosome, 9 on 2L, 5 on 2R, 15 on 3L, and 10 on 3R. The vast majority of all DCOs recovered were widely spaced, with an average distance between them of 10.5 Mb, significantly larger than expected by chance (P < 0.0001, expected average distance 8.1 Mb, binomial test based on 100,000 trials of randomly distributing 541 COs among the entire length of 980 chromosome arms). Of the 14 total DCOs recovered on the 2nd chromosome, 1 was the largest DCO observed in this study (19.9 Mb in male cs8.6; see Methods for an explanation of naming conventions) and 2 were among the smallest recovered (1.5 Mb in male w4.8; and 4.0 Mb in male w12.2) (Figure 1, Figure S3, Figure S4, and Table S6). Recovery of DCOs both as small as 1.5 Mb and as large as 19.9 Mb was unexpected. To determine how often we would expect to recover DCO events of these sizes by chance, we randomly distributed 52 DCO events across each of the 5 major chromosome arms and recovered a ≤2.0-Mb DCO in only 0.1% of 100,000 trials and a ≥19-Mb DCO in 0.2% of trials.
Interestingly, we also found that the strength of interference differed between chromosomes. Although the two arms of the 2nd chromosome had a similar, albeit slightly greater, number of SCO events as the other chromosome arms (X: 86, 2L: 88, 2R: 90, 3L: 84, 3R: 86), we observed proportionally fewer DCOs on the 2nd chromosome compared to the X and the 3rd chromosomes (P = 0.027, Fisher’s exact one-tailed test). If the strength of interference were equal across chromosome arms, we would expect all chromosomes to have a similar number of doubles. However, the number of doubles observed on the 2nd was proportionally about half that of the X and 3rd chromosomes, driven, in part, by the paucity of doubles observed on 2R.
These findings demonstrate the ability of crossover interference to influence the distribution of exchange events (Muller 1916; Lindsley and Sandler 1977; Berchowitz and Copenhaver 2010). They also may suggest that interference may not act equally among the five major chromosome arms. Indeed, the paucity of doubles observed on the 2nd compared to the X and 3rd suggest that interference may act differently on the 2nd chromosome than it does on the other chromosome arms (Figure 1).
To more quantitatively describe the distribution of chromosome arms that experienced no, one, or two CO events (denoted as E0, E1, and E2 bivalents, respectively), we employed the algebraic approach developed by Weinstein (1918) (Table S7). Our estimates of the frequencies of E1 and E2 bivalents are consistent with those obtained using data from much larger genetic studies of recombination on the X, 2nd, or 3rd chromosomes (Baker and Carpenter 1972; Parry 1973; Page et al. 2007; Collins et al. 2014). Specifically, our E2 values of 27% for the X, 18% for 2L, and 31% for 3L are similar to previously published datasets with much larger n values, and our E2 value of 10% for 2R is identical to a previously published study (Parry 1973) (Table S7). Taken together, these observations provide additional evidence that mechanisms of crossover control may differ between chromosome arms.
NCOs fail to show interference and are insensitive to the centromere effect
Although much is known about the distribution of COs in D. melanogaster, less is known about the genome-wide distribution of NCOs. Because of the challenge of identifying the precise location of large numbers of NCOs after a single meiosis, it has been unclear whether NCO events follow the same rules pertaining to interference and the centromere effect as COs.
The 291 NCOs identified in our study contained an average of 5.0 SNPs per event (minimum 1 and maximum 35; Table S3). The average maximum conversion tract length, defined as the distance between unconverted polymorphisms, was 1421 bp (Figure S5A, Table S3); the average minimum conversion tract length, defined as the distance between the first and last converted polymorphism, was 290 bp (Figure S5B, Table S3). The maximum likelihood estimate for the average tract length was found to be 440–442 bp (see Methods). This is consistent with previously reported estimates of conversion tract lengths, which range from 352 bp to 441 bp in studies using the rosy marker (Hilliker et al. 1994; Blanton et al. 2005) and from 476 bp to 518 bp in studies using WGS (Miller et al. 2012; Comeron et al. 2012). In addition, using maximum likelihood analysis, we find the conversion rate to be ∼2.1 × 10−8 per base pair per meiosis, consistent with a rate of ∼2.0 × 10−8 per base pair per meiosis reported for the rosy gene in two other studies (Hilliker et al. 1994; Blanton et al. 2005) and 1.8 × 10−8 per base pair per meiosis using WGS (Miller et al. 2012).
Determining the precise location of the observed NCOs on the genome sequence revealed 33 chromatids containing two or more conversion events, a number not significantly different than expected by chance (P = 0.8, based on 100,000 trials of randomly distributing 291 NCO events among 980 chromosome arms then counting the number of arms with two or more NCOs present), with 11 of the 33 instances occurring within 4 Mb of each other (P = 0.5, based on 100,000 trials of randomly distributing 291 NCO events, then counting those within 4 Mb of each other). Additionally, we found 128 instances where a conversion occurred on the same chromatid as either a single, double, or triple crossover event. Thirty-two of these conversions occurred within 4 Mb of a crossover, a number not significantly different from that expected by chance (P = 0.05, based on 100,000 trials of conservatively randomly distributing 541 CO and 291 NCO events, then counting those within 4 Mb of each other). Together, these data suggest that NCOs neither generate nor are subject to interference.
We next examined NCOs with respect to the centromere effect. On chromosome 2R, although none (0/90) of the SCOs fell within the centromere-proximal one-third of the chromosome arm, 18% (11/61) of the NCOs recovered were in this region (P < 0.0001, Fisher’s exact test) (Figure 1). In addition, only 2% (3/170) of the SCOs on the 3rd chromosome fell within the centromere-proximal one-third of either arm, whereas 27% (17/64) and 29% (19/66) of NCOs fell within that region on 3L and 3R, respectively (P < 0.0001, Fisher’s exact test). For each chromosome arm, we modeled a random distribution of conversion events and found that the number of NCO events in the proximal one-third of each chromosome arm was not different from those placed by random chance (see Methods for individual chromosome arm values and 95% confidence intervals). The only exception was chromosome 2R, in which we observed that significantly fewer NCOs occurred in the proximal one-third of the chromosome arm than expected by chance (P = 0.03). Note that although only 9% of the SNPs on chromosome 2R are found in the centromere-proximal one-third of the arm, all tests are based on the conservative assumption that SNPs are equally distributed along the chromosome arm, suggesting that this deviation is, in fact, explained by low SNP density in this region. These data therefore suggest that NCOs in D. melanogaster are insensitive to the centromere effect. Indeed, the paucity of SNPs in the most proximal region of each chromosome arm, including 2R, prevents us from determining exactly how close to the euchromatic/heterochromatic boundary conversions may occur (Figure 1); thus, we are likely underestimating the frequency of proximal conversion events.
To estimate the number of NCOs we may have missed due to decreased SNP density, we used data from the 291 NCOs we recovered to estimate the genome-wide NCO rate to be 2.1 × 10−8 conversions per base pair per meiosis. Applying this rate to the entire 132.5-Mb haploid genome (excluding the Y and 4th chromosomes and unmapped heterochromatic regions) yields 2.8 recoverable conversions per haploid meiosis (132.5 Mb × 2.1 × 10−8). Thus, in the 196 individual flies examined in this study, we should have recovered ∼549 NCO events had all events been equally detectable. Our observation that the NCOs we observe are insensitive to the centromere effect and to interference suggests that DSBs produced either near the centromere or in proximity to another DSB are preferentially repaired as NCOs. It is therefore likely that many of the 258 conversions we failed to detect (549 expected − 291 detected) occurred in regions of low SNP density, such as in the centromere-proximal euchromatic regions or in SNP deserts that occur randomly throughout the genome.
Recovery of complex NCO events
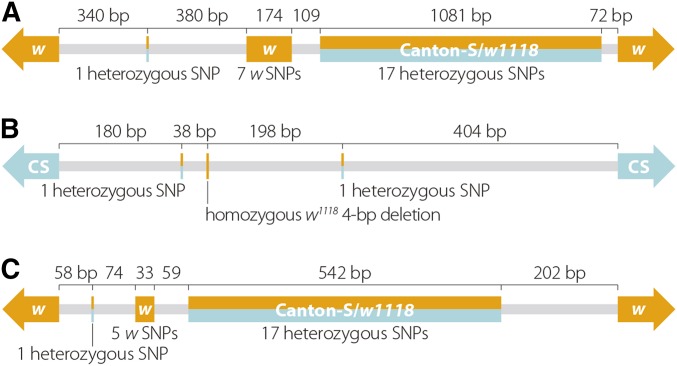
Unbiased recovery of NCO events on a genome-wide scale allows for the identification of unexpected meiotic repair products. We recovered three discontinuous NCOs on chromosome 2R that appear to be the result of either a mitotic repair event or a complex meiotic repair event. (When counting NCOs, we considered these three discontinuous tracts as single NCO events unless otherwise noted.) All three discontinuous repair events appear as two short conversion tracts with a nonconverted SNP between them (Figure 3). These events appear remarkably similar to a complex conversion event at rosy recovered by Carpenter (1982) and analyzed at the molecular level by Curtis and Bender (1991). There are several processes, including bidirectional repair or template switching during repair, that may have given rise to these events (Merker et al. 2003; Whitby 2005). Recovery and identification of more complex repair events such as this, perhaps by methods designed to enrich for them, would certainly contribute to the mechanistic understanding of the repair processes at play during D. melanogaster female meiosis.
Figure 3.
Recovery of complex NCO repair events. We recovered three instances of complex NCO repair similar to an event recovered by Carpenter (1982) and described by Curtis and Bender (1991). Exact coordinates for each NCO can be found in Table S3 and are based on D. melanogaster genome release 6 (dm6).
Transposable elements mediate copy number variation in Drosophila
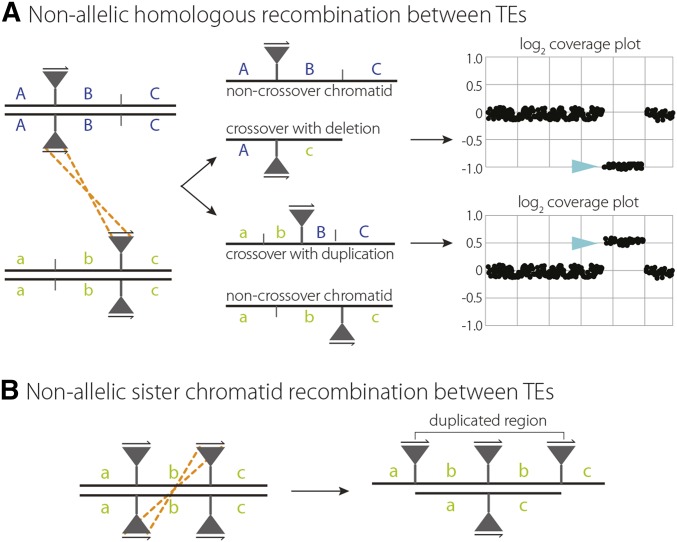
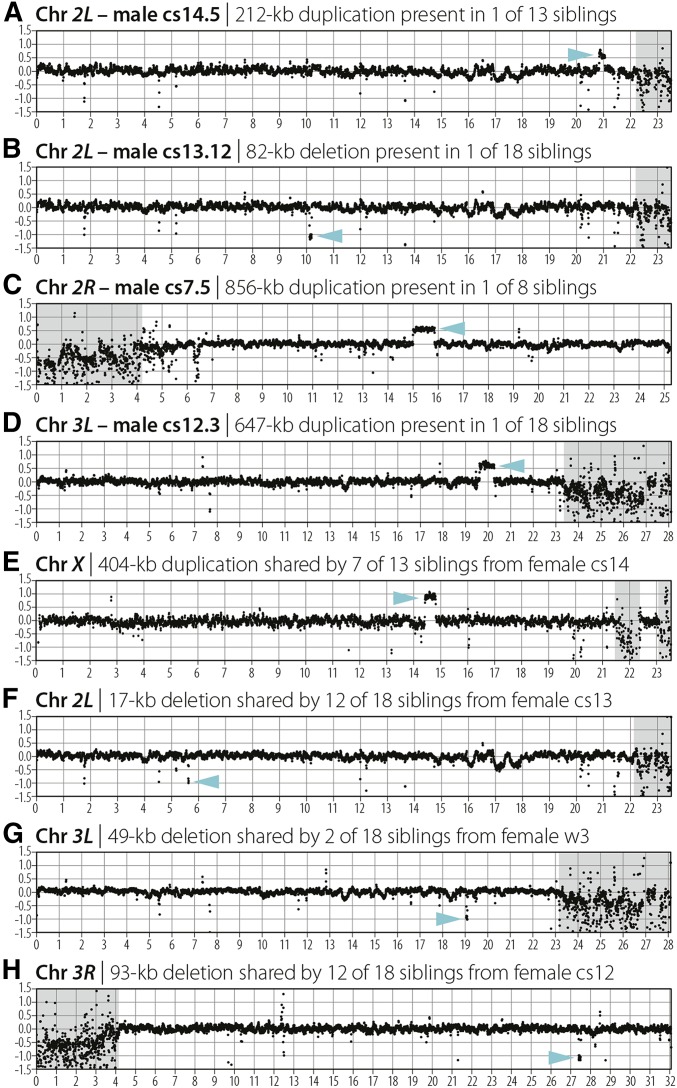
In addition to the recovery of complex meiotic repair events, whole-genome sequencing of individual flies also allowed us to observe evidence of ectopic exchange events mediated by TEs. TEs are mobile DNA elements that can replicate within a genome by moving into or near genes, sometimes with deleterious effects to the host. TEs have been shown to be an important component of genome evolution and are thought to cause large deletions or duplications through ectopic exchange or unequal crossing over either between homologs or sister chromatids (Figure 4) (Kaminker et al. 2002; Lee and Langley 2012). These CNVs may be visualized by plotting depth of coverage for an entire genome or region of interest (Figure 4A). We recovered one DCO (male cs14.5) on chromosome 2L in which the proximal of the two COs occurred at the same position as a TE present in the w1118 parental line but not in the Canton-S parental line (Figure 1, Table 1). The position of this proximal CO also defined a change in read depth, with ∼50% higher read depth on the distal side of the CO than on the proximal side (Figure 5A). Plotting read depth for the entire chromosome arm revealed a 212-kb duplication precisely defined by two TEs, with the distal TE present only in the Canton-S parental line and the proximal TE present only in the w1118 parental line (Table 1). We then created depth-of-coverage graphs for all 196 males sequenced in this study and identified three additional CNVs ≥10 kb in size that were present only in individual male offspring (Figure 5, B–D, Table 1), as well as four CNVs shared among multiple male siblings (Figure 5, E–H, Table 1).
Figure 4.
Model of unequal exchange between homologous chromosomes or sister chromatids. (A) Nonallelic homologous recombination between identical TEs on homologous chromosomes creates a CO with one chromatid carrying a duplication and another carrying a deletion. Expected log2 depth-of-coverage graphs are shown for autosomal duplications and deletions. (B) Unequal sister chromatid exchange between identical TEs creates one sister chromatid carrying a duplication and one with a deletion. Note that in these models TEs are oriented in the same direction.
Table 1. CNVs recovered in this study.
| Figure | CNV type | Chr | Proximal coordinate | Distal coordinate | Proximal featurea,b | TE orientationc | Distal feature | TE orientation |
|---|---|---|---|---|---|---|---|---|
| De novo CNVs | ||||||||
| 6A | Duplication | 2L | 20,845,594 | 21,057,582 | w: — | — | w: Roo | Unknown |
| cs: Roo | 3′–5′ | cs: — | — | |||||
| 6B | Deletion | 2L | 10,113,178 | 10,194,791 | w: — | — | w: — | — |
| cs: McClintock | Unknown | cs: — | — | |||||
| 6C | Duplication | 2R | 15,005,073 | 15,860,851 | w: hobo (DMHFL1) | 3′–5′ | w: hobo (DMHFL1) | 3′–5′ |
| cs: — | — | cs: — | — | |||||
| 6D | Duplication | 3L | 19,624,757 | 20,272,082 | w: DMIS297 | Unknown | w: — | — |
| cs: DMIS297 | Unknown | cs: DMIS297 | Unknown | |||||
| Inherited CNVs | ||||||||
| 6E | Duplication | X | 14,413,980 | 14,817,705 | w: Roo | Unknown | w: Roo | Unknown |
| cs: — | — | cs: — | — | |||||
| 6F | Deletion | 2L | 5,622,078 | 5,639,080 | w: — | — | w: — | — |
| cs: hobo (DMHFL1) | 3′–5′ | cs: hobo (DMHFL1) | 3′–5′ | |||||
| 6G | Deletion | 3L | 19,053,516 | 19,102,247 | w: — | — | w: — | — |
| cs: DMIS297 | Unknown | cs: DMIS297 | Unknown | |||||
| 6H | Deletion | 3R | 27,406,208 | 27,499,496 | w: — | — | w: — | — |
| cs: hobo (DMHFL1) | 3′–5′ | cs: — | — | |||||
Four CNVs (6A–6D) were recovered in only individual males and are thus likely to have arisen de novo. All four of these CNVs were defined by at least one TE in one of two parental genomes, and one CNV (6A) defined the proximal CO of a DCO event. Four CNVs (6E–6H) were shared among multiple individuals, and all four were defined by at least one TE present in one of the two parental genomes. The Figure column lists the panel in Figure 5 to which each event corresponds.
Roo, McClintock, hobo, and DMIS297 are different TE families; w = w1118; cs = Canton-S.
Em dash (—) indicates that no TE was observed.
TE, transposable element.
Figure 5.
Large de novo and inherited CNVs. Log2 depth of coverage for each chromosome arm is shown. Alignment of reads to heterochromatic regions (shaded in gray) is poor. Siblings are the number of males sequenced from an individual female. Arrowheads (blue) indicate the position of a CNV. Note that there appear to be many small CNVs (represented by single dots) along each chromosome arm that are simply differences between each stock and the D. melanogaster reference genome. (A–D) Candidate de novo events that were observed in only one male. (E–H) Representative CNVs that were inherited by more than one male from either their heterozygous mother or homozygous father.
The presence of a CNV in one male on the maternal haplotype that is not present in his siblings would suggest that the CNV is a de novo event; thus, it is likely that the CNVs in the four males represented in Figure 5, A–D are indeed de novo CNVs. Three of these four CNVs had identical TEs present at both sides of the CNV in either one or both of the parents (Table 1). The remaining de novo CNV was a deletion that contained a TE on only the distal side of the deletion in the Canton-S stock and no apparent parental TE or low-complexity sequence on the proximal side of the deletion (Table 1).
Because four of the CNVs observed were present in more than one sibling, it was presumed that these alleles were segregating in the parental germline. By analyzing TEs present in the w1118 or Canton-S stocks along with the flanking SNP profiles of the CNVs, we determined that three of the four inherited CNVs were likely sister chromatid recombination events mediated by identically oriented TEs (Figure 4B, Figure 5, E–H, Table 1). The remaining inherited CNV was a deletion defined on its proximal side by a TE present only in the Canton-S line but no apparent TE in either the w1118 or Canton-S stocks on its distal side. BLAST of read pairs from the distal side of the deletion revealed that all unmapped pairs matched a canonical hobo element (a TE family), suggesting that the deletion was mediated by the hobo element in the absence of an identical TE at the distal location.
Interestingly, four of the eight CNV events we observed lie in the proximal one-third of the chromosome arms, where crossing over is reduced. Previous studies have concluded that ectopic recombination is likely a major factor limiting the spread of TEs in natural populations (Charlesworth and Langley 1986). Our findings support this conclusion; however, our data also show that ectopic recombination occurs at a significant frequency in regions of the genome with lower recombination. This is surprising because the accumulation of TEs in centromere-proximal genomic regions has historically been thought to be caused by a low rate of ectopic recombination in these regions (Charlesworth and Langley 1989; Lee and Langley 2010). Our data suggest, rather, that the reduced efficacy of selection against TE-mediated CNV formation in regions of reduced recombination may contribute to the accumulation in these regions.
Discussion
Elucidating the properties controlling meiotic COs and NCOs has been of interest to Drosophila researchers for over a century. The present study helps to explain and clarify several observations that Drosophila researchers have made during that time. By examining 196 individual wild-type meiotic events, we are able to make accurate and precise observations about the number and position of COs and NCOs for the five major chromosome arms in D. melanogaster. We find, as expected, that COs are sensitive to the centromere effect and occur with less frequency in the centromere-proximal euchromatic regions of autosomal chromosome arms. NCOs, on the other hand, are not sensitive to the centromere effect and are often found in proximal euchromatic regions. NCOs also do not seem to be sensitive to or to generate interference and may occur close to crossover events, within double crossover events, and, surprisingly, even within close proximity to one another.
Crossover interference is evident in our dataset based on two observations. First, we recovered only 52 DCO events, significantly fewer than expected by random chance (Table S5), and only one TCO. Second, we find that DCOs are generally widely spaced, with an average distance of 10.5 Mb between the COs. Although interference is seen in many organisms from yeast to humans, the full mechanism remains a mystery (Berchowitz and Copenhaver 2010). D. mauritiana, a species closely related to D. melanogaster, appears to have about twice as many COs—with more centromere-proximal COs—per chromosome arm as D. melanogaster (True et al. 1995). It would be interesting to perform an experiment in D. mauritiana similar to the one described here to obtain detailed insight into the distribution and distance between these crossovers. Because these two species are ∼3 million years divergent (Lachaise et al. 1986), it may be possible to identify genes or polymorphisms that play an important role in this process.
In some organisms there are two main pathways for repairing DSBs as COs. These are referred to as the ZMM-dependent, or class I, pathway and the Mus81-dependent, or class II, pathway (Whitby 2005). Class II crossovers, which appear to act as a “backup” system in some organisms, are infrequent and are insensitive to interference (Novak et al. 2001; Hollingsworth and Brill 2004). Recent work in tomato (Solanum lycopersicum) using high-resolution microscopy to visualize class I and class II crossover events (Anderson et al. 2014) reported fewer class II events (18% of total COs) and found them much more often in the centromere-proximal euchromatin, suggesting that they may be less sensitive to the centromere effect than class I events. Perhaps then, it is possible that the 1.4-Mb DCO recovered on chromosome 2L, possibly the smallest DCO ever reported in D. melanogaster, is a product of the class II, noninterfering pathway, similar to the 3.0-Mb DCO observed on the X and the 4.0-Mb DCO observed on 2R. If these events are indeed class II events, they may be the first demonstration of this pathway in D. melanogaster.
Several studies have identified motifs associated with sites of crossing over in both D. melanogaster (Comeron et al. 2012; Miller et al. 2012; Singh et al. 2013) and other species of Drosophila (Cirulli et al. 2007; Kulathinal et al. 2008; Stevison and Noor 2010). Analyzing our current dataset, we detected no motifs enriched over background using 201 of the SCO events defined by two SNPs ≤500 bp apart (Figure S6). This finding suggests that either several motifs may be associated with COs, as reported by Comeron et al. (2012), or that crossovers in D. melanogaster may be associated with open chromatin and transcription in early meiosis (Adrian and Comeron 2013). Either one of these possibilities would make a true crossover-associated motif difficult to detect in a dataset of this size.
We recovered 541 total CO events and expected to recover 549 NCO events in our study (291 NCO events recovered, 258 NCO events not recovered). Early studies of NCO events at the ry locus recovered a significantly higher number of NCOs than COs (Chovnick et al. 1971), leading us to wonder whether we were underestimating the total number of NCO events we expected to recover. The increased ratio of NCO:CO events at ry in previous studies makes sense in light of our finding that NCO events are evenly distributed along the chromosome arm, while CO events are shifted to the distal two-thirds of the chromosome arm (Figure 1). Indeed, only four of 41 SCO events (9%) on 3R occurred between ry (3R: 13,032,528–13,038,020, cytological location 87D) and the centromere, demonstrating that few CO events are expected in this region. In addition, a smaller analysis of only the X chromosome from 30 individual males using the same genetic background recovered 15 COs, five NCOs, and calculated that seven additional NCO events were not recovered (Miller et al. 2012)—similar to the NCO:CO ratio observed in the current study.
During prophase I of meiosis I, ∼11–17 DSBs are produced per oocyte (Mehrotra and McKim 2006). Our likelihood analysis shows that 11 (2.8 per meiotic product × 4 haploid products of meiosis) of these DSBs will be repaired as NCO events. DSBs may also be repaired as COs and we recover an average of 2.8 COs per individual (541 COs/196 individuals) in this study. Because COs between homologs are apparent on only two chromatids, we estimate that 5.6 total COs (2.8 COs per individual × 2) are produced during a single meiosis, the same number reported in early estimates of 5.6 exchanges per meiosis (Lindsley and Grell 1967; Carpenter 1982). Therefore the observed number of CO events plus our estimate of the total number of NCO events likely account for nearly all the DSBs formed during meiosis.
The reported number of DSBs in repair-deficient mutants, 19–23, is somewhat higher than that observed in wild type (Jang et al. 2003; Mehrotra and McKim 2006; Lake et al. 2013). This difference may be because repair-deficient mutants may create more DSBs than wild type, as resolution of breaks as either COs or NCOs may provide feedback that limits the number of DSBs produced (Thacker et al. 2014). If this is the case, the absence of this feedback may artificially inflate the number of DSBs expected in a wild-type background and 19–23 may not be an accurate number of DSBs in D. melanogaster. If the true number of DSBs created during meiosis is indeed slightly higher than the 11–17 reported in wild type, then our analysis may be missing evidence of other repair events. Examples of these events include mismatch repair of a NCO that may mask an identifying SNP, causing an underestimation of the number of NCO events (Radford et al. 2007), or nonhomologous end joining or sister chromatid repair may resolve a DSB in a way that is undetectable using SNP or InDel polymorphisms (McVey et al. 2004; Johnson-Schlitz et al. 2007; Goldfarb and Lichten 2010).
Unexpectedly, we identified four large de novo and four large inherited CNVs in the 196 individual male genomes that we studied. Previous studies of TE-mediated copy number variation in Drosophila focused on assaying unequal exchange between one family of TEs, roo elements, near the white locus (Davis et al. 1987; Montgomery et al. 1991). Separately, a screen for de novo mutations resulting in eye color changes recovered five large deletions that were presumed to be the result of unequal exchange between TEs at different genomic positions (Watanabe et al. 2009). Using these five deletions, Watanabe et al. (2009) estimated the mutation rate for large deletions and duplications affecting multiple genes to be 1.7%, remarkably close to the 2% rate we observe for de novo CNVs in our study. The similar mutation rate observed in both studies supports the hypothesis that ectopic recombination is a common source of genetic variation in D. melanogaster and demonstrates the value of unbiased sequencing of individual meiotic products.
With one significant exception (Parry 1973), all previous genetic studies of recombination in Drosophila have focused on a single chromosome arm or studied offspring from recombinant inbred lines (Comeron et al. 2012). Ours is the first to characterize both NCOs and COs on all five major arms within a single meiosis. Our data show that the processes that position CO events are clearly distinct from those that position NCO events. It will be of considerable interest, as sequencing technology improves and declines in cost, to repeat this analysis in the presence of polar effect mutants or genotypes that elevate recombination. In addition, controlled crossing experiments such as ours should be repeated using recently isolated and characterized wild-type lines, which may carry polymorphisms affecting the distribution or number of meiotic repair events (Mackay et al. 2013; Lack et al. 2015), as it would be informative to identify lines in which these properties are significantly different from those described in this study or from each other. Much of this will involve repeating 20th century genetic assays with 21st century genomic approaches. But the goal will remain unchanged—to identify the mechanisms that ensure the proper number and position of exchanges and thus ensure the proper segregation of homologs at the first meiotic division.
Acknowledgments
We thank Angela Miller for editorial and figure preparation assistance; Kate Hall, Kendra Walton, Michael Peterson, Rhonda Egidy, and Anoja Perera for expert assistance with DNA sequencing; and members of the Hawley laboratory for helpful discussion and comments. R.S.H. and S.L.J. are supported by the Stowers Institute for Medical Research. R.S.H. is an American Cancer Society Research Professor supported by American Cancer Society award RP-05-086-06DDC. S.L.J. is supported by the American Cancer Society (RSG-11-030-01-CSM). J.P.B. is supported by National Science Foundation grants MCB-1022165 and MCB-1413532 and the National Institutes of Health (NIH-P20GM103638, principal investigator, Susan Lunte).
Footnotes
Communicating editor: M. P. Colaiácovo
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.115.186486/-/DC1.
Literature Cited
- Adrian A. B., Comeron J. M., 2013. The Drosophila early ovarian transcriptome provides insight to the molecular causes of recombination rate variation across genomes. BMC Genomics 14: 794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson L. K., Lohmiller L. D., Tang X., Hammond D. B., Javernick L., et al. , 2014. Combined fluorescent and electron microscopic imaging unveils the specific properties of two classes of meiotic crossovers. Proc. Natl. Acad. Sci. USA 111: 13415–13420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auton A., Rui Li Y., Kidd J., Oliveira K., Nadel J., et al. , 2013. Genetic recombination is targeted towards gene promoter regions in dogs. PLoS Genet. 9: e1003984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey T. L., Elkan C., 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 2: 28–36. [PubMed] [Google Scholar]
- Bailey T. L., Williams N., Misleh C., Li W. W., 2006. MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 34: W369–W373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker B. S., Carpenter A. T., 1972. Genetic analysis of sex chromosomal meiotic mutants in Drosophila melanogaster. Genetics 71: 255–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker B. S., Hall J. C., 1976. Meiotic mutants: genetic control of meiotic recombination and chromosome segregation, pp. 351–434 in The Genetics and Biology of Drosophila, Vol. 1a, edited by M. Ashburner and E. Novitski. Academic Press, New York.
- Baudat F., Buard J., Grey C., Fledel-Alon A., Ober C., et al. , 2010. PRDM9 is a major determinant of meiotic recombination hotspots in humans and mice. Science 327: 836–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beadle G. W., 1932. A possible influence of the spindle fibre on crossing-over in Drosophila. Proc. Natl. Acad. Sci. USA 18: 160–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berchowitz L. E., Copenhaver G. P., 2010. Genetic interference: don’t stand so close to me. Curr. Genomics 11: 91–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanton H. L., Radford S. J., McMahan S., Kearney H. M., Ibrahim J. G., et al. , 2005. REC, Drosophila MCM8, drives formation of meiotic crossovers. PLoS Genet. 1: e40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter A. T., 1982. Mismatch repair, gene conversion, and crossing-over in two recombination-defective mutants of Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 79: 5961–5965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan A. H., Jenkins P. A., Song Y. S., 2012. Genome-wide fine-scale recombination rate variation in Drosophila melanogaster. PLoS Genet. 8: e1003090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B., Langley C. H., 1986. The evolution of self-regulated transposition of transposable elements. Genetics 112: 359–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B., Langley C. H., 1989. The population genetics of Drosophila transposable elements. Annu. Rev. Genet. 23: 251–287. [DOI] [PubMed] [Google Scholar]
- Chovnick A., Ballantyne G. H., Holm D. G., 1971. Studies on gene conversion and its relationship to linked exchange in Drosophila melanogaster. Genetics 69: 179–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirulli E. T., Kliman R. M., Noor M. A. F., 2007. Fine-scale crossover rate heterogeneity in Drosophila pseudoobscura. J. Mol. Evol. 64: 129–135. [DOI] [PubMed] [Google Scholar]
- Collins K. A., Unruh J. R., Slaughter B. D., Yu Z., Lake C. M., et al. , 2014. Corolla is a novel protein that contributes to the architecture of the synaptonemal complex of Drosophila. Genetics 198: 219–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comeron J. M., Ratnappan R., Bailin S., 2012. The many landscapes of recombination in Drosophila melanogaster. PLoS Genet. 8: e1002905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis D., Bender W., 1991. Gene conversion in Drosophila and the effects of the meiotic mutants mei-9 and mei-218. Genetics 127: 739–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis P. S., Shen M. W., Judd B. H., 1987. Asymmetrical pairings of transposons in and proximal to the white locus of Drosophila account for four classes of regularly occurring exchange products. Proc. Natl. Acad. Sci. USA 84: 174–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Do, A. T., J. T. Brooks, M. K. Le Neveu, and J. R. LaRocque, 2014 Double-strand break repair assays determine pathway choice and structure of gene conversion events in Drosophila melanogaster. G3 (Bethesda) 4: 425–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobzhansky T., 1930. Translocations involving the third and the fourth chromosomes of Drosophila melanogaster. Genetics 15: 347–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldfarb T., Lichten M., 2010. Frequent and efficient use of the sister chromatid for DNA double-strand break repair during budding yeast meiosis. PLoS Biol. 8: e1000520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawley R. S., 1980. Chromosomal sites necessary for normal levels of meiotic recombination in Drosophila melanogaster. I. Evidence for and mapping of the sites. Genetics 94: 625–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heil, C. S. S., and M. A. F. Noor, 2012 Zinc finger binding motifs do not explain recombination rate variation within or between species of Drosophila. PLoS ONE 7: e45055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hey J., 2004. What’s so hot about recombination hotspots? PLoS Biol. 2: e190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilliker A. J., Chovnick A., 1981. Further observations on intragenic recombination in Drosophila melanogaster. Genet. Res. 38: 281–296. [DOI] [PubMed] [Google Scholar]
- Hilliker A. J., Harauz G., Reaume A. G., Gray M., Clark S. H., et al. , 1994. Meiotic gene conversion tract length distribution within the rosy locus of Drosophila melanogaster. Genetics 137: 1019–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingsworth N. M., Brill S. J., 2004. The Mus81 solution to resolution: generating meiotic crossovers without Holliday junctions. Genes Dev. 18: 117–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang J. K., Sherizen D. E., Bhagat R., Manheim E. A., McKim K. S., 2003. Relationship of DNA double-strand breaks to synapsis in Drosophila. J. Cell Sci. 116: 3069–3077. [DOI] [PubMed] [Google Scholar]
- Johnson-Schlitz D. M., Flores C., Engels W. R., 2007. Multiple-pathway analysis of double-strand break repair mutations in Drosophila. PLoS Genet. 3: e50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminker J. S., C. M. Bergman, B. Kronmiller, J. Carlson, R. Svirskas et al, 2002 The transposable elements of the Drosophila melanogaster euchromatin: a genomics perspective. Genome Biol. 3: RESEARCH0084. [DOI] [PMC free article] [PubMed]
- Kulathinal R. J., Bennett S. M., Fitzpatrick C. L., Noor M. A. F., 2008. Fine-scale mapping of recombination rate in Drosophila refines its correlation to diversity and divergence. Proc. Natl. Acad. Sci. USA 105: 10051–10056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachaise D., David J. R., Lemeunier F., Tsacas L., Ashburner M., 1986. The reproductive relationships of Drosophila sechellia with D. mauritiana, D. simulans, and D. melanogaster from the Afrotropical region. Evolution 40: 262–271. [DOI] [PubMed] [Google Scholar]
- Lack J. B., Cardeno C. M., Crepeau M. W., Taylor W., Corbett-Detig R. B., et al. , 2015. The Drosophila genome nexus: a population genomic resource of 623 Drosophila melanogaster genomes, including 197 from a single ancestral range population. Genetics 199: 1229–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lake C. M., Holsclaw J. K., Bellendir S. P., Sekelsky J., Hawley R. S., 2013. The development of a monoclonal antibody recognizing the Drosophila melanogaster phosphorylated histone H2A variant (γ-H2AV). G3 (Bethesda) 3: 1539–1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y. C. G., Langley C. H., 2010. Transposable elements in natural populations of Drosophila melanogaster. Philos. Trans. R. Soc. Lond. B Biol. Sci. 365: 1219–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y. C. G., Langley C. H., 2012. Long-term and short-term evolutionary impacts of transposable elements on Drosophila. Genetics 192: 1411–1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Durbin R., 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J. et al, 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libuda D. E., Uzawa S., Meyer B. J., Villeneuve A. M., 2013. Meiotic chromosome structures constrain and respond to designation of crossover sites. Nature 502: 703–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichten M., Goldman A., 1995. Meiotic recombination hotspots. Annu. Rev. Genet. [DOI] [PubMed] [Google Scholar]
- Lindsley D. L., Grell E. H., 1967. Genetic Variations of Drosophila melanogaster, Carnegie Institute of Washington, Washington, DC. [Google Scholar]
- Lindsley D. L., Sandler L., 1977. The genetic analysis of meiosis in female Drosophila melanogaster. Philos. Trans. R. Soc. Lond. B Biol. Sci. 277: 295–312. [DOI] [PubMed] [Google Scholar]
- Lindsley D. L., Zimm G. G., 1992. The Genome of Drosophila melanogaster, Academic Press, San Diego. [Google Scholar]
- Mackay T. F. C., Richards S., Stone E. A., Barbadilla A., Ayroles J. F., et al. , 2013. The Drosophila melanogaster genetic reference panel. Nature 482: 173–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzano-Winkler, B., S. E. McGaugh, and M. A. F. Noor, 2013 How hot are Drosophila hotspots? Examining recombination rate variation and associations with nucleotide diversity, divergence, and maternal age in Drosophila pseudoobscura. PLoS ONE 8: e71582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., et al. , 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20: 1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVey M., Radut D., Sekelsky J. J., 2004. End-joining repair of double-strand breaks in Drosophila melanogaster is largely DNA ligase IV independent. Genetics 168: 2067–2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehrotra S., McKim K. S., 2006. Temporal analysis of meiotic DNA double-strand break formation and repair in Drosophila females. PLoS Genet. 2: e200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merker J. D., Dominska M., Petes T. D., 2003. Patterns of heteroduplex formation associated with the initiation of meiotic recombination in the yeast Saccharomyces cerevisiae. Genetics 165: 47–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller D. E., Takeo S., Nandanan K., Paulson A., Gogol M. M., et al. , 2012. A whole-chromosome analysis of meiotic recombination in Drosophila melanogaster. G3 (Bethesda) 2: 249–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery E. A., Huang S. M., Langley C. H., Judd B. H., 1991. Chromosome rearrangement by ectopic recombination in Drosophila melanogaster: genome structure and evolution. Genetics 129: 1085–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller H. J., 1916. The mechanism of crossing-over. Am. Nat. 50: 193–221. [Google Scholar]
- Novak J. E., Ross-Macdonald P. B., Roeder G. S., 2001. The budding yeast Msh4 protein functions in chromosome synapsis and the regulation of crossover distribution. Genetics 158: 1013–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Offermann, C. A., and H. J. Muller, 1932 Regional differences in crossing over as a function of the chromosome structure. Proc. Sixth Int. Congress Genet. 2: 143–145. [Google Scholar]
- Page S. L., Hawley R. S., 2001. c(3)G encodes a Drosophila synaptonemal complex protein. Genes Dev. 15: 3130–3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page S. L., Nielsen R. J., Teeter K., Lake C. M., Ong S., et al. , 2007. A germline clone screen for meiotic mutants in Drosophila melanogaster. Fly (Austin) 1: 172–181. [DOI] [PubMed] [Google Scholar]
- Page S. L., Khetani R. S., Lake C. M., Nielsen R. J., Jeffress J. K., et al. , 2008. Corona is required for higher-order assembly of transverse filaments into full-length synaptonemal complex in Drosophila oocytes. PLoS Genet. 4: e1000194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry D. M., 1973. A meiotic mutant affecting recombination in female Drosophila melanogaster. Genetics 73: 465–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radford S. J., McMahan S., Blanton H. L., Sekelsky J., 2007. Heteroduplex DNA in meiotic recombination in Drosophila mei-9 mutants. Genetics 176: 63–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh N. D., Stone E. A., Aquadro C. F., Clark A. G., 2013. Fine-scale heterogeneity in crossover rate in the garnet-scalloped region of the Drosophila melanogaster X chromosome. Genetics 194: 375–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singhal S., Leffler E. M., Sannareddy K., Turner I., Venn O., et al. , 2015. Stable recombination hotspots in birds. Science 350: 928–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevison L. S., Noor M. A. F., 2010. Genetic and evolutionary correlates of fine-scale recombination rate variation in Drosophila persimilis. J. Mol. Evol. 71: 332–345. [DOI] [PubMed] [Google Scholar]
- Sturtevant A. H., 1913. A third group of linked genes in Drosophila ampelophila. Science 37: 990–992. [DOI] [PubMed] [Google Scholar]
- Sturtevant A. H., 1915. The behavior of the chromosomes as studied through linkage. Z. Vererbungsl. 13: 234–287. [Google Scholar]
- Sturtevant A. H., Beadle G. W., 1936. The relations of inversions in the X chromosome of Drosophila melanogaster to crossing over and disjunction. Genetics 21: 554–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sym M., Roeder G. S., 1994. Crossover interference is abolished in the absence of a synaptonemal complex protein. Cell 79: 283–292. [DOI] [PubMed] [Google Scholar]
- Thacker D., Mohibullah N., Zhu X., Keeney S., 2014. Homologue engagement controls meiotic DNA break number and distribution. Nature 510: 241–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorvaldsdottir H., Robinson J. T., Mesirov J. P., 2013. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinform. 14: 178–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- True J. R., Mercer J. M., Laurie C. C., 1995. Differences in crossover frequency and distribution among three sibling species of Drosophila. Genetics 142: 507–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincenten N., Kuhl L.-M., Lam I., Oke A., Kerr A. R., et al. , 2015. The kinetochore prevents centromere-proximal crossover recombination during meiosis. eLife 4: e10850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y., Takahashi A., Itoh M., Takano-Shimizu T., 2009. Molecular spectrum of spontaneous de novo mutations in male and female germline cells of Drosophila melanogaster. Genetics 181: 1035–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstein A., 1918. Coincidence of crossing over in Drosophila melanogaster (Ampelophila). Genetics 3: 135–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitby M. C., 2005. Making crossovers during meiosis. Biochem. Soc. Trans. 33: 1451. [DOI] [PubMed] [Google Scholar]
- Yamamoto M., Miklos G. L., 1977. Genetic dissection of heterochromatin in Drosophila: the role of basal X heterochromatin in meiotic sex chromosome behaviour. Chromosoma 60: 283–296. [DOI] [PubMed] [Google Scholar]
- Yamamoto M., Miklos G. L., 1978. Genetic studies on heterochromatin in Drosophila melanogaster and their implications for the functions of satellite DNA. Chromosoma 66: 71–98. [DOI] [PubMed] [Google Scholar]
- Zhang L., Espagne E., de Muyt A., Zickler D., Kleckner N. E., 2014. Interference-mediated synaptonemal complex formation with embedded crossover designation. Proc. Natl. Acad. Sci. USA 111: E5059–E5068. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Strains are available on request. Sequencing data from this project has been deposited at the National Center for Biotechnology Information (NCBI) under the following project numbers: PRJNA285112 (isogenized w1118 and Canton-S parental stocks) and PRJNA307070 (196 individual males). Scripts used to align genome data, to call SNPs, to identify CO and NCO sites, and to estimate NCO rate and tract length can be found at https://github.com/danrdanny/2016_CO_NCO_Paper.