Abstract
In response to replication stress, a phospho-signaling cascade is activated and required for coordination of DNA repair and replication of damaged templates (intra-S-phase checkpoint) . How phospho-signaling coordinates the DNA replication stress response is largely unknown. We employed state-of-the-art liquid chromatography tandem-mass spectrometry (LC-MS/MS) approaches to generate high-coverage and quantitative proteomic and phospho-proteomic profiles during replication stress in yeast, induced by continuous exposure to the DNA alkylating agent methyl methanesulfonate (MMS) . We identified 32,057 unique peptides representing the products of 4296 genes and 22,061 unique phosphopeptides representing the products of 3183 genes. A total of 542 phosphopeptides (mapping to 339 genes) demonstrated an abundance change of greater than or equal to twofold in response to MMS. The screen enabled detection of nearly all of the proteins known to be involved in the DNA damage response, as well as many novel MMS-induced phosphorylations. We assessed the functional importance of a subset of key phosphosites by engineering a panel of phosphosite mutants in which an amino acid substitution prevents phosphorylation. In total, we successfully mutated 15 MMS-responsive phosphorylation sites in seven representative genes including APN1 (base excision repair); CTF4 and TOF1 (checkpoint and sister-chromatid cohesion); MPH1 (resolution of homologous recombination intermediates); RAD50 and XRS2 (MRX complex); and RAD18 (PRR). All of these phosphorylation site mutants exhibited MMS sensitivity, indicating an important role in protecting cells from DNA damage. In particular, we identified MMS-induced phosphorylation sites on Xrs2 that are required for MMS resistance in the absence of the MRX activator, Sae2, and that affect telomere maintenance.
Keywords: mass spectrometry, phosphorylation, methyl methanesulfonate, DNA damage checkpoint, genetic interaction, homologous recombination, telomere, DNA damage response
CELLS utilize excision repair and DNA damage tolerance pathways without significant delay of the cell cycle to address low levels of DNA base damage (Hishida et al. 2009; Huang et al. 2013), while more extensive damage is hallmarked by the activation of additional checkpoints, prolonged cell cycle arrest, and utilization of additional repair mechanisms (Lazzaro et al. 2009). A classic example of an agent that elicits a profoundly different DNA damage response (DDR) at high and low doses is the monofunctional alkylating agent methyl methanesulfonate (MMS) (Friedberg and Friedberg 2006; Hanawalt 2015). At low doses, the MMS lesions are well tolerated by wild-type cells and do not elicit any discernible sensitivity (Huang et al. 2013); however, at higher concentrations, MMS-induced DNA damage present during the S phase leads to prolonged replication fork stall, a phenomenon termed “replication stress” (Shimada et al. 2002; Zeman and Cimprich 2013). As a result of replication stress, cells synchronize into a lengthened S phase due to a kinase-mediated checkpoint response (Paulovich and Hartwell 1995; Murakami-Sekimata et al. 2010).
Much of the known signaling in the DDR is mediated by a group of highly conserved checkpoint kinases (e.g., ATR/Mec1, ATM/Tel1, Chk2/Rad53, Chk1), which activate an extensive phospho-signaling network to enhance DNA repair capacity as well as induce cell cycle delay at G1, intra-S, or G2/M to allow additional time for cells to deal with higher doses of DNA damage (Weinert and Hartwell 1988; Siede et al. 1993; Paulovich and Hartwell 1995). In Saccharomyces cerevisiae, the intra-S-phase checkpoint is mediated by the serine/threonine protein kinases Mec1 and Tel1 (Paulovich and Hartwell 1995; Zeman and Cimprich 2013). Mec1 plays the predominant role in the activation of the intra-S-phase checkpoint, whereas Tel1 plays a backup role (Weinert et al. 1994; Greenwell et al. 1995). The long stretches of single-stranded DNA (ssDNA) exposed during replication fork stalling after DNA damage contribute to the activation of Mec1 and induction of the intra-S-phase checkpoint (Tercero et al. 2003; MacDougall et al. 2007).
Activation of the Mec1 kinase leads to activation of two well-known, bifurcated pathways: the Rad9-mediated DNA-damage checkpoint (DDC) pathway, and the Mrc1/Tof1/Ctf4/Csm3-mediated S phase-specific DNA-replication checkpoint (DRC) (Alcasabas et al. 2001; Katou et al. 2003; Uzunova et al. 2014). Similar to the Mec1/Tel1 relationship, the Rad9-mediated DDC pathway is required for MMS resistance, whereas the Mrc1-mediated DRC plays a backup role in MMS resistance (Foss 2001). Phosphorylations of Rad9 and Mrc1 in turn facilitate phosphorylation of the downstream checkpoint kinases (Rad53 and Chk1) (Vialard et al. 1998; Sanchez et al. 1999; Alcasabas et al. 2001), which, in turn, phosphorylate additional substrates, including Pds1 and Cdc5 polo-kinase, both of which contribute to cell cycle delay (Sanchez et al. 1999). Rad53 also phosphorylates and activates another kinase, Dun1, which contributes to the hyper-phosphorylation and inactivation of the transcriptional repressor Crt1 and leads to increased expression of genes related to DNA repair, including RNR (ribonucleotide-diphosphate reductase) genes (Huang et al. 1998).
Many downstream DNA repair proteins are reported to be phosphorylated during checkpoint activation, including proteins involved in post-replication repair (PRR), homologous recombination (HR), DNA replication, DNA repair, histone modification, and chromatin remodeling (Smolka et al. 2007; Chen et al. 2010; Bastos de Oliveira et al. 2015). For example, phosphorylation of Rev1 by Mec1 increases the proficiency of Polζ-mediated translesion synthesis (Pages et al. 2009), which together with the template-switch subpathways of PRR are important in dealing with replication stress, because the lesion-containing ssDNA resulting from a replication fork stall would not be subject to excision repair (Yang et al. 2010; Allen et al. 2011) and must be circumnavigated using PRR pathways. Much of the core PRR machinery is known to be phosphorylated, including Rad6, Rad18, Rev1, Mms2, and Rad5 (Chi et al. 2007; Albuquerque et al. 2008; Holt et al. 2009; Helbig et al. 2010), yet the physiological importance of most of these phosphorylations is unknown.
Over 100 checkpoint-induced phosphorylations have been identified in previous studies (Smolka et al. 2007; Chen et al. 2010; Bastos de Oliveira et al. 2015). However, because previous studies used a relatively high concentration (0.05%) of MMS, in which cells accumulate in the G1 phase and do not replicate the bulk of the genome (Murakami-Sekimata et al. 2010), phospho-signaling events that are exclusive to intra-S-phase checkpoint activation may be missing in these data sets. The identification and interpretation of these phospho-signaling events is of significance, as the key regulatory steps by which cells sense and respond to replication stress are poorly mapped out.
We utilized recent advances in proteomic technologies that enable near-comprehensive coverage of the yeast proteome to identify phosphorylation events during continuous treatment with a sublethal dose of MMS, which induces a replication stress response (Paulovich and Hartwell 1995; Zeman and Cimprich 2013). We identified many novel phosphorylation sites. We assessed the functional importance of a subset of key phosphosites by screening mutants in which an amino acid substitution prevents phosphorylation. All of these phosphorylation site mutants exhibited MMS sensitivity, indicating an important role for phosphorylation at these sites in protecting cells from DNA damage. Moreover, we performed a series of genetic and functional characterizations of phosphosite mutants, and we found that MMS-induced phosphorylation sites on Xrs2 are required for MMS resistance in the absence of the MRX activator, Sae2, and affect telomere maintenance.
Materials and Methods
Public access to the liquid chromatography tandem-mass spectrometry data
All mass spectrometry data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository (Vizcaíno et al. 2013) with the data-set identifier PXD002344 (reviewer account details: username: reviewer63953@ebi.ac.uk; password: s7u3qKaX).
Strains, medium, and growth conditions
S. cerevisiae strains used in this study are listed in Table 1. Strain BY4741 was obtained from Open Biosystems. All of the other strains used in this study are derived from BY4741. YPD medium contains 1% yeast extract, 2% peptone, and 2% glucose. SILAC (Stable Isotope Labeling by Amino acids in Cell culture) medium is synthetic defined (SD)-Lys-Arg (2% glucose) liquid medium supplemented with 40 mg/liter of lysine (light or heavy) and 20 mg/liter of arginine (light or heavy). Heavy lysine is L-lysine:2HCl (U-13C6, 99%; U-15N2, 99%); heavy arginine is l-arginine:HCl (U-13C6, 99%; U-15N4, 99%). All light and heavy lysine and arginine were purchased from Cambridge Isotope Laboratories Inc. MMS was purchased from Acros Organics (AC254609). YPD plates containing MMS were prepared ∼15 hr prior to use.
Table 1. S. cerevisiae strains.
| Strain | Genotype | Source |
|---|---|---|
| BY4741 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | Open Biosystems |
| yDH227 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 rad18ΔKANR | This study |
| yDH350 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 rad9ΔKANR | This study |
| yDH355 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 sgs1ΔKANR | This study |
| yDH357 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 xrs2ΔKANR | This study |
| yDH359 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 rad50ΔKANR | This study |
| yDH452 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 exo1ΔKANR | This study |
| yDH455 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 apn1ΔKANR | This study |
| yDH456 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 ctf4ΔKANR | This study |
| yDH457 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 ctf8ΔKANR | This study |
| yDH460 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 mph1ΔKANR | This study |
| yDH465 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 tof1ΔKANR | This study |
| yDH492 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 xrs2T675A yku80ΔKANR | This study |
| yDH513 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 dia2ΔKANR | This study |
| yDH567 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 xrs2S349A est3ΔKANR | This studya |
| yDH568 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 xrs2T675A est3ΔKANR | This studya |
| yDH569 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 xrs2S349A, T675A est3ΔKANR | This studya |
| yDH576 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 srs2ΔKANR | This study |
| yDH578 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 ctf4T401A, T411A | This study |
| yDH587 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 rad50T568A | This study |
| yDH599 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 apn1S350A, S356A | This study |
| yDH600 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 est3ΔKANR | This studya |
| yDH603 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 tof1S379A, S626A ctf8ΔKANR | This study |
| yDH604 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 tof1S379A, S626A rad9ΔKANR | This study |
| yDH606 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 xrs2S349A, T675A rad27ΔKANR | This study |
| yDH607 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 xrs2S349A, T675A pol32ΔKANR | This study |
| yDH608 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 xrs2S349A, T675A yku80ΔKANR | This study |
| yDH610 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 pol32ΔKANR | This study |
| yDH625 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 xrs2S349A, T675A srs2ΔNATR | This study |
| yDH627 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 rad27ΔKANR | This study |
| yDH628 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 clb2ΔKANR | This study |
| yDH629 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 yku80ΔKANR | This study |
| yDH638 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 xrs2S349A, T675A | This study |
| yDH641 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 xrs2S349A, T675A sgs1ΔKANR | This study |
| yDH650 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 rad18T155A, T282, S284A | This study |
| yDH654 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 xrs2S349A, T675A ctf4ΔKANR | This study |
| yDH664 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 mph1T540A, S542A, S543A | This study |
| yDH672 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 tof1S379A, S626A | This study |
| yDH685 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 xrs2S349A,T675A apn1ΔKANR | This study |
| yDH687 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 rtt109ΔKANR | This study |
| yDH690 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 xrs2S349A, T675A rtt109ΔKANR | This study |
| yDH693 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 tof1S379A, S626A srs2ΔKANR | This study |
| yDH698 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 xrs2S349A, T675A clb2ΔKANR | This study |
| yDH700 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 tof1S379A, S626A dia2ΔKANR | This study |
| yDH702 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 xrs2S349A, T675A rad17ΔKANR | This study |
| yDH706 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 xrs2S349A, T675A rad24ΔKANR | This study |
| yDH714 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 xrs2S349A, T675A exo1ΔKANR | This study |
| yDH730 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 sae2ΔKANR | This study |
| yDH741 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 xrs2S349A, T675A mec1ΔLEU2 sml1ΔHIS3 sae2ΔKANR | This study |
| yDH751 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 sae2ΔHIS3 | This study |
| yDH752 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 xrs2S349A sae2ΔHIS3 | This study |
| yDH753 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 xrs2T675A sae2ΔHIS3 | This study |
| yDH754 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 xrs2S349A T675A sae2ΔHIS3 | This study |
| yDH755 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 xrs2ΔURA3 sae2ΔHIS3 | This study |
| yDH794 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 xrs2S349A yku80ΔKANR | This study |
| yDH805 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 xrs2S349A T675A tel1ΔKANR sae2ΔHIS3 | This study |
| yDH806 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 tof1ΔURA3 sml1ΔHIS3 rad9ΔKANR | This study |
| yDH807 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 tof1ΔURA3 dia2ΔKANR | This study |
EST3 gene was deleted by PCR. The freshly made est3Δ transformants were grown in YPD for 14 hr (approximately eight generations) before being used for experiments and being frozen and stored at −80°C.
Quantitative MS screen for MMS-responsive phosphopeptides
Metabolic labeling of proteins, extraction, and digestion:
Wild-type cells were metabolically labeled for >20 generations in SILAC medium. The heavy (H)-SILAC-labeled cells were then continuously exposed to 0.01% MMS to induce replication stress, and the light (L)-SILAC-labeled cells were mock-exposed. After 3 hr, heavy and light cells were harvested and lysed using a previously described trichloroacetic acid (TCA) lysis method (Ziv et al. 2011). To ensure reproducibility, the entire experiment was repeated, and the labels were swapped such that the (L)-SILAC-labeled wild-type yeast cells were exposed to 0.01% MMS for 3 hr, and the (H)-SILAC-labeled wild-type yeast cells were mock-exposed. The protein pellets from TCA prep were resuspended in urea buffer [300 mM Tris, pH 8.0, 6 M Urea (Sigma U0631)]. Lysates from heavy and light cells were mixed 1:1 by protein mass. Five milligrams of each protein lysate was reduced in 100 mM Tris/20 mM TCEP (Thermo 77720) for 30 min at 37° with shaking, followed by alkylation with 50 mM iodoacetamide (Sigma I1149) in the dark at room temperature. Lysates were then diluted 1:10 with 100 mM Tris, pH 8, and trypsin was added at a 1:50 trypsin:protein ratio (by mass). After 2 hr, a second trypsin aliquot was added at a 1:100 trypsin:protein ratio. Digestion was carried out overnight at 37° with shaking. After 16 hr, the reaction was quenched with formic acid (FA) (Acros Organics 14793-2500) with a final concentration 1% by volume. Digests were desalted using Hydrophilic-Lipophilic-Balanced (HLB) cartridges (Waters WAT094225) with vacuum. HLB cartridges were washed with 3 vol of 0.1% FA in 80% acetonitrile (ACN) (Fisher A955-4) and then equilibrated with four washes of 0.1% FA. The digests were applied to the cartridge and then washed with 4 vol 0.1% FA before being eluted drop by drop with three washes of 0.1% FA in 80% ACN. The eluate was then aliquotted by volume, and digests were lyophilized and stored at −80° until use.
Fractionation of proteome and phosphoproteome samples:
The desalted tryptic digest was fractionated by high-pH reverse phase (RP) liquid chromatography, as follows: five milligrams of the protein digest were loaded onto an LC system consisting of an Agilent 1200 HPLC (Agilent, Santa Clara, CA) with mobile phases of 5 mM ammonium bicarbonate (NH4HCO3), pH 10 (A) and 5 mM NH4HCO3 in 90% ACN, pH 10 (B). The peptides were separated by a 10- × 250-mm XBridge C18 5-μm column (Waters catalog #186003256) over 50 min at a flow rate of 2.5 ml/min by the following time table: hold 5% B for 1 min, gradient from 5 to 40% B for 35 min, gradient from 40 to 60% B for 5 min, gradient from 60 to 90% B for 4 min, gradient from 90 to 5% B for 1 min, and re-equilibrate at 5% B for 4 min. Fractions were collected at 0.5-min intervals from 2 to 50 min by the shortest path by row in a 2-ml-deep well plate (Thermo 95040450). The high pH reverse phase fractions were concatenated into 12 samples by column (e.g., sample 1 contained fractions from wells A1, B1, C1, D1, etc.). For proteome analysis, 2% of each concatenated fraction was dried down (lyophilization) and re-suspended in 0.1% FA in 3% ACN for liquid chromatography tandem-mass spectrometry (LC-MS/MS) analysis. The remaining 98% was processed to enrich for phosphopeptides using immobilized metal affinity chromatography as previously described (Ficarro et al. 2009). Briefly, Ni-NTA-agarose beads (catalog #36113, Qiagen, Valencia, CA) were stripped with EDTA and incubated in a 10-mM FeCl3 solution to prepare magnetic Fe3+-NTA-agarose beads. Samples were reconstituted in 400 μl of 0.1% TFA in 80% ACN and incubated for 30 min with 75 μl of the 5% bead suspension, with mixing at 1400 × g at room temperature. After incubation, the beads were washed three times each with 150 μl of 0.1% TFA in 80% ACN and then once with 150 μl of 0.1% TFA. Phosphorylated peptides were eluted from the beads twice using 150 μl of 500 mM potassium phosphate, pH 7, after incubating for 3 min each time. Samples were desalted using StageTips loaded with reverse-phase material (Rappsilber et al. 2007), dried down, and resuspended in 0.1% FA and 3% ACN for LC-MS/MS analysis.
Mass-spectrometry-based analysis:
Global and phosphopeptide-enriched samples were analyzed by LC-MS/MS on a Thermo LTQ-Orbitrap Velos mass spectrometer. Peptides were loaded onto an LC system consisting of a nanoAcquity HPLC (Waters, Milford, MA) with mobile phases of 0.1% FA in water (A) and 0.1% FA in ACN (B). The peptides were separated on a 75-µm × 250-mm C18, 130 Å, 1.7-µm column (Waters catalog #186003545) over 152 min at a flow rate of 300 nl/min, with a gradient from 3 to 40% B for 120 min, a gradient from 40 to 90% B for 2 min, a hold of 90% B for 10 min, and re-equilibration at 3% B for 20 min. The HPLC was coupled to an LTQ-Orbitrap Velos hybrid mass spectrometer using an Advance CaptiveSpray source (Michrom Bioresources, Auburn, CA) operated in positive ion mode. A spray voltage of 1700 V was applied to the nanospray tip (catalog #559/25000/20). MS/MS analysis consisted of one full-scan MS from 300 to 2000 m/z at resolution 30,000, followed by 15 data-dependent MS/MS scans. Dynamic exclusion parameters included repeat count 1, exclusion list size 500, and exclusion duration 15 sec.
Analysis of LC-MS/MS data
Raw MS/MS spectra were searched against version 3.69 of the Yeast International Protein Index sequence database using three independent search engines (MaxQuant/Andromeda, Spectrum Mill, and xTandem) (Craig and Beavis 2004; Kapp et al. 2005; Cox and Mann 2008). All searches were performed with the tryptic enzyme constraint set for up to two missed cleavages, oxidized methionine set as a variable modification, and carbamidomethylated cysteine set as a static modification. For MaxQuant, the peptide MH+ mass tolerances were set at 20 ppm. For X!Tandem, the peptide MH+ mass tolerances were set at ±2.0 Da with post-search filtering of the precursor mass to 50 ppm, and the fragment MH+ mass tolerances were set at ±0.5 Da. For Spectrum Mill, peptide MH+ mass tolerances were set at 20 ppm and fragment MH+ mass tolerances were set at ±0.7 Da. The overall false discovery rate (FDR) was set at ≤0.03 based on a decoy database search. SILAC ratios and phosphosite localization probabilities are reported only in the MaxQuant results. Any site with a probability >0.8 was considered to be localized; ambiguous sites with a lower probability were manually examined to verify the location of the phosphorylation and ensure the quality of the reported results. For manual examination, the MS/MS spectra were examined to identify ions with the loss of phosphate, which is characteristic of Ser/Thr-phosphorylated peptides, and to confirm that all of the major ions were properly assigned and the assignment of the phosphate group to the specific site was correct. Quantification of the heavy:light ratios was performed using MaxQuant software, with a minimum ratio count of 2 and using unique + razor peptides for quantification. Functional enrichment analysis was conducted using the software package Funspec (Robinson et al. 2002).
Gene disruptions and integrations
All gene disruptions and integrations were achieved by homologous recombination at their respective chromosomal loci by standard PCR-based methods (Longtine et al. 1998). Briefly, a deletion cassette with a 0.5-kb region flanking the target open reading frame (ORF) was amplified by PCR from the corresponding xxxΔ::KANMX strain of the deletion array (Open Biosystems) and transformed into the target strain for gene knockout. The primers used in the gene disruptions were designed using 20-bp sequences that are 0.5 kb upstream and downstream of the target gene (Reid et al. 2002).
For gene disruptions utilizing the LEU2MX or HIS3MX cassette, the xxxΔ::KANMX strain from the deletion array was converted to xxxΔ::LEU2MX or xxxΔ::HIS3MX. The cassette conversion was achieved by amplifying the LEU2MX or HIS3MX cassette with primers MX-F (5′-ACATGGAGGCCCAGAATACCCT-3′) and MX-R (5′-CAGTATAGCGACCAGCATTCAC-3′) from plasmids pFA6a-Leu2MX6-GAL1 and pFA6a-His3MX6-pGAL1, respectively (Longtine et al. 1998), and the resulting PCR product was used to transform the xxxΔ::KANMX strain (the −MX cassettes each carry an identical 5′ Translational elongation factor EF-1 (TEF) promoter and 3′ terminator, which facilitates the KANMX::LEU2MX or KANMX::HIS3MX conversion).
To integrate the Myc-tag into the C terminus of the XRS2 gene, a region of plasmid pFA6a-13Myc-KanMX6 was amplified by PCR using primers that contain 55 bp of XRS2 gene sequence (55 nucleotides before and after the stop codon), followed by 20 bp homologous to plasmid pFA6a-13Myc-KanMX6 (F2 and R1) (Longtine et al. 1998). The PCR product was used to transform the indicated target yeast strains and integrated Myc-tag in the C terminus of the endogenous XRS2 with KanMX marker. The primers used were 5′-GGCGACGACGACGATGACGACGGTCCGAAGTTTACGTTCAAAAGAAGAAAAGGACGGATCCCCGGGTTAATTAA-3′ (XRS2-13myc F2) and 5′-ATGATAATGCAAAATATAATTTAATGAAATTGGAAATACTCGGAAAATTTATCAGAATTCGAGCTCGTTTAAAC-3′ (XRS2-13myc R1).
In vivo site-directed mutagenesis
We followed a protocol modified from a previously published yeast mutagenesis method (Storici et al. 2001) based on transformations of oligonucleotides that allow the rapid creation of site-directed DNA mutations in vivo. The protocol includes two steps: The first step involves the integration of a counterselectable reporter URA3 cassette into the target gene at the position of the codon where the change is desired, resulting in replacement of the three-nucleotide codon with URA3. (The URA3 cassette with a 50-bp region flanking the three-nucleotide codon was amplified by PCR from pRS406 and transformed into the target strain). The second step involves transformation with the mutation-containing oligonucleotides that eliminate the URA3 cassette. The two 93-bp “integrative recombinant oligonucleotides” are complementary to each other and contain the three-nucleotide mutated codon flanked by 45-bp sequences upstream and downstream of the URA3 cassette. Cells were transformed with these two complementary oligonucleotides, and loss of the URA3 cassette was counterselected using 5-fluoroorotic acid (TRC, F595000). The removal of URA3 and reinstatement of the continuous coding sequence was further confirmed by PCR. The acquired mutation was confirmed by sequencing of the entire gene. By repeating these processes, multiple mutations were made in a single gene.
Colony-based survival assays
Three independent, sequence-confirmed transformants were analyzed for each phospho-mutant, along with wild-type and deletion mutant controls. Log-phase cells were sonicated and counted using a Beckman Coulter Z1 particle counter. Cells were serially diluted in PBS and plated onto YPD plates ± appropriate concentrations of MMS. Viability was determined by scoring the number of colony-forming units (CFU) after 3–4 days at 30°. Viability was calculated as the number of CFUs on a MMS plate/the number of CFUs on an YPD plate.
Western blotting
Cell extracts were prepared from log-phase cells using a TCA lysis method (Ziv et al. 2011). Eighty micrograms of total protein were loaded on SDS-PAGE. Myc-tagged Xrs2 wild-type and mutant proteins were detected with anti-MYC-HRP (Fisher MA121316HRP). Rad53p was detected with the yC-19 anti-Rad53 antibody (Santa Cruz).
Southern blotting
Southern blotting for telomere lengths was carried out using a previously described DNA probe targeting telomeric Y′ regions (Singer et al. 1998). Digoxigenin (DIG)-labeled probe synthesis was carried out by PCR using the Roche DIG Probe Synthesis Kit following the manufacturer’s instructions. Genomic DNA was prepared using a Yeastar genomic DNA kit (Zymo Research D2002). Genomic DNA preparations were digested overnight with XhoI (NEB R0146S) and separated on 1% agarose gels. Separated DNA molecules were transferred onto nylon membranes via blot sandwich overnight in 20× SSC buffer (3.0 M NaCl and 0.3 M sodium citrate, at pH 7.0). DNA molecules were cross-linked onto the membrane using a UV cross-linker (Fisher Scientific) at 60 mJ/cm2, and the membrane was incubated with the Y′ telomeric DIG-labeled probe overnight. Antibody detection of the DIG probe was performed using a DIG luminescent detection kit (Roche 11363514910), and blots were imaged on a ChemiDoc XRS system (Bio-Rad).
Data availability
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.
Results
LC-MS/MS-based analysis of proteomic and phosphoproteomic responses associated with MMS-induced replication stress
To more comprehensively characterize the signaling events that comprise the replication stress response, we performed large-scale quantitative proteomic and phosphoproteomic profiling of yeast cells ± continuous exposure to 0.01% MMS. To do this, we metabolically labeled wild-type yeast cells in heavy SILAC medium (with Arg and Lys labeled with 13C and 15N) (Mann 2014) and exposed the cells to 0.01% MMS for 3 hr. Lysate from these cells was mixed 1:1 by protein mass with lysate from untreated yeast cells grown in light SILAC medium. The protein lysates were digested with trypsin, and the resulting peptides were subjected to fractionation via offline reverse-phase HPLC and split into aliquots for global proteome and phosphoproteome analyses. Phosphopeptides were enriched via iron metal affinity chromatography (Ficarro et al. 2009), and both the global and phosphopeptide-enriched samples were analyzed by LC-MS/MS (Figure 1A). Both data sets were analyzed using three search engines (MaxQuant/Andromeda, Spectrum Mill, and X!Tandem), and results for this combined analysis were reported for an FDR ≤ 0.03 (see Materials and Methods).
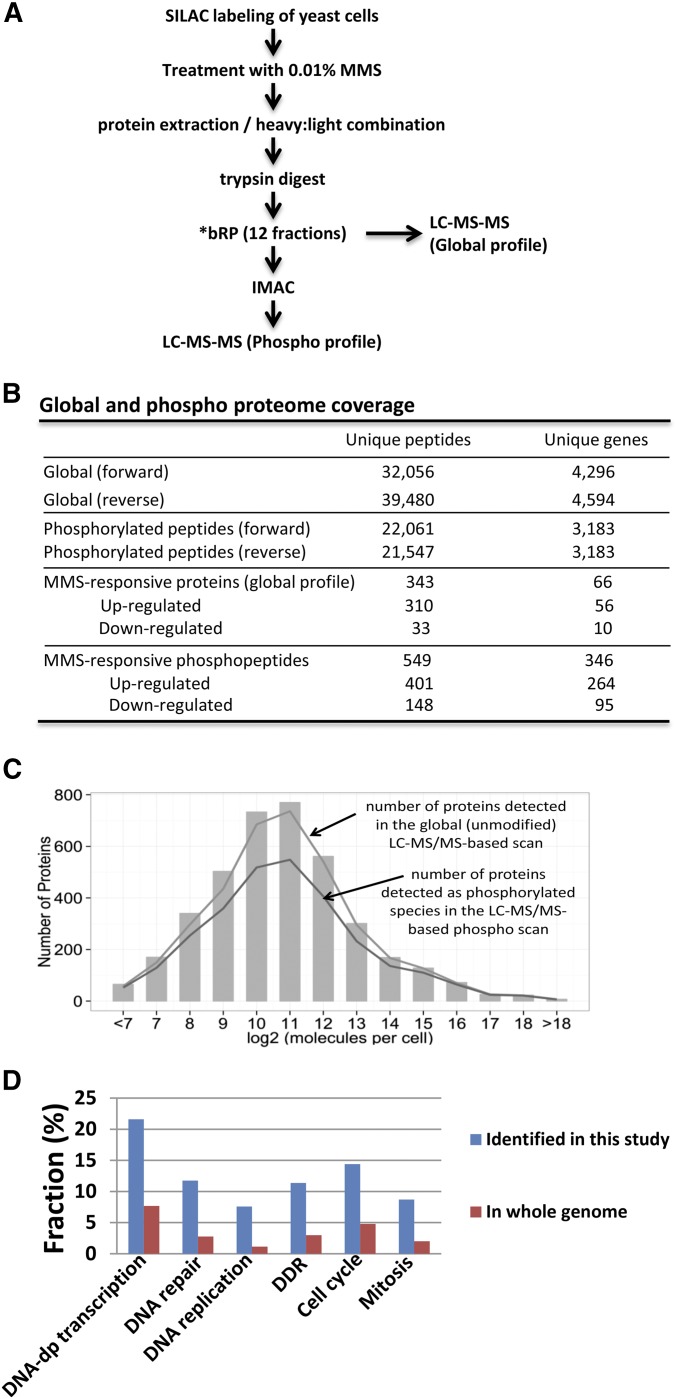
Figure 1.
Identification of MMS-responsive phospho-peptides by LC-MS/MS. (A) Strategy. We exposed heavy (H)-SILAC-labeled wild-type yeast cells to 0.01% MMS and mock-exposed light (L)-SILAC-labeled cells. After 3 hr, H- and L-labeled cells were mixed in equal amounts by protein mass. To ensure reproducibility, the entire experiment was repeated, and the labels were swapped such that the (L)-SILAC-labeled wild-type yeast cells were exposed to 0.01% MMS for 3 hr, and the (H)-SILAC-labeled wild-type yeast cells were mock-exposed. *bRP: basic reverse-phase (bRP) liquid chromatography (LC). (B) Global and phospho-proteome coverage. Note that “MMS-responsive” phospho-peptides include the peptides the phosphorylation of which increases (SILAC ratio ≥2) or decreases (SILAC ratio ≤0.5) after MMS treatment. (C) The high coverage of the yeast proteome achieved by LC-MS/MS is not biased. Bar graph is extracted from Ghaemmaghami et al. (2003) and represents the abundance distribution of 80% of the yeast proteome based on immuno-detection. Lines represent the abundance-based coverage achieved in our LC-MS/MS-based global and phospho scans. (D) The genes encoding MMS-inducible phosphoproteins are enriched in the functional categories of cell cycle regulation and DDR. Gene ontology enrichment analysis was conducted on the 264 MMS-induced phosphoproteins (≥2σ or twofold) using the Funspec software package (http://funspec.med.utoronto.ca/). After inputting the 264 gene names and setting the P-value cutoff as 10−6, the following processes were found to be enriched: DNA-dependent transcription (P = 1.1 × 10−13), DNA repair (P = 2.5 × 10−12), DNA replication (P = 4.1 × 10−12), DNA damage response (P = 9.2 × 10−11), cell cycle (P = 3.4 × 10−10), and mitosis (P = 4.1 × 10−9).
In the global proteome, we identified 32,056 unique peptides representing the products of 4296 genes (Supplemental Material, Table S1A), and in the phosphoproteome we identified 22,061 unique phosphopeptides representing the products of 3183 genes (Figure 1B and Table S1B). To ensure reproducibility, the entire experiment was repeated with the SILAC labels swapped (reverse experiment), with cells exposed to MMS cultured in light SILAC medium and the untreated cells cultured in heavy SILAC medium. The results for the reverse experiment were comparable to the initial forward experiments; we identified 39,480 unique peptides representing 4594 gene products in the global proteome (Table S1C), and 21,547 unique phosphopeptides representing 3183 gene products in the phosphoproteome (Figure 1B and Table S1D). Importantly, in our current study, we detected virtually all of the proteins known to be involved in the replication stress response (Figure S1). The coverage achieved for the yeast proteome was not significantly biased toward high-abundance proteins, as shown by comparing our LC-MS/MS results to the immunodetection-based global analysis of tagged protein expression in yeast from Ghaemmaghami et al. (2003) (Figure 1C).
We assessed for changes in protein abundance and phosphorylation after MMS treatment using the SILAC ratios. To minimize the noise present in the reported ratios (by focusing on only the most robust MS signals), peak areas (both heavy and light) in the lowest 20% of the results from the phospho and global proteome were removed from the analysis (Figure S2). To assess protein abundance changes, we considered proteins with at least two peptides quantified in both the forward and reverse experiments with reported ratios within 50%. Of the 1476 proteins that qualify based on our filtering criteria (Table S2), we found that 66 proteins exhibited a change in abundance of ≥2σ (or 1.5-fold) based on the median of all peptide ratios of a protein (Figure S3). Of the 66 MMS-responsive proteins, 56 were up-regulated and 10 were down-regulated after MMS treatment (Figure 1B and Table S3). The 56 MMS-induced proteins were highly biased for particular Gene Ontology (GO) biological processes such as oxidation-reduction processes (P < 10−14), deoxynucleotide biosynthesis (P = 1.3 × 10−10), and metabolic processes (P = 9.74 × 10−9) (Figure S4). These are consistent with previous reports that the expression of ribonucleotide-diphosphate reductase complex subunits (RNR genes) was stimulated by DNA damage checkpoint pathways (Gasch et al. 2001) and that exposure of cells to MMS triggers an oxidative stress response (Mizumoto et al. 1993; Gasch et al. 2001; Salmon et al. 2004).
For the analysis of the phosphoproteome, of the 6644 phosphopeptides quantified in the overlap of both the forward and reverse experiments (Table S4), 5524 phosphosites could be confidently localized to a specific residue (localization probability score >0.8) (Table S5), and 4449 of these sites were previously reported in the S. cerevisiae phosphorylation site database (PhosphoGRID) as well as in recently published results (Amoutzias et al. 2012; Sadowski et al. 2013; Bastos de Oliveira et al. 2015); as such, the remaining 1075 are novel site-of-phosphorylation identifications.
Based on the relative quantification measured by SILAC ratios, we identified 549 phosphopeptides mapping to 346 proteins that exhibited a change in abundance of ≥2σ (or twofold) in both forward and reverse experiments (Figure 1B and Table S6). Of these, 471 peptides contained a single phosphorylated residue, 77 were doubly phosphorylated, and 1 was triply phosphorylated. A total of 360 modifications were on serine residues, 51 on threonine, and 1 on tyrosine. With regards to directionality of the change in response to MMS, 401 phosphopeptides (420 phosphorylation sites in 264 proteins) increased in abundance after MMS treatment, indicating MMS-induced phosphorylation, while 148 decreased in abundance (Figure 1B and Table S6). In addition, the 264 genes showing MMS-induced phosphorylation were highly biased for genes with particular GO biological processes: DNA-dependent transcription (P = 1.1 × 10−13), DNA repair (P = 2.5 × 10−12), DNA replication (P = 4.1 × 10−12), DNA damage response (P = 9.2 × 10−11), cell cycle (P = 3.4 × 10−10), and mitosis (P = 4.1 × 10−9) (Figure 1D).
Among the 420 MMS-inducible phosphorylation sites, 376 were confident sites (with localization probability score >0.8) (Table S7). Of these, 271 sites were previously reported (Amoutzias et al. 2012; Sadowski et al. 2013; Bastos de Oliveira et al. 2015), and 105 were novel sites. Not surprisingly, 71/271 (26%) were previously annotated as “DNA damage induced,” and 34/271 (12%) were “cell cycle regulated.” However, the majority of previously identified phosphosites 164/271 (61%) had not been previously attributed to any biological condition (Table S7), suggesting that phosphoproteomic DNA damage signaling has to date been largely unmapped.
We next looked for motif enrichment among the 376 MMS-inducible phosphosites identified in our study. We found that 84/376 (22%) of the MMS-inducible phosphosites exhibited a consensus sequence for Mec1/Tel1 (S/T-Q), as compared to 188/5524 or 3% of all confident phosphorylation sites identified (P = 6.2 × 10−58). Therefore, Mec1/Tel1 (S/T-Q) consensus sequence is significantly enriched. Although other kinase motifs were represented among the MMS-induced phosphosites, none showed significant enrichment, such as an S/T-Ψ Rad53 motif (P = 0.495), an S/T-P minimal Cdk1 consensus sequence (P = 0.075), and an S/T-D/E minimal Casein kinase 2 consensus sequence (P = 0.366).
Selection of biologically important phosphorylation sites for further functional analysis
Our large-scale phosphoproteomic analyses yielded a significant number of novel MMS-dependent phosphorylation events. However, differential phosphoproteomics alone does not provide functional information regarding the importance of any specific phosphosite for surviving DNA damage. Thus we next chose a subset of previously uncharacterized MMS-induced phosphosites for further characterization via study of the effects of site-directed mutagenesis of the phosphosite of interest to nonphosphorylatable alanine. We chose evolutionally and functionally conserved DDR genes and avoided multiply phosphorylated proteins (due to the technical challenge of generating 3+ point mutations via site-directed mutagenesis). In total, we successfully mutated 15 MMS-inducible phosphorylation sites in seven representative genes including the following: APN1 (Base excision repair); CTF4 and TOF1 (checkpoint and sister-chromatid cohesion); MPH1 (resolution of HR intermediates); RAD50 and XRS2 (MRX complex); and RAD18 (PRR) (Table 2 and see also Figure S5 for MS1 quantification and MS/MS identification of Xrs2 and Tof1 peptides).
Table 2. Phosphorylation sites and genes selected for further characterization.
| Gene symbol | Target sequence | Modification position | Observed phenotype(s)a | Function |
|---|---|---|---|---|
| APN1 | ATAEPS(ph)DNDILSQMTK | S350 | MMS-sensitive | Base excision repair |
| APN1 | ATAEPSDNDILS(ph)QMTK | S356 | ||
| CTF4 | LFSDIT(ph)QEANAEDVFT(ph)QTHDGPSGLSEK | T401, T411 | MMS-sensitive | Checkpoint; Sister-chromatid cohesion |
| TOF1 | LTVSGS(ph)QALVDEK | S379 | MMS-sensitive; Interact with rad9Δ and dia2Δ. | |
| TOF1 | FNIS(ph)EGDITK | S626 | ||
| MPH1 | T(ph)GSSEEAQISGMNQK | T540 | MMS-sensitive | HR intermediate resolution |
| MPH1 | TGS(ph)S(ph)EEAQISGMNQK | S542 or S543b | ||
| RAD50 | QVFPLT(ph)QEFQR | T568 | MMS-sensitive | MRX |
| XRS2 | APEVEAS(ph)PVVSK | S349 | MMS-sensitive; Telomere maintenance; Interact with exo1Δ, yku80Δ, and sae2Δ | |
| XRS2 | NAAFLIT(ph)R | T675 | ||
| RAD18 | INFTSMT(ph)QS(ph)QIK | T282 or S284b | MMS-sensitive | PRR |
| RAD18 | SMT(ph)DILPLSSKPSK | T155 |
Since PRR genes play an essential role in the replication stress response, RAD18 was included even though the induction of phosphorylation by MMS for these proteins was detected in only one of the forward or label-swap experiments. The same is true for the phosphorylation of Mph1T540 and Apn1S356. The mass spectra of the detected phosphopeptides from these gene products (in either the forward or label-swap experiment) were manually inspected and confirmed.
The observed phenotypes reflect phenotypes of the phospho-mutants in which all detected MMS-inducible phosphosites are mutated.
The mass spectra were not able to distinguish between phosphorylation of Mph1 on S542 vs. S543 or phosphorylation of Rad18 on T282 vs. S284; in all ambiguous cases, both sites were mutated.
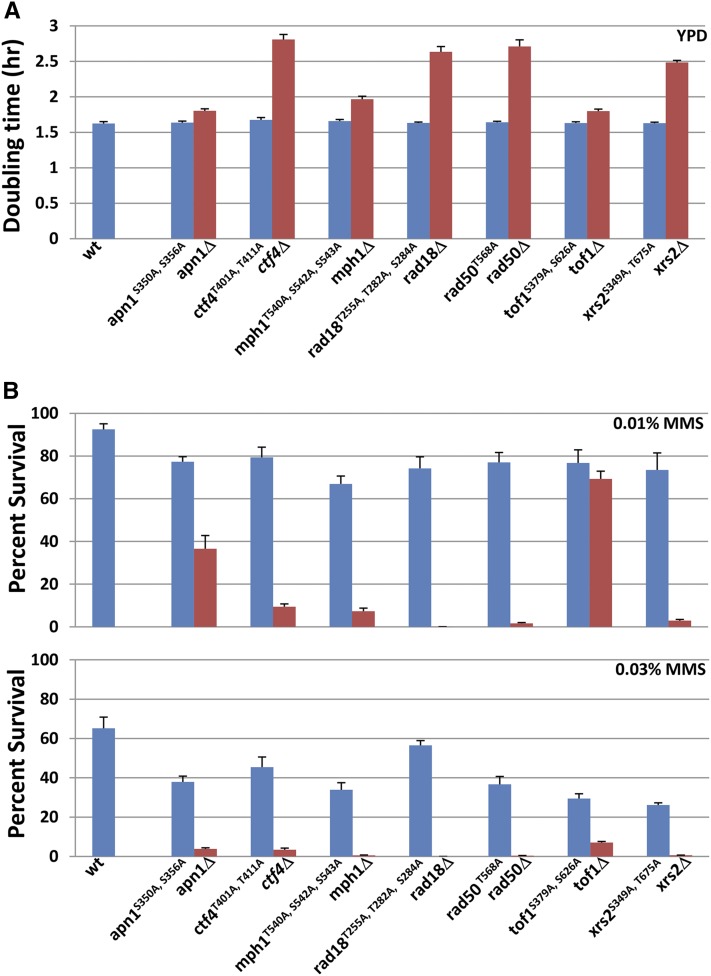
The vast majority of phosphorylation sites tend to locate in structurally disordered loop regions, and mutations in loop regions tend not to disrupt protein function (Iakoucheva et al. 2004; Gsponer et al. 2008). Eleven of the selected 15 phosphosites were predicted with high confidence to locate in loop regions (Phyre2 program, http://www.sbg.bio.ic.ac.uk/phyre2/), including apn1S350, ctf4T401, ctf4T411, mph1T540, mph1S542, rad18T155, rad18T282, tof1S379, tof1S626, xrs2S349, and xrs2T675. All of the nonphosphorylatable mutants demonstrate normal doubling time in rich medium, in contrast to many of their respective congenic deletion mutants (Figure 2A), suggesting that the mutant proteins are functional under normal growth conditions.
Figure 2.
Nonphosphorylatable alleles show mild but significant dose-dependent MMS sensitivity. (A) Doubling times of various phospho-mutants as compared to their respective deletion strains. Log-phase cultures were diluted in YPD such that all cultures started at a density of 5 × 105 cells/ml. The cell density of each culture was subsequently measured every 2 hr for 10 hr. The log numbers were then plotted. The doubling times were calculated from determining the slope of the straight line of each graph after linear regression. Three independent, sequence-verified isolates of each genotype were assayed, and the error bars represent the standard deviation for the three isolates. (B) Cell survival in two doses of MMS. For quantitative survival analyses in MMS, log-phase wild-type, nonphosphorylatable point mutants and deletion mutants were serially diluted in PBS and spread onto YPD, YPD + 0.01% MMS, or YPD + 0.03% MMS plates. Viable cells were determined by the number of CFUs after 3 days at 30°. Three independent sequence-verified, independent transformants of each strain were tested, and the error bars represent the standard deviation for the three isolates. Of note, a rad18Δ strain is highly sensitive to MMS so that zero CFUs were obtained from a 0.03% MMS plate.
We next assessed the functional importance of these mutated phospho-targets by examining the nonphosphorylatable amino acid substitution alleles for dose-dependent MMS sensitivity. We determined that all mutants conferred mild but significant MMS sensitivities in response to 0.01 and 0.03% MMS, which were intermediate between the sensitivity of the wild-type strain and a full deletion of the ORF (Figure 2B). From these data we conclude that each of the phosphosites contributes to its respective protein’s role in surviving MMS-mediated DNA damage; however, based on the intermediate phenotypes observed, none is the sole determinant of its protein’s respective role in the DNA damage response.
tof1S379A, S626A and xrs2S349A, T675A showed enhanced phenotypes in specific genetic backgrounds
The DDR collectively encompasses a wide array of both competing and collaborating repair and signaling mechanisms, with many proteins contributing multiple, sometimes-independent functions to more than one such pathway. As such, we hypothesized that the intermediate DDR phenotype conferred by each phospho-mutant may reflect the disabling of a subset of each respective protein’s complete repertoire of DDR functions. This hypothesis predicts that the nonphosphorylatable mutant alleles may show interactions with only a subset of the genes with which the corresponding deletion allele interacts. We tested this prediction by examining known interactions of tof1Δ and xrs2Δ in the respective nonphosphorylatable allele backgrounds, as described below.
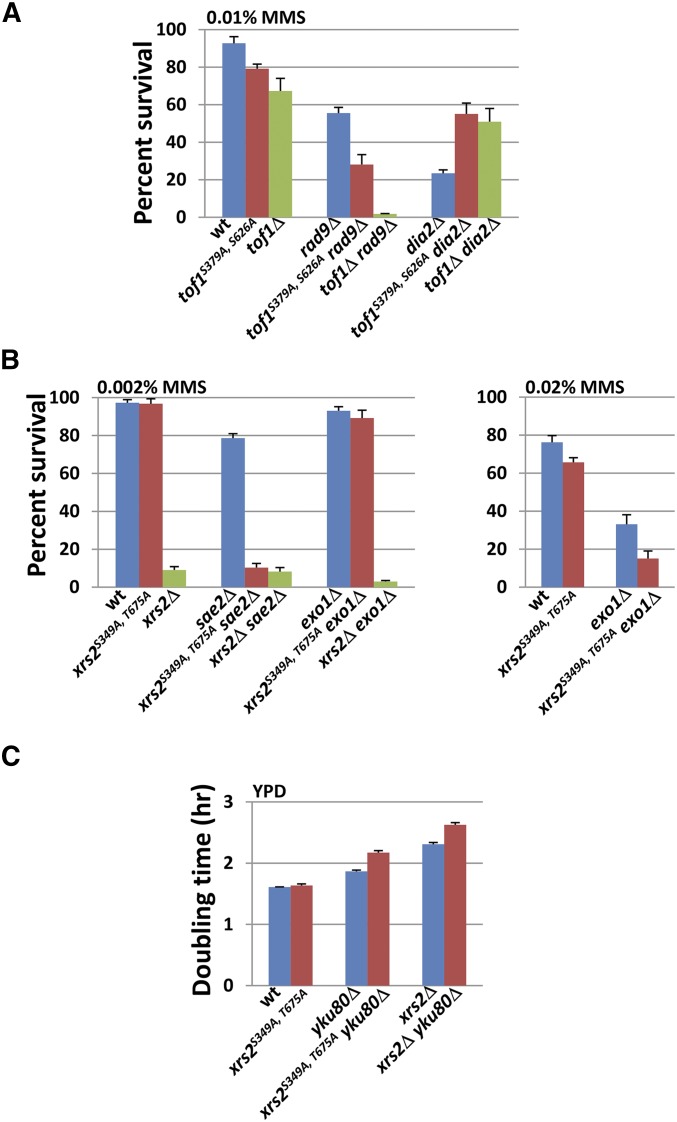
Tof1 has been implicated in sister-chromatid cohesion as well as in the activation of the DRC at stalled forks (Katou et al. 2003; Xu et al. 2004). These two known functions of Tof1 are independent of each other (Xu et al. 2004). Specifically, tof1Δ srs2Δ and tof1Δ ctf8Δ display synthetic lethality, which could result from sister-chromatid cohesion defects of tof1Δ (Xu et al. 2004). TOF1 also has a negative genetic interaction with the checkpoint mediator RAD9 (Foss 2001; Pan et al. 2006), as well as a positive interaction (suppression) with the checkpoint recovery mediator DIA2 (Fong et al. 2013) (Table 3 and Figure S6). When these interactions were tested in a nonphosphorylatable tof1 background, we found that tof1S379A, S626Arad9Δ cells displayed enhanced sensitivity as compared to either congenic single mutant. In addition, a tof1S379A, S626Adia2Δ strain exhibited less MMS sensitivity (suppression) than a dia2Δ single mutant (Figure S6). The phospho-mutant tof1S379A, S626Arad9Δ displayed intermediate MMS sensitivity vs. the tof1Δ rad9Δ double-deletion strain, indicating at least a partial role in survival in the absence of a fully functional checkpoint (Figure 3A), while tof1S379A, S626Adia2Δ fully recapitulated the suppression of loss of DIA2 as seen in tof1Δ (Figure 3A). While the exact biochemical nature of the suppression of MMS sensitivity by tof1Δ in a checkpoint-recovery-defective background is unknown, it has been hypothesized that loss of the Tof1-mediated replication checkpoint helps resumption of growth in dia2Δ cells in the presence of DNA damage (Fong et al. 2013), and these data suggest that tof1S379A, S626A may thus be defective in replication checkpoint activation. In contrast, the intermediate sensitivity phenotype exhibited by tof1S379A, S626Arad9Δ shows that the role of these two phosphosites in survival in a checkpoint-defective background is not absolute and suggests that additional biochemical properties not regulated by phosphorylation at these residues are still functional.
Table 3. Genetic interactions of tof1S379A, S626A and xrs2S349A, T675A.
| Genetic interactions testeda | Functions | Genetic interactions observed |
|---|---|---|
| tof1S379A, S626A | ||
| ctf8Δ | Required for sister-chromatid cohesion | No |
| srs2Δ | DNA helicase; involved in HR and sister-chromatid cohesion | No |
| rad9Δ | DNA damage-dependent checkpoint | Yes |
| dia2Δ | F-box protein; required for deactivation of Rad53 checkpoint kinase | Yes (suppression) |
| xrs2S349A, T675A | ||
| sgs1Δ | RecQ family DNA helicase; involved in HR | No |
| exo1Δ | 5′-3′ exonuclease; involved in HR | Yes |
| rtt109Δ | Histone acetyltransferase; involved in NHEJ | No |
| yku80Δ | Ku complex; involved in NHEJ and telomere function | Yesb |
| clb2Δ | B-type cyclin involved in cell cycle progression | No |
| pol32Δ | Third subunit of DNA polymerase delta | No |
| rad27Δ | Flap endonuclease; required for DNA replication and BER | No |
| apn1Δ | Apurinic/apyrimidinic endonuclease; required for BER | No |
| ctf4Δ | Required for sister-chromatid cohesion; DNA replication | No |
| srs2Δ | DNA helicase; involved in HR and sister-chromatid cohesion | No |
The genetic interactions with tof1S379A, S626A and xrs2S349A, T675A were selected from the manually curated genetic interactions with tof1Δ and xrs2Δ in the Saccharomyces Genome Database (http://www.yeastgenome.org/).
xrs2S349A, T675A yku80Δ showed enhanced growth defects (but not MMS sensitivity) as compared to yku80Δ.
Figure 3.
tof1S379A, S626A and xrs2S349A, T675A recapitulate a subset of the genetic interactions manifested by their respective deletion mutants. (A) tof1S379A, S626A shows negative interaction with rad9Δ and positive interaction with dia2Δ in the presence of MMS. The survival rates of wild-type and mutant strains in 0.01% MMS were determined as in Figure 2B. Three independent, PCR-confirmed gene knockout transformants of each strain were tested, and the error bars represent the standard deviation for the three isolates. [The tof1Δ rad9Δ strain is sml1Δ tof1Δ rad9Δ. The sml1Δ single mutation does not affect growth or survival at the tested MMS concentrations (data not shown)]. (B) xrs2S349A, T675A shows genetic interactions with sae2Δ and exo1Δ in the presence of MMS. The assay of survival rates of wild-type, single-, and double-mutant strains in indicated MMS concentrations were performed as in Figure 2B. Three independent, PCR-confirmed gene knockout transformants of each strain were tested, and the error bars represent the standard deviation for the three isolates. (C) xrs2S349A, T675A shows a synergistic growth interaction with yku80Δ. The wild type, xrs2S349A, T675A, and yku80Δ single- and double-mutant cells were grown in YPD to log phase at 30°. The cultures were diluted such that every culture started at a density of 106 cells/ml. The cell density of each culture was subsequently measured every 2 hr for 10 hr. The doubling time of each strain was calculated as in Figure 2A. Three independent, PCR-confirmed gene knockout transformants of each strain were tested, and the error bars represent the standard deviation for the three isolates.
While Tof1 is hypothesized to play multiple roles at stalled replication forks, Xrs2 has been implicated in even more varied DDR functions including checkpoint signaling (Nakada et al. 2003, 2004), recombination (Cejka 2015), and telomere maintenance (Nugent et al. 1998). As such, we chose 10 genes (known to interact with xrs2Δ and comprising a wide variety of DDR functions) to test the interaction of Xrs2 phospho-mutants with the loss of these functions. The genes chosen were SGS1, EXO1, RTT109, YKU80, CLB2, POL32, RAD27, APN1, CTF4, and SRS2. These genes are involved in many different cellular processes such as HR, nonhomologous end jointing (NHEJ), telomere maintenance, DNA replication, DNA repair, sister-chromatid cohesion, and cell cycle. Deletion of each of these 10 genes was tested for interaction effects with xrs2S349A, T675A (Table 3 and Figure S7). Of the 10 deletion mutants, two (exo1Δ and yku80Δ) exhibited interaction effects with xrs2S349A, T675A (Figure S7). xrs2S349A, T675Aexo1Δ cells displayed enhanced MMS sensitivity as compared to exo1Δ (at 0.02% MMS) (Figure 3B). Consistent with previously reported results (Nakada et al. 2004), complete deletion of the XRS2 ORF in combination with exo1Δ renders cells sensitive to MMS at very low concentrations (0.002%) (Figure 3B). At such dose, in contrast to the extreme MMS sensitivity of xrs2Δ exo1Δ, the xrs2S349A, T675Aexo1Δ interaction is nonevident, and the sensitivity of this strain is identical to exo1Δ. We conclude that phosphorylation of Xrs2 at Ser349 and Thr675 is important for Xrs2’s role in the DDR, and the nonexistent phenotype at low doses of MMS shows that some DDR functionality is retained.
In contrast to xrs2S349A, T675Aexo1Δ, we observed that the xrs2S349A, T675Ayku80Δ double mutant displayed no enhanced sensitivity to MMS vs. yku80Δ alone (Figure S7). However, while xrs2S349A, T675A did not exhibit a growth defect on its own, xrs2S349A, T675Ayku80Δ cells grew at a slower rate than yku80Δ alone (note that the full ORF deletion xrs2Δ exhibits a growth phenotype, in contrast to the phospho-mutant) (Figure 3C). From these data, we conclude that phosphorylation of Xrs2 on Ser-349 and/or Thr-675 is required for normal growth in an yku80Δ background, discussed further below.
Mutations of Ser349 and Thr675 of Xrs2 affect telomere maintenance and entry into senescence
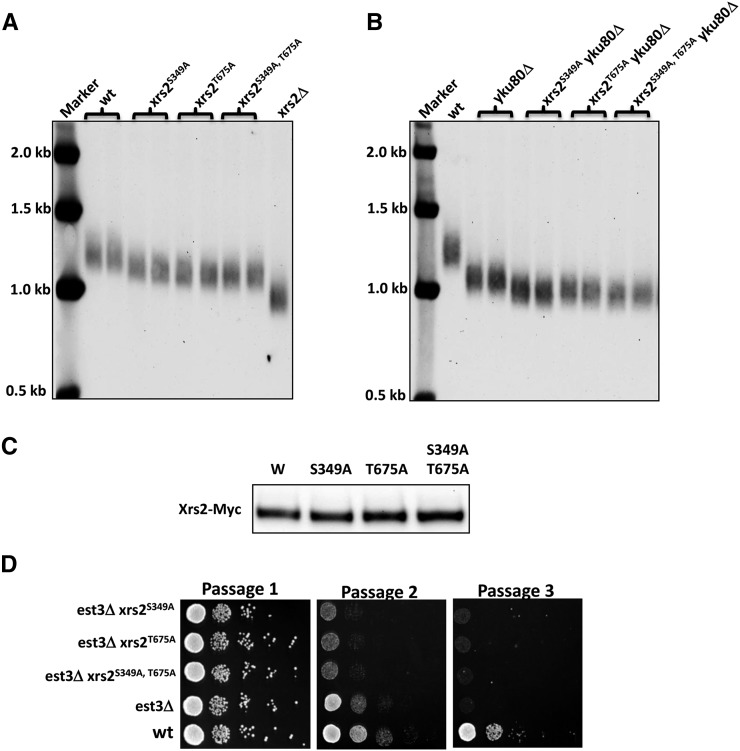
As described above, we observed that the phospho-mutant xrs2S349A, T675A exhibits a growth defect when combined with the end-capping mutant yku80Δ. One hypothesis for this genetic interaction is that xrs2S349A, T675Ayku80Δ could have an enhanced telomere defect, as loss of both genes was previously reported to result in synergistic telomere and growth defects (Nugent et al. 1998) (see also Figure 3C). Consistent with this hypothesis, all xrs2 phospho-mutants (xrs2S349A, xrs2T675A, and xrs2S349A, T675A) also showed mild (but significant) shortening of telomeres relative to wild type (Figure 4A), and there was no decrease in xrs2S349A, T675A expression (e.g., due to misfolding and degradation) as compared to the wild-type Xrs2p (Figure 4C). Furthermore, consistent with the genetic interaction between xrs2S349A, T675A and yku80Δ, the xrs2S349A, T675Ayku80Δ double mutants showed additive effects and exhibited telomere lengths shorter than yku80Δ (Figure 4B). In addition, the phospho-mutants xrs2S349Ayku80Δ and xrs2T675Ayku80Δ also displayed shorter telomere length than the yku80Δ single mutant (Figure 4B). From these data, we conclude that phosphorylations of S349 and T675 on Xrs2 are important for telomere maintenance independent of Yku80, supporting the hypothesis that the slow-growth phenotypes likely result from defective telomere maintenance.
Figure 4.
xrs2S349A and xrs2T675A mutants affect telomere maintenance and entry into senescence. (A and B) xrs2S349A and xrs2T675A strains exhibit a shortened telomere phenotype. XhoI-digested DNA was analyzed by Southern blot using a probe complementary to the Y′ subtelomere. (C) xrs2 phospho-mutant protein levels are unchanged as compared to the wild-type protein. Cells harboring Myc-tagged Xrs2 (wild-type or phospho-mutants) were grown in YPD to log phase and then harvested and lysed. The cell extracts were subjected to immunoblotting with anti-Myc antibodies. (D) Senescence assay on wild-type and xrs2 phospho-mutants. Initially, wild-type and freshly made est3Δ cells were resuspended in PBS to equal concentrations (107 cells/ml) and serial 10-fold dilutions were spotted on YPD plates. After 2 days at 30°, cells were scraped from the grown patches and resuspended in PBS to equal concentrations (107 cells/ml). Serial 10-fold dilutions were spotted on fresh YPD plates as above. To confirm the results, the entire process was repeated, and consistent results were obtained.
In telomerase-deficient cells, telomeres shorten over each round of DNA replication, leading to replicative senescence (“est” phenotype) (Gilson and Geli 2007; Palm and de Lange 2008), a phenotype that is accelerated by deletion of XRS2 (Chang et al. 2011). As such, we hypothesized that loss of phosphorylation of S349 and/or T675 on Xrs2 would also cause an accelerated senescence phenotype in the absence of telomerase. To test this, we generated a knockout of EST3 (an essential subunit of telomerase) in the xrs2 phospho-mutant backgrounds and examined the kinetics of entry into senescence. For this purpose, freshly made est3Δ mutants were serially spotted onto YPD plates for testing of survival (passage 1). After 2 days at 30°, cells from the grown patches were scraped and serially spotted on a fresh YPD plate (passage 2). The same process was repeated after another 2 days at 30° (passage 3). Initially, cell viability (passage 1) was equal among wild-type and est3Δ single and double mutants (Figure 4D). As expected, the est3Δ mutants entered senescence at passage 3, whereas wild-type cells remained viable (Figure 4D). However, when est3Δ was combined with nonphosphorylatable alleles of XRS2, entry into senescence was accelerated, as evidenced by a reduction in viability in passage 2 (Figure 4D). From these data we conclude that phosphorylation of S349 and T675 on Xrs2 prevents early entry into senescence in the absence of telomerase, with the failure to maintain normal-length telomeres (as described above) one likely cause.
Mutation of Xrs2 phosphorylation sites results in severe MMS sensitivity in the absence of the MRX activator Sae2
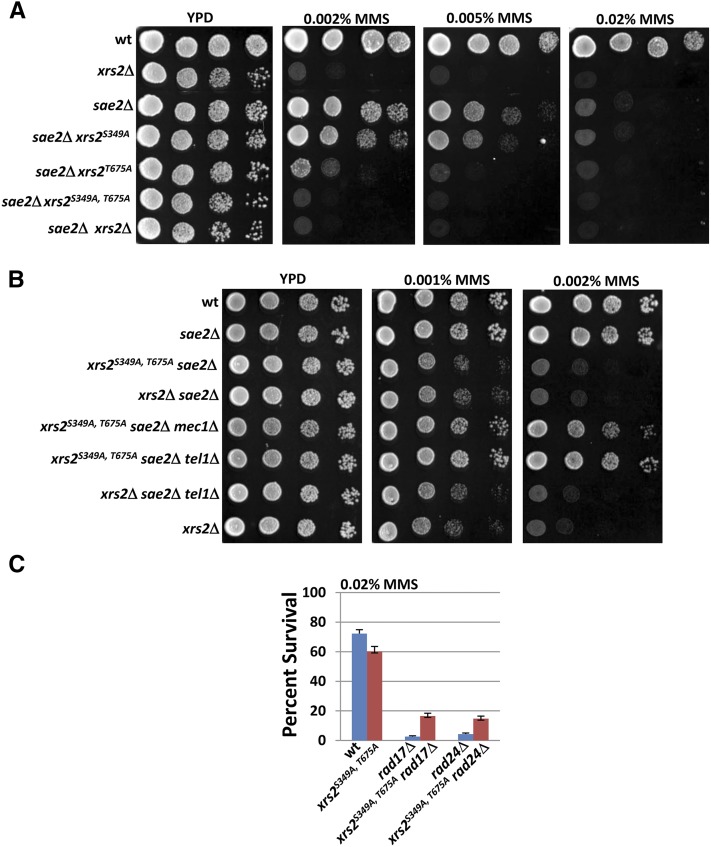
The Mre11 complex plays a parallel role with Exonuclease 1 (Exo1) in producing long-range ssDNA tails (Bernstein et al. 2013; Cannavo and Cejka 2014). The genetic interaction of xrs2S349A, T675A with exo1Δ raises the possibility that xrs2S349A, T675A may confer a defect in resection at sites of DNA damage. Since one of the functions of Xrs2 is to activate Mre11 nuclease activity (Trujillo et al. 2003), we hypothesized that the phenotype of xrs2S349A, T675A may be enhanced by deletion of another activator of Mre11 nuclease, Sae2 (Cannavo and Cejka 2014). Indeed, we observed that the xrs2S349A, T675Asae2Δ double-mutant strain was much more sensitive to MMS than the single mutants (Figure 3B and Figure 5A). To determine if only one of the two phosphorylation sites is responsible for the genetic interaction with sae2Δ, we further tested the MMS sensitivity of xrs2S349Asae2Δ and xrs2T675Asae2Δ (Figure 5A). We found that the xrs2T675Asae2Δ strain displayed strong sensitivity to MMS as compared to sae2Δ. In contrast, an xrs2S349Asae2Δ strain showed mild (if any) MMS sensitivity (Figure 5A). Thus, the phosphorylation sites differentially contribute to the MMS sensitivity in the absence of SAE2.
Figure 5.
xrs phospho-mutants show genetic interactions with sae2Δ and rescue MMS-sensitive phenotype caused by 9-1-1 mutants. (A) xrs phospho-mutant cells show severe sensitivity to MMS when combined with sae2Δ. The strains were grown in YPD overnight at 30°. Serial 10-fold dilutions were spotted onto YPD and YPD + MMS plates. The plates were incubated in 30° for 2 days. (B) mec1Δ and tel1Δ suppressed the severe sensitivity of xrs2S349A, T675A sae2Δ to MMS. The spot assay followed the same protocol as in A. (C) xrs2S349A, T675A suppressed the MMS sensitivity of 9-1-1 mutants. The cell survival rates of wild-type and mutant strains in the presence of MMS were determined as in Figure 2B. Three independent, PCR-confirmed gene knockout transformants of each genotype were assayed, and the error bars represent the standard deviations for the three isolates.
The Mre11 endonuclease activity (promoted by Sae2) is known to be required for removing the MRX complex from DSB ends to facilitate subsequent long-range resection (Clerici et al. 2006; Bernstein et al. 2013; Chen et al. 2015). Retention of MRX on the damage site results in prolonged checkpoint activation and growth defects (Clerici et al. 2006; Chen et al. 2015). One hypothesis to explain the strong interaction between xrs2S349A, T675A and sae2Δ is that xrs2S349A, T675A may also be defective in activating the endonuclease activity of Mre11 (without disruption of MRX); thus mutation of Ser349 and Thr675 on Xrs2 coupled with loss of Sae2 may cause drastic inactivation of Mre11 endonuclease activity, leading to retention of MRX and uncontrolled checkpoint activation. If this is the case, removing one of the checkpoint kinases Mec1 or Tel1 may alleviate the severe MMS-sensitivity phenotype at low concentrations of MMS when checkpoint functions are not critical for survival (Huang et al. 2013). To test this prediction, we deleted MEC1 or TEL1 in the xrs2S349A, T675Asae2Δ background. We found that both mec1Δ and tel1Δ suppressed the MMS-sensitivity phenotype of xrs2S349A, T675Asae2Δ (Figure 5B). Furthermore, although MMS-treated xrs2S349A, T675A cells exhibited only a modest increase in Rad53 phosphorylation in various genetic backgrounds tested (Figure S8), we found that when xrs2S349A, T675A was combined with deletion of the 9-1-1 checkpoint factors RAD17 or RAD24, xrs2S349A, T675A was able to suppress the MMS sensitivity of the 9-1-1 mutants (Figure 5C). Since the 9-1-1 complex and Tel1/Xrs2 act in parallel to activate the checkpoint (Piening et al. 2013), these observations suggest that the increased checkpoint activity in xrs2S349A, T675A may compensate for the loss of checkpoint activity caused by deletion of 9-1-1 genes. From these data, we conclude that the phosphorylation of residues Ser349 and Thr675 on XRS2 contributes to attenuation of MMS-triggered checkpoints.
Discussion
Among a panel of proteins in which we mutated S/T/Y sites identified in our screen to alanine, disruption of these sites resulted in DNA damage sensitivity and recapitulated known genetic interactions that have been previously observed via deletion of the entire ORF. Of note, in many cases the DDR phenotypes observed due to amino acid substitution were intermediate to that of the full ORF deletion; this could be due to a number of scenarios: (1) many DDR proteins are hyperphosphorylated, and removal of one to two sites may only partially abrogate the DDR phenotype; (2) some DDR proteins likely play multiple functional roles in the DDR, of which only a subset may be dependent on phosphorylation; or (3) one or more substitutions may destabilize the local or overall protein structure, potentially resulting in a null or hypomorphic allele. The majority of phosphorylation sites that we tested were predicted to be in structurally disordered loop regions, and none confers a growth defect when mutated; therefore, they are unlikely to disrupt protein structure (Iakoucheva et al. 2004; Gsponer et al. 2008; Dephoure et al. 2013). Nevertheless, we cannot entirely rule out this possibility. A common approach is to generate phospho-mimicking (aspartic or glutamic acid) amino acid substitutions of the site of interest and to determine if the observed phenotypes are reversed or neutralized. However, since the chemical environment introduced by phosphorylation is not completely equivalent to that of negatively charged residues, the failure rate of such studies is high, and the behavior of phosphomimetic mutations can be hard to interpret (Hunter 2012; Dephoure et al. 2013). In our studies, none of the phospho-mutants showed growth defects under normal conditions (in contrast to the full ORF deletion mutants, which exhibited mild-to-significant growth defects) (Figure 2A), suggesting that the engineered phosphosite substitutions behave similarly to wild type. Moreover, the expression level of Myc-tagged xrs2S349A, T675A is indistinguishable from wild-type Xrs2 (Figure 4B), suggesting that the mutant protein is not targeted for degradation.
Previous studies have demonstrated that members of the MRX complex are regulated by phosphorylation in response to DNA damage (Usui et al. 2001; Simoneau et al. 2014; Soriano-Carot et al. 2014). Our phospho-proteomic studies raise the possibility of novel phosphorylation mechanisms that act on Xrs2. The novel phosphosite Thr675 of Xrs2 plays a major role in cell survival in MMS when Sae2 is absent (Figure 5). SAE2 is required for activating Mre11 endonuclease activity (Cannavo and Cejka 2014). Given the strong genetic interaction with sae2Δ, we hypothesized that Thr675 on Xrs2 may function in a parallel pathway of activating Mre11 endonuclease activity, which is essential for timely removal of the MRX complex from DSB ends, and the retention of MRX results in prolonged checkpoint activation and growth defects (Clerici et al. 2006; Bernstein et al. 2013; Chen et al. 2015). Our genetic studies showed that tel1Δ and mec1Δ checkpoint mutants suppressed the severe sensitivity of an xrs2S349A, T675Asae2Δ strain to MMS (Figure 5B), consistent with the idea that prolonged checkpoint activation is responsible for the severe MMS-sensitivity phenotype. Furthermore, xrs2S349A, T675A by itself suppressed the MMS sensitivity of the 9-1-1 checkpoint mutants (Figure 5C). These phenotypes are consistent with the possibility that these phosphosites may promote MRX removal, possibly through activation of Mre11 in parallel with Sae2, and affect checkpoint reversal.
Alignment of Xrs2 with its human ortholog Nbs1 indicates that the phosphosite Thr675 occurs in a highly conserved C-terminal region and adjacent to an important ATM phosphorylated site Ser615 (http://www.ebi.ac.uk/Tools/psa/emboss_needle/). The C-terminal region contains highly conserved and functionally critical motifs, which are responsible for interaction with Mre11 and Tel1/ATM (Demuth and Digweed 2007; Schiller et al. 2012). In addition, the most common mutation responsible for Nijmegen breakage syndrome, 657Δ5, also locates in the C-terminal of Nbs1 (Demuth and Digweed 2007). It will be of interest to investigate whether mutation of Ser615 on Nbs1 exhibits a genetic interaction with the Sae2 ortholog CtIP.
The MRX complex also plays important roles in telomere maintenance. Cells lacking any one component of MRX (mrxΔ) have short and stable telomeres (Wellinger and Zakian 2015). In addition, mrxΔ also shows an accelerated senescence phenotype in the absence of telomerase (Chang et al. 2011). These phenotypes, albeit milder, are also reflected in both xrs2 phosphosite mutations, suggesting that phosphorylation on these residues may play important roles in telomere maintenance. Many genes involved in the DNA damage checkpoint response also affect telomere length (Wellinger and Zakian 2015). Tel1 is the master kinase that regulates telomere maintenance, and the kinase activity of Tel1 is required for such roles (Greenwell et al. 1995; Morrow et al. 1995). It was previously reported that Tel1 phosphorylates several proteins at telomeres including Xrs2 (Mallory et al. 2003; Tseng et al. 2006; Sridhar et al. 2014). However, it is unclear whether phosphorylation of Xrs2 by Tel1 is important for telomere maintenance (Mallory et al. 2003). In addition, the consensus sequences around the two MMS-induced phosphosites in this study do not contain S/T-Q, and therefore it is unlikely that Tel1 directly phosphorylates these sites. Another kinase that plays important roles in telomere maintenance is the cell cycle master regulator Cdc28 (Enserink and Kolodner 2010). Telomeres are actively maintained during late S phase, which is attributed mainly to the function of Cdk to promote telomere maintenance (Enserink and Kolodner 2010). Cdc28 has been recently implicated to be responsible for the phosphorylation of Ser349, studied here, on Xrs2 (Holt et al. 2009; Simoneau et al. 2014). It was found that mutation of this site and six other S/T-P sites on Xrs2 specifically stimulates NHEJ (Simoneau et al. 2014). Recent studies have shown that Nbs1 is also phosphorylated by Cdk1/2 to promote DSB repair in human cells (Wohlbold et al. 2012). The phosphorylation on Nbs1 occurs at Ser432, which is not far away from Ser349 on Xrs2, based on protein/protein alignments (http://www.ebi.ac.uk/Tools/psa/emboss_needle/). How the phosphorylation of Ser349 by Cdc28 affects telomere maintenance and NHEJ or whether there is a crosstalk between these two functions has yet to be investigated.
A wide variety of human diseases, including cancer, have been found to be associated with deregulated phosphorylation events and, in particular, mutated kinases (Cohen 2002). Thus, a careful reconstruction of the vast phospho-signaling networks that tightly regulate the DNA damage response, including the identification of all key points of failure for which a defect in phosphorylation has catastrophic consequences, will aid the development of novel anticancer therapies.
Acknowledgments
We thank Daniel Gottschling for plasmids used in this study and Travis Lorentzen and Regine Schoenherr for careful reading and helpful discussion of the manuscript. This work was supported by National Institutes of Health grant R01 CA129604.
Footnotes
Communicating editor: J. A. Nickoloff
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.115.185231/-/DC1.
Literature Cited
- Albuquerque C. P., Smolka M. B., Payne S. H., Bafna V., Eng J., et al. , 2008. A multidimensional chromatography technology for in-depth phosphoproteome analysis. Mol. Cell. Proteomics 7: 1389–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcasabas A. A., Osborn A. J., Bachant J., Hu F., Werler P. J., et al. , 2001. Mrc1 transduces signals of DNA replication stress to activate Rad53. Nat. Cell Biol. 3: 958–965. [DOI] [PubMed] [Google Scholar]
- Allen C., Ashley A. K., Hromas R., Nickoloff J. A., 2011. More forks on the road to replication stress recovery. J. Mol. Cell Biol. 3: 4–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amoutzias G. D., Y. He, K. S. Lilley, Y. Van de Peer, S. G. Oliver, 2012 Evaluation and properties of the budding yeast phosphoproteome. Mol. Cell Proteomics DOI: 10.1074/mcp.M111.009555. [DOI] [PMC free article] [PubMed]
- Bastos de Oliveira F. M., Kim D., Cussiol J. R., Das J., Jeong M. C., et al. , 2015. Phosphoproteomics reveals distinct modes of Mec1/ATR signaling during DNA replication. Mol. Cell 57: 1124–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein K. A., Mimitou E. P., Mihalevic M. J., Chen H., Sunjaveric I., et al. , 2013. Resection activity of the Sgs1 helicase alters the affinity of DNA ends for homologous recombination proteins in Saccharomyces cerevisiae. Genetics 195: 1241–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannavo E., Cejka P., 2014. Sae2 promotes dsDNA endonuclease activity within Mre11-Rad50-Xrs2 to resect DNA breaks. Nature 514: 122–125. [DOI] [PubMed] [Google Scholar]
- Cejka P., 2015. DNA end resection: nucleases team up with the right partners to initiate homologous recombination. J. Biol. Chem. 290: 22931–22938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang H. Y., Lawless C., Addinall S. G., Oexle S., Taschuk M., et al. , 2011. Genome-wide analysis to identify pathways affecting telomere-initiated senescence in budding yeast. G3 (Bethesda) 1: 197–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Donnianni R. A., Handa N., Deng S. K., Oh J., et al. , 2015. Sae2 promotes DNA damage resistance by removing the Mre11-Rad50-Xrs2 complex from DNA and attenuating Rad53 signaling. Proc. Natl. Acad. Sci. USA 112: E1880–E1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S. H., Albuquerque C. P., Liang J., Suhandynata R. T., Zhou H., 2010. A proteome-wide analysis of kinase-substrate network in the DNA damage response. J. Biol. Chem. 285: 12803–12812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi A., Huttenhower C., Geer L. Y., Coon J. J., Syka J. E., et al. , 2007. Analysis of phosphorylation sites on proteins from Saccharomyces cerevisiae by electron transfer dissociation (ETD) mass spectrometry. Proc. Natl. Acad. Sci. USA 104: 2193–2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clerici M., Mantiero D., Lucchini G., Longhese M. P., 2006. The Saccharomyces cerevisiae Sae2 protein negatively regulates DNA damage checkpoint signalling. EMBO Rep. 7: 212–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen P., 2002. Protein kinases: The major drug targets of the twenty-first century? Nat. Rev. Drug Discov. 1: 309–315. [DOI] [PubMed] [Google Scholar]
- Cox J., Mann M., 2008. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26: 1367–1372. [DOI] [PubMed] [Google Scholar]
- Craig R., Beavis R. C., 2004. TANDEM: matching proteins with mass spectra. Bioinformatics 20: 1466–1467. [DOI] [PubMed] [Google Scholar]
- Demuth I., Digweed M., 2007. The clinical manifestation of a defective response to DNA double-strand breaks as exemplified by Nijmegen breakage syndrome. Oncogene 26: 7792–7798. [DOI] [PubMed] [Google Scholar]
- Dephoure N., Gould K. L., Gygi S. P., Kellogg D. R., 2013. Mapping and analysis of phosphorylation sites: a quick guide for cell biologists. Mol. Biol. Cell 24: 535–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enserink J. M., Kolodner R. D., 2010. An overview of Cdk1-controlled targets and processes. Cell Div. 5: 11–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ficarro S. B., Adelmant G., Tomar M. N., Zhang Y., Cheng V. J., et al. , 2009. Magnetic bead processor for rapid evaluation and optimization of parameters for phosphopeptide enrichment. Anal. Chem. 81: 4566–4575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong C. M., Arumugam A., Koepp D. M., 2013. The Saccharomyces cerevisiae F-box protein Dia2 is a mediator of S-phase checkpoint recovery from DNA damage. Genetics 193: 483–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foss E. J., 2001. Tof1p regulates DNA damage responses during S phase in Saccharomyces cerevisiae. Genetics 157: 567–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedberg E. C., Friedberg E. C., 2006. DNA Repair and Mutagenesis. ASM Press, Washington, DC. [Google Scholar]
- Gasch A. P., Huang M., Metzner S., Botstein D., Elledge S. J., et al. , 2001. Genomic expression responses to DNA-damaging agents and the regulatory role of the yeast ATR homolog Mec1p. Mol. Biol. Cell 12: 2987–3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaemmaghami S., Huh W. K., Bower K., Howson R. W., Belle A., et al. , 2003. Global analysis of protein expression in yeast. Nature 425: 737–741. [DOI] [PubMed] [Google Scholar]
- Gilson E., Geli V., 2007. How telomeres are replicated. Nat. Rev. Mol. Cell Biol. 8: 825–838. [DOI] [PubMed] [Google Scholar]
- Greenwell P. W., Kronmal S. L., Porter S. E., J. Gassenhuber, B. Obermaier et al, 1995. TEL1, a gene involved in controlling telomere length in S. cerevisiae, is homologous to the human ataxia telangiectasia gene. Cell 82: 823–829. [DOI] [PubMed] [Google Scholar]
- Gsponer J., Futschik M. E., Teichmann S. A., Babu M. M., 2008. Tight regulation of unstructured proteins: from transcript synthesis to protein degradation. Science 322: 1365–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanawalt P. C., 2015. Historical perspective on the DNA damage response. DNA Repair (Amst.) 36: 2–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helbig A. O., Rosati S., Pijnappel P. W., van Breukelen B., Timmers M. H., et al. , 2010. Perturbation of the yeast N-acetyltransferase NatB induces elevation of protein phosphorylation levels. BMC Genomics 11: 685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hishida T., Kubota Y., Carr A. M., Iwasaki H., 2009. RAD6–RAD18–RAD5-pathway-dependent tolerance to chronic low-dose ultraviolet light. Nature 457: 612–615. [DOI] [PubMed] [Google Scholar]
- Holt L. J., Tuch B. B., Villén J., Johnson A. D., Gygi S. P., et al. , 2009. Global analysis of Cdk1 substrate phosphorylation sites provides insights into evolution. Science 325: 1682–1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D., Piening B. D., Paulovich A. G., 2013. The preference for error-free or error-prone postreplication repair in Saccharomyces cerevisiae exposed to low-dose methyl methanesulfonate is cell cycle dependent. Mol. Cell. Biol. 33: 1515–1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M., Zhou Z., Elledge S. J., 1998. The DNA replication and damage checkpoint pathways induce transcription by inhibition of the Crt1 repressor. Cell 94: 595–605. [DOI] [PubMed] [Google Scholar]
- Hunter T., 2012. Why nature chose phosphate to modify proteins. Philos. Trans. R. Soc. Lond. B Biol. Sci. 367: 2513–2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iakoucheva L. M., Radivojac P., Brown C. J., O’Connor T. R., Sikes J. G., et al. , 2004. The importance of intrinsic disorder for protein phosphorylation. Nucleic Acids Res. 32: 1037–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapp E. A., Schutz F., Connolly L. M., Chakel J. A., Meza J. E., et al. , 2005. An evaluation, comparison, and accurate benchmarking of several publicly available MS/MS search algorithms: sensitivity and specificity analysis. Proteomics 5: 3475–3490. [DOI] [PubMed] [Google Scholar]
- Katou Y., Kanoh Y., Bando M., Noguchi H., Tanaka H., et al. , 2003. S-phase checkpoint proteins Tof1 and Mrc1 form a stable replication-pausing complex. Nature 424: 1078–1083. [DOI] [PubMed] [Google Scholar]
- Lazzaro F., Giannattasio M., Puddu F., Granata M., Pellicioli A., et al. , 2009. Checkpoint mechanisms at the intersection between DNA damage and repair. DNA Repair (Amst.) 8: 1055–1067. [DOI] [PubMed] [Google Scholar]
- Longtine M. S., McKenzie A., III, Demarini D. J., Shah N. G., Wach A., et al. , 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14: 953–961. [DOI] [PubMed] [Google Scholar]
- MacDougall C. A., Byun T. S., C. Van, M. C. Yee, K. A. Cimprich, 2007. The structural determinants of checkpoint activation. Genes Dev. 21: 898–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory J. C., Bashkirov V. I., Trujillo K. M., Solinger J. A., Dominska M., et al. , 2003. Amino acid changes in Xrs2p, Dun1p, and Rfa2p that remove the preferred targets of the ATM family of protein kinases do not affect DNA repair or telomere length in Saccharomyces cerevisiae. DNA Repair (Amst.) 2: 1041–1064. [DOI] [PubMed] [Google Scholar]
- Mann M., 2014. Fifteen years of Stable Isotope Labeling by Amino Acids in Cell Culture (SILAC). Methods Mol. Biol. 1188: 1–7. [DOI] [PubMed] [Google Scholar]
- Mizumoto K., Glascott P. A., Jr, Farber J. L., 1993. Roles for oxidative stress and poly(ADP-ribosyl)ation in the killing of cultured hepatocytes by methyl methanesulfonate. Biochem. Pharmacol. 46: 1811–1818. [DOI] [PubMed] [Google Scholar]
- Morrow D. M., Tagle D. A., Shiloh Y., Collins F. S., Hieter P., 1995. TEL1, an S. cerevisiae homolog of the human gene mutated in ataxia telangiectasia, is functionally related to the yeast checkpoint gene MEC1. Cell 82: 831–840. [DOI] [PubMed] [Google Scholar]
- Murakami-Sekimata A., Huang D., Piening B. D., Bangur C., Paulovich A. G., 2010. The Saccharomyces cerevisiae RAD9, RAD17 and RAD24 genes are required for suppression of mutagenic post-replicative repair during chronic DNA damage. DNA Repair (Amst.) 9: 824–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakada D., Matsumoto K., Sugimoto K., 2003. ATM-related Tel1 associates with double-strand breaks through an Xrs2-dependent mechanism. Genes Dev. 16: 1957–1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakada D., Hirano Y., Sugimoto K., 2004. Requirement of the Mre11 complex and exonuclease 1 for activation of the Mec1 signaling pathway. Mol. Cell. Biol. 24: 10016–10025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nugent C. I., Bosco G., Ross L. O., Evans S. K., Salinger A. P., et al. , 1998. Telomere maintenance is dependent on activities required for end repair of double-strand breaks. Curr. Biol. 8: 657–660. [DOI] [PubMed] [Google Scholar]
- Pages V., Santa Maria S. R., Prakash L., Prakash S., 2009. Role of DNA damage-induced replication checkpoint in promoting lesion bypass by translesion synthesis in yeast. Genes Dev. 23: 1438–1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palm W., de Lange T., 2008. How shelterin protects mammalian telomeres. Annu. Rev. Genet. 42: 301–334. [DOI] [PubMed] [Google Scholar]
- Pan X., Ye P., Yuan D. S., Wang X., Bader J. S., et al. , 2006. A DNA integrity network in the yeast Saccharomyces cerevisiae. Cell 124: 1069–1081. [DOI] [PubMed] [Google Scholar]
- Paulovich A. G., Hartwell L. H., 1995. A checkpoint regulates the rate of progression through S phase in S. cerevisiae in response to DNA damage. Cell 82: 841–847. [DOI] [PubMed] [Google Scholar]
- Piening B. D., Huang D., Paulovich A. G., 2013. Novel connections between DNA replication, telomere homeostasis, and the DNA damage response revealed by a genome-wide screen for TEL1/ATM interactions in Saccharomyces cerevisiae. Genetics 193: 1117–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappsilber J., Mann M., Ishihama Y., 2007. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using Stage Tips. Nat. Protoc. 2: 1896–1906. [DOI] [PubMed] [Google Scholar]
- Reid R. J. D., Sunjevaric I., Keddache M., Rothstein R., 2002. Efficient PCR-based gene disruption in Saccharomyces strains using intergenic primers. Yeast 19: 319–328. [DOI] [PubMed] [Google Scholar]
- Robinson M. D., Grigull J., Mohammad N., Hughes T. R., 2002. FunSpec: a web-based cluster interpreter for yeast. BMC Bioinformatics 3: 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadowski, I., B.-J. Breitkreutz, C. Stark, T.-C. Su, M. Dahabieh et al., 2013 The PhosphoGRID Saccharomyces cerevisiae protein phosphorylation site database: version 2.0 update. Database (Oxford) 2013: bat026 [DOI] [PMC free article] [PubMed]
- Salmon T. B., Evert B. A., Song B., Doetsch P. W., 2004. Biological consequences of oxidative stress-induced DNA damage in Saccharomyces cerevisiae. Nucleic Acids Res. 32: 3712–3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez Y., Bachant J., Wang H., Hu F., Liu D., et al. , 1999. Control of the DNA damage checkpoint by chk1 and rad53 protein kinases through distinct mechanisms. Science 286: 1166–1171. [DOI] [PubMed] [Google Scholar]
- Schiller C. B., Lammens K., Guerini I., Coordes B., Feldmann H., et al. , 2012. Structure of Mre11-Nbs1 complex yields insights into ataxia-telangiectasia-like disease mutations and DNA damage signaling. Nat. Struct. Mol. Biol. 19: 693–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada K., Pasero P., Gasser S. M., 2002. ORC and the intra-S-phase checkpoint: a threshold regulates Rad53p activation in S phase. Genes Dev. 16: 3236–3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siede W., Friedberg A. S., Friedberg E. C., 1993. RAD9-dependent G1 arrest defines a second checkpoint for damaged DNA in the cell cycle of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 90: 7985–7989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simoneau A., Robellet X., Ladouceur A. M., D’Amours D., 2014. Cdk1-dependent regulation of the Mre11 complex couples DNA repair pathways to cell cycle progression. Cell Cycle 13: 1078–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer M. S., Kahana A., Wolf A. J., Meisinger L. L., Peterson S. E., et al. , 1998. Identification of high-copy disruptors of telomeric silencing in Saccharomyces cerevisiae. Genetics 150: 613–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolka M. B., Albuquerque C. P., Chen S., Zhou H., 2007. Proteome-wide identification of in vivo targets of DNA damage checkpoint kinases. Proc. Natl. Acad. Sci. USA 104: 10364–10369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriano-Carot M., Quilis I., Bañó M. C., Igual J. C., 2014. Protein kinase C controls activation of the DNA integrity checkpoint. Nucleic Acids Res. 42: 7084–7095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridhar A., Kedziora S., Donaldson A. D., 2014. At short telomeres Tel1 directs early replication and phosphorylates Rif1. PLoS Genet. 10: e1004691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storici F., Lewis L. K., Resnick M. A., 2001. In vivo site-directed mutagenesis using oligonucleotides. Nat. Biotechnol. 19: 773–776. [DOI] [PubMed] [Google Scholar]
- Tercero J. A., Longhese M. P., Diffley J. F., 2003. A central role for DNA replication forks in checkpoint activation and response. Mol. Cell 11: 1323–1336. [DOI] [PubMed] [Google Scholar]
- Trujillo K. M., Roh D. H., Chen L., Van Komen S., Tomkinson A., et al. , 2003. Yeast xrs2 binds DNA and helps target rad50 and mre11 to DNA ends. J. Biol. Chem. 278: 48957–48964. [DOI] [PubMed] [Google Scholar]
- Tseng S. F., Lin J. J., Teng S. C., 2006. The telomerase-recruitment domain of the telomere binding protein Cdc13 is regulated by Mec1p/Tel1p-dependent phosphorylation. Nucleic Acids Res. 34: 6327–6336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usui T., Ogawa H., Petrini J. H., 2001. A DNA damage response pathway controlled by Tel1 and the Mre11 complex. Mol. Cell 7: 1255–1266. [DOI] [PubMed] [Google Scholar]
- Uzunova S. D., Zarkov A. S., Ivanova A. M., Stoynov S. S., Nedelcheva-Veleva M. N., 2014. The subunits of the S-phase checkpoint complex Mrc1/Tof1/Csm3: dynamics and interdependence. Cell Div. 9: 4–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vialard J. E., Gilbert C. S., Green C. M., Lowndes N. F., 1998. The budding yeast Rad9 checkpoint protein is subjected to Mec1/Tel1-dependent hyperphosphorylation and interacts with Rad53 after DNA damage. EMBO J. 17: 5679–5688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizcaíno J. A., R. G. Côté, A. Csordas, J. A. Dianes, A. Fabregat et al, 2013. The PRoteomics IDEntifications (PRIDE) database and associated tools: status in 2013 Nucleic Acids Res. 41(Database issue): D1063–D1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinert T. A., Hartwell L. H., 1988. The RAD9 gene controls the cell cycle response to DNA damage in Saccharomyces cerevisiae. Science 241: 317–322. [DOI] [PubMed] [Google Scholar]
- Weinert T. A., Kiser G. L., Hartwell L. H., 1994. Mitotic checkpoint genes in budding yeast and the dependence of mitosis on DNA replication and repair. Genes Dev. 8: 652–665. [DOI] [PubMed] [Google Scholar]
- Wellinger R. J., Zakian V. A., 2015. Everything you ever wanted to know about Saccharomyces cerevisiae telomeres: beginning to end. Genetics 191: 1073–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohlbold L., Merrick K. A., De S., Amat R., Kim J. H., et al. , 2012. Chemical genetics reveals a specific requirement for Cdk2 activity in the DNA damage response and identifies Nbs1 as a Cdk2 substrate in human cells. PLoS Genet. 8: e1002935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H., Boone C., Klein H. L., 2004. Mrc1 is required for sister chromatid cohesion to aid in recombination repair of spontaneous damage. Mol. Cell. Biol. 24: 7082–7090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Gordenin D. A., Resnick M. A., 2010. A single-strand specific lesion drives MMS-induced hyper-mutability at a double-strand break in yeast. DNA Repair (Amst.) 9: 914–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeman M. K., Cimprich K. A., 2013. Causes and consequences of replication stress. Nat. Cell Biol. 16: 2–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziv, I., Y. Matiuhin, D. S. Kirkpatrick, Z. Erpapazoglou, S. Leon et al., 2011 A perturbed ubiquitin landscape distinguishes between ubiquitin in trafficking and in proteolysis. Mol Cell. Proteomics DOI: 10.1074/mcp.M111.009753. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.