Abstract
The Caenorhabditis elegans oxidative stress response transcription factor, SKN-1, is essential for the maintenance of redox homeostasis and is a functional ortholog of the Nrf family of transcription factors. The numerous levels of regulation that govern these transcription factors underscore their importance. Here, we add a thioredoxin, encoded by trx-1, to the expansive list of SKN-1 regulators. We report that loss of trx-1 promotes nuclear localization of intestinal SKN-1 in a redox-independent, cell non-autonomous fashion from the ASJ neurons. Furthermore, this regulation is not general to the thioredoxin family, as two other C. elegans thioredoxins, TRX-2 and TRX-3, do not play a role in this process. Moreover, TRX-1-dependent regulation requires signaling from the p38 MAPK-signaling pathway. However, while TRX-1 regulates SKN-1 nuclear localization, classical SKN-1 transcriptional activity associated with stress response remains largely unaffected. Interestingly, RNA-Seq analysis revealed that loss of trx-1 elicits a general, organism-wide down-regulation of several classes of genes; those encoding for collagens and lipid transport being most prevalent. Together, these results uncover a novel role for a thioredoxin in regulating intestinal SKN-1 nuclear localization in a cell non-autonomous manner, thereby contributing to the understanding of the processes involved in maintaining redox homeostasis throughout an organism.
Keywords: Caenorhabditis elegans, oxidative stress response, thioredoxin, ASJ neurons, cell non-autonomous signaling
THE ability of an organism to maintain redox homeostasis is critical for its survival. At the cellular level, exposure to oxidative insult can irreversibly damage DNA, proteins, and lipids, all of which can lead to cell apoptosis or necrosis (Ray et al. 2012; Thanan et al. 2014). At the organismal level, unresolved oxidative stress is considered a hallmark of numerous life-threatening diseases, including Alzheimer’s, Parkinson’s disease, atherosclerosis, and several forms of cancer (Hybertson et al. 2011; Thanan et al. 2014). To counteract oxidative insults, organisms have evolved specific pathways capable of sensing and responding to both endogenous and exogenous oxidative stress, termed “the oxidative stress response” (Lushchak 2011). This response is coordinated by oxidative stress response transcription factors, which activate the expression of detoxification and repair enzymes (McCord and Fridovich 1969; Anderson 1998; Lushchak 2011). In mammals, the major oxidative stress transcription factor is the nuclear factor erythroid 2-related factor, Nrf2, one of three Nrf paralogs (Hybertson et al. 2011). To ensure efficient surveillance of redox homeostasis, several mechanisms regulate Nrf2, including those that regulate its subcellular localization and protein turnover (Marinho et al. 2014).
The nematode Caenorhabditis elegans utilizes a functional ortholog of mammalian Nrf proteins, SKN-1, to coordinate its oxidative stress response (Walker et al. 2000; An and Blackwell 2003). More recently, a role for SKN-1 has been found in the regulation of the unfolded protein response and the maintenance of lipid homeostasis (Glover-Cutter et al. 2013; Lynn et al. 2015; Steinbaugh et al. 2015). Similar to Nrf2, SKN-1 regulation is also well studied, and overlapping mechanisms of regulation exist between mammals and worms. In general, both Nrf2 and SKN-1 seem to be regulated at the level of nuclear accumulation. Specifically, both mammals and worms employ cysteine-rich adaptor proteins, Keap1 and WDR-23, respectively, to facilitate the degradation of these transcription factors by the proteasome, thereby preventing their nuclear accumulation (Choe et al. 2009; Leung et al. 2014; Marinho et al. 2014). Furthermore, both mammalian and worm glycogen synthase kinase 3 phosphorylate Nrf2 and SKN-1, respectively, in a manner that impacts the subcellular localization of these transcription factors (An et al. 2005; Salazar et al. 2006). In C. elegans, additional mechanisms of SKN-1 regulation were elucidated. SKN-1 isoform C is antagonized by insulin/IGF-1-like signaling and is positively regulated by the p38 MAPK pathway via phosphorylation of Serines 74 and 340 (Inoue et al. 2005; Tullet et al. 2008). Exposure to oxidative stressors, such as sodium arsenite, impact these positive and negative regulators governing intestinal SKN-1, resulting in increased nuclear localization and transcriptional activation, thereby maintaining redox homeostasis (Inoue et al. 2005). However, while many factors and mechanisms of regulating SKN-1 are known, how these signaling pathways initially sense oxidative imbalance remains unclear.
Thioredoxins are small proteins that, due to their inherent amino acid chemistry, are redox reactive (Arner and Holmgren 2000; Powis and Montfort 2001; Buchanan et al. 2012). While thioredoxins can act as antioxidants via their ability to reduce oxidized proteins, they play a prominent role in the regulation of signaling pathways in several organisms (Fujino et al. 2006; Yoshioka et al. 2006). In mammals, thioredoxin 1, TRX1, serves as an allosteric inhibitor of apoptosis signal-regulating kinase 1, ASK1, by preventing dimerization at the N terminus of this MAPKKK, thereby inhibiting activation of p38 MAPK pathway signaling. Upon oxidation of TRX1 by reactive oxygen species (ROS), repression of ASK1 is relieved and ASK1 is able to homodimerize, activating its kinase activity and ultimately triggering the apoptotic response (Fujino et al. 2007). While the redox activity of thioredoxin is important for a majority of its cellular functions, thioredoxins have important, redox-independent cellular roles. For example, TRX1 promotes ASK1 ubiquitination and degradation irrespective of its redox activity (Liu and Min 2002). Moreover, a C. elegans thioredoxin, TRX-1, modulates DAF-28 signaling during dauer formation in a redox-independent fashion (Fierro-Gonzalez et al. 2011a).
In C. elegans, TRX-1 plays a role in life span, dauer formation, dietary restriction, and the oxidative stress response (Jee et al. 2005; Miranda-Vizuete et al. 2006; Fierro-Gonzalez et al. 2011a,b). However, no specific role for thioredoxins in signaling has been characterized in the worm. Given the general ability of thioredoxins to act as both redox-dependent and redox-independent regulators and for mammalian TRX1 to regulate the p38 MAPK pathway, we reasoned that a thioredoxin may regulate SKN-1 and/or the C. elegans oxidative stress response.
In this work, we explore whether thioredoxins are regulators of SKN-1 or one of the previously characterized SKN-1 regulatory components. Interestingly, we demonstrate that TRX-1, but not TRX-2 or TRX-3 (Cacho-Valadez et al. 2012; Jimenez-Hidalgo et al. 2014), affects the nuclear localization of intestinal SKN-1. Specifically, we observed that the nuclear localization of intestinal SKN-1 is increased in a trx-1(ok1449) null mutant and that this trx-1-dependent localization does not require redox activity but does require the p38 MAPK pathway. TRX-1 expression is restricted to the ASJ neurons (Jee et al. 2005; Miranda-Vizuete et al. 2006), indicating cell non-autonomous regulation and arguing against a direct interaction of TRX-1 with SKN-1 or its regulatory components. Nuclear localization of intestinal SKN-1 usually parallels activation of SKN-1-regulated genes, but we found by quantitative real time PCR (qRT-PCR) and RNA-Seq that loss of trx-1 did not increase the typical transcriptional activity of SKN-1 (An et al. 2005; Inoue et al. 2005; Tullet et al. 2008; Choe et al. 2009; Leung et al. 2014). Rather than an upregulation of the oxidative stress response transcriptional program, a downregulation of many genes was observed, particularly those encoding collagen and lipid transport and localization proteins. Interestingly, only three genes (lips-6, lips-11, and lbp-8), encoding two lipase-related proteins and a lipid chaperone, respectively, are up-regulated upon loss of trx-1. In summary, the data presented support a model in which nuclear localization, but not activation, of intestinal SKN-1 is regulated cell non-autonomously by TRX-1 in a redox-independent fashion.
Materials and Methods
Strains
C. elegans strains were grown and maintained as previously described (Hope 1999). C. elegans strains used in this study are listed in Supplemental Material, Table S4. The following bacterial strains were used in this study: Escherichia coli OP50, Enterococcus faecalis OG1RF, Pseudomonas aeruginosa PA14, and E. coli HT115.
Strain construction
In general, all mutant strains were backcrossed to wild-type N2 6x. To generate the VZ27, VZ26, and VZ157 strains, Is007 [skn-1b/c::gfp; rol-6(su1006)] worms were crossed with trx-1(ok1449), trx-2(tm2720), and trx-3(tm2820) mutants, respectively. To visualize TRX-1 localization, the OE3381 strain was generated by injecting the pPD95.77 plasmid containing trx-1 (1-kb promoter + gene) fused to gfp with the trx-1 3′ UTR (rather than the unc-54 3′ UTR native to this vector). To generate the VZ472, VZ458, and VZ461 strains, the trx-1 promoter was replaced with tissue-specific promoters (ssu-1; ASJ, ges-1; intestinal, daf-7; ASI) in the above-mentioned plasmid. These constructs were then injected into wild-type N2 worms along with the Punc-122::DsRed coinjection marker that causes red fluorescence in the coelomocytes. Finally, the arrays were transferred into the trx-1(ok1449); Is007 background to generate GF92, GF93, and GF94 strains, respectively. To determine p38 MAPK-signaling dependence, nsy-1(ok593), sek-1(km4), and pmk-1(km25) mutants were crossed into the trx-1(ok1449); Is007 background to generate the GF96, GF97, and GF98 strains, respectively. Ex060[skn-1(S74,340A)b/c::gfp; rol-6(su1006)] was also crossed into the trx-1(ok1449); Is007 background to further verify p38 MAPK-signaling dependence (GF95). To visualize gst-4 expression in the trx-1 background, the VZ433 strain was made by crossing trx-1(ok1449) mutants with dvIs19 [Pgst-4::gfp::NLS; rol-6(su1006)] worms. To generate GF99 and GF100, OE4064 and OE4067 were crossed into the trx-1(ok1449); Is007 background and maintained at 25° for four generations to remove the daf-28(sa191) mutation.
RNA interference
To knock down skn-1 expression, RNA interference (RNAi) was induced by feeding L1- to L4-stage worms with bacteria producing skn-1 double-stranded RNA. The skn-1 RNAi bacterial strain was made previously, as described (Hoeven et al. 2011).
Fluorescence microscopy
To visualize SKN-1B/C::GFP and TRX-1::GFP localization and gst-4::gfp expression, young adults were washed from plates and anesthetized with 1 mM levamisole. Anesthetized worms were mounted on 2% agarose pads and visualized and imaged using Olympus IX81 automated inverted microscope and Slidebook (version 5.0) software. For SKN-1B/C::GFP localization quantification, the percentage of intestinal SKN-1B/C::GFP nuclear localization was categorically scored as follows: none: no localization; low: posterior or anterior intestinal localization; medium: posterior and anterior intestinal localization; high: localization throughout the entire intestine (Inoue et al. 2005). For GF99 and GF100, percentage of intestinal SKN-1B/C::GFP nuclear localization was categorically scored in an area limited to the anterior intestine of animals as follows: none: no localization; low: weak fluorescence; medium: moderate fluorescence; or high: bright fluorescence. Chi square and Fisher’s exact tests (GraphPad Prism version 5.0) were used to calculate significance of SKN-1B/C::GFP nuclear localization averaged between three biological replicates of n ≥ 50 worms.
Western blotting
About 1000 worms were washed from NGM plates and collected in a 100-µl pellet using protein extraction buffer [50 mM Tris, pH 7.5, 50 mM NaCl, protease inhibitor cocktail (Roche, 11873580001), PhosStop (Roche, 04906845001)]. The pellet was sonicated (on ice) for 10 sec at level 5 and 50% duty. Suspensions were incubated in 1% SDS on ice for 5 min and then centrifuged in the cold at 14,000 × g for 10 min. Supernatants were transferred to a fresh Eppendorf tube, and total protein concentration was measured via BCA assay (Pierce, 23227). Sample buffer was added and protein lysates were boiled for 5 min. For SKN-1C detection, total protein was separated using a 10% SDS-PAGE gel with 15 µg of total protein per well. The gel was transferred to a nitrocellulose membrane for 60 min at 4°. The membrane was blocked in 5% milk + TBST for 1 hr at room temperature. The membrane was then incubated with 1:200 monoclonal SKN-1 antibody (FC4) overnight at 4° (Bowerman et al. 1993). The blot was washed eight times for 5-min intervals with TBST. The blot was then incubated with 1:1000 secondary HRP-conjugated anti-mouse antibody for 30 min and subsequently washed eight times for 5-min intervals. Blots were developed using SuperSignal West Dura Extended Duration Substrate (Pierce, 37071) and visualized using an ImageQuant LAS 4000 imager (GE Healthcare Life Sciences). Note that we were unable to detect SKN-1C::GFP and that there are presently no examples in the literature of successful detection of the fusion protein with this antibody. For phospho-NSY-1 detection, total protein was separated using an 8% SDS-PAGE gel with at least 70 µg of total protein per well. The gel was transferred to a nitrocellulose membrane for 75 min at 4°. The membrane was blocked in 5% BSA + TBST overnight at 4°. The membrane was then incubated with 1:1000 Phospho-ASK1 (Thr845) Antibody (Cell Signaling, #3765) or Phospho-p38 MAPK (Thr180/Tyr182) Antibody (Cell Signaling, #9211) for 5.5 hr at 4°. The blot was washed four times for 5-min intervals with TBST. The blot was then incubated with 1:3000 secondary HRP-conjugated anti-mouse antibody (for anti-ASK1 blot) or 1:5000 secondary HRP-conjugated anti-rabbit antibody (for anti-p38 blot) for 30 min and subsequently washed four times for 5-min intervals. Blots were developed as above. Both blots were repeated at least three times, obtaining similar results. Anti-α-tubulin was used as a loading control with a 1-hr incubation in 1:1000 primary antibody (Sigma, T9026) concentration and a 30-min incubation in 1:3000 goat anti-rabbit HRP-conjugated secondary antibody with similar washing procedures as described above.
Oxidative stress and killing assays
To assess sensitivity to oxidative stress, sodium arsenite (NaAsO2) was added to NGM plates to a final concentration of 10 mM. Overnight E. coli OP50 culture was generously seeded and incubated for growth (at 37°) on NaAsO2 plates. For microscopy, qRT-PCR, and Western blotting, ∼1000 worms were incubated on NaAsO2 plates for 5 hr at 20°. For survival assays, 90 worms were added to three replicate plates and scored hourly for survival. To assess sensitivity to pathogen stress, killing assays were performed as previously described, with slight modification (Mahajan-Miklos et al. 1999; Garsin et al. 2001). E. faecalis OG1RF grown in BHI for 5 hr was seeded onto BHI plates and grown overnight at 37°. P. aeruginosa PA14 grown in LB for 8 hr was seeded onto SK plates and grown overnight at 37°. A total of 90 worms were added to three replicate plates of each pathogen and scored for survival at various times points over the course of the assay. Kaplan–Meier log-rank analysis was used to compare the significance of the survival curves using median survival.
qRT-PCR analysis
Using TRIzol (Invitrogen), RNA was extracted (as directed by the manufacturer) from young adult worms exposed or left unexposed to sodium arsenite and analyzed via qRT-PCR, as previously described (Hoeven et al. 2011). The average gene expression of biological triplicates was graphed, and error bars represent the standard error of the mean (SEM). A paired Student’s t-test was used to determine significance where indicated. Primers used in this approach are listed in Table S5.
RNA sequencing
Young adult animals were incubated on NGM plates with and without 10 mM sodium arsenite for 5 hr, at which point total RNA was extracted (as described above) and sent for RNA sequencing (RNA-Seq). There were an average 101,140,446 reads/sample generated from the RNA-Seq. The average percentage of the bases with ≥Q30 reads was 90.1%. We used the C.elegans genome (version ce10) as the reference that can be downloaded from the iGenome databases (ftp://igenome:G3nom3s4u@ussd-ftp.illumina.com/Caenorhabditis_elegans/UCSC/ce10/Caenorhabditis_elegans_UCSC_ce10.tar.gz). The sequencing reads were mapped to the genome with the application TopHat (Trapnell et al. 2012; Trapnell et al. 2013), which utilizes the high-throughput sequence aligner bowtie (Langmead et al. 2009). The application Cufflinks further processed the transcripts assembly and the abundance estimation. The FPKMs (Fragments Per Kilobase of transcript per Million mapped reads) were calculated for the gene differential expression analysis. To visualize the results generated from Cufflinks, we used an R/Bioconductor package CummeRbund (http://bioconductor.org). Venn diagrams generated by R programming enabled the affected gene comparison between samples. To explore the functional groups for the differentially expressed genes, we further carried out the Gene Ontology (GO) analysis with the application Database for Annotation, Visualization and Integrated Discovery (Huang da et al. 2009a,b). The three subontologies (MF: molecular function; BP: biological process; and CC: cellular component) were assessed.
Data availability
All strains are available upon request. Table S4 contains genotypes for each strain used in this study. Table S5 contains nucleotide sequences for each primer used in qRT analysis. RNA-Seq gene expression data are available in Table S6 or at Gene Expression Omnibus under accession no. GSE77976.
Results
TRX-1 negatively regulates SKN-1 nuclear localization
Previously, we identified a role for SKN-1 in the C. elegans immune response. We found that the C. elegans dual oxidase, Ce-Duox1/BLI-3, is responsible for the purposeful production of ROS as a means of combatting bacterial infection (Chavez et al. 2009). In a follow-up study, we demonstrated that the ROS produced by BLI-3 during infection activates SKN-1-dependent expression of antioxidants in a p38 MAPK pathway-dependent manner, as a means to protect the host from the inadvertent consequences of protective ROS production (Hoeven et al. 2011). However, the mechanism(s) by which the p38 MAPK pathway is activated to stimulate SKN-1 remained elusive.
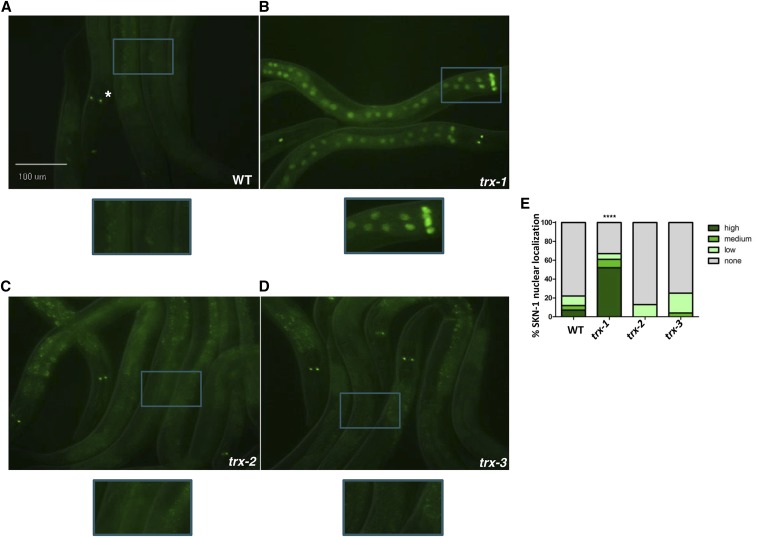
Given the ability of thioredoxins to regulate a variety of signaling pathways, and specifically p38 MAPK pathway activation in mammals, we wanted to determine whether thioredoxins also regulate SKN-1. Using fluorescence microscopy, we examined the ability of TRX-1, TRX-2, and TRX-3, three thioredoxins encoded by the C. elegans genome (Miranda-Vizuete et al. 2006; Cacho-Valadez et al. 2012; Jimenez-Hidalgo et al. 2014), to regulate SKN-1 nuclear localization using a well-characterized strain expressing GFP-tagged SKN-1 protein (SKN-1B/C::GFP), which has been shown to rescue skn-1-dependent functions (An and Blackwell 2003) Under normal conditions, with the exception of the ASI neurons where SKN-1::GFP is constitutively localized to the nuclei (white asterisk, Figure 1A), no specific expression of the GFP is detected (An and Blackwell 2003). Interestingly, however, SKN-1::GFP localizes to the nuclei of intestinal cells in trx-1 mutants, even in the absence of stress (Figure 1, A and B). This effect was specific to trx-1 since loss of either trx-2 or trx-3 does not affect intestinal SKN-1::GFP nuclear localization (Figure 1, A, C, and D). Quantification of nuclear localization of SKN-1::GFP demonstrated that loss of trx-1 results in a threefold increase in SKN-1::GFP nuclear localization (Figure 1E). The increase in nuclear localization of intestinal SKN-1::GFP is not due to a general increase in protein levels. We assayed the protein levels of intestinal SKN-1, isoform SKN-1C, via Western analysis and found that the SKN-1C levels remain unaltered in trx-1 null mutants compared to wild type (Figure S1). Moreover, the degree to which intestinal SKN-1::GFP nuclear localization increases upon loss of trx-1 is similar to that seen upon exposure to the oxidative stressor, sodium arsenite (Figure S2). From this, we conclude that TRX-1 suppresses the nuclear localization of intestinal SKN-1 and therefore may be a novel negative regulator of this transcription factor.
Figure 1.
TRX-1 negatively regulates nuclear localization of intestinal SKN-1. (A–D) Fluorescence microscopy was used to analyze the intestinal nuclear localization of SKN-1 (SKN-1B/C::GFP) upon the loss of trx-1, trx-2, or trx-3. Only upon the loss of trx-1 did SKN-1::GFP accumulate in intestinal nuclei. Asterisk in A depicts constitutive SKN-1B/C::GFP localization in the nucleus of the ASI neurons. Worms were visualized using a 20× objective. Blue boxes indicate the portion of the micrograph field that is magnified in the boxes below each micrograph. (E) Percentage of SKN-1::GFP nuclear localization was categorically scored and quantified as described in Materials and Methods. The percentage of SKN-1 nuclear localization increased threefold upon loss of trx-1 (P-value < 0.0001 as compared to wild type). Percentages are an average of three biological replicates (n = 100 worms per replicate).
TRX-1 regulates intestinal SKN-1 nuclear localization in a redox-independent fashion
Thioredoxins utilize a highly conserved CGPC (Cys-Gly-Pro-Cys) redox active site to reduce disulfide bonds of protein substrates (Holmgren and Lu 2010). C. elegans TRX-1 utilizes this redox reactive capability to reduce insulin in vitro (Jee et al. 2005; Miranda-Vizuete et al. 2006). However, thioredoxins also have redox-independent functions, notably in the promotion of protein folding and turnover (Berndt et al. 2008). Given the ability of thioredoxins to elicit both redox-dependent and redox-independent functions, we investigated whether the redox reactive residues of TRX-1 were required for its regulation of intestinal SKN-1 nuclear localization.
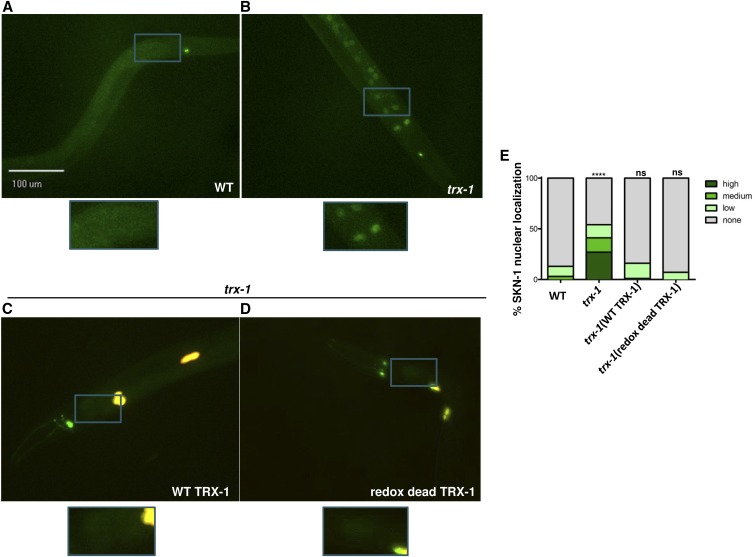
trx-1 mutants were complemented with either wild-type trx-1 or “redox dead” trx-1, in which the redox reactive cysteines of the TRX-1 active site (CGPC) were replaced with nonreactive serine residues (SGPS) (Fierro-Gonzalez et al. 2011a). The transgenes were tracked using Punc122::DsRed as a co-injection marker, which labels coelomocytes with red fluorescence (Loria et al. 2004). Some bleed-through into the green channel resulted in the marker appearing more yellow than red in the resulting pictures (Figure 2, C and D). Under nonstressed conditions, wild-type trx-1 restores proper SKN-1::GFP localization, similar to that seen in the skn-1b/c::gfp parent background (Figure 2, A, C, and E). Interestingly, complementation with redox dead trx-1 restores proper SKN-1::GFP localization as well, indicating that TRX-1 regulates intestinal SKN-1::GFP nuclear localization in a redox-independent fashion (Figure 2, D and E). Upon exposure to oxidative stress, SKN-1::GFP nuclear localization was observed in both the wild-type and redox dead complements of trx-1, indicating that intestinal SKN-1::GFP nuclear localization is indeed inducible in these strains (Figure S3). However, the stress-induced SKN-1::GFP nuclear localization was only partially complemented, as compared to the nontransgenic backgrounds. Since complementation with transgenes often leads to overexpression, this could indicate that overproduction of TRX-1 causes it to maintain its role as a negative regulator of intestinal SKN-1::GFP nuclear localization even during stress.
Figure 2.
TRX-1 regulates intestinal SKN-1 nuclear localization in a redox-independent fashion. Fluorescence microscopy was used to analyze the intestinal nuclear localization of SKN-1::GFP in (A) wild type, (B) trx-1 mutants, trx-1 mutants complemented with either (C) wild-type trx-1 or (D) redox dead trx-1. Worms were visualized using a 20× objective. Blue boxes indicate the portion of the micrograph field that is magnified in the boxes below each micrograph. (E) Percentage of SKN-1::GFP nuclear localization was categorically scored and quantified as described in Materials and Methods. While the percentage of SKN-1::GFP nuclear localization increased over twofold upon loss of trx-1 (P-value < 0.0001), complementation with either wild-type or redox dead trx-1 restored proper SKN-1 localization (P-value = 0.3843 and P-value = 0.1931, as compared to wild type, respectively). Percentages are an average of three biological replicates (n = 40 worms per replicate).
TRX-1 regulates SKN-1 localization cell non-autonomously from the ASJ neurons
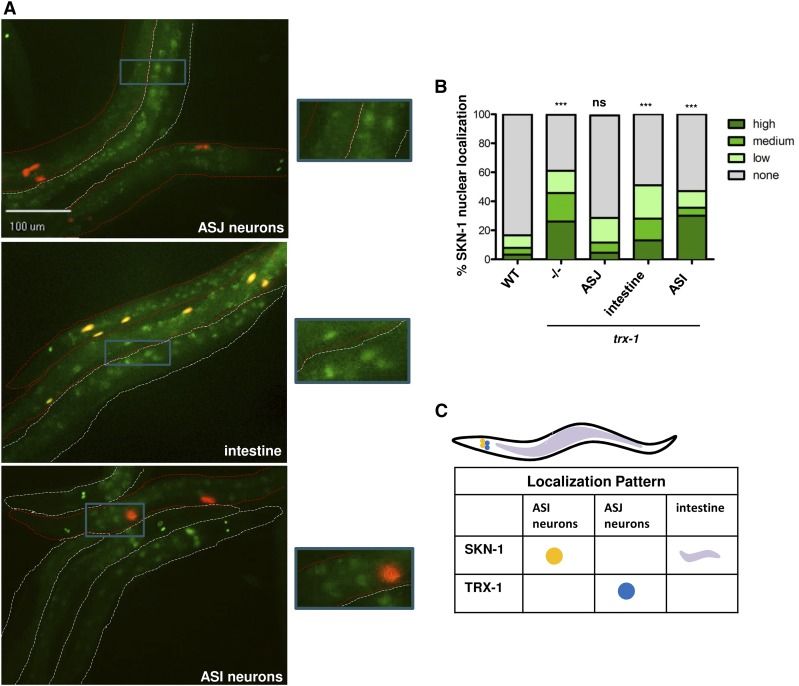
skn-1 is constitutively expressed and localized to the nuclei of the ASI neurons and conditionally becomes localized to intestinal nuclei when worms are exposed to stress (An and Blackwell 2003). trx-1 is expressed solely in the ASJ neurons and impacts worm longevity (Fierro-Gonzalez et al. 2011b; Gonzalez-Barrios et al. 2015). Given that the expression of trx-1 is restricted to the ASJ neurons, but impacts intestinal SKN-1::GFP localization, we wanted to address the possibility that TRX-1 regulates intestinal SKN-1 cell non-autonomously. To assess this, we complemented the trx-1; skn-1b/c::gfp strain with trx-1 expressed under the control of three separate tissue-specific promoters, ssu-1 (ASJ neurons), ges-1 (intestine), and daf-7 (ASI neurons) (Edgar and McGhee 1986; Schackwitz et al. 1996; Carroll et al. 2006). To track tissue-specific rescue throughout the population, Punc122::DsRed was again used as a co-injection marker (Loria et al. 2004). Some bleed-through into the green channel resulted in the marker appearing more yellow than red in the resulting pictures. In Figure 3A, worms outlined in red express trx-1 with the indicated tissue specificity, while worms outlined in white do not carry the tissue-specific rescue and display increased intestinal SKN-1 nuclear localization, thus serving as an internal control for the experiment. As evident in Figure 3A, the increased intestinal SKN-1 nuclear localization seen upon loss of trx-1 was abolished upon specific expression of trx-1 in the ASJ neurons, while rescue of trx-1 expression to the intestine or ASI neurons could not restore proper intestinal SKN-1 nuclear localization.
Figure 3.
TRX-1 regulates SKN-1 nuclear localization cell non-autonomously. (A) Fluorescence microscopy was used to analyze intestinal SKN-1 nuclear localization upon rescue of trx-1 expression in specific tissues in trx-1; skn-1b/c::gfp animals. Worms outlined in red expressed wild-type trx-1 under the regulation of a promoter specific for the designated tissue. Nontransgenic worms (outlined in white) had a trx-1; skn-1b/c::gfp genotype and served as internal controls. Only expression of trx-1 in the ASJ neurons rescued proper intestinal SKN-1::GFP nuclear localization. Worms were visualized using a 20× objective. Blue boxes indicate the portion of the micrograph field that is magnified in the boxes to the right of each micrograph. (B) Percentage of SKN-1::GFP nuclear localization was categorically scored and quantified as described in Materials and Methods. Percentages are an average of three biological replicates (n ≥ 35 worms per replicate). The trx-1 mutant exhibited a threefold increase in the percentage of intestinal SKN-1::GFP nuclear localization (P-value < 0.0001 as compared to wild type). The increased SKN-1::GFP nuclear localization seen upon loss of trx-1 could be fully rescued with specific expression of trx-1 in the ASJ neurons (P-value = 0.2377, as compared to wild type). Rescue of trx-1 expression in the intestine and ASI neurons did not restore proper SKN-1 nuclear localization (P-value < 0.0001 and P-value < 0.0001 as compared to wild type, respectively). (C) A schematic depicting the tissue localization of TRX-1 and SKN-1, emphasizing the absence of overlapping tissue expression between these two proteins. The ability of ASJ-expressed TRX-1 to regulate intestinal SKN-1 nuclear localization indicates a mechanism of cell non-autonomous regulation.
To more quantitatively assess these observations, the percentage of SKN-1 nuclear localization was categorically scored and analyzed (Figure 3B). Loss of trx-1 significantly increased SKN-1::GFP nuclear localization threefold, as shown in Figure 1E. Restoring trx-1 expression specifically in the ASJ neurons reduced SKN-1::GFP nuclear localization to levels seen in the skn-1b/c::gfp parent background. This suggests that TRX-1 regulates intestinal SKN-1::GFP nuclear accumulation cell non-autonomously from the ASJ neurons. To test whether intestinal SKN-1::GFP nuclear localization is inducible in the ASJ-specific complement of trx-1, the strain was exposed to oxidative stress. Intestinal SKN-1::GFP nuclear localization was partially restored (Figure S4). This finding further supports the observation that, when overexpressed, TRX-1 dampens the ability of oxidative stress to fully induce intestinal SKN-1::GFP nuclear localization (Figure S4). In contrast to the ASJ complement, restoring trx-1 expression to the intestine or the ASI neurons did not rescue the SKN-1::GFP nuclear localization caused by loss of trx-1. It is interesting that artificially driving expression of trx-1 in the intestine, the very same tissue in which SKN-1 localization is exhibited, could not restore proper SKN-1::GFP localization. This further suggests that a critical action required for TRX-1-dependent SKN-1 regulation must occur from the distal site of the ASJ neurons. Furthermore, while SKN-1::GFP is constitutively localized to the nuclei of the ASI neurons, expression of trx-1 in these neurons does not abrogate SKN-1::GFP nuclear localization. This suggests that TRX-1 specifically affects intestinal SKN-1 protein.
The model in Figure 3C summarizes the expression pattern of both trx-1 and skn-1. skn-1 is expressed in both the ASI neurons and intestine. trx-1 is expressed solely in the ASJ neurons. The cytoplasmic localization of SKN-1::GFP in the intestine occurs only when trx-1 mutants are complemented with ASJ-specific trx-1 expression. Therefore, we conclude that TRX-1 negatively impacts intestinal SKN-1 nuclear localization in a cell non-autonomous manner from the ASJ neurons.
TRX-1-dependent regulation of SKN-1 localization requires the p38 MAPK pathway
The p38 MAPK pathway is a critical regulator of SKN-1 localization and activation (Inoue et al. 2005). The p38 MAPK pathway is comprised of three kinases: NSY-1 (MAPKKK), SEK-1 (MAPKK), and PMK-1 (the p38 MAPK). One outcome of the stimulation of this pathway is the phosphorylation of SKN-1 at two serine residues, S74 and S340, leading to the nuclear translocation and transcriptional activation of this protein (Inoue et al. 2005). Highlighting the importance of this signaling pathway, in the absence of a functional p38 MAPK pathway or upon alanine substitution at these critical residues of SKN-1, intestinal SKN-1 nuclear localization and activation does not occur even during stress (Inoue et al. 2005). Given that TRX-1 negatively impacts intestinal SKN-1::GFP nuclear localization, we sought to address the dependence of this regulation on the p38 MAPK pathway.
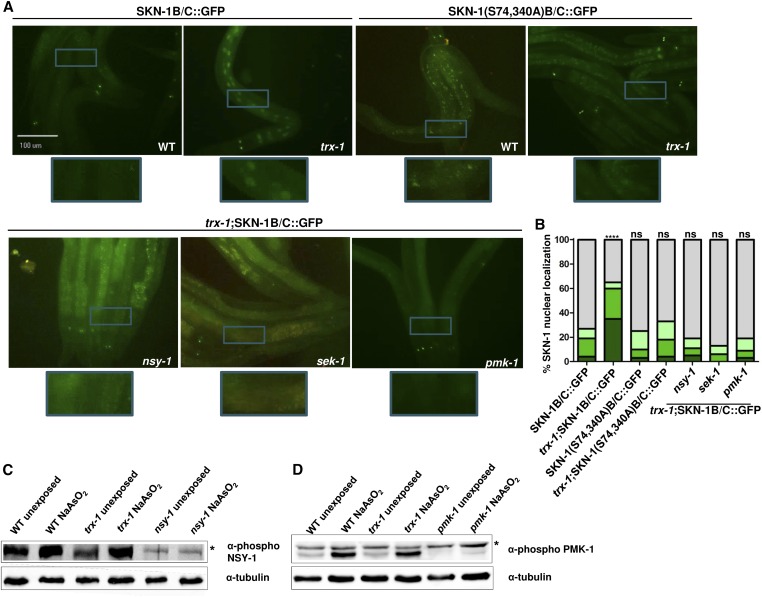
First, we examined the importance of SKN-1 phosphorylation in mediating the TRX-1-dependent intestinal SKN-1 nuclear localization. To do this, we generated a trx-1; skn-1(S74,340A)b/c::gfp strain. While this strain did manifest higher background fluorescence in the intestine, the inability to phosphorylate SKN-1 at S74 and 340 resulted in cytoplasmic retention of SKN-1::GFP even upon loss of trx-1 (Figure 4A), supported by the quantification of subcellular accumulation (Figure 4B). This suggests that TRX-1 functions to regulate SKN-1 nuclear translocation in a p38 phosphorylation-dependent manner. To further address the necessity of TRX-1-dependent SKN-1 regulation on the p38 MAPK pathway, we crossed the trx-1; skn-1b/c::gfp strain with null mutants of each p38 MAPK component [nsy-1(ok593), sek-1(km4), and pmk-1(km25)]. Fluorescent micrographs in Figure 4A and quantification in Figure 4B demonstrate that, while loss of trx-1 alone causes a significant, threefold increase in intestinal SKN-1::GFP nuclear localization, compared to wild type, the additional loss of any component of the p38 MAPK significantly abrogates this phenotype. These data suggest that TRX-1-dependent regulation of intestinal SKN-1::GFP nuclear localization requires p38 MAPK-dependent phosphorylation of SKN-1 at serine residues 74 and 340.
Figure 4.
TRX-1-dependent regulation of SKN-1 nuclear localization is dependent on the p38 MAPK pathway. (A) Fluorescence microscopy was used to analyze trx-1 worms that express a mutant form of SKN-1 [SKN-1(S74,340A)B/C::GFP], which cannot be phosphorylated by PMK-1. The increased percentage of intestinal SKN-1 nuclear localization seen upon loss of trx-1 was abrogated upon mutation of Serines 74 and 340 of SKN-1. Mutants of p38 MAPK pathway components (nsy-1, sek-1, and pmk-1) were crossed into trx-1; skn-1b/c::gfp animals and viewed by fluorescence microscopy. The increased percentage of intestinal SKN-1 nuclear localization seen upon loss of trx-1 was abrogated upon the additional loss of each p38 MAPK-signaling component. Wild-type and trx-1 mutants with a skn-1b/c::gfp background are shown as controls. Worms were visualized using a 20× objective. Blue boxes indicate the portion of the micrograph field that is magnified in the boxes below each micrograph. (B) Percentage of SKN-1::GFP nuclear localization was categorically scored and quantified as described in Materials and Methods. Percentages are an average of three biological replicates (n ≥ 50 worms per replicate). trx-1 mutants expressing the mutated form of SKN-1 did not exhibit increased nuclear SKN-1 localization (P-value = 0.4895, as compared to wild type). trx-1(ok1449); nsy-1(ok593), trx-1(ok1449); sek-1(km4) and trx-1(ok1449); pmk-1(km25) mutants did not exhibit the increased SKN-1 localization seen in trx-1 single mutants (P-value = 0.9629, P-value = 0.2322, and P-value = 0.8863, as compared to wild type, respectively). (C) Western blotting was used to analyze the level of phosphorylation (at residue Thr829) of NSY-1 in wild-type and trx-1 mutants with and without exposure to the oxidative stressor sodium arsenite. NSY-1 phosphorylation was not increased in trx-1 mutants, as compared to wild-type animals, regardless of sodium arsenite exposure. nsy-1 mutants served as a negative control and α-tubulin as a loading control. (D) Western blotting was used to analyze the level of phosphorylation (at residues Thr180 and Thr182) of PMK-1 in wild-type and trx-1 mutants with and without exposure to the oxidative stressor sodium arsenite. PMK-1 phosphorylation was not increased in trx-1 mutants, as compared to wild-type animals, regardless of sodium arsenite exposure. pmk-1 mutants served as a negative control and α-tubulin as a loading control. Black asterisks indicate nonspecific bands. Both of the blots shown are representative of three biological replicates.
Given that TRX-1-dependent regulation of intestinal SKN-1::GFP nuclear localization is dependent on the p38 MAPK pathway, we next assessed whether the dependence was direct or indirect, i.e., in the same or a parallel pathway. One mechanism by which TRX-1 could modulate SKN-1::GFP localization in a direct, p38 MAPK-dependent manner would be to regulate the level of activity of a p38 MAPK component. To address this, we sought to determine whether trx-1 mutants exhibit increased activation of NSY-1 and/or PMK-1, which was measured by assessing the levels of phosphorylation on the relevant residues in these kinases. In mammals, autophosphorylation of ASK1, the mammalian NSY-1 homolog, at Threonine 845 activates the kinase (Tobiume et al. 2002). In C. elegans, this threonine residue is found at position 829 of the amino acid sequence, and the surrounding amino acids are fully conserved between mammals and worms. We took advantage of a peptide-derived antibody specific for mammalian ASK1 phosphorylation at Threonine 845 that has previously been shown to cross-react with phosphorylated NSY-1 (Maruyama et al. 2014). To analyze PMK-1 activation, we obtained an antibody specific to PMK-1 phosphorylated at Threonine 180 and 182. (Schmeisser et al. 2013). Using Western blot analysis, we tested NSY-1 and PMK-1 phosphorylation in wild-type and trx-1 mutants, which demonstrated no increase in active NSY-1 (Figure 4C) or active PMK-1 (Figure 4D) in a trx-1 mutant, as compared to wild type. Exposure to sodium arsenite stress modestly increased NSY-1 activation and strongly increased PMK-1 phosphorylation, but to a similar extent in both wild type animals and trx-1 mutants (Figure 4, C and D). It was previously demonstrated that P. aeruginosa infection increases NSY-1 phosphorylation dramatically (Maruyama et al. 2014). Similarly, we saw a dramatic increase in NSY-1 phosphorylation upon P. aeruginosa infection; however, the level of increased NSY-1 phosphorylation of trx-1 mutants did not differ from that of wild-type animals (Figure S5). These data indicate that TRX-1 does not increase signaling through the p38 MAPK pathway, as measured by NSY-1 and PMK-1 phosphorylation levels, to cause the observed increase in SKN-1 nuclear localization. Overall, these experiments suggest that, while regulation of SKN-1 nuclear localization depends on p38 MAPK signaling, TRX-1 likely does not achieve this regulation by affecting the phosphorylation levels of the kinases.
TRX-1-dependent SKN-1 regulation does not result in the typical transcriptional activation of SKN-1 or an increase in any previously characterized protective responses
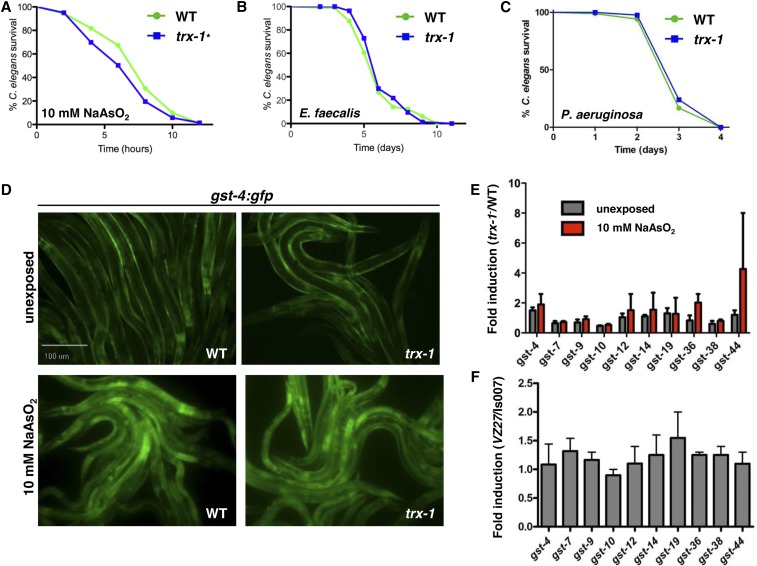
A classic hallmark of previously identified mechanisms of SKN-1 regulation is the correlation between intestinal SKN-1 nuclear localization and activation of the transcription factor, as exhibited by (i) resistance to both oxidative and pathogen stressors and (ii) the increased expression of phase II antioxidants (An and Blackwell 2003; An et al. 2005; Inoue et al. 2005; Tullet et al. 2008; Hoeven et al. 2011; Leung et al. 2014). Given that loss of trx-1 dramatically increases intestinal SKN-1 nuclear localization, we predicted that trx-1 mutants would exhibit increased expression of SKN-1-dependent antioxidants and demonstrate resistance to previously characterized oxidative and pathogen stressors (Wang et al. 1996; Oliveira et al. 2009; Hoeven et al. 2011). As previously shown with other oxidative stressors (Jee et al. 2005), trx-1 mutants were mildly sensitive to 10 mM sodium arsenite (oxidative stress). (Figure 5A). However, this may be explained by the intrinsic short-lived phenotype of trx-1 worms (Jee et al. 2005; Miranda-Vizuete et al. 2006). Furthermore, trx-1 mutants exhibited no significant increase in survival upon E. faecalis (Figure 5B) or P. aeruginosa (Figure 5C) infection (pathogen stress), indicating that loss of trx-1 does not impact previously characterized skn-1-dependent protective stress responses (An and Blackwell 2003; Hoeven et al. 2011).
Figure 5.
Loss of trx-1 does not promote previously characterized SKN-1-dependent protective responses. (A–C) Resistance to several stressors, including (A) 10 mM sodium arsenite, (B) E. faecalis infection, and (C) P. aeruginosa infection was examined. trx-1 mutants were mildly sensitive to oxidative stress induced by sodium arsenite (P-value = 0.02). Loss of trx-1 did not significantly alter the ability to resist either pathogen infection (P-value = 0.3261 and P-value = 0.1527, respectively). (D) Fluorescence microscopy was used to analyze gst-4::gfp expression in wild-type and trx-1 mutant animals with (bottom) and without (top) 5 hr of exposure to 10 mM sodium arsenite. gst-4 expression was not induced in the intestine of trx-1 mutants as compared to wild type. Worms were visualized using a 10× objective. (E) qRT-PCR was used to determine the fold change in SKN-1-dependent gene expression in trx-1 mutants (as compared to wild-type animals) with (red bars) and without (black bars) 5 hr of exposure to 10 mM sodium arsenite. Expression of SKN-1-dependent genes did not significantly change in trx-1 mutants, regardless of stress. (F) qRT-PCR was used to determine the fold change in SKN-1-dependent gene expression in trx-1;skn-1b/c::gfp (VZ27) animals as compared to skn-1b/c::gfp (Is007) control animals. The expression of SKN-1-dependent genes did not significantly change upon the loss of trx-1 in the skn-1b/c::gfp parent background. The average gene expression of biological triplicates is shown, and the error bars represent SEM.
Next, we examined the transcriptional activity of SKN-1 in wild-type and trx-1 mutants under both stressed and unstressed conditions. As previously mentioned, increased intestinal SKN-1::GFP nuclear localization typically results in an enhancement of its transcriptional activity (An et al. 2005; Inoue et al. 2005; Tullet et al. 2008; Leung et al. 2014). A transcriptional gst-4::gfp reporter strain, which acts as a readout of SKN-1-dependent transcriptional activation (Park et al. 2009), was crossed with the trx-1 mutant and analyzed via fluorescence microscopy. Loss of trx-1 did not significantly increase gst-4 expression, as determined by GFP fluorescence in intestinal cells under unstressed conditions (Figure 5D). Furthermore, upon exposure to sodium arsenite, induction of this reporter in the intestine of trx-1 mutants was comparable to that of wild type (Figure 5D). To enable the examination of additional SKN-1-regulated genes in a quantitative manner, qRT-PCR was utilized. In agreement with the gst-4::gfp reporter strain, loss of trx-1 did not significantly increase gene expression of any of the 10 SKN-1-dependent genes analyzed (Oliveira et al. 2009), as compared to wild-type worms, under unstressed conditions or after exposure to sodium arsenite (Figure 5E). Furthermore, there is no significant difference in SKN-1-dependent transcripts in trx-1; skn-1b/c::gfp worms, as compared to skn-1b/c::gfp animals (Figure 5F). These data suggest that, while intestinal nuclear localization of SKN-1 is increased, loss of trx-1 may not affect the classical antioxidant transcriptional activity of SKN-1.
Transcriptome analysis reveals changes in cuticle components and lipid localization and transport upon loss of trx-1
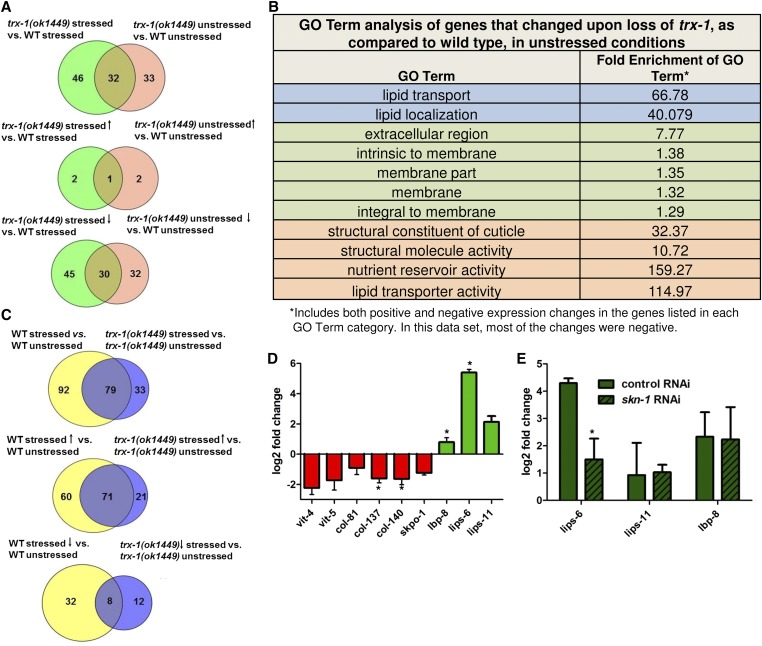
Because TRX-1-dependent SKN-1 nuclear localization did not appear to activate expression of the SKN-1 regulon in a limited analysis (Figure 5, D and E), we utilized a more unbiased approach to identify SKN-1-regulated genes in a trx-1 mutant background. Furthermore, we wanted to better understand how loss of trx-1 impacts the transcriptome as a whole. We used RNA-Seq to look at global transcriptional changes in trx-1 mutants as compared to wild-type worms with and without exposure to sodium arsenite. Interestingly, loss of trx-1 results in a decrease in gene expression, with 75 (with stress) and 62 (without stress) genes being down-regulated, while only 3 genes are up-regulated regardless of stress (Figure 6A). Specifically, upon loss of trx-1, there is a significant enrichment in the down-regulation of genes encoding cuticle components and lipid localization and transport (Figure 6B). Furthermore, RNA-Seq also validated that the oxidative stress transcriptional response in trx-1 animals does not greatly differ from that of wild-type animals (Figure 6C). While trx-1 animals have an overall lower gene expression changes than wild-type animals, regardless of stress, the enrichment levels of glutathione transferase activity do not differ between stressed trx-1 and wild-type animals (Table S1, Table S2, and Table S3). While thioredoxins are commonly regarded as general, cellular antioxidants, these data suggest that this is not the major role of TRX-1. The RNA-Seq results were validated using qRT-PCR to verify the changes in gene expression of 6 down-regulated and 3 up-regulated genes (Figure 6D). Overall, we conclude that loss of trx-1 causes a general down-regulation of the expression of cuticle components and lipid localization and transport genes. Furthermore, the transcriptional oxidative stress response does not seem to be impaired in trx-1 animals, as compared to wild-type animals.
Figure 6.
Transcriptome analysis of trx-1 mutants. (A) (Top) Number of shared genes significantly changed in trx-1 animals, as compared to wild-type animals, under both stressed (10 mM sodium arsenite) and unstressed conditions. (Middle) Number of shared, overexpressed genes. (Bottom) Number of shared, underexpressed genes. (B) GO term analysis of genes that changed upon loss of trx-1, as compared to wild type, under unstressed conditions. Blue: biological processes; green: cellular component; orange: molecular function. (C) (Top) Number of shared genes significantly changed under stressed (10 mM sodium arsenite), as compared to unstressed, conditions in both wild-type and trx-1 animals. (Middle) Number of shared, overexpressed genes. (Bottom) Number of shared, underexpressed genes. (D) qRT-PCR validation of select genes found to be differentially expressed using RNA-Seq upon loss of trx-1, as compared to wild type. The average gene expression of biological triplicates was graphed. Error bars represent the standard error of the mean (SEM), and an asterisk indicates a P-value < 0.05. (E) qRT-PCR was used to measure the log2-fold change in expression of the three up-regulated genes (lips-6, lips-11, lbp-8) upon loss of trx-1, as compared to wild type, after exposure to either skn-1 RNAi or control RNAi. Expression of lips-6 is partially dependent on skn-1 (P-value = 0.032), while lips-11 and lbp-8 expression appears independent (P-value = 0.446 and P-value = 0.453, respectively). The average gene expression of biological triplicates was graphed, and error bars represent SEM.
Three genes, lips-6, lips-11, and lbp-8, are up-regulated upon loss of trx-1. Interestingly, these three genes are related to lipid metabolism as they encode two lipase-like proteins and one lipid-binding protein, respectively. We were interested in addressing whether SKN-1 was important for the up-regulation of these genes. Using qRT-PCR, we examined the fold change of these genes in trx-1 animals, as compared to wild type, after knockdown of skn-1 via RNAi. Only lips-6 expression is significantly reduced after knockdown of skn-1 (Figure 6E), although it is not a complete reduction, suggesting that, while SKN-1 has an effect, other factors may be involved in lips-6 regulation.
Discussion
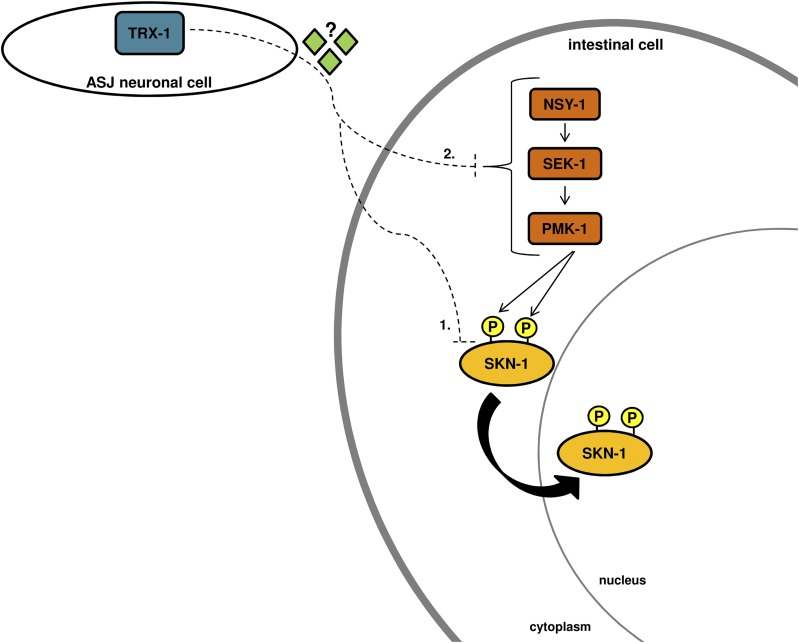
Here, we demonstrate a novel mechanism of SKN-1 regulation in which TRX-1 specifically regulates intestinal SKN-1 nuclear localization in a redox-independent, cell non-autonomous fashion from the ASJ neurons (Figure 1; Figure 2; Figure 3). This method of regulation requires the p38 MAPK pathway, although whether this dependence is direct or indirect requires further study (Figure 4). While TRX-1 regulates nuclear accumulation of intestinal SKN-1, it does not impact SKN-1 transcriptional activity or previously characterized SKN-1-dependent protective responses (Figure 5). RNA-Seq revealed that loss of trx-1 impacts the expression of both cuticle components and lipid localization and transport genes, indicating that TRX-1 may directly influence these processes (Figure 6). Figure 7 depicts a schematic summary our findings.
Figure 7.
TRX-1 regulates SKN-1 nuclear localization cell non-autonomously. TRX-1 expressed in the ASJ neurons negatively controls nuclear localization of intestinal SKN-1, independently of its redox activity, probably through an unknown signaling intermediate. Two possible models by which ASJ-localized TRX-1 could cell non-autonomously direct SKN-1 nuclear localization in the intestine are shown. (1) A novel modification of SKN-1 could occur that results in p38 MAPK pathway-dependent nuclear localization of SKN-1, but not activation. (2) An alteration in signaling through the p38 MAPK pathway activity could cause SKN-1 localization without concurrent transcriptional activation.
The ability of TRX-1 to impact intestinal SKN-1 nuclear localization is not shared by all thioredoxins, as loss of TRX-2 and TRX-3 had no effect (Figure 1). TRX-2 is a component of the mitochondrial thioredoxin system in several tissues and is 54% similar and 32% identical to TRX-1 (Cacho-Valadez et al. 2012). TRX-3 is localized to the cytoplasm and nuclei of intestinal cells, is important for resistance to certain bacterial and fungal pathogens, and is 42% similar and 26% identical to TRX-1 (Jimenez-Hidalgo et al. 2014). Given the differences in cellular expression patterns and subcellular localization, it is not surprising that the regulation of intestinal SKN-1 nuclear localization is not common to the thioredoxin family as a whole. However, given that TRX-3 is localized to the same tissue as SKN-1, we were surprised to find that loss of trx-3 does not affect intestinal SKN-1 nuclear localization. These data suggest that a critical step in TRX-1-dependent SKN-1 regulation occurs from the ASJ neurons specifically and/or that regions of sequence unique to trx-1 may be critical for its ability to regulate SKN-1 nuclear localization.
Thioredoxins utilize a pair of conserved, redox reactive cysteine residues at their active site to fulfill a variety of oxidoreductase-related functions, including maintenance of cellular homeostasis and regulation of transcription factors (reviewed in Holmgren 1985 and Holmgren and Lu 2010). However, thioredoxins also facilitate important cellular processes, such as chaperone-like functions, independently of their oxidoreductase functions (Du et al. 2015). Specifically in C. elegans, TRX-1 has been shown to modulate DAF-28 signaling during dauer formation in a redox-independent fashion (Fierro-Gonzalez et al. 2011a). “Redox dead” complementation of trx-1 demonstrated that TRX-1 regulates SKN-1 in a manner independent of its redox status, expanding upon the list of redox-independent functions of TRX-1. This is further supported by the fact that there is no significant difference in the ability of stressed trx-1 mutants, as compared to both stressed wild-type animals and unstressed trx-1 mutants, to impact intestinal SKN-1 nuclear localization. These data suggest that TRX-1 negatively regulates intestinal SKN-1 nuclear localization regardless of its redox ability or the presence of an oxidative stressor.
Cell non-autonomous regulation is a classic method of coordinating an organism-wide response in a predictive, adaptive fashion. In C. elegans, cell non-autonomous signaling regulates several stress responses, including the heat-shock response, the unfolded protein response, pathogen stress response, and longevity (Prahlad et al. 2008; Sun et al. 2011; Taylor and Dillin 2013; Zhang et al. 2013; van Oosten-Hawle and Morimoto 2014a,b). In this work, we expand the list of stress pathways governed cell non-autonomously to include the oxidative stress transcription factor SKN-1. A variety of molecules, ranging from microRNAs to neurohormones and neuropeptides, signal between various tissues, including between the neurons and the intestine (van Oosten-Hawle and Morimoto 2014a,b). RNA-Seq of trx-1 mutants revealed a change in gene expression of several lipid localization and transport genes. For example, lbp-8, an intestinal lipid chaperone that impacts two nuclear receptors, NHR-49 and NHR-80, to promote longevity (Folick, Oakley et al. 2015) was one of the genes with increased expression in the trx-1 background. Lipids, such as yolk proteins, are implicated as signaling molecules (Grant and Hirsh 1999). Moreover, SKN-1 was recently implicated in the maintenance of lipid homeostasis. Specifically, certain lipids and the activity of specific lipases were shown to activate SKN-1 in the absence of germline stem cells. In addition, SKN-1 was shown to regulate lipid metabolism in the absence of germline stem cells (Steinbaugh et al. 2015). In light of these recent findings and the work herein, we postulate that loss of trx-1 affects intestinal SKN-1 nuclear localization via modulation of lipid homeostasis.
The requirement of the p38 MAPK pathway for TRX-1-dependent SKN-1 regulation was not surprising, given that the p38 MAPK-signaling pathway is essential in several previously identified mechanisms of SKN-1 regulation, including daf-2, gsk-3, and wdr-23 (An et al. 2005; Tullet et al. 2008; Leung et al. 2014). Whether this is a direct or indirect dependence is still unknown, and several models can be postulated. First, it is possible that TRX-1 acts in the same pathway as p38 MAPK signaling to regulate SKN-1 localization. While we ruled out the possibility that loss of trx-1 increases NSY-1 or PMK-1 phosphorylation, other post-translational modifications of either kinase may facilitate proper SKN-1 regulation in a TRX-1-dependent manner. Another possibility is that TRX-1 regulates intestinal SKN-1 nuclear localization in a pathway parallel to, but still dependent on, the p38 MAPK pathway. For example, TRX-1 may be able to regulate SKN-1 only after it has been phosphorylated by PMK-1 at serines 74 and 340 (Figure 7).
One of the more intriguing findings of this study was the severance of SKN-1 nuclear localization and transcriptional activation in the trx-1 background. As previously mentioned, other mechanisms that affect nuclear localization of SKN-1 such as regulation via gsk-3, wdr-23, and the insulin and p38 MAPK-signaling pathways, are characterized by congruence between the degree of nuclear localization of intestinal SKN-1 and the degree of SKN-1-associated gene expression (An et al. 2005; Inoue et al. 2005; Tullet et al. 2008; Choe et al. 2009; Leung, Hasegawa et al. 2014). Dissociation of these phenotypes has been previously described, however. For example, several proteasome regulatory subunits and ubiquitin hydrolases result in increased intestinal SKN-1 nuclear localization but not increased gst-4. Furthermore, loss of certain chaperonins and a proteasomal protease, pas-6, elicits an opposite effect, in which increased gst-4 expression does not result in detectable intestinal SKN-1 nuclear localization (Kahn et al. 2008). Additionally, loss of TOR signaling or treatment with rapamycin, which results in increased autophagy and decreased translation, causes an increase in SKN-1-associated gene expression without concomitant nuclear localization (Robida-Stubbs et al. 2012). In conclusion, TRX-1 is now one of several factors that affect intestinal SKN-1 nuclear localization in a manner independent of its degree of transcriptional activation. Understanding the basis of SKN-1 activity, or lack of activity, under these incongruous circumstances is an area ripe for future investigation.
Using RNA-Seq, we were able to look globally at the transcriptional impact of the loss of trx-1. This method was powerful for several reasons. First, it allowed us to verify that the SKN-1-dependent oxidative stress transcriptional response is not highly activated in trx-1 animals. Additionally, the response to oxidative stress is unimpaired; it was activated in a relatively normal manner under oxidative stress conditions, as compared to wild-type animals (Table S1 and Table S2). Second, this approach allowed us to take an unbiased look at the classes of genes affected by the loss of trx-1. One prominent class of genes down-regulated upon loss of trx-1 was the collagen gene family. Collagen is a critical structural component of the cuticle and the extracellular matrix (Page and Johnstone 2007). While no obvious cuticle defects are apparent in the trx-1 mutant, it is possible that the structural integrity of the animals is affected. Furthermore, SKN-1-dependent collagen production is a key factor in maintaining a healthy extracellular matrix, ultimately promoting longevity (Ewald et al. 2014). This is of particular interest given that loss of trx-1 results in a significant longevity defect (Jee et al. 2005; Miranda-Vizuete et al. 2006), which could potentially be explained by the down-regulation of collagen gene expression seen in this background. Finally, this approach allowed us to identify potential modulators of the TRX-1-dependent mechanism of intestinal SKN-1 nuclear localization. For example, the most enriched class of genes whose expression changes upon loss of trx-1 are the lipid localization and transport genes. Interestingly, lipid gene regulators play a role in cell non-autonomous signaling in C. elegans (Zhang et al. 2013). Therefore, it is possible that TRX-1 present in the ASJ neurons uses its ability to modulate lipid localization and transport genes to affect intestinal SKN-1 nuclear localization cell non-autonomously.
In conclusion, we report that in C. elegans the major oxidative stress transcription factor, SKN-1, is cell non-autonomously regulated by the ASJ neurons via the thioredoxin TRX-1. This further expands the list of stress responses that are modulated from distant tissues at the organismal level. Additionally, we uncovered another example in which nuclear localization and activation of SKN-1 are not synonymous. Finally, the large number of collagen genes repressed in the trx-1 mutant, based on our RNA-Seq data, may provide an explanation for why these animals are short-lived. While these findings increase the complexity of intestinal SKN-1 regulation, they highlight the importance of maintaining its proper regulation.
Acknowledgments
We thank T. K. Blackwell and the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health (NIH) National Center for Research Resources, for the C. elegans strains. T. K. Blackwell is also acknowledged for sharing an aliquot of the FC4 antibody. We also thank V. Garcia and J. Hourihan for helpful advice regarding the use of phospho-specific antibodies for Western blotting. The research reported in this publication was supported by the National Institute of Allergy and Infectious Diseases and the National Institute of General Medical Sciences of the NIH under award nos. R01AI076406 (to D.A.G.) and R01GM98200 (to S.A.). S.A. was also supported by the American Cancer Society RSG014-044-DDC. A.M.-V. is a member of the EU-ROS (BM1203) and GENiE (BM1408) COST Actions, and his work was financed by grants from the Junta de Andalucía (Projects P07-CVI-02697 and P08-CVI-03629). P.S. was supported by the Swedish Research Council, the Torsten Söderberg and Åhlén Foundations, and by the Karolinska Institute Strategic Neurosciences Program. The content is solely the responsibility of the authors and does not necessarily represent the official views of the granting agencies.
Footnotes
Communicating editor: M. V. Sundaram
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.115.185272/-/DC1.
Literature Cited
- An J. H., Blackwell T. K., 2003. SKN-1 links C. elegans mesendodermal specification to a conserved oxidative stress response. Genes Dev. 17: 1882–1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An J. H., Vranas K., Lucke M., Inoue H., Hisamoto N., et al. , 2005. Regulation of the Caenorhabditis elegans oxidative stress defense protein SKN-1 by glycogen synthase kinase-3. Proc. Natl. Acad. Sci. USA 102: 16275–16280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson M. E., 1998. Glutathione: an overview of biosynthesis and modulation. Chem. Biol. Interact. 111–112: 1–14. [DOI] [PubMed] [Google Scholar]
- Arnér E. S., Holmgren A., 2000. Physiological functions of thioredoxin and thioredoxin reductase. Eur. J. Biochem. 267: 6102–6109. [DOI] [PubMed] [Google Scholar]
- Berndt, C., C. H. Lillig, and A. Holmgren, 2008 Thioredoxins and glutaredoxins as facilitators of protein folding. Biochim. Biophys. Acta 1783: 641–650. [DOI] [PubMed]
- Bowerman B., Draper B. W., Mello C. C., Priess J. R., 1993. The maternal gene skn-1 encodes a protein that is distributed unequally in early C. elegans embryos. Cell 74: 443–452. [DOI] [PubMed] [Google Scholar]
- Buchanan, B. B., A. Holmgren, J. P. Jacquot, and R. Scheibe, 2012 Fifty years in the thioredoxin field and a bountiful harvest. Biochim. Biophys. Acta 1820: 1822–1829. [DOI] [PubMed]
- Cacho-Valadez B., Muñoz-Lobato F., Pedrajas J. R., Cabello J., Fierro-González J. C., et al. , 2012. The characterization of the Caenorhabditis elegans mitochondrial thioredoxin system uncovers an unexpected protective role of thioredoxin reductase 2 in β-amyloid peptide toxicity. Antioxid. Redox Signal. 16: 1384–1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll B. T., Dubyak G. R., Sedensky M. M., Morgan P. G., 2006. Sulfated signal from ASJ sensory neurons modulates stomatin-dependent coordination in Caenorhabditis elegans. J. Biol. Chem. 281: 35989–35996. [DOI] [PubMed] [Google Scholar]
- Chávez V., Mohri-Shiomi A., Garsin D. A., 2009. Ce-Duox1/BLI-3 generates reactive oxygen species as a protective innate immune mechanism in Caenorhabditis elegans. Infect. Immun. 77: 4983–4989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe K. P., Przybysz A. J., Strange K., 2009. The WD40 repeat protein WDR-23 functions with the CUL4/DDB1 ubiquitin ligase to regulate nuclear abundance and activity of SKN-1 in Caenorhabditis elegans. Mol. Cell. Biol. 29: 2704–2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du H., Kim S., Hur Y. S., Lee M. S., Lee S. H., et al. , 2015. A cytosolic thioredoxin acts as a molecular chaperone for peroxisome matrix proteins as well as antioxidant in peroxisome. Mol. Cells 38: 187–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar L. G., McGhee J. D., 1986. Embryonic expression of a gut-specific esterase in Caenorhabditis elegans. Dev. Biol. 114: 109–118. [DOI] [PubMed] [Google Scholar]
- Ewald, C. Y., J. N. Landis, J. P. Abate, C. T. Murphy, and T. K. Blackwell, 2014 Dauer-independent insulin/IGF-1-signalling implicates collagen remodelling in longevity. Nature 519: 97–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fierro-González J. C., Cornils A., Alcedo J., Miranda-Vizuete A., Swoboda P., 2011a The thioredoxin TRX-1 modulates the function of the insulin-like neuropeptide DAF-28 during dauer formation in Caenorhabditis elegans. PLoS One 6: e16561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fierro-González J. C., González-Barrios M., Miranda-Vizuete A., Swoboda P., 2011b The thioredoxin TRX-1 regulates adult lifespan extension induced by dietary restriction in Caenorhabditis elegans. Biochem. Biophys. Res. Commun. 406: 478–482. [DOI] [PubMed] [Google Scholar]
- Folick A., Oakley H. D., Yu Y., Armstrong E. H., Kumari M., et al. , 2015. Aging. Lysosomal signaling molecules regulate longevity in Caenorhabditis elegans. Science 347: 83–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujino G., Noguchi T., Takeda K., Ichijo H., 2006. Thioredoxin and protein kinases in redox signaling. Semin. Cancer Biol. 16: 427–435. [DOI] [PubMed] [Google Scholar]
- Fujino G., Noguchi T., Matsuzawa A., Yamauchi S., Saitoh M., et al. , 2007. Thioredoxin and TRAF family proteins regulate reactive oxygen species-dependent activation of ASK1 through reciprocal modulation of the N-terminal homophilic interaction of ASK1. Mol. Cell. Biol. 27: 8152–8163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garsin D. A., Sifri C. D., Mylonakis E., Qin X., Singh K. V., et al. , 2001. A simple model host for identifying Gram-positive virulence factors. Proc. Natl. Acad. Sci. USA 98: 10892–10897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover-Cutter K. M., Lin S., Blackwell T. K., 2013. Integration of the unfolded protein and oxidative stress responses through SKN-1/Nrf. PLoS Genet. 9: e1003701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Barrios M., Fierro-González J. C., Krpelanova E., Mora-Lorca J. A., Pedrajas J. R., et al. , 2015. Cis- and trans-regulatory mechanisms of gene expression in the ASJ sensory neuron of Caenorhbditis elegans. Genetics 200: 123–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant B., Hirsh D., 1999. Receptor-mediated endocytosis in the Caenorhabditis elegans oocyte. Mol. Biol. Cell 10: 4311–4326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeven Rv., McCallum K. C., Cruz M. R., Garsin D. A., 2011. Ce-Duox1/BLI-3 generated reactive oxygen species trigger protective SKN-1 activity via p38 MAPK signaling during infection in C. elegans. PLoS Pathog. 7: e1002453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmgren, A., 1985 Thioredoxin. Annu. Rev. Biochem. 54: 237–271. [DOI] [PubMed] [Google Scholar]
- Holmgren A., Lu J., 2010. Thioredoxin and thioredoxin reductase: current research with special reference to human disease. Biochem. Biophys. Res. Commun. 396: 120–124. [DOI] [PubMed] [Google Scholar]
- Hope, I. A., 1999 C. elegans: A Practical Approach. The Practical Approach Series, Oxford University Press, Oxford. [Google Scholar]
- Huang da W., Sherman B. T., Lempicki R. A., 2009a. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W., Sherman B. T., Lempicki R. A., 2009b. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4: 44–57. [DOI] [PubMed] [Google Scholar]
- Hybertson B. M., Gao B., Bose S. K., McCord J. M., 2011. Oxidative stress in health and disease: the therapeutic potential of Nrf2 activation. Mol. Aspects Med. 32: 234–246. [DOI] [PubMed] [Google Scholar]
- Inoue H., Hisamoto N., An J. H., Oliveira R. P., Nishida E., et al. , 2005. The C. elegans p38 MAPK pathway regulates nuclear localization of the transcription factor SKN-1 in oxidative stress response. Genes Dev. 19: 2278–2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jee C., Vanoaica L., Lee J., Park B. J., Ahnn J., 2005. Thioredoxin is related to life span regulation and oxidative stress response in Caenorhabditis elegans. Genes Cells 10: 1203–1210. [DOI] [PubMed] [Google Scholar]
- Jiménez-Hidalgo M., Kurz C. L., Pedrajas J. R., Naranjo-Galindo F. J., González-Barrios M., et al. , 2014. Functional characterization of thioredoxin 3 (TRX-3), a Caenorhabditis elegans intestine-specific thioredoxin. Free Radic. Biol. Med. 68: 205–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn N. W., Rea S. L., Moyle S., Kell A., Johnson T. E., 2008. Proteasomal dysfunction activates the transcription factor SKN-1 and produces a selective oxidative-stress response in Caenorhabditis elegans. Biochem. J. 409: 205–213. [DOI] [PubMed] [Google Scholar]
- Langmead B., Trapnell C., Pop M., Salzberg S. L., 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung C. K., Hasegawa K., Wang Y., Deonarine A., Tang L., et al. , 2014. Direct interaction between the WD40 repeat protein WDR-23 and SKN-1/Nrf inhibits binding to target DNA. Mol. Cell. Biol. 34: 3156–3167. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Liu Y., Min W., 2002. Thioredoxin promotes ASK1 ubiquitination and degradation to inhibit ASK1-mediated apoptosis in a redox activity-independent manner. Circ. Res. 90: 1259–1266. [DOI] [PubMed] [Google Scholar]
- Loria P. M., Hodgkin J., Hobert O., 2004. A conserved postsynaptic transmembrane protein affecting neuromuscular signaling in Caenorhabditis elegans. J. Neurosci. 24: 2191–2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lushchak, V. I., 2011 Adaptive response to oxidative stress: bacteria, fungi, plants and animals. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 153: 175–190. [DOI] [PubMed]
- Lynn D. A., Dalton H. M., Sowa J. N., Wang M. C., Soukas A. A., et al. , 2015. Omega-3 and -6 fatty acids allocate somatic and germline lipids to ensure fitness during nutrient and oxidative stress in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 112: 15378–15383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan-Miklos S., Tan M. W., Rahme L. G., Ausubel F. M., 1999. Molecular mechanisms of bacterial virulence elucidated using a Pseudomonas aeruginosa-Caenorhabditis elegans pathogenesis model. Cell 96: 47–56. [DOI] [PubMed] [Google Scholar]
- Marinho H. S., Real C., Cyrne L., Soares H., Antunes F., 2014. Hydrogen peroxide sensing, signaling and regulation of transcription factors. Redox Biol. 2: 535–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama T., Araki T., Kawarazaki Y., Naguro I., Heynen S., et al. , 2014. Roquin-2 promotes ubiquitin-mediated degradation of ASK1 to regulate stress responses. Sci. Signal. 7: ra8. [DOI] [PubMed] [Google Scholar]
- McCord J. M., Fridovich I., 1969. Superoxide dismutase. An enzymic function for erythrocuprein (hemocuprein). J. Biol. Chem. 244: 6049–6055. [PubMed] [Google Scholar]
- Miranda-Vizuete A., Fierro González J. C., Gahmon G., Burghoorn J., Navas P., et al. , 2006. Lifespan decrease in a Caenorhabditis elegans mutant lacking TRX-1, a thioredoxin expressed in ASJ sensory neurons. FEBS Lett. 580: 484–490. [DOI] [PubMed] [Google Scholar]
- Oliveira R. P., Porter Abate J., Dilks K., Landis J., Ashraf J., et al. , 2009. Condition-adapted stress and longevity gene regulation by Caenorhabditis elegans SKN-1/Nrf. Aging Cell 8: 524–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page, A. P., and I. L. Johnstone, 2007 The cuticle. (March 19, 2007), WormBook, ed. The C. elegans Research Community, WormBook, /10.1895/wormbook.1.138.1, http://www.wormbook.org.
- Park S. K., Tedesco P. M., Johnson T. E., 2009. Oxidative stress and longevity in Caenorhabditis elegans as mediated by SKN-1. Aging Cell 8: 258–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powis G., Montfort W. R., 2001. Properties and biological activities of thioredoxins. Annu. Rev. Biophys. Biomol. Struct. 30: 421–455. [DOI] [PubMed] [Google Scholar]
- Prahlad V., Cornelius T., Morimoto R. I., 2008. Regulation of the cellular heat shock response in Caenorhabditis elegans by thermosensory neurons. Science 320: 811–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray P. D., Huang B. W., Tsuji Y., 2012. Reactive oxygen species (ROS) homeostasis and redox regulation in cellular signaling. Cell. Signal. 24: 981–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robida-Stubbs S., Glover-Cutter K., Lamming D. W., Mizunuma M., Narasimhan S. D., et al. , 2012. TOR signaling and rapamycin influence longevity by regulating SKN-1/Nrf and DAF-16/FoxO. Cell Metab. 15: 713–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salazar M., Rojo A. I., Velasco D., de Sagarra R. M., Cuadrado A., 2006. Glycogen synthase kinase-3beta inhibits the xenobiotic and antioxidant cell response by direct phosphorylation and nuclear exclusion of the transcription factor Nrf2. J. Biol. Chem. 281: 14841–14851. [DOI] [PubMed] [Google Scholar]
- Schackwitz W. S., Inoue T., Thomas J. H., 1996. Chemosensory neurons function in parallel to mediate a pheromone response in C. elegans. Neuron 17: 719–728. [DOI] [PubMed] [Google Scholar]
- Schmeisser S., Priebe S., Groth M., Monajembashi S., Hemmerich P., et al. , 2013. Neuronal ROS signaling rather than AMPK/sirtuin-mediated energy sensing links dietary restriction to lifespan extension. Mol. Metab. 2: 92–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinbaugh M. J., Narasimhan S. D., Robida-Stubbs S., Moronetti Mazzeo L. E., Dreyfuss J. M., et al. , 2015. Lipid-mediated regulation of SKN-1/Nrf in response to germ cell absence. eLife 4: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J., Singh V., Kajino-Sakamoto R., Aballay A., 2011. Neuronal GPCR controls innate immunity by regulating noncanonical unfolded protein response genes. Science 332: 729–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor R. C., Dillin A., 2013. XBP-1 is a cell-nonautonomous regulator of stress resistance and longevity. Cell 153: 1435–1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanan R., Oikawa S., Hiraku Y., Ohnishi S., Ma N., et al. , 2014. Oxidative stress and its significant roles in neurodegenerative diseases and cancer. Int. J. Mol. Sci. 16: 193–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobiume K., Saitoh M., Ichijo H., 2002. Activation of apoptosis signal-regulating kinase 1 by the stress-induced activating phosphorylation of pre-formed oligomer. J. Cell. Physiol. 191: 95–104. [DOI] [PubMed] [Google Scholar]
- Trapnell C., Roberts A., Goff L., Pertea G., Kim D., et al. , 2012. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7: 562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Hendrickson D. G., Sauvageau M., Goff L., Rinn J. L., et al. , 2013. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol. 31: 46–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tullet J. M., Hertweck M., An J. H., Baker J., Hwang J. Y., et al. , 2008. Direct inhibition of the longevity-promoting factor SKN-1 by insulin-like signaling in C. elegans. Cell 132: 1025–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Oosten-Hawle P., Morimoto R. I., 2014a Organismal proteostasis: role of cell-nonautonomous regulation and transcellular chaperone signaling. Genes Dev. 28: 1533–1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Oosten-Hawle P., Morimoto R. I., 2014b Transcellular chaperone signaling: an organismal strategy for integrated cell stress responses. J. Exp. Biol. 217: 129–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker A. K., See R., Batchelder C., Kophengnavong T., Gronniger J. T., et al. , 2000. A conserved transcription motif suggesting functional parallels between Caenorhabditis elegans SKN-1 and Cap’n’Collar-related basic leucine zipper proteins. J. Biol. Chem. 275: 22166–22171. [DOI] [PubMed] [Google Scholar]
- Wang T. S., Kuo C. F., Jan K. Y., Huang H., 1996. Arsenite induces apoptosis in Chinese hamster ovary cells by generation of reactive oxygen species. J. Cell. Physiol. 169: 256–268. [DOI] [PubMed] [Google Scholar]
- Yoshioka J., Schreiter E. R., Lee R. T., 2006. Role of thioredoxin in cell growth through interactions with signaling molecules. Antioxid. Redox Signal. 8: 2143–2151. [DOI] [PubMed] [Google Scholar]
- Zhang P., Judy M., Lee S. J., Kenyon C., 2013. Direct and indirect gene regulation by a life-extending FOXO protein in C. elegans: roles for GATA factors and lipid gene regulators. Cell Metab. 17: 85–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All strains are available upon request. Table S4 contains genotypes for each strain used in this study. Table S5 contains nucleotide sequences for each primer used in qRT analysis. RNA-Seq gene expression data are available in Table S6 or at Gene Expression Omnibus under accession no. GSE77976.