Abstract
Diversity in insect pigmentation, encompassing a wide range of colors and spatial patterns, is among the most noticeable features distinguishing species, individuals, and body regions within individuals. In holometabolous species, a significant portion of such diversity can be attributed to the melanin synthesis genes, but this has not been formally assessed in more basal insect lineages. Here we provide a comprehensive analysis of how a set of melanin genes (ebony, black, aaNAT, yellow, and tan) contributes to the pigmentation pattern in a hemipteran, Oncopeltus fasciatus. For all five genes, RNA interference depletion caused alteration of black patterning in a region-specific fashion. Furthermore, the presence of distinct nonblack regions in forewings and hindwings coincides with the expression of ebony and aaNAT in these appendages. These findings suggest that the region-specific phenotypes arise from regional employment of various combinations of the melanin genes. Based on this insight, we suggest that melanin genes are used in two distinct ways: a “painting” mode, using predominantly melanin-promoting factors in areas that generally lack black coloration, and, alternatively, an “erasing” mode, using mainly melanin-suppressing factors in regions where black is the dominant pigment. Different combinations of these strategies may account for the vast diversity of melanin patterns observed in insects.
Keywords: Oncopeltus fasciatus, insect pigmentation, melanin patterning, melanin-promoting factors, melanin suppressors
PIGMENT patterns are among the most striking and variable features of insect morphology. An extraordinary diversity in coloration distinguishes species, populations within species, individuals within populations, and different body regions (Wittkopp and Beldade 2009). Most insights into the mechanisms underlying such diversity have come from studies on melanization in Drosophila (Wittkopp et al. 2003; Wittkopp and Beldade 2009). Melanization is the pigmentation process wherein precursors (catecholamines) are converted into pigment molecules that are incorporated into the cuticle (Wittkopp and Beldade 2009). These studies have helped to identify a network of melanin genes and their roles in body color patterning (Wright 1987; Wittkopp et al. 2003; Wittkopp and Beldade 2009). The core part of this proposed pathway is shown in Figure 1. The pathway begins with the conversion of tyrosine to dihydroxyphenylalanine (DOPA). DOPA can then be used in two different manners: to produce DOPA melanin (black) or to be converted to dopamine, another precursor of black melanin. In the conversions from DOPA/dopamine to black melanin, the yellow gene is thought to play an essential role in promoting these processes. However, it is still unclear whether yellow plays a role in producing DOPA melanin, dopamine melanin, or both (question marks in Figure 1). Alternatively, production of dopamine melanin can be suppressed by converting dopamine to N-β-alanyldopamine (NBAD) or N‐acetyldopamine (NADA). The NBAD branch contains three main genes: ebony, black, and tan. Black catalyzes the production of β-alanine, which binds to dopamine by the activity of Ebony, thus forming NBAD, the precursor of yellow sclerotin. It is also possible to convert NBAD back to dopamine by the activity of Tan. The dopamine produced under such circumstance is then converted to dopamine melanin, thus promoting dark coloration (Wittkopp et al. 2002b; True et al. 2005). In Heliconius, Tan also has been recognized as an additional factor that promotes dark melanization (Ferguson et al. 2011). The NADA branch relies on the function of arylalkylamine-N-acetyltransferase (AANAT), which converts dopamine to NADA, the precursor of colorless sclerotin. These five genes, including three melanin-suppressing factors (ebony, black, and aaNAT) and two melanin-promoting factors (tan and yellow), are thought to be the main genes in the mechanism responsible for producing different melanin patterns (Wright 1987; Wittkopp et al. 2003; Wittkopp and Beldade 2009).
Figure 1.
A summary of the melanin pathway in insects. [Redrawn from Wright (1987).] Black melanin originates from either DOPA or dopamine, which are converted from tyrosine by TH and DDC. While yellow is recognized as a melanin-promoting factor, it is still unclear whether it converts DOPA melanin, dopamine melanin, or both (hence the question marks next to it). Dopamine can be converted to NBAD sclerotin (yellow pigment), which requires the functions of both ebony and black. This conversion can be reversed by tan, another melanin-promoting factor. Dopamine also can be converted to NADA sclerotin, which is colorless and catalyzed by aaNAT. All conversions from precursors (DOPA, dopamine, NBAD, and NADA) to the final pigments require multistep biochemical reactions, as illustrated by dashed lines.
Assays in Drosophila have provided functional evidence associating ebony, yellow, and tan with body and wing pigmentation (Walter et al. 1996; Wittkopp et al. 2002b; Gompel et al. 2005; True et al. 2005; Jeong et al. 2008). In addition, functional studies in Tribolium have also reported the essential involvement of ebony, black, and yellow (Arakane et al. 2009; Tomoyasu et al. 2009; Arakane et al. 2010), whereas the pigmented phenotype of aaNAT was reported only in Bombyx (Zhan et al. 2010; Osanai-Futahashi et al. 2012). A systematic functional profile of these melanin genes is lacking in any of the more basal hemimetabolous insect lineages. In this mode of development, the embryo hatches as a miniature adult that undergoes a succession of molts, with melanization occurring at each stage. Thus, as a complement to the studies already performed in holometabolous insects, it is important to begin evaluating the roles of melanization genes in the pigmentation of hemimetabolous species. This also establishes a foundation for understanding the evolution of the melanin pathway in insects.
Accordingly, we performed the first systematic functional analysis of ebony, black, tan, yellow, and aaNAT in a hemimetabolous species, the milkweed bug O. fasciatus. This insect is well suited for the study of pigmentation because it features striking aposematic black/orange warning colorations and color patterns that are distinct in different body regions. The black coloration has been confirmed recently to be produced via the melanin pathway, whereas most of the orange coloration comes from another source (Liu et al. 2014). The RNA interference (RNAi) approach in Oncopeltus is also highly efficient and generates phenotypes displaying systemic responses (Liu and Kaufman 2004a, b, 2005; Angelini and Kaufman 2005). This allows for the functional analysis of pigmentation genes at a whole-body scale, as was highlighted by a previous study (Liu et al. 2014). The depletion of master synthesis genes (tyrosine hydroxylase and dopa decarboxylase, which initialize the melanin pathway) (Figure 1) caused a great reduction in black coloration throughout the whole body (Liu et al. 2014). In the present study, RNAi knockdown of the five melanin genes caused different regional pigment phenotypes. More specifically, the phenotypes observed in the abdomen and hindwings of RNAi adults were different from those in the head, thorax, and forewings. These observations are unexpected because RNAi depletion of ebony, black, or aaNAT in holometabolous species (Tribolium and Bombyx) caused alterations in color patterns over the entire body (Tomoyasu et al. 2009; Arakane et al. 2010; Zhan et al. 2010; Osanai-Futahashi et al. 2012). Results presented here from Oncopeltus suggest that different melanin genes are used to form black patterns in distinct body regions. This finding provides novel insights regarding how pigment diversity is created in different body regions by mechanisms that could be general to other insects with complex color patterns, including other aposematic species whose color patterns play critical roles in interspecies communication.
Materials and Methods
Cloning and sequence analysis of complementary DNA (cDNA) fragments
Extraction of total RNA from embryos of O. fasciatus, as well as the follow-up cDNA generation, nested RT-PCR, and cloning were carried out according to Liu et al. (2014) and Li and Popadić (2004). cDNA fragments of ebony and aaNAT were obtained from Periplaneta americana embryos (another hemimetabolous species used here for comparison and confirmation) following an established protocol from Chesebro et al. (2009). Multiple reactions of nested PCR were performed to amplify the gene fragments. The primers used for PCR cloning, as well as the lengths of the cDNA fragments obtained, are listed in Supplemental Material, Table S1. All these cDNA fragments cover most of the coding region of each gene. The orthologies of these Oncopeltus genes were confirmed by phylogenetic analysis (Figure S1 and Figure S2). In the case of Oncopeltus Ubx, a previously described clone was used (Medved et al. 2015).
RNA interference (RNAi)
For all genes in this assay, double-stranded RNA (dsRNA) was generated using the entire cDNA fragment to ensure the specificity of RNAi depletion. The prepared dsRNA was then injected into the abdomens of Oncopeltus nymphs following established protocols (Chesebro et al. 2009; Liu et al. 2014). Approximately 2 µl of dsRNA was injected at a concentration of 2–3 µg/µl into each individual. Injections were performed around the beginning of the last nymphal stage (fifth nymphs). Ubx RNAi was performed following an established protocol (Medved et al., 2015). For ebony RNAi, 20 nymphs were injected, and 11 successfully molted into adults. For tan RNAi, 45 nymphs were injected, and 37 survived to adult stage. For black RNAi, 32 nymphs were injected, and 20 molted to adults. For yellow RNAi, 45 nymphs were injected, and 38 molted to adults. For aaNAT RNAi, 25 nymphs were injected, and 17 molted to adults. All images were chosen to show the representative phenotypes. For Ubx RNAi, 17 nymphs were injected, all of which were dissected at the fifth nymphal stage for in situ hybridization.
The RNAi analysis in Periplaneta was carried out following Chesebro et al. (2009). dsRNA was prepared and injected into the ventral abdomens of late nymphs of Periplaneta. Approximately 3 μl of dsRNA was injected at a concentration of 3–4 μg/μl into each nymph. Of the 10 nymphs injected with ebony, six successfully molted into adults. For aaNAT RNAi, of the 10 nymphs injected, seven successfully molted into adults. All images were chosen to show the representative phenotypes.
RT-PCR analysis
To determine the efficiency of our RNAi approach, we performed independent RT-PCR analyses on ebony, black, tan, yellow, aaNAT, and Ubx as controls (shown in Figure S3). Total RNA was extracted from the whole bodies (with internal organs removed). Two individuals were used for RNA extraction in each group. The procedures of total RNA extraction and cDNA synthesis were carried out following Liu et al. (2014). Primers used for RT-PCR are listed in Table S1. Complementary RT-PCR analyses on ebony and aaNAT were performed in P. americana (Figure S4).
In addition, to determine whether the RNAi depletion was consistent throughout the body, we performed RT-PCR analyses on isolated body regions of black RNAi individuals that showed the greatest contrast in pigmented phenotypes between the abdomen and anterior body regions (Figure S5). Two wild-type or black RNAi fifth nymphs (day 8 of development) were used. The epidermis of head, thorax, forewing, hindwing, and abdomen were dissected and separated. Each regional sample was used for total RNA extraction and cDNA synthesis following Liu et al. (2014).
Image processing
Microscopic images were obtained using a SZX16 Microscope (Olympus) and DP72 Camera (Olympus). To minimize the possible variation in captured images, the background and all the microscope and camera settings were standardized: the images under a particular magnification were taken under the same light conditions, aperture, exposure time, and white balance.
In addition, to quantify the changes in black intensity observed in yellow and tan RNAi adults, we measured the mean grayscales (at a scale of 0–255) of the black subregions using ImageJ in separate microscopic images of the head, thorax, forewing, hindwing, and ventral abdomen. The measurements were carried from 10 individuals of wild-type, yellow RNAi, or tan RNAi Oncopeltus adults. The grayscale values were converted to brightness level as grayscale value/255 × 100. The converted brightness levels from these three groups were then compared to each other using group t-tests (the level of significance was 0.05). As a control, the same measurements and analyses were also applied to the orange subregions in the dorsal abdomens.
In situ hybridization
The digoxigenin-labeled antisense RNA probes of ebony and aaNAT were synthesized as described in Li and Popadić (2004). For each gene, both sense and antisense probes were used. The formation of wing tissue is completed at day 5 of the fifth nymphal stage, after which the cuticle starts forming, which, in turn, restricts the use of postembryonic in situ hybridization analysis. Before day 5, the wing tissue is not completely formed and cannot be fixed intact. After day 6, the cuticle prevents the riboprobes from penetrating into the wing tissue. Therefore, in this assay, the wings were dissected from wild-type and Ubx RNAi fifth nymphs that were exactly 5–6 days old. In each group, 16–22 wings were analyzed, and the images shown represent the consensus expression patterns. The in situ hybridization was modified from Tomoyasu et al. (2009). Details will be provided on request.
Data availability
The authors state that all data necessary for confirming the conclusions presented in this article are represented fully within the article. The accession numbers of the O. fasciatus sequences from GenBank are as follows: KX023894 (ebony), KX023895 (black), KX023896 (yellow), KX023897 (tan), and KX023898 (aaNAT).
Results
Functions of melanin genes on black patterns of body pigmentation in Oncopeltus
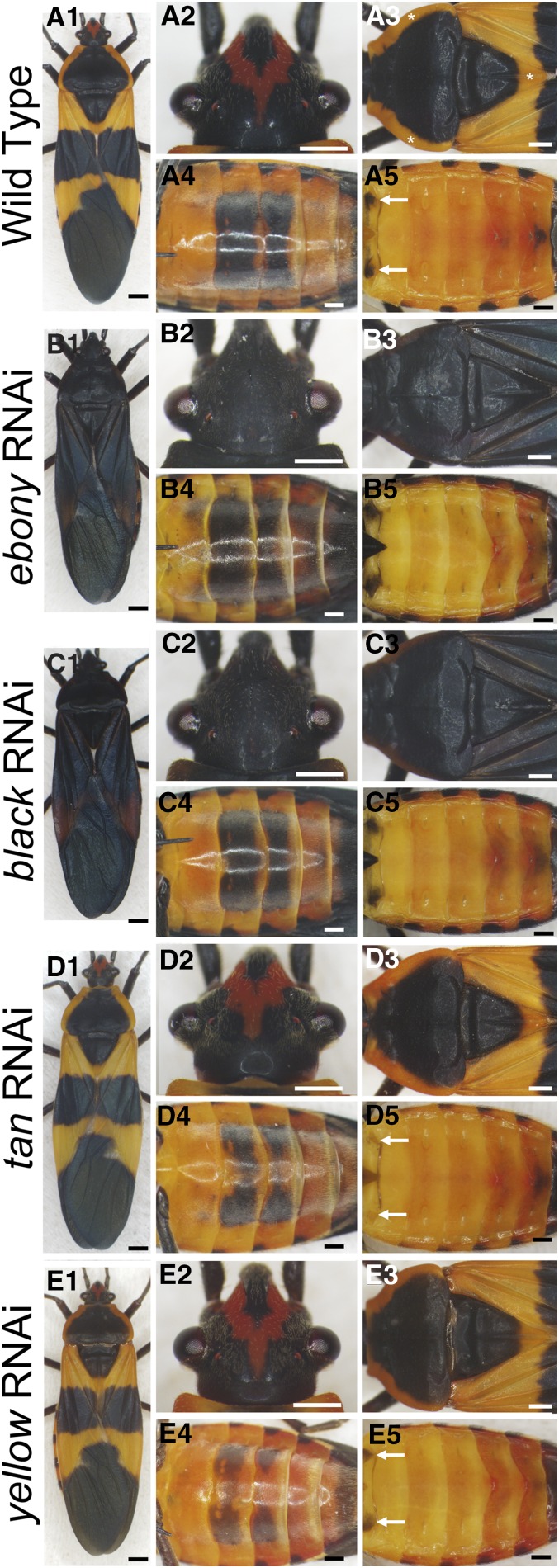
The Oncopeltus body has warning coloration consisting of alternating black and orange subregions and patches (Figure 2A1). The dorsal head features a V-shaped orange stripe flanked by black pigmentation covering the rest of the head (Figure 2A2). Dorsal plates of the thoracic segments are primarily black, with the exception of orange at the lateral edges on the prothorax (T1) and posterior tip of the mesothorax (T2; asterisks, Figure 2A3). In contrast, most of the abdomen is orange, with the exception of two rectangular black patches on the ventral side of A3–A4 segments (Figure 2A4) and two small spots on the dorsal A1 segment (white arrows, Figure 2A5).
Figure 2.
Functions of ebony, black, tan, and yellow in different body regions of Oncopeltus adults. (A1–A5) Wild-type Oncopeltus shows alternating black-orange patterning. Black pigmentation is present in the head (A2), thorax (A3), ventral abdomen (A4), and dorsal abdomen (A5). (B1–B5) The ebony RNAi adult phenotypes showed the expansion of black pigment in the head (B2) and thorax (B3), whereas such an expansion was only moderate in the ventral abdomen (B4) and barely noticeable in dorsal abdomen (B5). (C1–C5) The black RNAi adults showed a phenotype similar to the ebony RNAi individuals described earlier. (D1–D5) The tan RNAi adult phenotypes showed a differential defect of black pigment across the body. The defect of black was subtle in the head (D2) and thorax (D3), while it was quite significant in ventral abdomen (D4). Also, on the dorsal A1 segment, the two black patches were reduced (arrows, D5). (E1–E5) The yellow RNAi adults showed varying levels of reduction in black pigment between body regions. Reduction was moderate in the head (E2) and thorax (E3) whereas quite significant in ventral abdomen (E4). The two black patches on the dorsal A1 segment were also significantly reduced (arrows, E5). Scale bars, 1 mm (A1, B1, C1, D1, and E1) and 500 µm (A2–A5, B2–B5, C2–C5, D2–D5, and E2–E5).
To examine the role of the melanin pathway in Oncopeltus color patterning, we cloned five putative core melanin genes: ebony, black, tan, yellow, and aaNAT. Phylogenetic analyses confirmed that the clones obtained are indeed Drosophila orthologs (Figure S1 and Figure S2). To test the efficiency of RNAi, we performed RT-PCR analyses for each gene, which showed that in all instances the original transcription level was significantly reduced throughout the body, including the epidermis (Figure S3). These levels of depletion are consistent with previous RNAi analyses in Oncopeltus showing changes in coloration encompassing the entire body (Liu et al. 2014). These findings, combined with an additional RT-PCR result showing a significant reduction in the level of black transcripts in all body regions (Figure S5), support the conclusion that the observed phenotypes resulted from a systemic response.
Studies in Drosophila and Tribolium have shown that ebony plays the central role in suppressing black pigmentation (Figure 1). On loss of function of ebony, a global darkening of body pigmentation was observed in both species (Wittkopp et al. 2002b; Takahashi et al. 2007; Tomoyasu et al. 2009). Based on these observations, we expected ebony RNAi Oncopeltus adults to exhibit black coloration throughout the body. Among 11 ebony RNAi adults, all but one exhibited a significant expansion of black melanization in the anterior body regions. As illustrated in Figure 2, B1–B3, all nonblack anterior subregions, including the orange V-shaped stripe on the head, the lateral edges on T1, and the posterior tip on T2, became black. However, in these 10 individuals, the expansion of black pigmentation was only moderate in posterior body regions. The black rectangle on the third abdominal segment (A3) expanded anteriorly into the A2 segment, whereas the A4 rectangle expanded posteriorly into A5 and A6 segments (Figure 2B4). The remainder of the ventral abdomen and the non-black-pigmented dorsal abdomen were generally unaltered (Figure 2B5). Therefore, the different outcomes observed between anterior and posterior body regions suggested that while ebony was critical for nonblack patches in the head and thorax, it played only a minor role in the majority of nonblack subregions in the abdomen.
Ebony is the core enzyme in the conversion of dopamine to NBAD, catalyzing the binding of β-alanine to dopamine (Figure 1). Therefore, the synthesis of β-alanine, which is catalyzed by the product of the black gene, is also critical for the NBAD branch (Wright 1987; Arakane et al. 2009). Among 20 black RNAi adults, 18 showed an expansion of black coloration in the head and thorax that covered the original orange pigmentation (Figure 2, C1–C3). In the ventral abdomen, the effects were highly consistent among these 18 individuals, showing little of the melanin expansion that was observed in ebony knockdowns (compare Figure 2C4 with Figure 2B4). The dorsal abdomen and most of the nonblack portions of the ventral abdomen also remained unchanged (Figure 2, C4 and C5). These observations show that black was required for the nonblack patches in the anterior body regions but not for the abdomen. Combined with ebony RNAi insights, these results establish that suppression of melanin by the NBAD branch is used differently between the anterior and posterior body regions of Oncopeltus.
While these results focus on the mechanism that creates nonblack subregions, it is equally important to understand the complementary process—the generation of black areas across the insect body. Studies in Drosophila have shown that other than tyrosine hydroxylase (TH) and dopa decarboxylase (DDC), the enzymes required for the production of DOPA and dopamine, additional promoting enzymes are required for melanization (Wright 1987; Wittkopp et al. 2002b; True et al. 2005; Jeong et al. 2008). One of the essential enzymes is Tan, which counteracts Ebony in the NBAD branch (Figure 1), thus promoting melanin production (True et al. 2005; Jeong et al. 2008). Functional analyses on tan have been reported only in Drosophila, which showed that its loss causes a global reduction of melanin patterns (True et al. 2005; Jeong et al. 2008). Of 37 tan RNAi Oncopeltus adults, 34 exhibited a consistent phenotype (Figure 2D1). Black patterns in the head and thorax were generally unaltered (Figure 2, D2 and D3). In contrast, the melanin patterns showed noticeable changes in the abdomen (Figure 2, D4 and D5). In particular, the two black spots on the dorsal A1 segment were significantly reduced (arrows in Figure 2D5). The same trend also was observed in the ventral A3 and A4 segments, exhibiting a reduction in the middle portion of each black rectangle (Figure 2D4). In three individuals, the degree of reduction was more pronounced, with either the left or right half of the A4 rectangle disappearing completely. It is worth noting that the intensity of black coloration within the remaining A3 and A4 rectangles was not significantly different from wild type (Figure S6, P > 0.05). These observations suggest that tan is required for proper patterning of black pigmentation in the Oncopeltus abdomen but not for its intensity. Such a role for tan in abdominal melanin patterning is consistent with previous studies of Drosophila species (True et al. 2005; Jeong et al. 2008).
As shown earlier, the depletions of ebony, black, and tan did not affect the majority of the nonblack subregions in the abdomen. These observations suggest that the NBAD branch of the melanin pathway is not involved in the generation of these subregions. Another possible candidate is arylalkylamine-N-acetyltransferase (aaNAT), which is the core gene in the NADA branch of the melanin pathway (Figure 1). Studies in Drosophila have shown that aaNAT is responsible for converting dopamine to NADA as a way of depleting melanin and creating colorless sclerotin (Wright 1987; Hintermann et al. 1995; Brodbeck et al. 1998). Consistent with this, the depletion of aaNAT causes an increase in melanization across the body in Bombyx (Zhan et al., 2010). In order to determine whether this mechanism also can explain the nonblack subregions in the abdomen of Oncopeltus, we depleted aaNAT in fifth instar nymphs. The consequent adults, however, showed no effect in color patterns in the head, thorax, or abdomen (Figure S7). This observation indicates that the NADA branch of the melanin pathway is not required for black patterns in these body regions.
In addition to the preceding four genes within the NBAD and NADA branches, we also tested the function of yellow, another important gene that promotes melanin production (Wright 1987; Wittkopp et al. 2002a, b; Jeong et al. 2008; Tomoyasu et al. 2009; Arakane et al. 2010). Although this gene is not within the NBAD branch, its depletion in Drosophila causes a severe reduction in melanization across the whole body (Wittkopp et al. 2002b; Jeong et al. 2008). Among the 38 yellow RNAi adults, 32 displayed a significant reduction in the intensity of black pigments (Figure S6, P < 0.05) in ventral A3 and A4 rectangles (Figure 2, E4 and E5). In addition, in 30 of 32 individuals, a reduction in the middle portions of the A3 and A4 rectangles also was observed (Figure 2E4). These pigmented phenotypes in the abdomen are similar to those reported in Drosophila melanogaster (Wittkopp et al. 2002b; Jeong et al. 2008). While abdomens showed an increase in average brightness of 51%, the reduction in black intensity in the head and thorax was more moderate (Figure 2, E1 and E3), with an increase in average brightness of 28 and 15%, respectively (Figure S6). These findings indicate that yellow was involved in regulating both the extent of melanin patterns and their intensity in the abdomen but had much less effect in the thorax and abdomen. Based on yellow and tan RNAi results, we speculate that these melanin-promoting factors may be critical for the melanin patterns in the posterior body regions.
Distinct black patterns between the forewing and hindwing are generated by different branches of the melanin pathway in Oncopeltus
The forewing and hindwing in Oncopeltus also have distinct melanin patterns. The forewing has an alternating black and orange pattern (Figure 3A1), whereas the hindwing is colorless at the proximal end and black throughout the distal region (Figure 3A2). To determine whether the NBAD branch regulates the melanin patterns in both pairs of wings, we examined the wings of ebony and black RNAi adults. In both instances, black pigmentation greatly expanded into the orange subregions on the forewing (Figure 3, B1 and C1). However, the depletion of either gene generated no noticeable changes in color patterns in the hindwing (Figure 3, B2 and C2). These observations indicate that the NBAD branch was required for repressing melanization in the nonmelanized areas on the forewing, whereas similar nonmelanized regions on the hindwing used a different mechanism.
Figure 3.
Functions of ebony, black, aaNAT, yellow, and tan in the forewing and hindwing of Oncopeltus adults. (A1–A2) Pigmentation patterns of wild-type wings. (B1–B2) The ebony RNAi adults showed an expansion of black pigmentation into the orange subregion of the forwing (B1), while the hindwing remained unaffected (B2). (C1–C2) The black RNAi adult wings showed a similar phenotype to the ebony RNAi adults, as described earlier. (D1–D2) The aaNAT RNAi adult phenotypes show no effect on the forewing (D1), while melanization expanded into the anal region of the hindwing (asterisk in D2). (E1–E2) The yellow RNAi adult wing phenotype showed a reduction of black pigment on the forewing (E1) and the hindwing (E2). (F1–F2) The tan RNAi adult phenotype showed no noticeable pigmentation effect in the forewing and hindwing. FW, forewing; HW, hindwing. Scale bars, 1 mm.
Candidate genes responsible for suppressing melanization in the proximal hindwing include the NADA branch of the melanin pathway (Figure 1). Because aaNAT, the core gene within this branch, is reported to be responsible for converting dopamine to colorless NADA sclerotin in Drosophila (Wright 1987; Hintermann et al. 1995; Brodbeck et al. 1998), it is possible that such a mechanism also can generate the colorless pattern in the hindwing of Oncopeltus. To test this hypothesis, we observed the hindwing coloration in aaNAT RNAi adults. In all 17 resulting RNAi individuals, the anal lobe region of the hindwing became melanized (Figure 3D2), whereas the black pigmentation of the forewing was not affected (Figure 3D1). These observations indicate that aaNAT was involved in suppression of melanin formation in the colorless anal lobe region of the hindwing. However, there was no indication that this role was required for proper pigmentation of the forewing. In summary, the Oncopeltus forewing and hindwing seem to use distinct mechanisms to generate nonblack subregions: the NBAD branch is applied to suppress melanization in the orange areas of the forewing, whereas the hindwing employs the NADA branch to generate the colorless anal lobe.
In addition to the melanin-suppressing factors ebony, black, and aaNAT, we tested the function of the melanin-promoting factors yellow and tan in the black subregions of Oncopeltus wings. In yellow RNAi adults, there was a significant reduction in black intensity in both pairs of wings (Figure 3, E1 and E2, and Figure S6, P < 0.05 in forewing and P 0.05 in hindwing). This effect was much greater in the hindwing (average brightness increased by 89%) than the forewing (19%) (Figure S6). In contrast, the depletion of tan did not generate any noticeable effect on the black patterns of either the forewing or hindwing (Figure 3, F1 and F2) nor significant reduction in their black intensity (Figure S6, P > 0.05), indicating that this gene was not essential for wing melanin patterns. These observations suggest that the roles in wing melanization are distinct between different melanin-promoting factors: yellow is required for the proper intensity of black melanin in the wings, especially the hindwing, whereas tan may not be involved at all in wing pigmentation in Oncopeltus.
Differential involvement of pigmentation genes between forewing and hindwing correlates with their expression patterns
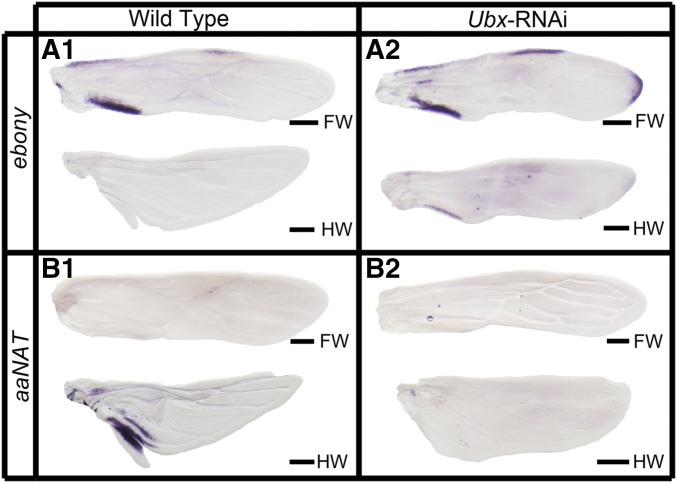
Previous studies in the wings of Drosophila and Heliconius have shown strong correlations between melanin patterns and the expression patterns of melanin genes (Wittkopp et al. 2002b; Gompel et al. 2005; Ferguson et al. 2011; Hines et al. 2012). In particular, such correlations have been seen in the D. melanogaster abdomen (Rebeiz et al. 2009; Camino et al. 2015) and forewing (Gompel et al. 2005) and occur in diverse Drosophila species with divergent patterns of melanic pigmentation (Werner et al. 2010; Arnoult et al. 2013; Ordway et al. 2014; Camino et al. 2015), indicating that these correlations are functionally meaningful. Thus, we hypothesized that the difference in mechanisms regulating nonblack patterns between forewing (NBAD branch) and hindwing (NADA branch) comprise differential expressions of the relevant core genes. To test this hypothesis, we used in situ hybridization to detect in developing wing pads of Oncopeltus the expression patterns of ebony and aaNAT, the two essential genes in the NBAD and NADA branches. Note that because formation of the cuticle precludes the expression analysis later in development (see Materials and Methods), we can only study early stages during which initial patterns are beginning to be laid out and do not fully correspond to the final patterns. As shown in Figure 4A1, ebony expression was observed in three distinct patches on the forewing: two are located on the anterior and posterior edges of the proximal portion and the other in the middle of the anterior edge. These locations correspond with the anterior and posterior margins of orange subregions on the forewing (Figure 3A1). The expression of ebony on the hindwing, however, was not detectable (Figure 4A1), consistent with the fact that ebony RNAi displayed no phenotype. However, aaNAT expression was not observed in the forewing but was present in the anal lobe region of the hindwing (Figure 4B1). This expression pattern of aaNAT was strongly correlated with its RNAi phenotype, where the colorless anal lobe region of the hindwing became melanized (Figure 3A2). Hence, the expression patterns of ebony and aaNAT correlated with their functions in generating the nonblack patches on the forewing and hindwing.
Figure 4.
In situ hybridization of ebony and aaNAT in developing wild-type and Ubx RNAi Oncopeltus wings. (A1) In wild-type wings, ebony is expressed exclusively in the forewing. (A2) In Ubx RNAi wings, the hindwing exhibits expression of ebony. (B1) In wild-type wings, aaNAT is expressed only in the hindwing. (B2) In Ubx RNAi wings, aaNAT is no longer expressed in the hindwing. FW, forewing; HW, hindwing. Scale bars, 200 µm.
These differences in RNAi phenotypes between forewing and hindwing pigmentation may be explained by the differential activations of ebony and aaNAT. If so, this regional regulation of melanin genes would require specific selector genes (Wittkopp et al. 2003; Wittkopp and Beldade 2009). In the butterfly Junonia coenia, the Hox gene Ultrabithorax (Ubx) plays such a role by differential regulation of hindwing and forewing color patterns (Weatherbee et al. 1999). Recently, Ubx was shown to also control the identity of hindwing in Oncopeltus (Medved et al. 2015), allowing us to test the generality of the selector gene’s role in regional regulation. Therefore, on depletion of Ubx, we would expect ebony to be expressed in the developing hindwing, whereas aaNAT should be absent. As shown in Figure 4A2, the hindwing expresses ebony at the proximal margins, which resembles the patterns observed in the wild-type forewing. However, the expression of aaNAT is lost in the hindwing, even in a moderate phenotype showing an intermediate shape transformation (Figure 4B2). These findings indicate that Ubx governs the differential activation of melanin genes between the wings. This further suggests that the different pigmentation roles of melanin genes across different Oncopeltus body regions, as shown by the present RNAi analyses, may be due to the functions of other region-specific regulatory genes.
The expression patterns of black, yellow, and tan, however, could not be detected using in situ hybridization. Note that the wing tissue is amenable to this procedure only during the middle part of the fifth nymphal stage (see Materials and Methods) when these three genes may not be active. To test this possibility, we examined the expression of black, yellow, and tan during the entire fifth nymphal stage (Figure S8). As predicted, expression of these genes was noticeable only during the latter half of the stage (Figure S8). These observations suggest that during the development of Oncopeltus wings, enzymes such as Ebony and AANAT, which use dopamine as direct substrates (Figure 1), perform their patterning roles at early-middle fifth nymphal stage. In contrast, Black, Yellow, and Tan are turned on at a later stage.
Functions of melanin-suppressing factors in Periplaneta
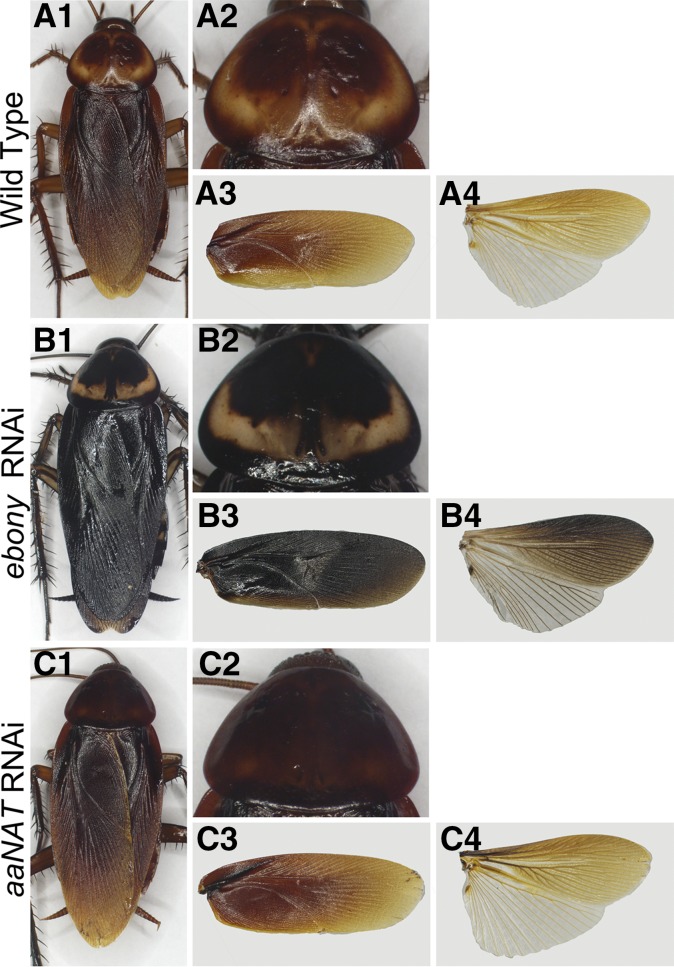
The preceding sections have shown that in Oncopeltus, melanin-suppressing factors (ebony and aaNAT) play critical roles in the appearance of light patches of color in body regions that are mostly dark in coloration (e.g., the head, thorax, and wings). To determine whether such a mechanism is applicable to nonaposematic hemimetabolous insects, we examined the American cockroach, P. americana (Figure 5A1), whose uniform brown body coloration is also generated by the melanin pathway (Lemonds 2015). In the middle of the dorsal T1 plate, however, there are paired oval regions created by clear sclerotin (Figure 5A2). The Periplaneta forewing is dark brown (Figure 5A3), whereas the hindwing features a brown anterior distal half and clear sclerotin in the posterior proximal half (Figure 5, A3 and A4). To determine how the melanin-suppressing factors can generate the color patterns in Periplaneta, we performed RNAi depletion of ebony and aaNAT using previously reported gene fragments (Bembenek et al. 2005; Blenau and Baumann 2005).
Figure 5.
The functions of ebony and aaNAT in P. americana adults. (A1–A4) Pigmentation patterns of wild-type adult Periplaneta. (A1) Most of the body is covered by dark brown pigments. (A2) The T1 plate has a clear sclerotin patch in the shape of a horseshoe and two brown oval patches in the middle. (A3) The forewing displays a uniform brown coloration. (A4) The hindwing has a distinct pigmentation pattern with a brown pigment on the entire anterior region but is unpigmented in the posterior region, except for the veins, which are also brown in color. (B1–B4) The ebony RNAi adults established a significant increase in melanization. (B1) The increase in melanization resulted in a black appearance. (B2) Most of the T1 plate became black pigmented, with the exception of clear patches. (B3) The forewing showed a similar increase in melanization throughout the wing. (B4) The hindwing appeared darker compared to wild type, especially the anterior region. (C1–C4) The aaNAT RNAi phenotypes showed similar brown coloration to those observed in wild type, except for the T1 plate (C2), which lost the clear patches and is completely brown. FW, forewing; HW, hindwing.
Adults from ebony RNAi treatments showed a significant increase in overall melanization, with black pigment covering most of their bodies (Figure 5B1). However, the clear sclerotin on the dorsal T1 plate remained (Figure 5B2). The wings also showed a substantial increase in pigmentation (Figure 5, B3 and B4), especially the forewing, which became black in color (Figure 5B3). These observations indicate that in Periplaneta, ebony plays an essential role in maintaining the proper intensity of melanization in areas that are already melanized, but it has no effect on clear sclerotin regions. In other words, ebony can regulate the darkness levels of the already-established melanin pattern, but it cannot alter the pattern. This finding is similar to that of a previous report in Tribolium, where the colorless patch on the hindwing was not altered by ebony RNAi (Tomoyasu et al. 2009).
The aaNAT RNAi adults retained wild-type brown pigmentation (Figure 5C1). However, the clear sclerotin areas in the T1 plate became melanized (Figure 5C2), with the overall coloration in the forewing and hindwing remaining similar to wild type (Figure 5, C3 and C4). These results reveal that aaNAT is responsible for creating some (T1 plate), but not all, colorless regions (posterior half of the hindwing). Alternatively, it is possible that these latter regions of clear sclerotin are produced by putative aaNAT paralogs (Mehere et al. 2011; Barbera et al. 2013; Long et al. 2015). These findings suggest that in both Oncopeltus and Periplaneta, some aspects of melanin patterning may be conserved (e.g., creating clear sclerotin by using AANAT in hindwing in Oncopeltus and in T1 in Periplaneta).
Discussion
Pigmentation functions of melanin genes in Oncopeltus
In this study, we performed a comprehensive functional analysis of the putative core melanin genes using RNAi. Although there has been limited success in RNAi silencing by direct body cavity injection in Drosophila, this approach has proven to be highly effective in other insects (Li et al. 2015). This is especially the case in Oncopeltus, which is characterized by a strong systemic gene depletion following RNAi treatment (Liu and Kaufman, 2004a, b, 2005; Angelini and Kaufman 2005). The recent RNAi analysis of the essential enzymes for the production of black melanin in this species also showed a systemic reduction in coloration encompassing the entire body (Liu et al. 2014). In this study, the whole-body RNAi response is documented by RT-PCR results showing similar reduction in the amount of transcript across different body regions in black RNAi individuals (Figure S5). Hence, the observed region-specific effects of RNAi against melanin genes are likely true phenotypes resulting from systemic responses rather than low-penetrance phenotypes. These results suggest distinct regional utilization of melanin genes.
The NBAD branch of the melanin pathway has been reported to be essential for melanin patterning in Drosophila (Wright 1987; Wittkopp et al. 2002b). In this pathway, both black and ebony can suppress melanin formation via the production of β-alanine and NBAD, respectively (Figure 1). In Oncopeltus, RNAi knockdown of these two genes caused the nonblack subregions to become black in the head (Figure 2, B2 and C2), thorax (Figure 2, B3 and C3), and forewing (Figure 3, B1 and C1), whereas most of the nonblack patches in the hindwing (Figure 3, B2 and C2) and abdomen (Figure 2, B4 and B5 and C4 and C5) were not affected. These findings suggest that the melanin-suppressing role of ebony and black is crucial in the head, thorax, and forewing but not necessary for proper coloration in the hindwing and abdomen. This is in contrast with findings in Drosophila and Tribolium, where loss of function of either ebony or black results in an overall darkening of body pigmentation (Wright 1987; Wittkopp et al. 2002b; Arakane et al. 2009; Tomoyasu et al. 2009).
Another gene of the NBAD branch, tan, counteracts the function of ebony and promotes black pigmentation by converting NBAD back to dopamine (Wright 1987; True et al. 2005; Jeong et al. 2008). tan RNAi phenotypes in Oncopeltus indicate that it is essential for the black patterns present in the abdomen (Figure 2, D4 and D5) but is not a significant player in forming the melanin patterning in the head (Figure 2D2), thorax (Figure 2D3), and wings (Figure 3, F1 and F2). Again, these results are different from those in Drosophila, where tan mutants display a reduction in melanization spanning the entire body (True et al. 2005; Jeong et al. 2008).
Despite the fact that the reactions catalyzed by tan and ebony appear as a circuit from the biochemical perspective (Figure 1), in Oncopeltus, they are employed differently in distinct body regions. It is possible that in hemimetabolous insects, only half of this circuit is essential for melanization in one specific body region, whereas the other half might be active but not required for pigmentation. Instead, they might be involved in other biological processes such as behavior (Wittkopp and Beldade 2009). Subsequently, in more derived groups such as dipterans, the entire tan-ebony circuit (both reactions) became fully involved in melanization (True et al. 2005). Extending future studies of tan to additional hemimetabolous species may show how the NBAD branch of the melanin pathway has changed during insect evolution.
Finally, the NADA branch also can suppress melanin production by transforming dopamine to NADA, thus creating colorless tissue (Wright 1987). The key gene in this pathway is aaNAT (Wright 1987; Hintermann et al. 1995; Brodbeck et al. 1998). Here we observed that the role of aaNAT is restricted to the anal lobe region of the hindwing (Figure 3D2), which correlates with its expression profile (Figure 4B1). This observation is in contrast to the depletion of aaNAT in the silkworm Bombyx, which causes a global darkening of the adult body (Zhan et al. 2010; Osanai-Futahashi et al. 2012). Thus, Oncopeltus appears to preferentially activate aaNAT expression in wings to create the colorless tissue in the anal lobe region. It is worth noting that aaNAT may not be the only gene used in this region because the anterior portion of the proximal half of the hindwing remains colorless when aaNAT is depleted by RNAi (Figure 3D2). Future studies will need to assess whether other members of the AANAT family are involved in generating colorless patches in this region (Mehere et al. 2011; Long et al. 2015).
The pigmented phenotypes of yellow have been reported in both Drosophila and Tribolium, with distinct results. Loss of yellow causes global reduction of melanin in Drosophila (Wittkopp et al. 2002b), whereas only the hindwing is affected in Tribolium (Tomoyasu et al. 2009; Arakane et al. 2010). In Oncopeltus, yellow RNAi adults displayed a mix of features previously observed in Drosophila and Tribolium: black coloration in the abdomen and hindwing is greatly reduced, but other body regions were only moderately affected (Figure S6). Therefore, the restriction of yellow function appears to have changed during insect evolution. At present, we cannot rule out the possibility that in regions where yellow RNAi was observed to have a moderate effect, other yellow family members might play additional roles in melanization (Han et al. 2002). However, currently the functional roles of most of the yellow family genes are yet to be determined. Even for the yellow gene itself, despite the fact that it is known to be required for melanization in Drosophila and Tribolium (Wright 1987; Wittkopp et al. 2002a, b; Jeong et al. 2008; Tomoyasu et al. 2009; Arakane et al. 2010; Bray et al. 2014), its enzymatic activity has not yet been established. In terms of the position of yellow in the melanin pathway, two different hypotheses have been proposed (Figure 1): it may be essential for the production of either DOPA melanin (Wright 1987; Walter et al. 1996) or dopamine melanin (Wittkopp et al. 2002b, 2003). Our present results and another recent report (Liu et al. 2014) show that black pigment in the hindwing is dopamine melanin, suggesting that yellow may be involved only in dopamine melanin production.
Overall, our results show that the pigmentation functions of the five genes under study are regionalized in Oncopeltus. This is in contrast with previous studies in Drosophila, Tribolium, and Bombyx, in which depletion of ebony, black, tan, aaNAT, or yellow resulted in alteration of melanin patches throughout the body (Wright 1987; Walter et al. 1996; Wittkopp et al. 2002b; True et al. 2005; Gibert et al. 2007; Jeong et al. 2008; Arakane et al. 2009; Tomoyasu et al. 2009; Zhan et al. 2010; Osanai-Futahashi et al. 2012). Hence, the insight from Oncopeltus is that the entire melanin pathway can be split into different sections that are used in different body regions.
In this study, the regional patterns of melanin genes are observed from analyses at a whole-body scale. To obtain a deeper comprehension of how complex melanin patterns are generated will require analyzing pigmentation genes at a finer morphologic scale (e.g., a subregion of a segment). Classical studies in Drosophila have shown that selector genes involved in a general anteroposterior axis determination, such as hedgehog (hh), engrailed (en), and optomotor-blind (omb), also regulate melanin patterning in the abdominal segments (Kopp and Duncan 1997; Kopp et al. 1997). It is tempting to speculate that similar mechanisms may account for the presence of centrally positioned black rectangles on A3 and A4 segments in Oncopeltus. Also, other axis-determination mechanisms, especially mediolateral, may also be involved in generating the black spots located on the lateral edges of each abdominal segment (Figure 2A5). Furthermore, it is interesting to note that hh and en regulate melanin genes in a cell-autonomous manner in Drosophila (Kopp et al. 1997). In contrast, melanin genes themselves can function nonautonomously, semiautonomously, or autonomously in different body regions (Hanna 1953; Hotta and Benzer 1970; Borycz et al. 2002; Wittkopp et al. 2002b; True et al. 2005). At present, we do not know whether such region-specific cell autonomy of melanin genes also may account for production of the regional color patterns observed in Oncopeltus. Extending future studies in this direction will be required to gain a better understanding of the regulatory mechanisms that generate species-specific melanin patterns in insects in general.
Region-specific employment of melanin genes
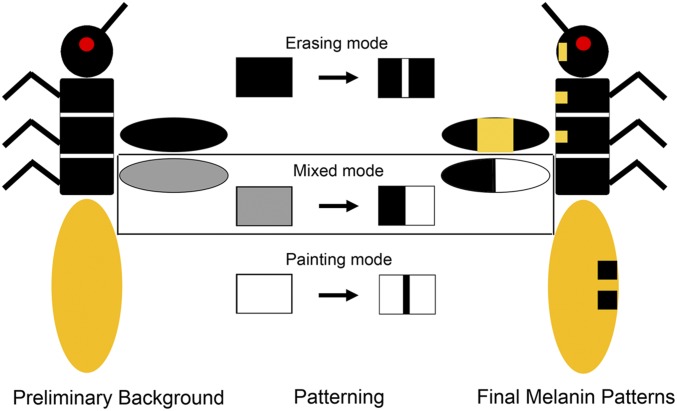
To better understand the regional utilization of melanin genes, we provide a summary of the general principles that appear to guide melanin patterning (Figure 6). In this summary, we refer to a putative case as the “preliminary background,” in which only the basic enzymes required for melanin production, such as TH and DDC, would be active. In other words, none of the melanin-promoting or melanin-suppressing factors function in such a background. Under the first scenario, the “painting” mode would be used in a body region that lacks melanin patterns, such as the Oncopeltus abdomen, which is predominantly orange. The preliminary background in this case would be nonmelanin because most of the abdomen may not be capable of producing black pigments. The dark melanin is “painted” onto specific areas of this background, which requires melanin-promoting factors (tan and yellow) to ensure the proper boundary and intensity of black patterns. Under a second scenario, the “erasing” mode would apply to a body region where dark melanin is the predominant pigment (as observed in the head, thorax, and forewing in Oncopeltus, as well as the body and wings of Periplaneta). In this instance, the preliminary background would be fully melanized, and the melanin suppressors such as ebony, black, and aaNAT are used for “erasing” melanin production in a specific area or for lowering the overall melanin intensity. In addition to situations where one of these two modes is used exclusively, there are also regions in which the melanin-promoting factors and suppressors can both play active roles, such as the hindwing in Oncopeltus, where melanin and nonmelanin subregions are equally distributed. Under this scenario, the preliminary background is defined as a region undergoing melanization but lacking a proper level of intensity. Finalizing the melanin patterns requires both the melanin-suppressing factor (aaNAT), which generates the nonmelanin patches, and the melanin-promoting factor (yellow), which intensifies the dark color within melanin patches.
Figure 6.
Three proposed modes of insect melanin patterning, as illustrated by pigmentation in Oncopeltus. The final melanin patterns (right) are generated from the preliminary background (left) via different modes of melanin patterning. In the head, thorax, and forewing, where the background is fully black, the nonmelanin patches are generated by “erasing” the background. In the abdomen, where the background is nonblack, the melanin patches are “painted” onto the background. A mixture of these two modes may be used in the hindwing, in which black is intensified in the melanin subregions, whereas melanin is suppressed in the nonblack subregions.
In terms of their generality, these three scenarios can account for most of the functional results in the previously studied species. The painting mode may be evidenced in the dark-colored pterostigma on the hindwing of Tribolium. In those studies, this specific black pattern becomes lighter in yellow RNAi individuals, whereas ebony RNAi has no effect on the flanking nonblack subregions (Tomoyasu et al. 2009; Arakane et al. 2010). The erasing mode may be evidenced in the butterfly Papilio, where the default fully melanized forewing was observed in ebony mutant adults (Koch et al. 2000). Another example is the depletion of ebony or black in Tribolium, which results in the general blackening of the whole body (Arakane et al. 2009; Tomoyasu et al. 2009). In addition to the present results in Oncopeltus, the mixed mode has been fully confirmed only in D. melanogaster, where functional analyses revealed that ebony and yellow contribute equally to both wing and body pigmentation (Wittkopp et al. 2002b; Gompel et al. 2005). In general, our summary of the principles of melanin patterning can serve as a practical framework explaining the diversity in melanin coloration observed in previously reported insects. A broader taxonomic sampling in basal groups, from which we can infer the ancestral melanin patterning, will be required to determine whether this framework can be applied to a wider range of insect species.
Acknowledgments
We thank Victor Medved for help with cloning gene fragments in Oncopeltus. We also thank Mark VanBerkum, Patricia Wittkopp, William Branford, and two anonymous reviewers for helpful comments that greatly improved this manuscript. This work was supported in part by National Institutes of Health grant GM-071927 to A.P. and a WSU Rumble Fellowship to J.L. J.H.M.’s participation was facilitated by National Science Foundation grants IOS-0950416 and IOS-1354667 and a HITS grant from the Huck Institutes of the Life Sciences.
Footnotes
Communicating editor: D. M. Parichy
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.115.186684/-/DC1.
Literature Cited
- Angelini D. R., Kaufman T. C., 2005. Functional analyses in the milkweed bug Oncopeltus fasciatus (Hemiptera) support a role for Wnt signaling in body segmentation but not appendage development. Dev. Biol. 283: 409–423. [DOI] [PubMed] [Google Scholar]
- Arakane Y., Lomakin J., Beeman R. W., Muthukrishnan S., Gehrke S. H., et al. , 2009. Molecular and functional analyses of amino acid decarboxylases involved in cuticle tanning in Tribolium castaneum. J. Biol. Chem. 284: 16584–16594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arakane Y., Dittmer N. T., Tomoyasu Y., Kramer K. J., Muthukrishnan S., et al. , 2010. Identification, mRNA expression and functional analysis of several yellow family genes in Tribolium castaneum. Insect Biochem. Mol. Biol. 40: 259–266. [DOI] [PubMed] [Google Scholar]
- Arnoult L., Su K. F., Manoel D., Minervino C., Magrina J., et al. , 2013. Emergence and diversification of fly pigmentation through evolution of a gene regulatory module. Science 339: 1423–1426. [DOI] [PubMed] [Google Scholar]
- Barbera M., Mengual B., Collantes-Alegre J. M., Cortes T., Gonzalez A., et al. , 2013. Identification, characterization and analysis of expression of genes encoding arylalkylamine N-acetyltransferases in the pea aphid Acyrthosiphon pisum. Insect Mol. Biol. 22: 623–634. [DOI] [PubMed] [Google Scholar]
- Bembenek J., Sakamoto K., Takeda M., 2005. Molecular cloning of a cDNA encoding arylalkylamine N-acetyltransferase from the testicular system of Periplaneta americana: primary protein structure and expression analysis. Arch. Insect Biochem. Physiol. 59: 219–229. [DOI] [PubMed] [Google Scholar]
- Blenau W., Baumann A., 2005. Molecular characterization of the ebony gene from the American cockroach, Periplaneta americana. Arch. Insect Biochem. Physiol. 59: 184–195. [DOI] [PubMed] [Google Scholar]
- Borycz J., Borycz J. A., Loubani M., Meinertzhagen I. A., 2002. tan and ebony genes regulate a novel pathway for transmitter metabolism at fly photoreceptor terminals. J. Neurosci. 22: 10549–10557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray M. J., Werner T., Dyer K. A., 2014. Two genomic regions together cause dark abdominal pigmentation in Drosophila tenebrosa. Heredity 112: 454–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodbeck D., Amherd R., Callaerts P., Hintermann E., Meyer U. A., et al. , 1998. Molecular and biochemical characterization of the aaNAT1 (Dat) locus in Drosophila melanogaster: differential expression of two gene products. DNA Cell Biol. 17: 621–633. [DOI] [PubMed] [Google Scholar]
- Camino E. M., Butts J. C., Ordway A., Vellky J. E., Rebeiz M., et al. , 2015. The evolutionary origination and diversification of a dimorphic gene regulatory network through parallel innovations in cis and trans. PLoS Genet. 11: e1005136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesebro J., Hrycaj S., Mahfooz N., Popadic A., 2009. Diverging functions of Scr between embryonic and post-embryonic development in a hemimetabolous insect, Oncopeltus fasciatus. Dev. Biol. 329: 142–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson L. C., Maroja L., Jiggins C. D., 2011. Convergent, modular expression of ebony and tan in the mimetic wing patterns of Heliconius butterflies. Dev. Genes Evol. 221: 297–308. [DOI] [PubMed] [Google Scholar]
- Gibert J. M., Peronnet F., Schlotterer C., 2007. Phenotypic plasticity in Drosophila pigmentation caused by temperature sensitivity of a chromatin regulator network. PLoS Genet. 3: e30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gompel N., Prud’homme B., Wittkopp P. J., Kassner V. A., Carroll S. B., 2005. Chance caught on the wing: cis-regulatory evolution and the origin of pigment patterns in Drosophila. Nature 433: 481–487. [DOI] [PubMed] [Google Scholar]
- Han Q., Fang J., Ding H., Johnson J. K., Christensen B. M., et al. , 2002. Identification of Drosophila melanogaster yellow-f and yellow-f2 proteins as dopachrome-conversion enzymes. Biochem. J. 368: 333–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna A., 1953. Non-autonomy of yellow in gynandromorphs of Drosophila melanogaster. J. Exp. Zool. 123: 523–560. [Google Scholar]
- Hines H. M., Papa R., Ruiz M., Papanicolaou A., Wang C., et al. , 2012. Transcriptome analysis reveals novel patterning and pigmentation genes underlying Heliconius butterfly wing pattern variation. BMC Genomics 13: 288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hintermann E., Jeno P., Meyer U. A., 1995. Isolation and characterization of an arylalkylamine N-acetyltransferase from Drosophila melanogaster. FEBS Lett. 375: 148–150. [DOI] [PubMed] [Google Scholar]
- Hotta Y., Benzer S., 1970. Genetic dissection of the Drosophila nervous system by means of mosaics. Proc. Natl. Acad. Sci. USA 67: 1156–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong S., Rebeiz M., Andolfatto P., Werner T., True J., et al. , 2008. The evolution of gene regulation underlies a morphological difference between two Drosophila sister species. Cell 132: 783–793. [DOI] [PubMed] [Google Scholar]
- Koch P. B., Behnecke B., ffrench-Constant R. H., 2000. The molecular basis of melanism and mimicry in a swallowtail butterfly. Curr. Biol. 10: 591–594. [DOI] [PubMed] [Google Scholar]
- Kopp A., Duncan I., 1997. Control of cell fate and polarity in the adult abdominal segments of Drosophila by optomotor-blind. Development 124: 3715–3726. [DOI] [PubMed] [Google Scholar]
- Kopp A., Muskavitch M. A., Duncan I., 1997. The roles of hedgehog and engrailed in patterning adult abdominal segments of Drosophila. Development 124: 3703–3714. [DOI] [PubMed] [Google Scholar]
- Lemonds, T. R., 2015 The contribution of the melanin pathway to overall body pigmentation changes during ontogenesis of Periplaneta Americana. Master of Science Thesis, Wayne State University, Detroit. [DOI] [PubMed] [Google Scholar]
- Li H., Popadic A., 2004. Analysis of nubbin expression patterns in insects. Evol. Dev. 6: 310–324. [DOI] [PubMed] [Google Scholar]
- Li Z., Zeng B., Ling L., Xu J., You L., et al. , 2015. Enhancement of larval RNAi efficiency by over-expressing Argonaute2 in Bombyx mori. Int. J. Biol. Sci. 11: 176–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Lemonds T. R., Popadic A., 2014. The genetic control of aposematic black pigmentation in hemimetabolous insects: insights from Oncopeltus fasciatus. Evol. Dev. 16: 270–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P. Z., Kaufman T. C., 2004a Kruppel is a gap gene in the intermediate germband insect Oncopeltus fasciatus and is required for development of both blastoderm and germband-derived segments. Development 131: 4567–4579. [DOI] [PubMed] [Google Scholar]
- Liu P. Z., Kaufman T. C., 2004b hunchback is required for suppression of abdominal identity, and for proper germband growth and segmentation in the intermediate germband insect Oncopeltus fasciatus. Development 131: 1515–1527. [DOI] [PubMed] [Google Scholar]
- Liu P. Z., Kaufman T. C., 2005. even-skipped is not a pair-rule gene but has segmental and gap-like functions in Oncopeltus fasciatus, an intermediate germband insect. Development 132: 2081–2092. [DOI] [PubMed] [Google Scholar]
- Long Y., Li J., Zhao T., Li G., Zhu Y., 2015. A new arylalkylamine N-acetyltransferase in silkworm (Bombyx mori) affects integument pigmentation. Appl. Biochem. Biotechnol. 175: 3447–3457. [DOI] [PubMed] [Google Scholar]
- Medved V., Marden J. H., Fescemyer H. W., Der J. P., Liu J., et al. , 2015. Origin and diversification of wings: Insights from a neopteran insect. Proc. Natl. Acad. Sci. USA 112: 15946–15951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehere P., Han Q., Christensen B. M., Li J., 2011. Identification and characterization of two arylalkylamine N-acetyltransferases in the yellow fever mosquito, Aedes aegypti. Insect Biochem. Mol. Biol. 41: 707–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ordway A. J., Hancuch K. N., Johnson W., Wiliams T. M., Rebeiz M., 2014. The expansion of body coloration involves coordinated evolution in cis and trans within the pigmentation regulatory network of Drosophila prostipennis. Dev. Biol. 392: 431–440. [DOI] [PubMed] [Google Scholar]
- Osanai-Futahashi M., Ohde T., Hirata J., Uchino K., Futahashi R., et al. , 2012. A visible dominant marker for insect transgenesis. Nat. Commun. 3: 1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebeiz M., Pool J. E., Kassner V. A., Aquadro C. F., Carroll S. B., 2009. Stepwise modification of a modular enhancer underlies adaptation in a Drosophila population. Science 326: 1663–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi A., Takahashi K., Ueda R., Takano-Shimizu T., 2007. Natural variation of ebony gene controlling thoracic pigmentation in Drosophila melanogaster. Genetics 177: 1233–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomoyasu Y., Arakane Y., Kramer K. J., Denell R. E., 2009. Repeated co-options of exoskeleton formation during wing-to-elytron evolution in beetles. Curr. Biol. 19: 2057–2065. [DOI] [PubMed] [Google Scholar]
- True J. R., Yeh S. D., Hovemann B. T., Kemme T., Meinertzhagen I. A., et al. , 2005. Drosophila tan encodes a novel hydrolase required in pigmentation and vision. PLoS Genet. 1: e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter M. F., Zeineh L. L., Black B. C., McIvor W. E., Wright T. R., et al. , 1996. Catecholamine metabolism and in vitro induction of premature cuticle melanization in wild type and pigmentation mutants of Drosophila melanogaster. Arch. Insect Biochem. Physiol. 31: 219–233. [DOI] [PubMed] [Google Scholar]
- Weatherbee S. D., Nijhout H. F., Grunert L. W., Halder G., Galant R., et al. , 1999. Ultrabithorax function in butterfly wings and the evolution of insect wing patterns. Curr. Biol. 9: 109–115. [DOI] [PubMed] [Google Scholar]
- Werner T., Koshikawa S., Williams T. M., Carroll S. B., 2010. Generation of a novel wing colour pattern by the Wingless morphogen. Nature 464: 1143–1148. [DOI] [PubMed] [Google Scholar]
- Wittkopp P. J., Beldade P., 2009. Development and evolution of insect pigmentation: genetic mechanisms and the potential consequences of pleiotropy. Semin. Cell Dev. Biol. 20: 65–71. [DOI] [PubMed] [Google Scholar]
- Wittkopp P. J., Vaccaro K., Carroll S. B., 2002a Evolution of yellow gene regulation and pigmentation in Drosophila. Curr. Biol. 12: 1547–1556. [DOI] [PubMed] [Google Scholar]
- Wittkopp P. J., True J. R., Carroll S. B., 2002b Reciprocal functions of the Drosophila yellow and ebony proteins in the development and evolution of pigment patterns. Development 129: 1849–1858. [DOI] [PubMed] [Google Scholar]
- Wittkopp P. J., Carroll S. B., Kopp A., 2003. Evolution in black and white: genetic control of pigment patterns in Drosophila. Trends Genet. 19: 495–504. [DOI] [PubMed] [Google Scholar]
- Wright T. R., 1987. The genetics of biogenic amine metabolism, sclerotization, and melanization in Drosophila melanogaster. Adv. Genet. 24: 127–222. [PubMed] [Google Scholar]
- Zhan S., Guo Q., Li M., Li J., Miao X., et al. , 2010. Disruption of an N-acetyltransferase gene in the silkworm reveals a novel role in pigmentation. Development 137: 4083–4090. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors state that all data necessary for confirming the conclusions presented in this article are represented fully within the article. The accession numbers of the O. fasciatus sequences from GenBank are as follows: KX023894 (ebony), KX023895 (black), KX023896 (yellow), KX023897 (tan), and KX023898 (aaNAT).