Abstract
The Drosophila egg chamber provides a useful model for examining mechanisms by which cell fates are specified and maintained in the context of a complex tissue. The egg chamber is also an excellent model for understanding the mechanism by which cytoskeletal filaments are organized and the critical interplay between cytoskeletal organization, polarity establishment, and cell fate specification. Previous work has shown that Egalitarian (Egl) is required for specification and maintenance of oocyte fate. Mutants in egl either completely fail to specify an oocyte, or if specified, the oocyte eventually reverts back to nurse cell fate. Due to this very early role for Egl in egg chamber maturation, it is unclear whether later stages of egg chamber development also require Egl function. In this report, we have depleted Egl at specific stages of egg chamber development. We demonstrate that in early-stage egg chambers, Egl has an additional role in organization of oocyte microtubules. In the absence of Egl function, oocyte microtubules completely fail to reorganize. As such, the localization of microtubule motors and their cargo is disrupted. In addition, Egl also appears to function in regulating the translation of critical polarity determining messenger RNAs (mRNAs). Finally, we demonstrate that in midstage egg chambers, Egl does not appear to be required for microtubule organization, but rather for the correct spatial localization of oskar, bicoid, and gurken mRNAs.
Keywords: RNA localization, molecular motor, cell polarity, microtubule organization
THE proper formation of a tissue or organ requires the precise specification of cell fates and the subsequent maintenance of these differentiated fates. Often, these processes take place during embryonic development. In many instances, disruption of tissue morphogenesis results in organismal lethality, complicating mechanistic studies. In this regard, the Drosophila melanogaster egg chamber is a useful tool for study. The ovary is a nonessential organ. Thus, genes with essential roles in formation of the mature Drosophila egg can be studied using adult animals. In addition, abundant genetic and molecular tools are available in Drosophila, facilitating mechanistic analysis of tissue morphogenesis (Sullivan et al. 2000).
Each Drosophila ovary is composed of 16–20 ovarioles (Spradling 1993). The germline stem cells and their associated somatic niche are found at the anterior tip of the ovariole in a region known as the germarium (Spradling 1993) (Figure 1, A and B). Division of a germline stem cell produces a daughter cell known as a cystoblast (Spradling et al. 2001; Gonzalez-Reyes 2003). The cystoblast undergoes four rounds of cell division to produce a cyst containing 16 germ cells. The cyst is eventually surrounded by a layer of somatic cells known as follicle cells. As the cyst progresses through the germarium, oocyte fate is specified (Deng and Lin 2001; Riechmann and Ephrussi 2001; Huynh and St Johnston 2004). Thus, once the cyst emerges from the germarium as an egg chamber, it contains 15 nurse cells and one oocyte (Spradling 1993) (Figure 1, A and B). The egg chamber progresses through 14 stages of morphogenesis before it is competent for fertilization. During these stages, the cyst grows in size and distinct fates are specified in the follicle cells (Spradling 1993). Although 14 different stages of egg chamber maturation can be identified, based on morphological features and the cell fates that are specified, egg chamber maturation represents a developmental continuum. Thus, not every stage of egg chamber development is observed in all ovarioles.
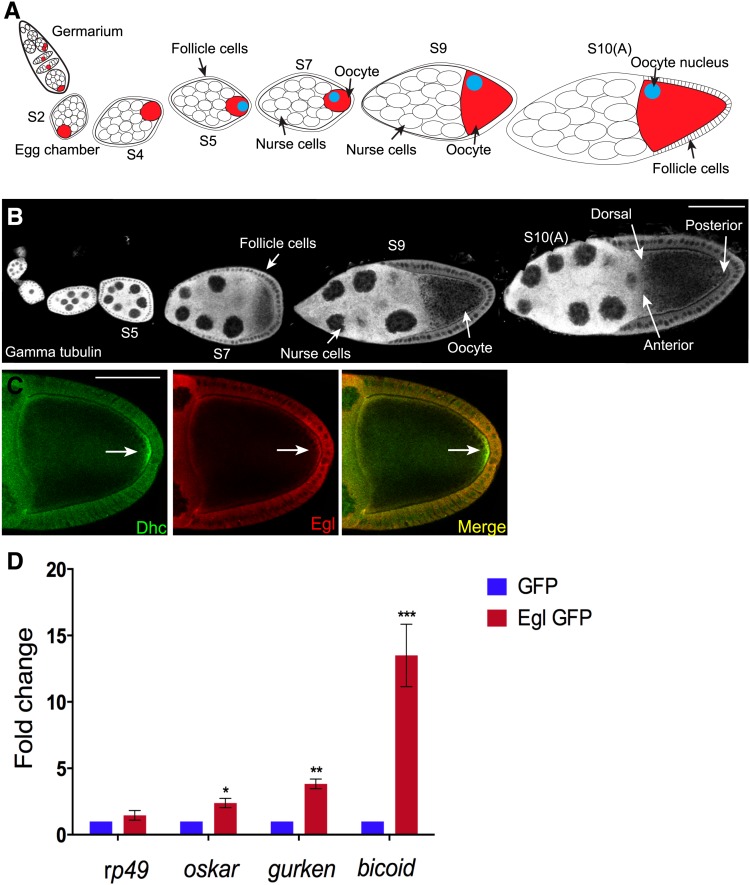
Figure 1.
The localization and mRNA binding properties of Egl. (A) Schematic of a Drosophila ovariole. The germline stem cells and their niche reside at the anterior tip of the ovariole in a region known as the germarium. The stem cell divides to produce a daughter cell known as a cystoblast. The cystoblast undergoes four rounds of cell division with incomplete cytokinesis to produce a cyst containing 16 germ cells. One of these 16 germ cells will become the oocyte (red cell); the rest will assume nurse cell fate. During maturation within the germarium, the oocyte comes to reside at the posterior of the cyst. Also within the germarium, the cyst becomes surrounded by a layer of somatic cells known as follicle cells. This structure is now referred to as an egg chamber. The egg chamber progresses through 14 stages of morphogenesis before it is competent for fertilization. Egg chambers from the following stages are indicated in the schematic: stage 2 (S2), stage 4 (S4), stage 5 (S5), stage 7 (S7), stage 9 (S9), and stage 10A (S10A). Between stages 5 and 7, signaling events between the oocyte and the overlying follicle cells result in reorganization of oocyte microtubules. As a consequence, the oocyte nucleus migrates from the posterior to the dorsal–anterior margin (blue circle). (B) Representative Drosophila ovariole from a wild-type strain. The egg chambers were stained with an antibody against γ-tubulin. (C) Ovaries were dissected from wild-type flies and fixed and processed for immunofluorescence using antibodies against Dynein heavy chain (Dhc, green) and Egl (red). The arrow indicates enrichment of Dhc at the posterior pole. (D) Ovaries were dissected from flies expressing either GFP or Egl-GFP in the female germline. Lysates were prepared and incubated with GFP-Trap beads. The coprecipitating RNAs were reverse transcribed and processed for qPCR using primers against the indicated genes. The level of coprecipitating rp49, osk, bcd, and grk was normalized to the level of coprecipitating γ-tubulin mRNA. rp49 and γ-tubulin represent mRNAs that are not asymmetrically localized within the egg chamber. The entire experiment was done in triplicate. The error bars represent standard deviation. *P = 0.033, **P = 0.0013, and ***P = 0.0009, unpaired t-test. Bar, 50 μm.
Between stages 2 and 6 there is a buildup of gurken (grk) messenger RNA (mRNA) and protein within the oocyte (Neuman-Silberberg and Schupbach 1993, 1996). This pool of Grk signals to the overlying follicle cells to adopt posterior cell fates (Gonzalez-Reyes et al. 1995; Roth et al. 1995). Once posterior fate has been specified, the follicle cells signal back to the oocyte. Although the nature of this signal is still unknown, the Notch and Hippo pathways are thought to contribute to the process (Ruohola et al. 1991; Meignin et al. 2007; Polesello and Tapon 2007; Yu et al. 2008; Yan et al. 2011). In addition, proteins within the extracellular matrix and factors that link the oocyte to the follicle cells are also involved (Deng and Ruohola-Baker 2000; Frydman and Spradling 2001; MacDougall et al. 2001). This signaling cascade initiates a reorganization of oocyte microtubules. Prior to this, oocyte microtubules are arranged with minus ends enriched at the posterior pole (Theurkauf et al. 1992). However, the redistribution of microtubules that takes place in response to Grk signaling results in minus ends enriched along the anterior and lateral cortex and plus ends that display a slight enrichment at the posterior pole of the oocyte (Theurkauf et al. 1992; Clark et al. 1994, 1997; Parton et al. 2011; Sanghavi et al. 2012). This reorganization is required for migration of the oocyte nucleus from the posterior to the anterior of the oocyte (Gonzalez-Reyes et al. 1995; Roth et al. 1995) (Figure 1A). In addition, this microtubule organization is required for the posterior localization of oskar (osk) mRNA, the anterior localization of bicoid (bcd) mRNA, and the localization of grk mRNA at the dorsal–anterior margin (St Johnston 2005). The precise localization of these mRNAs and their resulting protein products is essential for polarizing the oocyte and the resulting embryo (Berleth et al. 1988; Ephrussi and Lehmann 1992; Gavis and Lehmann 1992; Neuman-Silberberg and Schupbach 1993).
Given the complex patterning events that are required for producing a mature Drosophila egg, it is not surprising that many genes have been implicated in these various processes. Among these genes is egalitarian (egl). egl null mutants fail to specify an oocyte (Theurkauf et al. 1993; Carpenter 1994). Instead, egg chambers in these mutants contain 16 nurse cells (Theurkauf et al. 1993; Carpenter 1994). Egl is thought to function in this pathway by restricting meiosis to a single cell and by promoting the microtubule-dependent transport of factors to the presumptive oocyte (Mach and Lehmann 1997; Huynh and St Johnston 2000; Bolivar et al. 2001). Milder mutants in egl that are defective in binding Dynein light chain (Dlc) initially specify an oocyte. However, oocyte fate is not maintained in these mutants, eventually resulting in cysts containing 16 nurse cells (Navarro et al. 2004). In embryos, Egl has been shown to function as an adaptor for Dynein, by linking this microtubule motor to mRNAs destined for localization (Dienstbier et al. 2009). Thus, Egl is required at very early stages of Drosophila egg chamber morphogenesis, and much later, during development of the embryo.
The role of Egl in oocyte specification and maintenance has precluded a detailed analysis of Egl function during mid and late stages of egg chamber development. Using a short hairpin RNA (shRNA)-mediated depletion strategy, we have been able to specifically deplete Egl at defined stages of egg chamber development. We demonstrate that in early-stage egg chambers, Egl is required for organization of oocyte microtubules and translational regulation of specific mRNAs. In mid- and late-stage egg chambers, Egl does not appear to play a role in microtubule organization, but rather, functions in the localization of osk, bcd, and grk mRNAs. Thus, Egl function is required at multiple stages in the development of a mature Drosophila egg.
Materials and Methods
Fly stocks
The following fly stocks were used: Oregon-R-C (used as wild type; Bloomington Stock Center, no. 5); GFP-Stau (Zimyanin et al. 2008); eb1 shRNA (Bloomington Stock Center, no. 36680; donor TRiP); egl shRNA-1 (Bloomington Stock Center, no. 43550; donor TRiP); khc shRNA (Bloomington Stock Center, no. 35409; donor TRiP); osk84 (Kim-Ha et al. 1991); Df(3R)p-XT103 (Deficiency for osk; Bloomington Stock Center, no. 1962; donor Thom Kaufman); P{w[+mC]=UASp-Act5C.mRFP}13, w[*] (Bloomington Stock Center, no. 24777; donor Susan Parkhurst); pnt-LacZ (Becalska and Gavis 2010), Par1GFP N1S isoform (Doerflinger et al. 2006); and Dp(2;3)BAC.cnn+eGFP::Ex1A (Eisman et al. 2015).
The expression of the shRNA and the rescue transgene was driven using either P{w[+mC]=matalpha4-GAL-VP16}V37 (referred to in the Results section as the early-stage driver; Bloomington Stock Center, no. 7063; donor Andrea Brand) or w[*]; P{w[+mC]=matalpha4-GAL-VP16}V2H (referred to in the Results section as the mid-stage driver; Bloomington Stock Center, no. 7062; donor Andrea Brand).
The eglR-GFP transgene was constructed by first cloning the coding sequence of egl into pUC19. Site-directed mutagenesis was used to mutate the following nucleotides within egl: A1599C and G1602C. This was done using the Q5 Site-Directed Mutagenesis Kit (New England Biolabs). Once the desired mutations were confirmed by sequencing, the entire coding sequence of eglR was cloned into pUASp-attB-K10 (Koch et al. 2009). The EGFP coding sequence was subcloned immediately downstream of eglR. This resulted in the eglR-GFP transgene that encoded wild-type Egl protein, yet was not targeted by egl shRNA-1. Transgenic flies were generated by BestGene. To test for rescue, crosses were set up to recombine eglR-GFP and egl shRNA-1 onto the same chromosome.
Egl shRNA-2 flies were generated by cloning the following sequence into the NheI and EcoR1 sites of the Valium 22 vector (Ni et al. 2011): 5′-ACCGAGGTGTCCGATCTCAAA-3′. The insert was confirmed by sequencing. Transgenic flies containing this shRNA construct were generated by BestGene. The egl shRNA-2 construct was inserted at the attP40 site on chromosome 2.
Antibodies
Unless specifically stated, the indicated dilutions are for immunofluorescence. The following antibodies were used: rabbit anti-Egl (1:2000); rabbit anti-Staufen (D. St Johnston; 1:2500); mouse anti-Dhc (Developmental Studies Hybridoma Bank, 1:150; donor J. Scholey); rabbit anti-Khc (Cytoskeleton, 1:150); mouse anti-BicD (Developmental Studies Hybridoma Bank, 1:150; donor Ruth Steward); chicken anti-Oskar (1:50 after blocking with osk protein null ovaries); mouse anti-Oskar (D. Chen, 1:1000); rabbit anti-Oskar (A. Ephrussi, 1:2000); FITC-conjugated mouse anti-α-tubulin (Sigma, 1:100); rabbit anti-Eb1 (S. Rogers, 1:500); mouse anti-β-galactosidase (Promega, 1:2000); mouse anti-γ-tubulin (Sigma, 1:100); mouse anti-Grk (Developmental Studies Hybridoma Bank, 1:175; donor T. Schüpbach); rat anti-GFP (Nacalai USA, 1:600); mouse anti-Orb (Developmental Studies Hybridoma Bank, 1:300; donor P. Schedl); mouse anti-LaminDmO (Developmental Studies Hybridoma Bank, clone ADL84.12; 1:200; donor P. A. Fisher); goat antirabbit Alexa 594 and 488 (Life Technologies, 1:400 and 1:200, respectively); goat antimouse Alexa 594 and 488 (Life Technologies, 1:400 and 1: 200); and goat antichicken 594 and 488 (Life Technologies, 1:400 and 1:200, respectively).
The chicken anti-Osk antibody was prepared by injecting the following peptide into chicken (Aves Labs): ESNYISVREEYPDIDSEVR. Yolk was collected and antibody specific to this peptide was purified by Aves Labs.
Immunofluorescence
Prior to dissection, flies were fattened on yeast paste for 3 days. Ovaries were dissected as previously described (Liu et al. 2015) and fixed in either 8% formaldehyde for 10 min (for the α-tubulin staining experiment) or 4% formaldehyde for 20 min (for the remaining experiments). The fixative was diluted in PBS. For the experiments involving mouse and rabbit antibodies, the immunofluorescence staining was performed as previously described (Liu et al. 2015). For experiments involving the chicken anti-Osk antibody, the oocytes were blocked in BlokHen II (Aves Labs) diluted 1:10 in PBS for 30 min. The primary antibody was incubated in 1× PBST (PBS + 0.1% Triton X-100) + 0.2% BSA overnight at 4°. The next day, the samples were washed three times in PBST. The ovaries were then incubated in goat antichicken Alexa 594 (Life Technologies). The secondary antibody was diluted in 1× PBST + 0.2% BSA and incubated overnight at 4°. The following day, the ovaries were washed three times in PBST, mounted on slides, and imaged.
In situ hybridization
osk, bcd, and grk mRNAs were visualized as previously described (Sanghavi et al. 2013).
Microscopy
Images were captured on a Zeiss LSM 780 upright or a Zeiss LSM 510 upright confocal microscope. Images were processed for presentation using Fiji, Adobe Photoshop, and Adobe Illustrator.
Quantification
Localization phenotypes were quantified by scoring egg chambers of the indicated genotype from at least three different experiments. The level of Egl in stage 10 egg chamber (see Figure 2E) was quantified using the Zen software provided by Zeiss. The level of fluorescence intensity in a 50-μm-square area in the nurse cells of control and Egl-depleted egg chambers was determined. This was done on a single confocal slice of 25 different egg chambers for each genotype. The level of Grk oocyte enrichment (see Figure 7E and Supplemental material, Figure 5E) was determined using the same software. For this analysis, the fluorescence intensity within the oocyte was compared to the fluorescence intensity within the remainder of the egg chamber. A single confocal slice in the middle plane of the egg chamber was used for this analysis. For these experiments, 15 egg chambers from each genotype were quantified. The statistical calculators provided by GraphPad were used for calculating significance. An unpaired t-test was performed using standard deviation, mean, and the n value.
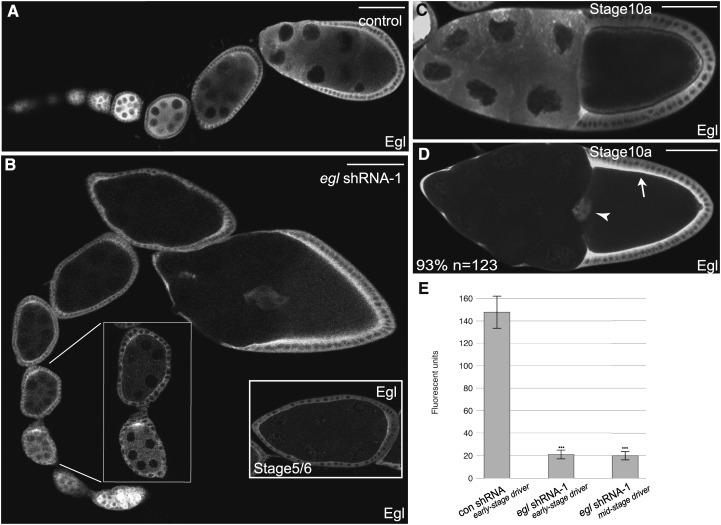
Figure 2.
Germline-specific depletion of Egl. (A–D) Egg chambers expressing either a control shRNA (A and C) or egl shRNA-1 (B and D) were fixed and processed for immunofluorescence using an antibody against Egl. The control used in this and in following experiments corresponds to a strain expressing shRNA against eb1. Expression of shRNAs was under control of a maternal α-tubulin driver that is active from early-stage egg chambers onwards (see Figure S1A). The left inset in B represents a magnified view of the area indicated by the white lines. The right inset in B shows a stage 5/6 egg chamber from a different ovariole from flies expressing egl shRNA-1. The arrow in D indicates the overlying main body follicle cells. The arrowhead in D indicates the border cells. Also indicated in D is the percentage of stage 10 egg chamber displaying convincing depletion of Egl within the germline. (E) The level of Egl within the germline was quantified using the following strains: a strain expressing a control shRNA, a strain expressing egl shRNA-1 using the early-stage driver, and a strain expressing egl shRNA-1 using the midstage driver (see Figure S1B). Ovaries were fixed and processed for immunofluorescence using an antibody against Egl. The fluorescence intensity of Egl immunostaining was measured within the germline of 25 different egg chambers from each strain. Error bars represent standard deviation. ***P < 0.0001, unpaired t-test. Bar, 50 μm.
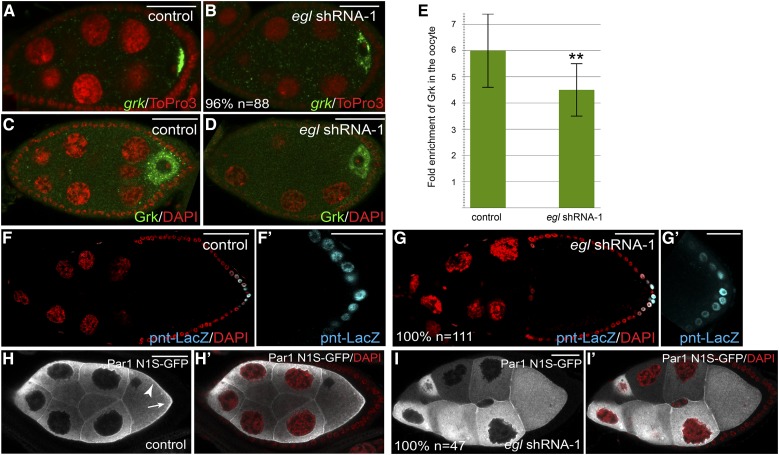
Figure 7.
The role of Egl in microtubule organization. (A and B) Ovaries from flies expressing a control shRNA (A) or egl shRNA-1 (B) using the early-stage driver were fixed and processed for in situ hybridization using an antisense probe against grk mRNA (green). The egg chambers were counterstained with ToPro3 (red). (C and D) Ovaries from flies expressing a control shRNA (A) or egl shRNA-1 (B) using the early-stage driver were fixed and processed for immunofluorescence using an antibody against Grk (green). The egg chambers were counterstained with DAPI (red). (E) The enrichment of Grk within the oocyte of control egg chambers was compared to that from Egl-depleted egg chambers. For each genotype, 15 egg chambers were quantified. **P = 0.0022, unpaired t-test. (F and G) Ovaries from flies expressing pnt-LacZ (F) or from flies expressing pnt-LacZ and egl shRNA-1 using the early-stage driver (G) were fixed and processed for immunofluorescence using an antibody against β-galactosidase (cyan). The ovaries were also counterstained with DAPI (red). F′ and G′ represent a magnified view of the posterior follicle cells in F and G, respectively. (H and I) Ovaries from flies expressing Par1-GFP (H) or from flies expressing Par1-GFP and egl shRNA-1 using the early-stage driver (I) were fixed and processed for immunofluorescence using an antibody against GFP (gray). The ovaries were also counterstained with DAPI (red). In H′ and I′, the DAPI and GFP signals are overlaid. The arrowhead in H indicates the membrane localization of Par-GFP in the control strain. The arrow in H indicates the posterior enrichment of Par1-GFP in control egg chambers. The penetrance of each phenotype and the number of egg chambers scored is indicated. Bar in F and G, 50 μm; in all other panels, 25 μm.
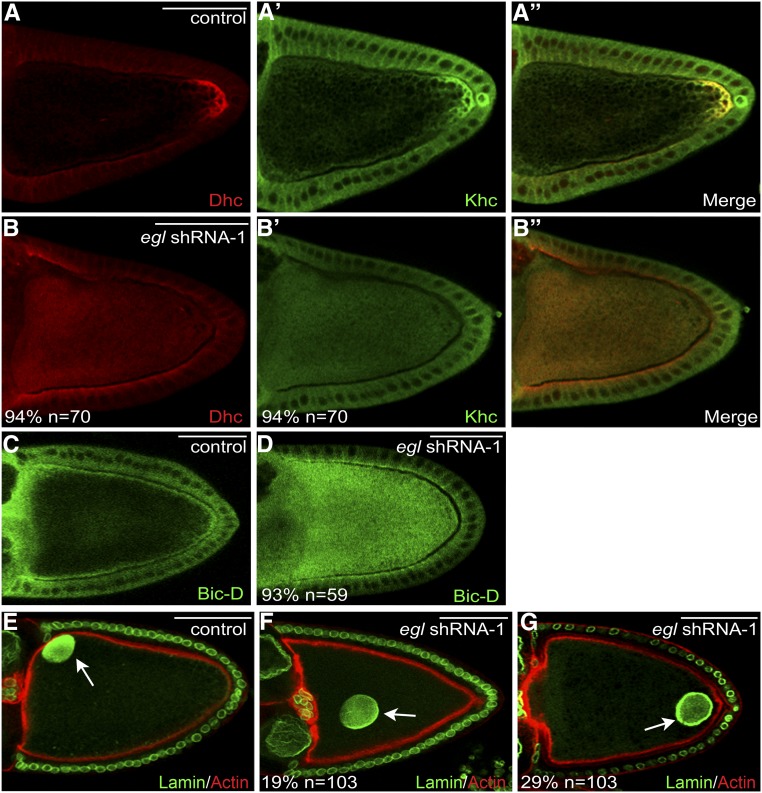
Figure 5.
Delocalization of Dynein, Kinesin, BicD, and the oocyte nucleus in Egl-depleted egg chambers. (A and B) Ovaries from flies expressing a control shRNA (A) or egl shRNA-1 (B) were fixed and processed for immunofluorescence using antibodies against Dhc (red) and Khc (A′ and B′, green). A merged image is also shown (A′′ and B′′). (C and D) Ovaries from flies expressing a control shRNA (C) or egl shRNA-1 (D) were fixed and processed for immunofluorescence using an antibody against BicD. (E–G) Ovaries from flies expressing a control shRNA (E) or egl shRNA-1 (F and G) were fixed and processed for immunofluorescence using an antibody against Lamin DmO (green). The egg chambers were also counterstained with Phalloidin (red). The arrow in E indicates the normal localization of the oocyte nucleus. The arrows in F and G indicate the mislocalized oocyte nucleus in Egl-depleted egg chambers. The number of egg chambers scored, as well as the penetrance of each phenotype, are indicated. Bar, 50 μm.
Analyzing mRNA association
Ovaries were dissected from flies expressing GFP or Egl-GFP. A total of 600 µg of each lysate was then incubated with GFP-Trap beads (Chromotek) to purify GFP and Egl-GFP. The procedure for immunoprecipitation followed by RT-PCR was previously described (Sanghavi et al. 2013). The RNA that coprecipitated with GFP and Egl-GFP was reverse transcribed using random hexamers and Superscript III (Life Technologies) using the directions provided by the manufacturer. The concentration of the cDNA was determined and 60 ng of cDNA was used in each quantitative PCR (qPCR) reaction. The SsoAdvanced Universal SYBR Green Supermix from Bio-Rad was used for qPCR. The reaction was run using a Bio-Rad CFX96 Real-Time PCR System. The following primers were used in the qPCR reactions:
-
osk mRNA
forward: 5′-CCCGAGGGTACTCAGATCAT-3′
reverse: 5′-GCGGTGCAAGATTTGTTAGA-3′
-
bcd mRNA
forward: 5′-AATCGGATCAGCACAAGGAC-3′
reverse: 5′-GCGTTGAATGACTCGCTGTA-3′
-
grk mRNA
forward: 5′-ATCCGATGGTGAACAACACA-3′
reverse: 5′-CGACGACAGCATGAGGAGTA-3′
-
rp49 mRNA
forward: 5′-CCAGTCGGATCGATATGCTA-3′
reverse: 5′-GGGCATCAGATACTGTCCCT-3′
-
γ-tubulin mRNA
forward: 5′-CCACCATCATGAGTCTGAGC-3′
reverse: 5′-ACCGATGAGGTTGTTGTTCA-3′.
To determine mRNA association, the ct values were compared to γ-tubulin. rp49 represents an unlocalized mRNA and serves as a further control. The entire experiment was done in triplicate.
Analyzing mRNA levels
Ovaries were dissected from flies expressing a control shRNA against eb1 or from flies expressing egl shRNA-1. RNA was extracted using TRIzol (Life Technologies), following the directions provided by the manufacturer. cDNA was generated using 5 µg of total RNA from each sample. Random hexamers and Superscript III (Life Technologies) was used for the reverse transcription. A total of 15 ng of cDNA was used in each qPCR reaction. The primer sequences are shown above. The geometric mean was determined for γ-tubulin and rp49 and this was used to compare to the levels of osk, bcd, and grk. The entire experiment was done in triplicate.
Data availability
All strains used in this work will be made available upon request. The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.
Results
The localization and RNA binding properties of Egalitarian
Egalitarian (Egl) is a Dynein adaptor that has been shown to link this motor with localized mRNAs in the Drosophila embryo (Navarro et al. 2004; Dienstbier et al. 2009). Within the ovary, the majority of studies have focused on the role of Egl in specification and maintenance of oocyte fate (Theurkauf et al. 1993; Mach and Lehmann 1997; Huynh and St Johnston 2000; Bolivar et al. 2001). The goal of the present study was to examine the function of Egl during midstages of egg chamber maturation.
Dynein heavy chain (Dhc), the motor subunit of the Dynein complex, was enriched at the posterior pole of the oocyte in stage 10 egg chambers (Figure 1C, arrow). This localization pattern is consistent with published findings (McGrail et al. 1995). Partial colocalization was observed between Egl and Dhc at the posterior pole and along the oocyte cortex (Figure 1C). It should be noted, however, that whereas Dhc displayed an obvious enrichment at the posterior pole, the posterior enrichment of Egl was slightly less apparent (Figure 1C).
We next determined whether Egl preferentially associated with mRNAs destined for localization within the oocyte. Although numerous mRNAs are specifically localized within the Drosophila egg chamber (Jambor et al. 2015), the localization pathways of gurken (grk), bicoid (bcd), and oskar (osk) mRNAs have been most extensively characterized (Gonsalvez and Long 2012; Weil 2014). Lysates expressing GFP or Egl-GFP were subjected to immunoprecipitation using GFP antibody beads. The coprecipitating mRNAs were extracted and analyzed using reverse transcriptase (RT) followed by quantitative PCR. The values were normalized to the level of γ-tubulin mRNA detected in each pellet. The nonlocalizing mRNA, rp49, was not specifically enriched in the Egl pellet (Figure 1D). By contrast, a significant enrichment of bcd mRNA was detected in the Egl pellet (Figure 1D). grk mRNA, and to a lesser extent, osk mRNA, were also enriched in Egl pellets (Figure 1D). Thus, as observed in Drosophila embryos (Dienstbier et al. 2009; Vazquez-Pianzola et al. 2014), Egl associates with mRNAs that are destined for localization.
Depletion of Egl in midstage egg chambers
Classical mutant alleles of egl have not been very useful in examining the role of this protein during midstages of egg chamber maturation. Egl performs an essential function in oocyte specification. Consequently, loss-of-function mutants in egl never specify an oocyte (Theurkauf et al. 1993; Carpenter 1994). Mutants in egl that are defective in binding Dynein light chain (Dlc) initially specify an oocyte. However, oocyte fate is not maintained in these mutants, and the oocyte eventually reverts to a nurse cell fate (Navarro et al. 2004). Occasionally, these mutants produce morphologically normal stage 10 egg chambers, and within these, osk mRNA is often delocalized (Navarro et al. 2004). These results suggest an important role for Egl in localizing mRNAs in midstage egg chambers.
To more directly examine the role of Egl in mRNA localization, we employed an shRNA-mediated depletion strategy. We have successfully used this approach to analyze the role of Dhc and the Dynein regulator, Dynamitin, in mRNA localization and endocytosis in midstage egg chambers (Sanghavi et al. 2013; Liu et al. 2015). A strain expressing an shRNA against egl (egl shRNA-1) was obtained from the Bloomington Drosophila Stock Center. In addition, we generated an independent shRNA targeting a different region of egl (egl shRNA-2) (see Materials and Methods for details). The shRNAs were expressed using a maternal α-tubulin driver that is only active after the oocyte has been specified (Supplemental Material, Figure S1A). This expression profile enables us to bypass the requirement for Egl in oocyte specification and to study the role of this protein during later stages of oogenesis.
We next determined the efficacy and specificity of Egl depletion using fly strains expressing a control shRNA, egl shRNA-1, or egl shRNA-2. Egg chambers from these flies were fixed and processed for immunofluorescence using an antibody against Egl. In control flies, Egl was abundantly expressed in the germline and soma during all stages of oogenesis (Figure 2, A and C). By contrast, Egl was significantly depleted from the germline of flies expressing egl shRNA-1 (Figure 2, B and D). Typically, depletion was observed from stage 5 onwards (Figure 2, B and D). Consistent with the expression profile of the driver, the level of Egl in the somatic follicle cells was unaffected in these flies (Figure 2D, arrow and arrowhead). Quantification of Egl depletion in stage 10 egg chambers is shown in Figure 2E. Similar results were obtained by Western blot analysis of whole ovary lysates (Figure S2A). It should be noted, however, that this analysis underrepresents the germline-specific depletion of Egl. These lysates contain follicle cells and stages 1–4 egg chambers. As shown by immunofluorescence analysis, Egl is not depleted in somatic cells and in these very-early-stage egg chambers (Figure 2B). In contrast to egl shRNA-1, expression of egl shRNA-2 was not capable of depleting endogenous Egl (Figure S2B). Therefore, most of the remaining experiments were carried out using flies expressing egl shRNA-1.
Egl is required for localization of osk, grk, and bcd mRNAs
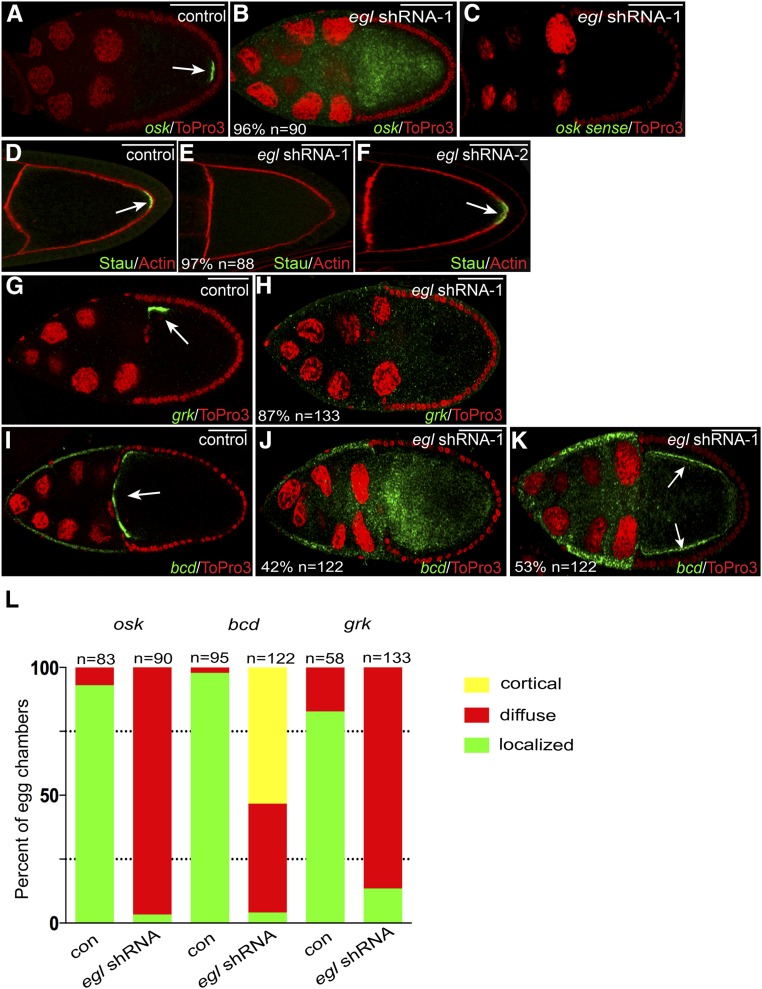
osk mRNA localizes to the posterior pole of the oocyte in wild-type stage 10 egg chambers (Ephrussi et al. 1991; Kim-Ha et al. 1991). The localization of osk mRNA plays an important role in restricting the expression of Oskar (Osk) protein to this site. The same posterior localization pattern was observed for osk mRNA in egg chambers expressing a control shRNA or egl shRNA-2 (Figure 3A, arrow; data not shown). By contrast, osk mRNA was completely delocalized in egg chambers expressing egl shRNA-1 (Figure 3, B and L). The delocalized osk mRNA was not only present in the oocyte, but also in the nurse cell cytoplasm (Figure 3B).
Figure 3.
mRNA localization defects in Egl-depleted egg chambers. (A and B) Egg chambers expressing either a control shRNA (A) or egl shRNA-1 (B) using the early-stage driver were fixed and processed for in situ hybridization using an antisense probe against osk mRNA (green). The egg chambers were counterstained with ToPro3 to reveal nuclei (red). The arrow in A indicates the normal localization of osk mRNA at the posterior pole. (C) To examine the specificity of the in situ hybridization signal, ovaries from egl shRNA-1-expressing flies were fixed and processed for in situ hybridization using a sense probe against osk mRNA (green). The egg chambers were also counterstained with ToPro3 (red). (D–F) Egg chambers expressing a control shRNA (D), egl shRNA-1 (E), or egl shRNA-2 (F) were fixed and processed for immunofluorescence using an antibody against Staufen (Stau, green). The egg chambers were also counterstained with phalloidin to reveal F-actin (red). The arrows in D and F indicate the posterior localization of Stau in control and egl shRNA-2-expressing egg chambers. Also indicated in E is the penetrance of the phenotype and the number of egg chambers that were scored. (G and H) Egg chambers expressing a control shRNA (G) or egl shRNA-1 (H) using the early-stage driver were fixed and processed for in situ hybridization using an antisense probe against grk mRNA (green). The samples were counterstained with ToPro3 (red). The arrow in G indicates the normal localization of grk mRNA at the dorsal–anterior corner of the oocyte. (I–K) The same two strains used in the above panels were also examined using an antisense probe against bcd mRNA (green). ToPro3 was used to visualize nuclei (red). The arrow in I indicates the normal localization of bcd mRNA at the anterior margin of the oocyte. In 42% of egg chambers expressing egl shRNA-1, bcd mRNA was delocalized throughout the entire egg chamber (J). In 53% of egg chambers expressing egl shRNA-1, bcd mRNA accumulated along the cortex (K, arrows). (L). The graph represents quantification of mRNA localization phenotypes. The two strains analyzed were ones expressing either a control shRNA or egl shRNA-1 using the early-stage driver. The green bars represent a wild-type localization pattern (examples in A, G, and I). The red bars represent egg chambers containing mRNA that was diffusely distributed throughout the entire egg chamber (examples are B, H, and J). The yellow bar represents mRNA that was detected along the cortex (K). The number of egg chambers scored for each genotype is indicated. Bar, 50 μm.
Imaging the delocalized osk mRNA required a higher gain setting than what was used for control egg chambers. This result is expected because in control egg chambers, most molecules of osk mRNA are highly enriched at the posterior pole, confined to a relatively small three-dimensional volume. When osk mRNA is delocalized, the same number of osk molecules is now distributed over a much larger volume. Thus, visualizing the delocalized osk mRNA requires a higher gain. However, an alternative interpretation of this result is that the level of osk mRNA is reduced upon depletion of Egl. To address this latter possibility, we performed quantitative analysis using total RNA extracted from control and egl shRNA-1-expressing ovaries. This analysis revealed no statistical difference in the level of osk mRNA between these two strains (Figure S2C).
To verify that the diffuse signal corresponds to delocalized osk mRNA, these strains were processed for in situ hybridization using a sense probe against osk. The egg chambers were imaged using the same high-gain setting. Despite using 10 times more sense probe in comparison to antisense probe, we did not detect any signal in Egl-depleted oocytes (Figure 3C). Thus, the in situ hybridization signal is specific for delocalized osk mRNA. As a final test, control and Egl-depleted egg chambers were processed for immunofluorescence using an antibody against Staufen, a core component of the osk mRNP (St Johnston et al. 1991; Zimyanin et al. 2008). Consistent with the in situ hybridization result, Staufen was absent from the oocyte posterior in Egl-depleted oocytes (Figure 3, D and E). As expected, expression of egl shRNA-2 had no effect on the localization of Staufen (Figure 3F, arrow).
We next examined the localization of grk and bcd mRNAs. grk mRNA localizes to the dorsal–anterior corner of stage 10 wild-type egg chambers (Neuman-Silberberg and Schupbach 1993). This localization pattern is required for concentrating Gurken (Grk) protein at this site. In contrast to egg chambers expressing a control shRNA, grk mRNA was completely delocalized in Egl-depleted egg chambers (Figure 3G, arrow, H, and L). bcd mRNA localizes to the anterior margin of stage 10 egg chambers (Berleth et al. 1988). This results in a high concentration of bcd mRNA at the anterior pole of wild-type embryos (Berleth et al. 1988). As with osk and grk mRNAs, bcd mRNA was completely delocalized in Egl-depleted egg chambers (Figure 3, I–L). In ∼45% of egg chambers, bcd mRNA was diffusely localized throughout the entire egg chamber (Figure 3, J and L). In the remainder, bcd mRNA was localized along the oocyte cortex (Figure 3, K and L). As noted with osk, the level of grk and bcd mRNAs was unchanged upon depletion of Egl (Figure S2C).
A potential caveat of these results is that these phenotypes might be nonspecific and caused due to off-target depletion of unknown factors. To address this issue, we attempted to rescue the mRNA localization defect using a wild-type Egl-GFP transgene. An shRNA-resistant Egl transgene was generated by introducing mutations at the wobble position within the shRNA-1 recognition site of egl (Figure S3A). The net result was an eglR-GFP transgene that encoded wild-type Egl protein, yet was not targeted by the shRNA. This transgene was then introduced into the egl shRNA-1 background. The expression of egl shRNA-1 as well as transgenic EglR-GFP was under the control of the same driver. Thus, as the level of endogenous Egl is depleted, transgenic EglR-GFP is expressed. For simplicity, we will refer to these as “rescue” flies.
We first confirmed that EglR-GFP was expressed in the rescue flies and that it localized in a similar manner to endogenous Egl (Figure S3B). We next examined the localization of osk, grk, and bcd mRNAs. The localization pattern of all three mRNAs resembled wild type (Figure S3, C–F). Based on these results, we conclude that the mRNA delocalization phenotype is caused due to specific depletion of Egl.
Egl regulates the translation of osk and grk mRNAs
In wild-type egg chambers, translation of osk mRNA is repressed during its transport to the posterior pole (Kim-Ha et al. 1995; Johnstone and Lasko 2001). Once osk mRNA is correctly localized, translational repression is relieved, and the mRNA is activated for translation (Johnstone and Lasko 2001). Several factors have been identified that are required for precisely regulating the translation of osk mRNA (Wilhelm and Smibert 2005). Osk protein performs an essential function in maintaining the anterior–posterior polarity of the oocyte and embryo (Ephrussi and Lehmann 1992; Smith et al. 1992). Thus, ectopic expression of Osk results in oocytes with defective polarity and embryos that are not able to establish proper cell fates. As shown in the preceding section, depletion of Egl results in complete delocalization of osk mRNA. We therefore wished to determine whether this delocalized message was translated.
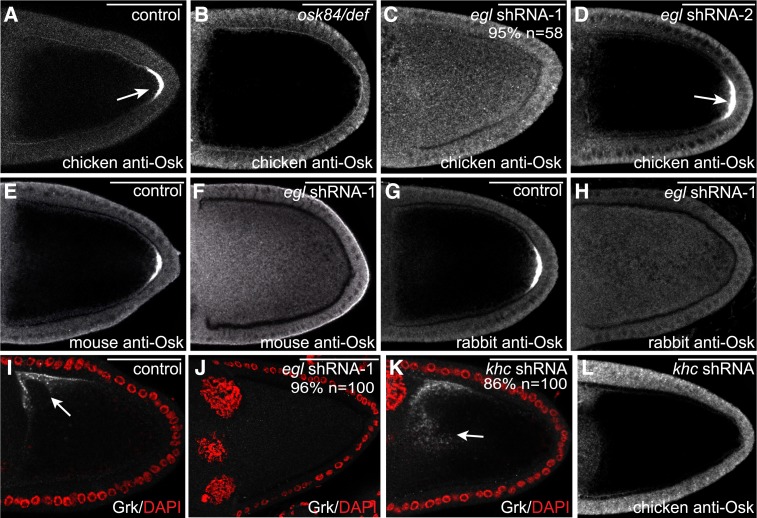
To examine the expression and localization of Osk protein, we developed a chicken anti-Osk antibody. As expected, this antibody recognized the posterior restricted Osk protein in wild-type egg chambers (Figure 4A, arrow; Figure S2D). By contrast, the posterior crescent of Osk protein was not observed in osk protein-null egg chambers (Figure 4B). Thus, this antibody is capable of detecting endogenous Osk. It should be noted, however, that variable nonspecific staining of follicle cells was observed using this antibody. The follicle cell staining was seen in both wild-type and osk protein-null egg chambers (Figure 4, A and B).
Figure 4.
The role of Egl in translation regulation. (A–D) Egg chambers from the following strains were fixed and processed for immunofluorescence using a chicken anti-Osk antibody: a strain expressing a control shRNA (A), osk 84/osk Def (B, an osk protein-null strain), a strain expressing egl shRNA-1 using the early-stage driver (C), and a strain expressing egl shRNA-2 using the early-stage driver (D). Arrows in A and D indicate the normal posterior localization of Osk protein. (E–H) Ovaries from flies expressing a control shRNA (E and G) or egl shRNA-1 (F and H) were fixed and processed for immunofluorescence using a mouse anti-Osk antibody (E and F) or a rabbit anti-Osk antibody (G and H). (I–K) Ovaries from flies expressing a control shRNA (I), egl shRNA-1 (J), or khc shRNA (K) were fixed and processed for immunofluorescence using an antibody against Grk (gray-scale image). The egg chambers were also counterstained with DAPI to visualize nuclei (red). The arrow in I indicates the normal localization of Grk protein at the dorsal–anterior corner of the oocyte. The arrow in K represents the delocalized Grk protein in Khc-depleted oocytes. (L) Ovaries from flies expressing khc shRNA were fixed and processed for immunofluorescence using a chicken anti-Osk antibody. The number of egg chambers scored, as well as the penetrance of each phenotype, are indicated. Bar, 50 μm.
Egg chambers from flies expressing a control shRNA or egl shRNA-1 were fixed and processed for immunofluorescence using this chicken anti-Osk antibody. Surprisingly, this analysis revealed that in the absence of Egl, the delocalized osk mRNA was translated into protein (Figure 4C, Figure S2E). As a control for specificity, we examined egg chambers from flies expressing egl shRNA-2. As noted previously, this shRNA is not capable of depleting endogenous Egl (Figure S2B). Consequently, osk mRNA remains localized in this strain, and Osk protein is restricted to the posterior pole (Figure 4D, data not shown).
To verify that our antibody was specifically detecting Osk protein, we repeated the experiment using two additional anti-Osk antibodies, one generated in mouse and another generated in rabbit (Markussen et al. 1995; Huang et al. 2014). The same result was obtained using both antibodies (Figure 4, E–H; Figure S2, F and G). Based on these findings, we conclude that in the absence of Egl, the delocalized osk mRNA is no longer maintained in a translationally repressed state.
We next examined the expression and localization of Grk protein. The enrichment of grk mRNA at the dorsal–anterior corner of the oocyte contributes to the localization of Grk protein at this site. Polarized secretion of Grk, and its signaling to the overlying follicle cells, is necessary for establishing the dorsal–ventral axis of the oocyte (Neuman-Silberberg and Schüpbach 1993). In contrast to egg chambers expressing a control shRNA, dorsal–anterior signal for Grk protein was not observed in Egl-depleted oocytes (Figure 4, I and J). In fact, specific signal that corresponds to Grk protein was almost absent in Egl-depleted egg chambers (Figure 4J). The net result of this phenotype is that the overlying follicle cells do not receive a signal to establish dorsal cell fates. Consequently, most mature eggs from these strains display a ventralized phenotype reflected by a loss of dorsal appendages (97%, n = 65; data not shown).
To validate this result, we examined the localization and expression of Grk protein in Khc-depleted egg chambers. In khc null oocytes, grk mRNA is delocalized and the delocalized mRNA is translated into protein (Brendza et al. 2002; Duncan and Warrior 2002; Januschke et al. 2002). Similar results were obtained in Khc-depleted oocytes; grk mRNA was delocalized, and some of the delocalized mRNA was translated into protein (Figure S4, C and D; Figure 4K). Importantly, in contrast to grk mRNA, delocalized osk mRNA was not translated in Khc-depleted oocytes (Figure S4, A and B; Figure 4L).
Based on these results we conclude that in the absence of Egl, delocalized osk mRNA is inappropriately translated into protein. By contrast, the delocalized grk mRNA present in these same oocytes appears to be translated much less efficiently. These results reveal an important distinction in the translational regulation of osk and grk mRNAs.
As shown in the previous section, expression of an shRNA-refractory eglR-GFP transgene was able to rescue the osk and grk mRNA localization defects (Figure S3, C and E). The same flies were also examined using anti-Osk and anti-Grk antibodies. This revealed that Osk protein was restricted to the posterior pole in the rescue flies, and Grk protein was restricted to the dorsal–anterior corner of the oocyte (Figure S3G; data not shown). Thus, expression of the shRNA-refractory eglR-GFP transgene was able to rescue the localization and translational regulation of osk and grk mRNAs.
bcd mRNA is translated in the embryo by a mechanism that involves lengthening of the poly(A) tail (Salles et al. 1994). Thus, wild-type oocytes express very little Bicoid (Bcd) protein (Driever and Nusslein-Volhard 1988) (data not shown). A similar situation was observed in Egl-depleted oocytes. The delocalized bcd mRNA was not precociously translated into protein in these egg chambers (data not shown).
Egl is required for the normal localization of Dhc, Khc, and BicD
Egl is considered an adaptor of the Dynein motor. Egl binds directly to Dlc, a component of the Dynein complex (Navarro et al. 2004), and in Drosophila embryos, Egl serves to link Dynein to mRNAs destined for localization (Dienstbier et al. 2009). We therefore examined the localization of Dynein in Egl-depleted egg chambers using an antibody against Dhc. In stage 10 egg chambers expressing a control shRNA, Dhc was enriched at the posterior pole of the oocyte and also along the anterior and lateral cortex (Figure 5A). A similar pattern was observed in wild-type egg chambers (data not shown) (McGrail et al. 1995). In the absence of Egl, Dhc was no longer enriched at the oocyte posterior or along the cortex in stage 10 egg chambers. Rather, Dhc was diffusely distributed throughout the cytoplasm of the oocyte (Figure 5B). A similar distribution was observed for Khc (Figure 5, A′, A′′, B′, and B′′).
We next examined the localization of Bicaudal-D (BicD), an interacting partner of Egl (Mach and Lehmann 1997). In wild-type stage 10 egg chambers, and in strains expressing a control shRNA, BicD localized along the oocyte cortex (Figure 5C; data not shown). As with Dynein and Kinesin, loss of Egl resulted in diffuse distribution of BicD throughout the oocyte with no obvious enrichment along the cortex (Figure 5D).
Disruption of the Dynein motor is associated with defects in the localization of the oocyte nucleus (Swan et al. 1999; Brendza et al. 2002; Duncan and Warrior 2002; Januschke et al. 2002). In stage 10 wild-type egg chambers, or in egg chambers expressing a control shRNA, the oocyte nucleus was anchored at the dorsal–anterior corner of the oocyte (Figure 5E, arrow; data not shown). By contrast, 48% of Egl-depleted egg chambers contained a mislocalized oocyte nucleus. In some egg chambers, the oocyte nucleus was present close to the anterior, but was not anchored adjacent to the cortex (Figure 5F, arrow). In addition, in several Egl-depleted egg chambers, the oocyte nucleus was observed at the posterior pole, having completely failed to migrate toward the anterior (Figure 5G, arrow). Thus, depletion of Egl disrupts the localization, and possibly the activity, of the Dynein motor complex. The normal localization of Dhc, Khc, BicD, and the oocyte nucleus, was restored upon expression of the shRNA-refractory eglR-GFP transgene (Figure S3, H and I; data not shown).
Microtubule organization is disrupted upon depletion of Egl
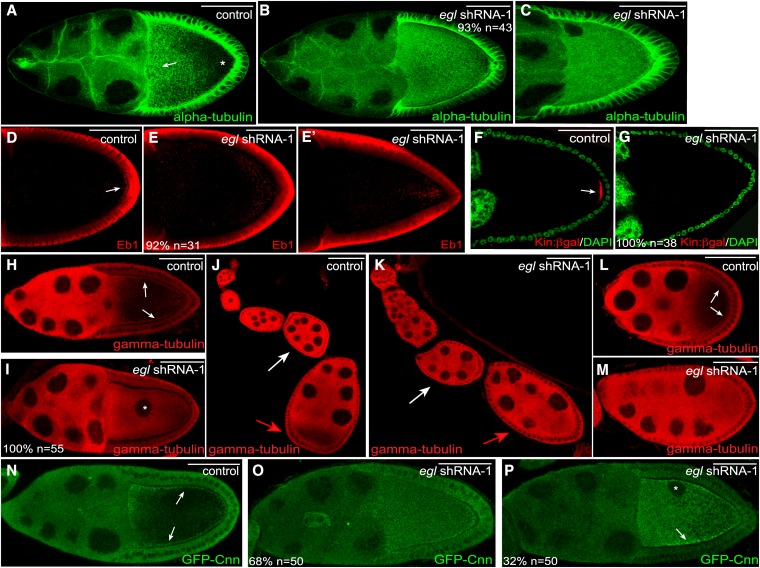
The severe delocalization phenotype observed for Dhc, Khc, and BicD upon depletion of Egl suggests that a defect in microtubule organization might underlie these phenotypes. To directly test this possibility, we first examined the overall organization of microtubules using an antibody against α-tubulin, a core component of the microtubule polymer. Stage 10 control egg chambers contained a dense array of microtubules along the anterior portion of the oocyte (Figure 6A, arrow), whereas the posterior was characterized by an overall lower density of microtubules (Figure 6A, asterisk). By contrast, a uniformly high level of α-tubulin staining was observed throughout the entire oocyte in Egl-depleted egg chambers (Figure 6, B and C). Thus, depletion of Egl disrupts the normal organization of oocyte microtubules.
Figure 6.
Microtubule organization in Egl-depleted egg chambers. (A–C) Ovaries from flies expressing a control shRNA (A) or egl shRNA-1 (B and C) were fixed and processed for immunofluorescence using an antibody against α-tubulin. There is a high density of microtubules at the anterior margin of control oocytes (A, arrow). The posterior of control oocytes contains a much lower density of microtubules (A, asterisk). (D–E′) Ovaries from flies expressing a control shRNA (D) or egl shRNA-1 (E and E′) were fixed and processed for immunofluorescence using an antibody against Eb1. The arrow in D indicates the posterior enrichment of Eb1 foci in control egg chambers. (F and G) Ovaries from control flies expressing Kinesin β-gal (F, Kin:β-gal, red) or Kinesin β-gal along with egl shRNA-1 (G) were fixed and processed for immunofluorescence using an antibody against β-galactosidase. The egg chambers were also counterstained with DAPI (green). The arrow in F indicates the posterior localization of Kin:β-gal in control egg chambers. (H–M) Ovaries from flies expressing a control shRNA (H, J, and L) or egl shRNA-1 (I, K, and M) were fixed and processed for immunofluorescence using an antibody against γ-tubulin. The arrows in H indicate the cortical enrichment of γ-tubulin in control stage 10 egg chambers. The asterisk in I indicates the mislocalized oocyte nucleus. The white arrows in J and K indicate a stage 5 egg chamber and the red arrows in these panels indicate a stage 7 egg chamber. The arrows in L indicate the cortical enrichment of γ-tubulin in an early stage 9 egg chamber. (N–P) Ovaries from flies expressing GFP-Cnn (N) or from flies expressing GFP-Cnn and egl shRNA-1 (O and P) were fixed and processed for immunofluorescence using an antibody against GFP. Expression of the shRNA was under control of the early-stage maternal α-tubulin driver. Arrows in N indicate the cortical enrichment of GFP-Cnn in the control strain. The arrow in P indicates the residual cortical enrichment of GFP-Cnn in a subset of Egl-depleted egg chambers. The asterisk indicates the mislocalized oocyte nucleus. The number of egg chambers scored, as well as the penetrance of each phenotype, are indicated. Bar, 50 μm.
To more precisely define the organization of microtubules in Egl-depleted oocytes, we examined these egg chambers using markers for microtubule plus ends. The first marker used was an antibody against End binding protein 1 (Eb1), a highly conserved plus-end binding protein (Schuyler and Pellman 2001). In wild-type egg chambers, Eb1 foci were detected throughout the oocyte, with enrichment at the posterior pole (Sanghavi et al. 2012) (Figure 6D). In Egl-depleted egg chambers, Eb1 foci were scattered throughout the oocyte with no obvious enrichment at the posterior pole (Figure 6E). Occasionally, a cloud of Eb1 foci could be detected close to the posterior (Figure 6E′).
We validated these results using another plus-end marker, a strain expressing the Kin:β-gal transgene. This transgene represents a fusion between the motor domain of Khc and β-galactosidase (Clark et al. 1994). Kin:β-gal localizes to the posterior pole of stage 9 and 10a egg chambers (Clark et al. 1994) (Figure 6F, arrow). This was not observed in Egl-depleted oocytes. Instead, diffuse signal for Kin:β-gal was observed throughout the oocyte (Figure 6G). Thus, upon depletion of Egl, microtubule plus ends, which are normally enriched at the posterior pole, become scattered within the oocyte.
We next examined the localization of microtubule minus ends. An antibody against γ-tubulin, a core component of microtubule organizing centers, was used for this analysis. γ-Tubulin was enriched along the oocyte cortex in stages 9 and 10 control egg chambers (Figure 6H) (Cha et al. 2002). The interior of control oocytes was characterized by a lower level of γ-tubulin staining (Figure 6H) (Cha et al. 2002). In Egl-depleted oocytes, the cortical enrichment of γ-tubulin was not detected. Instead, signal for γ-tubulin was detected throughout the entire oocyte (Figure 6I). The normal distribution of microtubule plus and minus ends was restored upon expression of the shRNA-refractory eglR-GFP transgene (Figure S3, J–L), thus demonstrating the specificity of these phenotypes.
During the course of these experiments, we noticed a stage-specific redistribution of γ-tubulin in control egg chambers. In early stage egg chambers, γ-tubulin was uniformly distributed throughout the entire egg chamber (Figure 6J, white arrow). At stage 7, there was a transition in the localization of γ-tubulin within the oocyte. From stage 7 onwards, γ-tubulin became concentrated along the oocyte cortex, and very little signal was observed within the oocyte cytoplasm (Figure 6J, red arrow, and L). In contrast to the control, this transition did not occur in Egl-depleted egg chambers, and γ-tubulin remained diffusely distributed (Figure 6, K and M). Thus, Egl appears to be required for this oocyte-specific reorganization of microtubule minus ends.
Similar results were obtained using a strain expressing GFP-tagged Centrosomin (Cnn), another component of microtubule organizing centers (Megraw et al. 1999). In control egg chambers, GFP-Cnn was enriched around the cortex of the oocyte (Figure 6N). This pattern was disrupted upon depletion of Egl. In 68% of egg chambers, GFP-Cnn was diffusely distributed throughout the entire oocyte (Figure 6O). In the remaining egg chambers, GFP-Cnn signal was still detected within the interior of Egl-depleted oocytes. However, residual cortical enrichment of GFP-Cnn could also be detected (Figure 6P, arrow).
The role of Egl in microtubule organization
The reorganization of oocyte microtubules that occurs in stage 7 egg chambers is initiated by Grk protein. During early stages of egg chamber maturation, grk mRNA and protein become selectively enriched within the oocyte (Neuman-Silberberg and Schüpbach 1996). The oocyte-enriched Grk protein signals the overlying follicle cells to establish posterior cell fate (Gonzalez-Reyes et al. 1995; Roth et al. 1995). Once established, posterior follicle cells signal back to the oocyte by a mechanism that remains poorly understood. The net result of this process is a reorganization of oocyte microtubules such that microtubule density is highest at the anterior margin, microtubule minus ends are enriched along the cortex, and microtubule plus ends are enriched at the posterior pole (Steinhauer and Kalderon 2006).
As noted above, this pattern of microtubule organization was completely disrupted when Egl was depleted from stage 5 onwards. We therefore examined grk mRNA and protein localization in these egg chambers. In control egg chambers, grk mRNA was highly enriched at the posterior of the oocyte (Figure 7A). grk mRNA was still enriched within oocytes of Egl-depleted stage 5 egg chambers. However, the mRNA was not restricted to the posterior pole (Figure 7B). Grk protein preferentially accumulated within the oocyte of control egg chambers (Figure 7, C and E). A similar qualitative localization pattern was observed for Grk protein in Egl-depleted oocytes (Figure 7D). However, quantification revealed that the oocyte enrichment of Grk was modestly reduced in Egl-depleted egg chambers (Figure 7E). Thus, depletion of Egl in stage 5 egg chambers results in subtle defects in the localization of grk mRNA and in the oocyte enrichment of Grk protein.
We next determined whether posterior cell fates could be specified in Egl-depleted egg chambers. The pointed-LacZ (pnt-LacZ) enhancer trap strain was used for this analysis. LacZ signal from this strain is induced in posterior follicle cells in a Grk-dependent manner (Gonzalez-Reyes and St Johnston 1998; Becalska and Gavis 2010). Control egg chambers as well as Egl-depleted egg chambers contained posterior LacZ signal (Figures 7, F and G; Figure S2, H and I). Thus, despite a subtle defect in the oocyte enrichment of Grk protein, posterior follicle cell fate is specified in Egl-depleted egg chambers.
Once specified, the posterior follicle cells signal back to the oocyte. This results in the recruitment of Par1 kinase to the posterior of stage 7 egg chambers (Doerflinger et al. 2006). Par1, along with additional members of the Par complex, function within the oocyte to reorganize oocyte microtubules (St Johnston and Ahringer 2010). We therefore examined the localization of Par1-GFP in control and Egl-depleted stage 7 egg chambers. As expected, Par1-GFP was localized on the plasma membrane and enriched at the posterior of the oocyte in control egg chambers (Figure 7H). By contrast, in Egl-depleted oocytes, Par1-GFP was neither localized on the plasma membrane nor enriched at the posterior pole (Figure 7I).
Collectively, these results suggest that Egl is required for orchestrating the reorganization of microtubules that takes place in stage 7 egg chamber. At least one aspect of this function involves the posterior recruitment of Par1.
A specific role for Egl in mRNA localization in stage 10 egg chambers
The maternal α-tubulin driver used in the preceding sections is located on the third chromosome. This driver is not expressed in the germarium, but is turned on in early-stage egg chambers after oocyte specification (Figure S1A). For simplicity, we will refer to this strain as the “early-stage driver.” As shown previously, expression of egl shRNA-1 using this driver results in depletion of endogenous Egl from stage 5 onward (Figure 2B).
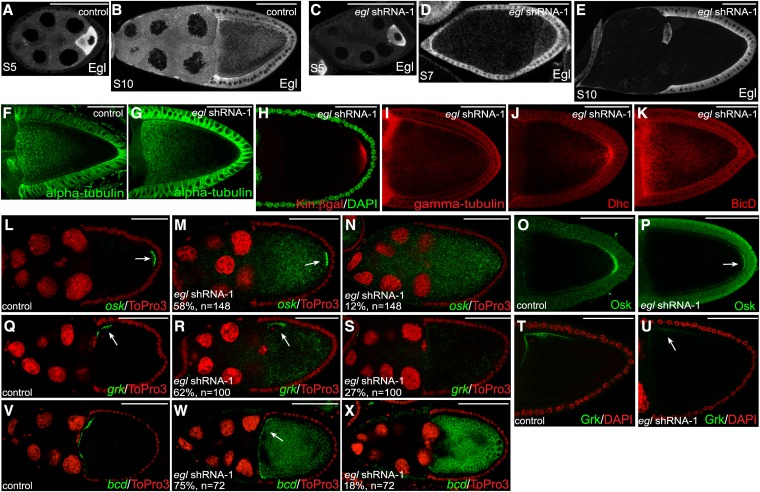
An additional maternal α-tubulin driver is available from the Bloomington Drosophila Stock Center. This driver is present on the second chromosome, and is expressed at later stages of egg chamber maturation, from stages 5 or 6 onwards (Sanghavi et al. 2013) (Figure S1B). For simplicity, we will refer to this strain as the “mid-stage driver.” The difference in expression profile between these drivers most likely results from position effects determined by their respective genomic locations. Nevertheless, expression of egl shRNA-1 using the midstage driver resulted in stage 10 egg chambers that were almost completely devoid of endogenous Egl within the germline (Figure 8, B and E). Quantification revealed that the level of depletion was similar to what was observed using the early-stage driver (Figure 2E). By contrast, the level of endogenous Egl in stages 5 and 7 egg chambers was only modestly reduced using the midstage driver (Figure 8, A, C, and D). Consistent reduction in the level of Egl was observed between stages 8 and 9 (data not shown).
Figure 8.
Egl is required in stage 10 egg chambers for mRNA localization. (A–E) Ovaries from flies expressing a control shRNA (A and B) or egl shRNA-1 (C–E) were fixed and processed for immunofluorescence using an antibody against Egl. A and C represent stage 5 egg chambers. D represents a stage 7 egg chamber and B and E represent stage 10 egg chambers. Expression of the shRNAs was driven using the midstage driver (see Figure S1B). (F and G) Ovaries from flies expressing a control shRNA (F) or egl shRNA-1 (G) using the midstage driver were fixed and processed for immunofluorescence using an antibody against α-tubulin (green). (H) Egg chambers from flies coexpressing Kin:β-gal and egl shRNA-1 using the midstage driver were fixed and processed for immunofluorescence using an antibody against β-galactosidase (green). The egg chambers were also counterstained with DAPI (red). (I–K) Ovaries from flies expressing egl shRNA-1 (I–K) using the midstage driver were fixed and processed for immunofluorescence using antibodies against γ-tubulin (I), Dhc (J), and BicD (K). (L–N) Ovaries from flies expressing a control shRNA (L) or egl shRNA-1 (M and N) using the midstage driver were fixed and processed for in situ hybridization using an antisense probe against osk mRNA (green). The egg chambers were counterstained with ToPro3 (red). The arrow in L indicates the posterior localization of osk mRNA in control strains. The arrow in M indicates the residual posterior enrichment of osk mRNA in a subset of Egl-depleted egg chambers. (O and P) The same strains used in the preceding panels were fixed and processed for immunofluorescence using an antibody against Osk. The arrow in P indicates the residual posterior Osk observed in Egl-depleted egg chambers. (Q–S) Ovaries from flies expressing a control shRNA (Q) or egl shRNA-1 (R and S) using the midstage driver were fixed and processed for in situ hybridization using an antisense probe again grk mRNA (green). The egg chambers were counterstained with ToPro3 (red). The arrow in Q indicates the dorsal–anterior localization of grk mRNA in control strains. The arrow in R indicates the residual enrichment of grk at the dorsal–anterior corner in a subset of Egl-depleted egg chambers. (T and U) The same strains used in the preceding panels were fixed and processed for immunofluorescence using an antibody against Grk. The arrow in U indicates the residual Grk protein at the dorsal–anterior corner of Egl-depleted egg chambers. (V–X) Ovaries from flies expressing a control shRNA (V) or egl shRNA-1 (W and X) using the midstage driver were fixed and processed for in situ hybridization using an antisense probe again bcd mRNA (green). The egg chambers were also counterstained with ToPro3 (red). The arrow in V indicates the anterior localization of bcd mRNA in control strains. The arrow in W indicates the residual anterior enrichment of bcd in a subset of Egl-depleted egg chambers. The number of egg chambers scored and the penetrance of the phenotypes are indicated. Bar, 50 μm.
We first examined microtubule organization in these Egl-depleted egg chambers. As noted previously, the initiating event in reorganization of oocyte microtubules is the oocyte enrichment of grk mRNA and protein in stage 5 egg chambers. In contrast to what was observed using the early-stage driver, grk mRNA localization and Grk protein accumulation within the oocyte was unaffected upon expression of egl shRNA-1 using the midstage driver (Figure S5, A–E). Consistent with this finding, the overall microtubule organization in stage 10 egg chambers resembled wild type (Figure 8, F and G). Furthermore, microtubule plus ends were enriched at the posterior pole and microtubule minus ends were detected around the cortex in these Egl-depleted egg chambers (Figure 8, H and I). In addition, the cortical localization of BicD was restored and Dhc was enriched at the posterior pole in egg chambers expressing egl shRNA-1 using the midstage driver (Figure 8, J and K). Thus, despite the fact that these stage 10 egg chambers are significantly depleted of Egl, their microtubule organization appears unaffected. Based on these results, we conclude that Egl is required between stages 5 and 7 for establishing the proper organization of oocyte microtubules. However, once this organization has been established, Egl does not appear to be required for maintaining microtubule organization in stage 10 egg chambers. One potential limitation of this conclusion, however, is that subtle defects might be present in the organization of oocyte microtubules that are not detected by our assays.
We next examined the localization of osk mRNA in stage 10 egg chambers expressing egl shRNA-1 using the midstage driver. In 58% of these egg chambers, osk mRNA was delocalized throughout the oocyte with residual enrichment at the posterior pole (Figure 8, L and M). Additionally, in 12% of egg chambers, osk mRNA was completely delocalized within the entire egg chamber with no residual posterior enrichment (Figure 8N). Thus, Egl is required in stage 10 egg chambers for the correct posterior localization of osk mRNA. We next examined these egg chambers using the anti-Osk antibody. A reduced level of Osk protein was detected at the posterior pole of these Egl-depleted egg chambers in comparison to the control (Figure 8, O and P). However, signal for Osk was not detected within the interior of the oocyte (Figure 8P). Thus, the delocalized osk mRNA present in these oocytes is not translated. This result suggests that the role of Egl in regulating the translation of osk mRNA is either stage specific or that the translation of delocalized osk mRNA is indirectly caused by defects in microtubule organization. Although both scenarios are equally plausible, the latter scenario is consistent with the findings of Becalska and Gavis (2010). Becalska and Gavis (2010) recently demonstrated a link between the translational regulation of osk mRNA and microtubule organization.
In addition to osk mRNA, Egl is also required in stage 10 egg chambers for the correct localization of grk and bcd mRNAs. In 62% of stage 10 egg chambers expressing egl shRNA-1 using the midstage driver, grk mRNA was delocalized within the oocyte with residual enrichment along the dorsal–anterior margin (Figure 8, Q and R). In a smaller percentage of egg chambers (27%), grk mRNA was completely delocalized throughout the entire egg chamber with no obvious dorsal–anterior localization (Figure 8S). Consistent with the delocalization of grk mRNA, a significantly reduced level of Grk protein was detected at the dorsal–anterior corner of the oocyte (Figure 8, T and U). Similar finding were obtained for bcd mRNA; 75% of egg chambers displayed residual enrichment of bcd mRNA at the anterior cortex, whereas in 18% of egg chambers the mRNA was completely delocalized (Figure 8, V–X).
Collectively, our results suggest that the efficient localization of osk, grk, and bcd mRNAs in stage 10 egg chambers requires Egl.
Discussion
Previous work has shown that Egl performs an essential function in specification and maintenance of oocyte fate (Theurkauf et al. 1993; Carpenter 1994; Navarro et al. 2004). Egl performs these functions within the germarium and early stage (stages 1–3) egg chambers, respectively. Using targeted depletion of Egl at various stages of egg chamber maturation, we have expanded on these studies. We demonstrate that Egl is required for the reorganization of oocyte microtubules that takes place in stage 7 egg chambers. By contrast, if Egl is present within early-stage egg chambers, but depleted during stages 9 and 10, microtubule organization appears unaffected. However, localization of osk, bcd, and grk mRNAs is disrupted. The localization of these mRNAs is critical for polarization of late stage oocytes and resulting embryos. Thus, formation of the mature Drosophila egg involves multiple functions of Egl at various stages of egg chamber maturation.
Egl and oocyte microtubules
Egg chambers from loss-of-function egl mutants fail to specify an oocyte, and instead contain 16 nurse cells (Theurkauf et al. 1993; Carpenter 1994). In fact, this phenotype is the source of the name “Egalitarian” (all are equal, no special cell has been set aside as the oocyte). The precise mechanism by which Egl functions to establish oocyte fate is unknown. However, the available data suggest that this function likely involves regulation of the microtubule cytoskeleton (Theurkauf et al. 1993). Milder mutants in egl that are defective in binding Dynein light chain (Dlc) initially specify an oocyte (Navarro et al. 2004). However, oocyte fate is not maintained in these early egg chambers, and the oocyte ultimately reverts to a nurse cell fate (Navarro et al. 2004).
In this study, we demonstrate that Egl has additional roles in development of the Drosophila egg chamber. Depletion of Egl from stage 5 onwards has no effect on oocyte maintenance. The oocyte marker, Orb, is selectively localized within a single cell, the DNA within the oocyte nucleus retains its condensed morphology, and border cell migration occurs normally (Figure S2, J–M; data not shown). Border cells are a group of follicle cells that migrate from the anterior of the egg chamber toward the oocyte by a mechanism that involves sensing chemoattractive cues produced by the oocyte (Montell 2006). Thus, if border cells migrate normally, it stands to reason that the oocyte is present and is functioning properly within this context. Despite this, several factors that are normally asymmetrically sorted become delocalized in the absence of Egl. These include mRNAs such as osk, bcd, and grk (Figure 3). In addition, the localization of motors and motor-associated factors are also affected (Figure 5). How might this occur? One possibility is that Egl is directly involved in the transport of each of these molecules. An alternative, and more plausible explanation, is that the delocalization of these factors is brought about by the severely disorganized microtubules present in Egl-depleted oocytes.
Oocyte microtubules undergo a reorganization at stage 7 such that microtubule density becomes highest at the anterior of the oocyte, minus ends become enriched around the cortex, and plus ends become enriched at the posterior pole (Steinhauer and Kalderon 2006). This process appears to be completely disrupted if Egl is depleted from stage 5 egg chambers onward (Figure 6). We envision two scenarios by which this could occur. One possibility is that Egl somehow directly functions to organize the arrangement and/or nucleation of oocyte microtubules. Consistent with this scenario is the finding that Egl is required within the germarium for directing the localization of the microtubule organizing center to the presumptive oocyte (Theurkauf et al. 1993). Also consistent with this notion is our observation that microtubule density within the oocyte is significantly lower in stage 5 egg chambers that have been depleted of Egl (Figure S2, N and O).
Another possibility is that Egl is required for the enrichment of grk mRNA and protein within the oocyte and a defect in these processes indirectly affects microtubules. Indeed, grk mRNA is not correctly localized in Egl-depleted stage 5 egg chambers. Although grk mRNA in enriched within these oocytes, unlike the control, it is not localized at the oocyte posterior (Figure 7, A and B). In addition, the oocyte enrichment of Grk protein is modestly reduced in Egl-depleted stage 5 egg chambers (Figure 7, C–E). However, these deficits do not appear to affect the Grk-dependent induction of posterior follicle cell fate (Figure 7, F and G). Thus, it is unlikely that Egl contributes to microtubule organization by affecting this arm of the pathway. Once posterior follicle cell fate has been established, these cells signal back to the oocyte, resulting in the recruitment of Par1 kinase to the posterior of the oocyte (Doerflinger et al. 2006). This process is severely affected in Egl-depleted egg chambers (Figure 7, H and I). Thus, Egl functions within the oocyte to facilitate the reorganization of microtubules in response to the posterior follicle cells signal.
It is worth noting that depletion of Egl produces a unique phenotype with respect to microtubule organization. As stated previously, grk mutants fail to reorganize oocyte microtubules at stage 7. However, in grk mutants, osk mRNA is delocalized to a central focus within the oocyte and bcd mRNA is present at both poles (Gonzalez-Reyes et al. 1995; Roth et al. 1995). By contrast, osk mRNA is uniformly distributed throughout the entire egg chamber in Egl-depleted oocytes (Figure 3B). In addition, bcd mRNA is either uniformly distributed or enriched along the oocyte cortex in Egl-depleted egg chambers (Figure 3, J and K). The common phenotype between Egl depletion and grk mutants is the defective migration of the oocyte nucleus away from the posterior pole (Figure 5G) (Gonzalez-Reyes et al. 1995; Roth et al. 1995).
Furthermore, in par1 and baz/par3 mutants, osk mRNA is delocalized to a central focus (Shulman et al. 2000; Tomancak et al. 2000; Becalska and Gavis 2010; Doerflinger et al. 2010). In some mutant alleles of these genes, bcd mRNA is relatively unaffected (Shulman et al. 2000), whereas in others, bcd mRNA localizes along the lateral cortex in a similar manner to Egl depletion (Becalska and Gavis 2010; Doerflinger et al. 2010). In some mutant alleles of baz, the oocyte nucleus is retained at the posterior (Becalska and Gavis 2010; Doerflinger et al. 2010), whereas in some mutants of par-1, migration of the oocyte nucleus is unaffected (Shulman et al. 2000). These varying results highlight the need for caution in interpreting the role of these genes in microtubule reorganization. The strength and nature of the phenotype will likely depend on the strength and nature of the mutant allele that was used. In addition, with an shRNA-based approach, the phenotypes will depend of the efficacy and timing of knockdown. Thus, we cannot conclude with certainly that the phenotype we describe in this report arises from complete depletion of Egl from stages 5 onward. A residual level of Egl might persist in these egg chambers and produce a milder phenotype than what would be observed if Egl could be completely eliminated.
An important future question will be to determine the molecular mechanism by which Egl functions in microtubule organization.
Egl and mRNA localization
As noted previously, Egl has been implicated in the Dynein-dependent localization of mRNAs in the Drosophila embryo (Dienstbier et al. 2009). We therefore investigated whether Egl might perform a similar function in the oocyte. Consistent with a role for Egl in mRNA localization, Egl-GFP was found to preferentially associate with localized mRNAs (Figure 1D). In addition, depletion of Egl from stage 5 onwards resulted in complete delocalization of osk, bcd, and grk mRNAs (Figure 3). However, as noted in the previous section, microtubules were severely disorganized in this background. Thus, from this experiment, we cannot conclude whether Egl is specifically required for mRNA localization. By contrast, depletion of Egl from stage 8 onwards does not appear to affect the organization of oocyte microtubules (Figure 8, F–I). Despite this, osk, bcd, and grk mRNAs were all significantly delocalized in these egg chambers (Figure 8). Our results therefore suggest that, much like in the embryo, Egl functions in mRNA localization within the oocyte. Because the efficient localization of all three mRNAs requires the Dynein motor (Duncan and Warrior 2002; Januschke et al. 2002; MacDougall et al. 2003; Weil et al. 2006; Sanghavi et al. 2013), it is likely that Egl performs this function via its association with Dynein (Navarro et al. 2004).
Egl and translation regulation
An unexpected finding was that Egl was required for the translational regulation of osk and grk mRNAs. Depletion of Egl from stage 5 onwards resulted in translation of delocalized osk mRNA (Figure 4C). By contrast, delocalized grk mRNA in these same oocytes appeared to be inefficiently translated (Figure 4J). How might Egl function in regulating the translation of these mRNAs? One possibility is a direct role for Egl in this process. A potential role for Egl in translation regulation was postulated by Huynh and St Johnston (2000). It is also possible that Egl participates indirectly in this process. As noted previously, depletion of Egl from stage 5 onwards resulted in profound defects in microtubule organization (Figure 6). Interestingly, Becalska and Gavis (2010) have demonstrated a correlation between microtubule organization and translation of osk mRNA. In general, mutants that affect microtubule organization cause ectopic translation of osk mRNA (Becalska and Gavis 2010). Similarly, mutants with defective microtubule organization have been shown to affect the translation of grk mRNA. In these mutants, grk mRNA is inefficiently translated (Abdu et al. 2006; Klattenhoff et al. 2007; Pane et al. 2007). It is therefore possible that the aberrant microtubule organization present in these Egl-depleted egg chambers underlies the defective translational regulation of osk and grk mRNAs. Consistent with this notion, depletion of Egl from stage 8 onwards using the midstage driver resulted in significant delocalization of osk mRNA. However, microtubules were correctly organized in these egg chambers, and the delocalized osk mRNA remained translationally repressed (Figure 8, M–P).
A model for Egl function in the Drosophila female germline
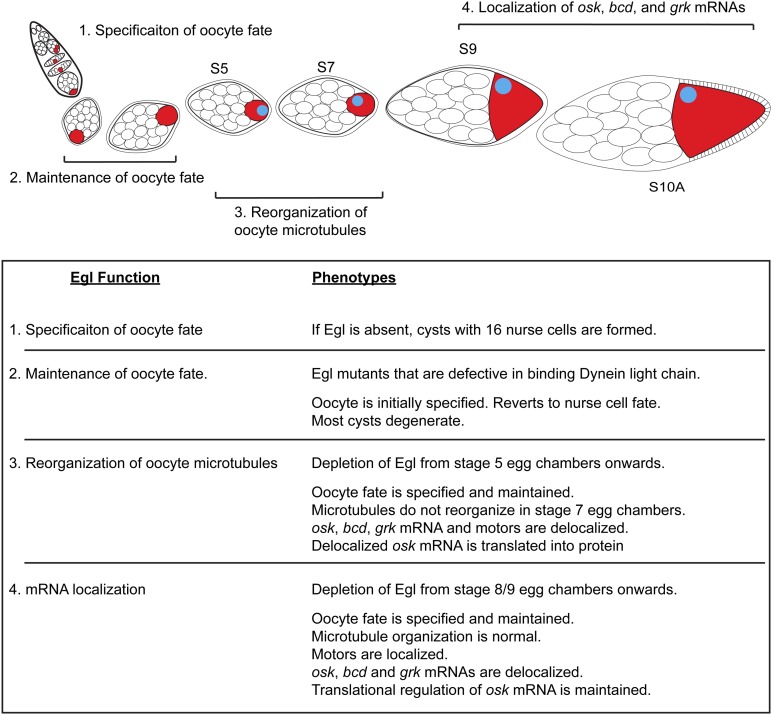
Published results have indicated a role for Egl within the germarium in establishment of oocyte fate (Figure 9) (Theurkauf et al. 1993; Carpenter 1994; Mach and Lehmann 1997; Huynh and St Johnston 2000; Bolivar et al. 2001). In early-stage egg chambers (stages 1–3), Egl is required for maintaining oocyte fate (Figure 9) (Navarro et al. 2004). During midstages of oogenesis (stages 5–7), we demonstrate that Egl is required within the oocyte to facilitate the reorganization of microtubules in response to the signal from posterior follicle cells (Figure 9). Finally, during stages 9 and 10, Egl is required for the efficient localization of osk, bcd, and grk mRNAs (Figure 9). The precise localization of these mRNAs is critical for establishing and maintaining the polarity of the oocyte and the future embryo. Thus, Egl performs essential functions at multiple stages during development of the Drosophila egg.
Figure 9.
Model for Egl function in maturation of the Drosophila egg chamber. Based on the findings in this report, as well as published results, we conclude that Egl has at least four separate functions during egg chamber maturation: (1) Within the germarium, Egl is required for oocyte specification (Theurkauf et al. 1993; Carpenter 1994). (2) In early-stage egg chambers, Egl is also required for maintaining oocyte fate (Navarro et al. 2004). (3) Between stages 5 and 7, Egl is required for reorganization of oocyte microtubules. (4) In stages 9 and 10 egg chambers, Egl is required for the correct spatial localization of osk, bcd, and grk mRNAs.
Acknowledgments
We thank S. Bullock, D. Chen, A. Ephrussi, V. Gelfand, L. Gavis, P. MacDonald, S. Roger, D. St Johnston, and T. Kaufman for antibodies and fly strains. We are grateful to the Bloomington Stock Center for providing fly strains and to the Developmental Studies Hybridoma bank for antibodies. This work was supported by a grant from the National Institutes of Health to G.B.G. (1R01GM100088-01).
Footnotes
Communicating editor: L. Cooley
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.115.184622/-/DC1.
Literature Cited
- Abdu U., Bar D., Schüpbach T., 2006. spn-F encodes a novel protein that affects oocyte patterning and bristle morphology in Drosophila. Development 133: 1477–1484. [DOI] [PubMed] [Google Scholar]
- Becalska A. N., Gavis E. R., 2010. Bazooka regulates microtubule organization and spatial restriction of germ plasm assembly in the Drosophila oocyte. Dev. Biol. 340: 528–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berleth T., Burri M., Thoma G., Bopp D., Richstein S., et al. , 1988. The role of localization of bicoid RNA in organizing the anterior pattern of the Drosophila embryo. EMBO J. 7: 1749–1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolivar J., Huynh J. R., Lopez-Schier H., Gonzalez C., St Johnston D., et al. , 2001. Centrosome migration into the Drosophila oocyte is independent of BicD and egl, and of the organisation of the microtubule cytoskeleton. Development 128: 1889–1897. [DOI] [PubMed] [Google Scholar]
- Brendza R. P., Serbus L. R., Saxton W. M., Duffy J. B., 2002. Posterior localization of dynein and dorsal-ventral axis formation depend on kinesin in Drosophila oocytes. Curr. Biol. 12: 1541–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter A. T., 1994. Egalitarian and the choice of cell fates in Drosophila melanogaster oogenesis. Ciba Found. Symp. 182: 223–246, discussion 246–254. [PubMed] [Google Scholar]
- Cha B. J., Serbus L. R., Koppetsch B. S., Theurkauf W. E., 2002. Kinesin I-dependent cortical exclusion restricts pole plasm to the oocyte posterior. Nat. Cell Biol. 4: 592–598. [DOI] [PubMed] [Google Scholar]
- Clark I., Giniger E., Ruohola-Baker H., Jan L. Y., Jan Y. N., 1994. Transient posterior localization of a kinesin fusion protein reflects anteroposterior polarity of the Drosophila oocyte. Curr. Biol. 4: 289–300. [DOI] [PubMed] [Google Scholar]
- Clark I. E., Jan L. Y., Jan Y. N., 1997. Reciprocal localization of Nod and kinesin fusion proteins indicates microtubule polarity in the Drosophila oocyte, epithelium, neuron and muscle. Development 124: 461–470. [DOI] [PubMed] [Google Scholar]
- Deng W., Lin H., 2001. Asymmetric germ cell division and oocyte determination during Drosophila oogenesis. Int. Rev. Cytol. 203: 93–138. [DOI] [PubMed] [Google Scholar]
- Deng W. M., Ruohola-Baker H., 2000. Laminin A is required for follicle cell-oocyte signaling that leads to establishment of the anterior-posterior axis in Drosophila. Curr. Biol. 10: 683–686. [DOI] [PubMed] [Google Scholar]
- Dienstbier M., Boehl F., Li X., Bullock S. L., 2009. Egalitarian is a selective RNA-binding protein linking mRNA localization signals to the dynein motor. Genes Dev. 23: 1546–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doerflinger H., Benton R., Torres I. L., Zwart M. F., St Johnston D., 2006. Drosophila anterior-posterior polarity requires actin-dependent PAR-1 recruitment to the oocyte posterior. Curr. Biol. 16: 1090–1095. [DOI] [PubMed] [Google Scholar]
- Doerflinger H., Vogt N., Torres I. L., Mirouse V., Koch I., et al. , 2010. Bazooka is required for polarisation of the Drosophila anterior-posterior axis. Development 137: 1765–1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driever W., Nusslein-Volhard C., 1988. A gradient of bicoid protein in Drosophila embryos. Cell 54: 83–93. [DOI] [PubMed] [Google Scholar]
- Duncan J. E., Warrior R., 2002. The cytoplasmic dynein and kinesin motors have interdependent roles in patterning the Drosophila oocyte. Curr. Biol. 12: 1982–1991. [DOI] [PubMed] [Google Scholar]
- Eisman R. C., Phelps M. A., Kaufman T., 2015. An amino-terminal Polo kinase interaction motif acts in the regulation of centrosome formation and reveals a novel function for centrosomin (cnn) in Drosophila. Genetics 201: 685–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ephrussi A., Lehmann R., 1992. Induction of germ cell formation by oskar. Nature 358: 387–392. [DOI] [PubMed] [Google Scholar]
- Ephrussi A., Dickinson L. K., Lehmann R., 1991. oskar organizes the germ plasm and directs localization of the posterior determinant nanos. Cell 66: 37–50. [DOI] [PubMed] [Google Scholar]
- Frydman H. M., Spradling A. C., 2001. The receptor-like tyrosine phosphatase lar is required for epithelial planar polarity and for axis determination within drosophila ovarian follicles. Development 128: 3209–3220. [DOI] [PubMed] [Google Scholar]
- Gavis E. R., Lehmann R., 1992. Localization of nanos RNA controls embryonic polarity. Cell 71: 301–313. [DOI] [PubMed] [Google Scholar]
- Gonsalvez G. B., Long R. M., 2012. Spatial regulation of translation through RNA localization. F1000 Biol. Rep. 4: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Reyes A., 2003. Stem cells, niches and cadherins: a view from Drosophila. J. Cell Sci. 116: 949–954. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Reyes A., St Johnston D., 1998. Patterning of the follicle cell epithelium along the anterior-posterior axis during Drosophila oogenesis. Development 125: 2837–2846. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Reyes A., Elliott H., St Johnston D., 1995. Polarization of both major body axes in Drosophila by gurken-torpedo signalling. Nature 375: 654–658. [DOI] [PubMed] [Google Scholar]
- Huang H., Li Y., Szulwach K. E., Zhang G., Jin P., et al. , 2014. AGO3 Slicer activity regulates mitochondria-nuage localization of Armitage and piRNA amplification. J. Cell Biol. 206: 217–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh J. R., St Johnston D., 2000. The role of BicD, egl, orb and the microtubules in the restriction of meiosis to the Drosophila oocyte. Development 127: 2785–2794. [DOI] [PubMed] [Google Scholar]
- Huynh J. R., St Johnston D., 2004. The origin of asymmetry: early polarisation of the Drosophila germline cyst and oocyte. Curr. Biol. 14: R438–R449. [DOI] [PubMed] [Google Scholar]
- Jambor H., Surendranath V., Kalinka A. T., Mejstrik P., Saalfeld S., et al. , 2015. Systematic imaging reveals features and changing localization of mRNAs in Drosophila development. eLife 4 10.7554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Januschke J., Gervais L., Dass S., Kaltschmidt J. A., Lopez-Schier H., et al. , 2002. Polar transport in the Drosophila oocyte requires Dynein and Kinesin I cooperation. Curr. Biol. 12: 1971–1981. [DOI] [PubMed] [Google Scholar]
- Johnstone O., Lasko P., 2001. Translational regulation and RNA localization in Drosophila oocytes and embryos. Annu. Rev. Genet. 35: 365–406. [DOI] [PubMed] [Google Scholar]
- Kim-Ha J., Smith J. L., Macdonald P. M., 1991. oskar mRNA is localized to the posterior pole of the Drosophila oocyte. Cell 66: 23–35. [DOI] [PubMed] [Google Scholar]
- Kim-Ha J., Kerr K., Macdonald P. M., 1995. Translational regulation of oskar mRNA by bruno, an ovarian RNA-binding protein, is essential. Cell 81: 403–412. [DOI] [PubMed] [Google Scholar]
- Klattenhoff C., Bratu D. P., McGinnis-Schultz N., Koppetsch B. S., Cook H. A., et al. , 2007. Drosophila rasiRNA pathway mutations disrupt embryonic axis specification through activation of an ATR/Chk2 DNA damage response. Dev. Cell 12: 45–55. [DOI] [PubMed] [Google Scholar]
- Koch R., Ledermann R., Urwyler O., Heller M., Suter B., 2009. Systematic functional analysis of Bicaudal-D serine phosphorylation and intragenic suppression of a female sterile allele of BicD. PLoS One 4: e4552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G., Sanghavi P., Bollinger K. E., Perry L., Marshall B., et al. , 2015. Efficient endocytic uptake and maturation in Drosophila oocytes requires Dynamitin/p50. Genetics 201: 631–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDougall N., Lad Y., Wilkie G. S., Francis-Lang H., Sullivan W., et al. , 2001. Merlin, the Drosophila homologue of neurofibromatosis-2, is specifically required in posterior follicle cells for axis formation in the oocyte. Development 128: 665–673. [DOI] [PubMed] [Google Scholar]
- MacDougall N., Clark A., MacDougall E., Davis I., 2003. Drosophila gurken (TGFalpha) mRNA localizes as particles that move within the oocyte in two dynein-dependent steps. Dev. Cell 4: 307–319. [DOI] [PubMed] [Google Scholar]
- Mach J. M., Lehmann R., 1997. An Egalitarian-BicaudalD complex is essential for oocyte specification and axis determination in Drosophila. Genes Dev. 11: 423–435. [DOI] [PubMed] [Google Scholar]
- Markussen F. H., Michon A. M., Breitwieser W., Ephrussi A., 1995. Translational control of oskar generates short OSK, the isoform that induces pole plasma assembly. Development 121: 3723–3732. [DOI] [PubMed] [Google Scholar]
- McGrail M., Gepner J., Silvanovich A., Ludmann S., Serr M., et al. , 1995. Regulation of cytoplasmic dynein function in vivo by the Drosophila Glued complex. J. Cell Biol. 131: 411–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megraw T. L., Li K., Kao L. R., Kaufman T. C., 1999. The centrosomin protein is required for centrosome assembly and function during cleavage in Drosophila. Development 126: 2829–2839. [DOI] [PubMed] [Google Scholar]
- Meignin C., Alvarez-Garcia I., Davis I., Palacios I. M., 2007. The Salvador-Warts-Hippo pathway is required for epithelial proliferation and axis specification in Drosophila. Curr. Biol. 17: 1871–1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montell D. J., 2006. The social lives of migrating cells in Drosophila. Curr. Opin. Genet. Dev. 16: 374–383. [DOI] [PubMed] [Google Scholar]
- Navarro C., Puthalakath H., Adams J. M., Strasser A., Lehmann R., 2004. Egalitarian binds dynein light chain to establish oocyte polarity and maintain oocyte fate. Nat. Cell Biol. 6: 427–435. [DOI] [PubMed] [Google Scholar]
- Neuman-Silberberg F. S., Schüpbach T., 1993. The Drosophila dorsoventral patterning gene gurken produces a dorsally localized RNA and encodes a TGF alpha-like protein. Cell 75: 165–174. [PubMed] [Google Scholar]
- Neuman-Silberberg F. S., Schüpbach T., 1996. The Drosophila TGF-α-like protein Gurken: expression and cellular localization during Drosophila oogenesis. Mech. Dev. 59: 105–113. [DOI] [PubMed] [Google Scholar]
- Ni J. Q., Zhou R., Czech B., Liu L. P., Holderbaum L., et al. , 2011. A genome-scale shRNA resource for transgenic RNAi in Drosophila. Nat. Methods 8: 405–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pane A., Wehr K., Schüpbach T., 2007. zucchini and squash encode two putative nucleases required for rasiRNA production in the Drosophila germline. Dev. Cell 12: 851–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parton R. M., Hamilton R. S., Ball G., Yang L., Cullen C. F., et al. , 2011. A PAR-1-dependent orientation gradient of dynamic microtubules directs posterior cargo transport in the Drosophila oocyte. J. Cell Biol. 194: 121–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polesello C., Tapon N., 2007. Salvador-Warts-Hippo signaling promotes Drosophila posterior follicle cell maturation downstream of notch. Curr. Biol. 17: 1864–1870. [DOI] [PubMed] [Google Scholar]
- Riechmann V., Ephrussi A., 2001. Axis formation during Drosophila oogenesis. Curr. Opin. Genet. Dev. 11: 374–383. [DOI] [PubMed] [Google Scholar]
- Roth S., Neuman-Silberberg F. S., Barcelo G., Schüpbach T., 1995. cornichon and the EGF receptor signaling process are necessary for both anterior-posterior and dorsal-ventral pattern formation in Drosophila. Cell 81: 967–978. [DOI] [PubMed] [Google Scholar]
- Ruohola H., Bremer K. A., Baker D., Swedlow J. R., Jan L. Y., et al. , 1991. Role of neurogenic genes in establishment of follicle cell fate and oocyte polarity during oogenesis in Drosophila. Cell 66: 433–449. [DOI] [PubMed] [Google Scholar]
- Salles F. J., Lieberfarb M. E., Wreden C., Gergen J. P., Strickland S., 1994. Coordinate initiation of Drosophila development by regulated polyadenylation of maternal messenger RNAs. Science 266: 1996–1999. [DOI] [PubMed] [Google Scholar]
- Sanghavi P., Lu S., Gonsalvez G. B., 2012. A functional link between localized Oskar, dynamic microtubules, and endocytosis. Dev. Biol. 367: 66–77. [DOI] [PubMed] [Google Scholar]
- Sanghavi P., Laxani S., Li X., Bullock S. L., Gonsalvez G. B., 2013. Dynein associates with oskar mRNPs and is required for their efficient net plus-end localization in Drosophila oocytes. PLoS One 8: e80605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuyler S. C., Pellman D., 2001. Microtubule “plus-end-tracking proteins”: the end is just the beginning. Cell 105: 421–424. [DOI] [PubMed] [Google Scholar]
- Shulman J. M., Benton R., St Johnston D., 2000. The Drosophila homolog of C. elegans PAR-1 organizes the oocyte cytoskeleton and directs oskar mRNA localization to the posterior pole. Cell 101: 377–388. [DOI] [PubMed] [Google Scholar]
- Smith J. L., Wilson J. E., Macdonald P. M., 1992. Overexpression of oskar directs ectopic activation of nanos and presumptive pole cell formation in Drosophila embryos. Cell 70: 849–859. [DOI] [PubMed] [Google Scholar]
- Spradling A., Drummond-Barbosa D., Kai T., 2001. Stem cells find their niche. Nature 414: 98–104. [DOI] [PubMed] [Google Scholar]
- Spradling A. C., 1993. Developmental genetics of oogenesis, The Development of Drosophila melanogaster, edited by M. Bate, Martinez Arias A. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Steinhauer J., Kalderon D., 2006. Microtubule polarity and axis formation in the Drosophila oocyte. Dev. Dyn. 235: 1455–1468. [DOI] [PubMed] [Google Scholar]
- St Johnston D., 2005. Moving messages: the intracellular localization of mRNAs. Nat. Rev. Mol. Cell Biol. 6: 363–375. [DOI] [PubMed] [Google Scholar]
- St Johnston D., Ahringer J., 2010. Cell polarity in eggs and epithelia: parallels and diversity. Cell 141: 757–774. [DOI] [PubMed] [Google Scholar]
- St Johnston D., Beuchle D., Nusslein-Volhard C., 1991. staufen, a gene required to localize maternal RNAs in the Drosophila egg. Cell 66: 51–63. [DOI] [PubMed] [Google Scholar]
- Sullivan W., Ashburner M., Hawley R. S., 2000. Drosophila Protocols, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Swan A., Nguyen T., Suter B., 1999. Drosophila Lissencephaly-1 functions with Bic-D and dynein in oocyte determination and nuclear positioning. Nat. Cell Biol. 1: 444–449. [DOI] [PubMed] [Google Scholar]
- Theurkauf W. E., Smiley S., Wong M. L., Alberts B. M., 1992. Reorganization of the cytoskeleton during Drosophila oogenesis: implications for axis specification and intercellular transport. Development 115: 923–936. [DOI] [PubMed] [Google Scholar]
- Theurkauf W. E., Alberts B. M., Jan Y. N., Jongens T. A., 1993. A central role for microtubules in the differentiation of Drosophila oocytes. Development 118: 1169–1180. [DOI] [PubMed] [Google Scholar]
- Tomancak P., Piano F., Riechmann V., Gunsalus K. C., Kemphues K. J., et al. , 2000. A Drosophila melanogaster homologue of Caenorhabditis elegans par-1 acts at an early step in embryonic-axis formation. Nat. Cell Biol. 2: 458–460. [DOI] [PubMed] [Google Scholar]
- Vazquez-Pianzola P., Adam J., Haldemann D., Hain D., Urlaub H., et al. , 2014. Clathrin heavy chain plays multiple roles in polarizing the Drosophila oocyte downstream of Bic-D. Development 141: 1915–1926. [DOI] [PubMed] [Google Scholar]
- Weil T. T., 2014. mRNA localization in the Drosophila germline. RNA Biol. 11: 1010–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weil T. T., Forrest K. M., Gavis E. R., 2006. Localization of bicoid mRNA in late oocytes is maintained by continual active transport. Dev. Cell 11: 251–262. [DOI] [PubMed] [Google Scholar]
- Wilhelm J. E., Smibert C. A., 2005. Mechanisms of translational regulation in Drosophila. Biol. Cell 97: 235–252. [DOI] [PubMed] [Google Scholar]
- Yan Y., Denef N., Tang C., Schüpbach T., 2011. Drosophila PI4KIIIalpha is required in follicle cells for oocyte polarization and Hippo signaling. Development 138: 1697–1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Poulton J., Huang Y. C., Deng W. M., 2008. The hippo pathway promotes Notch signaling in regulation of cell differentiation, proliferation, and oocyte polarity. PLoS One 3: e1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimyanin V. L., Belaya K., Pecreaux J., Gilchrist M. J., Clark A., et al. , 2008. In vivo imaging of oskar mRNA transport reveals the mechanism of posterior localization. Cell 134: 843–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All strains used in this work will be made available upon request. The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.