Abstract
Detecting the molecular basis of adaptation is one of the major questions in population genetics. With the advance in sequencing technologies, nearly complete interrogation of genome-wide polymorphisms in multiple populations is becoming feasible in some species, with the expectation that it will extend quickly to new ones. Here, we investigate the advantages of sequencing for the detection of adaptive loci in multiple populations, exploiting a recently published data set in cattle (Bos taurus). We used two different approaches to detect statistically significant signals of positive selection: a within-population approach aimed at identifying hard selective sweeps and a population-differentiation approach that can capture other selection events such as soft or incomplete sweeps. We show that the two methods are complementary in that they indeed capture different kinds of selection signatures. Our study confirmed some of the well-known adaptive loci in cattle (e.g., MC1R, KIT, GHR, PLAG1, NCAPG/LCORL) and detected some new ones (e.g., ARL15, PRLR, CYP19A1, PPM1L). Compared to genome scans based on medium- or high-density SNP data, we found that sequencing offered an increased detection power and a higher resolution in the localization of selection signatures. In several cases, we could even pinpoint the underlying causal adaptive mutation or at least a very small number of possible candidates (e.g., MC1R, PLAG1). Our results on these candidates suggest that a vast majority of adaptive mutations are likely to be regulatory rather than protein-coding variants.
Keywords: FST, domestication, linkage disequilibrium, next-generation sequencing, selective sweeps
DETECTING the molecular basis of adaptation in natural species is one of the major questions in population genetics. With the spectacular progress of genotyping and sequencing technologies, genome-wide scans for positive selection have been performed in multiple species and populations within the last decade. Livestock species provide a considerable resource for these selection scans, because they have been subjected to strong artificial selection since their initial domestication, leading to a large variety of breeds with distinct morphology, coat color, or specialized production. In addition, the economic value of these species and the need to improve them has motivated the development of standardized single-nucleotide polymorphism (SNP) chips and the genotyping of millions of animals using these chips, providing considerable data for population genetics analyses. For instance, in taurine cattle, at least 21 genomic scans for selection have already been published and were reviewed in Gutierrez-Gil et al. (2015). Numerous genomic scans for selection have also been published in other livestock species; see de Simoni Gouveia et al. (2014) for a review.
The regions detected by these studies are generally convincing, because they contain interesting positional and functional candidate genes (e.g., Fariello et al. 2014) and/or are statistically enriched with genes from regulation pathways related to production traits (e.g., Flori et al. 2009). Nevertheless, these regions often span several megabases and typically include up to tens of genes, so determining the exact gene(s) under selection in each region, and even more the causal mutation(s), remains difficult from these studies. This might change with the recent advent of next-generation sequencing (NGS) technologies, which allow one to characterize a very large proportion of the variants in the genome of a species. However, although several examples of genomic scans for selection based on large sequencing samples are already available in livestock species such as pig (Rubin et al. 2012; Li et al. 2013), cattle (Qanbari et al. 2014), or chicken (Rubin et al. 2010; Roux et al. 2015), the advantage of using sequencing data for detecting selection signatures has still not been widely discussed.
Another interesting question is the small overlap observed between studies, even when these studies focus on similar populations (Biswas and Akey 2006; Qanbari et al. 2011). This can largely be explained by the fact that many different detection methods have been applied, whose sensitivity depends on the type of selection event: recent or old, complete or ongoing, from a new variant (i.e., “hard”) or standing variation (i.e., “soft”), etc. These differences between methods have been described by several studies (Biswas and Akey 2006; Sabeti et al. 2006; de Simoni Gouveia et al. 2014). However, the practical implications of these differences when comparing the regions detected by different studies, or by different approaches within the same study, are rarely discussed.
Here we detect selection signatures in four European taurine cattle breeds (Angus, Fleckvieh, Holstein, and Jersey), using large samples of sequencing data that have been recently published by the 1000 bull genomes project (Daetwyler et al. 2014). We use two different statistical approaches: a within-breed approach detecting genomic regions with low genetic diversity (Boitard et al. 2009) and a between-population approach detecting genomic regions with large allele (Bonhomme et al. 2010) or haplotype (Fariello et al. 2013) frequency differences between breeds. These two analyses provide regions whose genomic features are significantly different from what would be expected under neutrality, even when accounting for the effects of past population size changes, population structure, and gene flow on the four breeds of our study. We show that applying the above methods to sequencing data improves the detection power of selection signatures and reduces considerably the length of detected regions. In some particular situations, it even leads to identifying the exact mutation under selection. We also provide a detailed characterization of the regions that are detected only by the within-population approach (in one or several populations), only by the between-population approach, or jointly by the two approaches.
Materials and Methods
Samples and sequencing
A total of 234 genome sequences were obtained from the 1000 bull genomes project, run II (Daetwyler et al. 2014). These included 129 Holsteins (125 Black and 4 Red), 43 Fleckviehs, 47 Angus, and 15 Jerseys. We considered all these sequences except the 4 Red Holsteins. We based our analyses on the phased and corrected autosomal data produced by the 1000 bull genomes project (Daetwyler et al. 2014). These data included 27,535,425 biallelic single nucleotide polymorphisms (SNPs) and 1,507,728 biallelic indels.
Choice of unrelated animals
To remove potential biases arising from sample size heterogeneity between breeds and inbreeding within breeds, we selected a subset of 25 unrelated animals in Holstein, Fleckvieh, and Angus breeds, while keeping all 15 Jersey animals. Within each breed, we computed the genetic relationship matrix (GRM) of all available animals based on SNPs from chromosome 1 with minor allele frequency (MAF) >10%, using the GCTA 1.04 software (Yang et al. 2011). We then selected unrelated animals as follows. First, we removed all animals with inbreeding value (the diagonal term of the GRM) >1.5 (this threshold was chosen because inbreeding values >1.5 clearly appeared as outliers compared to the rest of the distribution; Supplemental Material, Figure S1). Second, we considered all animal pairs with genetic relationship >0.3 in absolute value and removed the most inbred animal of each pair. Third, we performed a hierarchical clustering of the remaining animals based on the distance where is the genetic relationship between animals i and j and M is the maximum value of the GRM, and sampled the 25 most distant animals.
Estimation of a demographic model for the joint history of the four breeds
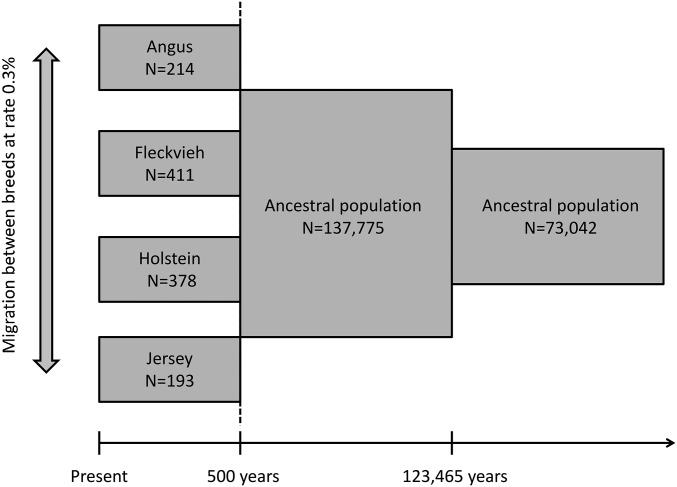
To model the demographic history of the four breeds under study, we assumed that these breeds diverged simultaneously from a common ancestral population, generations ago. Although the population tree estimated by FLK (Bonhomme et al, 2010) would suggest a slightly more recent divergence between Fleckvieh and Jersey (Figure 4), we considered that this difference was negligible and preferred reducing the number of parameters to be estimated. Based on previous studies suggesting that effective population size in taurine cattle strongly declined since domestication (MacLeod et al. 2013; Boitard et al. 2016), we allowed one population size change from (the ancestral population size) to (the “domestic” population size) in the ancestral population, generations ago. The divergence between breeds implied a second population size change: after this event, each breed i was assumed to have a specific population size which possibly differed from Finally, we assumed that each breed received, every generation, a proportion m of migrants from any of the other breeds.
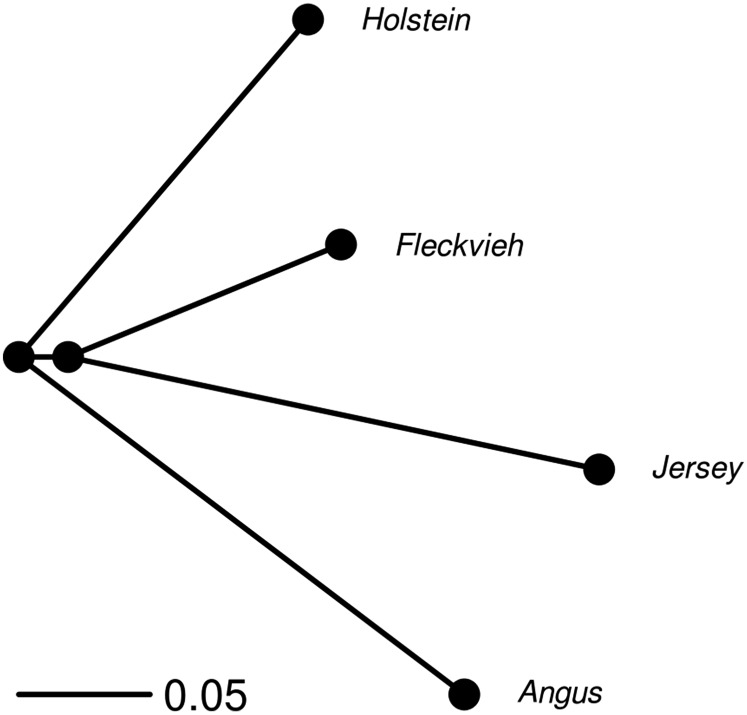
Figure 4.
Population tree estimated from the 1000 bull genomes data. Branch length is measured in units of drift (t/2N, where t is the time in generations and N the effective population size).
We estimated the parameters of this model using the composite-likelihood approach implemented in fastsimcoal2 (Excoffier et al. 2013), which is based on the joint site frequency spectrum (SFS). In our data set, this joint SFS had a very high dimension (51 51 51 31). Consequently, we instead considered the collection of joint SFS obtained for all six population pairs (Angus Fleckvieh, Angus Holstein, …), following the recommendations of the authors. We computed these observed joint SFS from our data using home-made scripts and provided them as input to fastsimcoal2, version 5.2.8 (May 2015). We performed 50 independent EM estimations and selected the one with the highest composite likelihood. For each estimation, we used the folded SFS (option -m) and the default settings −n100000 −N100000 −M0.001 −l10 −L40. In fastsimcoal2, time is scaled in generations. Time in years was obtained by assuming a generation time of 5 years.
Detection of genomic regions with low within-breed diversity
Model:
We looked for hard-sweep signatures within each breed, using the hidden Markov model (HMM) of Boitard et al. (2009). In this model, only biallelic variants are considered and the derived allele frequency at variant i, denoted is taken as the observed state of the HMM at this position. Each variant i is also assumed to have a hidden state which can take three different values: “selection,” for variants that are very close to a swept site; “neutral,” for variants that are far away from any swept site; and “intermediate,” for variants in between. These three values are associated with different allele frequency distributions. The neutral allele frequency distribution is estimated using all variants in the genome, assuming most of them have indeed evolved under neutrality. Allele frequency distributions in the intermediate and selection states are deduced from this neutral distribution using the derivations in Nielsen et al. (2005) and are typically more skewed toward extreme allele frequencies. The hidden states form a Markov chain along the genome with a per base pair probability p of switching state, so that close variants tend to be in the same hidden state. Under this HMM, the most likely sequence of hidden states can be predicted from the sequence of observed states, using the Viterbi algorithm. Each set of consecutive variants with predicted state selection is called a sweep window. The method of Boitard et al. (2009) is implemented in the freq-hmm program, available at https://forge-dga.jouy.inra.fr/projects/pool-hmm.
Implementation:
Ancestral Bovinae alleles at 448,289 SNPs included in the Illumina BovineHD BeadChip were obtained from Utsunomiya et al. (2013). To check this information we also aligned the bovine sequence against the sequence of three Bovidae species (Rocha et al. 2014). For 365,146 SNPs, the ancestral allele in our alignment was consistent with that reported by Utsunomiya et al. (2013) so we used this information for sweep detection. For all other SNPs, we used a folded allele frequency distribution; i.e., allele frequencies and were considered as the same observed state. Indels were not included at this stage of the analysis (they were considered only when looking at candidate polymorphisms within the region).
To reduce computation time, we estimated the neutral allele frequency distributions in each breed, using only 5% of the SNPs from each chromosome, which were selected at random. These SFS are shown in Figure S2.
The type I error of the above method, i.e., the probability that it detects a sweep window in a population that has evolved under neutrality, depends on parameter p (see Boitard et al. 2009 for more details). To control the genome-wide number of false positives, we simulated 5000 samples of length 500 kb under neutral evolution using ms (Hudson 2002) and adjusted p so that sweeps were detected in only 0.1% of these samples. We performed this calibration for each breed, using the same sample size and proportion of unfolded sites as in the data used for sweep detection. Parameter θ was also estimated from these data using Waterson’s estimator (Watterson 1975) and ρ was taken equal to which is rather low compared to current estimates in cattle (Sandor et al. 2012; Ma et al. 2015). As the detection sensitivity of the HMM method increases when decreases (Boitard et al. 2009), our adjusted value of p should be conservative. Assuming a 2.5-Gb genome as that of bovine (focusing on autosomes) is equivalent to 5000 windows of 500 kb, we expected no more than five false positive signals over the genome with this value of p.
Influence of demography on sweep detection:
We evaluated the robustness of the above detection approach under two neutral demographic scenarios: the multipopulation model estimated by fastsimcoal2 (Figure 1) and the single-population model estimated in Boitard et al. (2016). For each model, we simulated 20,000 samples of length 500 kb using ms, assuming a mutation rate and a recombination rate of 1e-8 per base pair and generation, and we applied the HMM procedure to these samples. For the multipopulation model, each simulated sample included genomes from the four breeds, which were split to apply the HMM within each population. For the single-population scenario, breed-specific samples were directly simulated independently of each other, each breed being associated to a different population size history. For each scenario and breed, the total length of simulated samples was equivalent to that of four cattle genomes. Consequently, the estimated proportion m of false positive signals per genome was given by the total number of sweeps detected in the simulations, divided by four, and the variance of this estimation was equal to The confidence interval of this estimation was approximated by
Figure 1.
Joint demography of the four cattle breeds, estimated from our data using the approach of Excoffier et al. (2013). Population sizes correspond to the number of haploid individuals. Parameter values correspond to the EM iteration with the highest likelihood, but similar values were obtained from the second- and third-best EM iterations.
Comparing hard-sweep signatures detected in different populations:
For a given variant i, the evidence for a hard-sweep window occurring around this variant in population j was measured by the statistic where was the posterior probability of hidden state selection returned by the backward–forward algorithm applied to the HMM in population j. When considering a given region of the genome, the evidence for a hard sweep in population j was quantified by the median of over the variants of the region. To detect selection signatures that are really breed specific, we computed for each breed the distribution of in three different classes of regions: (i) those where a hard sweep was detected in this breed, (ii) those where a hard sweep was detected in another breed, and (iii) those where no hard sweep was detected (Figure S3). Obviously, class i and class iii regions lead to very different distributions, with lower values in the latter (i.e., in the completely neutral regions). In addition, the distribution of in class ii regions was not similar to that found in class iii regions, but was shifted toward that of class i regions. Based on these observations, we therefore considered that a hard sweep was breed specific when, in all other breeds, the value of of the region was below a given quantile q of the class iii distribution. For 55 breed-specific sweeps were detected (listed in File S1) and for 12 breed-specific regions were detected.
Detection of genomic regions with large differentiation between breeds
We applied two methods for the detection of genomic regions exhibiting large genetic differentiation between populations: FLK (Bonhomme et al. 2010), a single-marker approach based on allele frequency differences, and its haplotypic extension hapFLK (Fariello et al. 2013), which exploits the linkage disequilibrium information to capture differences of haplotype frequencies.
Kinship matrix:
In contrast to the statistic, FLK and hapFLK account for the population history through a kinship matrix, which captures (i) differences in effective population sizes between populations and (ii) possible shared ancestry between populations. The kinship matrix is inferred from a population tree, with branch length expressed in units of drift, i.e., measured in fixation indexes where t is the number of generations from the root and N the effective population size. We estimated the population tree, using neighbor joining on the Reynold’s genetic distances between populations (see Bonhomme et al. 2010 for details). We used the ancestral allele reconstruction of Utsunomiya et al. (2013) to root the population tree and estimate the population kinship matrix.
FLK:
For the single-marker analysis, we performed the FLK test on all variants and computed P-values using the theoretical distribution, which was a good fit to the observed distribution (Figure S4).
hapFLK:
For the hapFLK test, to save computation time, we removed variants that had low minor allele frequency () in all breeds. Note that as the analysis looks for signals of differentiation, removing these variants does not preclude detection. In subsequent reanalysis of small genomic regions, we kept all variants in the analysis. hapFLK makes use of the local clustering approach in Scheet and Stephens (2006) to model haplotype diversity. This model requires specifying a number of haplotype clusters as the input parameter. Using the cross-validation procedure implemented in the fastPHASE software, we found that 15 clusters provided the smallest imputation error rate. hapFLK can be computed on unphased or phased genotype data. Genotype calling made used of imputation approaches, and data were therefore already phased (Daetwyler et al. 2014). We computed hapFLK both on the haplotype data and on the genotype (but imputed) data and found the two analyses provided similar results (not shown).
The distribution of hapFLK is not known, but, from theoretical arguments hapFLK is a deviance statistic. However, the variance parameter of this deviance is not known. If this parameter was known, then hapFLK should follow a distribution, where N is the number of populations and K the number of haplotype clusters. Building on this fact, we compared a set of quantiles (from 0.05 to 0.95 every 0.05) of the distribution to the observed quantiles of the hapFLK statistic We found that the relationship between and was very close to linear (Figure S5). Thus, we used the parameter of the linear model to scale the hapFLK statistic to a distribution that was then used to compute P-values.
Extracting significantly differentiated regions:
To call significant regions, we applied the approach of Storey and Tibshirani (2003), aimed at controlling the false discovery rate (FDR), at the 15% level; i.e., we called significant variants with q-values and selection signatures regions where more than one variant was called significant. We note, however, that our level of control of the FDR for the regions themselves may not be well calibrated as the tests are correlated. This procedure still provides a significant threshold so that among marker discoveries there is a sixfold enrichment in favor of the number of statistics under the alternative.
Influence of demography on hapFLK:
To evaluate the robustness to demography of hapFLK and our scaling approach, we performed 10,000 simulations of 50-kb windows under the population model estimated with fastsimcoal2. We applied the same testing procedure as with the real data, including filtering out SNPs with low minor allele frequencies in all breeds. The resulting hapFLK distribution was different from the one observed on real data; in particular it showed a depletion of low hapFLK values compared to real data; i.e., on real data some regions look more similar between breeds than on simulated data. The reasons for this are not clear, but we note that (i) fastsimcoal2 estimation does not use haplotype or linkage disequilibrium information so the haplotype patterns simulated are not expected to necessarily fit the data; (ii) simulations assume homogeneous recombination and mutation rates, which does not hold on real data; and (iii) common background/purifying selection between breeds might reduce differentiation in some genome regions. Despite the lesser hapFLK variance in simulations, scaling the hapFLK distribution to a with 14 d.f. provided a very good fit (see Figure S6). Applying the Storey and Tibshirani (2003) approach to estimate the proportion of alternative hypotheses in the resulting P-value distribution led to an estimate of 0 (i.e., ). A python script to perform the scaling of hapFLK to distributions is now available on the hapFLK webpage: https://forge-dga.jouy.inra.fr/projects/hapflk/documents.
Data availability
All data necessary for confirming the conclusions presented in the article are represented fully within the article or cited references.
Results
Genomic regions with low within-population diversity: hard-sweep signatures
We looked for hard-sweep signatures within each breed, using the method of Boitard et al. (2009), as described in Materials and Methods. This method detects regions showing an excess of low- and high-frequency derived alleles compared to the rest of the genome. Although this pattern is typically expected under a hard-sweep scenario, i.e., when a new mutation appears in the population and goes to fixation due to positive selection, it may also arise from purely demographic events, in particular bottlenecks. To test whether our analysis could be influenced by such false positive signals, we first simulated genomic samples under two different neutral demographic models, which both allow to reproduce the genetic diversity of the breeds under study, and applied the method of Boitard et al. (2009) to these samples.
Analysis of neutral samples:
In the first demographic model, we considered the joint history of the four breeds and accounted for several important features of this history: (i) the shared ancestry of the four breeds, which diverged recently from an ancestral pool of European domestic animals; (ii) the population size differences between breeds since their divergence; and (iii) the possible existence of gene flow between breeds. Moreover, because recent studies suggested that effective population size in taurine cattle strongly declined since domestication (MacLeod et al. 2013; Boitard et al. 2016), we allowed one population size change in the ancestral population. We estimated the parameters of this model from the joint allele frequency spectra observed in our data for all breed pairs, using the approach of Excoffier et al. (2013) (see Materials and Methods for more details), and obtained the demography shown in Figure 1.
In this estimated demography, the ancestral population size change was not a decline related to domestication, but an older expansion occurring in the wild population, ∼120,000 years before present. However, a very strong population decline was found at the time where the four breeds diverged, from an order of 100,000 individuals to an order of 100 individuals. Interestingly, the estimated divergence time (500 years before present) was consistent with a geographic isolation process starting a few hundred years before the strict separation of these populations, induced by the creation of modern breeds (Felius et al. 2011). In addition, the order of magnitude of estimated recent effective sizes (100) and the ranking of breeds according to these sizes were consistent with previous studies (Bovine HapMap Consortium 2009; Leroy et al. 2013; MacLeod et al. 2013; Boitard et al. 2016). Thus, this simple model seemed to provide a reasonable approximation of the demography of the four breeds under study. When genomic samples were simulated from this model, the average number of sweeps detected per genome was equal to 0 in Fleckvieh and Holstein, 3.75 (±1.93) in Angus, and 17.25 (±4.15) in Jersey.
We performed the same test using a second demographic model, which was estimated in another study (Boitard et al. 2016) based on the same data set as that considered here. This model treats each breed independently of the others, so it does not account for shared ancestry or gene flow. However, it accounts for the variations of population size over time more accurately than the previous model, because population size in each breed is modeled as a stepwise process with 21 time windows (figure 6 in Boitard et al. 2016). The population size within each time window is estimated from the allele frequency and linkage disequilibrium patterns observed in the breed, using an approximate Bayesian computation approach. When genomic samples were simulated from this second model, the average number of sweeps detected per genome was even lower than with the first model: 0 in Fleckvieh, Holstein, and Angus and 0.75 (±0.87) in Jersey.
Overall, these results indicate that the number of false hard-sweep signals detected by the method of Boitard et al. (2009) should be negligible in Angus, Fleckvieh, and Holstein and relatively small in Jersey, even when accounting for the demography of these breeds.
Overview of the detected signals:
When analyzing the cattle data with the same approach, we detected 1057 hard-sweep signals: 226, 384, 316, and 131 in Holstein, Angus, Jersey, and Fleckvieh, respectively. According to the simulation results presented above, the false discovery rate associated with this analysis should be <7% in Jersey () and close to 0 in the other breeds. The size of detected regions ranged from 8.2 to 948 kb, with a median of 78.7 kb. Some signals were overlapping between breeds so that after merging them we obtained 798 sweeps that were unique to one of the breeds (159, 297, 249, and 93, respectively) and 118 that were shared between at least two breeds. Overall this provided 916 regions covering ∼4.3% of the autosomal genome. Among these 916 regions, 450 included no (protein-coding) gene, 268 included a single gene, 154 included between 2 and 5 genes, and 44 included >5 genes, with a maximum of 19 genes. Overall, 1088 genes were included in sweeps windows, which represents ∼5.7% of all annotated genes in the bovine genome, so there was a slight enrichment of protein-coding genes within sweep regions. The list of all detected regions and of genes included in these regions is given in File S2.
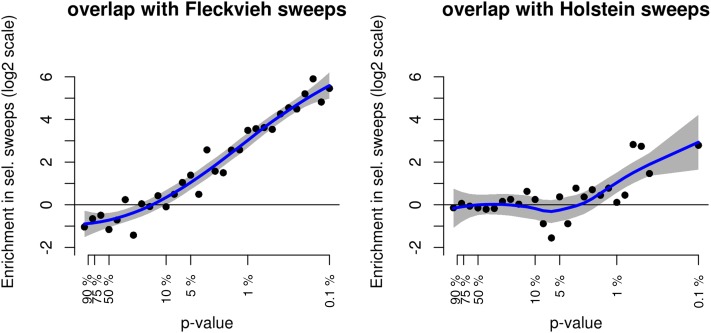
In a recent genome scan for selection focusing on the Fleckvieh breed (Qanbari et al. 2014), the 43 Fleckvieh sequences considered in our study were analyzed using the composite likelihood ratio (CLR) method (Nielsen et al. 2005) and the integrated haplotype score (iHS) method (Voight et al. 2006). Seventy-three hard-sweep signals were found with the former approach and 67 with the latter. Since the HMM approach used in this study aims at capturing the same allele frequency patterns as in the CLR method, we checked whether our results in Fleckvieh were consistent with those in Qanbari et al. (2014). To this end, we compared the distributions of CLR P-values within regions associated with selective sweeps to their distribution on the rest of the genome. Figure 2 (left) plots the ratio of the two densities (on a log2 scale) for increasing levels of significance of the CLR test. It shows a very strong enrichment of low CLR P-values in the sweep windows we detected in Fleckvieh, compared to the rest of the genome. We performed the same analysis with iHS P-values and found a similarly strong enrichment (Figure S7), which can be explained by the fact that iHS also tries to detect hard-sweep patterns, even if the information used (the length of haplotypes) is different.
Figure 2.
Comparison between HMM and CLR results. Shown is the proportion of Fleckvieh CLR P-values (obtained from Qanbari et al. 2014) in sweep windows vs. other windows in the genome. On the left, the sweep windows considered were those detected in Fleckvieh, independently of what happened in other breeds. On the right, the sweep windows considered were those detected only in Holstein.
We also observed an enrichment, albeit of lower intensity, of CLR and iHS low P-values in the sweep windows detected in other breeds than the Fleckvieh. For example, Figure 2 (right) shows the enrichment in low Fleckvieh CLR P-values in selective sweep regions detected only in the Holstein breed. A similar trend was observed in selective sweep regions specific to the Jersey and Angus breeds (not shown). Hence some of the hard sweeps detected in one breed also have probably taken place in the other breeds, but to a slightly lower extent that did not lead to a significant signal. These signatures must be related to favorable alleles that either started to increase in frequency before the divergence of the breeds or were selected in parallel in different breeds. However, selection signatures that are specific to one breed are interesting because they illustrate the importance of this breed for cattle functional diversity (Gutierrez-Gil et al. 2015). We therefore derived a way of finding clear breed-specific sweeps as follows.
Hard-sweep regions specific to one population:
The HMM approach “tags” a region as selected by reconstructing a hidden state at each position of the genome. Based on the observation above, we suspected that some regions were not tagged as selected, but might still have a nonnegligible probability of being adaptive under the HMM model, explaining the enrichment patterns observed in Figure 2. To investigate this possibility, we derived a statistic () quantifying the strength of evidence for selection in a breed, measured as the log odds ratio of selection over neutrality in a region (see Materials and Methods for details). As expected, the in one breed showed clearly different distributions in regions tagged as selected in this breed and in regions where no selection was detected in any breed (Figure S3). In regions where selection was detected in another breed, the distribution of was skewed toward higher values compared to clearly neutral regions (Figure S3). We exploited this to call breed-specific sweeps regions where was unambiguously consistent with the neutral density in all other breeds (see Materials and Methods for details). Fifty-five breed-specific regions were detected, and we could check that this time the sweeps specific to Holstein did not show any enrichment in low Fleckvieh CLR P-values (Figure S8). The 12 sweeps exhibiting the most contrasted patterns for are listed in Table S1.
Hard-sweep regions shared by all populations:
We also looked for sweep signals shared by all breeds, as they might correspond to older selection events, anterior to the divergence of the four breeds considered here and possibly related to initial cattle domestication. We found only one region with a sweep detected in all four breeds, but we also considered regions where a sweep was detected in three breeds and where allele frequencies in the fourth one were almost consistent with a sweep. This provided eight candidate regions, four of which include a single gene (Table 1).
Table 1. Sweep regions shared among all breeds.
| Chromosome | Start (Mb) | End (Mb) | Genes |
|---|---|---|---|
| 1 | 1.781 | 1.818 | lincRNA, Polled locus (Allais-Bonnet et al. 2013) |
| 1 | 107.452 | 107.557 | PPM1L |
| 1 | 107.571 | 107.749 | ARL14 |
| 5 | 68.675 | 68.751 | SLC41A2 |
| 7 | 4.574 | 4.745 | FKBP8, ELL, ISYNA1, SSBP4, LRRC25, GDF15 |
| 10 | 59.148 | 59.338 | CYP19A1 |
| 16 | 44.672 | 44.956 | CLSTN1, PIK3CD, TMEM201, SLC25A33 |
| 16 | 45.644 | 45.903 | RERE, SLC45A1 |
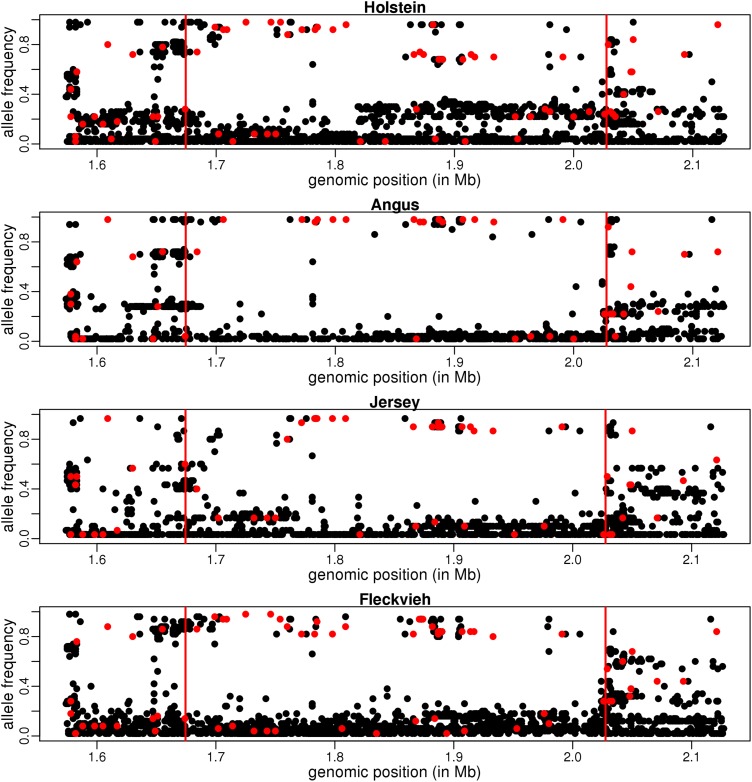
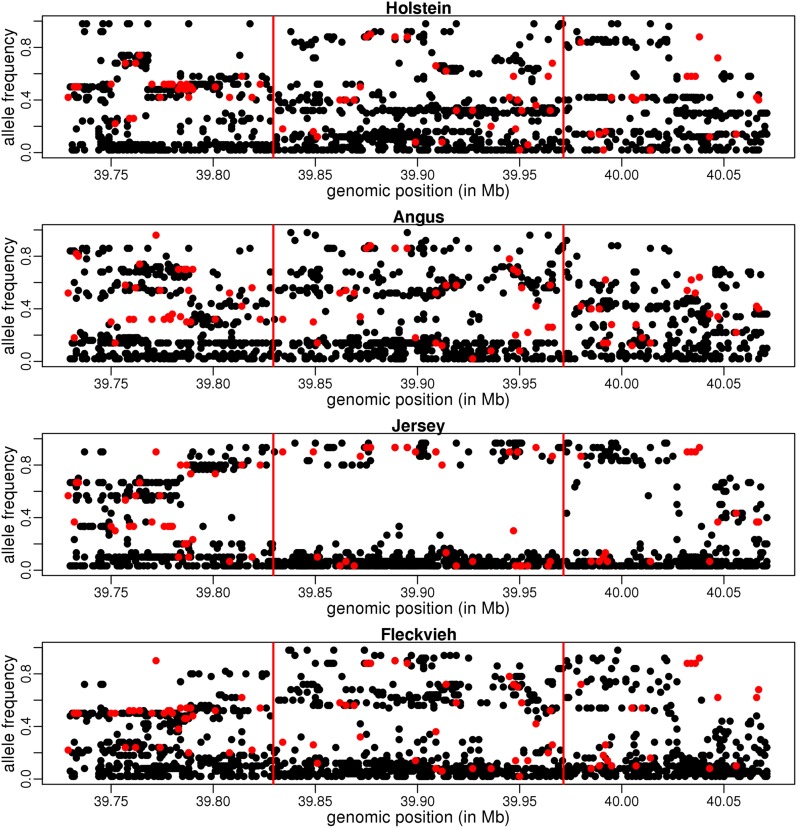
Several of these genes represent natural selection targets in cattle, as they are related to husbandry, metabolism, or fertility. On chromosome 1, we found evidence for selection in a region 10 kb upstream the OLIG1 gene, encompassing a lincRNA, orthologous to the human gene LINC00945, whose expression has been shown to be associated with polledness in Holstein and Fleckvieh (Allais-Bonnet et al. 2013). PPM1L is a protein phosphatase that has been shown to be involved in the response to exercise in humans (Tonevitsky et al. 2013). SLC25A33, located in the middle of one of the shared sweeps windows, encodes for mitochondrial pyrimidine nucleotide transporters and is essential for mitochondrial DNA and RNA metabolism in humans (Di Noia et al. 2014). CYP19A1 encodes the key enzyme for estrogen biosynthesis. Many studies have documented its role during the development of bovine follicles, and it has been found more abundant in bovine cells of twinners vs. controls (Echternkamp et al. 2012). To illustrate the allele frequency patterns observed in such regions, allele frequencies in the polled locus region are provided in Figure 3.
Figure 3.
Allele frequencies in the sweep region at the polled locus. For SNPs where the ancestral allele is known (in red), the frequency is that of the derived allele. For other SNPs (in black) the frequency is that of the minor allele (among all breeds). Vertical red bars delimit the union of detected regions among the four breeds.
Genome regions with large genetic differentiation between breeds
We applied two approaches to detect genome regions that exhibit outlying divergence in single-site (Bonhomme et al. 2010) or haplotype (Fariello et al. 2013) frequencies between populations. The first step in these two approaches is to estimate a population tree summarizing the neutral history of the populations under study. For our data set, this population tree (Figure 4) was approximately star shaped, (i.e., breeds essentially evolved in parallel from an ancestral population), although we estimated a small shared history between the Fleckvieh and Jersey populations. Fixation indexes, represented by the branch length from the root to the tips of the tree, had similar values in all populations, the Jersey’s one being slightly higher. The genome-wide level of differentiation between populations was rather large, with between-population values ranging from 0.22 to 0.4.
When genetic drift is large, single-marker tests are expected to have low power because even large allele frequency differences can be explained by drift alone. This was indeed the case here for the single-marker differentiation analysis (FLK): the smallest observed P-value was which corresponded to an FDR of ∼10% when applying the approach of Storey and Tibshirani (2003). Although it is not clear how to correct P-values for correlation between markers in such a setting (genome-wide differentiation-based tests), even this smallest P-value did not provide clear evidence of selection. This does not mean that there is no selection in these data, only that genuine selection signatures cannot be discriminated from background noise provoked by drift when looking at sites independently. However, as illustrated later in this study, given a region where a selection signature has been found, FLK can help in identifying the mutations that have likely been under selection.
The hapFLK method (Fariello et al. 2013) is similar to FLK, but it incorporates linkage disequilibrium (LD) information through the exploitation of a multilocus LD model (Scheet and Stephens 2006). Because hapFLK combines information across multiple sites, it has been shown to have better detection power than single-site statistics (Fariello et al. 2013, 2014). This was confirmed when applied to this data set, as we could confidently find 67 significant regions using a FDR threshold of 15%. hapFLK has been shown to be robust to bottlenecks and to a certain extent to gene flow (Fariello et al. 2013). We confirmed this by simulating haplotypes under the demographic model estimated by the approach of Excoffier et al. (2013) (Figure 1) and calculating hapFLK on the simulated data (see Materials and Methods). While the fixation indexes computed from the simulated samples were very close to the ones computed from our data (Table S2), hapFLK P-values obtained from simulated samples did not lead to any signal called significant with the Storey and Tibshirani (2003) approach (Figure S6).
Ten of the significant regions detected by hapFLK likely resulted from assembly errors and were thus not considered in the rest of our analysis (Table S3). The cumulated length of the remaining 57 regions, listed and annotated in Table S4, was 9.1 Mb, 0.36% of the total autosome length. Detected regions spanned from a few hundred base pairs to >1 Mb with a median length of ∼20 kb (Figure S9). The median size of detected regions was thus considerably smaller than that obtained in previous studies where hapFLK was applied to 60K data in sheep (Fariello et al. 2014; Kijas 2014). Nineteen of the regions encompassed at least one gene while 38 contained no gene. In total, 82 genes were included in hapFLK regions, which represents ∼0.4% of the total number of genes in the genome, corresponding to a small enrichment in protein-coding genes in hapFLK signatures.
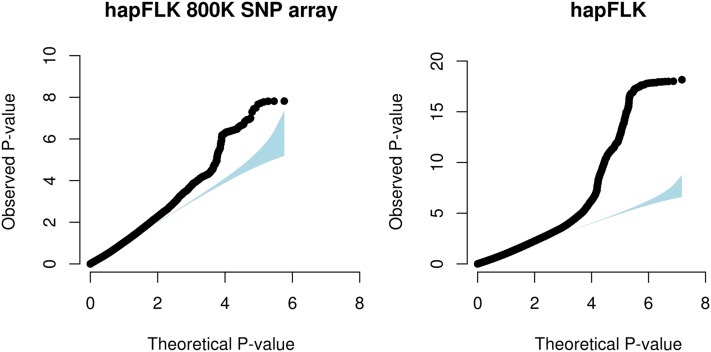
To investigate whether sequencing improves detection power with hapFLK, we thinned the data set by considering only SNPs present on the Illumina BovineHD BeadChip. We found (Figure 5) that the excess of small P-values was much larger when applying hapFLK to all sites identified in the 1000 bull genomes project than when applying it only to the SNPs that are included in the SNP chip. Note that this was not the case with FLK, where detection power was low with both sequencing and SNP chip data due to the amount of drift, as already discussed above (Figure S10).
Figure 5.
Influence of NGS on hapFLK detection power. Shown is a probabiliy probability (PP) plot of the hapFLK test applied to SNPs of the Illumina BovineHD SNP chip (left) or to all sites of the 1000 bull genomes project (right).
Hard-sweep regions showing a strong differentiation signal
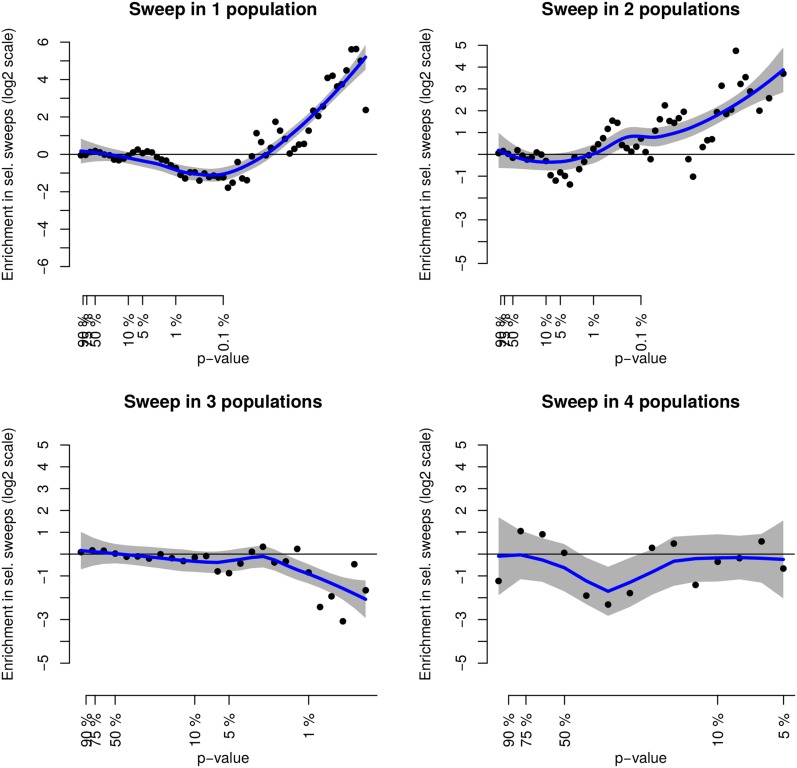
Eight selection signatures were found with both the HMM and the hapFLK analyses, pointing out regions that combine a low diversity within at least one breed and a strong haplotypic differentiation between breeds (Table 2). Although this is significantly more than expected if the two kinds of signals were independent (), it is somewhat surprising to observe that many hard sweeps were not detected with hapFLK.
Table 2. Hard-sweep selection signatures associated with significant differentiation signals.
| Hard-sweep region | hapFLK | ||||||
|---|---|---|---|---|---|---|---|
| BTA | Start | End | Population | Start | End | P-value | Candidate genes |
| 6 | 71.439 | 71.558 | F | 70.332 | 71.607 | KIT | |
| 18 | 14.755 | 14.963 | F | 14.305 | 14.872 | MC1R | |
| 14 | 24.805 | 25.076 | H, A | 24.937 | 25.070 | PLAG1 | |
| 10 | 5.736 | 5.843 | A | 5.736 | 5.782 | Intergenic | |
| 13 | 64.149 | 64.197 | A | 63.879 | 64.546 | ASIP | |
| 20 | 22.923 | 23.203 | J | 23.110 | 23.125 | ANKRD55 | |
| 7 | 43.436 | 43.542 | A, J | 43.473 | 43.474 | OR cluster | |
| 24 | 14.006 | 14.040 | F | 14.021 | 14.023 | Intergenic | |
Signatures are ordered by decreasing hapFLK P-values. Region coordinates are expressed in megabases on assembly UMD 3.1. Population abbreviations: A, Angus; F, Fleckvieh; H, Holstein; J, Jersey.
A prevalent reason for this is the large genome-wide level of differentiation between the four breeds, which reduces the power of differentiation-based tests (Fariello et al. 2013). Indeed, hard-sweep regions detected in one or two populations showed a clear enrichment in low hapFLK P-values (Figure 6). This indicates that many hard sweeps exhibit a mild differentiation signal, although the power to detect them with hapFLK is not sufficient. In addition, some hard-sweep regions did not show any differentiation signal, because the same haplotype was fixed or at least increased in frequency in all populations. This is typically the case of hard-sweep regions detected in three or four populations, for which there was a depletion of low hapFLK P-values (Figure 6). This may also concern regions where a hard sweep was detected in one or two populations, but where the swept haplotype was also at quite high frequency in other populations, as already discussed above.
Figure 6.
Distribution of hapFLK within hard-sweep regions. Shown is a comparison of the distribution of hapFLK P-values in hard-sweep regions identified within populations and in the rest of the genome. The plot shows the ratio of the P-values distribution in hard sweeps detected in one to four populations to their distribution on the part of the genome where no hard sweep is detected. y-axis is on a log2 scale and x-axis is on a log10 scale.
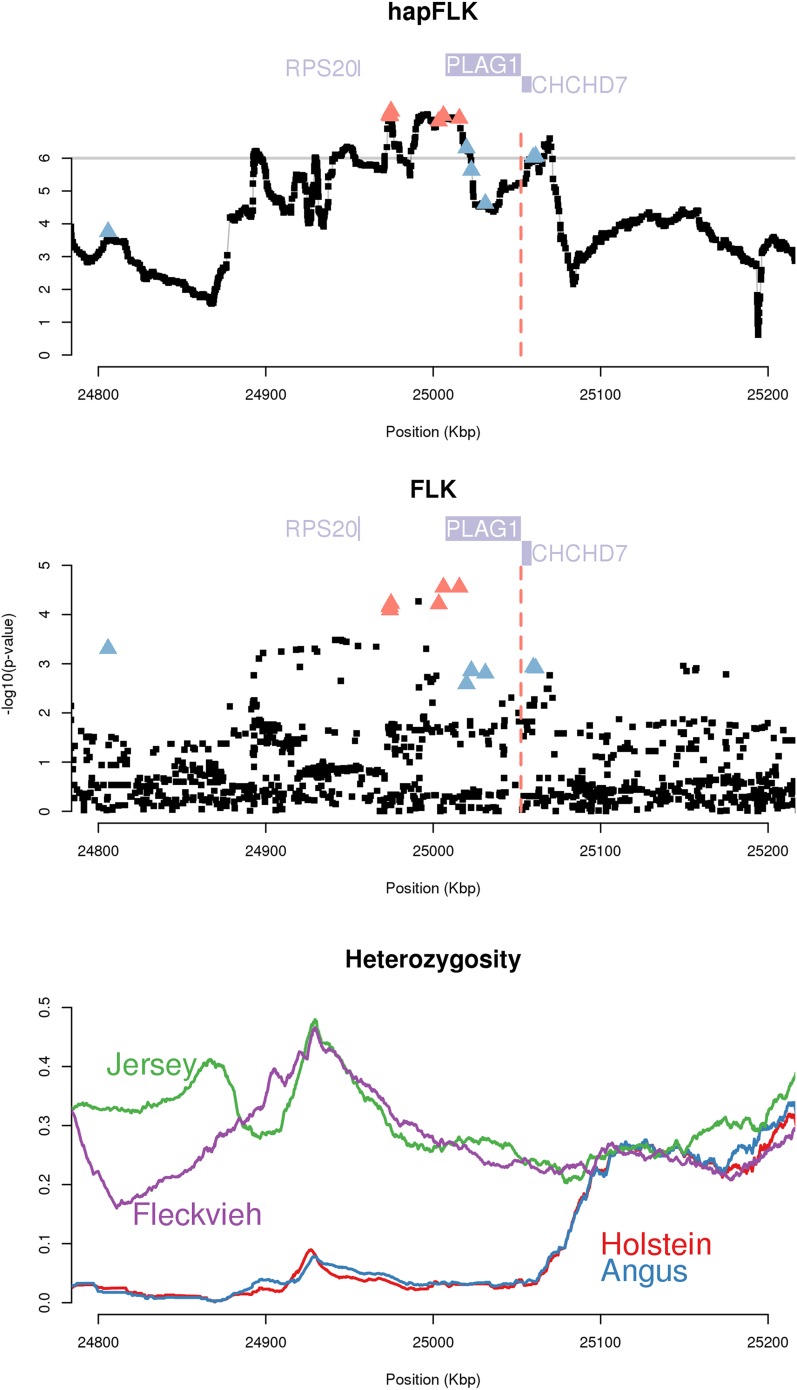
Among the regions with evidence for both a hard-sweep and an extreme differentiation signature, the top three corresponded to genes and mutations of known phenotypic effects that recapitulate the most obvious phenotypic divergence of the four breeds in this data set. The most differentiated region corresponded to the KIT gene, which has been shown to be associated with white spotting patterns in the Holstein (Hayes et al. 2010) and the Fleckvieh (Qanbari et al. 2014), while the Jersey and the Angus are nonspotted breeds. The next most differentiated region harbored the MC1R gene, for which previous studies have identified two causal polymorphisms for coat color (Klungland et al. 1995). Finally, the third signature was a small genomic region comprising the PLAG1 gene (Figure 7, top). Hard sweeps identified in this region indicate a past selection event affecting the Angus and the Holstein breeds, whereas no selection was evidenced in the Fleckvieh and the Jersey breeds (Table 2), as can also be seen from heterozygosity patterns in the region (Figure 7, bottom). The region surrounding PLAG1 was previously demonstrated to harbor a QTL for calving ease in the Fleckvieh (Pausch et al. 2011) and one for stature in a Holstein Jersey cross (Karim et al. 2011). Our results are consistent with these studies and suggest that the allele favoring high stature was selected in Holstein and Angus, but not in Jersey and Fleckvieh.
Figure 7.
Selection signature around PLAG1. Shown are hapFLK (top) and FLK (middle) P-values (log10 scale) for the selection signature around PLAG1 and local heterozygosity in the four breeds (bottom) for the same region. In the top and middle panels, genes are indicated by purple solid rectangles, and red solid triangles correspond to the candidate QTNs of Karim et al. (2011).
The other common regions include the ASIP gene, which plays a crucial role in adipocyte development and seems to be expressed in a wide set of tissues in different cattle breeds (Albrecht et al. 2012), and the ANKRD55 region that is strongly associated with autoimmune disorders in humans [in particular, multiple sclerosis (Stahl et al. 2010) and type 2 diabetes (Morris et al. 2012).
hapFLK signatures of soft or incomplete sweeps
Apart from the eight signatures above, none of the other hapFLK signatures matched hard sweeps detected by the HMM approach. These signatures most likely resulted from incomplete sweeps for which the selected allele did not reach fixation or soft sweeps where selection targeted an allele that was already at intermediate frequency in the population. Indeed, such signals do not lead to the skewed allele frequency patterns that are looked for by the approach of Boitard et al. (2009). This hypothesis was confirmed by examining haplotype diversity patterns in hapFLK signatures, which typically show haplotypes of large but not fixed frequency in at least one population (e.g., Figure S11 for region 1 in Table S4).
Two signatures are located close to the homologous region of a human promoter region, near ROBO1 on one hand and the prolactin receptor gene PRLR on the other, hinting that the causal mutation is likely regulatory in nature. ROBO1 has been shown to be involved in early follicular development in sheep (Dickinson and Duncan 2010; Dickinson et al. 2010), while the prolactin gene and its receptor are involved in a large range of biological functions (development, metabolism, immunology, reproduction, etc.) (Bole-Feysot et al. 1998).
The PRLR gene lies 6 Mb from the growth hormone receptor (GHR) gene, itself close (140 kb) to another hapFLK signature. It has been evidenced that two QTL affecting milk, fat, and protein yield segregate near these two genes in a Finnish dairy cattle population (Viitala et al. 2006). Our results suggest that these QTL might be in regulatory regions of these genes and that they have responded to selection in populations from the 1000 bull genomes. We found another selection signature, in Jersey, within a gene potentially involved in milk production (Figure S12), ARL15. Seven highly differentiated variants (FLK P-value ) were located in an ARL15 intron. ARL15 is a protein of unknown function that has been shown to be strongly associated with adiponectin levels in humans (Richards et al. 2009). Adiponectin is a hormone involved in glucose metabolism, with a low concentration of adiponectin being associated with insulin resistance. In dairy cows, insulin resistance is maintained in early lactation, favoring mammary glucose uptake. Giesy et al. (2012) showed that the reduction of plasma adiponectin concentration in early lactating cows was not associated with changes in the adiponectin expression itself. Our results could suggest a potential role of ARL15 in this process, with a particular adaptation of the Jersey dairy cattle at this gene.
Apart from the PLAG1 signature, several others include genes involved in morphology and growth: RUNX3 (Yoshida et al. 2004; Soung do et al. 2007), STARD3NL (Rivadeneira et al. 2009), and RASSF2 (Song et al. 2012) are involved in bone development; NCAPG and/or LCORL match a large QTL for many growth traits in cattle, horses, and sheep (Eberlein et al. 2009; Weikard et al. 2010; Lindholm-Perry et al. 2011; Bongiorni et al. 2012; Makvandi-Nejad et al. 2012; Signer-Hasler et al. 2012; Lindholm-Perry et al. 2013; Metzger et al. 2013; Tetens et al. 2013; Kijas 2014; Randhawa et al. 2015; Sahana et al. 2015; Xu et al. 2015); and CTNNBL1 (Liu et al. 2008) is associated with obesity traits in humans.
Identifying causal adaptive polymorphisms
As demonstrated above, genomic scans for selection based on sequencing data have a higher detection power than those based on genotyping chip data and locate the selection signatures with higher precision. This is an expected outcome of the higher marker density, and the same could be said when comparing high-density to medium-density chip data. But a more fundamental difference between dense SNP chip data and sequencing data is that, with the latter, one can reasonably expect the causal polymorphism under selection to be included in the observed data. Clearly, not all selection signatures can be related to a single polymorphism, and even in this case this polymorphism might be absent in our data due to insufficient coverage or remaining calling issues. Still, the favorable situation where a single polymorphism leads to a selective advantage and is present in the data should also occur. One natural question is thus to determine whether such variants can be identified only from genetic diversity patterns. We show below that this is indeed possible.
HapFLK signals
Haplotype frequency differences detected by hapFLK typically result from the increase in frequency of one particular allele in a population due to positive selection, which implied the increase in frequency of one or several haplotypes carrying this allele by genetic hitchhiking. Thus, in regions detected by hapFLK, the causal polymorphism under selection should be the one with the largest allele frequency differentiation, and a natural strategy to detect this polymorphism is to look at the variants with the largest FLK value. Two of the regions identified by hapFLK validate this strategy.
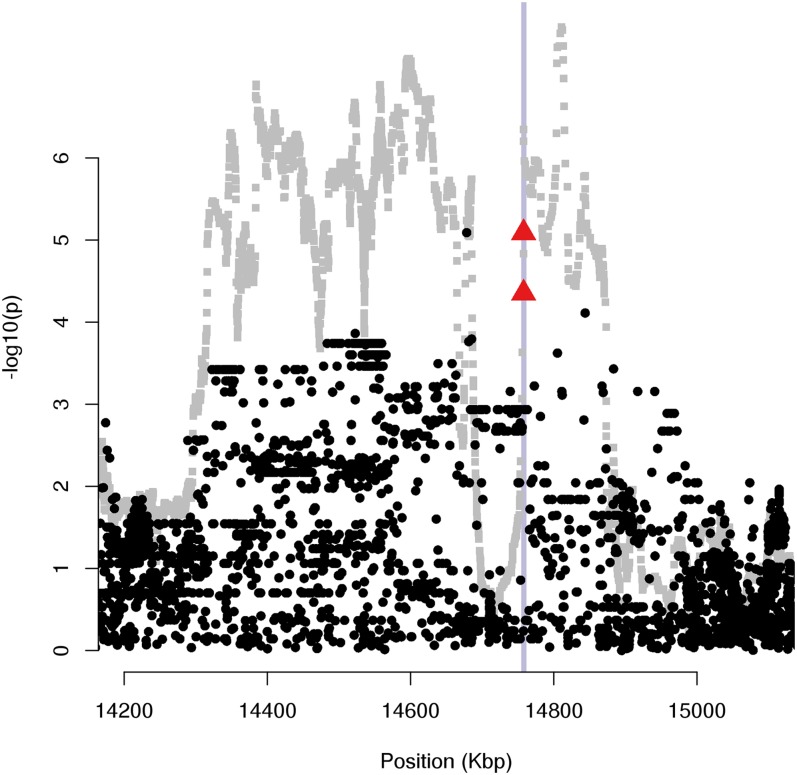
Within the MC1R selection signature, we found three polymorphisms with clear outlying FLK values (Figure 8), and two of these corresponded to the known causal mutations mentioned previously: a single-base mutation at position 14,757,910 (rs109688013), responsible for the black pigmentation in Holstein and Angus breeds, and a single-base deletion at position 14,757,924 (rs110710422), responsible for the red pigmentation in the Fleckvieh breed (Klungland et al. 1995). The third outlying polymorphism (rs110494166) in the region was located at position 14,678,403, within an intron of the nearby FANCA gene, and exhibited the same allele frequencies as rs110710422.
Figure 8.
Selection signature around MC1R. Shown are hapFLK (gray) and FLK (black) P-values (log10 scale) for the selection signature around MC1R (purple vertical line). Red triangles highlight the two known causal mutations for red and black coat color in cattle.
In the PLAG1 region, the causal mutation was shown to be one of eight candidate quantitative trait nucleotides (QTNs) (Karim et al. 2011). We found seven polymorphisms exhibiting a high level of differentiation between breeds in this region based on the FLK statistic (P-value <) (Figure 7, middle, and Table S5). Of these seven candidates, six were common with the candidate QTNs listed in table 2 of Karim et al. (2011). rs134215421 and rs109815800 showed particularly extreme allele frequency differences between Holstein and Angus on the one hand and Jersey and Simmental on the other hand. These two mutations are located at the 3′ end of the PLAG1 gene, rs134215421 being 1 kb downstream and rs109815800 within an intron of the PLAG1 gene. One SNP (rs134029466) was not identified as a potential QTN in Karim et al. (2011) but showed a strong signal for differentiation. Two of the mutations in Karim et al. (2011), rs209821678 and rs210030313, which are considered by the authors as the most serious candidates because they affect a highly conserved element, were not available in the 1000 bull genomes project. One is a variable number tandem repeat (VNTR), a kind of polymorphism that is hardly callable from short-read sequence data, and the other one is a SNP that lies 44 bp from the VNTR, which exhibited very low-quality scores and was therefore not called in the 1000 bull genomes data. While these polymorphisms cannot be considered as disqualified based on our study, their positions, highlighted in Figure 7, lie in a region where the differentiation signal was not significantly elevated.
Hard-sweep signals
Hard-sweep signals are expected when an allele goes from very low frequency to almost fixation in a population due to positive selection. In these regions, detecting the causal selected variant only from genetic data of the swept population is impossible, because all physical positions show either extreme allele frequencies or no polymorphism at all. However, if we assume that other sampled populations evolved neutrally in the region, alleles that were initially at low frequency in the swept population have likely remained at relatively low frequency in these other populations. This should result in high genetic differentiation at and around the selected polymorphism, which again can be detected using the FLK statistic.
For all hard-sweep regions except the ones shared by all populations, we thus tried to identify the selected site by looking for polymorphisms with a high FLK value (P ). To ensure that the detected polymorphisms could indeed be the causal ones, we further required a high allele frequency () in the swept population(s) and only in this (these) one(s). We found only 12 sweep regions exhibiting such causal candidates (Table 3 and Table S6). Again, this small proportion is related to the fact that in most cases the selected allele must actually be at relatively high frequency even in nonswept populations, due to undetected ongoing sweeps or just random drift.
Table 3. Private hard-sweep regions including candidate mutations.
| Chr | Start (Mb) | End (Mb) | Sel pop | Nb mut | Pval FLK | Sel freq | Genes |
|---|---|---|---|---|---|---|---|
| 6 | 71.440 | 71.560 | F | 2 | 0.92 | Intergenic | |
| 7 | 25.430 | 26.000 | J | 8 | 0.77 | CHSY3, KIAA1024L, ADAMTS19 SLC27A6, FBN2, SLC12A2 | |
| 7 | 26.280 | 27.060 | J | 16 | 0.83 | ||
| 14 | 24.810 | 25.080 | H, A | 7 | 0.04 | LYN, RPS20, PLAG1, CHCHD7 ENSBTAG00000039031, MOS | |
| 18 | 14.760 | 14.960 | F | 3 | 0.95 | TUBB6, TUBB3, DEF8, LOC532875 DBNDD1, GAS8, SHCBP1, MC1R | |
| 20 | 24.230 | 24.740 | J | 1 | 0.77 | LOC530348, SNX18, COX8A, LOC783202, HSPB3 | |
| 20 | 25.030 | 25.390 | J | 21 | 0.80 | Intergenic | |
| 20 | 26.640 | 27.230 | J | 2 | 0.77 | Intergenic | |
| 20 | 39.830 | 39.970 | J | 1 | 0.80 | SLC45A2, ADAMTS12, RXFP3 | |
| 22 | 35.680 | 35.790 | J | 1 | 0.77 | Intergenic | |
| 22 | 35.960 | 36.030 | J | 2 | 0.90 | MAGI1 | |
| 27 | 4.140 | 4.230 | J | 1 | 0.90 | Intergenic |
Horizontal spaces are used to group closely related sweep windows, which might result from the same selection event. Abreviations A, F, H, and J are defined in Table 2 legend. Chr, chromosome; Sel pop, population(s) under selection; Nb mut, Number of candidate mutations; Pval FLK, lowest P-value of the FLK test; Sel freq, Allele frequency at the position with lowest P-value.
The regions detected by this approach include those of KIT, PLAG1, and MC1R. In all these regions, a very limited number of potentially causal polymorphisms were detected, and in the case of MC1R these candidates included the true causal variants. This provides a validation of the detection strategy considered here and suggests that other regions in Table 3 should also contain interesting putative causal polymorphisms.
Of particular interest, a sweep region on chromosome 20 included a single causal candidate at position 39,872,347 (Figure 9 and Figure S13). This polymorphism is located downstream of SLC45A2, a gene that has been associated with fertility (Killeen et al. 2014) and residual feed intake (Karisa et al. 2013) in cattle and with pigmentation in several other species including humans (Sturm 2009; Stefanaki et al. 2013; Morice-Picard et al. 2014), dogs (Wijesena and Schmutz 2015), and tigers (Xu et al. 2013). It is also located downstream of RXFP3, a gene known to be involved in food intake regulation and body weight in mice (Ganella et al. 2012; Smith et al. 2014)
Figure 9.
Allele frequencies in the sweep region including SLC45A2 and RXFP3. For SNPs where the ancestral allele is known (in red), the frequency is that of the derived allele. For other SNPs (in black) the frequency is that of the minor allele (among all breeds).
Another interesting region was found on chromosome 22, where two strong candidate polymorphisms were located 30 kb upstream of MAGI1 (Figure S14 and Figure S15). Although not reported in Table 3 due to P-values slightly > five other suggestive polymorphisms are located within MAGI1 introns. One of these, at position 36,011,838, is located within a highly conserved region according to a multiple alignment of 36 eutherian mammals (www.ensembl.org). At this position, all considered species carry allele G, which is also the most frequent allele in Angus, Fleckvieh, and Holstein, while allele A swept in Jersey. MAGI1 is a scaffolding protein present in tight junctions of epithelial cells and might be implied in nervous system functions. Adaptive selection around this gene was already reported in some West African cattle breeds (Gautier et al. 2009).
A more complex situation was observed on chromosome 7, where 8 and 16 candidate polymorphisms were found in two sweep windows distant by 300 kb. Among all these candidates, the highest FLK value was reached at position 26,459,812, in an intergenic region between SLC27A6 and FBN2. Interestingly, this polymorphism was located in a very conserved region, and it was the only one in this case among all candidates in the sweep window. Allele T was found in almost all species at this position and was almost fixed in Angus, Fleckvieh, and Holstein, while allele A swept in Jersey. SLC27A6 has been shown related to fatty acid metabolism in cattle (Bionaz and Loor 2008; Nafikov et al. 2013), while FBN2 has been related to several development processes, including bone formation in mice (Nistala et al. 2010).
Strong candidate mutations were also found in two regions without protein-coding genes on the current bovine annotation, and in general we note that all candidate polymorphisms were located in intergenic or regulatory regions. This implies that validating the effect of these polymorphisms will be difficult, but this also outlines the potential of selective sweep studies to improve genome annotation.
Hard-sweep signals shared by all breeds
In regions where hard-sweep signals were shared by all breeds (Table 1), identifying causal polymorphisms only from our data set is impossible. Indeed, the majority of polymorphisms have similar diversity patterns, with a low minor allele frequency in all four breeds. In addition, positions that were monomorphic in our data set are also good candidates, since they might result from the complete fixation of a favorable allele in the four breeds.
To identify potential causal variants in these regions, one possible approach can be to use data from related species, looking for highly conserved positions for which the major (or the only) allele of our bovine data set is absent in other mammals. To illustrate this approach, we implemented the following procedure. Based on a multiple alignment of the bovine reference sequence with 10 other mammal reference sequences (Rocha et al. 2014), we selected the positions where (i) the bovine allele (or the major bovine allele in the case of polymorphic positions) was distinct from the yak allele, (ii) the bovine allele was unobserved among the 10 other species, and (iii) the yak allele was observed in >7 (of 9) other species, including at least buffalo or sheep (the two closest species). Such positions represent convincing candidates for two reasons. First, alleles that are observed in other mammal species must have been segregating in the bovine for a very long time and are thus very unlikely to produce a hard-sweep pattern, even if they became positively selected at some point in the bovine history. Second, highly conserved positions are more likely functional, and thus subject to selection, than less conserved ones.
Among the 1,382,681 positions included in the regions in Table 1, only 91 satisfied the three conditions above. Further removing 7 positions for which the minor bovine allele was at quite high frequency, we finally obtained 84 causal candidates (Table S7). All sweep regions except one exhibited convincing causal variants located in coding or regulatory regions, but more work will be needed to determine the exact causal variants in these regions.
Discussion
We performed in this study a genomic scan for selection in European taurine cattle, based on large samples of sequencing data from four different breeds. We used two detection approaches, based respectively on the genetic diversity within breeds and on the genetic differentiation between breeds, and compared the signals detected by these two approaches.
One important conclusion from our analysis is that sequencing data represent a great opportunity in the context of genomic scans for selection. Indeed, the detection power of hapFLK was higher when applied to sequencing data than when applied to high-density chip data (Figure 5). This was consistent with previous studies looking for selection signatures in cattle (Ma et al. 2015) and humans (Liu et al. 2014), which also found, based on computer simulations, that higher power could be expected from sequencing data compared to SNP chip data. The localization of selection signatures was also found more accurate with sequencing data, both for hapFLK and for the within-population approach, as the median size of detected regions (a few tens of kilobases) was considerably lower than with SNP chip data. Note that the reduced size of detected regions does not mean that we detected older selection events. It just comes from the fact that, in many regions, only the most significant part of the sweep was identified. For instance, the selection events detected by hapFLK are quite recent, because they must have occurred more recently than breed divergence (∼500 years before present according to our demographic analysis). Still, the region with excess differentiation, which is captured by hapFLK, is generally smaller than the sweep itself, because part of the swept haplotype can be shared between breeds. An example of such a signature can be seen in the ARL15 signature (Figure S12). Importantly, the higher precision provided by the use of NGS data reduces the number of genes included in each window, allowing potentially an easier exploration of the molecular functions driving selection.
Since sequencing data capture a large proportion of the SNPs and small indels segregating in a sample, genomic scans for selection based on such data can be expected to identify even the causal polymorphism under selection in a given sweep region. We demonstrated that this is indeed the case in some regions, where the few variants with the highest allele frequency differentiation between breeds, measured by the FLK statistic, included the causal variant. For instance, in the MC1R region, the two mutations that have been shown to affect coat color in the breeds of our study were included in the top three FLK values. Similarly, in the PLAG1 region, a good overlap was observed between variants with top FLK values and the QTNs found by Karim et al. (2011) in a genome-wide association study analysis on stature. In several other regions, variants with top FLK values also lead to promising candidate polymorphisms (Table 3 and Table S6), which were often located in highly conserved regions or/and in the vicinity of genes with interesting metabolic functions. However, it is important to point out that identifying the selected variant from FLK values will essentially be possible in population-specific hard-sweep scenarios, where the favorable allele has almost fixed in the selected population(s) and has remained absent or at very low frequency in the other sampled populations. In all other situations, and for instance those where the favorable allele is at intermediate frequency also in neutral populations, the FLK value of the causal variant will be reached just by chance by many neutral variants, so identifying the causal variant will require additional data, for instance phenotypes related to the selection constraint.
Our study also illustrated the small overlap between the selection signatures detected by the within- and the between-population approach and allowed us to better characterize the regions detected by each of these approaches. Among the 916 hard-sweep regions detected by the within-population approach, only 8 were detected by hapFLK. One reason is that, in many of the regions where a hard sweep was detected, the swept allele was likely at quite high frequency also in the other breeds, reducing the differentiation signal. Indeed, in regions where a sweep was detected in one of the breeds, the distribution in the other breeds was shifted toward that of swept regions, even if this signal was not strong enough to be detected by the HMM (Figure S3). This can be explained by the long shared history between the four breeds considered in this study, which have a common geographic origin and have been strictly isolated only in the last 200 years. Convergent selection of similar alleles in different breeds may also have occurred, as the same traits were selected in these breeds, but this is not the most parsimonious hypothesis. Finally, the small proportion of hard sweeps detected by hapFLK was also likely related to a power issue, because none of the most breed-specific hard sweeps listed in Table S1 were detected by this approach. Although such regions clearly showed some signal of differentiation, this signal was not strong enough to be detected by hapFLK, because the very high levels of genetic drift in cattle breeds imply that only extremely differentiated regions can be considered as significantly under selection. Actually, we can see from Table 2 that in most hard-sweep regions detected by hapFLK, there was not one hard sweep in a single population, but at least two sweeps (either complete or incomplete) implying distinct haplotypes in the four breeds, which represents an even stronger differentiation signal (Figure S16). We expect the 1000 bull genomes data set to grow by inclusion of new populations, which will most likely increase power of differentiation approaches such as FLK and hapFLK by adding information on breeds more closely related to each other than on the initial release.
On the other hand, among the 57 selection signatures detected by hapFLK, 49 were not detected by the within-population approach. This comes from the fact that this approach is specifically designed to detect hard-sweep signals, while hapFLK also detects incomplete or soft sweeps (Fariello et al. 2013, 2014). For the 49 regions detected by hapFLK and not by the within-breed approach, the evidence of a hard sweep, measured by the statistic was always very low in all breeds, indicating that the signal detected by hapFLK had nothing to do with a hard sweep in one of the breeds. For example, in the ARL15 signature, the swept haplotype clearly segregates at moderate frequency (∼85%) in the Jersey population (Figure S12). This ability of hapFLK to detect incomplete and soft sweeps is extremely interesting for the study of livestock species. Indeed, recent intensive selection in modern livestock breeds has mainly targeted polygenic traits such as milk or meat production, which is much more likely to produce soft- and incomplete-sweep patterns than hard-sweep patterns (Pritchard et al. 2010; Hernandez et al. 2011).
In conclusion, our study illustrates how sequencing data offer great advantages for the detection of adaptive loci: higher power and better precision in localizing adaptive genes and even in some rare cases causal mutations. However, we found that many of our strongest signals and mutations lie in noncoding regions of the genome, hinting that a majority of adaptive mutations are regulatory in nature. A better annotation of genomes, such as obtained by high-throughput postgenomic approaches, in combination with phenotyping in large population samples will be key in uncovering the biological basis of adaptation.
Acknowledgments
Data analyzes were performed on the computer cluster of the Genotoul bioinformatics platform Toulouse Midi-Pyrénées (www.bioinfo.genotoul.fr). The cattle genomes were obtained from the 1,000 bull genomes project (www.1000bullgenomes.com). We would like to thank Didier Boichard for useful discussions on this project and two anonymous reviewers for constructive comments on the manuscript.
Footnotes
Communicating editor: R. Nielsen
Supplemental material is available online at http://www.genetics.org/cgi/data/genetics.115.181594/DC1.
Literature Cited
- Albrecht E., Komolka K., Kuzinski J., Maak S., 2012. Agouti revisited: transcript quantification of the asip gene in bovine tissues related to protein expression and localization. PLoS One 7: e35282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allais-Bonnet A., Grohs C., Medugorac I., Krebs S., Djari A., et al. , 2013. Novel insights into the bovine polled phenotype and horn ontogenesis in bovidae. PLoS One 8: e63512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bionaz M., Loor J. J., 2008. Acsl1, agpat6, fabp3, lpin1, and slc27a6 are the most abundant isoforms in bovine mammary tissue and their expression is affected by stage of lactation. J. Nutr. 138: 1019–1024. [DOI] [PubMed] [Google Scholar]
- Biswas S., Akey J. M., 2006. Genomic insights into positive selection. Trends Genet. 22: 437–446. [DOI] [PubMed] [Google Scholar]
- Boitard S., Schlotterer C., Futschik A., 2009. Detecting selective sweeps: a new approach based on hidden Markov models. Genetics 181: 1567–1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boitard S., Rodríguez W., Jay F., Mona S., Austerlitz F., 2016. Inferring population size history from large samples of genome-wide molecular data - an approximate Bayesian computation approach. PLoS Genet. 12: 1–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bole-Feysot C., Goffin V., Edery M., Binart N., Kelly P. A., 1998. Prolactin (prl) and its receptor: actions, signal transduction pathways and phenotypes observed in prl receptor knockout mice. Endocr. Rev. 19: 225–268. [DOI] [PubMed] [Google Scholar]
- Bongiorni S., Mancini G., Chillemi G., Pariset L., Valentini A., 2012. Identification of a short region on chromosome 6 affecting direct calving ease in Piedmontese cattle breed. PLoS One 7: e50137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonhomme M., Chevalet C., Servin B., Boitard S., Abdallah J., et al. , 2010. Detecting selection in population trees: the Lewontin and Krakauer test extended. Genetics 186: 241–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bovine HapMap Consortium , 2009. Genome-wide survey of SNP variation uncovers the genetic structure of cattle breeds. Science 324: 528–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daetwyler H. D., Capitan A., Pausch H., Stothard P., van Binsbergen R., et al. , 2014. Whole-genome sequencing of 234 bulls facilitates mapping of monogenic and complex traits in cattle. Nat. Genet. 46: 858–865. [DOI] [PubMed] [Google Scholar]
- de Simoni Gouveia, J. J., M. V. G. B. da Silva, S. R. Paiva, and S. M. P. de Oliveira, 2014. Identification of selection signatures in livestock species. Genet. Mol. Biol. 37: 330–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Noia M. A., Todisco S., Cirigliano A., Rinaldi T., Agrimi G., et al. , 2014. The human slc25a33 and slc25a36 genes of solute carrier family 25 encode two mitochondrial pyrimidine nucleotide transporters. J. Biol. Chem. 289: 33137–33148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson R. E., Duncan W. C., 2010. The SLIT-ROBO pathway: a regulator of cell function with implications for the reproductive system. Reproduction 139: 697–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson R. E., Hryhorskyj L., Tremewan H., Hogg K., Thomson A. A., et al. , 2010. Involvement of the SLIT/ROBO pathway in follicle development in the fetal ovary. Reproduction 139: 395–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberlein A., Takasuga A., Setoguchi K., Pfuhl R., Flisikowski K., et al. , 2009. Dissection of genetic factors modulating fetal growth in cattle indicates a substantial role of the non-SMC condensin I complex, subunit G (NCAPG) gene. Genetics 183: 951–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echternkamp S., Aad P., Eborn D., Spicer L., 2012. Increased abundance of aromatase and follicle stimulating hormone receptor mRNA and decreased insulin-like growth factor-2 receptor mRNA in small ovarian follicles of cattle selected for twin births. J. Anim. Sci. 90: 2193–2200. [DOI] [PubMed] [Google Scholar]
- Excoffier L., Dupanloup I., E. Huerta-Sánchez, V. C. Sousa, and M. Foll, 2013. Robust demographic inference from genomic and SNP data. PLoS Genet. 9: e1003905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fariello M. I., Boitard S., Naya H., SanCristobal M., Servin B., 2013. Detecting signatures of selection through haplotype differentiation among hierarchically structured populations. Genetics 193: 929–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fariello M. I., Servin B., Tosser-Klopp G., Rupp R., Moreno C., et al. , 2014. Selection signatures in worldwide sheep populations. PLoS One 9: e103813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felius M., Koolmees P. A., Theunissen B., European Cattle Genetic Diversity Consortium, andLenstra J. A., 2011. On the breeds of cattle––historic and current classifications. Diversity 3: 660–692. [Google Scholar]
- Flori L., Fritz S., Jaffrézic F., Boussaha M., Gut I., et al. , 2009. The genome response to artificial selection: a case study in dairy cattle. PLoS One 4: e6595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganella D. E., Ryan P. J., Bathgate R. A., Gundlach A. L., 2012. Increased feeding and body weight gain in rats after acute and chronic activation of rxfp3 by relaxin-3 and receptor-selective peptides: functional and therapeutic implications. Behav. Pharmacol. 23: 516–525. [DOI] [PubMed] [Google Scholar]
- Gautier M., Flori L., Riebler A., Jaffrezic F., Laloe D., et al. , 2009. A whole genome Bayesian scan for adaptive genetic divergence in West African cattle. BMC Genomics 10: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giesy S. L., Yoon B., Currie W. B., Kim J. W., Boisclair Y. R., 2012. Adiponectin deficit during the precarious glucose economy of early lactation in dairy cows. Endocrinology 153: 5834–5844. [DOI] [PubMed] [Google Scholar]
- Gutierrez-Gil B., Arranz J. J., Wiener P., 2015. An interpretive review of selective sweep studies in Bos taurus cattle populations: identification of unique and shared selection signals across breeds. Front. Genet. 6: 167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes B. J., Pryce J., Chamberlain A. J., Bowman P. J., Goddard M. E., 2010. Genetic architecture of complex traits and accuracy of genomic prediction: coat colour, milk-fat percentage, and type in Holstein cattle as contrasting model traits. PLoS Genet. 6: e1001139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez, R. D., J. L. Kelley, E. Elyashiv, S. C. Melton, A. Auton et al., 2011 Classic selective sweeps were rare in recent human evolution. Science 331: 920–924. [DOI] [PMC free article] [PubMed]
- Hudson R., 2002. Generating samples under a Wright-Fisher neutral model of genetic variation. Bioinformatics 18: 337–338. [DOI] [PubMed] [Google Scholar]
- Karim L., Takeda H., Lin L., Druet T., Arias J. A., et al. , 2011. Variants modulating the expression of a chromosome domain encompassing plag1 influence bovine stature. Nat. Genet. 43: 405–413. [DOI] [PubMed] [Google Scholar]
- Karisa B., Thomson J., Wang Z., Stothard P., Moore S., et al. , 2013. Candidate genes and single nucleotide polymorphisms associated with variation in residual feed intake in beef cattle. J. Anim. Sci. 91: 3502–3513. [DOI] [PubMed] [Google Scholar]
- Kijas J. W., 2014. Haplotype-based analysis of selective sweeps in sheep. Genome 57: 433–437. [DOI] [PubMed] [Google Scholar]
- Killeen A. P., Morris D. G., Kenny D. A., Mullen M. P., Diskin M. G., et al. , 2014. Global gene expression in endometrium of high and low fertility heifers during the mid-luteal phase of the estrous cycle. BMC Genomics 15: 234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klungland H., Vage D., Gomez-Raya L., Adalsteinsson S., Lien S., 1995. The role of melanocyte-stimulating hormone (msh) receptor in bovine coat color determination. Mamm. Genome 6: 636–639. [DOI] [PubMed] [Google Scholar]
- Leroy G., Mary-Huard T., Verrier E., Danvy S., Charvolin E., et al. , 2013. Methods to estimate effective population size using pedigree data: examples in dog, sheep, cattle and horse. Genet. Sel. Evol. 45: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Tian S., Jin L., Zhou G., Li Y., et al. , 2013. Genomic analyses identify distinct patterns of selection in domesticated pigs and Tibetan wild boars. Nat. Genet. 45: 1431–1438. [DOI] [PubMed] [Google Scholar]
- Lindholm-Perry A. K., Sexten A. K., Kuehn L. A., Smith T. P., King D. A., et al. , 2011. Association, effects and validation of polymorphisms within the NCAPG - LCORL locus located on BTA6 with feed intake, gain, meat and carcass traits in beef cattle. BMC Genet. 12: 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindholm-Perry A. K., Kuehn L. A., Oliver W. T., Sexten A. K., Miles J. R., et al. , 2013. Adipose and muscle tissue gene expression of two genes (NCAPG and LCORL) located in a chromosomal region associated with cattle feed intake and gain. PLoS One 8: e80882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Saw W. Y., Ali M., Ong R. T. H., Teo Y. Y., 2014. Evaluating the possibility of detecting evidence of positive selection across Asia with sparse genotype data from the Hugo pan-Asian SNP consortium. BMC Genomics 15: 332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y. J., Liu X. G., Wang L., Dina C., Yan H., et al. , 2008. Genome-wide association scans identified ctnnbl1 as a novel gene for obesity. Hum. Mol. Genet. 17: 1803–1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L., O’Connell J. R., VanRaden P. M., Shen B., Padhi A., et al. , 2015. Cattle sex-specific recombination and genetic control from a large pedigree analysis. PLoS Genet. 11: e1005387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLeod I. M., Larkin D. M., Lewin H. A., Hayes B. J., Goddard M. E., 2013. Inferring demography from runs of homozygosity in whole-genome sequence, with correction for sequence errors. Mol. Biol. Evol. 30: 2209–2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makvandi-Nejad S., Hoffman G. E., Allen J. J., Chu E., Gu E., et al. , 2012. Four loci explain 83% of size variation in the horse. PLoS One 7: e39929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger J., Schrimpf R., Philipp U., Distl O., 2013. Expression levels of LCORL are associated with body size in horses. PLoS One 8: e56497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morice-Picard F., Lasseaux E., Cailley D., Gros A., Toutain J., et al. , 2014. High-resolution array-cgh in patients with oculocutaneous albinism identifies new deletions of the tyr, oca2, and slc45a2 genes and a complex rearrangement of the oca2 gene. Pigment Cell Melanoma Res. 27: 59–71. [DOI] [PubMed] [Google Scholar]
- Morris A. P., Voight B. F., Teslovich T. M., Ferreira T., Segre A. V., et al. , 2012. Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat. Genet. 44: 981–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nafikov R., Schoonmaker J., Korn K., Noack K., Garrick D., et al. , 2013. Association of polymorphisms in solute carrier family 27, isoform a6 (slc27a6) and fatty acid-binding protein-3 and fatty acid-binding protein-4 (fabp3 and fabp4) with fatty acid composition of bovine milk. J. Dairy Sci. 96: 6007–6021. [DOI] [PubMed] [Google Scholar]
- Nielsen R., Williamson L., Kim Y., Hubisz M., Clark A., et al. , 2005. Genomic scans for selective sweeps using SNP data. Genome Res. 15: 1566–1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nistala H., Lee-Arteaga S., Smaldone S., Siciliano G., Carta L., et al. , 2010. Fibrillin-1 and -2 differentially modulate endogenous tgf-β and bmp bioavailability during bone formation. J. Cell Biol. 190: 1107–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pausch H., Flisikowski K., Jung S., Emmerling R., Edel C., et al. , 2011. Genome-wide association study identifies two major loci affecting calving ease and growth-related traits in cattle. Genetics 187: 289–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard J. K., Pickrell J. K., Coop G., 2010. The genetics of human adaptation: hard sweeps, soft sweeps, and polygenic adaptation. Curr. Biol. 20: R208–R215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qanbari S., Gianola D., Hayes B., Schenkel F., Miller S., et al. , 2011. Application of site and haplotype-frequency based approaches for detecting selection signatures in cattle. BMC Genomics 12: 318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qanbari S., Pausch H., Jansen S., Somel M., Strom T. M., et al. , 2014. Classic selective sweeps revealed by massive sequencing in cattle. PLoS Genet. 10: e1004148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randhawa, I. A., M. S. Khatkar, P. C. Thomson, and H. W. Raadsma, 2015 Composite selection signals for complex traits exemplified through bovine stature using multi-breed cohorts of European and African Bos taurus. G3 5: 1391–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards J. B., Waterworth D., O’Rahilly S., Hivert M. F., Loos R. J., et al. , 2009. A genome-wide association study reveals variants in ARL15 that influence adiponectin levels. PLoS Genet. 5: e1000768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivadeneira F., Styrkarsdottir U., Estrada K., Halldorsson B. V., Hsu Y. H., et al. , 2009. Twenty bone-mineral-density loci identified by large-scale meta-analysis of genome-wide association studies. Nat. Genet. 41: 1199–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha D., Billerey C., Samson F., Boichard D., Boussaha M., 2014. Identification of the putative ancestral allele of bovine single-nucleotide polymorphisms. J. Anim. Breed. Genet. 131: 483–486. [DOI] [PubMed] [Google Scholar]
- Roux, P. F., S. Boitard, Y. Blum, B. Parks, A. Montagner et al., 2015 Combined QTL and selective sweep mappings with coding SNP annotation and cis-eQTL analysis revealed park2 and jag2 as new candidate genes for adiposity regulation. G3 5: 517–529. [DOI] [PMC free article] [PubMed]
- Rubin C. J., Zody M. C., Eriksson J., Meadows J. R., Sherwood E., et al. , 2010. Whole-genome resequencing reveals loci under selection during chicken domestication. Nature 464: 587–591. [DOI] [PubMed] [Google Scholar]
- Rubin C. J., Megens H. J., Barrio A. M., Maqbool K., Sayyab S., et al. , 2012. Strong signatures of selection in the domestic pig genome. Proc. Natl. Acad. Sci. USA 109:19529–19536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabeti P., SSchaffner, BFry, JLohmueller, PVarilly et al, 2006. Positive natural selection in the human lineage. Science 312: 1614–1620. [DOI] [PubMed] [Google Scholar]
- Sahana G., Hoglund J. K., Guldbrandtsen B., Lund M. S., 2015. Loci associated with adult stature also affect calf birth survival in cattle. BMC Genet. 16: 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandor C., Li W., Coppieters W., Druet T., Charlier C., et al. , 2012. Genetic variants in rec8, rnf212, and prdm9 influence male recombination in cattle. PLoS Genet. 8: e1002854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheet P., Stephens M., 2006. A fast and flexible statistical model for large-scale population genotype data: applications to inferring missing genotypes and haplotypic phase. Am. J. Hum. Genet. 78: 629–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Signer-Hasler H., Flury C., Haase B., Burger D., Simianer H., et al. , 2012. A genome-wide association study reveals loci influencing height and other conformation traits in horses. PLoS One 7: e37282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C. M., Chua B. E., Zhang C., Walker A. W., Haidar M., et al. , 2014. Central injection of relaxin-3 receptor (rxfp3) antagonist peptides reduces motivated food seeking and consumption in c57bl/6j mice. Behav. Brain Res. 268: 117–126. [DOI] [PubMed] [Google Scholar]
- Song H., Kim H., Lee K., Lee D. H., Kim T. S., et al. , 2012. Ablation of Rassf2 induces bone defects and subsequent haematopoietic anomalies in mice. EMBO J. 31: 1147–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soung do, Y., Y. Dong, Y. Wang, M. J. Zuscik, E. M. Schwarz et al, 2007. Runx3/AML2/Cbfa3 regulates early and late chondrocyte differentiation. J. Bone Miner. Res. 22: 1260–1270. [DOI] [PubMed] [Google Scholar]
- Stahl E. A., Raychaudhuri S., Remmers E. F., Xie G., Eyre S., et al. , 2010. Genome-wide association study meta-analysis identifies seven new rheumatoid arthritis risk loci. Nat. Genet. 42: 508–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanaki I., Panagiotou O. A., Kodela E., Gogas H., Kypreou K. P., et al. , 2013. Replication and predictive value of SNPs associated with melanoma and pigmentation traits in a southern European case-control study. PLoS One 8: e55712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey J. D., Tibshirani R., 2003. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA 100: 9440–9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturm R. A., 2009. Molecular genetics of human pigmentation diversity. Hum. Mol. Genet. 18: R9–R17. [DOI] [PubMed] [Google Scholar]
- Tetens J., Widmann P., Kuhn C., Thaller G., 2013. A genome-wide association study indicates LCORL/NCAPG as a candidate locus for withers height in German Warmblood horses. Anim. Genet. 44: 467–471. [DOI] [PubMed] [Google Scholar]
- Tonevitsky A. G., Maltseva D. V., Abbasi A., Samatov T. R., Sakharov D. A., et al. , 2013. Dynamically regulated miRNA-mRNA networks revealed by exercise. BMC Physiol. 13: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utsunomiya Y.T., A. M. Perez O’Brien, T. S. Sonstegard, C. P. Van Tassell, A. S. do Carmo et al, 2013. Detecting loci under recent positive selection in dairy and beef cattle by combining different genome-wide scan methods. PLoS One 8: e64280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viitala S., JSzyda, SBlott, NSchulman, MLidauer et al, 2006. The role of the bovine growth hormone receptor and prolactin receptor genes in milk, fat and protein production in Finnish Ayrshire dairy cattle. Genetics 173: 2151–2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voight B. F., Kudaravalli S., Wen X., Pritchard J. K., 2006. A map of recent positive selection in the human genome. PLoS Biol. 4: e72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watterson G., 1975. On the number of segregating sites in genetical models without recombination. Theor. Popul. Biol. 7: 256–276. [DOI] [PubMed] [Google Scholar]
- Weikard R., Altmaier E., Suhre K., Weinberger K. M., Hammon H. M., et al. , 2010. Metabolomic profiles indicate distinct physiological pathways affected by two loci with major divergent effect on Bos taurus growth and lipid deposition. Physiol. Genomics 42A: 79–88. [DOI] [PubMed] [Google Scholar]
- Wijesena H. R., Schmutz S. M., 2015. A missense mutation in slc45a2 is associated with albinism in several small long haired dog breeds. J. Hered. 106: 285–288. [DOI] [PubMed] [Google Scholar]
- Xu L., Bickhart D. M., Cole J. B., Schroeder S. G., Song J., et al. , 2015. Genomic signatures reveal new evidences for selection of important traits in domestic cattle. Mol. Biol. Evol. 32: 711–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Dong G. X., Hu X. S., Miao L., Zhang X. L., et al. , 2013. The genetic basis of white tigers. Curr. Biol. 23: 1031–1035. [DOI] [PubMed] [Google Scholar]
- Yang J., Lee S. H., Goddard M. E., Visscher P. M., 2011. Gcta: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 88: 76–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida C. A., Yamamoto H., Fujita T., Furuichi T., Ito K., et al. , 2004. Runx2 and Runx3 are essential for chondrocyte maturation, and Runx2 regulates limb growth through induction of Indian hedgehog. Genes Dev. 18: 952–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data necessary for confirming the conclusions presented in the article are represented fully within the article or cited references.