Abstract
Deleterious alleles can reach high frequency in small populations because of random fluctuations in allele frequency. This may lead, over time, to reduced average fitness. In this sense, selection is more “effective” in larger populations. Recent studies have considered whether the different demographic histories across human populations have resulted in differences in the number, distribution, and severity of deleterious variants, leading to an animated debate. This article first seeks to clarify some terms of the debate by identifying differences in definitions and assumptions used in recent studies. We argue that variants of Morton, Crow, and Muller’s “total mutational damage” provide the soundest and most practical basis for such comparisons. Using simulations, analytical calculations, and 1000 Genomes Project data, we provide an intuitive and quantitative explanation for the observed similarity in genetic load across populations. We show that recent demography has likely modulated the effect of selection and still affects it, but the net result of the accumulated differences is small. Direct observation of differential efficacy of selection for specific allele classes is nevertheless possible with contemporary data sets. By contrast, identifying average genome-wide differences in the efficacy of selection across populations will require many modeling assumptions and is unlikely to provide much biological insight about human populations.
Keywords: genetic drift, genetic load, human genetics, purging, selection
ONE of the best-known predictions of population genetics is that smaller populations harbor less diversity at any one time but accumulate a higher number of deleterious variants over time (Kimura et al. 1963). Considerable subsequent theoretical effort has been devoted to the study of fitness differences at equilibrium in populations of different sizes (e.g., Glémin 2003) and in subdivided populations (e.g., Whitlock 2002; Roze and Rousset 2003). The reduction in diversity has been observed in human populations that have undergone strong population bottlenecks. For example, heterozygosity decreased in populations that left Africa and further decreased with successive founder events (Tishkoff et al. 1996; Ramachandran et al. 2005; 1000 Genomes Project Consortium 2012; Casals et al. 2013). The effect of demography on the accumulation of deleterious variation has been more elusive in both humans and nonhuman species. In conservation genetics, where fitness can be measured directly and effective population sizes are small, a modest correlation between population size and fitness was observed (Reed and Frankham 2003). In humans, the first estimates of the fitness cost of deleterious mutations were obtained through the analysis of census data (Crow 1958), but recent studies have focused on bioinformatic prediction using genomic data (Davydov et al. 2010; Adzhubei et al. 2013). Lohmueller et al. (2008) found that sites that were variable among Europeans were more likely to be deleterious than sites that were variable among African Americans and attributed the finding to a reduced efficacy of selection in Europeans because of the out-of-Africa (OOA) bottleneck. However, recent studies (Do et al. 2014; Simons et al. 2014) suggest that there has not been enough time for substantial differences in fitness to accumulate in these populations, at least under an additive model of dominance. By contrast Peischl et al. (2013) and, more recently, Henn et al. (2016) have claimed significant differences among populations under range expansion models, and Fu et al. (2014) claim a slight excess in the number of deleterious alleles in European Americans compared to African Americans. These apparent contradictions have sparked a heated debate as to whether the efficacy of selection has indeed been different across human populations (Fu et al. 2014; Lohmueller 2014). Part of the apparent discrepancy stems for disagreement about how we should measure the effect of selection.
What does it mean for selection to be “effective”? Some genetic variants increase the expected number of offspring by carriers. As a result, these variants tend to increase in frequency in the population. This correlation between the fitness of a variant and its fate in the population—i.e., natural selection—holds independent of the biology and history of the population. However, the rate at which deleterious alleles are removed from a population depends on mutation, dominance, linkage, and demography and can vary across populations. Multiple metrics have been proposed to quantify the action of selection in human populations and verify the classical population genetics predictions, leading to apparent discrepancies between studies.
In this article, we first review different metrics used in recent empirical work to quantify the action of selection in human populations. We show that many commonly used metrics implicitly rely on “steady state” or “equilibrium” assumptions, wherein genetic diversity within populations is independent of time. This condition is not met in human populations. We discuss two measures of the efficacy of selection that are appropriate for the study of human populations and other out-of-equilibrium populations.
We then seek to provide an intuitive but quantitative understanding of the effect of mutation, selection, and drift on the efficacy of selection in out-of-equilibrium populations. This is done via a combination of extensive simulation and analytical work describing differentiation between populations after a split from a common ancestor. Using this information, we discuss how the classical predictions concerning the effect of demography on selection could be verified in empirical data from human populations.
Methods
Measuring selection in out-of-equilibrium populations
We consider large panmictic populations whose size may change over time and whose reproduction follows the Wright-Fisher model (Ewens 2012). Given alleles a and A, we assume that genotype has fitness 1, has fitness , and has fitness . We suppose that A is the least favorable allele () and that . In a random-mating population, an allele A at frequency adds an average of to individual fitness compared to the optimal genotype. We compute the expected fitness over multiple loci as under the assumption that the individual selection coefficients are small. Finally, we define the genetic load as the total relative fitness reduction compared to the optimal genotype . This yields
To study the effect of selection over short time spans and in out-of-equilibrium populations, we want to define instantaneous measures of the effect of selection on the genetic load and the frequency of deleterious alleles. In this article, the rate of adaptation refers to the instantaneous rate of fitness increase (or load decrease) in a population. It has contributions from selection, mutation, and drift. The contribution of selection has been the object of considerable theoretical attention: it is the object of the fitness increase theorem (FIT) (see, e.g., Ewens 2012). We will refer to the contribution of selection to the rate of adaptation as the FIT efficacy of selection.
We also wish to study the effect of selection on the frequency of deleterious alleles. There are multiple ways to combine frequencies across loci to obtain a single, genome-wide metric: any linear function of the allele frequencies, with a weight assigned to locus i, provides an equally acceptable metric. A natural option, which weights alleles according to their selection coefficient, is Morton, Crow, and Muller’s total mutational damage (Morton et al. 1956), which is equivalent to the additive genetic load that would be observed if all dominance coefficients were replaced by 1/2, i.e., . Mutation and selection systematically affect , but genetic drift does not. We define the Morton efficacy of selection as the contribution of selection to . In simulations, where all alleles have equal fitness, we use . Another common choice, in empirical studies, is to set for all sites annotated as deleterious by a prediction algorithm and zero otherwise (Do et al. 2014; Fu et al. 2014; Simons et al. 2014). Because different empirical studies use different , direct comparisons of the results can be challenging.
Because Morton and FIT efficacies are instantaneous measures of the effect of selection, they can be integrated over time to measure the effect of selection over arbitrary periods. Their integrals over long periods are directly related to classical steady-state metrics such as the rate of fixation of deleterious alleles and the average genetic load in a population.
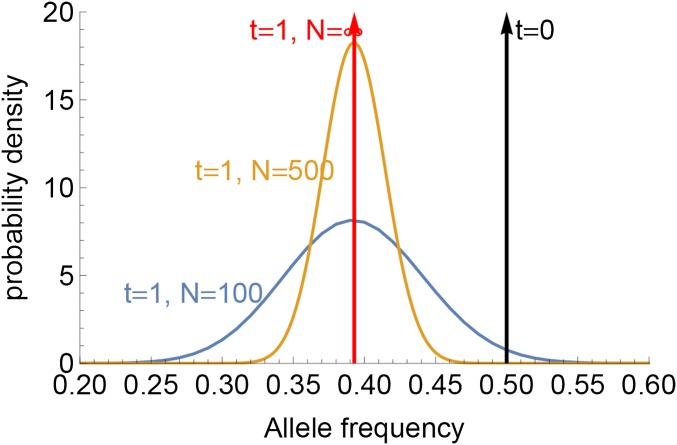
To understand how genetic drift affects the FIT and Morton efficacies of selection, consider an allele with parental frequency x, selection coefficient and no dominance (). In the descending population, this allele is drawn with probability . Figure 1 shows the resulting distribution in offspring allele frequency for , , and . The average frequency is independent of N; hence, the expected FIT and Morton efficacies are equal in all populations. Genetic drift does not instantaneously change the effect of selection.
Figure 1.
Frequency distribution of deleterious alleles with initial frequency 0.5 after one generation, assuming population sizes of and ; a selection coefficient ; and no dominance (). The average effect of selection after one generation does not depend on N.
If we let these populations evolve further, however, we will eventually find that deleterious allele frequencies decrease more slowly in smaller populations. This happens because natural selection acts on fitness differences and therefore requires genetic variation. By dispersing allele frequencies and reducing diversity, genetic drift also reduces the subsequent effect of selection (Figure 2). Drift accumulated during one generation can change the efficacy of selection for many future generations. Conversely, the current average efficacy of selection depends on the drift accumulated in many previous generations. This delay between the action of drift and its impact on selection can be ignored in steady-state populations but not in out-of-equilibrium populations. For this reason, measures of the effect of selection that have been developed for populations of constant size can be misleading or biased when applied to populations that are out of equilibrium.
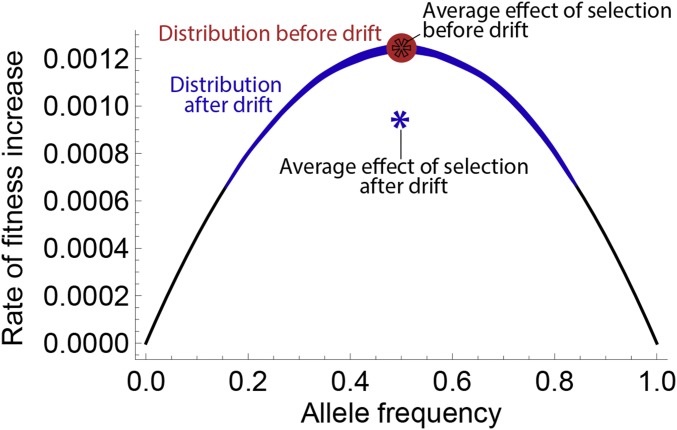
Figure 2.
Effect of drift on the rate of adaptation for . We consider two populations with initial allele frequency 0.5 and selection coefficient . The red population does not undergo drift, and the blue population undergoes one generation of neutral drift, leading to increased variance in allele frequency. The reduced diversity at each locus leads to a lower average rate of fitness increase per generation (blue star). Because the rate of fitness increase is a concave function of allele frequency, drift always reduces the future effect of selection when .
Other measures of the effect of selection
The rate at which deleterious mutations are eradicated from a population, for example, is an intuitive metric for the effect of selection that has been recently applied to out-of-equilibrium populations (Gazave et al. 2013). Over long time scales or in the steady state, this rate of eradication is indeed equivalent to Morton’s efficacy of selection. However, in out-of-equilibrium populations, the rate of eradication is a biased measure of the effect of selection. In Figure 1, the smaller population has a higher rate of eradication of deleterious alleles, but this reflects the action of drift rather than the effect of selection. This effect of drift on the rate of eradication of deleterious alleles is short-lived on phylogenetic time scales, but it can be the dominant effect for time scales relevant to human populations.
Classical work on the efficacy of selection in steady-state populations has emphasized the role of the combined parameter in the dynamics of deleterious alleles. The importance of this combined parameter has led some authors to argue that it should be used as a metric for the efficacy of selection even outside the steady state (Do et al. 2014; Lohmueller 2014). This is problematic for practical and fundamental reasons. On the practical side, the parameter is a function of time and does not allow for comparison between populations over finite times: is not a rate, and its time integral is meaningless. At a more fundamental level, an instantaneous difference between two populations in the product simply indicates a difference in effective population sizes. The interesting biological question is not whether the population sizes are different but whether these differences lead to differential action of selection by the process illustrated in Figure 2.
More generally, it is commonly proposed that the effect of selection should be measured relative to the effect of drift (Lohmueller 2014) because the classical parameter is a ratio between a selection term s and a drift term . Such a relative measure is not necessary: Morton and FIT efficacies are absolute measures of the effect of selection, and they do capture the classical interaction between selection and genetic drift. In populations of constant size, these efficacies do depend on the relative magnitude of selection and drift coefficients through the classical parameter . In out-of equilibrium populations, however, they depend on a more complex function of s and In other words, the classical parameter does not measure the effect of selection as compared to the effect of drift but rather the effect of selection as modulated by past genetic drift.
Finally, even though most classical work has focused on the effect of selection on fitness or allele frequency, Henn et al. (2016) recently proposed to measure the effect of selection on diversity, defining a “reduction in heterozygosity” () statistic that compares the heterozygosity of selected and neutral sites. We show in Section S2 of Supplemental Material, File S1 that is robust to the effect of genetic drift, but it can be biased by recent mutations.
Asymptotic Results
To study the effect of selection after a population split, we calculate the moments of the expected allele frequency distribution under the diffusion approximation. In this formulation, represents the expected number of alleles with frequency between x and at time t. In a randomly mating population of size and constants s and h, the evolution of approximately follows the diffusion equation (Ewens 2012)
| (1) |
where u is the total mutation rate. The first term describes the effect of drift; the second term, the effect of selection; and the third term, the influx of new mutations: δ is Dirac’s delta distribution. From this equation, we can easily calculate evolution equations for moments of the expected allele frequency distribution . For example, the rate of change in allele frequencies is driven by mutation and selection, i.e.,
| (2) |
where is a function of the diversity in the population that generalizes the heterozygosity [see the Appendix for detailed calculations and Evans et al. (2007) and Balick et al. (2015) for other applications of the moment approach]. We can define the contributions of selection and mutation to changes in allele frequency as and . Morton’s efficacy of selection at a locus is simply . Whereas the effect of mutation is constant and independent of population size, Morton’s efficacy depends on the history of the population through :
| (3) |
Similarly, changes in the expected fitness W can be decomposed into contributions from mutation, drift, and selection:
| (4) |
Favorable mutations increase fitness, drift increases fitness when fitness of the heterozygote is below the mean of the homozygotes, and selection always increases average fitness.
The FIT efficacy is therefore
| (5) |
The right-hand side is the additive variance in fitness, and Equation 5 is an expression of the FIT [see, e.g., equations 1.9 and 1.42 in Ewens (2012)]. Importantly, the FIT efficacy describes only one of three genetic contributions to the rate of adaptation. Interpreting changes in fitness in terms of FIT efficacy requires picking apart the effects of drift and mutation from those of selection. In addition to these genetic effects, changes in the environment can directly affect fitness, introducing a further confounder (Mustonen and Lassig 2010).
Now consider an ancestral population that splits into two isolated randomly mating populations with initial sizes and at time . The populations may experience continuous population size fluctuations. If we expand the moments of the allele frequency distribution in Taylor series around we can easily solve the diffusion equation to study the differentiation between the two populations right after the bottleneck. Here we provide an overview of the main results. Detailed derivations are provided in the Appendix.
The difference in fitness between the two populations grows linearly in time under dominance
| (6) |
where t is measured in generations, is the expected heterozygosity in the source population, and represents terms at least quadratic in t. This rapid, linear differentiation is driven by drift coupled with dominance. The smaller population has higher fitness when for : drift hides dominant, deleterious alleles from the action of selection.
If the source population is large and , we have (Crow and Kimura 2009), and the rate of fitness differentiation is independent of s. This generalizes Haldane’s observation that load is insensitive to the selection coefficient in large populations (Haldane 1937). By contrast to the constant-size population case, however, the observation does not hold when . The initial response to the bottleneck is independent of fitness for (Figure S2 and Figure S3) but not for or (Figure 3 and Figure S1).
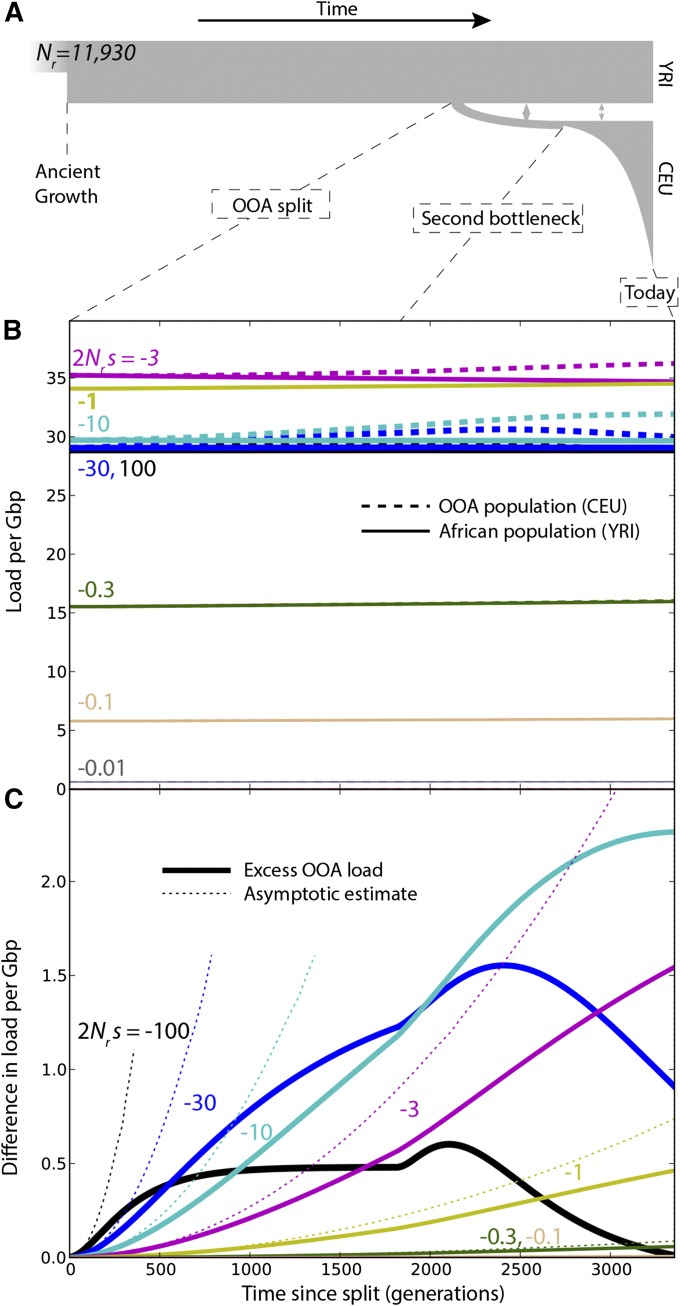
Figure 3.
(A) Changes in load after the OOA split model as a function of s, given ancestral population size and no dominance (). (B) Load evolution after the split. We subtracted the load due to variants fixed in all populations. (C) Difference in load between populations. Because , this is equivalent to the difference in additive load. Dotted lines show asymptotic results from Equations 8 and 10. Load is given per Gbp of variants at the specified selection coefficient. The total amount of variation under strong selection in the human genome is likely much less than 1 Gbp.
The effect of selection on fitness differences grows only quadratically
| (7) |
where is a measure of diversity that reduces to when (see Appendix).
This slower response is the mathematical consequence of the intuition provided by Figure 1 and Figure 2: right after the split, the fitnesses are identical, and the efficacy of selection is the same in both populations. It takes time for drift to increase the variance in allele frequency and cause differences in the efficacy of selection, accounting for a factor . It then takes time for differences in the efficacy of selection to accumulate and produce differences in fitness, accounting for an additional factor .
Combining Equations 6 and 7, we get an asymptotic result for the load differentiation:
| (8) |
This expression describes the leading differentiation in fitness in all the simulations that follow. It is straightforward to refine this asymptotic result by computing higher-order corrections. However, the number of terms in the expansion increases rapidly. Some of these terms are of particular interest, such as the contribution of new mutations. Since the direct effect of mutation on load is independent of demography (Equation 4), we must wait for mutations to accumulate before load differentiation can begin. This leads to an additional factor of compared to the case of standing variation. The contribution of drift acting on new recessive mutations is therefore quadratic:
| (9) |
The effect of selection on new mutations is only cubic in time: we must wait for mutations to appear (contributing a factor of ), then wait for drift to cause differences on the frequency distribution of the new mutations {contributing a factor of }, and finally wait for selection to act on these frequency distribution differences (contributing a factor of ). The leading contribution of selection is therefore
Finally, since drift alone does not produce differences in average allele frequencies, the rate of differentiation in deleterious allele frequencies is always quadratic in time, i.e.,
| (10) |
Simulations
We simulated the evolution of using dadi (Gutenkunst et al. 2009) and the OOA demographic model illustrated in Figure 3A. This model begins with an ancestral population of size with frequency distribution following the quasi-stationary distribution of Kimura (1964) and features population splits and size changes that were inferred from synonymous polymorphism from the 1000 Genomes Project data set (Gravel et al. 2011). We estimated the probability that a variant is at frequency i in a finite sample of size for each population given a mutation rate of bp−1 generation−1 (Gravel et al. 2013) in an infinite genome. We used the finite-sample predictions to estimate the expected genetic load and the contributions of drift, selection, and mutation. Finally, to ensure that the results were not model dependent, we repeated each simulation using a different demographic model described in Do et al. (2014), featuring a single deeper but shorter OOA bottleneck.
We simulated all combinations of selection coefficients and dominance coefficients . The contributions of selection and drift were obtained using Equation 4. To emphasize the long-term effects of the OOA bottleneck even after drift is suppressed, simulations also were carried to future times assuming large population sizes () and no migrations (Figure S6). In all cases, Equations 8–10 capture the initial increase in load (Figure 3, Figure S1, Figure S2, and Figure S7).
Genic selection: h = 1/2
Simulated differences in load are modest and limited to intermediate-effect variants (, ). Assuming the distribution of fitness effects inferred from European-American data by Boyko et al. (2008), the excess load in the OOA population is 0.49 per Gbp of amino-acid-changing variants, in addition to a total accumulated load of 24 Gbp−1 in the African population (this accumulated load does not include variation that was fixed at the time of the split). If we consider the 24 Mb of exome covered by the 1000 Genomes Project and assume that 70% of mutations are coding in that region (Hwang and Green 2004), the model predicts a nonsynonymous load difference of 0.008. The total estimated nonsynonymous load, excluding mutations fixed in the ancestral state, is 0.4 in the African-American population. In this model, the reduced efficacy of selection caused by the OOA bottleneck leads to a relative increase in nonrecessive load of 2%. Since we did not consider fixed ancestral deleterious alleles in the total load, this figure is an overestimate of the relative increase in load due to the bottleneck. The relative increase reaches a maximum of 8% for mutations with . The results are similar if we use the distribution of fitness effects inferred from African-American data (Boyko et al. 2008).
Using the simple bottleneck demographic model of Do et al. (2014), we find very similar load (24 Gbp−1) and load differences across populations (2% of the total load).
Partial and complete dominance
The picture changes dramatically when we consider recessive deleterious variants (). Reactions to changes in population size are linear rather than quadratic, and they are more substantial than in the additive case (Figure S1). The OOA load due to segregating variants with almost doubles after 500 generations. This excess load in the OOA population is due entirely to drift and leads to an increased efficacy of selection in the OOA population because a higher proportion of deleterious alleles is now visible to selection. The difference in load for the most deleterious variants is therefore not sustained. Both the number of very deleterious variants and the associated genetic load eventually become higher in the simulated Yoruba population. By contrast, weak-effect deleterious variants contribute more load in the simulated European population.
Even though a bottleneck inexorably leads to increased load when no dominance is present, the additional exposure of recessive variants therefore leads to “purging,” a reduction in the frequency of deleterious alleles [see Glémin (2003) and references therein]. Simulations show that the increase in recessive load can last hundreds or thousands of generations for weakly deleterious variants. Glémin (2003) argued that the purging effect is suppressed in constant-sized population when is much less than “2 to 5.” This also holds in a nonequilibrium setting in recessive alleles going through a bottleneck (Figure S1) (see also Wang et al. 1999). The time required for purging to compensate the initial fitness loss increases rapidly as the magnitude of the selection coefficients decreases: whereas our model predicts a reduced load in present-day OOA populations for alleles with , it would take over 20,000 generations of continued isolation in a large constant-sized population to see purging in alleles with (Figure S6).
Opposite effects are observed for dominant deleterious variants (Figure S7). Drift tends to increase fitness by combining more of the deleterious alleles into homozygotes, reducing their average effect on fitness. The difference between populations is much less pronounced and less sustained than in the recessive case. Equation 6 shows that the reduced magnitude is caused by reduced ancestral heterozygosity : dominant deleterious alleles are much less likely to reach appreciable allele frequencies before the split. Here again, the population with the highest load depends on the selection coefficient, with a higher load in the simulated European population for strongly deleterious variants and a higher load in the simulated Yoruba population for the weakly deleterious variants.
The distribution of dominance coefficients for mutations in humans is largely unknown, but nonhuman studies suggest that partial recessive may be the norm [see, e.g., Henn et al. (2015) and references therein]. Under models with , we find that the genetic load is elevated in OOA populations for most selection coefficients , whereas the additive genetic load is mostly reduced (Figure S3, B and C, and Figure S4, B and C). These simulations suggest that the rate of adaptation was reduced in OOA populations (i.e., the genetic load is higher in OOA population), while the efficacy of selection was higher in the OOA population, whether it is measured by the Morton efficacy or the FIT efficacy (Figure S5). Thus, unless most nearly neutral variation has , we do not expect an overall elevated number of deleterious variants in OOA populations. As we move closer to additive selection, e.g., at the contributions of alleles with larger and weaker selection coefficients are of comparable magnitude and opposite direction. Because of our limited ability to estimate selection coefficients in humans, this might explain why observing differences in the overall frequency of deleterious alleles between populations has been so difficult. This also suggests that any claim for an across-the-board difference in the efficacy of selection between two populations will have to rely on a number of assumptions about fitness coefficients in human populations.
Present-Day Differences in the Efficacy of Selection
The Wright-Fisher predictions for the instantaneous Morton and FIT efficacies of selection (Equations 3 and 5) depend on the present-day allele frequency distribution, on the dominance coefficient h, and on the selection coefficient s. However, s is a multiplicative factor in both equations and cancels out when we consider the relative rate of adaptation across populations. We can therefore use Equations 3 and 5 to estimate differences in the efficacy of selection between populations based on the present-day distribution of allele frequencies. For nearly neutral alleles, the present-day frequency distribution is similar to the neutral frequency spectrum and largely independent of h. We can therefore use the present-day frequency spectrum for synonymous variation to estimate the relative efficacy of selection for all nearly neutral alleles at different values of h (Figure 4). Figure S9 and Figure S10 show similar results for nonsynonymous and predicted deleterious alleles (for the most deleterious classes, the assumption that the present-day frequency spectra depend weakly on h is less accurate).
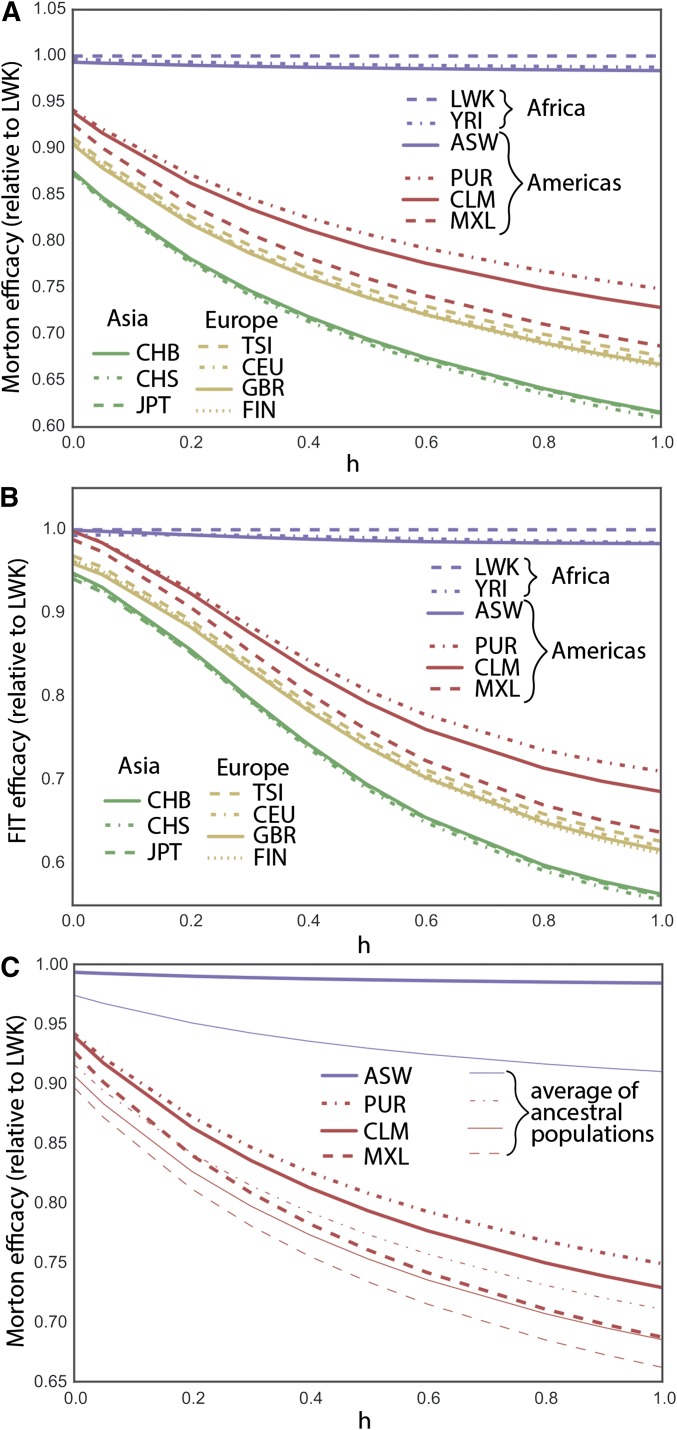
Figure 4.
(A) Present-day Morton efficacy of selection for nearly neutral variants, estimated from 1000 Genomes Project synonymous variation. (B) FIT efficacy for the same variants and (C) Morton efficacy in admixed populations are increased compared to the average efficacies in ancestral populations.
In the nearly neutral case, the Luhya population (LWK) shows the highest Morton and FIT efficacies of selection for most dominance parameters and is used as a basis of comparison. The estimated FIT efficacy of selection is higher in African population for all dominance coefficients, as is the Morton efficacy, except for completely recessive alleles. The reduction in Morton efficacy of selection for nearly neutral variation in OOA populations is 25–39% for dominant variants, 19–31% for additive variants, and 6–13% for fully recessive variants. The reduction in the FIT efficacy in OOA populations is 29–44% for dominant variants, 19–31% for additive variants, and 0.2–6% for fully recessive variants. This is also consistent with the interpretation of Glémin (2003) that purging, measured as the reduction in the frequency of recessive alleles caused by a bottleneck, is not expected for nearly neutral variants. By contrast, estimates using sites with high predicted pathogenicity according to combined annotation-dependent depletion (CADD) (Kircher et al. 2014) do suggest that purging of deleterious variation by drift is still ongoing in OOA populations (Figure S9 and Figure S10).
Admixed populations from the Americas with the highest African ancestry proportion also show elevated predicted efficacy of selection: African Americans (75.9% African ancestry) (Baharian et al. 2015), Puerto Rican (14.8% African ancestry) (Gravel et al. 2013), Colombians (7.8% African ancestry) (Gravel et al. 2013), and Mexican Americans (5.4% African ancestry) (Gravel et al. 2013). The predicted Morton efficacy of selection in admixed populations is much larger than the weighted average of source populations would suggest (Figure 4C, which uses CHB, CEU, and YRI as ancestral population proxies for native, European, and African ancestries). By averaging out some of the genetic drift experienced by the source populations since their divergence, admixture increases the overall amount of additive variance in the population and therefore leads to a substantial and rapid increase in the predicted efficacy of selection for nearly neutral alleles.
Data availability
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.
Discussion
Selection affects evolution in many ways. It tends to increase the frequency of favorable alleles and the overall fitness of a population, and it often reduces diversity. The rates at which it performs these tasks vary across populations, and population geneticists like to frame these differences in terms of the efficacy of selection. The word “efficacy” implies a measure of achievement, but there are many ways to define achievement for selection. We considered two measures of achievement: the change in deleterious allele frequency (i.e., the Morton efficacy) and the change in load caused by selection (i.e., the FIT efficacy). Even though the two quantities are closely related and are equal for additive selection, the Morton efficacy is much easier to measure: systematic differences in the frequency of deleterious alleles are robust to drift and to modest changes in the environment. By contrast, the FIT efficacy is impossible to observe directly and requires picking apart the contributions of selection, drift, and the environment. Given the long-standing controversy about how this should be done in the context of Fisher’s fundamental theorem (Ewens and Lessard 2015), we would advise against using it.
We have argued that other popular measures for the efficacy of selection (Lohmueller et al. 2008; Casals et al. 2013; Lohmueller 2014; Henn et al. 2016) are biased in out-of-equilibrium populations studied over short time scales. Many previous claims that selection acted differentially in human populations (Lohmueller et al. 2008; Casals et al. 2013) could be explained by these biases. Confirming this interpretation, Fu et al. (2014) found no differences in the average frequency of deleterious alleles between African Americans and European Americans in the ESP 6500 data set (NHLBI GO Exome Sequencing Project 2012). However, they did report a slight but extremely significant difference in the average number of deleterious alleles per individual for a set of putatively deleterious SNPs. The contrasting results are surprising because the two statistics are equal up to a multiplicative constant: the average number of deleterious alleles per genome equals the mean frequency of deleterious alleles multiplied by the number of loci. We could reproduce the results from Fu et al. (2014) but found that the statistical test used did not account for variability introduced by genetic drift in a finite genome: results remained significant if allele frequencies were randomly permuted between African Americans and European Americans (see Section S1 of File S1 for details). This emphasizes that an empirical observation of differences in genetic load must be robust to both finite sample size and finite genome to be attributed to differences in the efficacy of selection.
Figure 4, Figure S9, and Figure S10 strongly suggest that the OOA bottleneck still influences the present-day efficacy of selection. By extension, they also suggest that the efficacy of selection did differ and will differ among populations. Importantly, the differences in frequency distributions across populations that provide this support are not a consequence of past differences in the efficacy of selection but a possible cause for such differences in the present and future. We have shown that some of the future differences are not inevitable and can be attenuated by demographic processes including admixture. Therefore, measuring actual differences in the efficacy of selection can only be achieved by measuring actual differences in the average frequency or effect of deleterious alleles.
The simulations presented here, together with the results of Simons et al. (2014) and Do et al. (2014), do suggest that the classical prediction on the differential efficacy of selection in small populations can be verified if only we can accurately isolate variants of specific selective effect and dominance coefficients. By picking apart variants of different selection and dominance coefficients, we should soon be able to convincingly and directly observe the consequences of differences in the efficacy of selection. The recent results of Henn et al. (2016) using a version of the Morton efficacy suggest such differences for a subset of variants and therefore provide important experimental validation for a classical population genetics prediction. By contrast, the observation of genome-wide differences in the efficacy of selection across populations depends on the cancellation of effects across different variant classes and therefore can depend sensitively on the particular choice of a metric. For this reason, overall differences in load among populations may not be particularly informative about the fundamental processes governing human evolution.
Acknowledgments
I thank S. Baharian, M. Barakatt, B. Henn, D. Nelson, and S. Lessard for useful comments on this manuscript and W. Fu and J. Akey for help in reproducing their results. This research was undertaken, in part, thanks to funding from the Canada Research Chairs Program and a Sloan Research Fellowship.
Appendix
Background
To derive the asymptotic results in the text, we start with the diffusion approximation for the distribution of allele frequencies x over time t in an infinite-sites model (see Crow and Kimura 2009, section 8.6):
| (A1) |
where N is the effective population size, h is the dominance coefficient, s is the selection coefficient, and u is the mutation rate. In this model, new mutations are constantly added via Dirac’s delta function δ. Because there are no back-mutations in this model, the proportion of fixed mutations increases over time without bound. Because we are only interested in population differences accumulating over a short time span, however, we can simply ignore the (infinite) number of deleterious alleles that fixed before the population split. The time scales that we will consider are short enough that back-mutations and multiple mutations contribute little to changes in allele frequencies.
A complete solution of this problem can be expressed as a superposition of Gegenbauer polynomials (Kimura 1964). However, here we are looking for simple asymptotic results that will help us to understand the dynamics of the problem. We will consider the evolution of moments of the allele frequency distribution . Similar moment approaches have been used in Evans et al. (2007) and Balick et al. (2015). Because there is a potentially infinite number of fixed sites at frequencies 0 and 1, it is often convenient to distinguish contributions from segregating sites and fixed sites, i.e.,
where is the number of sites fixed at frequency 0, is the number of sites fixed at frequency 1, and is Kronecker’s delta. Both and can be infinite in this model, but this will not be a problem because we will ultimately consider only differences or rates of change in the moments, and these remain finite. In this notation, is the (possibly infinite) number of sites, and is the expected number of alternate alleles per haploid genome.
To obtain evolution equations for the moments, we integrate both sides of Equation A1 using . The left-hand side gives
| (A2) |
and the right-hand side can be integrated by parts. For , this yields
where and are defined by continuity from the open interval and do not include fixed sites. Because the number of sites is constant () and the diffusion equation is continuous, we require
| (A3) |
These equations are equivalent to equations 3.18 and 3.19 in Kimura (1964).
To obtain an evolution equation for at arbitrary k, we return to the integration of Equation A1 with . We use the left-hand-side expression obtained in Equation A2 and once again integrate the right-hand side by parts. This yields
| (A4) |
where
| (A5) |
These are functions of the moments μ and therefore can be thought of as measures of the shape of the frequency distribution φ.
The first term in Equation A4 represents the effect of drift; the second term, the effect of selection; and the third term, the effect of mutation. For example, if and , we get
The frequency of damaging alleles can decrease because of selection or increase because of mutation.
Response in Allele Frequencies
Solving Equation A4 in general is challenging because can depend on and , leading to an infinite number of coupled equations. However, it can be used to calculate the response in allele frequency to a sudden change in demographic or selective conditions. Consider a population of size that experiences a change in size to at time . We can expand for short times, i.e.,
| (A6) |
where is the kth moment prior to the population size change, and represent terms at least cubic in t. The coefficients can be evaluated by expanding both sides of Equation A4 using Equation A6 and then collecting powers of t. For example, we get
The frequency of variants can increase even in a steady-state regime with because our model assumes a constant supply of irreversible mutations. However, this linear term is independent of and does not contribute to differences across populations that share a common ancestor. Differences in the number of segregating sites between two populations with sizes and appear at the next order in t. Computations are elementary but a bit more cumbersome. Matching terms linear in t in Equation A4, we find Equation 10:
where is the moment computed for the common ancestral population.
Response in Genetic Load
To compute the fitness in the diploid case, we write
| (A7) |
Using Equation A4, we get
| (A8) |
where
| (A9) |
are the instantaneous contributions of selection, mutation, and drift to changes in fitness. The mutation term is constant in time and independent of population size; it does not contribute directly to differences across populations. The drift term, by contrast, has an explicit dependence on the population size; this leads to differentiation between populations that grow linearly in time. To see this, we compute the load using Equation A7 and the time dependence computed in Equation A6 as in the section Response in Allele Frequencies:
| (A10) |
This reduction in load is driven by drift, i.e., the third term in Equation A9. It is not caused by selection, in the sense that it does not result from differential reproductive success between individuals. As expected, the contribution of drift vanishes for additive variants ().
For arbitrary h, the change in fitness caused by selection is
where is a statistic of the ancestral frequency distribution, i.e.,
| (A11) |
which reduces to the heterozygosity when The statistic depends only on the ancestral frequency distribution and the dominance coefficient.
Genetic drift also contributes to the changes in load at second order in t through . In addition to the linear term from Equation A10, we find three quadratic contributions that vanish when : a second-order contribution of genetic drift, a contribution from the rate of change in population size and drift, and a contribution from new mutations and drift. Even though these terms can be comparable in magnitude to the contribution of selection in Equation A11 when , they are subdominant to Equation A10. We only consider the contribution of new mutations in some detail because this contribution tells us whether population differentiation in the genetic load is due to old, shared variation or to new, population-specific variation.
Effect of New Mutations
If we set in the preceding equations, we can calculate the impact of new mutations on the genetic load. The leading term is again due to drift and dominance:
| (A12) |
while the leading term describing the efficacy of selection is now cubic in t:
When , drift also contributes terms to . These are reasonably straightforward to compute but are subdominant to Equation A12. We therefore use the asymptotic result
| (A13) |
Comparisons with simulated data are shown in Figure S8.
Footnotes
Communicating editor: R. Nielsen
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.115.184630/-/DC1.
Literature Cited
- 1000 Genomes Project Consortium , 2012. An integrated map of genetic variation from 1,092 human genomes. Nature 491: 56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adzhubei I., Jordan D. M., Sunyaev S. R., 2013. Predicting functional effect of human missense mutations using PolyPhen-2. Curr. Protoc. Hum. Genet. 76: 7.20.1–7.20.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baharian S., Barakatt M., Gignoux C. R., Shringarpure S., 2015. The Great Migration and African-American genomic diversity. bioRxiv (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balick D. J., Do R., Cassa C. A., Reich D., Sunyaev S. R., 2015. Dominance of deleterious alleles controls the response to a population bottleneck. PLoS Genet. 11: e1005436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyko A. R., Williamson S. H., Indap A. R., Degenhardt J. D., Hernandez R. D., et al. , 2008. Assessing the evolutionary impact of amino acid mutations in the human genome. PLoS Genet. 4: e1000083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casals F., Hodgkinson A., Hussin J., Idaghdour Y., Bruat V., et al. , 2013. Whole-exome sequencing reveals a rapid change in the frequency of rare functional variants in a founding population of humans. PLoS Genet. 9: e1003815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crow J. F., 1958. Some possibilities for measuring selection intensities in man. Hum. Biol. 30: 1–13. [PubMed] [Google Scholar]
- Crow J. F., Kimura M., 2009. An Introduction to Population Genetics Theory. Blackburn Press, Caldwell, NJ. [Google Scholar]
- Davydov E. V., Goode D. L., Sirota M., Cooper G. M., Sidow A., et al. , 2010. Identifying a high fraction of the human genome to be under selective constraint using GERP++. PLOS Comput. Biol. 6: e1001025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Do R., Balick D., Li H., Adzhubei I., Sunyaev S., et al. , 2014. No evidence that selection has been less effective at removing deleterious mutations in Europeans than in Africans. Nat. Genet. 47: 126–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans S. N., Shvets Y., Slatkin M., 2007. Non-equilibrium theory of the allele frequency spectrum. Theor. Popul. Biol. 71: 109–119. [DOI] [PubMed] [Google Scholar]
- Ewens W. J., 2012. Mathematical Population Genetics 1: Theoretical Introduction. Springer Science & Business Media, New York. [Google Scholar]
- Ewens W. J., Lessard S., 2015. On the interpretation and relevance of the fundamental theorem of natural selection. Theor. Popul. Biol. 104: 59–67. [DOI] [PubMed] [Google Scholar]
- Fu W., Gittelman R. M., Bamshad M. J., Akey J. M., 2014. Characteristics of neutral and deleterious protein-coding variation among individuals and populations. Am. J. Hum. Genet. 95: 421–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazave E., Chang D., Clark A. G., Keinan A., 2013. Population growth inflates the per-individual number of deleterious mutations and reduces their mean effect. Genetics 195: 969–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glémin S., 2003. How are deleterious mutations purged? Drift vs. nonrandom mating. Evolution 57: 2678–2687. [DOI] [PubMed] [Google Scholar]
- Gravel S., Henn B. M., Gutenkunst R. N., Indap A. R., Marth G. T., et al. , 2011. Demographic history and rare allele sharing among human populations. Proc. Natl. Acad. Sci. USA 108: 11983–11988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gravel S., Zakharia F., Moreno-Estrada A., Byrnes J. K., Muzzio M., et al. , 2013. Reconstructing Native American migrations from whole-genome and whole-exome data. PLoS Genet. 9: e1004023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutenkunst R. N., Hernandez R. D., Williamson S. H., Bustamante C. D., 2009. Inferring the joint demographic history of multiple populations from multidimensional SNP frequency data. PLoS Genet. 5: e1000695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldane J., 1937. The effect of variation of fitness on fitness. Am. Nat. 71: 337–349. [Google Scholar]
- Henn B. M., Botigué L. R., Bustamante C. D., Clark A. G., Gravel S., 2015. Estimating the mutation load in human genomes. Nat. Rev. Genet. 16: 333–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henn B. M., Botigué L. R., Peischl S., Dupanloup I., Lipatov M., et al. , 2016. Distance from sub-Saharan Africa predicts mutational load in diverse human genomes. Proc. Natl. Acad. Sci. USA 113: E440–E449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang D. G., Green P., 2004. Bayesian Markov chain Monte Carlo sequence analysis reveals varying neutral substitution patterns in mammalian evolution. Proc. Natl. Acad. Sci. USA 101: 13994–14001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M., 1964. Diffusion models in population genetics. J. Appl. Probab. 1: 177. [Google Scholar]
- Kimura M., Maruyama T., Crow J. F., 1963. The mutation load in small populations. Genetics 48: 1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher M., Witten D. M., Jain P., O’Roak B. J., Cooper G. M., et al. , 2014. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 46: 310–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohmueller K. E., 2014. The distribution of deleterious genetic variation in human populations. Curr. Opin. Genet. Dev. 29: 139–146. [DOI] [PubMed] [Google Scholar]
- Lohmueller K. E., Indap A. R., Schmidt S., Boyko A. R., Hernandez R. D., et al. , 2008. Proportionally more deleterious genetic variation in European than in African populations. Nature 451: 994–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton N. E., Crow J. F., Muller H. J., 1956. An estimate of the mutational damage in man from data on consanguineaous marriages. Proc. Natl. Acad. Sci. USA 42: 855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustonen V., Lassig M., 2010. Fitness flux and ubiquity of adaptive evolution. Proc. Natl. Acad. Sci. USA 107: 4248–4253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NHLBI GO Exome Sequencing Project, 2012 Exome variant server. Available at: evs.gs.washington.edu/EVS/. Accessed October 2015.
- Peischl S., Dupanloup I., Kirkpatrick M., Excoffier L., 2013. On the accumulation of deleterious mutations during range expansions. Mol. Ecol. 22: 5972–5982. [DOI] [PubMed] [Google Scholar]
- Ramachandran S., Deshpande O., Roseman C. C., Rosenberg N. A., Feldman M. W., et al. , 2005. Support from the relationship of genetic and geographic distance in human populations for a serial founder effect originating in Africa. Proc. Natl. Acad. Sci. USA 102: 15942–15947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed D., Frankham R., 2003. Correlation between fitness and genetic diversity. Conserv. Biol. 17: 230–237. [Google Scholar]
- Roze D., Rousset F., 2003. Selection and drift in subdivided populations: a straightforward method for deriving diffusion approximations and applications involving dominance, selfing and local extinctions. Genetics 165: 2153–2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons Y. B., Turchin M. C., Pritchard J. K., Sella G., 2014. The deleterious mutation load is insensitive to recent population history. Nat. Genet. 46: 220–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tishkoff S. A., Dietzsch E., Speed W., Pakstis A. J., Kidd J. R., et al. , 1996. Global patterns of linkage disequilibrium at the CD4 locus and modern human origins. Science 271: 1380–1387. [DOI] [PubMed] [Google Scholar]
- Wang J., Hill W. G., Charlesworth D., Charlesworth B., 1999. Dynamics of inbreeding depression due to deleterious mutations in small populations: mutation parameters and inbreeding rate. Genet. Res. 74: 165–178. [DOI] [PubMed] [Google Scholar]
- Whitlock M. C., 2002. Selection, load and inbreeding depression in a large metapopulation. Genetics 160: 1191–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.