Abstract
VEGF and angiopoietin-1 (Ang-1) are essential factors to promote angiogenesis through regulation of a plethora of signaling events in endothelial cells (ECs). Although pathways activated by VEGF and Ang-1 are being established, the unique signaling nodes conferring specific responses to each factor remain poorly defined. Thus, we conducted a large-scale comparative phosphoproteomic analysis of signaling pathways activated by VEGF and Ang-1 in ECs using mass spectrometry. Analysis of VEGF and Ang-1 networks of regulated phosphoproteins revealed that the junctional proteins ZO-1, ZO-2, JUP and p120-catenin are part of a cluster of proteins phosphorylated following VEGF stimulation that are linked to MAPK1 activation. Down-regulation of these junctional proteins led to MAPK1 activation and accordingly, increased proliferation of ECs stimulated specifically by VEGF, but not by Ang-1. We identified ZO-1 as the central regulator of this effect and showed that modulation of cellular ZO-1 levels is necessary for EC proliferation during vascular development of the mouse postnatal retina. In conclusion, we uncovered ZO-1 as part of a signaling node activated by VEGF, but not Ang-1, that specifically modulates EC proliferation during angiogenesis.
The concerted action of VEGF and angiopoietin-1 (Ang-1)1 on endothelial cells (ECs) regulates the process of new blood vessel formation, called angiogenesis (1). During vascular development, VEGF and Ang-1 have complementary roles to form mature blood vessels. VEGF plays a key role in vessel sprouting and initiation of new vessels, whereas Ang-1 is required for subsequent vessel maturation (2–4). Pathological angiogenesis leads to aberrant blood vessel formation in diseases such as cancer progression and metastasis or in vascular retinopathies (5, 6). Targeting intracellular signaling events elicited by VEGF and Ang-1 in ECs therefore holds promise for the treatment of angiogenic diseases (7).
Through activation of their cognate tyrosine kinase receptors, VEGFR2 and Tie2, VEGF and Ang-1 trigger phosphorylation of multiple intracellular effectors to induce proliferation, survival and migration of ECs (8, 9). When examined individually, it is appreciated that both receptors activate common signaling pathways in ECs such as ERK/MAPK (10, 11), PI3K/Akt (12–14), and p38 MAPK (11, 15) to induce angiogenesis. However, VEGF and Ang-1 must signal differently to cell–cell junctions to respectively augment or decrease endothelial permeability to macromolecules (16–19). This shows that, in order to induce angiogenesis, VEGF and Ang-1 must activate overlapping and diverging signaling pathways in ECs. There are numerous studies on the implication of individual intracellular signaling pathways that are activated by VEGF and Ang-1 to control angiogenesis. However, a global comparison and analysis of signaling pathways activated in ECs by these growth factors is needed to uncover novel interrelations between specific intracellular signaling events that control the angiogenic response.
The endothelial junctions have long been associated with barrier functions, however they also receive and transmit signals that regulate cell communication, differentiation and proliferation (20–22). Proteins that form endothelial intercellular junctions integrate signaling events that are important for angiogenesis. For instance, genetic deletion of VE-cadherin, β-catenin, or ZO-1 in mice leads to embryonic lethally because of vascular defects (23–26). In addition, it is well established that signals transmitted from intercellular junctions to the nucleus control contact-mediated inhibition of cell proliferation. In ECs, the adherens junction proteins β-catenin and p120-catenin are known to elicit signaling pathways that induce proliferation when junctions are disrupted (21, 27). Both proteins can translocate to the nucleus and act as modulator of gene expression through interaction with the TCF/LEF transcription factors for β-catenin or by relieving the repressor activity of the transcription factor Kaiso for p120-catenin (28, 29). The tight junction protein ZO-1 was recently shown in ECs to function as a major cytoskeletal organizer that orchestrates adherens junctions to control barrier function, cell migration, and angiogenesis (30). However, the role of ZO-1 in the regulation of EC proliferation is undefined.
Herein, the phosphoproteomes of ECs treated with VEGF or Ang-1 were systematically compared to profile the activation of intracellular signaling pathways. Network analysis of the phosphoproteins regulated by VEGF and Ang-1 uncovered a cluster of cell-cell junction proteins unique to VEGF treatment, which is linked to activation of MAPK1 and promotion of EC proliferation. We demonstrate that ZO-1 is the central regulator of this cluster of cell junction proteins to promote MAPK1 activation. Furthermore, we observed that reduction of the cellular levels of ZO-1 correlates with cell proliferation during retinal vascular development in mice. Collectively, our comparative phosphoproteomic analyses identified a regulatory signaling node, differentially engaged by VEGF over Ang-1, that controls EC proliferation.
EXPERIMENTAL PROCEDURES
Cell Culture and Reagents
Bovine aortic endothelial cells (BAECs), obtained from VEC Technologies (Rensselaer, NY), were cultured in Dulbecco Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum (HyClone), 2 mm l-glutamine, 100 U/ml penicillin, and 100 μg/ml streptomycin. BAECs were treated with the recombinant human VEGF-A and recombinant human Ang-1 obtained from R&D System. The primary antibodies used were: Anti-phospho-p44/42 MAPK (Thr202/Tyr204) (monoclonal antibody [mAb]), p42/44 MAPK (polyclonal antibody [pAb]), phospho-Ser1179-eNOS (pAb), eNOS (mAb), beta-actin (mAb), phospho-Ser252 p120-catenin (pAb), and BrdU (mAb) from Cell Signaling Technology, Danvers, MA. Anti-p120-catenin (mAb), anti-VE-cadherin (pAb), and anti-ZO-1 (pAb) were from Santa Cruz Biotechnology, Santa Cruz, CA. Anti-JUP (mAb) and anti-β-catenin (mAb) were from BD Transduction Laboratories, San Jose, CA. Anti-phospho-Ser268 p120-catenin was from Novus Biologicals, Littleton, CO. Anti-phospho-histone 3 (mAb) was from Abcam, Cambridge, UK and Rhodamine conjugated Lectin I was from Vector Laboratories, Burlingame, CA.
Trypsin Digestion
For phosphoproteomics experiments, cells were washed three times with ice-cold PBS and scraped into lysis buffer containing 8 m urea, 50 mm Tris-HCl pH 7.6, 1 mm ethylenediaminetetraacetic (EDTA), 1 mm ethyleneglycoltetraacetic (EGTA), 10 mm sodium fluoride (NaF), 1 mm sodium orthovanadate (Na3VO4), 1 mm sodium pyrophosphate (Na4P2O7) and 50 mm NaCl. The lysates were sonicated and cleared by centrifugation at 3,000 × g for 10 min. Protein concentration was carried out using BCA protein assays. β-casein phosphoprotein (50 ng; Sigma) was spiked in all samples as an internal standard to normalize for phosphopeptide recovery between conditions. For in-solution digestion, 2 mg of protein was reduced at 37 °C using dithiothreitol (DTT) for one hour and alkylated by iodoacetaminde for 60 min at room temperature in the dark. Then, the mixture was digested using trypsin (ratio enzyme/total protein of 1:50) followed by incubation at 37 °C overnight. The tryptic digestion was quenched by adding 1% trifluoroacetic acid (TFA).
Desalting and Phosphopeptides Enrichment with TiO2
The resultant peptides were desalted using Oasis HLB extraction plate 30 μm (Waters UK). Briefly, the wells were equilibrated with 500 μl of 100% methanol and washed with 500 μl of H2O. Afterward, wells were loaded with the peptide mixture, washed with 500 μl of 5% methanol and eluted with 400 μl of 100% methanol. The recovered peptides were lyophilized and subjected to phosphopeptides enrichment as follow (31–33). Peptides were resuspended in 200 μl of 80% Acetonitrile (ACN)/3% TFA/300 mg DHB (dihydroxybenzoic acid). TiO2 beads (GL sciences) were resuspended in the same buffer and 20 μl of this slurry was added to each (1:2 peptide to beads ratio). Samples were incubated for 30 min on a rotator at room temperature and centrifuged at 5000 × g for 1 min. Phosphopeptides-bound TiO2 beads were washed 3 times with 30% ACN, 3% TFA on a StageTip C8 material (ThermoFisher Scientific) and then 3 times with 80% ACN, 0.3% TFA. Phosphopeptides were eluted twice using a C8 StageTip with 75 μl 40% ACN, 15% NH4OH. Eluted phosphopeptides samples were vacuum-dried prior to LC-MS/MS analyses.
Liquid Chromatography-tandem Mass Spectrometry
Phosphopeptides enriched samples were resolubilized under agitation for 15 min in 21 μl of 1% ACN/1% formic acid. The LC column was a PicoFrit fused silica capillary column (17 cm × 75 μm i.d; New Objective, Woburn, MA), self-packed with C-18 reverse-phase material (Jupiter 5 μm particles, 300 Å pore size; Phenomenex, Torrance, CA) using a high-pressure packing cell. This column was installed on the Easy-nLC II system (Proxeon Biosystems, Odense, Denmark) and coupled to the LTQ Orbitrap Velos (ThermoFisher Scientific, Bremen, Germany) equipped with a Proxeon nanoelectrospray Flex ion source. The buffers used for chromatography were 0.2% formic acid (buffer A) and 100% ACN/0.2% formic acid (buffer B). Peptides were loaded on-column at a flow rate of 600 nL/minute and eluted with a 2-slopes gradient at a flow rate of 250 nL/minute. Buffer B first increased from 1 to 40% in 110 min and then from 40 to 80% B in 50 min.
LC-MS/MS data was acquired using a data-dependent top8 method combined with a MS3 scanning upon detection of a neutral loss of phosphoric acid (48.99, 32.66, or 24.5 Th) in MS2 scans. The mass resolution for full MS scan was set to 60,000 (at m/z 400) and lock masses were used to improve mass accuracy. Mass over charge ratio range was from 375 to 1800 for MS scanning with a target value of 1,000,000 charges and from ∼1/3 of parent m/z ratio to 1800 for MSn scanning in the linear ion trap analyzer with a target value of 10,000 charges. The data-dependent scan events used a maximum ion fill time of 100 milliseconds and target ions already selected for MS/MS were dynamically excluded for 30 sec after two repeat counts. Nanospray and S-lens voltages were set to 1.5 kV and 50 V, respectively. The normalized collision energy used was of 27 with an activation q of 0.25 and activation time of 10 milliseconds. Capillary temperature was 250 °C.
Experimental Design and Statistical Rationale
Raw mass spectrometry data were processed using the MaxQuant software (version 1.3.0.5) (34). Database searching was performed using the Andromeda search engine (version 1.3.0.5) integrated into MaxQuant against the bovine NCBI and UniProt database (89,075 entries downloaded on May 2012 and 27,028 entries downloaded on June 2012, respectively) and against the human UniProt database (86,749 entries downloaded on June 2012) (35). Enzyme specificity was set to trypsin and up to two missed cleavages was allowed. Cysteine carbamidomethylation (C) was set as fixed modification and oxidation (M) and phosphorylation (STY) were set as variable modification. The minimum required peptide length was six amino acids. Mass tolerances for precursor ions and fragment ions were set to 20 ppm and 0.5 Da, respectively. The “matching between runs” algorithm in MaxQuant was enabled with a time window of 3 min to transfer identifications between adjacent replicates. The false discovery rate (FDR) was estimated by searching against the databases with the reversed sequences. For protein and peptide identification, the maximum FDR was set to 1%. For each peptide, the modification sites and the localization probabilities were assigned and calculated by MaxQuant. Phosphorylation events with a localization probability above 0.75 were considered localized on the respective S/T/Y residue. Three biological replicates of nontreated, VEGF or Ang-1 treated cells were performed. In addition, one technical replicate for nontreated and Ang-1 and two technical replicates for VEGF treated cells were performed to validate reproducibility of the MS analysis. Correlations between the technical replicates are shown in supplemental Fig. S1A. Moreover, a phosphopeptide had to be identified in at least two biological replicates per conditions in order to be considered for further statistical analyses. The technical replicates for each condition were averaged and used for phosphopeptide quantification. First, in order to determine phosphopeptides that were statistically significant following VEGF or Ang-1 treatment, we performed an analysis of variance where the intensities of each treatment were compared with nontreated. Phosphopeptides with a p value below 0.05 were retained. Secondly, to identify the regulated phosphopeptides, the ratio of the intensities from VEGF or Ang-1 treatments to nontreated was calculated for each experiment to determine the fold-changes. The fold-changes of all phosphopeptides from the three biological replicates were log2 transformed and then averaged. Finally, the average log2 fold-change of each phosphopeptide was normalized using a β-casein phosphopeptide (FQpSEEQQQTEDELQDK) that was added to all samples as an internal standard to correct for phosphopeptide recovery between the different LC-MS/MS runs (correction factor VEGF: 0.11; Ang-1: −0.15; See supplemental Table S1). A cut-off of > 1.4-fold increase (log2 fold-change > 0.5) and < 0.7-fold decrease (log2 fold-change < 0.5) was applied to define regulated phosphopeptides. The MS proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) (36) via the PRIDE partner repository with the data set identifier PXD002456. MaxQuant viewer version 1.3.0.5 was used to view the annotated spectra.
Bioinformatic Analysis
Gene ontology annotations for biological processes were obtained from the Gene Ontology integrated in STRING database (version 9.1) (37). Data was visualized using Perseus 1.3.0.4. Ingenuity pathway analysis (IPA, Qiagen) was used to identify pathways that were significantly enriched. Only terms or pathways that were significantly enriched with a p value of less than 0.05 were used in the analysis. To generate protein interactions network, STRING interactions database was used. The published or informatic-predicted interactions were first determined using standard STRING-defined confidence (medium confidence 0.4). The obtained STRING network data were imported into the Cytoscape software (version 3.0.1) (http://www.cytoscape.org) (38). For clustering analysis, MCODE plugin in Cytoscape was used to identify functional protein clusters within the networks. Cluster enrichment was manually annotated based on the GO biological process enrichment.
Plasmids and Transfections
p120-catenin, ZO-1, ZO-2, JUP, VE-cadherin, α-catenin, β-catenin small interfering RNA (siRNA), as well as nonsilencing control (CT) siRNA were obtained from IDT technology, Coralville, IA. si-CT AUGAACGUGAAUUGCUCAAUU, si-VE-cadherin ACAAAGAACUGGACAGAGAUU, si-p120-catenin-(1) CAAGAAAGGCAAAGGGAAAUU, si-p120-catenin-(2) GGACAGAAAGAUUCGGAUAUU, si-β-catenin AAGUAGCUGAUAUUGAUGGACUU, si-α-catenin GGGCAAUGCUGGACGUAAAUU, si-ZO-1-(1) GCAGAGAGGAAGAGAGAAUUU, siZO-1-(2) CAGCAAAGGUGUACAGGAAUU, si-ZO-2 GGUUAAAUACCGUGAGACAUU, si-JUP GCAACAACAGCAAGAACAAUU. Human ZO-1 tagged myc was obtained from Addgene. Murine p120-catenin was a generous gift from Dr. Albert Reynolds (Vanderbilt University Medical Center). p120-catenin was inserted in a pCMV-myc tag. BAECs were transfected with plasmids or siRNAs using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) according to manufacturer's instructions.
Immunoblotting
For immunoblotting, cells were solubilized with a lysis buffer containing 1% Nonidet P-40, 0.1% sodium dodecyl sulfate (SDS), 0.1% deoxycholic acid, 50 mm Tris (pH 7.4), 0.1 mm EGTA, 0.1 mm EDTA, 20 mm NaF, 1 mm sodium Na4P2O7, and 1 mm Na3VO4. Lysate were incubated for 30 min at 4 °C, centrifuged at 14,000 × g for 10 min, boiled in SDS sample buffer, separated by SDS-polyacrylamide gel electrophoresis, transferred onto a nitrocellulose membrane (Hybond ECL; Amersham Biosciences, Mississauga, ON, Canada), and Western blotted. Antibody detection was performed by a LI-COR Odyssey infrared imaging system (LI-COR Biosciences, Lincoln, NE) using Alexa-680 and Alex-800-labeled secondary antibodies (Invitrogen) or by a chemiluminescence-based detection system (ECL; GE healthcare). The quantifications of band intensities presented are means of at least three independent experiments.
RNA Extraction and Quantitative (q)RT-PCR
Total RNA was extracted with an RNeasy Mini kit (Qiagen, Hilden, Germany), and reversed-transcribed into complementary DNA by using SuperScript II Reverse Transcriptase (Invitrogen) according to the manufacturer's instructions. Real-time PCR was performed with the SYBR Select Master Mix (Applied Biosystems, Foster City, CA) and conducted in triplicate for each sample with RNA preparations from at least two independent experiments. Gene expression levels were analyzed on an Eco™ Real-Time PCR System (Illumina) and normalized based on β-actin mRNA expression. Bovine cyclin D1 forward primer is TTACACTGACAACTCCATCCG and its reverse primer is CATCTTGGAGAGGAAGTGCTC. Bovine β-actin forward primer is GACAGGATGCAGAAAGAGATCA and its reverse primer is AATCCACACGGAGTACTTGC.
Immunofluorescence
BAECs were transfected and then cultured on 0.1% gelatin-coated coverslips. Cells were washed with cold PBS and fixed for 20 min in Methanol/Acetone (1:1) at −20 °C. Fixed cells were rinsed with PBS and blocked with 1% BSA for 1 h at room temperature. After blocking, cells were incubated with primary antibodies overnight in 0.1% BSA in PBS. Bound primary antibodies were visualized after 1 h of incubation using Alexa Fluor 488-labeled donkey anti-goat, Alexa Fluor 568-labeled donkey anti-mouse, Alexa Fluor 568-labeled donkey anti-rabbit, Alexa Fluor 488 goat anti-mouse. Coverslips were mounted using Fluoromount (Sigma-Aldrich) and observed using an Olympus FV-300 confocal laser-scanning microscope. Samples were viewed with a 60 × oil objective and a 1.5 × digital zoom. Images were assembled via ImageJ and Photoshop CS4 (Adobe Systems, Mountain View, CA).
BrdU Immunostaining
For basal proliferation, BAECs were transfected and then cultured on 0.1% gelatin-coated coverslips for 24 h in 10% FBS. For VEGF or Ang-1 treatment, cells were cultured and treated in 1% FBS overnight. Cells were incubated with 0.03 mg/ml BrdU at 37 °C for 30 min and then fixed with 70% ethanol for 5 min. Cells were rinsed with PBS and denatured with 1.5 m HCl for 30 min at room temperature. After incubation with 1% BSA to block nonspecific staining for 60 min, they were incubated with BrdU antibody overnight at 4 °C. After three washes with PBS, cells were incubated with Alexa-Fluor 568-labeled goat anti-mouse for one hour. DAPI was used to stain the nuclei.
Flow Cytometry
BAECs were treated with 0.03 mg/ml BrdU at 37 °C for 30 min. Cells were then fixed with 70% ethanol for 1 h at 4 °C. After fixation, cells were denatured and permeabilized with 1.5 m HCl and 0.03% Triton X-100 for 30 min, washed with PBS and incubated with BrdU (dilution 1:50) and ZO-1 (dilution 1:50) antibodies for 2 h at room temperature. After one wash with PBS, cells were incubated with Alexa-Fluor 647-labeled goat anti-mouse and Alexa-Fluor 488-labeled donkey anti-rabbit for 45 min. Stained cells were processed by flow cytometry FACSCanto II (BD biosciences), and the results were analyzed using FlowJo (www.flowjo.com).
Retina Immunostaining
Dissection and whole mount staining of postnatal retinas at the stage P5 were performed as described previously (39). Retinas were fixed for 2 h on ice in 4% paraformaldehyde (PFA). Dissected retinas were blocked overnight in 1% BSA, 0.3% Triton X-100 in PBS. For lectin I staining, retinas were equilibrated with Pblec buffer containing 1 mm CaCl2, 1 mm MgCl2, 1% Triton X-100 in PBS (pH 6.8) and then stained with Rhodamine conjugated LectinI (dilution 1:100) overnight at 4 °C. For ZO-1 and phospho-histone 3 (pH3) staining, retinas were incubated with rabbit ZO-1 (Life Technologies, dilution 1:100) and mouse pH3 (Abcam, dilution 1:100) in blocking buffer overnight at 4 °C. After primary incubation, retinas were labeled with Alexa-Fluor 488-labeled goat anti-rabbit, Alexa-Fluor 647-labeled goat anti-mouse for 2 h at room temperature. Stained retinas were flat-mounted using Fluoromount G (Electron Microscopy Sciences, Hatfield, PA). Z-stack confocal imaging was performed on Zeiss LSM700 confocal laser-scanning microscope using a 63× oil objective and a 2.5× digital zoom. All quantifications were performed on z-stack confocal images. Images were analyzed using Fiji software.
Statistical Analyses
Data are represented as the means ± S.E. Two-tailed independent Student's tests were used when comparing two groups. Comparisons between multiple groups were made using one-way ANOVA followed by post-hoc Bonferroni's multiple comparisons test among groups. p value < 0.05 was considered statistically significant.
RESULTS
Phosphoproteome Profiling of VEGF or Ang-1 Stimulated ECs
To analyze in a comprehensive manner the phosphorylation events regulated by VEGF and Ang-1, we performed a phosphoproteomic profiling of BAECs stimulated with VEGF or Ang-1. Because VEGF and Ang-1 activate numerous signaling pathways implicated in cell survival, proliferation and migration through the phosphorylation of MAPK, Akt and eNOS, we first established that 10 min of stimulation was the optimal time point of these signaling intermediates in BAECs (Fig. 1A).
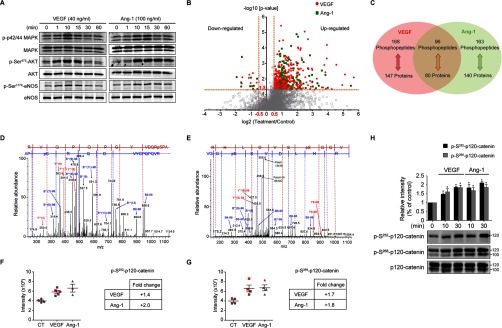
Fig. 1.
Analysis of the phosphoproteome in VEGF or Ang-1 treated BAECs. A, Activation of Akt, MAPK and eNOS, using the indicated phosphospecific antibodies, was monitored in BAECs treated with VEGF (40 ng/ml) or Ang-1 (100 ng/ml) for the indicated times. Equal protein loading was confirmed by reprobing the membranes with antibodies against total Akt, MAPK and eNOS. These experiments were repeated at least three times with similar results. B, Volcano plot showing the distribution of phosphopeptides abundance of VEGF or Ang-1 over control (fold-change, x axis) as a function of statistical significance (-log10 of p value, y axis). Red circles correspond to 255 significantly up-regulated peptides and 9 significantly down-regulated peptides upon VEGF stimulation. Green squares correspond to 248 significantly up-regulated peptides upon Ang-1 stimulation and 11 significantly down-regulated peptides to Ang-1 treatment. A cut-off of log2 fold-change > 0.5 and log2 fold-change < 0.5 was applied to define regulated phosphopeptides. -log10 of p value > 1.3 (p value < 0.05) represents significant peptides. C, Venn diagram showing the number of phosphopeptides and their corresponding phosphoproteins significantly regulated by VEGF and/or Ang-1. Data represents four to five replicates per condition. D, E, A representative mass spectrum of the phosphopeptide corresponding to phosphorylation of Ser252 (D) and Ser268 (E) of p120-catenin. F, G, The intensities of the phosphopeptide corresponding to phosphorylation of Ser252 (F) and Ser268 (G) of p120-catenin were used to calculate the fold-change ratio in VEGF or Ang-1 treatment. Data are represented as mean ± S.E. Red represents the intensity of the technical replicates. H, Immunoblotting validation of p120-catenin phosphorylation sites identified by mass spectrometry. BAECs were treated with VEGF (40 ng/ml) or Ang-1 (100 ng/ml) for the indicated times. Phosphorylation of Ser252 and Ser268 of p120-catenin was monitored using site-specific antibodies. Histogram represents the ratio of the phosphorylation levels over total protein measured by densitometry of three independent experiments. Data are represented as mean ± S.E.
To profile the phosphoproteomes of BAECs treated with VEGF or Ang-1, we used a label-free approach based on the comparison of phosphopeptide intensities between control and treated cells together with statistical analysis of replicates. First, whole BAEC lysates were enriched for phosphopeptides with TiO2 followed by LC-MS/MS analysis using an LTQ Orbitrap mass spectrometer. In total, three biological replicates of nontreated, VEGF or Ang-1 treated cells and one technical replicate of Ang-1 and two technical replicates of VEGF treated cells were performed. All replicates were combined for statistical analysis. We identified a total of 2426 unique phosphopeptides corresponding to 1001 individual proteins (FDR ≤ 1%) (supplemental Table S1). Phosphopeptides enrichment by TiO2 was 64.3% (3772 total peptides). To evaluate the reproducibility of our label-free approach, linear correlations of raw data for any two replicates were performed. Correlation coefficients of biological and technical replicates were between 0.64 and 0.96 (supplemental Fig. S1A, S1B). Analysis of the overall distribution of identified phosphorylation sites revealed that most phosphopeptides were singly phosphorylated (67.1%), whereas the others had either double (24.8%) or multiple (8.1%) phosphorylation sites. In addition, 87.9% of the phosphorylated sites identified were on serine, 10.2% on threonine and 1.8% on tyrosine (supplemental Fig. S2A, S2B). As expected, we identified less tyrosine phosphorylated than serine or threonine phosphorylated peptides because of the low affinity of TiO2 for phosphotyrosine residues and the lower numbers of tyrosine phosphorylation events in cells (40, 41).
To identify phosphopeptides that were up- or down-regulated in response to VEGF or Ang-1 stimulation of BAECs, the average intensities of the replicates was calculated and the treated/nontreated ratio for each phosphopeptide was determined. The distribution of the log2 ratio for all phosphopeptides following normalization for the β-casein phosphopeptide standard (see Methods) shows a median distribution of 0.40 and 0.65 for VEGF and Ang-1 phosphopeptides, respectively (supplemental Fig. S2C). Distribution of the significantly modulated phosphopeptides (p value < 0.05) shows that more than 95% were above a log2 fold-change of ± 0.5 (supplemental Fig. S2D). Thus, a cut-off of > 1.4-fold increase (log2 fold-change > 0.5) and < 0.7 fold-decrease (log2 fold-change < 0.5) was applied. All significant phosphopeptides were identified in at least two biological replicates. The correlation coefficients between the log2 fold-changes of the VEGF and Ang-1 treatment conditions were determined for all quantified phosphopeptides (supplemental Fig. S3A, S3B) and for significantly regulated phosphopeptides (supplemental Fig. S3C, S3D). As anticipated, the correlation coefficients were markedly improved when applied only to phosphopeptides that were significantly regulated by VEGF or Ang-1 treatments. The correlation coefficients of significantly regulated phosphopeptides for the biological replicates of the VEGF treatment were 0.61, 0.62, and 0.66 and were 0.66, 0.71, and 0.75 for the Ang-1 condition (supplemental Fig. S3C, S3D). Thus, a total of 427 phosphopeptides were significantly modulated upon VEGF and Ang-1 stimulation (Fig. 1B, supplemental Fig. S2D). In VEGF-treated cells, 255 phosphopeptides were found up-regulated and nine were down-regulated whereas in Ang-1-treated cells, 248 phosphopeptides were up-regulated and 11 were down-regulated (Fig. 1B). In sum, our analyses identified 168 phosphopeptides, corresponding to 147 phosphoproteins, regulated solely by VEGF; 163 phosphopeptides, corresponding to 140 phosphoproteins, regulated solely by Ang-1, and 96 phosphopeptides, corresponding to 80 phosphoproteins, regulated by both VEGF and Ang-1 (Fig. 1C).
To validate our mass spectrometry analyses, we confirmed that peptides corresponding to the well-established phosphorylation sites Thr145 and Tyr147 of bovine MAPK1 (Thr185 and Tyr187 in human MAPK1) shown in immunoblots of Fig. 1A were present in VEGF and Ang-1 samples. Indeed, we found a peptide with both residues phosphorylated that was increased by an average of 4.9- and 7.5-folds in VEGF and Ang-1 samples, respectively (supplemental Table S1 and Fig. 2C). In addition, to confirm the accuracy of peptide identification by MaxQuant, we manually searched the MS/MS spectra corresponding to peptides of phosphorylated Ser252 and Ser268 of the cell junction protein p120-catenin (CTNND1) (Fig. 1D, 1E). In order to quantify the phosphopeptide abundance in VEGF or Ang-1 treatment, we used the intensities of the different replicates calculated by MaxQuant to determine the average log2 fold-change of each treatment. As shown in Fig. 1F and 1G, the phosphorylated peptides pSer252 and pSer268 of p120-catenin were identified in at least three out of four replicates and were increased in BAECs treated with VEGF and Ang-1. These significant increases in phosphopeptide intensities compared with control cells were confirmed by Western blots using commercially available phosphorylation site-specific antibodies against pSer252 and pSer268 of p120-catenin. Consistent with the quantification by MS, the phosphorylation levels of Ser252 and Ser268 of p120-catenin were increased in BAECs treated with VEGF and Ang-1 at 10 min of stimulation (Fig. 1H).
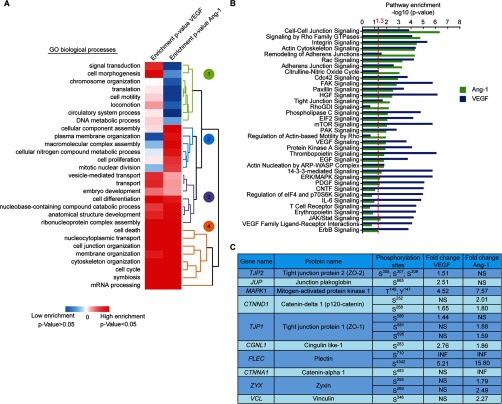
Fig. 2.
Comparative analysis of the phosphoproteomes of VEGF and Ang-1 treated cells. A, A hierarchical clustering heatmap showing three different clusters of biological processes enrichment of VEGF or Ang-1 regulated phosphoproteins. Phosphoproteins regulated by VEGF or Ang-1 were analyzed using the GO biological process annotation built in STRING database. Red corresponds to GO terms significantly enriched (p value<0.05) and blue represents nonsignificant biological processes (p value > 0.05). B, VEGF and Ang-1 regulated proteins were applied into IPA and the top pathways significantly enriched of each condition were represented and compared. A -log10 of p value > 1.3 correspond to the pathway significantly enriched calculated by Fisher's exact test. C, Phosphorylation sites and fold-change of selected phosphoproteins that are linked to the regulation of cell junctions. NS: not significant. INF: infinity indicates the ratio of phosphopeptides that were only identified in VEGF or Ang-1 samples but not in control samples.
Comparative Analysis of VEGF and Ang-1 Phosphoproteomes
To determine the biological processes and signaling pathways that emerged as being enriched in VEGF or Ang-1-regulated phosphoproteomes, we performed Gene Ontology analyses of phosphoproteins identified against the entire human database and biological processes annotations with a p value less than 0.05 were retained (hypergeometric test) (supplemental Table S2). Hierarchical clustering of biological processes resulted into three groups (Fig. 2A). The first cluster (green), comprised of biological processes enriched only in VEGF-treated cells, includes processes such as signal transduction (GO:0007165), translation (GO:0006412), and cell morphogenesis (GO:0000902). The second cluster (blue) encloses mainly biological processes weakly enriched in VEGF and Ang-1 treatments such as cellular component assembly (GO:0022607) and cell proliferation (GO:0008283). Finally, the third and fourth cluster (purple and orange) corresponds to high enrichments in both VEGF and Ang-1 conditions and includes biological processes associated with cell cycle (GO:0007049), transport (GO:0006810), mRNA processing (GO:0006397), cell differentiation (GO:0030154), membrane organization (GO:0061024), and notably cell junction organization (GO:0034330) and cytoskeleton organization (GO:0007010) (Fig. 2A). To characterize the signaling pathways modulated by VEGF and Ang-1, a pathway enrichment analysis was conducted using IPA. The top significantly enriched pathways modulated by VEGF and Ang-1 were determined (Fig. 2B, supplemental Table S2). Signaling pathways such as ERK/MAPK, mTOR, and PDGF signaling were mostly enriched in the VEGF condition. However, Cdc42 signaling, Rac signaling, actin cytoskeleton and cell-cell junction signaling pathways were among the most enriched in both VEGF and Ang-1 conditions. This is consistent with the known modulatory roles of VEGF and Ang-1 on EC junctions. Moreover, several phosphorylation sites on proteins involved in the regulation of cell–cell junctions that are regulated by VEGF or Ang-1 were identified (Fig. 2C). For example, pSer252 and pSer268 on p120-catenin (CTNND1), and pThr145 and pSer147 on MAPK1 were phosphorylated under VEGF and Ang-1 conditions. In contrast, pSer453 on catenin-α1 (CTNNA1) and pSer346 on Vinculin (VCL) were phosphorylated only by Ang-1. pSer305/307/309 on tight junction protein 2 (ZO-2) and pSer665 on junction plakoglobin (JUP) were phosphorylated only with VEGF treatment. Finally, pSer586 on tight junction protein 1 (ZO-1) was phosphorylated only with VEGF whereas pSer881 and pSer895 were only found in Ang-1 samples. The function of many of these phosphorylation sites remains unknown (www.phosphosite.org) (42).
Potential Link Between MAPK-induced Proliferation and Endothelial Junctions
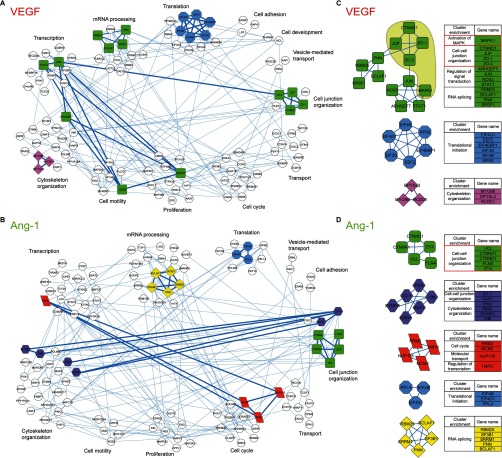
Based on our phosphoproteomic data, we generated a protein interaction network of phosphoproteins that are significantly regulated by VEGF or Ang-1 using the STRING database and assembled by Cytoscape. A network of 110 interacting nodes and 189 connecting edges for VEGF-regulated proteins was determined (Fig. 3A). The Ang-1 interaction network showed 120 nodes and 201 edges (Fig. 3B). The proteins were grouped based on their biological functions using GO annotations and the Uniprot database. These interaction network highlighted subsets of proteins associated with cell junction organization, cell motility, cell proliferation, cytoskeleton organization, cell cycle, translation, transcription, RNA splicing, and transport. Based on these networks, we performed a protein interaction clustering using MCODE in order to regroup highly interconnected proteins based on known protein-protein interactions (37, 43). The parameters used in MCODE to identify a cluster based on connectivity are shown in supplemental Table S3. We identified three subnetworks in VEGF-treated cells (Fig. 3C) and six subnetworks in Ang-1-treated cells (Fig. 3D). The STRING database provides a protein–protein interaction network in which each edge represents a known link between two proteins based on the literature. Therefore, proteins of similar functions tend to be regrouped within a cluster. For example, the blue cluster (Fig. 3C) contains a group of proteins all involved in translational initiation and the yellow cluster encloses proteins all involved in RNA splicing (Fig. 3D). In contrast, the green clusters link together phosphoproteins of various functions such as RNA splicing, regulation of signal transduction, activation of MAPK and cell-cell junction organization (Fig. 3C, 3D).
Fig. 3.
Phosphoprotein interaction network of VEGF and Ang-1 treated cells. Protein interaction networks generated with VEGF (A) and Ang-1 (B) regulated phosphoproteins using STRING database version 9.1 with medium confidence and visualized by Cytoscape. The proteins that did not interact with any other proteins are not shown. The interaction network was subjected to a cluster analysis using the algorithm MCODE based on the protein interaction level. Clusters are represented with different colors. Phosphoproteins in white are not part of any cluster. The tables contain the gene names and the functional enrichment of VEGF (C) and Ang-1 (D) clusters. The functional enrichment was manually annotated using GO terms of the biological process category and Uniprot database. The green cluster shows the junctional proteins in the same cluster with MAPK1 in VEGF but not in Ang-1 interaction network.
Ang-1 and VEGF act in opposite manner on EC junctions and because our phosphoproteomic analysis yielded a high enrichment of proteins involved in the regulation of junctions, we focused on clusters in which junctional phosphoproteins were identified. In VEGF-treated cells, ZO-1, ZO-2, JUP, and p120-catenin were found in the same cluster (green cluster) as MAPK1 (Fig. 3C). In Ang-1-treated cells, p120-catenin and α-catenin formed a cluster with vinculin, zyxin and filamin A. Moreover, ZO-1 clustered separately with cytoskeleton organization proteins such as proteins paxillin, vimentin, and drebrin 1. Remarkably in the Ang-1 interaction network, none of the junctional phosphoproteins identified clustered with MAPK1 (Fig. 3D). Notably, the peptides identified for MAPK1 were phosphorylated on Thr145 and Tyr147, well-known indicators of MAPK activation and cell proliferation (Fig. 1A). This suggests that in order to induce MAPK1 activation, VEGF signaling may be more reliant on junctional proteins than Ang-1-induced activation of MAPK1. Thus, these findings revealed a potential link between MAPK-induced proliferation and p120-catenin, JUP, ZO-1, and ZO-2 junctional proteins specifically in VEGF-stimulated ECs (Fig. 3C).
Down-regulation of p120-catenin, ZO-1, ZO-2, and JUP Induces Proliferation of ECs
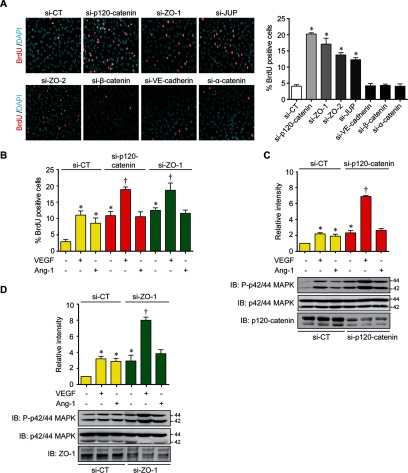
Because the formation cell-cell contacts between ECs is involved in the inhibition of proliferative signals, we investigated further the potential link that we uncovered between proteins involved in the regulation of endothelial junctions and MAPK-induced proliferation. To explore this in more details, we used siRNAs to down-regulate p120-catenin, ZO-1, ZO-2, and JUP in BAECs and monitored cell proliferation by measuring BrdU labeling (Fig. 4A). Interestingly, we found that down-regulation of p120-catenin, ZO-1, ZO-2, and JUP significantly increased BrdU incorporation by ∼20, 17, 13, and 12%, respectively (Fig. 4A). These results were confirmed by the use of a second siRNA against p120-catenin and ZO-1 that showed similar results (supplemental Fig. S4A). The knockdown efficiency of all siRNAs was confirmed by immunoblotting (supplemental Fig. S4A, S4B). Similarly, we observed increased Cyclin D1 mRNA levels in ZO-1 down-regulated cells confirming cell cycle progression and proliferation (supplemental Fig. S4C). To determine if this increase in cell proliferation was specific to junction phosphoproteins identified in our cluster analyses, we down-regulated other components of adherens junctions. Down-regulation of VE-cadherin, β-catenin or α-catenin did not increase proliferation of ECs (Fig. 4A). In the network analysis, MAPK1 clustered with junction phosphoproteins only for the VEGF-stimulated condition although Ang-1-stimulation can also promote MAPK activation and EC proliferation (Fig. 3C, 3D). The effect of down-regulation of p120-catenin and ZO-1 on VEGF and Ang-1-stimulated proliferation of BAECs was compared. Down-regulation of p120-catenin and of ZO-1 increased basal cell proliferation that was increased further in VEGF-stimulated cells. In contrast, Ang-1 stimulation did not stimulate further proliferation in cells where p120-catenin or ZO-1 is down-regulated (Fig. 4B). However, down-regulation of VE-cadherin or β-catenin did not affect proliferation induced by VEGF or Ang-1 treatment (supplemental Fig. S4D). To confirm the link between MAPK1 activation and cell junction proteins, we monitored MAPK phosphorylation (P-p42/44 MAPK) in p120-catenin or ZO-1 down-regulated BAECs. In agreement with the proliferation results, VEGF-stimulated phosphorylation of MAPK was increased significantly in p120-catenin and ZO-1 down-regulated cells (Fig. 4C, 4D). Our results indicate that cellular levels of p120-catenin, ZO-1, ZO-2, and JUP influence MAPK-mediated proliferation of ECs.
Fig. 4.
Endothelial cell junction proteins regulate cell proliferation. A, Representative images (left) and quantification (right) of BrdU incorporation in BAECs cultured in 10% serum and transfected with siRNA against p120-catenin, JUP, ZO-1, ZO-2, α-catenin, β-catenin or VE-cadherin. Data are represented as mean ± S.E. of at least three different experiments (*p < 0.05 compared with si-CT). B, Quantification of the percentage of BrdU incorporation in p120-catenin or ZO-1 siRNA transfected BAECs in response to VEGF (40 ng/ml) or Ang-1 (100 ng/ml) treatment in 1% serum overnight. Data are represented as mean ± S.E. of three different experiments (*p < 0.05 as compared with CT, †p < 0.05 as compared with VEGF treatment). C, D, BAECs were transfected with p120-catenin (C) or ZO-1 (D) siRNA and phosphorylation of p42/44 MAPK was monitored by immunoblotting. Total p42/44 MAPK was used as loading control. Data are represented as mean ± S.E. of at least three different experiments (*p < 0.05 as compared with CT, †p < 0.05 as compared with VEGF treatment).
ZO-1 is Central in the Modulation of Endothelial Cell Proliferation
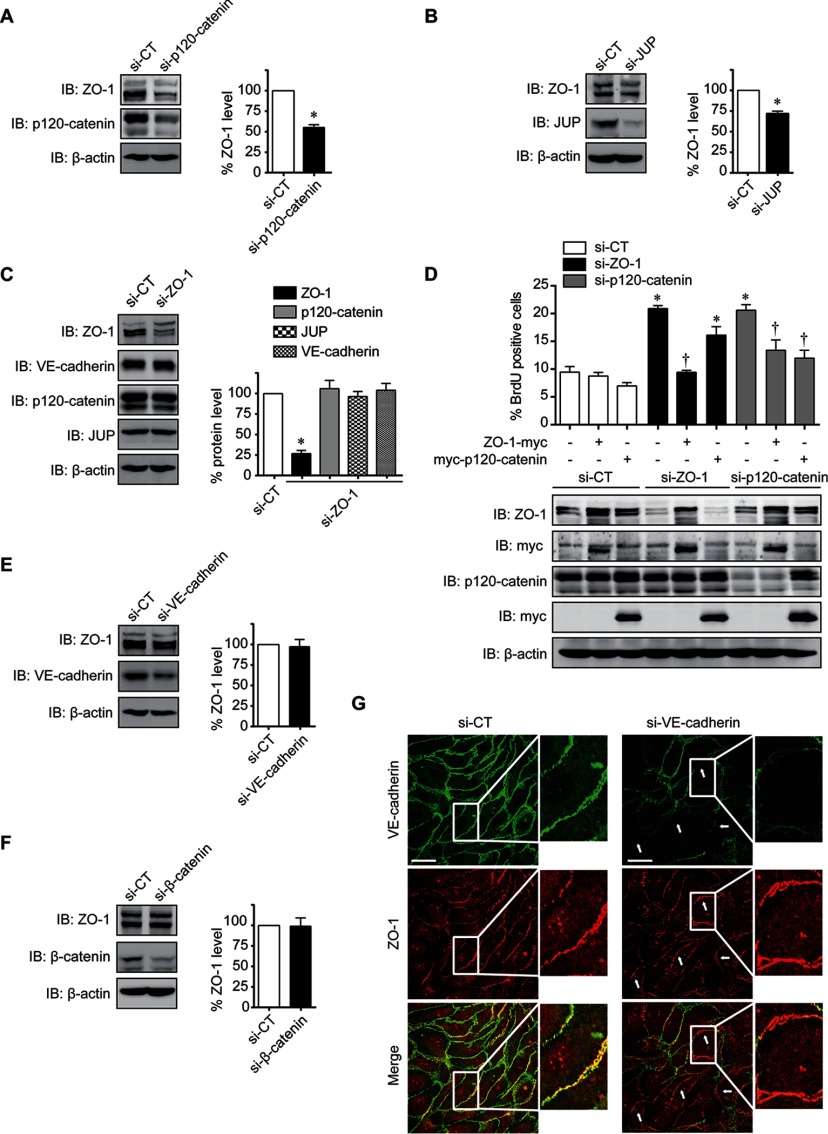
When investigating the relationship between junctional proteins within the VEGF network cluster, we observed that transfection of BAECs with siRNAs against p120-catenin or JUP also reduced ZO-1 protein levels (Fig. 5A, 5B). However, siRNA against ZO-1 did not decrease the protein levels of p120-catenin, JUP or VE-cadherin (Fig. 5C). Moreover, the localization of p120-catenin, JUP, or VE-cadherin at the cell membrane was not affected in BAECs transfected with ZO-1 siRNA (supplemental Fig. S5A, S5B, S5C). Thus, these results indicate that to induce EC proliferation, down-regulation of a cell junction protein must consequently provoke a reduction of the cellular levels of ZO-1. Furthermore, expression of myc-tagged human ZO-1 reduced proliferation of BAECs transfected with siRNA against bovine ZO-1. Similarly, proliferation induced by transfection of p120-catenin siRNA is reduced when myc-tagged p120-catenin is expressed (Fig. 5D). However, expression of myc-tagged ZO-1 in p120-catenin down-regulated cells did block proliferation induced by p120-catenin siRNA. In contrast, expression of myc-tagged p120-catenin did not significantly affect proliferation induced by ZO-1 siRNA (Fig. 5D). Interestingly, down-regulation of the adherens junction proteins VE-cadherin and β-catenin, which are not part of the ZO-1/MAPK1 cluster, did not induce cell proliferation and did not affect ZO-1 levels or its localization at cell–cell junctions (Fig. 5E, 5F, 5G). Thus, we identified ZO-1 as the central regulator of this cluster of junctional proteins and that modulation of the cellular levels of ZO-1 could control proliferation of ECs.
Fig. 5.
ZO-1 is a major regulator of endothelial cell proliferation. A, B Immunoblot showing ZO-1 expression levels in p120-catenin (A) or JUP (B) siRNA transfected BAECs. Quantification of ZO-1 protein levels of three different experiments was represented as mean ± S.E. (*p < 0.05 as compared with CT). C, BAECs were transfected with ZO-1 siRNA and the expression levels of p120-catenin, ZO-1, VE-cadherin and JUP was monitored by immunoblotting. Histograms represent the mean ± S.E. of three different experiments (*p < 0.05 as compared with CT, †p < 0.05 as compared with VEGF treatment). White bar represents the basal level of each protein. D, Quantification of BrdU incorporation in BAECs transfected with siRNA against bovine ZO-1 or p120-catenin and expression plasmids for human ZO-1-myc or for myc-p120-catenin. Data are presented as mean ± S.E. of at least three experiments (*p < 0.05 compared with si-CT, †p < 0.05 compared with si-ZO-1). Representative immunoblots, showing ZO-1 and p120-catenin protein levels, from at least three experiments with similar results. E, F Immunoblot showing ZO-1 protein levels in BAECs transfected with siRNA against VE-cadherin (E) or β-catenin (F). Histograms show the quantification of ZO-1 protein levels in VE-cadherin (top panel) or β-catenin (bottom panel) siRNA transfected cells. Data are represented as mean ± S.E. (*p < 0.05 as compared with CT). G, Representative confocal images of immunofluoresence staining of ZO-1 (red) and VE-cadherin (green) in BAECs transfected with control or VE-cadherin siRNA. White arrowheads point to staining at cell-cell junctions and higher magnification view of the boxed region is shown. Scale bar represents 20 μm.
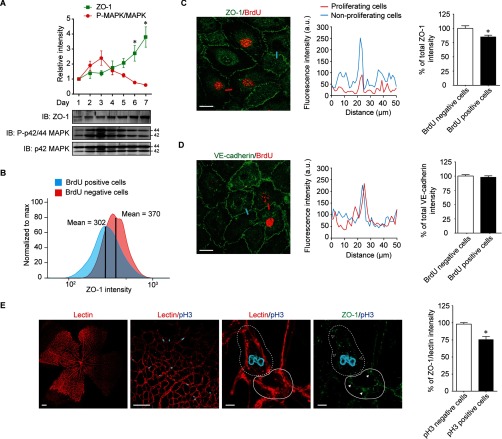
To further investigate the relationship between ZO-1 levels, MAPK, and EC proliferation, we hypothesized that the cellular ZO-1 levels are inversely correlated with cell proliferation in a cell density-dependent manner. To verify this, BAECs were seeded at low cell density and ZO-1 levels were determined until cells proliferated to confluence (7 days). We found that ZO-1 levels increased with cell density and reduced phosphorylation of MAPK was observed in parallel (Fig. 6A). This correlation between ZO-1 levels and cell proliferation has been observed in other cell systems and indicated a possible role for ZO-1 in contact-mediated inhibition of EC proliferation (44). Next, we examined ZO-1 levels in proliferating ECs. Thus, we performed flow cytometry analysis of confluent BAECs stained for BrdU and ZO-1. Cells were treated with BrdU for 30 min at 37 °C, then fixed and labeled for BrdU and ZO-1. BrdU positive cells showed an overall 19% decrease in ZO-1 intensity compared with BrdU negative cells (Fig. 6B, supplemental Fig. S6). Furthermore, we visualized the decrease of ZO-1 levels in proliferating cells by immunofluorescence microscopy. Indeed, proliferating BrdU positive BAECs displayed a 20% decrease in ZO-1 staining compared with nonproliferating cells (Fig. 6C). However, the levels of VE-cadherin were identical in proliferating and nonproliferating cells (Fig. 6D). In the mouse retina, the development of the vascular plexus forms during the first week after birth and the growth vessels reaches the retinal edges at approximately postnatal day (P) 8 (45). In order to determine ZO-1 levels in proliferating ECs in vivo, we examined mouse retina at P5 where growth of the vascular plexus is not fully completed. To identify proliferating ECs in the retina, phospho-histone 3 (pH3) staining was performed and ZO-1 levels were quantified in ECs (lectin I positive/red). At day P5, a significant proportion of ECs were proliferating (Fig. 6E). In agreement with our results in cultured ECs, ZO-1 levels, when normalized to the staining of the EC marker lectin I, was decreased by 23% in proliferating cells (pH3 positive cells) compared with nonproliferating cells (Fig. 6E) in the developing vasculature of the mouse postnatal retina.
Fig. 6.
ZO-1 levels correlate with endothelial cell proliferation in vitro and in vivo. A, Equal amount of BAECs were grown at low density at day 1 and cultured until they reached maximal cell density at day 7. Cells were harvested each day and equal amount of protein were analyzed by immunoblotting. Representative immunoblot of three different experiments was shown. The relative intensity of ZO-1 (green) and P-p42/44 MAPK (red) were quantified and the data are represented as mean ± S.E. (*p < 0.05 compared with day 1). B, Flow cytometry analysis showing overlay histograms of ZO-1 intensity in BrdU positive and negative confluent BAECs. Samples are scaled to the percentage of its maximum signal. The mean of ZO-1 intensity in BrdU positive and negative cells was shown. C, D Immunfluoresence staining of BAECs showing ZO-1 (C) or VE-cadherin (D) levels (green) in the proliferative cells stained with BrdU (red) versus nonproliferating cells. Representative fluorescence intensity profiles of ZO-1 or VE-cadherin in proliferating and nonproliferating cells (blue) measured along the lines drawn across cell-cell junctions is shown. Scale bar represents 20 μm. Quantification of ZO-1 or VE-cadherin levels in the BrdU positive cells was expressed as percentage of decrease relative to nonproliferating cells. Each column represents at least 50 measurements. Values are represented as mean ± S.E. (*p < 0.05 compared with BrdU negative cells). E, Low magnification confocal micrographs of a P5 mouse retina show lectin I (panel 1) staining to indicate retinal vessels in which the proliferating cells were stained with pH3 (panel 2). Scale bar represents 200 μm. Panel 3 and 4 show higher magnification images of lectin I (panel 3) or ZO-1 (panel 4) staining. Empty arrows indicate ZO-1 staining in the proliferating cell (dashed line). Full arrows indicate ZO-1 staining in the nonproliferating cell (solid line). Quantification of the percentage of total ZO-1 intensity normalized to lectin I intensity in the pH3 positive cells compared with pH3 negative cells. Each column represents the average of 25 measurements. Data are represented as mean ± S.E. (n = 6, *p < 0.05 compared with BrdU negative cells). Scale bar represents 5 μm.
DISCUSSION
VEGF and Ang-1, through their respective receptors, activate overlapping signaling cascades such that the molecular mechanisms explaining their unique biological activities remain largely undefined. Herein, we demonstrate that by comparing and analyzing the phosphoproteomes of ECs subjected to stimulation by angiogenic factors, we can uncover molecular networks of phosphoproteins that are determinants of basic cellular functions such as proliferation. We identified a link between MAPK1-induced proliferation and the junctional proteins ZO-1, ZO-2, JUP (junction plakoglobin), and p120-catenin. We found that ZO-1 is the central modulator of the actions of these proteins on MAPK activation and cell proliferation. Importantly, our results define a role for ZO-1 in the regulation of EC proliferation during development of the vasculature in the mouse retina.
The use of a label-free proteomics approach based on the comparison of phosphopeptide intensities, the use of multiple replicates and straightforward statistical analysis provided us with accurate identifications and reliable quantifications to determine the phosphoproteomes of VEGF or Ang-1 stimulated ECs (46, 47). Moreover, the identification of phosphorylation sites allowed us to profile the proteins modulated by VEGF and Ang-1 treatment in order to define molecular networks that could regulate ECs during and important for angiogenesis. This method also allowed us to confirm several previously reported phosphorylation sites regulated by VEGF or Ang-1. Notably, we identified regulated phosphopeptides in VEGF and Ang-1 conditions corresponding to the well-established phosphorylation sites on Thr185 and Tyr187 on human MAPK1 (ERK2/p44 MAPK) that are linked to its activation and cell proliferation (10). In addition, phosphorylation of Ser82/78 (human/bovine) on HSPB1 (heat shock protein β1 or HSP27) was previously shown to be phosphorylated by Protein Kinase D and to participate in VEGF-stimulated EC migration (48). Other phosphorylation sites identified highlight important signaling pathways that are responsible for the effects of VEGF and/or Ang-1 in ECs. For example, we identified a group of phosphoproteins, EIF4B (Ser406) (49), EIF4EBP1 (Ser65) (50, 51), EIF4G1 (Ser1077/1104; human/bovine) (52), and RPS6 (Ser235, Ser236) (53) that are all linked to the regulation of mRNA translation. Furthermore, phosphorylation sites on proteins known to be involved in cell migration and cytoskeletal reorganization such as Dock1 (Ser1857/1860; human/bovine) (54, 55), Dock6 (Ser1343) (56), ARHGEF7 (Ser694/673, Ser518/497; human/bovine) (57, 58) and eNOS (Ser633/635; human/bovine) (59) were also identified in our analyses. Analysis of pathway enrichment and GO terms indicate that cell junctions, cytoskeleton reorganization processes are highly modulated by VEGF and Ang-1 (Fig. 2). Interestingly, the phosphoproteins of VEGF-stimulated ECs showed high enrichment of mTOR, MAPK and 14–3-3 signaling pathways. These are all consistent with the known effects of VEGF in angiogenesis (10, 60).
In VEGF-treated cells, our bioinformatics analyses revealed a link between MAPK1 activation and the cell junction proteins ZO-1, ZO-2, JUP, and p120-catenin. Signaling at cell junctions is well known to modulate cell density and proliferation. However, we show that the induction of EC proliferation by down-regulation of these proteins is accomplished independently of adherens junction integrity. Indeed, we show that down-regulation of VE-cadherin or β-catenin does not provoke an increase in EC proliferation in contrast to down-regulation of ZO-1, ZO-2, JUP, and p120-catenin. Interestingly, the concomitant reduction in the cellular levels of ZO-1 is necessary for increased cell proliferation (Fig. 4, 5). Few studies have directly linked these proteins to the regulation of cell proliferation. Deletion of ZO-1 or ZO-2 in mice is embryonic lethal because of reduced yolk sac angiogenesis or to an arrest in early gastrulation and decrease in cell proliferation (23, 61). Furthermore, ZO-1 was found to participate in cell cycle regulation and cell density in epithelial cells via the Y-box transcription factor ZO-1-associated nucleic acid binding protein (ZONAB) (44). ZONAB was found to associate with cell division kinase (CDK) 4 and regulate its nuclear accumulation. On the other hand, the translocation of p120-catenin to the nucleus was reported to promote proliferation of ECs through the transcription factor Kaiso (29). Finally, expression of plakoglobin under the control of the keratin K14 promoter in mice reduced proliferation of epithelial cells of the epidermis and hair follicles, in agreement with our observations in ECs (62).
The comparison of interaction networks showed that MAPK1 clustered with junctional proteins only in VEGF-treated cells. This differential clustering led us to investigate further the implication of these proteins in the regulation of VEGF-stimulated proliferation (Fig. 3). It is established that the cellular levels of junctional proteins increase with cell density to control contact mediated inhibition of cell proliferation (21, 29, 44). Here, we reveal that ZO-1 is a central regulator of this process. Notably, we show that down-regulation of ZO-1 specifically enhances VEGF-mediated MAPK activation and cell proliferation, which is not observed under Ang-1 stimulation (Fig. 4). Thus, it is tempting to propose that because VEGF is known to disrupt EC junctions, in contrast to Ang-1, this contributes to facilitation and activation of the proliferative signals in ECs, which require a decrease in the cellular levels of ZO-1. Our results suggest that ECs must reduce ZO-1 levels in order to enter the proliferative program. Importantly, we found that proliferating cells display decreased ZO-1 levels compared with nonproliferating cells. Moreover, we demonstrate that ZO-1 levels are decreased in ECs during retinal vascular development in mice. Further studies will need to determine how ZO-1 directly affects cell proliferation.
Interestingly, we identified Ser617/Ser586 (human/bovine) of ZO-1 to be phosphorylated in response to VEGF and found Ser912/Ser881 and Ser926/Ser895 (human/bovine) to be phosphorylated under Ang-1 conditions. This suggests that these phosphorylation events regulate differentially the function of ZO-1. Because phosphorylation of Ser617 is positively regulated by VEGF and that depletion of ZO-1 has a similar effect as VEGF treatment, the possibility that phosphorylation of this residue is somehow inactivating ZO-1 is currently being investigated. Alternatively, it has been previously reported that VEGF and Ang-1 modulate differently the expression of ZO-1 or ZO-2. For example, in brain microvascular endothelial cells Ang-1 was found to counteract VEGF-induced permeability by increasing expression of ZO-2 (63). Moreover, in human peritoneal mesothelial cells, VEGF was reported to decrease the expression levels of ZO-1 in response to glucose degradation products (64).
In conclusion, by comparing the phosphoproteomes and signaling networks of ECs stimulated with well-defined growth factors, VEGF and Ang-1, we revealed a major role for ZO-1 in the control of cell proliferation during angiogenesis. This indicates that global phosphoproteomics analyses may serve to delineate interrelations between signaling pathways that act as regulators of basic cell function that are important in physiological and pathological settings.
Supplementary Material
Acknowledgments
We thank Dr. Audrey Claing (Université de Montréal, Montreal, Canada) and Dr. Jean-François Côté (IRCM, Montreal, Canada) for critical reading of the manuscript.
Footnotes
Author contributions: R.C. designed and performed the experiments, analyzed the data, prepared the figures, and wrote the manuscript; Y.Z., S.T. and C.D. performed the experiments and analyzed the data, D.F. participated in designing the experiments and analyzed the data; J.-P.G. designed and supervised the experiments, analyzed the data, prepared the figures, and wrote the manuscript.
* This work was supported by grants from the Canadian Institutes of Health Research to J.-P.G. (MOP-86464 and MOP-111031). J.-P.G. holds a Université de Montréal research Chair and was in receipt of a Fonds de recherche du Québec - Santé (FRQS) Senior career award.
 This article contains supplemental materials.
This article contains supplemental materials.
Conflict of interest: The authors have no conflict of interest to declare.
1 The abbreviations used are:
- Ang-1
- angiopoietin-1
- BAEC
- bovine aortic endothelial cell
- EC
- endothelial cell
- FDR
- false discovery rate
- GO
- gene ontology
- LC-MS/MS
- liquid chromatography-tandem mass spectrometry
- MAPK
- mitogen-activated protein kinase
- siRNA
- small interfering RNA
- VEGF
- vascular endothelial growth factor
- DMEM
- Dulbecco Modified Eagle Medium
- pAb
- polyclonal antibody
- mAb
- monoclonal antibody
- EDTA
- ethylenediaminetetraacetic
- EGTA
- ethyleneglycoltetraacetic
- TFA
- trifluoroacetic acid
- ANOVA
- Analysis of variance
- CT
- control
- DTT
- dithiothreitol
- ACN
- Acetonitrile
- (q)RT-PCR
- quantitative reverse transcriptase – polymerase chain reaction
- BrdU
- bromodeoxyuridine
- pH3
- phospho-histone 3
- P5
- postnatal day 5
- IB
- immunoblot
- IPA
- ingenuity pathway analysis.
REFERENCES
- 1. Chung A. S., Lee J., and Ferrara N. (2010) Targeting the tumour vasculature: insights from physiological angiogenesis. Nat. Rev. Cancer 10, 505–514 [DOI] [PubMed] [Google Scholar]
- 2. Ferrara N., Carver-Moore K., Chen H., Dowd M., Lu L., O'Shea K. S., Powell-Braxton L., Hillan K. J., and Moore M. W. (1996) Heterozygous embryonic lethality induced by targeted inactivation of the VEGF gene. Nature 380, 439–442 [DOI] [PubMed] [Google Scholar]
- 3. Jain R. K. (2003) Molecular regulation of vessel maturation. Nat. Med. 9, 685–693 [DOI] [PubMed] [Google Scholar]
- 4. Suri C., Jones P. F., Patan S., Bartunkova S., Maisonpierre P. C., Davis S., Sato T. N., and Yancopoulos G. D. (1996) Requisite role of angiopoietin-1, a ligand for the TIE2 receptor, during embryonic angiogenesis. Cell 87, 1171–1180 [DOI] [PubMed] [Google Scholar]
- 5. Carmeliet P. (2003) Angiogenesis in health and disease. Nat. Med. 9, 653–660 [DOI] [PubMed] [Google Scholar]
- 6. Tremolada G., Del Turco C., Lattanzio R., Maestroni S., Maestroni A., Bandello F., and Zerbini G. (2012) The role of angiogenesis in the development of proliferative diabetic retinopathy: impact of intravitreal anti-VEGF treatment. Exp. Diabetes Res. 2012, 728325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Adams R. H., and Alitalo K. (2007) Molecular regulation of angiogenesis and lymphangiogenesis. Nat. Rev. Mol. Cell Biol. 8, 464–478 [DOI] [PubMed] [Google Scholar]
- 8. Huang H., Bhat A., Woodnutt G., and Lappe R. (2010) Targeting the ANGPT-TIE2 pathway in malignancy. Nat. Rev. Cancer 10, 575–585 [DOI] [PubMed] [Google Scholar]
- 9. Olsson A. K., Dimberg A., Kreuger J., and Claesson-Welsh L. (2006) VEGF receptor signalling - in control of vascular function. Nat. Rev. Mol. Cell Biol. 7, 359–371 [DOI] [PubMed] [Google Scholar]
- 10. D'Angelo G., Struman I., Martial J., and Weiner R. I. (1995) Activation of mitogen-activated protein kinases by vascular endothelial growth factor and basic fibroblast growth factor in capillary endothelial cells is inhibited by the antiangiogenic factor 16-kDa N-terminal fragment of prolactin. Proc. Natl. Acad. Sci. U.S.A. 92, 6374–6378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Harfouche R., Gratton J. P., Yancopoulos G. D., Noseda M., Karsan A., and Hussain S. N. (2003) Angiopoietin-1 activates both anti- and proapoptotic mitogen-activated protein kinases. FASEB J. 17, 1523–1525 [DOI] [PubMed] [Google Scholar]
- 12. Gerber H. P., McMurtrey A., Kowalski J., Yan M., Keyt B. A., Dixit V., and Ferrara N. (1998) Vascular endothelial growth factor regulates endothelial cell survival through the phosphatidylinositol 3′-kinase/Akt signal transduction pathway. Requirement for Flk-1/KDR activation. J. Biol. Chem. 273, 30336–30343 [DOI] [PubMed] [Google Scholar]
- 13. Kim I., Kim H. G., So J. N., Kim J. H., Kwak H. J., and Koh G. Y. (2000) Angiopoietin-1 regulates endothelial cell survival through the phosphatidylinositol 3′-Kinase/Akt signal transduction pathway. Circ. Res. 86, 24–29 [DOI] [PubMed] [Google Scholar]
- 14. Papapetropoulos A., Fulton D., Mahboubi K., Kalb R. G., O'Connor D. S., Li F., Altieri D. C., and Sessa W. C. (2000) Angiopoietin-1 inhibits endothelial cell apoptosis via the Akt/survivin pathway. J. Biol. Chem. 275, 9102–9105 [DOI] [PubMed] [Google Scholar]
- 15. Rousseau S., Houle F., Landry J., and Huot J. (1997) p38 MAP kinase activation by vascular endothelial growth factor mediates actin reorganization and cell migration in human endothelial cells. Oncogene 15, 2169–2177 [DOI] [PubMed] [Google Scholar]
- 16. Gavard J., Patel V., and Gutkind J. S. (2008) Angiopoietin-1 prevents VEGF-induced endothelial permeability by sequestering Src through mDia. Dev. Cell 14, 25–36 [DOI] [PubMed] [Google Scholar]
- 17. Jho D., Mehta D., Ahmmed G., Gao X. P., Tiruppathi C., Broman M., and Malik A. B. (2005) Angiopoietin-1 opposes VEGF-induced increase in endothelial permeability by inhibiting TRPC1-dependent Ca2 influx. Circ. Res. 96, 1282–1290 [DOI] [PubMed] [Google Scholar]
- 18. Wang Y., Pampou S., Fujikawa K., and Varticovski L. (2004) Opposing effect of angiopoietin-1 on VEGF-mediated disruption of endothelial cell-cell interactions requires activation of PKC beta. J. Cell. Physiol. 198, 53–61 [DOI] [PubMed] [Google Scholar]
- 19. Oubaha M., and Gratton J. P. (2009) Phosphorylation of endothelial nitric oxide synthase by atypical PKC zeta contributes to angiopoietin-1-dependent inhibition of VEGF-induced endothelial permeability in vitro. Blood 114, 3343–3351 [DOI] [PubMed] [Google Scholar]
- 20. Matter K., and Balda M. S. (2003) Signalling to and from tight junctions. Nat. Rev. Mol. Cell Biol. 4, 225–236 [DOI] [PubMed] [Google Scholar]
- 21. Wallez Y., and Huber P. (2008) Endothelial adherens and tight junctions in vascular homeostasis, inflammation and angiogenesis. Biochim. Biophys. Acta 1778, 794–809 [DOI] [PubMed] [Google Scholar]
- 22. Perez-Moreno M., Jamora C., and Fuchs E. (2003) Sticky business: orchestrating cellular signals at adherens junctions. Cell 112, 535–548 [DOI] [PubMed] [Google Scholar]
- 23. Katsuno T., Umeda K., Matsui T., Hata M., Tamura A., Itoh M., Takeuchi K., Fujimori T., Nabeshima Y., Noda T., Tsukita S., and Tsukita S. (2008) Deficiency of zonula occludens-1 causes embryonic lethal phenotype associated with defected yolk sac angiogenesis and apoptosis of embryonic cells. Mol. Biol. Cell 19, 2465–2475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cattelino A., Liebner S., Gallini R., Zanetti A., Balconi G., Corsi A., Bianco P., Wolburg H., Moore R., Oreda B., Kemler R., and Dejana E. (2003) The conditional inactivation of the beta-catenin gene in endothelial cells causes a defective vascular pattern and increased vascular fragility. J. Cell Biol. 162, 1111–1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gory-Faure S., Prandini M. H., Pointu H., Roullot V., Pignot-Paintrand I., Vernet M., and Huber P. (1999) Role of vascular endothelial-cadherin in vascular morphogenesis. Development 126, 2093–2102 [DOI] [PubMed] [Google Scholar]
- 26. Vittet D., Buchou T., Schweitzer A., Dejana E., and Huber P. (1997) Targeted null-mutation in the vascular endothelial-cadherin gene impairs the organization of vascular-like structures in embryoid bodies. Proc. Natl. Acad. Sci. U.S.A. 94, 6273–6278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Beckers C. M., Garcia-Vallejo J. J., van Hinsbergh V. W., and van Nieuw Amerongen G. P. (2008) Nuclear targeting of beta-catenin and p120ctn during thrombin-induced endothelial barrier dysfunction. Cardiovasc Res. 79, 679–688 [DOI] [PubMed] [Google Scholar]
- 28. Nelson W. J., and Nusse R. (2004) Convergence of Wnt, beta-catenin, and cadherin pathways. Science 303, 1483–1487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhu Y. T., Chen H. C., Chen S. Y., and Tseng S. C. (2012) Nuclear p120 catenin unlocks mitotic block of contact-inhibited human corneal endothelial monolayers without disrupting adherent junctions. J. Cell Sci. 125, 3636–3648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tornavaca O., Chia M., Dufton N., Almagro L. O., Conway D. E., Randi A. M., Schwartz M. A., Matter K., and Balda M. S. (2015) ZO-1 controls endothelial adherens junctions, cell-cell tension, angiogenesis, and barrier formation. J. Cell Biol. 208, 821–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Thingholm T. E., Jorgensen T. J., Jensen O. N., and Larsen M. R. (2006) Highly selective enrichment of phosphorylated peptides using titanium dioxide. Nat. Protoc. 1, 1929–1935 [DOI] [PubMed] [Google Scholar]
- 32. Larsen M. R., Thingholm T. E., Jensen O. N., Roepstorff P., and Jorgensen T. J. (2005) Highly selective enrichment of phosphorylated peptides from peptide mixtures using titanium dioxide microcolumns. Mol. Cell. Proteomics 4, 873–886 [DOI] [PubMed] [Google Scholar]
- 33. Cantin G. T., Shock T. R., Park S. K., Madhani H. D., and Yates J. R. 3rd (2007) Optimizing TiO2-based phosphopeptide enrichment for automated multidimensional liquid chromatography coupled to tandem mass spectrometry. Anal. Chem. 79, 4666–4673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cox J., and Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 35. Cox J., Neuhauser N., Michalski A., Scheltema R. A., Olsen J. V., and Mann M. (2011) Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 10, 1794–1805 [DOI] [PubMed] [Google Scholar]
- 36. Vizcaino J. A., Deutsch E. W., Wang R., Csordas A., Reisinger F., Rios D., Dianes J. A., Sun Z., Farrah T., Bandeira N., Binz P. A., Xenarios I., Eisenacher M., Mayer G., Gatto L., Campos A., Chalkley R. J., Kraus H. J., Albar J. P., Martinez-Bartolome S., Apweiler R., Omenn G. S., Martens L., Jones A. R., and Hermjakob H. (2014) ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat. Biotechnol. 32, 223–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Von Mering C., Huynen M., Jaeggi D., Schmidt S., Bork P., and Snel B. (2003) STRING: a database of predicted functional associations between proteins. Nucleic Acids Res. 31, 258–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Shannon P., Markiel A., Ozier O., Baliga N. S., Wang J. T., Ramage D., Amin N., Schwikowski B., and Ideker T. (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pitulescu M. E., Schmidt I., Benedito R., and Adams R. H. (2010) Inducible gene targeting in the neonatal vasculature and analysis of retinal angiogenesis in mice. Nat. Protoc. 5, 1518–1534 [DOI] [PubMed] [Google Scholar]
- 40. Hunter T., and Sefton B. M. (1980) Transforming gene product of Rous sarcoma virus phosphorylates tyrosine. Proc. Natl. Acad. Sci. U.S.A. 77, 1311–1315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sharma K., D'Souza R. C., Tyanova S., Schaab C., Wisniewski J. R., Cox J., and Mann M. (2014) Ultradeep human phosphoproteome reveals a distinct regulatory nature of Tyr and Ser/Thr-based signaling. Cell Rep. 8, 1583–1594 [DOI] [PubMed] [Google Scholar]
- 42. Hornbeck P. V., Chabra I., Kornhauser J. M., Skrzypek E., and Zhang B. (2004) PhosphoSite: A bioinformatics resource dedicated to physiological protein phosphorylation. Proteomics 4, 1551–1561 [DOI] [PubMed] [Google Scholar]
- 43. Bader G. D., and Hogue C. W. (2003) An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics 4, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Balda M. S., Garrett M. D., and Matter K. (2003) The ZO-1-associated Y-box factor ZONAB regulates epithelial cell proliferation and cell density. J. Cell Biol. 160, 423–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Stahl A., Connor K. M., Sapieha P., Chen J., Dennison R. J., Krah N. M., Seaward M. R., Willett K. L., Aderman C. M., Guerin K. I., Hua J., Lofqvist C., Hellstrom A., and Smith L. E. (2010) The mouse retina as an angiogenesis model. Invest Ophthalmol Vis. Sci. 51, 2813–2826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Cox J., and Mann M. (2011) Quantitative, high-resolution proteomics for data-driven systems biology. Annu. Rev. Biochem. 80, 273–299 [DOI] [PubMed] [Google Scholar]
- 47. Olsen J. V., Blagoev B., Gnad F., Macek B., Kumar C., Mortensen P., and Mann M. (2006) Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell 127, 635–648 [DOI] [PubMed] [Google Scholar]
- 48. Evans I. M., Britton G., and Zachary I. C. (2008) Vascular endothelial growth factor induces heat shock protein (HSP) 27 serine 82 phosphorylation and endothelial tubulogenesis via protein kinase D and independent of p38 kinase. Cell Signal 20, 1375–1384 [DOI] [PubMed] [Google Scholar]
- 49. van Gorp A. G., van der Vos K. E., Brenkman A. B., Bremer A., van den Broek N., Zwartkruis F., Hershey J. W., Burgering B. M., Calkhoven C. F., and Coffer P. J. (2009) AGC kinases regulate phosphorylation and activation of eukaryotic translation initiation factor 4B. Oncogene 28, 95–106 [DOI] [PubMed] [Google Scholar]
- 50. Gingras A. C., Gygi S. P., Raught B., Polakiewicz R. D., Abraham R. T., Hoekstra M. F., Aebersold R., and Sonenberg N. (1999) Regulation of 4E-BP1 phosphorylation: a novel two-step mechanism. Genes Dev. 13, 1422–1437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Shin S., Wolgamott L., Tcherkezian J., Vallabhapurapu S., Yu Y., Roux P. P., and Yoon S. O. (2014) Glycogen synthase kinase-3beta positively regulates protein synthesis and cell proliferation through the regulation of translation initiation factor 4E-binding protein 1. Oncogene 33, 1690–1699 [DOI] [PubMed] [Google Scholar]
- 52. Dobrikov M. I., Shveygert M., Brown M. C., and Gromeier M. (2014) Mitotic phosphorylation of eukaryotic initiation factor 4G1 (eIF4G1) at Ser1232 by Cdk1:cyclin B inhibits eIF4A helicase complex binding with RNA. Mol. Cell. Biol. 34, 439–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Roux P. P., Shahbazian D., Vu H., Holz M. K., Cohen M. S., Taunton J., Sonenberg N., and Blenis J. (2007) RAS/ERK signaling promotes site-specific ribosomal protein S6 phosphorylation via RSK and stimulates cap-dependent translation. J. Biol. Chem. 282, 14056–14064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Cote J. F., and Vuori K. (2002) Identification of an evolutionarily conserved superfamily of DOCK180-related proteins with guanine nucleotide exchange activity. J. Cell Sci. 115, 4901–4913 [DOI] [PubMed] [Google Scholar]
- 55. Laurin M., and Cote J. F. (2014) Insights into the biological functions of Dock family guanine nucleotide exchange factors. Genes Dev. 28, 533–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Shaheen R., Faqeih E., Sunker A., Morsy H., Al-Sheddi T., Shamseldin H. E., Adly N., Hashem M., and Alkuraya F. S. (2011) Recessive mutations in DOCK6, encoding the guanidine nucleotide exchange factor DOCK6, lead to abnormal actin cytoskeleton organization and Adams-Oliver syndrome. Am. J. Hum. Genet. 89, 328–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kuo J. C., Han X., Hsiao C. T., Yates J. R. 3rd, and Waterman C. M. (2011) Analysis of the myosin-II-responsive focal adhesion proteome reveals a role for beta-Pix in negative regulation of focal adhesion maturation. Nat. Cell Biol. 13, 383–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Yu H. W., Chen Y. Q., Huang C. M., Liu C. Y., Chiou A., Wang Y. K., Tang M. J., and Kuo J. C. (2015) beta-PIX controls intracellular viscoelasticity to regulate lung cancer cell migration. J. Cell Mol. Med. 19, 934–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Morales-Ruiz M., Fulton D., Sowa G., Languino L. R., Fujio Y., Walsh K., and Sessa W. C. (2000) Vascular endothelial growth factor-stimulated actin reorganization and migration of endothelial cells is regulated via the serine/threonine kinase Akt. Circ. Res. 86, 892–896 [DOI] [PubMed] [Google Scholar]
- 60. Zhuang G., Yu K., Jiang Z., Chung A., Yao J., Ha C., Toy K., Soriano R., Haley B., Blackwood E., Sampath D., Bais C., Lill J. R., and Ferrara N. (2013) Phosphoproteomic analysis implicates the mTORC2-FoxO1 axis in VEGF signaling and feedback activation of receptor tyrosine kinases. Sci. Signal 6, ra25. [DOI] [PubMed] [Google Scholar]
- 61. Xu J., Kausalya P. J., Phua D. C., Ali S. M., Hossain Z., and Hunziker W. (2008) Early embryonic lethality of mice lacking ZO-2, but Not ZO-3, reveals critical and nonredundant roles for individual zonula occludens proteins in mammalian development. Mol. Cell. Biol. 28, 1669–1678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Charpentier E., Lavker R. M., Acquista E., and Cowin P. (2000) Plakoglobin suppresses epithelial proliferation and hair growth in vivo. J. Cell Biol. 149, 503–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Lee S. W., Kim W. J., Jun H. O., Choi Y. K., and Kim K. W. (2009) Angiopoietin-1 reduces vascular endothelial growth factor-induced brain endothelial permeability via upregulation of ZO-2. Int. J. Mol. Med. 23, 279–284 [PubMed] [Google Scholar]
- 64. Leung J. C., Chan L. Y., Li F. F., Tang S. C., Chan K. W., Chan T. M., Lam M. F., Wieslander A., and Lai K. N. (2005) Glucose degradation products downregulate ZO-1 expression in human peritoneal mesothelial cells: the role of VEGF. Nephrol. Dial Transplant 20, 1336–1349 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.