Abstract
Hundreds of genes have been associated with respiratory chain disease (RCD), the most common inborn error of metabolism so far. Elimination of the respiratory electron chain by depleting the entire mitochondrial DNA (mtDNA, ρ0 cells) has therefore one of the most severe impacts on the energy metabolism in eukaryotic cells. In this study, proteomic data sets including the post-translational modifications (PTMs) phosphorylation and ubiquitination were integrated with metabolomic data sets and selected enzyme activities in the osteosarcoma cell line 143B.TK−. A shotgun based SILAC LC-MS proteomics and a targeted metabolomics approach was applied to elucidate the consequences of the ρ0 state. Pathway and protein–protein interaction (PPI) network analyses revealed a nonuniform down-regulation of the respiratory electron chain, the tricarboxylic acid (TCA) cycle, and the pyruvate metabolism in ρ0 cells. Metabolites of the TCA cycle were dysregulated, such as a reduction of citric acid and cis-aconitic acid (six and 2.5-fold), and an increase of lactic acid, oxalacetic acid (both twofold), and succinic acid (fivefold) in ρ0 cells. Signaling pathways such as GPCR, EGFR, G12/13 alpha, and Rho GTPases were up-regulated in ρ0 cells, which could be indicative for the mitochondrial retrograde response, a pathway of communication from mitochondria to the nucleus. This was supported by our phosphoproteome data, which revealed two main processes, GTPase-related signal transduction and cytoskeleton organization. Furthermore, a general de-ubiquitination in ρ0 cells was observed, for example, 80S ribosomal proteins were in average threefold and SLC amino acid transporters fivefold de-ubiquitinated. The latter might cause the observed significant increase of amino acid levels in ρ0 cells. We conclude that an elimination of the respiratory electron chain, e.g. mtDNA depletion, not only leads to an uneven down-regulation of mitochondrial energy pathways, but also triggers the retrograde response.
The mitochondrial energy metabolism is necessary for the generation of more than 90% of cellular energy in form of adenosine triphosphate (ATP) (1, 2). Human mitochondria contain ∼1500–2000 proteins (3) and have an own genome (mtDNA) encoding 37 genes, including 13 proteins of the oxidative phosphorylation (OXPHOS), 22 tRNAs, and two rRNAs (4). Mitochondria store the majority of cellular calcium, play an important role during apoptosis, heat production, membrane potential, and harbor important catabolic and anabolic pathways such as TCA cycle, beta oxidation, amino acid, and heme synthesis pathways (5). Respiratory chain diseases (RCD)1 represent a large subset of mitochondrial disorders and are biochemically characterized by defective OXPHOS, leading predominantly to neurological and muscular degeneration. They occur at an estimated prevalence of one in 5000 live births and are collectively the most common inborn error of metabolism (6). Human cells lacking mtDNA (ρ0 cells) were originally obtained from the human osteosarcoma cell line 143B.TK− (7) by chronic exposure to the DNA intercalating dye ethidium bromide. Since then, ρ0 cell lines have been established from various tissues and species applying additional methods such as application of the anticancer drug ditercalinium (8) or restriction enzymes specifically targeting mitochondria (9, 10). However, ρ0 cells are viable in culture, provided appropriate conditions are met (11), e.g. supplementation with uridine to compensate for impaired pyrimidine biosynthesis, and pyruvate to provide electron acceptors for anaerobic glycolysis. The mitochondria of ρ0 cells still maintain an electrochemical gradient across the inner membrane by a mechanism coupled to ATP hydrolysis (12), thus making them net consumers of ATP. Our study was performed with the ρ0 cell line generated from 143B.TK− cells, as it is one of the best characterized ρ0 cell lines available (7, 9, 13, 14).
The progression of mitochondrial diseases has a broad spectrum with variable clinical manifestations and can originate by mutations either in the mitochondrial or the nuclear genome (15, 16). More than 250 gene defects have been reported to date and this number continues to grow (17). MtDNA depleted cells can be used to investigate the pathogenesis of specific mtDNA mutations, and for developing a better understanding of interactions between nuclear and mitochondrial genomes in mitochondrial disease (12).
However, little is known about protein abundance changes, the influence and regulation of post-translational modifications (PTMs) or metabolites in ρ0 cells. Based on a previous proteomics study from ρ0 mitochondria separated by 2-D electrophoresis, others could identify an uneven down-regulation of subunits of the respiratory electron chain and of mitochondrial ribosomes (18).
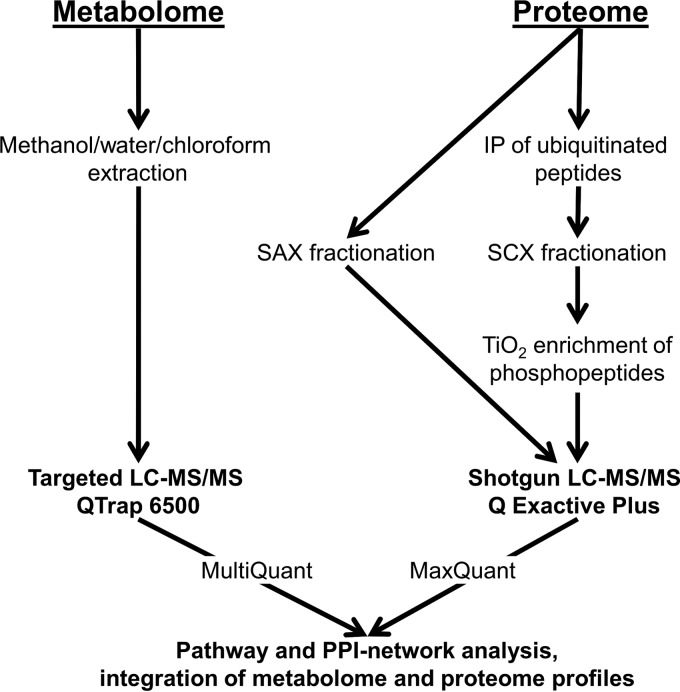
In this study, metabolome and proteome profiles of the parental cell line 143B.TK− versus ρ0 were integrated, including PTM analyses such as phosphorylation and ubiquitination to characterize the impact of the absence of mtDNA for the entire cell (Fig. 1). For quantitative proteome profiling, a shotgun LC-MS/MS approach including the classical SILAC labeling was performed. For comprehensive metabolome profiling, a targeted LC-MS approach, based on multiple reaction monitoring (MRM) (19) was applied.
Fig. 1.
A schematic workflow indicating applied methods, instruments, and software tools for an integrated metabolome and proteome profiling.
Our study revealed that mtDNA depletion leads to a nonuniform down-regulation of the mitochondrial energy metabolism in ρ0 cells on the proteome level. Metabolites of the TCA cycle were highly dysregulated which in turn had an impact on the amino acid levels, which were up-regulated. Perturbation of the mitochondrial energy metabolism could be indicative for an activation of the retrograde response, supported by proteome data and phosphorylation patterns in GTPase signaling pathways and the cytoskeleton as well as a general de-ubiquitination in ρ0 cells.
EXPERIMENTAL PROCEDURES
Cell Culture
The thymidine kinase deficient (TK−) osteosarcoma cell line 143B.TK− (ATCC-CRL-8303), with a bromodeoxyuridine resistance was obtained from LGC Standards and is the parental line of recently generated ρ0 cells (13) by the protocol from (7). The wild-type cell line 143B.TK− and the according ρ0 cells were cultivated in SILAC DMEM (Silantes, Munich, Germany, without l-lysine and l-arginine) containing 4.5 g/L glucose, 1 mm pyruvate, supplemented with 5% dialyzed FBS (Silantes, Munich, Germany), 1% Penicillin-Streptomycin-Neomycin (Invitrogen, Carlsbad, CA), 100 μg/ml bromodeoxyuridine (Sigma-Aldrich) and 50 μg/ml uridine (Sigma-Aldrich) at 37 °C in a humidified atmosphere of 5% CO2. Cells were labeled with light (L) or heavy (H) isotopes of arginine (12C614N4, 13C615N4;30 mg/L) and lysine (12C614N2, 13C615N2; 80 mg/L; both: Silantes) and grown to confluency in one 300 cm2 polystyrene flask per replicate. Proteomic experiments were done in biological quadruplicates, including a label-switch. For metabolome profiling and enzymatic measurements, light SILAC labeled cultures were grown in biological triplicates.
Verification of the ρ0 Status by PCR
Full depletion of the mtDNA was verified by PCR according to (13). In brief, genomic DNA was isolated using the QIAmp DNA Mini Kit for amplification of a 399-bp mtDNA product with following primers: 5′TTCACAAAGCGCCTTCCCCCGT and 5′GCGATGGTGAGAGCTAAGGTCGGG, which span a region of nt 3153–nt 3551 (accession number NC_012920.1). For the 238-bp nuclear DNA product amplification, primer 5′AGTGTCTTAAGAGTAAAGCTGGCCACA and 5′ TTGCCTTTGTTGCATTTTCTACAG, spanning a region of exon 5 of the gene USMG5 (accession number NT_030059.13, nt 55953445 - nt 55953207) were used. PCR was performed with 50 ng of genomic DNA as template, 250 μm dNTPs (Invitrogen), 0.5 μm of each primer, and 2.5 U of Taq polymerase (Invitrogen) were used.
PCR conditions were as following: initial denaturation at 94 °C for 2 min, denaturation at 94 °C for 30 s, primer annealing 30 s at 60 °C, elongation 60 s at 72 °C, 30 cycles in a thermocycler (Mastercycler personal 5332, Eppendorf, Hamburg, Germany). PCR products and mass calibration ladder (New England Biolabs, Ipswich, MA) were loaded and separated on a 2% agarose/TBE gel.
Measurement of TCA Cycle and Respiratory Electron Chain Enzyme Activities
Sample preparation for spectrophotometric detection of selected enzyme activities was done as reported previously (20). Measurements were performed in biological triplicates from independent culture dishes and normalized for total protein content. Malate dehydrogenase (MDH) and fumarase (FH) activities were measured according to the manufactures' protocol (Biovision Technologies, Golden, CO), citrate synthase (CS), and isocitrate dehydrogenase (IDH2) according to (21).
Metabolite Extraction and Profiling by Targeted LC-MS
Metabolite extraction was done as reported previously by us (19). Protein containing pellets of the first extraction-step were used to determine the protein concentration by a BC assay (Sigma-Aldrich), which was used for sample normalization. Additionally, an internal standard, containing chloramphenicol and C13-labeled l-glutamine, l-arginine, l-proline, l-valine, and uracil (3.5 μm final concentration) was added to each sample. Samples were cleaned by iso-disc filters (iso-disc filters PTFE 13 mm × 0.2 mm, Supelco, Bellafonte, PA), to avoid column clogging. Dry residuals were suspended in 50 μl ACN, 0.1% FA, and 50 μl MeOH, 0.1% FA for analysis by according HILIC mode, in 50 μl H2O, 0.1% FA for RPLC mode and centrifuged at 17,500 × g for 5 min at 4 °C. The supernatants were transferred to microvolume inserts, 5 μl per run were injected for LC-MS/MS analysis.
264 metabolites, such as amino acids, nucleic acids, bile acids, carbohydrates, vitamins, hormones, nucleotides, and biogenic amines beside others, were selected to cover most of the important metabolic pathways in mammals. Metabolites are chemically very diverse, therefore several different LC columns have been used for metabolite separation: A Reprosil-PUR C18-AQ (1.9 μm, 120 Å, 150 × 2 mm ID; Dr. Maisch; Ammerbuch, Germany) column and a zicHILIC (3.5 μm, 100 Å, 150 × 2.1 mm ID; di2chrom; Marl, Germany). LC-MS instrument (1290 series UHPLC; Agilent, Santa Clara, CA) conditions online coupled to a QTrap 6500 (Sciex, Foster City, CA) were reported previously (19).
A list of all metabolites including MRM ion ratios, retention times and KEGG or HMDB metabolite identifiers can be found in supplemental Table S1. The mass spectrometry metabolomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository (22) with the data set identifier PXD002425.
Relative quantification was performed using MultiQuantTM software v.2.1.1 (Sciex, Foster City, CA). Integration settings were a Gaussian smooth width of two points and a peak splitting factor of two. Peak integrations were reviewed manually and normalized according to the protein content and subsequently by internal standards.
Cell Harvesting and Sample Preparation for Proteomics
Cells were harvested and lysed under denaturing conditions in a buffer containing 4% SDS, 0.1 m DTT, and 0.1 m Tris, pH 8.0. Equal amounts of differently labeled ρ0 and parental samples were mixed, ∼17 mg of protein for each replicate in total. Lysates were sonicated for 1 min, boiled at 95 °C for 5 min and precipitated with acetone at −20 °C overnight. Lyophilized proteins were dissolved in 8 m urea, 10 mm Tris, pH 8, alkylated with a final concentration of 5.5 mm chloroacetamide for 30 min and Lys-C digested (1:2000) for 4 h at room temperature followed by a trypsin digestion (1:1000) overnight in 2 m urea at 37 °C (23). Subsequent, the peptides were purified with C18 columns. For whole proteome profiling 100 μg of each sample was further separated using six pH fractions (pH levels 11, 8, 6, 5, 4, and 3) of strong anion exchange chromatography (SAX, 3 m Purification, Meriden, CT) according to (24). The remaining sample amount of each C18 purified peptide mixture was used for immunoprecipitation of ubiquitinated peptides using the PTMScan Ubiquitin Remnant Motif Kit (Cell Signaling, Cambridge, UK) (25). Peptides were immunoprecipitated with 40 μl of α-diGly coupled to protein A agarose beads over night at 4 °C on a rotation wheel. The beads were washed three times in ice-cold immunoprecipitation buffer followed by three washes in water. Subsequently, the enriched peptides were purified and desalted with C18 StageTips. The peptides which did not bind to the immunoaffinity beads were used for subsequent phosphoproteome profiling. Therefore, samples were fractionated by strong cation exchange (SCX) chromatography. Five μg of each fraction were again used for proteome profiling, whereas the remaining sample was used for TiO2 (GL Sciences, Torrance, CA) enrichment of phosphorylated peptides, according to (23). All SAX and SCX fractions, as well as the PTM scans, were allocated to the corresponding replicate and analyzed jointly by MaxQuant (Fig. 1).
LC-MS Instrument Settings for Shotgun Proteome Profiling and Data Analysis
LC-MS/MS was carried out by nanoflow reverse phase liquid chromatography (Dionex Ultimate 3000, Thermo Scientific, Waltham, MA) coupled online to a Q-Exactive Plus Orbitrap mass spectrometer (Thermo Scientific, Waltham, MA). Briefly, the LC separation was performed using a PicoFrit analytical column (75 μm ID × 25 cm long, 15 μm Tip ID (New Objectives, Woburn, MA) in-house packed with 3-μm C18 resin (Reprosil-AQ Pur, Dr. Maisch, Ammerbuch-Entringen, Germany). Peptides were eluted using a nonlinear gradient from 2 to 40% solvent B 79.9% acetonitrile, 20% H2O, 0.1% formic acid). 3 kV were applied for nanoelectrospray over 210 min at a flow rate of 266 nL/min (solvent A: 99.9% H2O, 0.1% formic acid; solvent B: generation. A cycle of one full FT scan mass spectrum (300–1750 m/z, resolution of 70,000 at m/z 200, AGC target 1e6) was followed by 12 data-dependent MS/MS scans (resolution of 35,000, AGC target 5e5) with normalized collision energy of 25 eV. To avoid repeated sequencing of the same peptides a dynamic exclusion window of 30 s was used. In addition, only the peptide charge states between two to eight were sequenced, in the case of ubiquitinated peptide samples, only charge states three to eight were allowed.
Raw MS data were processed with MaxQuant software (v1.5.0.0) (26) with the Andromeda search engine (27) and searched against the human proteome database UniProtKB with 69,714 entries released in 06/2015. Additionally, the “requantify” and “match between runs” features were implemented to increase the number of peptides which can be used for quantification. A false discovery rate (FDR) of 0.01 for proteins and peptides, a minimum peptide length of seven amino acids, a mass tolerance of 4.5 ppm for precursor, and 20 ppm for fragment ions were required. A minimum Andromeda score of 0 and 40 (delta score 0 and 9) for unmodified peptides and modified peptides was applied. A maximum of two missed cleavages was allowed for the tryptic digest, except for ubiquitination where three missed cleavages were allowed. Following SILAC modifications were used: 13C615N4-arginine and 13C615N2-lysine. Cysteine carbamidomethylation was set as fixed modification, whereas N-terminal acetylation, methionine oxidation, diGly modification of lysine and phosphorylation of serine, threonine, and tyrosine were set as variable modifications. The two later PTM's were only used for the according enriched fractions. MaxQuant processed output files can be found in supplemental Table S2, showing peptide and protein identification, accession numbers, % sequence coverage of the protein, posterior error probability (PEP) values, and normalized SILAC ratios. To correct for mixing errors of total protein amounts, the SILAC ratios were normalized for each LC-MS run separately, according to (26). Contaminants as well as proteins identified by site modification and proteins derived from the reversed part of the decoy database were strictly excluded from further analysis. For PTM analysis, only high confidence sites, defined by a localization probability higher than 0.75 for phosphorylation sites and 0.9 for ubiquitination sites, PEP score smaller than 0.01 and an Andromeda score difference between the best and second best peptide match larger than five, were considered (28). All PTM analyses were performed with these high confidence criteria.
Statistical, Pathway, and PPI Network Analyses
For metabolome data sets, a two-sample t test, and for the proteome data sets, a one sample t test was performed. Multiple test correction was done by Benjamini-Hochberg (BH) with a FDR of 0.05. Significantly regulated metabolites and proteins were marked by a plus sign in according (supplemental Tables S2 and S3).
For comprehensive proteome data analyses, gene set enrichment analysis (GSEA, v2.1.0) (29) was applied to see if a priori defined sets of proteins show statistically significant, concordant differences between ρ0 and parental state. Only proteins with valid values in all replicates were averaged and used for GSEA analysis and log2 transformation (supplemental Table S2). GSEA default settings were used, except the minimum size exclusion was set to five and reactome v5.0 was used as gene set database. The cut off for significantly regulated pathways was set to ≤ 0.05 p value and ≤ 0.25 FDR. Only pathways with significant values in merged replicates were extracted, average p- and FDR values are shown in Table I.
Table I. Significantly regulated Reactome pathways in ρ0 versus parental cells, analyzed by GSEA (thresholds: p value ≤ 0.05; q-value ≤ 0.25).
| Reactome pathway | GSEA |
Globaltest |
|||
|---|---|---|---|---|---|
| Protein entries | p-Value | q-Value | p-Value | q-Value | |
| > down-regulated pathways in ρ0 cells | |||||
| Respiratory electron transport | 41 | 0.00 | 0.00 | 0.00 | 0.00 |
| TCA cycle and respiratory electron transport | 77 | 0.00 | 0.00 | 0.00 | 0.00 |
| Respiratory electron transport ATP synthesis by chemiosmotic coupling and heat production by uncoupling proteins | 51 | 0.00 | 0.00 | 0.00 | 0.00 |
| Pyruvate metabolism and citric acid TCA cycle | 28 | 0.00 | 0.04 | 0.00 | 0.00 |
| TCA cycle | 17 | 0.01 | 0.24 | 0.00 | 0.00 |
| > up-regulated pathways in ρ0 cells | |||||
| G12/13 α signaling events | 24 | 0.00 | 0.02 | 0.02 | 0.04 |
| Deposition of new CENPA containing nucleosomes at the centromere | 15 | 0.00 | 0.03 | 0.03 | 0.06 |
| Signaling by Rho GTPases | 40 | 0.00 | 0.03 | 0.04 | 0.07 |
| Meiosis | 34 | 0.00 | 0.03 | 0.00 | 0.00 |
| Meiotic synapsis | 19 | 0.00 | 0.05 | 0.00 | 0.00 |
| GPCR downstream signaling | 46 | 0.00 | 0.08 | 0.00 | 0.01 |
| Chromosome maintenance | 53 | 0.00 | 0.16 | 0.00 | 0.01 |
| Signaling by GPCR | 74 | 0.00 | 0.16 | 0.00 | 0.00 |
| Mitotic prometaphase | 61 | 0.00 | 0.19 | 0.00 | 0.01 |
| Hormone sensitive lipase HSL mediated triacylglycerol hydrolysis | 7 | 0.00 | 0.14 | 0.00 | 0.00 |
| Apoptosis induced DNA fragmentation | 11 | 0.00 | 0.04 | 0.06 | 0.10 |
| Packaging of telomere ends | 7 | 0.01 | 0.15 | 0.00 | 0.00 |
| Cell cell communication | 38 | 0.01 | 0.19 | 0.05 | 0.08 |
| Regulation of insulin secretion by acetylcholine | 5 | 0.01 | 0.18 | 0.01 | 0.03 |
| Rap1 signaling | 7 | 0.01 | 0.15 | 0.02 | 0.04 |
| Gap junction trafficking | 8 | 0.01 | 0.18 | 0.01 | 0.02 |
| EGFR downregulation | 19 | 0.01 | 0.18 | 0.03 | 0.05 |
| Recycling pathway of L1 | 19 | 0.01 | 0.19 | 0.01 | 0.02 |
| Gap junction degradation | 8 | 0.01 | 0.14 | 0.01 | 0.02 |
| Meiotic recombination | 20 | 0.02 | 0.14 | 0.00 | 0.01 |
| Glucagon signaling in metabolic regulation | 10 | 0.02 | 0.19 | 0.07 | 0.10 |
| GPCR ligand binding | 7 | 0.02 | 0.19 | 0.00 | 0.00 |
| Regulation of insulin secretion by glucagon like peptide1 | 14 | 0.02 | 0.18 | 0.07 | 0.10 |
| Factors involved in megakaryocyte development and platelet production | 55 | 0.02 | 0.23 | 0.00 | 0.01 |
| RNA Pol I promoter opening | 9 | 0.03 | 0.18 | 0.06 | 0.10 |
| Gi α signaling events | 12 | 0.04 | 0.22 | 0.00 | 0.00 |
To cope with the strong abundance of down-regulated as compared with up-regulated genes, we additionally employed a self-contained test. The test was performed on the normalized intensity ratios (or their inverse for the label-switched experiments, respectively) using the “globaltest” R package (30). Gene symbols were mapped to Entrez Gene IDs using the “org.Hs.eg.db” annotation package. The Broad gene sets (msigdb_v5.0.xml) were downloaded from http://www.broadinstitute.org/gsea/downloads.jsp and filtered for the subset of 509 Reactome pathway annotations that have at least five gene members. After performing the global test, the p values were corrected for multiple testing using the Benjamini-Hochberg method and are displayed additionally in Table I.
For protein–protein interaction (PPI) network analyses, the software tool String v.10 has been used to visualize networks of significantly down-regulated proteins with a confidence level of 0.7 (31). Protein nodes which were not integrated into a network were removed.
For PTM analyses, regulated phosphorylation and ubiquitination sites were defined by two standard deviations from the median of unmodified peptides. Resulting lists were subjected to the gene set enrichment analysis tool of the DAVID Bioinformatics Resources (32, 33), and analyzed by Gene Ontology (GO) biological process (GOTERM_BP_FAT), or molecular function (GOTERM_MF_FAT), respectively. Additionally, the overrepresentation analysis feature offered by the ConsensusPathDB (CPDB) (34) was used to find enriched pathways among regulated sites. The default parameters were used (all database resources checked, minimum overlap of five, p value cutoff = 0.01). In both analyses, lists of all phosphorylated or ubiquitinated sites were used as background sets.
Multiple data sets and software tools were used to elucidate as much information as possible, as every database has another focus and sets of pathways defined. Further data analyses were performed using Perseus (v1.5.1.6), a post data acquisition package of MaxQuant. GraphPad Prism 5.03 was used for graphing.
RESULTS
Verification of the mtDNA Depletion in ρ0 Cells
To verify complete mtDNA depletion in ρ0 cells, a PCR of nuclear and mtDNA encoded genes was executed. Nuclear PCR products were detected in both, 143B.TK− and ρ0 cells, whereas mtDNA PCR products could exclusively be amplified in 143B.TK− cells, confirming the mtDNA depletion of ρ0 cells (supplemental Fig. S1). Furthermore, no evidence of mtDNA encoded proteins could be found by LC-MS/MS in ρ0 cells.
Metabolome Profiling
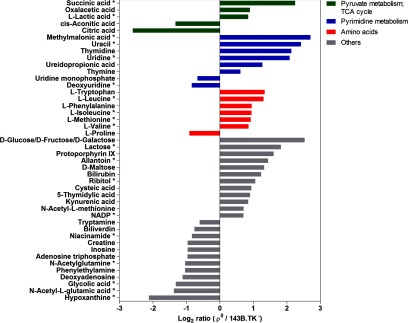
To elucidate metabolic alterations of mtDNA depleted cells, a MRM based approach was used for identification and relative quantification. In total, 103 metabolites in ρ0 versus 143B.TK− cells were quantified, 44 of them were ≥ 1.5-fold and 19 of them were significantly regulated (Fig. 2). Metabolites of the TCA cycle showed a striking dysregulation, for example, lactic acid was twofold elevated in ρ0 cells, acetyl CoA was unchanged and citric acid and cis-aconitic acid were the highest down-regulated metabolites (six and 2.5-fold, respectively) in ρ0 cells. In clear contrast, succinic acid and oxalacetic acid were five and twofold increased in ρ0 cells (Fig. 2). Most of the detected amino acids were significantly up-regulated (two to threefold) in ρ0 cells, except for the twofold down-regulation of proline. Compounds of the pyrimidine metabolism were highly regulated (two to sevenfold) in ρ0 cells (Fig. 2), like precursors or intermediates of uridine, such as uracil, ureidopropionic acid, methylmalonic acid, and deoxyuridine. Both, ρ0 and parental cells were grown under equal conditions, but as ρ0 cells are dependent on the supplementation of uridine, we conclude that this can lead to a bias in the pyrimidine metabolism. An entire list of all identified and quantified metabolites can be found in (supplemental Table S3).
Fig. 2.
Profile of regulated metabolites in ρ0versus parental cells. Only ≥ 1.5-fold and/or significantly (* adjusted p value < 0.05) changed metabolites are shown. Amino acids, metabolites of the TCA cycle and pyrimidine metabolism and others are grouped.
Proteome Profiling
For comprehensive proteome profiling, tryptic peptides were fractionated using strong anion exchange (SAX) and strong cation exchange (SCX) chromatography. Thus, four biological replicates for ρ0 and parental cells, including the SILAC label switches were analyzed in a total of 124 LC-MS/MS runs. 1.4 million MS2 spectra were identified, belonging to more than 8000 protein groups with at least one peptide per protein group in total. Only proteins with ratios, based on at least two peptides in all replicates were used for further data analysis. This stringent criterion resulted in a final protein group list of 4815 entries (n = 4), of which 2708 were found to be significantly regulated after BH correction. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository (22) with the data set identifier PXD002425.
MaxQuant processed output files can be found in supplemental Table S2. The reproducibility of the biological replicates was tested by Pearson correlation and visualized in a multiscatter plot for all proteome-, phosphorylation-, and ubiquitination profiles (supplemental Fig. S2A–S2C).
Pathway and PPI Network Analyses
The pathway enrichment tool GSEA was applied to reveal if a priori defined sets of proteins show statistically significant, concordant differences between ρ0 and 143B.TK− states. Pathways with significant p- and q-values are listed in Table I. As we observed an abundance of down-regulated genes as opposed to up-regulated genes, we also performed a self-contained test for enriched pathways, confirming the result from GSEA (Table I).
Additionally, a PPI network analysis of significantly down-regulated proteins was performed.
Significantly Down-regulated Pathways in ρ0 Cells
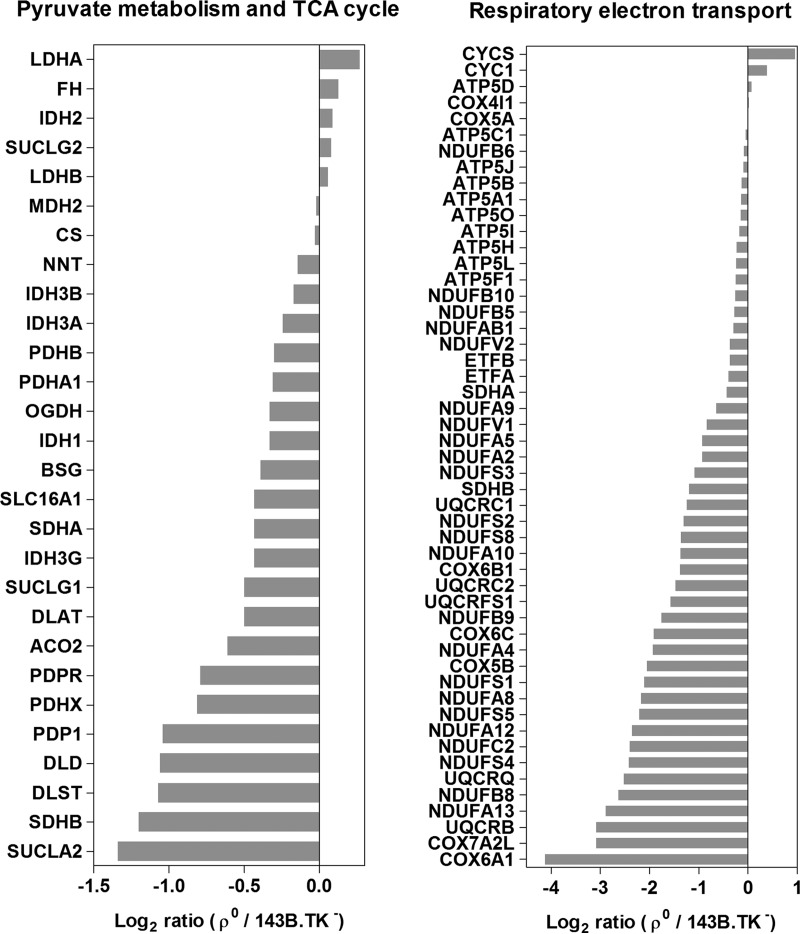
Significantly down-regulated pathways were detected exclusively within the mitochondrial energy metabolism, such as the pyruvate metabolism, the TCA cycle, and the respiratory electron chain (Table I, Fig. 3). Within the pyruvate metabolism, an up-regulation of the anaerobic part in ρ0 cells, the lactate dehydrogenases LDHA and LDHB (Fig. 3A) was detected. Both are catalyzing the interconversion of pyruvate and lactate by simultaneously interconverting NADH and NAD+, to restore reducing equivalents for anaerobic glycolysis.
Fig. 3.
Significantly down-regulated Reactome pathways and individual protein ratios between ρ0 and parental cells. An unequal regulation of complexes and their subunits is shown in A pyruvate metabolism and TCA cycle and B respiratory electron transport and ATP synthesis by chemiosmotic coupling and heat production by uncoupling.
The same observation was made for proteins involved in the TCA cycle, the entire pathway was significantly down-regulated in ρ0 cells (Table I), but individual enzymes were unregulated such as citrate synthase (CS), or showed even an up-regulation, such as fumarase (FH, Fig. 3A). Interestingly, this dysregulation was also observable within enzyme complexes, such as for succinate-CoA ligase and isocitrate dehydrogenase, where some subunit abundancies were decreased (SUCLA2, IDH3G) and others instead increased (SUCLG2, IDH2) in ρ0 cells.
As expected, a significant down-regulation of the entire respiratory electron transport chain (Table I) was observed, as all mitochondrial encoded (core) subunits were missing in ρ0 cells. As previously observed in our study of rotenone (specific complex I inhibitor) treated HeLa cells (19), subunits were not uniformly down-regulated in ρ0 cells (Fig. 3B). Most of the respiratory electron transport chain subunits were indeed up to 17-fold decreased, but unchanged or slightly up-regulation of the complex III subunit cytochrome c1 (1.3-fold; CYC1) and the electron carrier cytochrome c (twofold; CYCS) was observed. ATP synthase subunits, necessary to assemble the protein complex even without mitochondrial encoded subunits for coupled ATP hydrolysis were unchanged in ρ0 cells (12, 13).
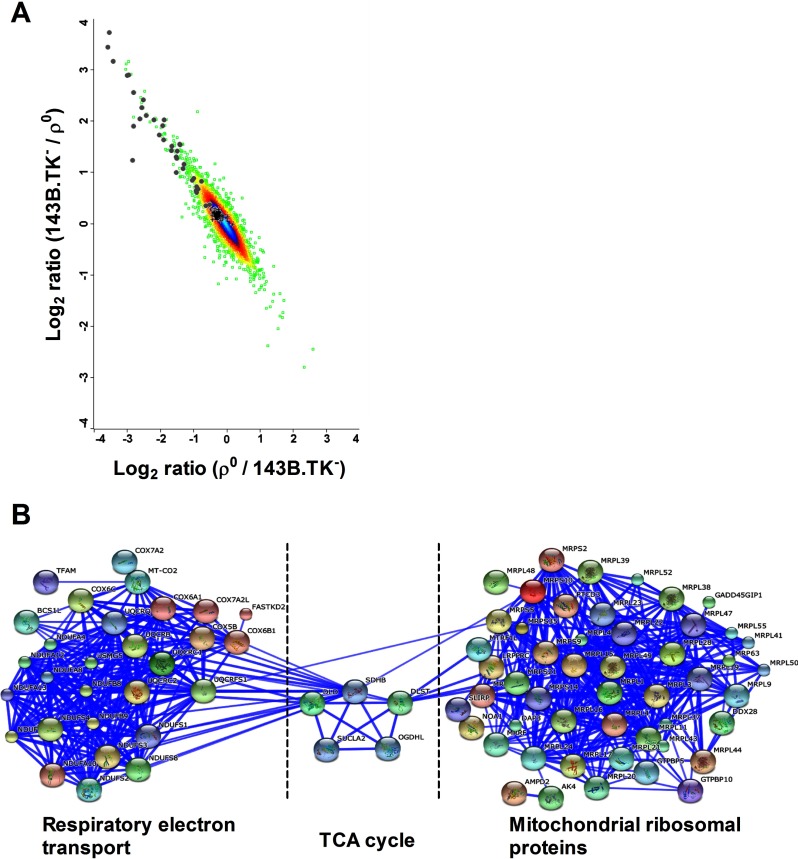
Cytoplasmic ribosomal subunits were unchanged (on average 1.1-fold down-regulated in ρ0 cells, Fig. 4A). In clear contrast, mitochondrial ribosomal proteins were on average threefold down-regulated in ρ0 cells (Fig. 4A). As mitochondrial ribosomal proteins are not defined as an independent pathway, this striking down-regulation was visualized by a PPI network (Fig. 4B). One of the highest down-regulated proteins in the ρ0 state was mitochondrial transcription factor A (TFAM, eightfold). TFAM binds to the mitochondrial DNA and functions in mitochondrial transcription regulation and mtDNA maintenance (35).
Fig. 4.
Intensity scatter plot of protein abundances and a PPI network of down-regulated proteins in ρ0versus parental cells. A, Shown are unregulated cytoplasmic 80S ribosomal proteins, indicated by black crosses and mitochondrial 55S ribosomal proteins, which were down-regulated in ρ0 cells, indicated by gray dots. B, Protein–protein interaction (PPI) network analysis of significantly down-regulated proteins in ρ0 cells (average of valid values), featuring subunits of the respiratory electron chain, proteins involved in the TCA cycle and mitochondrial 55S ribosomal proteins. The proteins that are known to interact with each other are linked with a blue line. The cut off for regulated proteins was determined using two standard deviations from the median.
Significantly Up-regulated Pathways in ρ0 Cells
Surprisingly, most of the significantly regulated pathways were indeed up-regulated in ρ0 cells (Table I). Many of these pathways play a role in the cell cycle and cell signaling. For example, numerous signaling pathways, such as G12/13 alpha signaling, Rho GTPases, G-protein-coupled receptor (GPCR) signaling, and Gi α signaling were found to be significantly up-regulated in ρ0 cells. Among Rho GTPases, the well-studied proteins RhoA, Rac1 and Cdc42 were found to be core enriched within the pathway. Furthermore, within the meiosis pathway, proteins involved in the telomere nucleoprotein complex, lamin, and histone proteins were up-regulated. Lamin proteins are thought to be involved in nuclear stability, chromatin structure, and gene expression.
Measurement of Enzyme Activities of the TCA Cycle
Proteome and metabolome profiling revealed an uneven regulation of proteins and metabolites involved in the TCA cycle (Fig. 2 and Fig. 3). Thus, enzyme activities were determined for individual TCA cycle enzymes to explore, how these activities are correlated to according protein abundances and metabolite levels (Table II).
Table II. Enzyme activities of the TCA cycle in ρ0 and 143B.TK−. CS, citrate synthase; IDH, isocitrate dehydrogenase; MDH, malate dehydrogenase; FH, fumarase.
| Enzyme | ρ0 [mU/mg of protein] ± S.D. | 143B.TK− [mU/mg of protein] ± S.D. | p value |
|---|---|---|---|
| CS | 31.3 ± 3 | 27.7 ± 2 | 0.16 |
| IDH2 | 13 ± 1 | 15.7 ± 2.1 | 0.12 |
| MDH2 | 504.4 ± 91.5 | 161.1 ± 18.8 | 0.003 |
| FH | 25 ± 1.5 | 13.4 ± 4.2 | 0.01 |
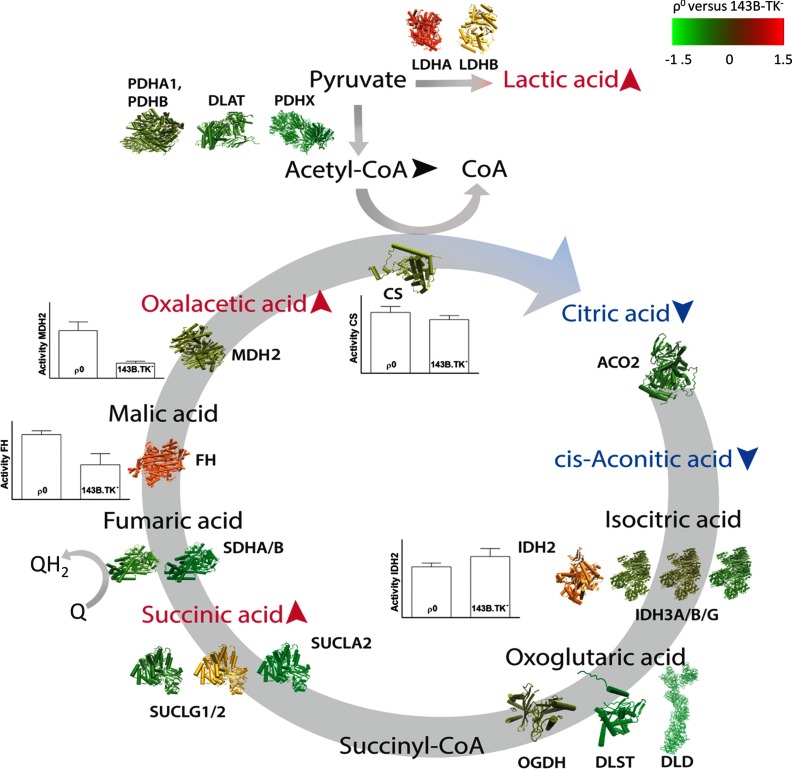
The enzyme activities of CS and IDH2 were unaltered, whereas MDH2 and FH showed significantly increased enzyme activities in ρ0 cells. Because mitochondrial fractions were used for enzyme measurements, only the activity of IDH2 was determined. All enzyme activities correlated well with protein abundances, except for MDH2. For a better overview, protein abundances, metabolite levels, and enzyme activities of the TCA cycle between ρ0 and parental cells were integrated and visualized in Fig. 5.
Fig. 5.
Integration of protein abundances, metabolite levels, and enzyme activities of TCA cycle compounds between ρ0 and parental cells. Symbolic 3D protein structures of enzymes and their subunits are colored according to the heat map, red = up-regulated, green = down-regulated in ρ0 cells. Enzyme activities are taken from Table II. Accumulation or reduction of metabolites in ρ0 versus parental cells is indicated by adjacent arrows.
Phosphoproteome Profiling
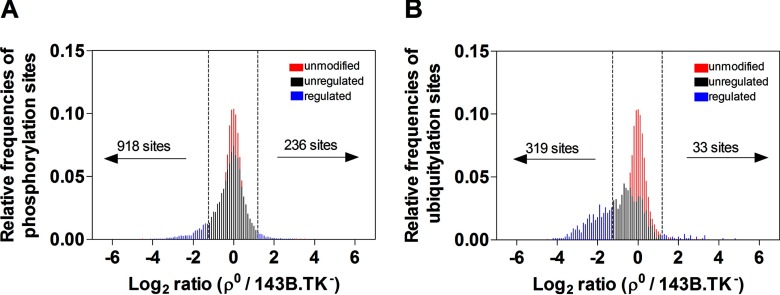
The phosphoproteome was analyzed by sequentially applying SCX separation of peptides and enrichment by TiO2 beads to elucidate the impact of the mtDNA depletion on the phosphorylation status. In total, we identified 14,905 phosphorylation sites, 9051 of them with a high confidence and ratios, belonging to a total of 3371 proteins. Nine hundred and eighteen phosphorylation sites were down-regulated, 236 up-regulated in ρ0 cells (Fig. 6A). A curated list of 1158 genes with high confidence of mitochondrial localization derived from the Human MitoCarta2.0 (36) was used to identify 609 (13%) verified mitochondrial proteins in our data set. Only 4% (124 proteins) of all 3371 phosphorylated proteins were localized in the mitochondrion, showing a fivefold underrepresentation of phosphorylation events in mitochondria compared with the total proteome.
Fig. 6.
Distribution of relative frequencies of phosphorylation and ubiquitination sites in ρ0versus parental cells (average of replicates; n = 1 - 4). A, Log2 ratios of unmodified peptides (red), unregulated phosphorylation sites (black) and regulated phosphorylation sites (blue) and B, for regulated ubiquitination sites (blue). The cut off value for significantly regulated PTM sites was based on two standard deviations from the median of unmodified peptides (dotted lines; −1.2; 1.2).
Regulated phosphoproteins were submitted to DAVID as well as CPDB for computing enrichment with existing lists created from prior knowledge organized into gene-set libraries. The resulting lists of the GO category “biological process” and the CPDB pathways revealed two main processes, GTPase-related signal transduction and cytoskeleton organization, as down-regulated phosphorylation dependent pathways in ρ0 versus parental cells (supplemental Table S4).
Ubiquitylome Profiling
A specific diGly antibody, recognizing the remnant glycine after tryptic digestion was used for detection of ubiquitinated proteins. It has to be mentioned that the applied methodology cannot distinguish between ubiquitination, neddylation, or ISGylation, as the same diglycine motif is present on lysines after tryptic digestion, the target of the diGly specific antibody. Ubiquitination can affect proteins in many different ways, such as signal for degradation, alter cellular locations, affect the enzyme activity, and promote or prevent protein interactions (37–39). In total, 1398 ubiquitinated sites, 900 of them with a high confidence and ratios, belonging to 551 proteins were identified. MG132, a potent inhibitor of the proteasome was not used for accumulating ubiquitinated proteins, because this could have a significant impact on other PTMs, the metabolome and proteome in general.
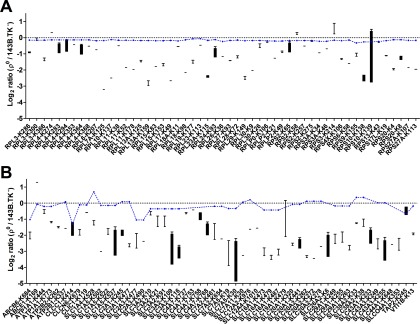
In clear contrast to the more equal distribution of up- and down-regulated phosphorylation sites (Fig. 6A), a severe de-ubiquitination was observed in ρ0 cells (Fig. 6B). In total, 319 ubiquitinated sites were down- and just 33 were up-regulated in the ρ0 state. Again, enrichment was computed using DAVID as well as CPDB, the resulting list of de-ubiquitinated proteins of the GO category “molecular function” displayed the following altered pathways, among others: amino acid transporter activities, structural constituent of the cytoskeleton and ribosomes (supplemental Table S4). For instance, all 80S ribosomal proteins were on average threefold de-ubiquitinated in ρ0 cells (Fig. 7A). Most striking, SLC transporter proteins, most of them amino acid transporters, were on average fivefold de-ubiquitinated (Fig. 7B).
Fig. 7.
De-ubiquitinated target proteins in ρ0 cells. Proteins and their specific de-ubiquitination sites are shown for A, cytosolic ribosomal proteins and B, SLC transporter proteins and other transporter or channel proteins. The log2 ratios of all replicates (n = 1 - 4) are shown (reversed values for label switches) in a box plot. For comparison, the mean of according log2 protein ratios is indicated by connected blue dots.
OTU domain-containing protein 7B (OTUD7B) was one of the highest de-ubiquitinated proteins (18-fold) in ρ0 cells. OTUD7B has a de-ubiquitinating activity toward Lys11, Lys48 or Lys63-linked polyubiquitin and mediates de-ubiquitination of epidermal growth factor receptor (EGFR), which can lead to recycling of EGFR to the plasma membrane (40). EGFR itself was eightfold de-ubiquitinated in ρ0 cells and is known to activate several signaling cascades (41). Adenylate cyclase type 10 (ADCY10), catalyzing the formation of the signaling molecule cAMP was more than 100-fold de-ubiquitinated in ρ0 cells, showing the importance of retrograde signaing again (42).
Furthermore, another protein involved in EGFR signaling, coiled-coil domain-containing protein 50 (CCDC50), was on average sixfold de-ubiquitinated in ρ0 cells. Several proteins of the cytoskeleton, such as keratin 18 (KRT18) with seven distinct ubiquitination sites were on average 11-fold; integrin beta-1 (ITGB1), a receptor for structural elements was on average fourfold, and synaptopodin (SYNPO), an actin-associated protein that may play a role in modulating actin-based shape and motility was 18-fold de-ubiquitinated in ρ0 cells. CD44 antigen, mediating cell–cell and cell–matrix interactions was ninefold de-ubiquitinated in ρ0 cells.
Thus, de-ubiquitination plays a major role in ρ0 cells by altering 80S ribosomal subunits, SLC transporters, as well as proteins involved in the cytoskeleton.
An increase of ubiquitination was for example observed in four different ATP synthase subunit beta (ATP5B) sites (average 15-fold). This catalytic subunit of the ATPase (F1 part) is essential for ATP consumption in ρ0 cells.
Many proteins with increased ubiquitination patterns were involved in cell cycle or DNA replication, such as cancer susceptibility candidate 5 (CASC5), essential for spindle-assembly checkpoint signaling and for correct chromosome alignment; eukaryotic translation elongation factor 1 alpha 1 (EEF1A1); anaphase-promoting complex subunit 1 (ANAPC1), a cell cycle-regulated E3 ubiquitin ligase that controls progression through mitosis and the G1 phase of the cell cycle; DNA polymerase subunit gamma-1 (POLG), involved in the replication of mitochondrial DNA; DNA-directed RNA polymerase II subunit RPB1 and 2 (POLR2A, POLR2B), RNA polymerases catalyzes the transcription of DNA into RNA. The increase of ubiquitination did not alter the protein abundancies, but might have an effect on their activities.
DISCUSSION
The impact of a nonfunctional respiratory electron chain was investigated by comparing the osteosarcoma cell line 143B.TK− to thereof derived mtDNA depleted cells, ρ0 cells. Molecular consequences of this depletion were studied by an integrated metabolic and proteomic approach. Nuclear DNA mutations, because of EtBr treatment, cannot be entirely excluded for ρ0 cells, but so far, no indications were reported in the literature. Nevertheless, nuclear DNA mutations may be responsible for some proteomic changes in nonmitochondrial proteins.
A severely, but unevenly down-regulation of the mitochondrial energy metabolism (pyruvate metabolism, TCA cycle and respiratory electron chain) was observed in ρ0 cells, accompanied by a down-regulation of mitochondrial ribosomal proteins. Only the anaerobic part of the pyruvate metabolism was up-regulated in ρ0 cells, LDHA as well as its product lactic acid, as ρ0 cells can only rely on anaerobic glycolysis and lactic acid formation.
Interpretation of dysregulated proteins and metabolites in the TCA cycle is a complex task, as many other metabolites such as fatty acids, cholesterol, and amino acids are entering or exiting the TCA cycle on different places. Moreover, the regulation of the TCA cycle is largely determined by allosteric mechanisms and substrate availability, for example, NADH inhibits the pyruvate dehydrogenase, isocitrate dehydrogenase, α-ketoglutarate dehydrogenase, and citrate synthase. Two distinct break points of the TCA cycle in ρ0 cells were identified: First, citric acid and cis-acontic acid levels were six and 2.5-fold decreased in ρ0 cell. Second, the electron carrier ubiquinol (QH2) of the respiratory chain, might accumulate, because CIII is failing to oxidize QH2 to ubiquinone (Q). Therefore, succinic acid cannot be oxidized to fumaric acid because of a lack of Q. The metabolites before these two break points, oxalacetic acid and succinic acid, were found to be two and fivefold increased, most likely caused by a congestion introduced by the break points. Even within enzyme complexes of the TCA cycle, we discovered an unequal regulation. For example, succinyl-CoA ligase [ADP/GDP-forming] subunit alpha (SUCLG1) and [ADP-forming] subunit beta (SUCLA2) were down-regulated, whereas the [GDP-forming] subunit beta (SUCLG2) was up-regulated in ρ0 cells.
Hence, we investigated enzymatic activities to elucidate functionality. FH and MDH2 enzyme activities were significantly increased in ρ0 cells, CS and IDH2 were unchanged, matching to according protein abundances. It is not clear why increased levels of oxalacetic acid and a normal CS activity result in reduced citric acid levels. The Vmax of CS was measured in vitro, but steric inhibition could considerably decrease the turnover rate of CS in vivo.
Beside the general down-regulation of proteins in the respiratory electron chain in ρ0 cells, an up-regulation of the heme containing proteins cytochrome c (CYCS) and cytochrome c1 (CYC1) was observed. CYCS is an electron carrier in the mitochondrial intermembrane space and plays a major role in triggering apoptosis (43) and was actually one of the highest down-regulated proteins in our previous study, were an artificial complex I deficiency was induced by rotenone (19). These cytochromes might therefore be differently regulated. Unchanged ratios of ATP synthase subunits between ρ0 and parental cells can be explained by the maintenance of the ATP hydrolysis function. Together with the adenine nucleotide translocator (ANT) that exchanges ATP4− against ADP3− as an electrogenic transport, this pathway contributes to mitochondrial membrane potential in ρ0 cells (44). As complex I subunits are assembled stepwise to finally form a functional complex (45), these subcomplex-intermediates have differences in their regulation, stability and half-life, which can result in this observed uneven down-regulation.
A significant up-regulation of cell signaling pathways was observed, such as G12/13 alpha signaling, Rho GTPases, G-protein-coupled receptor (GPCR) signaling, and Gi α signaling. Rho GTPases are best known for their ability to induce dynamic rearrangements of the plasma membrane-associated actin cytoskeleton (46) and have been implicated in many important cell biological processes, including cell growth control, actomyosin contractility, and microtubule dynamics, cytokinesis, cell motility, cell–cell and cell–extracellular matrix adhesion, cell transformation and invasion, and development (46–54). The cAMP-dependent pathway is a G protein-coupled receptor-triggered signaling cascade used in cell communication, proliferation and differentiation (55), and breakdown of glycogen and fat. This conspicuous increase of signaling pathways in ρ0 cells could be explained by the mitochondrial retrograde response, a pathway of communication from mitochondria to the nucleus that influences many cellular and organismal activities under both, normal and pathophysiological conditions (56). These retrograde responses are for the most part adaptive in that they represent cellular adjustments to altered mitochondrial states. The observed alterations of signaling pathways, that are regulators of the cellular cytoskeleton, fit perfectly to described morphological adjustments in ρ0 cells (35, 37). It has already been reported half a century ago, that mitochondrial morphology is dependent on the respiration status (57). Furthermore, immunolabeling studies revealed that ρ0 cells have severe cytoskeletal alterations, such as a collapsed vimentin network, a dense accumulation of mitochondria around the nucleus and only a few mitochondria distributed throughout the cytoplasm (58). In ρ0 cells, the reticulum appears disrupted, that yields in a distribution of small individual organelles instead of reticular networks with tubular cristae (59–61). The total amount of mitochondrial volume does not appear altered between ρ0 and parental cells, only its morphology (59). Additionally, ρ0 mitochondria were shown to be less mobile (60).
To further elucidate effects of the ρ0 state, phosphorylation and ubiquitination events were investigated and quantified. Post-translational modifications provide a powerful mechanism to rapidly and temporarily alter protein functions and locations in the cell, and they are capable of providing information to regulate proteins by creating docking sites for PTM-recognition domains (62). GO analyses of the phosphoproteome data set revealed GTPase-related signaling pathways and cytoskeleton organization to be down-regulated. Additionally, many regulated key phosphorylation sites of Rho GTPases and proteins involved in the cytoskeleton were identified in this study.
In contrast, the ubiquitylome revealed a clear de-ubiquitination in ρ0 cells (Fig. 6 and Fig. 7). The most interesting and largest groups of de-ubiquitinated proteins were cytosolic ribosomal and SLC transporter proteins. The total amount of these proteins was unchanged in ρ0 versus parental cells (Fig. 7A, 7B), only the modification status changed. A de-ubiquitination of SLC transporters could be indicative for an increased transporter activity or a relocalization (63), but the ubiquitin linkage type, which determines the specific function, cannot be investigated by our experimental setup.
Obvious is the accumulation of amino acid levels in our ρ0 samples (Fig. 2), most likely induced by an increased SLC transporter activity as a result of the observed de-ubiquitination (most of them were amino acid transporters localized in the plasma membrane) and as a response to the interrupted respiratory electron chain. The amino acid metabolism is tightly linked to the TCA cycle and amino acids can serve as alternative substrates for generating energy.
Ubiquitination of ribosomes is a hot topic, but not well understood yet. It is discussed, dependent on the exact position of the modification that the ribosomal function, altered via ubiquitin specific binding proteins and ubiquitination dependent degradation of mature ribosomes by autophagy, is a regulated mechanism (64).
A few connections between the regulation of transporters and ubiquitination have been shown so far. Variations in the E3 ubiquitin-protein ligase NEDD4-like (NEDD4L) gene for example were associated with sodium lithium counter transport activity, based on a combined gene expression profiling and linkage analysis (65). Kelch-like protein 3 (KLHL3), a E3 ubiquitin ligase complex that acts as a regulator of ion transport in the distal nephron has been shown to increase the paracellular chloride permeability in a KLHL3 knockdown experiment, indicating that the paracellular pathway is physiologically regulated through the ubiquitination pathway (66). There have been increasing indications that ubiquitylation of small GTPases occurs in a regulated fashion, primarily upon activation, and is an important means to control signaling output (67). Beside ribosomes and SLC transporters, proteins involved in regulating the actin cytoskeleton and the proteasome were found to be de-ubiquitinated. The actin cytoskeleton is regulated through signaling by Rho-like GTPases, such as RhoA, which stimulates myosin-based contractility, and CDC42 and Rac1, which promote actin polymerization and protrusion (47–52). It has been shown that Rac1 ubiquitylation regulates the dynamics of Rac1 at the periphery of the cell (68). Furthermore, it was shown that RhoA is a direct target of Cul3-based ubiquitin ligase complexes, and cells lacking Cul3 show impaired ubiquitination and degradation of RhoA and exhibit remarkable abnormal actin stress fibers. De-ubiquitination of RhoA finally inhibits migration potential of cultured mouse and human cells and leads to defective RhoA-mediated convergent extension movements (69). The proteome and PTM analyses are supporting our hypothesis that ρ0 cells trigger the retrograde response. Thus, phosphorylation events and de-ubiquitination in ρ0 cells are regulating and tuning the retrograde response, cytoskeleton rearrangements, and SLC transport activities, which leads to the known ρ0 phenotype, featuring an altered mitochondrial morphology such as a collapsed vimentin network and a dense accumulation of mitochondria around the nucleus.
In summary, a significant, but uneven down-regulation of the entire mitochondrial energy pathways such as pyruvate metabolism, TCA cycle, and respiratory electron chain was observed in ρ0 cells. Signaling pathways were significantly up-regulated on the proteome level in ρ0 cells, further supported by our phospho-proteomics data showing a striking regulation of proteins involved in GTPase signaling and the cytoskeleton organization. The most surprising finding was the remarkable de-ubiquitination in ρ0 cells, especially of proteins involved in the cytoskeleton, cytosolic ribosomes, and SLC amino acid transporters. Moreover, the observed de-ubiquitylation of Rho GTPases and EGFR signaling is an important aspect of regulating signaling cascades. This indicates a tight regulation of ubiquitination in response to mitochondrial energy alterations. Increased amino acid levels in ρ0 cells could be the effect of de-ubiquitinated and thereby activated SLC transporter proteins to adapt to new energy sources. Such an integrated omics approach including metabolites, proteins, and their PTMs can elucidate important connections and interactions within a cell, leading to the discovery of new molecular mechanisms and features of RCD.
Supplementary Material
Acknowledgments
We thank Ralf Herwig for his help with statistical analyses and Monica Shevack for her contribution in figure preparation.
Footnotes
Author contributions: D.M. designed research; I.A. performed research; I.A. and D.M. analyzed data; I.A., I.W., and D.M. wrote the paper; C.H. performed global test, david, cpdb.
* This work is part of the Ph.D. thesis of I.A. Our work is supported by the Max Planck Society and by the DFG to I.W. (Sonderforschungsbereich 815, project Z1).
 This article contains supplemental materials.
This article contains supplemental materials.
1 The abbreviations used are:
- RCD
- Respiratory chain disease
- 143B.TK−
- 143B Thymidine kinase deficient
- 2-DE
- 2-D electrophoresis
- BC assay
- Bicinchoninic acid assay
- BH
- Benjamini-Hochberg
- DMEM
- Dulbecco's Modified Eagle Medium
- dNTP
- Deoxynucleotide
- FA
- Formic acid
- FBS
- Fetal bovine serum
- FDR
- False discovery rate
- GO
- Gene ontology
- GSEA
- Gene set enrichment analysis
- HILIC
- Hydrophilic interaction chromatography
- MeOH
- Methanol
- MRM
- Multiple reaction monitoring
- mtDNA
- Mitochondrial DNA
- PEP
- Posterior error probability
- PPI
- Protein–protein interaction
- PTM
- Post-translational modification
- RP
- Reversed phase
- SAX
- Strong anion exchange chromatography
- SCX
- Strong cation exchange chromatography
- SILAC
- Stable isotope labeling by amino acids in cell culture
- TBE
- TRIS-Borat-EDTA
- TiO2
- Titanium dioxide
- ρ0
- Rho 0.
REFERENCES
- 1. Ernster L., and Schatz G. (1981) Mitochondria: a historical review. J. Cell Biol. 91, 227s–255s [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Watt I. N., Montgomery M. G., Runswick M. J., Leslie A. G. W., and Walker J. E. (2010) Bioenergetic cost of making an adenosine triphosphate molecule in animal mitochondria. Proc. Natl. Acad. Sci. U.S.A. 107, 16823–16827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Prokisch H., Andreoli C., Ahting U., Heiss K., Ruepp A., Scharfe C., and Meitinger T. (2006) MitoP2: the mitochondrial proteome database – now including mouse data. Nucleic Acids Res. 34, D705–D711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Anderson S., Bankier A. T., Barrell B. G., de Bruijn M. H., Coulson A. R., Drouin J., Eperon I. C., Nierlich D. P., Roe B. A., Sanger F., Schreier P. H., Smith A. J., Staden R., and Young I. G. (1981) Sequence and organization of the human mitochondrial genome. Nature 290, 457–465 [DOI] [PubMed] [Google Scholar]
- 5. Ruiz-Romero C., and Blanco F. J. (2009) Mitochondrial proteomics and its application in biomedical research. Mol. Biosyst. 5, 1130–1142 [DOI] [PubMed] [Google Scholar]
- 6. DiMauro S., Hirano M., and Schon E. A. (2006) Approaches to the treatment of mitochondrial diseases. Muscle Nerve 34, 265–283 [DOI] [PubMed] [Google Scholar]
- 7. King M. P., and Attardi G. (1996) Isolation of human cell lines lacking mitochondrial DNA. Methods Enzymol. 264, 304–313 [DOI] [PubMed] [Google Scholar]
- 8. Inoue K., Takai D., Hosaka H., Ito S., Shitara H., Isobe K., LePecq J. B., Segal-Bendirdjian E., and Hayashi J. (1997) Isolation and characterization of mitochondrial DNA-less lines from various mammalian cell lines by application of an anticancer drug, ditercalinium. Biochem. Biophys. Res. Commun. 239, 257–260 [DOI] [PubMed] [Google Scholar]
- 9. Kukat A., Kukat C., Brocher J., Schäfer I., Krohne G., Trounce I. A., Villani G., and Seibel P. (2008) Generation of rho0 cells utilizing a mitochondrially targeted restriction endonuclease and comparative analyses. Nucleic Acids Res. 36, e44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Schubert S., Heller S., Löffler B., Schäfer I., Seibel M., Villani G., and Seibel P. (2015) Generation of rho zero cells: visualization and quantification of the mtDNA depletion process. Int. J. Mol. Sci. 16, 9850–9865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. King M. P., and Attardi G. (1989) Human cells lacking mtDNA: repopulation with exogenous mitochondria by complementation. Science 246, 500–503 [DOI] [PubMed] [Google Scholar]
- 12. Chandel N. S., and Schumacker P. T. (1999) Cells depleted of mitochondrial DNA (rho0) yield insight into physiological mechanisms. FEBS Lett. 454, 173–176 [DOI] [PubMed] [Google Scholar]
- 13. Wittig I., Meyer B., Heide H., Steger M., Bleier L., Wumaier Z., Karas M., and Schägger H. (2010) Assembly and oligomerization of human ATP synthase lacking mitochondrial subunits a and A6L. Biochim. Biophys. Acta 1797, 1004–1011 [DOI] [PubMed] [Google Scholar]
- 14. Chae S., Ahn B. Y., Byun K., Cho Y. M., Yu M.-H., Lee B., Hwang D., and Park K. S. (2013) A systems approach for decoding mitochondrial retrograde signaling pathways. Sci. Signal. 6, rs4. [DOI] [PubMed] [Google Scholar]
- 15. DiMauro S. (2004) The many faces of mitochondrial diseases. Mitochondrion 4, 799–807 [DOI] [PubMed] [Google Scholar]
- 16. Nunnari J., and Suomalainen A. (2012) Mitochondria: in sickness and in health. Cell 148, 1145–1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mayr J. A., Haack T. B., Freisinger P., Karall D., Makowski C., Koch J., Feichtinger R. G., Zimmermann F. A., Rolinski B., Ahting U., Meitinger T., Prokisch H., and Sperl W. (2015) Spectrum of combined respiratory chain defects. J. Inherit. Metab. Dis., 38, 629–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chevallet M., Lescuyer P., Diemer H., van Dorsselaer A., Leize-Wagner E., and Rabilloud T. (2006) Alterations of the mitochondrial proteome caused by the absence of mitochondrial DNA: a proteomic view. Electrophoresis 27, 1574–1583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gielisch I., and Meierhofer D. (2015) Metabolome and proteome profiling of complex I deficiency induced by rotenone. J. Proteome Res. 14, 224–235 [DOI] [PubMed] [Google Scholar]
- 20. Mayr J. A., Meierhofer D., Zimmermann F., Feichtinger R., Kögler C., Ratschek M., Schmeller N., Sperl W., and Kofler B. (2008) Loss of complex I due to mitochondrial DNA mutations in renal oncocytoma. Clin. Cancer Res. 14, 2270–2275 [DOI] [PubMed] [Google Scholar]
- 21. Meierhofer D., Mayr J. A., Foetschl U., Berger A., Fink K., Schmeller N., Hacker G. W., Hauser-Kronberger C., Kofler B., and Sperl W. (2004) Decrease of mitochondrial DNA content and energy metabolism in renal cell carcinoma. Carcinogenesis 25, 1005–1010 [DOI] [PubMed] [Google Scholar]
- 22. Vizcaíno J. A., Côté R. G., Csordas A., Dianes J. A., Fabregat A., Foster J. M., Griss J., Alpi E., Birim M., Contell J., O'Kelly G., Schoenegger A., Ovelleiro D., Pérez-Riverol Y., Reisinger F., Ríos D., Wang R., and Hermjakob H. (2013) The PRoteomics IDEntifications (PRIDE) database and associated tools: status in 2013. Nucleic Acids Res. 41, D1063–1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Meierhofer D., Weidner C., Hartmann L., Mayr J. A., Han C.-T., Schroeder F. C., and Sauer S. (2013) Protein sets define disease states and predict in vivo effects of drug treatment. Mol. Cell. Proteomics 12, 1965–1979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wiœniewski J. R., Zougman A., and Mann M. (2009) Combination of FASP and StageTip-based fractionation allows in-depth analysis of the hippocampal membrane proteome. J. Proteome Res. 8, 5674–5678 [DOI] [PubMed] [Google Scholar]
- 25. Wagner S. A., Beli P., Weinert B. T., Schölz C., Kelstrup C. D., Young C., Nielsen M. L., Olsen J. V., Brakebusch C., and Choudhary C. (2012) Proteomic analyses reveal divergent ubiquitylation site patterns in murine tissues. Mol. Cell. Proteomics 11, 1578–1585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cox J., and Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 27. Cox J., Neuhauser N., Michalski A., Scheltema R. a, Olsen J. V., and Mann M. (2011) Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 10, 1794–1805 [DOI] [PubMed] [Google Scholar]
- 28. Iesmantavicius V., Weinert B. T., and Choudhary C. (2014) Convergence of ubiquitylation and phosphorylation signaling in rapamycin-treated yeast cells. Mol. Cell. Proteomics 13, 1979–1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Subramanian A., Tamayo P., Mootha V. K., Mukherjee S., Ebert B. L., Gillette M. A., Paulovich A., Pomeroy S. L., Golub T. R., Lander E. S., and Mesirov J. P. (2005) Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Goeman J. J., van de Geer S. A., de Kort F., and van Houwelingen H. C. (2004) A global test for groups of genes: testing association with a clinical outcome. Bioinformatics 20, 93–99 [DOI] [PubMed] [Google Scholar]
- 31. Franceschini A., Szklarczyk D., Frankild S., Kuhn M., Simonovic M., Roth A., Lin J., Minguez P., Bork P., von Mering C., and Jensen L. J. (2013) STRING v9.1: protein–protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 41, D808–D815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Huang D. W., Sherman B. T., and Lempicki R. A. (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 [DOI] [PubMed] [Google Scholar]
- 33. Huang D. W., Sherman B. T., and Lempicki R. A. (2009) Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kamburov A., Stelzl U., Lehrach H., and Herwig R. (2013) The ConsensusPathDB interaction database: 2013 update. Nucleic Acids Res. 41, D793–D800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hansson A., Hance N., Dufour E., Rantanen A., Hultenby K., Clayton D. A., Wibom R., and Larsson N.-G. (2004) A switch in metabolism precedes increased mitochondrial biogenesis in respiratory chain-deficient mouse hearts. Proc. Natl. Acad. Sci. U.S.A. 101, 3136–3141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Calvo S. E., Clauser K. R., and Mootha V. K. (2015) MitoCarta2.0: an updated inventory of mammalian mitochondrial proteins. Nucleic Acids Res., gkv1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Glickman M. H., and Ciechanover A. (2002) The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol. Rev. 82, 373–428 [DOI] [PubMed] [Google Scholar]
- 38. Mukhopadhyay D., and Riezman H. (2007) Proteasome-independent functions of ubiquitin in endocytosis and signaling. Science 315, 201–205 [DOI] [PubMed] [Google Scholar]
- 39. Schnell J. D., and Hicke L. (2003) Nontraditional functions of ubiquitin and ubiquitin-binding proteins. J. Biol. Chem. 278, 35857–35860 [DOI] [PubMed] [Google Scholar]
- 40. Mizuno E., Iura T., Mukai A., Yoshimori T., Kitamura N., and Komada M. (2005) Regulation of epidermal growth factor receptor down-regulation by UBPY-mediated deubiquitination at endosomes. Mol. Biol. Cell 16, 5163–5174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Avraham R., and Yarden Y. (2011) Feedback regulation of EGFR signaling: decision making by early and delayed loops. Nat. Rev. Mol. Cell Biol. 12, 104–117 [DOI] [PubMed] [Google Scholar]
- 42. Schmid A., Sutto Z., Nlend M.-C., Horvath G., Schmid N., Buck J., Levin L. R., Conner G. E., Fregien N., and Salathe M. (2007) Soluble adenylyl cyclase is localized to cilia and contributes to ciliary beat frequency regulation via production of cAMP. J. Gen. Physiol. 130, 99–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Liu X., Kim C. N., Yang J., Jemmerson R., and Wang X. (1996) Induction of apoptotic program in cell-free extracts: requirement for dATP and cytochrome c. Cell 86, 147–157 [DOI] [PubMed] [Google Scholar]
- 44. Buchet K., and Godinot C. (1998) Functional F1-ATPase essential in maintaining growth and membrane potential of human mitochondrial DNA-depleted rho degrees cells. J. Biol. Chem. 273, 22983–22989 [DOI] [PubMed] [Google Scholar]
- 45. Vogel R. O., Smeitink J. A. M., and Nijtmans L. G. J. (2007) Human mitochondrial complex I assembly: a dynamic and versatile process. Biochim. Biophys. Acta 1767, 1215–1227 [DOI] [PubMed] [Google Scholar]
- 46. Govek E.-E., Newey S. E., and Van Aelst L. (2005) The role of the Rho GTPases in neuronal development. Genes Dev. 19, 1–49 [DOI] [PubMed] [Google Scholar]
- 47. Bosco E. E., Mulloy J. C., and Zheng Y. (2009) Rac1 GTPase: a “Rac” of all trades. Cell. Mol. Life Sci. 66, 370–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Didsbury J., Weber R., Bokoch G., Evans T., and Snyderman R. (1989) rac, a novel ras-related family of proteins that are botulinum toxin substrates. J. Biol. Chem. 264, 16378–16382 [PubMed] [Google Scholar]
- 49. Hall A. (1990) The cellular functions of small GTP-binding proteins. Science 249, 635–640 [DOI] [PubMed] [Google Scholar]
- 50. Kozma R., Sarner S., Ahmed S., and Lim L. (1997) Rho family GTPases and neuronal growth cone remodeling: relationship between increased complexity induced by Cdc42Hs, Rac1, and acetylcholine and collapse induced by RhoA and lysophosphatidic acid. Mol. Cell. Biol. 17, 1201–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ridley A. J., Paterson H. F., Johnston C. L., Diekmann D., and Hall A. (1992) The small GTP-binding protein rac regulates growth factor-induced membrane ruffling. Cell 70, 401–410 [DOI] [PubMed] [Google Scholar]
- 52. Ridley A. J., and Hall A. (1992) The small GTP-binding protein rho regulates the assembly of focal adhesions and actin stress fibers in response to growth factors. Cell 70, 389–399 [DOI] [PubMed] [Google Scholar]
- 53. Alblas J., Ulfman L., Hordijk P., and Koenderman L. (2001) Activation of Rhoa and ROCK are essential for detachment of migrating leukocytes. Mol. Biol. Cell 12, 2137–2145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Worthylake R. A., and Burridge K. (2001) Leukocyte transendothelial migration: orchestrating the underlying molecular machinery. Curr. Opin. Cell Biol. 13, 569–577 [DOI] [PubMed] [Google Scholar]
- 55. Hanoune J., and Defer N. (2001) Regulation and role of adenylyl cyclase isoforms. Annu. Rev. Pharmacol. Toxicol. 41, 145–174 [DOI] [PubMed] [Google Scholar]
- 56. Butow R. A., and Avadhani N. G. (2004) Mitochondrial signaling: the retrograde response. Mol. Cell 14, 1–15 [DOI] [PubMed] [Google Scholar]
- 57. Hackenbrock C. R. (1968) Ultrastructural bases for metabolically linked mechanical activity in mitochondria. II. Electron transport-linked ultrastructural transformations in mitochondria. J. Cell Biol. 37, 345–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Annunen-Rasila J., Ohlmeier S., Tuokko H., Veijola J., and Majamaa K. (2007) Proteome and cytoskeleton responses in osteosarcoma cells with reduced OXPHOS activity. Proteomics 7, 2189–2200 [DOI] [PubMed] [Google Scholar]
- 59. Gilkerson R. W., Margineantu D. H., Capaldi R. A., and Selker J. M. (2000) Mitochondrial DNA depletion causes morphological changes in the mitochondrial reticulum of cultured human cells. FEBS Lett. 474, 1–4 [DOI] [PubMed] [Google Scholar]
- 60. Jazayeri M. (2003) Inducible expression of a dominant negative DNA polymerase-gamma depletes mitochondrial DNA and produces a rho 0 phenotype. J. Biol. Chem. 278, 9823–9830 [DOI] [PubMed] [Google Scholar]
- 61. Garcia J. J. (2000) Structure, functioning, and assembly of the ATP synthase in cells from patients with the T8993G mitochondrial DNA mutation. Comparison with the enzyme in rho0 cells completely lacking mtDNA. J. Biol. Chem. 275, 11075–11081 [DOI] [PubMed] [Google Scholar]
- 62. Deribe Y. L., Pawson T., and Dikic I. (2010) Post-translational modifications in signal integration. Nat. Struct. Mol. Biol. 17, 666–672 [DOI] [PubMed] [Google Scholar]
- 63. Brenkman A. B., de Keizer P. L. J., van den Broek N. J. F., Jochemsen A. G., and Burgering B. M. T. (2008) Mdm2 induces mono-ubiquitination of FOXO4. PLoS One 3, e2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Kraft C., Deplazes A., Sohrmann M., and Peter M. (2008) Mature ribosomes are selectively degraded upon starvation by an autophagy pathway requiring the Ubp3p/Bre5p ubiquitin protease. Nat. Cell Biol. 10, 602–610 [DOI] [PubMed] [Google Scholar]
- 65. Zheng X., Morrison A. C., Feingold E., Turner S. T., and Ferrell R. E. (2011) Association between NEDD4L gene and sodium lithium countertransport. Am. J. Hypertens. 24, 145–148 [DOI] [PubMed] [Google Scholar]
- 66. Gong Y., Wang J., Yang J., Gonzales E., Perez R., and Hou J. (2015) KLHL3 regulates paracellular chloride transport in the kidney by ubiquitination of claudin-8. Proc. Natl. Acad. Sci. U.S.A. 112, 4340–4345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Nethe M., and Hordijk P. L. (2010) The role of ubiquitylation and degradation in RhoGTPase signaling. J. Cell Sci. 123, 4011–4018 [DOI] [PubMed] [Google Scholar]
- 68. Nethe M., Anthony E. C., Fernandez-Borja M., Dee R., Geerts D., Hensbergen P. J., Deelder A. M., Schmidt G., and Hordijk P. L. (2010) Focal-adhesion targeting links caveolin-1 to a Rac1-degradation pathway. J. Cell Sci. 123, 1948–1958 [DOI] [PubMed] [Google Scholar]
- 69. Chen Y., Yang Z., Meng M., Zhao Y., Dong N., Yan H., Liu L., Ding M., Peng H. B., and Shao F. (2009) Cullin mediates degradation of RhoA through evolutionarily conserved BTB adaptors to control actin cytoskeleton structure and cell movement. Mol. Cell 35, 841–855 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.