Abstract
Exosomes, derived from multivesicular bodies (MVBs), contain proteins and genetic materials from their cell of origin and are secreted from various cells types, including kidney epithelial cells. In general, it is thought that protein cargo is ubiquitylated but that ubiquitin is cleaved by specific deubiquitylases during the process of cargo incorporation into MVBs. Here, we provide direct evidence that, in vivo, deubiquitylation is not essential. Ubiquitin was detected within human MVBs and urinary exosomes by electron microscopy. Of the >6000 proteins identified in human urinary exosomes was mass spectrometry, 15% were ubiquitylated with various topologies (Lys63>Lys48> Lys11>Lys6>Lys29>Lys33>Lys27). A significant preference for basic amino acids upstream of ubiquitylation sites suggests specific ubiquitylation motifs. The current studies demonstrate that, in vivo, deubiquitylation of proteins is not necessary for their incorporation into MVBs and highlight that urinary exosomes are an enriched source for studying ubiquitin modifications in physiological or disease states.
Exosomes are extracellular nanovesicles (20–100 nm) that are secreted from various cells types in the body (1). In the urinary system, exosomes are secreted from epithelial cells from all segments of the kidney tubule and urinary tract (2). Urinary exosomes contain proteins and genetic materials e.g. mRNA and miRNA, from their cell of origin that may represent physiology or pathophysiology of a variety of renal and systemic diseases (3).
In general, exosomes originate in late endosomal compartments called multivesicular bodies (MVBs). An essential posttranslational modification that marks membrane proteins for incorporation into MVBs is ubiquitylation (4). Ubiquitin is a small 76 amino acid protein that requires reversible enzymatic activity of E1, an activation enzyme; E2, a conjugation enzyme; and E3, a ligation enzyme, for binding to a specific substrate. Ubiquitin acts as a signal for mediating a range of cellular functions, including protein trafficking, DNA repair, endocytosis, proteasomal and lysosomal degradation, and transcriptional regulation. The role of ubiquitin in various cellular functions can be directly related to the type of ubiquitin modifications on a specific substrate, such as monoubiquitylation, multimonoubiquitylation, and polyubiquitylation (5). Various topologies of polyubiquitin chains also play different roles in biology. For example, Lys48-linked chains can target proteins for proteasomal degradation, whereas Lys63-linked chains can target proteins for lysosomal degradation, aid in DNA repair, or play a role in transcriptional regulation (5).
During the process of protein trafficking, ubiquitylated membrane protein cargo can be recognized by the endosomal-sorting complex required for transport (ESCRT) apparatus on the outer surface of MVBs. Through a cascade of protein interactions, internal luminal vesicles (ILVs) are formed inside the MVB that can be released into the extracellular environment as exosomes upon the fusion of the MVB outer membrane with the plasma membrane (6). ESCRTs (0-II) contain ubiquitin-binding domains, and it is widely accepted that these domains on the ESCRT complex are responsible for recognizing mono- and polyubiquitylated (especially Lys63-linked chain) proteins and sequestering them to MVBs (7). What is unclear is what happens to the ubiquitin molecule during cargo incorporation to MVBs. Several studies suggest that deubiquitylases are recruited to the ESCRT complex and cleave ubiquitin from cargo proteins before they are incorporated into the ILVs (8–11). However, we previously provided biochemical evidence of ubiquitylated proteins in urinary exosomes (2).
Here, we conclusively identify ubiquitylated proteins in renal epithelial ILVs and in urinary exosomes. Using protein mass spectrometry coupled with extensive bioinformatics, we demonstrate that in human urinary exosomes numerous proteins are ubiquitylated, with quantification of polyubiquitin chain topologies highlighting an enrichment of Lys63-linked ubiquitin molecules in exosomes. Our studies demonstrate that, in human epithelial cells, deubiquitylation of protein cargo is not an essential step in ILV and exosome formation. Furthermore, urinary exosomes may provide an enriched source for a noninvasive approach to study ubiquitin modifications in physiological or disease states.
EXPERIMENTAL PROCEDURES
Urine Collection and Exosome Isolation
Urine was collected from healthy volunteers ages 19–45 years following the Danish guidelines for collection of biological materials according to the Act on Research Ethics Review of Health Research Projects, Act number 593 of 14 July 2011, section 14(3). Low-density membrane fractions containing exosomes (“exosome fraction”) were isolated as described (2) and resuspended in an appropriate buffer, depending on the subsequent experiment. For size distribution analysis of the membrane-bound structures using a NanoSight (Malvern, UK), exosome samples were diluted 400 times in particle-free PBS. Five videos of 60 s were recorded at camera level 13, and no aggregates were observed. Videos were analyzed at detection threshold 4 on the NanoSight Software 3.0.
Immunoblotting
For an individual experiment, urine from six healthy volunteers was pooled into three samples. Each pooled sample (G1–3) contained urine from one female and one male to generalize the findings of the study. 10 ml of urine were subject to precipitation by addition of 40 ml of prechilled 25% trichloroacetic acid in 100% acetone followed by overnight incubation at -20°C. Exosomes were isolated from the remaining urine as described (2), with the 17,000 × g pellets also collected. Immunoblots of samples were generated by standard techniques and probed with a ubiquitin (P4D1) mouse monoclonal primary antibody (#3936, Cell Signaling Technology, Danvers, MA) at 1:250 dilution, followed by chemiluminescence detection. The experiment was repeated three independent times in different laboratories, with replicate experiments yielding similar results.
Immunogold Electron Microscopy
Exosomes were isolated from urine collected from six healthy volunteers, combined and resuspended in 200 μl PBS. Samples were mixed (1:1, v:v) with 4% paraformaldehyde in PBS and incubated for 20 min at room temperature. 20 μl of the sample were added to a mesh nickel formvar/carbon-coated grid, and the droplet incubated for 10 min at room temperature and washed in PBS (2 x) for 5 min before processing for immunogold electron microscopy as previously described (12). Primary antibodies were rabbit polyclonal anti-AQP2 (13) or Na-Cl cotransporter (NCC) (12), and mouse monoclonal anti-ubiquitin (P4D1, Cell Signaling Technology or P4G7, Covance). Grids were imaged using an FEI Morgagni electron microscope.
LC-MS/MS sample preparation using in-gel digestion
Exosome fractions were reduced and alkylated as described (14). 50 μg of proteins were separated by SDS-PAGE, stained with Coomassie (Imperial Protein Stain, Pierce) and fractionated into 32 pieces before in-gel digestion and mass spectrometry (MS) sample preparation as described (2).
In-Solution Digestion Followed by Di-Glycine Remnant (K-ε-GG) Immunoaffinity Purification
Exosome fractions were resuspended in 8 m urea/50 mm Tris-HCl/75 mm NaCl. Samples were reduced, alkylated using 55 mm chloroacetamide at room temperature for 30 min in the dark, quenched using 40 mm DTT/50 mm NH4HCO3, and digested with Lys-C (1:100 (w/w) of protease:protein) at room temperature for 4 h, followed by trypsin digestion at 37°C overnight (1:25 (w/w) of protease:protein). After desalting, peptides were immunoaffinity purified using the PTMScan® Ubiquitin Remnant Motif (K-ε-GG) Kit (#5562, Cell Signaling Technology). Reactions were performed using 1 mg of exosome peptides, 4 μl of immunoaffinity purification (IAP) beads and incubation of the mixture at 4°C for 4 h with rotation. After washing, peptides were eluted twice each with 50 μl of 0.15% trifluoroacetic acid, desalted using C18 StageTips, dried, and stored at -20°C until MS analysis.
Nano-Liquid Chromatography, Mass Spectrometry Analysis, and Database Searches
Exosome samples for LC-MS/MS analysis were prepared from urine pooled from 20 biological replicates (10 males and 10 females with various ethnic backgrounds). LC-MS/MS analysis was performed in two technical replicates for all samples. Peptides were separated by nano-liquid chromatography (UltiMate® 3000 RSLCnano system, Thermo Fisher Scientific) coupled to a mass spectrometer (Q Exactive Hybrid Quadrupole-Orbitrap, Thermo Fisher Scientific) through an EASY-Spray nano-electrospray ion source (Thermo Fisher Scientific). The MS methods included a full MS scan at a resolution of 70,000 followed by 10 data-dependent MS2 scans at a resolution of 17,500. For in-gel digested samples, the full MS scan range of 200 to 2000 m/z was selected and precursor ions with the charge states of +1 or greater than +8 were excluded. For immunoaffinity purified samples, due to the higher charge states of the enriched ubiquitylated peptides, the full MS scan range of 300 to 1500 m/z was selected and the additional exclusion of +2 precursor ions was implemented. Normalized collision energy of HCD fragmentation was set at 35%. A dynamic exclusion of 60 s was used. Peaklist-generating software used was Thermo Xcalibur 2.2 (August 12, 2011). Raw LC-MS/MS files were searched by X! Tandem (CYCLONE, 2013.2.01), Andromeda (MaxQuant, version 1.3.0.5), and SEQUEST (Proteome Discoverer, version 1.4) algorithms against human databases (ENSEMBL GRCh37.70: 104,785 sequences and NCBI RefSeq: 35,930 sequences) plus common contaminants concatenated with their reversed sequences. A target–decoy approach was used to limit a false discovery rate of the identified peptides to less than 2% (15). For each MS2 scan, peptide sequences from the three search algorithms that passed the false discovery rate threshold were compared, and ambiguous identification was discarded. Parent and fragment monoisotopic mass errors were set at 10 ppm. Carbamidomethyl at cysteine was used as a fixed modification mass. Variable modifications were oxidation at methionine, di-glycine (+114.042927, Unimod Accession #: 121) at lysine, and LeuArgGlyGly (LRGG, +383.228103, Unimod Accession #: 535) at lysine. A maximum of three missed cleavage sites were allowed.
Quantification of Polyubiquitin Chains Using Ubiquitin-Absolute Protein Quantification (AQUA)
Lys11-, Lys48-, and Lys63-linked polyubiquitin AQUA peptides (Cell Signaling Technology) were used (16). After in-solution digestion, 30 μg of sample were spiked with an AQUA peptide mixture containing 2.5 pmol of each GG-linked ubiquitin-AQUA peptide of interest (Lys11: TLTGK[GG]TITLEVEPSDTIENVK, Lys48: LIFAGK[GG]QLEDGR, and Lys63: TLSDYNIQK[GG]ESTLHLVLR, where L is the heavy isotope labeled amino acid residue) before LC-MS/MS. Supplemental Table 2 shows the inclusion list used to identify the GG-linked endogenous (light) and AQUA (heavy) peptides. Absolute quantification of the GG-linked endogenous peptides was performed using extracted ion chromatograms from the endogenous peptide compared with the AQUA peptide in five biological replicates.
Bioinformatics Analysis
Functional annotation of proteins were performed by DAVID Bioinformatics Resources 6.7 (http://david.abcc.ncifcrf.gov/) (17). Conserved domain analysis was performed by the Batch Web CD-Search Tool (http://www.ncbi.nlm.nih.gov/Structure/bwrpsb/bwrpsb.cgi) (18). CPhos was utilized to assess the conservation of ubiquitylated sites (http://helixweb.nih.gov/CPhos/) (19). Annotations of ubiquitylated sites were extracted from the UniProt Knowledgebase (http://www.uniprot.org/). Novelty of ubiquitylated sites were determined by comparison with ubiquitylation datasets from PhosphoSitePlus (http://www.phosphosite.org/) (20). Motif analysis was performed by motif-x (http://motif-x.med.harvard.edu/) (21) using a background of all identified peptides prealigned using K as the central residue (to correct for any bias introduced in the analysis via the use of trypsin digest) and default parameters (width = 13, occurrences = 20, and significance = 0.000001).
Determination of Ubiquitylation Forms of Proteins in Urinary Exosomes
Only ubiquitylated proteins with at least 10 identified peptides (ubiquitylated and unmodified peptides) were analyzed. A distribution of molecular masses corresponding to the identified locations of ubiquitylated and unmodified peptides of each protein from a 1D SDS-PAGE gel was plotted. A fifth-order polynomial fitting assigned a molecular mass range for each gel fraction. The unmodified mass of a protein was derived from the mode of its nonubiquitylated peptide distribution. Molecular masses of the ubiquitylated forms of each ubiquitylated protein were derived from the corresponding locations on the gel of its ubiquitylated peptides. The mass difference between the ubiquitylated and unmodified form of each ubiquitylated protein was calculated.
Data Deposition
The mass spectrometry proteomics data, including annotated spectra for all modified peptides and proteins identified on the basis of a single peptide, have been deposited to: 1) the ProteomeXchange Consortium via the PRoteomics IDEntifications (PRIDE) partner repository with the dataset identifier PXD002645 (Username: reviewer33476@ebi.ac.uk, Password: eAMpXajt) and 2) the MS-Viewer (http://prospector2.ucsf.edu/prospector/cgi-bin/msform.cgi?form=msviewer) with the following keys: 5r4leji5o5, c5pjqvljul, w6lbkewo30, niolgjxqzv, and ke8clhonnl.
RESULTS
Ubiquitylated Proteins Are Enriched in a Low-Density Membrane Fraction Isolated from Urine
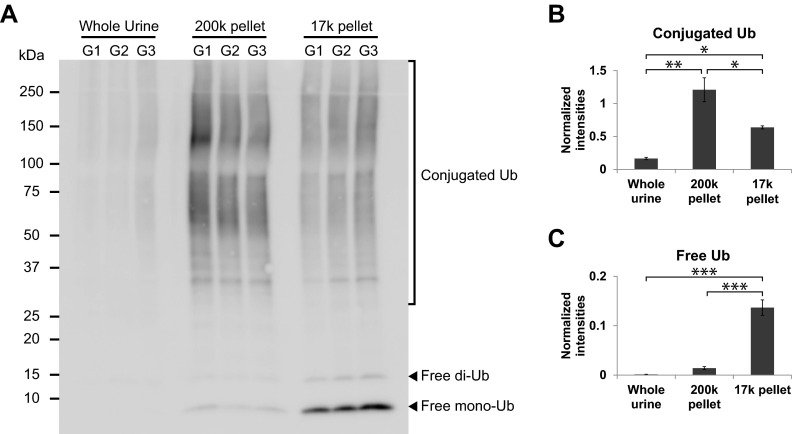
Quantitative assessment of the relative abundances of ubiquitylated proteins in different urine fractions was performed by comparing ubiquitin signal intensities between whole urine precipitate, 17,000 × g pellet (containing larger cellular membranes, larger organelles e.g. nuclei/mitochondria and other forms of fragmented cellular debris), and 200,000 × g pellet (containing low-density membranes e.g. exosomes and exosome-like vesicles) by immunoblotting (Fig. 1A). The signal intensities of conjugated ubiquitin (Ub) molecules were significantly greater in the 200,000 × g pellet samples relative to 17,000 × g pellet samples and were lowest in precipitated whole urine samples (Fig. 1B). However, free Ub signals (mainly mono-Ub) were almost exclusively observed in the 17,000 × g pellet samples (Fig. 1C) in agreement with the notion that this fraction contains nonspecific cellular materials and whole cells (containing soluble free mono-Ub). These data indicate that ubiquitylated proteins (indicated by conjugated Ub signal), but not free Ub molecules, are recruited to and concentrated in low-density membrane fractions of urine.
Fig. 1.
Enrichment of ubiquitylated proteins in a low-density membrane fraction of human urine. (A) Immunoblotting of ubiquitin in whole urine, 200,000 × g (200k) “low-density membrane” pellet, and 17,000 × g (17k) pellet of human urine from three groups of pooled samples (G1–3). Each lane is loaded with an equal quantity of total protein. Conjugated Ub signal (observed at >25 kDa) is predominant in the 200k pellet (B), whereas free mono- and di-Ub signal is predominant in the 17k pellet (C). Values are S.E. and *, **, and *** indicate p values of less than 0.05, 0.01, and 0.001, respectively, by one-way analysis of variance (ANOVA) followed Tukey's multiple comparison tests. Data are representative of three individual experiments (see Methods).
Ubiquitin Is Detected in Both MVBs and in Urinary Exosomes
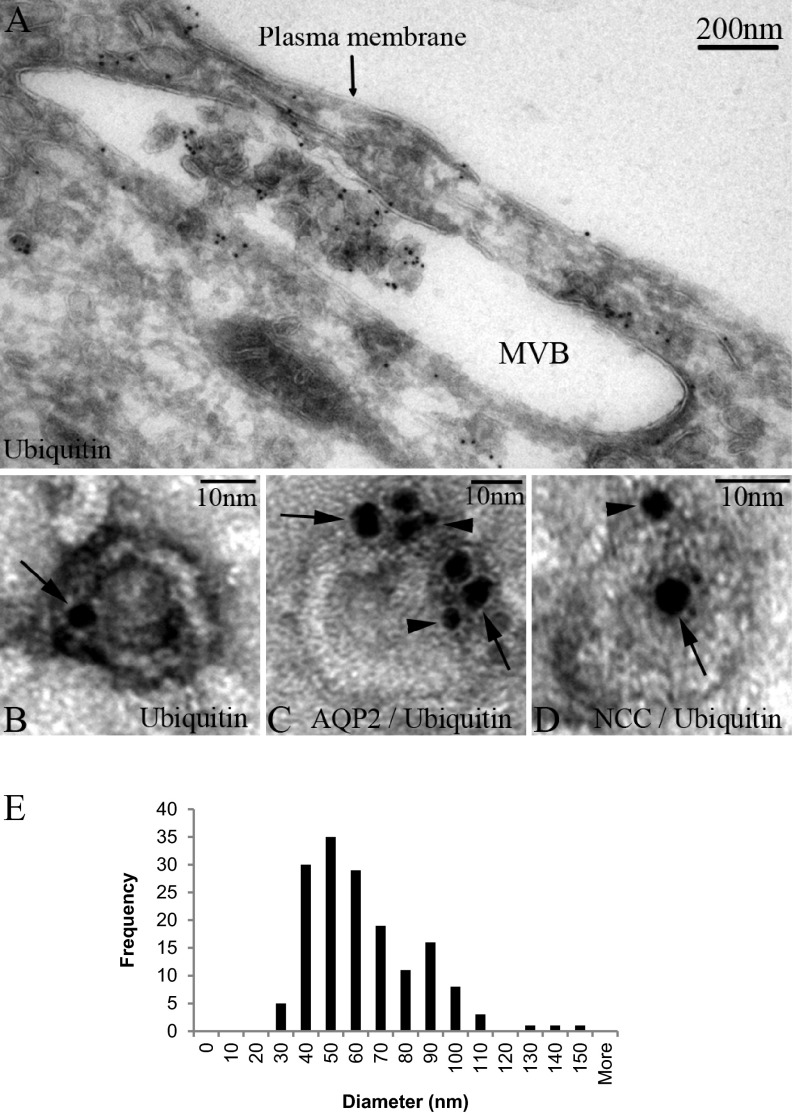
Detection of ubiquitylated proteins in the low-density membrane fractions from urine supports a mechanism where ubiquitylated proteins are recruited to MVBs, where a proportion are sequestered into ILVs without prior deubiquitylation before release into the urinary space as exosomes. To support such a mechanism, immunogold electron microscopy was performed to identify Ub within MVBs within epithelial cells of the human kidney and within human urinary exosomes. MVBs were identified in cells from various segments of the kidney tubule (Fig. 2A and Supplemental Fig. 1), the majority of which contained ILVs immuno-positive for Ub. In urine low-density membrane fractions, exosomes were also positive for Ub (Fig. 2B). Size distribution analysis of the membrane-bound structures in this fraction using transmission electron microscopy demonstrated that they have a mean diameter of 58.2 ± 1.8 nm (S.E.) (Fig. 2E). Nanosight analysis of various different purifications of low-density membrane fractions supported their classification as exosomes with a mean diameter of 89.4 ± 2.52 nm (S.E.) (Supplemental Fig. 2). A cohort of exosomes or MVBs (Figs. 2C and 2D and Supplemental Fig. 1) were colabeled for Ub and the common urinary exosome cargo proteins AQP2 and SLC12A3 (NCC); however, based on these data, whether the ubiquitylation occurs directly on AQP2/NCC or other cargo proteins was unclear.
Fig. 2.
Identification of ubiquitin in human MVBs and urinary exosomes by immunogold electron microscopy. (A) Ubiquitin is readily detected in association with internal luminal vesicles of an MVB within a human kidney epithelial cell. Exosomes isolated from human urine labeled with ubiquitin (B) or ubiquitin plus AQP2 (C) or NCC (D); arrows indicate ubiquitin (5 nm gold particles) and arrowheads indicate AQP2 or NCC (2 nm gold particles). (E) Size distribution of low-density membrane vesicles from 200,000 × g pellet fraction (n = 159).
Large-Scale Identification of Ubiquitylated Proteins in Urinary Exosomes
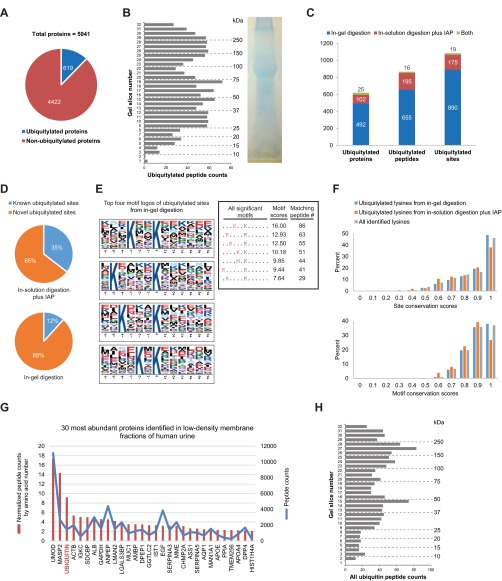
To determine if ubiquitylation of cargo proteins within exosomes is a common event, large-scale analysis of specific ubiquitylation sites on urinary exosome cargo from humans was performed. Two techniques prior to LC-MS/MS analysis were performed to both maximize coverage of membrane-associated proteins and to specifically enrich for ubiquitylated peptides (Supplemental Fig. 3). Overall, 5041 proteins were identified; among these proteins were 619 unique ubiquitylated proteins (Fig. 3A). All proteins identified and their associated information are listed in a publicly accessible online database at http://interpretdb.au.dk/database/UbExo/. This database is the largest urinary exosomal proteome from normal humans to date. Ubiquitylated peptides were identified throughout the molecular weight range of the 1D SDS-PAGE gel (Fig. 3B), corresponding well with the low-density membrane fraction immunoblot (Fig. 1A). A low correlation between the numbers of ubiquitylated proteins, peptides, and sites identified using the two techniques (Fig. 3C) highlighted their complementary nature in the detection of ubiquitylation events. In total, 1084 sites from 866 peptides were identified. With reference to the human ubiquitylation site dataset in the PhosphoSitePlus online resource (version date: 050415), the majority of ubiquitylated sites identified herein are novel, in particular those identified following in-gel digestion (88% novel, Fig. 3D). Motif analysis of the immunoaffinity purification (IAP) results revealed no significant motif surrounding ubiquitylated lysine residues. In contrast, several significant motifs were identified from the in-gel digestion sites, the majority of which contained K or R on the N-terminal side (within -6 position) of the ubiquitylated lysine residue (Fig. 3E). Comparing among various eukaryotic species in the HomoloGene database (http://www.ncbi.nlm.nih.gov/homologene), ubiquitylated sites from the in-gel digestion were slightly more conserved than those from the IAP at both ubiquitylated lysine residues and their surrounding motifs (upper and lower panels of Fig. 3F, respectively), although the overall conservation of these ubiquitylated sites was not higher than the background “all lysines” identified. Consistent with the high number of ubiquitylated proteins identified, ubiquitin was the third most abundant protein in low-density membrane fractions of human urine (Fig. 3G) and was observed with a similar distribution as the ubiquitylated peptides along the 1D SDS-PAGE gel (Fig. 3H, r2 = 0.4993, p value < 0.0001 in comparison to the distribution in Fig. 3B).
Fig. 3.
Large-scale identification of ubiquitylated proteins in urinary exosomes. (A) Total ubiquitylated and nonubiquitylated proteins identified in human urinary exosomes. (B) Quantity of ubiquitylated peptides from 1D SDS-PAGE gel. (C) Total ubiquitylated proteins, peptides, and sites identified from in-gel digestion, in-solution digestion with IAP, or both. (D) Novel versus known ubiquitylated sites identified from either in-solution digestion with IAP or in-gel digestion. (E) Top four motif logos of ubiquitylated sites identified from in-gel digestion display strong preferences for lysine at various positions N-terminal to the ubiquitylated lysine residue. Seven significant motifs with their scores and matching peptide numbers are shown in a table. The logos and scores of the overrepresented ubiquitylation motifs were generated by motif-x program. (F) Distributions of site and motif conservation scores of ubiquitylated lysines from in-gel digestion, in-solution digestion plus IAP, and all identified lysines. (G) Thirty most abundant proteins identified in low-density membrane fractions of human urine based on normalized and total peptide counts (see supplemental data 1 for the complete list). A gene symbol for each protein is displayed, except for ubiquitin (encoded by four different genes i.e. RPS27A, UBA52, UBB, and UBC). (H) Instances of ubiquitin peptide identification from 1D SDS-PAGE gel.
Bioinformatic Analysis of Ubiquitylated Proteins in Urinary Exosomes
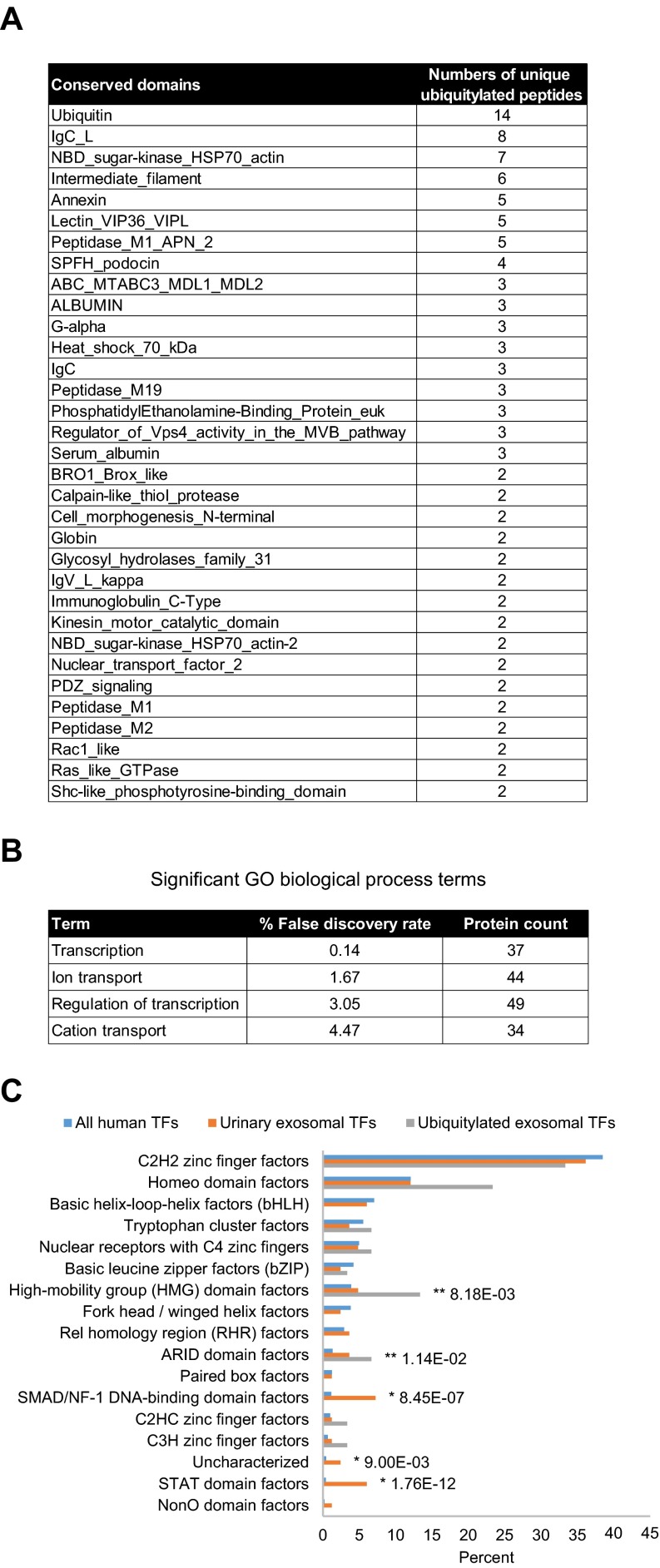
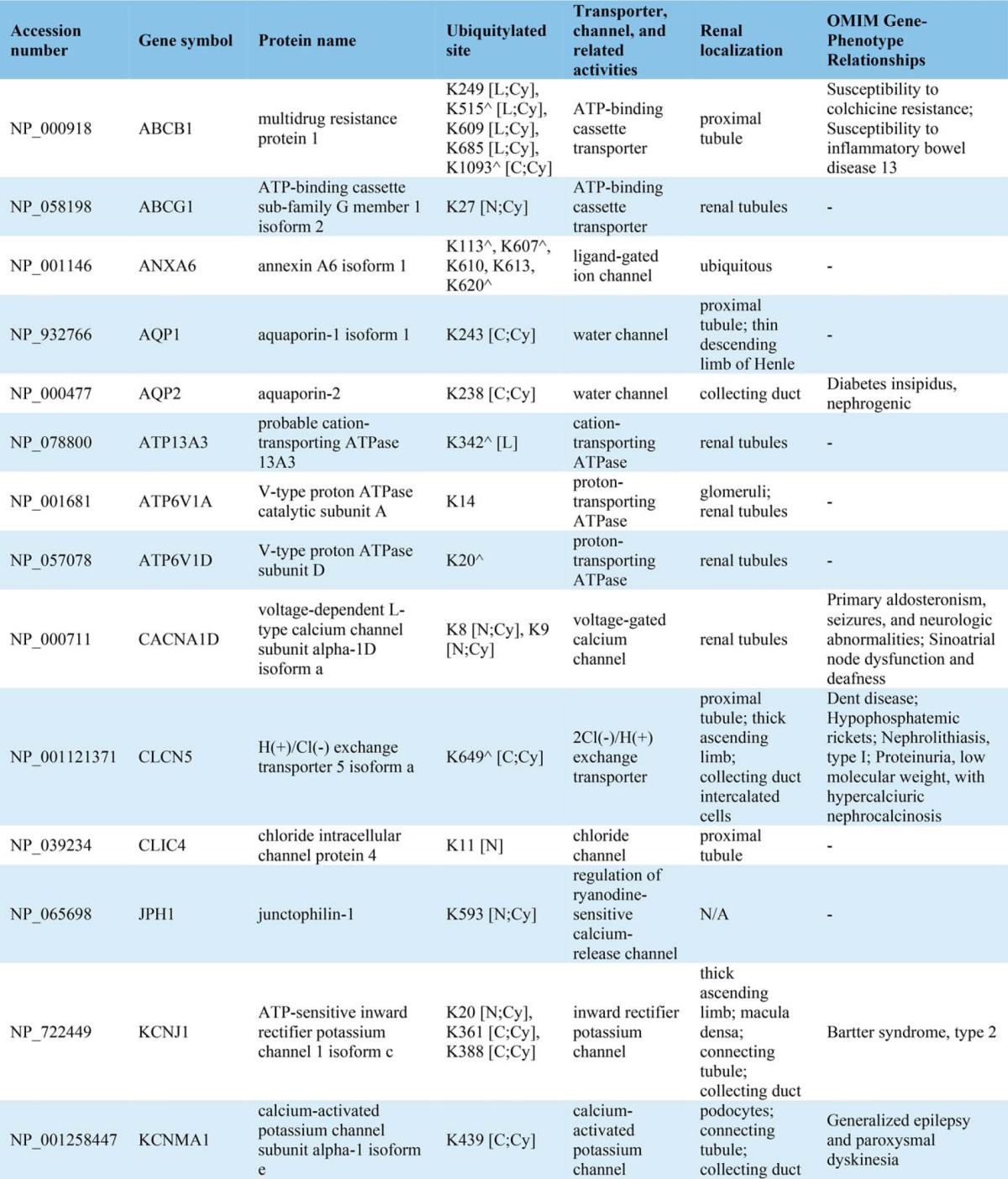
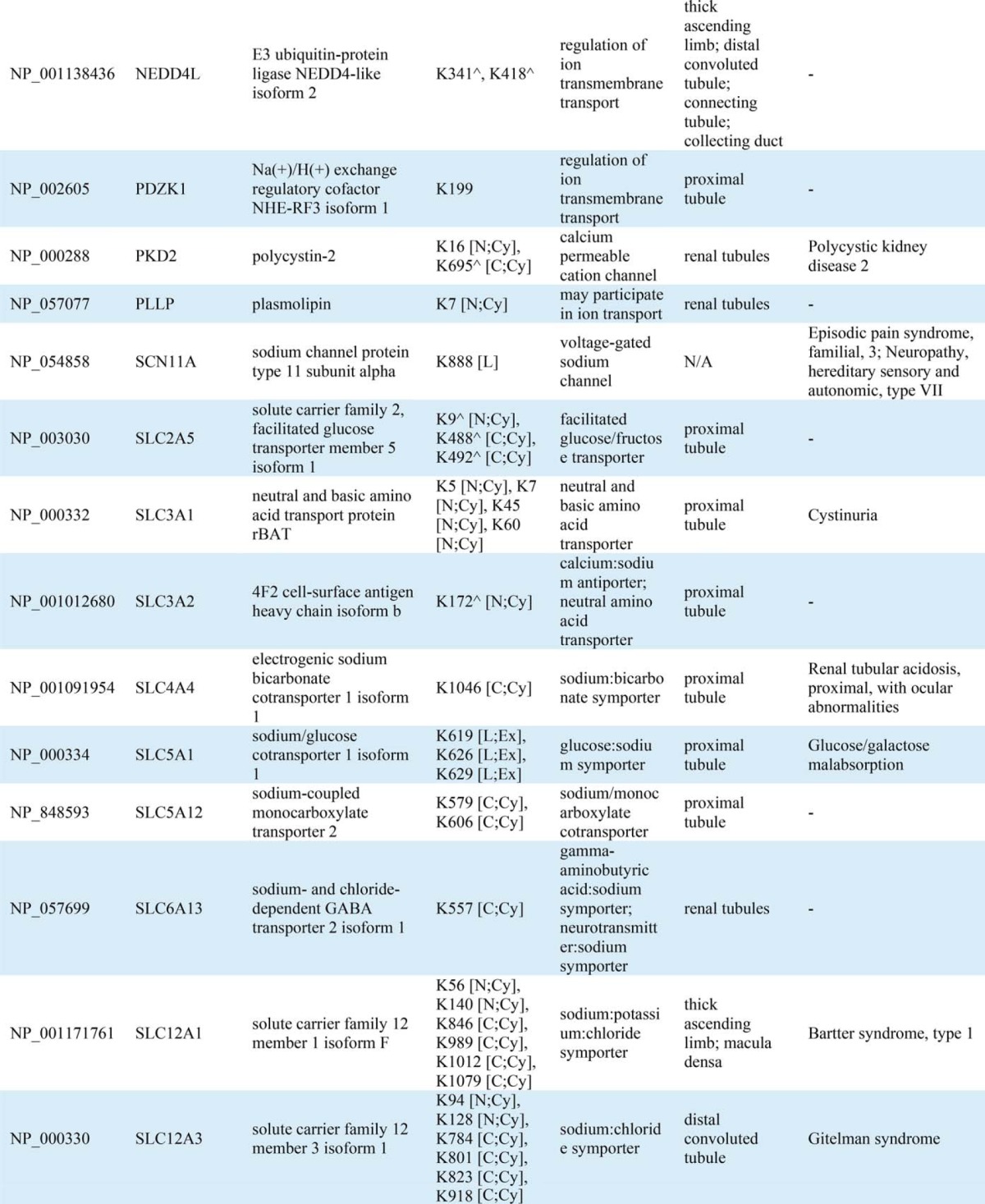
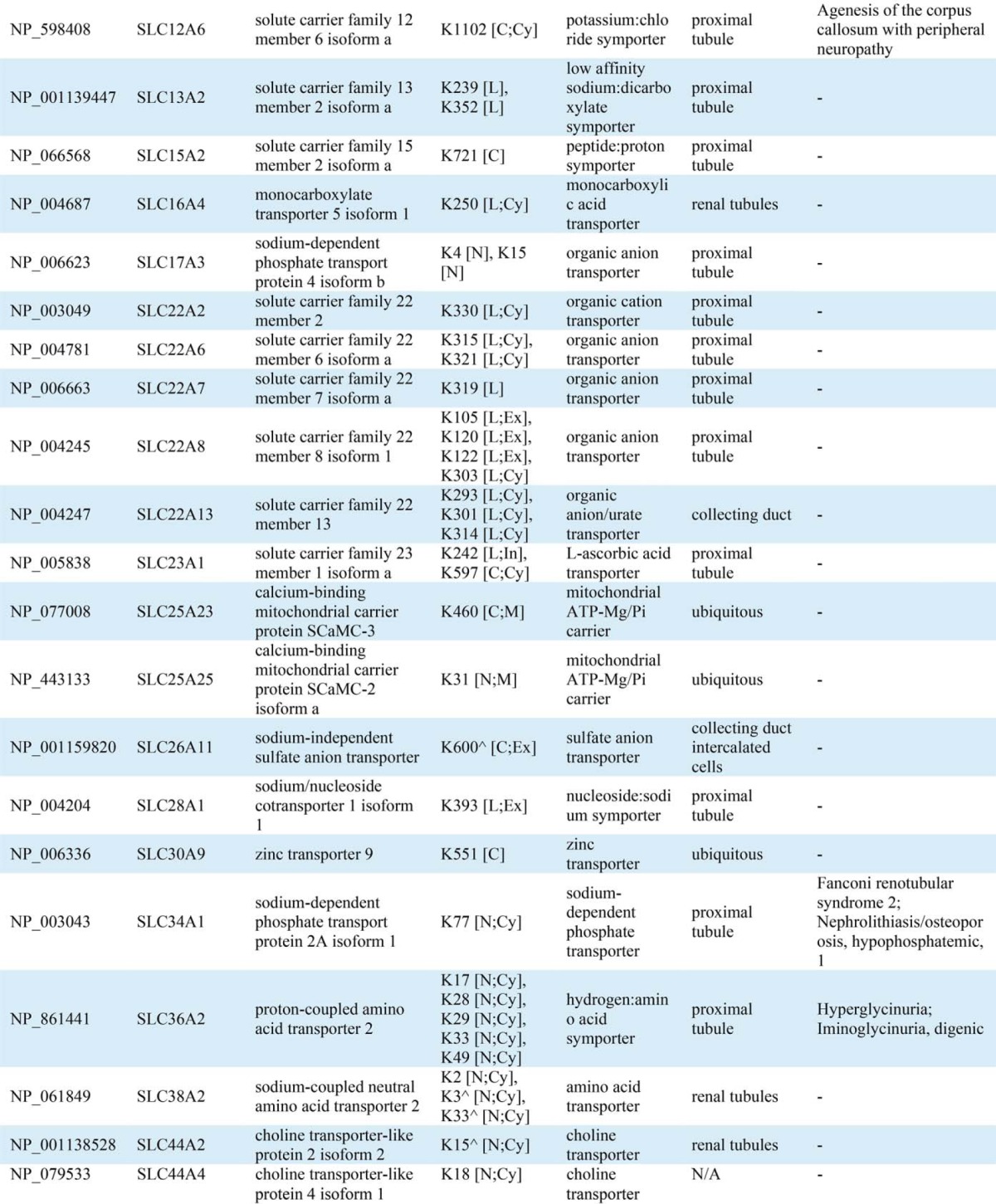
Conserved domain analysis identified ubiquitylated sites in a variety of protein functional domains (Fig. 4A). The domain “Ubiquitin” was highly conserved with the highest instance of ubiquitylated peptides and sites (14 unique peptides and all seven side chain linkages of ubiquitin i.e. Lys6, Lys11, Lys27, Lys29, Lys33, Lys48, and Lys63) indicating that, in addition to total ubiquitin, polyubiquitin chains are highly enriched in urinary exosomes. Though ubiquitylation is known to play a role in multiple processes, gene-annotation enrichment analysis revealed that in urinary exosomes, ubiquitylated proteins involved in transcription and transcriptional regulation as well as ion or solute transport were significantly overrepresented relative to the general distribution of protein classes (Fig. 4B). 83 transcription factors (TFs) were discovered in urinary exosomes; among these TFs, 30 (36%) were novel ubiquitylated TFs in human (Table IA). Fig. 4C demonstrates the relative percentages of transcription factor classes between all human TFs and urinary exosomal TFs or ubiquitylated exosomal TFs. Several classes of TFs in urinary exosomes were significantly overrepresented against the background of all human TFs, including high-mobility group domain factors, AT-Rich Interaction Domain (ARID) domain factors, small mothers against decapentaplegic/nuclear factor 1 (SMAD/NF-1) DNA-binding domain factors, and STAT domain factors. Several of these TFs are known to play a role in modulating kidney development, cell proliferation, autophagy, the cell cycle, cell morphology, transcriptional activation by nuclear hormone receptors, and innate immune responses. Multiple ubiquitylated ion or solute transport proteins were detected in urinary exosomes, with 80% of the identified ubiquitylation sites previously unknown (Table IB). The ubiquitylated ion/solute transport proteins identified in human urinary exosomes was representative of the whole renal tubule. Additional features of the ubiquitylated peptides identified are highlighted in Supplemental Fig. 4.
Fig. 4.
Bioinformatic analysis of ubiquitylated proteins in urinary exosomes. (A) Top conserved domains found to be associated with the ubiquitylated peptides identified (only conserved domains with at least two unique ubiquitylated peptides are shown (see supplemental data 2 for all conserved domains detected). (B) Significantgene ontology biological process terms determined by gene-annotation enrichment analysis. (C) Percentage distributions of transcription factor (TF) classes of all human TFs, urinary exosomal TFs, and ubiquitylated exosomal TFs. * and ** indicate significantly overrepresented TF classes of urinary exosomal TFs and ubiquitylated exosomal TFs, respectively, compared against the background of all human TFs by chi-squared tests.
Table IA. Ubiquitylated transcription factors discovered in urinary exosomes (all of their ubiquitylated sites are novel in human).
| Accession Number | Gene Symbol | Protein Name | Ubiquitylated Site | Class |
|---|---|---|---|---|
| NP_001177 | BACH1 | Transcription regulator protein BACH1 | K657 | Basic leucine zipper factors (bZIP) |
| NP_006456 | ARID3B | AT-rich interactive domain-containing protein 3B | K388 | ARID domain factors |
| NP_001253969 | JARID2 | Protein Jumonji isoform 2 | K507 | ARID domain factors |
| NP_001417 | EN1 | Homeobox protein engrailed-1 | K9 | Homeo domain factors |
| NP_000513 | HOXA13 | Homeobox protein Hox-A13 | K192, K209 | Homeo domain factors |
| NP_078777 | HOXD1 | Homeobox protein Hox-D1 | K216, K217, K219 | Homeo domain factors |
| NP_055436 | HOXD4 | Homeobox protein Hox-D4 | K11, K16 | Homeo domain factors |
| NP_006553 | LBX1 | Transcription factor LBX1 | K148 | Homeo domain factors |
| NP_001193955 | POU2F2 | POU domain, class 2, transcription factor 2 isoform 3 | K21 | Homeo domain factors |
| NP_055850 | ZHX3 | Zinc fingers and homeoboxes protein 3 | K858 | Homeo domain factors |
| NP_006485 | ERF | ETS domain-containing transcription factor ERF | K465 | Tryptophan cluster factors |
| NP_001070729 | NCOR2 | Nuclear receptor corepressor 2 isoform 2 | K1150, K1158 | Tryptophan cluster factors |
| NP_001136040 | BBX | HMG box transcription factor BBX isoform 1 | K338, K341 | High-mobility group (HMG) domain factors |
| NP_055798 | HMGXB3 | HMG domain-containing protein 3 | K74 | High-mobility group (HMG) domain factors |
| NP_005977 | SOX1 | Transcription factor SOX-1 | K45 | High-mobility group (HMG) domain factors |
| NP_579878 | WHSC1 | Histone-lysine N-methyltransferase NSD2 isoform 1 | K6, K17, K20, K22 | High-mobility group (HMG) domain factors |
| NP_722520 | ZFPM1 | Zinc finger protein ZFPM1 | K5 | C2H2 zinc finger factors |
| NP_001243208 | ZNF26 | Zinc finger protein 26 isoform 1 precursor | K550 | C2H2 zinc finger factors |
| NP_003406 | ZNF268 | Zinc finger protein 268 isoform a | K261, K274 | C2H2 zinc finger factors |
| NP_003416 | ZNF45 | Zinc finger protein 45 | K5, K11 | C2H2 zinc finger factors |
| NP_001001668 | ZNF470 | Zinc finger protein 470 | K140 | C2H2 zinc finger factors |
| NP_444270 | ZNF518B | Zinc finger protein 518B | K2, K25, K1010, K1012, K1014, K1015 | C2H2 zinc finger factors |
| NP_694951 | ZNF524 | Zinc finger protein 524 | K47 | C2H2 zinc finger factors |
| NP_001001411 | ZNF676 | Zinc finger protein 676 | K17 | C2H2 zinc finger factors |
| NP_057727 | ZNF771 | Zinc finger protein 771 | K24 | C2H2 zinc finger factors |
| NP_001264220 | ZNF891 | Zinc finger protein 891 | K495 | C2H2 zinc finger factors |
| Q9UL68 | MYT1L | myelin transcription factor 1-like [Source:HGNC Symbol;Acc:7623] | K479 | C2HC zinc finger factors |
| NP_940983 | ZC3H6 | Zinc finger CCCH domain-containing protein 6 | K819 | C3H zinc finger factors |
| NP_005117 | NR1D2 | Nuclear receptor subfamily 1 group D member 2 isoform 1 | K77, K80, K88 | Nuclear receptors with C4 zinc fingers |
| NP_995582 | NR5A2 | Nuclear receptor subfamily 5 group A member 2 isoform 1 | K92 | Nuclear receptors with C4 zinc fingers |
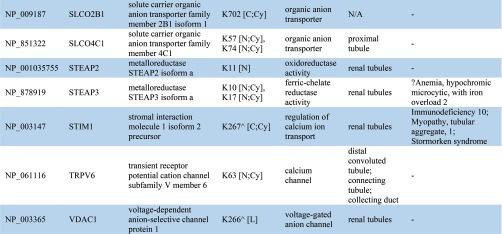
Table IB. Ubiquitylated transporter, channel, and related proteins identified in urinary exosomes. ^ indicates known ubiquitylated sites. Ubiquitylated site topological domain is reported as the following notation: [NH2-tail (N), COOH-tail (C), loop (L); Cytoplasmic (Cy), Extracellular (Ex), Intramembrane (In), Mitochondrial intermembrane (M)].
Quantification of Various Ubiquitylation Forms and Ubiquitin Topologies
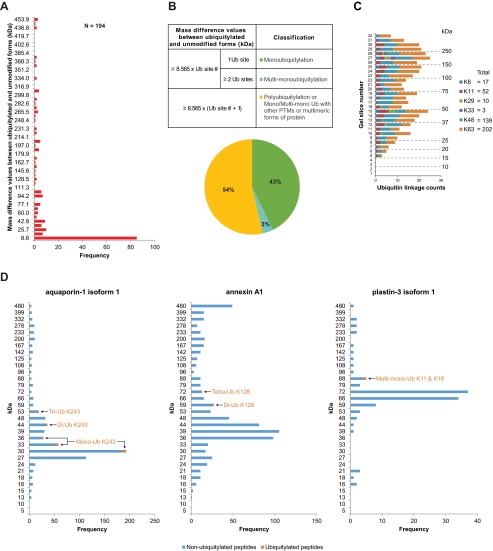
Monoubiquitylation, multimonoubiquitylation, or polyubiquitylation, in addition to the type of ubiquitin linkage utilized, are essential factors that can determine the role of ubiquitylation in various biological processes, which may include a mechanism for cargo protein incorporation into MVBs. To determine the proportion of these ubiquitylation forms on proteins in urinary exosomes, we calculated a mass difference between ubiquitylated and unmodified forms of each ubiquitylated protein based on its peptide distribution pattern on a 1D SDS-PAGE gel (Fig. 5A). A classification scheme was implemented on these mass difference values in reference to an expected mass increase by a single ubiquitylation event (as a function of the ubiquitylated site number multiplied by the molecular weight of a ubiquitin molecule, 8.565 kDa, Fig. 5B). This classification resulted in three different groups of ubiquitylated proteins (Supplemental Table 1) corresponding with monoubiquitylation (43%), multimonoubiquitylation (3%), and equivocal events e.g. polyubiquitylation and mono/multimonoubiquitylation with other PTMs or multimeric protein forms (54%). For the latter group, the presence of polyubiquitylated proteins was reinforced by the identification of many polyubiquitin chains with a combined pattern on the 1D SDS-PAGE gel comparable to that of the ubiquitylated peptides except in the < 15 kDa region (Fig. 5C, r2 = 0.6891, p value < 0.0001 in comparison to the distribution in Fig. 3B). Furthermore, a specific configuration of polyubiquitylation on many proteins can be determined by an unambiguous mass distribution pattern of their ubiquitylated peptides over the background of their nonubiquitylated peptides (Fig. 5D and supplemental data 3 for the molecular weight distribution plots of all the classified proteins). Using this approach, some ubiquitylated peptides were found at or below the expected molecular weight of their target protein (Supplemental Fig. 5), which may represent degradation products or proteins in a modified e.g. cleaved, form.
Fig. 5.
Quantification of ubiquitylation forms in urinary exosomes. (A) Frequency distribution of mass differences between the ubiquitylated and unmodified forms of ubiquitylated proteins. 194 events satisfying the criterion in Materials and Methods were evaluated. (B) Upper panel: scheme to classify ubiquitylation forms into three groups. Lower panel: percentages of the different groups of ubiquitylated proteins. (C) Instances of different polyubiquitin linkages in urinary exosomes. (D) Paradigms of specific configurations of ubiquitylation determined by an unambiguous mass distribution pattern of ubiquitylated peptides over the background of nonubiquitylated peptides. Left panel: aquaporin-1 isoform 1 (unmodified form = 28 kDa; K243 ubiquitylated peptides were identified from the corresponding gel locations of mono-, di-, and tri-ubiquitylated (Ub) forms of the protein). Middle panel: annexin A1 (unmodified form = 39 kDa; K128 ubiquitylated peptides were identified from the corresponding gel locations of di- and tetra-Ub forms of the protein). Right panel: plastin-3 isoform 1 (unmodified form = 71 kDa; a doubly ubiquitylated peptide at K11 and K18 was identified from the corresponding gel location of multimono-Ub form of the protein).
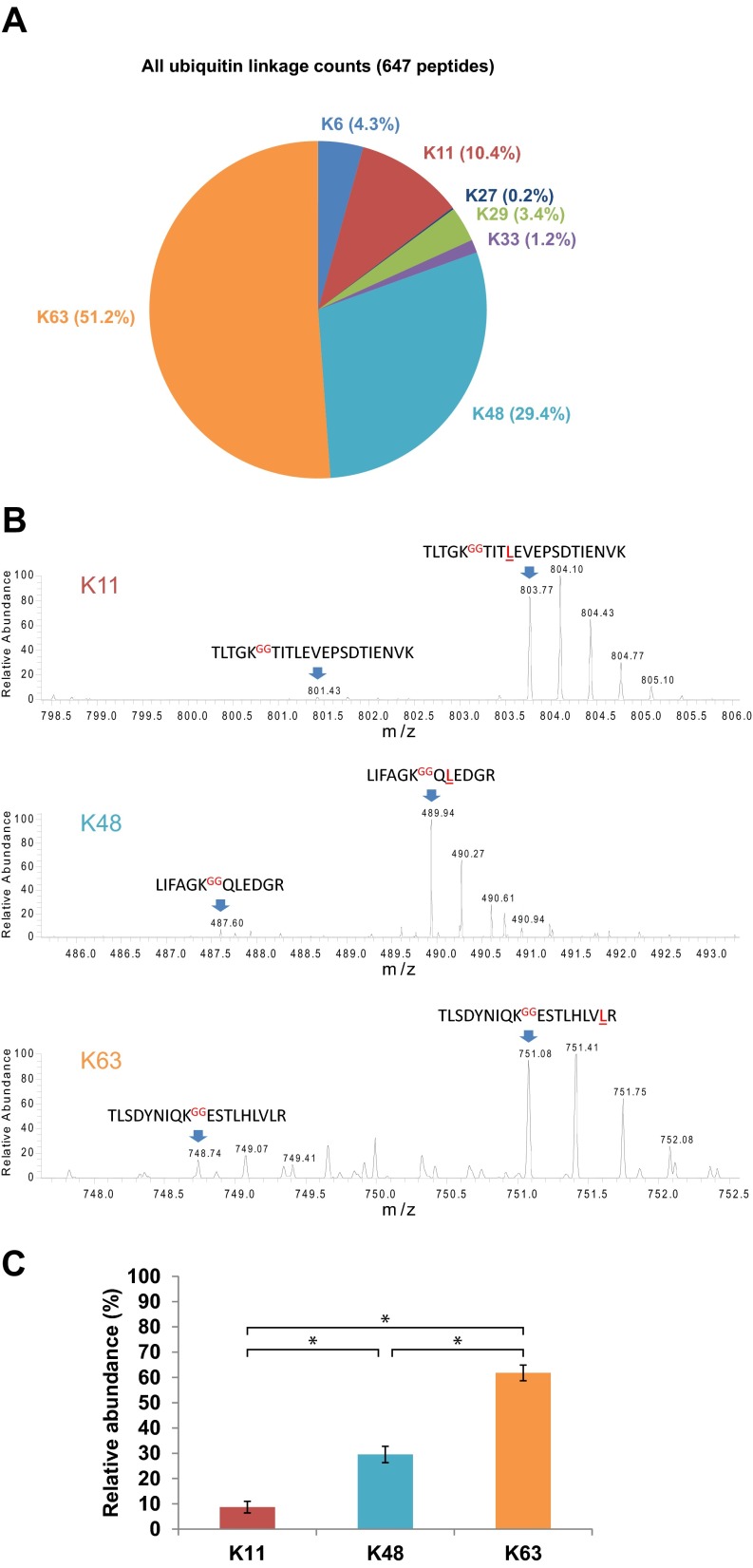
To initially quantify polyubiquitin chain topologies, peptide counting was utilized (Fig. 5C from the 1D SDS-PAGE data and Fig. 6A from all the data combined). This indicated that K63-linked polyubiquitin chains (Lys63) were most frequently observed, followed by Lys48, Lys11, Lys6, Lys29, Lys33, and Lys27. The estimated relative abundance of K11, K48, and K63 polyubiquitin chains provided by peptide counting was confirmed using an absolute protein quantification (AQUA) technique (Figs. 6B and 6C). Consistent with the peptide counting data, Lys63 polyubiquitin chains were verified by AQUA to be significantly more abundant in human urinary exosomes than K48 or K11 chains.
Fig. 6.
Quantification of polyubiquitin chain topologies in human urinary exosomes. (A) Relative abundance of seven polyubiquitin linkages quantified by peptide counting. (B) Representative mass spectra demonstrate relative peak heights of the GG-linked endogenous (light) and AQUA (heavy) peptides of K11-, K48-, and K63-linked polyubiquitin chains. An equal amount (2.5 pmol) of each GG-linked ubiquitin-AQUA peptide was spiked into each sample. (C) Relative abundance of K11, K48, and K63 polyubiquitin chains quantified using the AQUA technique (n = 5). * p < 0.0001 by Tukey's multiple comparison tests.
DISCUSSION
Urinary extracellular nanovesicles (exosomes) are predominantly derived from the epithelial cells that line the kidney tubule and urinary tract. Alterations in their protein or nucleic acid cargo have been proposed to reflect the physiological and pathophysiological state of cells within the renal tubule, the genitourinary tract, or elsewhere in the body (3). In general (22), it is thought that protein cargo to be incorporated into urinary exosomes is ubiquitylated, recognized by the ESCRT apparatus on MVBs, and deubiquitylated by deubiquitylases during the process of cargo incorporation into MVBs (8–11). In this study, we provide direct evidence that deubiquitylation of protein cargo is not an essential step in internal luminal vesicle and urinary exosome formation and a large proportion of exosome protein cargo is ubiquitylated with various topologies. Our studies demonstrate that urinary exosomes are an enriched source for a noninvasive approach to study ubiquitin modifications in physiological or disease states and that analysis of cargo ubiquitylation status (rather than abundance) may be better suited to discovery of new biomarkers for renal and systemic diseases. Furthermore, the large number of ubiquitylation sites on human proteins identified in this study will be a major resource to the cell biology community and be of broad utility for targeted analysis of individual ubiquitylation site function.
Advancing our previous studies (2), biochemical analysis indicated that ubiquitylated proteins, but not free ubiquitin, are in low-density membrane fractions of human urine. Direct detection of ubiquitin within ILVs of MVBs in kidney tubule epithelial cells and urinary exosomes by immunogold electron microscopy supports a mechanism where ubiquitylated proteins are recruited to MVBs, and a proportion are sequestered into ILVs without prior deubiquitylation before release into the urinary space as exosomes. Consistent with this mechanism, ubiquitin was the third most abundant protein in low-density membrane fractions of human urine and ∼13% of all the proteins identified in exosomes were ubiquitylated. Although the centrifugation technique we used to isolate the low-density membrane fractions of human urine is considered the “gold standard,” as with all biochemical techniques, we cannot completely exclude that some small membrane-bound vesicles make up a very small percentage of the analyzed sample. However, the vesicle size distribution by transmission electron microscopy and Nanosight analysis, coupled with the immunogold detection of ubiquitin in membrane-bound vesicles <100 nm in diameter, strongly supports our conclusions of ubiquitylated proteins within exosomes. Furthermore, although this is the first study of ubiquitylated proteins from exosomes derived directly from human subjects, detection of ubiquitin in exosomes isolated from culture media of myeloid-derived suppressor cells (23), human B-cells (HLA-DR15), or mouse immature splenic dendritic cells (D1) (24) further supports a model where deubiquitylation of protein cargo does not always occur during MVB formation. In addition to ubiquitylated cargo, our mass spectrometry data identified numerous nonubiquitylated peptides in low-density membrane fractions of human urine. This has three possible explanations. First, nonubiquitylated peptides may be identified, although they arise from a ubiquitylated cargo protein. Secondly, nonubiquitylated proteins may be targeted by ESCRT-interaction domains for entry into ILVs. In line with this, ubiquitin was proposed not as a unique sorting signal that is essential for MVB recruitment but rather as a temporary ESCRT-interaction domain that can be added and removed from proteins to regulate their interaction with the ESCRT machinery (25). Thirdly, we cannot exclude a model where specific cargos, for example, particular proteins classes, are deubiquitylated during ILV formation.
Our application of in-gel digestion or in-solution immunoaffinity isolation techniques prior to large-scale ubiquitylation profiling using LC-MS/MS resulted in complimentary datasets that allowed in-depth analysis of ubiquitylated exosomal proteins. Although previous studies, the majority performed in yeast or cell culture models, have indicated that monoubiquitylation is important for marking membrane proteins to be incorporated into MVBs e.g. (4, 26), we detected proteins with profiles corresponding with monoubiquitylation, multimonoubiquitylation, and polyubiquitylation in low-density membrane fractions of human urine. Although a proportion of these ubiquitin forms cannot be exactly determined due to unknown parameters that can also alter a protein mass e.g. other PTMs, multimer formation, and endogenous cleavage, it supports our model that deubiquitylation of protein cargo during ILV formation is not necessary. Combined with our observations that all classes of polyubiquitin chains (K6, K11, K27, K29, K33, K48, and K63) are detected on cargo from urinary exosomes with variable frequencies, these data support the growing body of evidence that multimonoubiquitylation and polyubiquitylation are necessary to overcome the low-affinity binding of ubiquitin to ubiquitin interaction motifs on adapter proteins and facilitate rapid internalization and endosomal sorting of particular classes of membrane proteins e.g. (27, 28). Furthermore, the high abundance of K63-linked ubiquitin chains we detected on urinary exosome cargo is in line with studies in yeast and cultured mammalian cells that K63-linkage is a better signal than monoubiquitylation during endosomal sorting and may even be important for MVB biogenesis (29). One question that our studies cannot address, but would be an interesting avenue of future research, is whether deubiquitylation of cargo proteins actually alters the rate of exosome formation and secretion. For example, experiments examining the secretion rate and/or concentration of exosomes into cell culture media following treatment of cells with broad spectrum deubiquitylase or E3 ligase inhibitors may provide novel information on this aspect.
Defining a valid ubiquitylation motif has been the subject of various studies using multiple, potentially biased, enrichment strategies (30–33). Despite identification of, for example, a significant preference for hydrophobic residues such as Phe, Tyr, Trp, Leu, Ile, and Val adjacent to ubiquitylated lysines (30, 33, 34), the majority of studies have been unsuccessful in identifying specific ubiquitylation recognition motifs (32). Our use of in-gel digestion of exosomal protein cargo prior to LC-MS/MS, without a specific enrichment strategy (e.g. K-ε-GG immunoaffinity purification), provided a nonbiased platform for analysis of overrepresented motifs surrounding ubiquitylation sites. We determined that there was a unique and strong preference for an additional lysine within six amino acids upstream of the ubiquitylated lysine residue. Although not a specific “ubiquitylation motif” per se, this clear preference for lysine surrounding the ubiquitylated residue is in line with a previous study highlighting a significant preference for positively charged residues (Lys and Arg) upstream of the ubiquitylation site (35). The current datasets will provide additional information to build computational models for ubiquitylation-binding motifs (33, 34).
Another observation from the current study is that we observed several instances of DiGly modification on C-terminal lysine residues, despite the use of chloroacetamide during alkylation steps. We have also observed this phenomenon ourselves in other studies (not published), supporting that this is a valid identification and not an artifact. Furthermore, DiGly modification on C-terminal lysine residues has been reported by others, who observed that following trypsin digest of a poly-Ub standard, 23% of identified ubiquitylation sites were on the terminal lysine (36). This fits well with our study that observed 20% of ubiquitylation sites on C-terminal lysines. However, it is still plausible that this phenomenon could be a result of chloroacetamide treatment under some certain conditions (albeit less common than with iodoacetamide). Therefore, the C-terminal ubiquitylation sites identified in this study should be interpreted with caution.
In addition to being an enriched source of ubiquitylated proteins for studying the characteristics of ubiquitin modifications, the urinary exosomal proteome we generated is the largest from normal humans to date. The majority of the ubiquitylated sites identified herein are novel in human, thus the datasets will serve as a major resource for targeted analysis of individual ubiquitylation site function. In total, 21% of ubiquitylated proteins in exosomes were classified as transmembrane proteins (a similar percentage to total number of transmembrane proteins identified in exosomes), supporting the idea that in mammalian cells ILV formation can occur via both ESCRT-dependent and -independent mechanisms (37–39) or that cytosolic components are randomly loaded into ILVs upon intraluminal budding. Within the large pool of ubiquitylated cargo in urinary exosomes, proteins involved in transcription/transcriptional regulation were significantly overrepresented. Although previous studies have identified mRNA and miRNAs in urinary microvesicles (40–42), the current study suggests that urinary exosomes may be able to vectorally influence other cells in the renal epithelia and urinary tract systems not only by delivering RNA cargo but also by delivering a wide range of TFs and regulatory proteins that may control/contribute to gene expression.
In conclusion, this study provides a new model for cargo recruitment to MVBs and ILV formation prior to urinary exosome secretion (Supplemental Fig. 6). The lack of deubiquitylation of cargo proteins proposed in this model is likely to be similar for exosomes formed and secreted from other cell types. Furthermore, analysis of ubiquitylated cargo within urinary exosomes can be used to study the molecular physiology and pathophysiology of ubiquitylation events in renal epithelial and urinary tract systems and potentially for the discovery of new biomarkers for renal and systemic diseases.
Supplementary Material
Acknowledgments
Ahmed Basim Abduljabar, Helle Høyer, and Else-Merete Locke are thanked for expert technical assistance. Michael Lykke Hvam is thanked for help with Nanosight analysis. Peter Baker and Robert Chalkey helped with mass spectrometry data conversion and upload of spectra to Protein Prospector.
Footnotes
Author contributions: R.A.F. and T.P. designed the research; A.R.H., L.C., P.S., R.A.F., and T.P. performed the research; M.A.K., R.A.F., and T.P. analyzed data; and R.A.F. and T.P. wrote the paper.
* This work was supported by The Danish Council for Independent Research Medical Research Council (4004–00043) and Natural Science Council (4002–00364), the Lundbeck Foundation, the Novo Nordisk Foundation, the Carlsberg Foundation, and the Aarhus University Research Foundation. TP is supported by the Chulalongkorn Academic Advancement into Its 2nd Century Project.
 This article contains supplemental material.
This article contains supplemental material.
The authors declare no competing financial interests.
1 The abbreviations used are:
- AQP2
- aquaporin 2
- HCD
- higher-energy C-trap dissociation
- PTM
- post-translational modification
- Ub
- ubiquitin.
REFERENCES
- 1. Colombo M., Raposo G., and Théry C. (2014) Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu. Rev. Cell Dev. Biol. 30, 255–289 [DOI] [PubMed] [Google Scholar]
- 2. Pisitkun T., Shen R.F., and Knepper M.A. (2004) Identification and proteomic profiling of exosomes in human urine. Proc. Natl. Acad. Sci. U.S.A. 101, 13368–13373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. van Balkom B.W., Pisitkun T., Verhaar M.C., and Knepper M.A. (2011) Exosomes and the kidney: prospects for diagnosis and therapy of renal diseases. Kidney Int. 80, 1138–1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Katzmann D.J., Babst M., and Emr S.D. (2001) Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell 106, 145–155 [DOI] [PubMed] [Google Scholar]
- 5. Komander D., and Rape M. (2012) The ubiquitin code. Annu. Rev. Biochem. 81, 203–229 [DOI] [PubMed] [Google Scholar]
- 6. Shields S. B., and Piper R. C. (2011) How ubiquitin functions with ESCRTs. Traffic 12, 1306–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ren X., and Hurley J. H. (2010) VHS domains of ESCRT-0 cooperate in high-avidity binding to polyubiquitinated cargo. EMBO J. 29, 1045–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Amerik A. Y., Nowak J., Swaminathan S., and Hochstrasser M. (2000) The Doa4 deubiquitinating enzyme is functionally linked to the vacuolar protein-sorting and endocytic pathways. Mol. Biol. Cell 11, 3365–3380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. McCullough J., Clague M. J., and Urbé S. (2004) AMSH is an endosome-associated ubiquitin isopeptidase. J. Cell Biol. 166, 487–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Agromayor M., and Martin-Serrano J. (2006) Interaction of AMSH with ESCRT-III and deubiquitination of endosomal cargo. J. Biolog. Chem. 281, 23083–23091 [DOI] [PubMed] [Google Scholar]
- 11. Mizuno E., Kobayashi K., Yamamoto A., Kitamura N., and Komada M. (2006) A deubiquitinating enzyme UBPY regulates the level of protein ubiquitination on endosomes. Traffic 7, 1017–1031 [DOI] [PubMed] [Google Scholar]
- 12. Pedersen N. B., Hofmeister M. V., Rosenbaek L. L., Nielsen J., and Fenton R. A. (2010) Vasopressin induces phosphorylation of the thiazide-sensitive sodium chloride cotransporter in the distal convoluted tubule. Kidney Int. 78, 160–169 [DOI] [PubMed] [Google Scholar]
- 13. Hoffert J. D., Fenton R. A., Moeller H. B., Simons B., Tchapyjnikov D., McDill B. W., Yu M. J., Pisitkun T., Chen F., and Knepper M. A. (2008) Vasopressin-stimulated increase in phosphorylation at Ser269 potentiates plasma membrane retention of aquaporin-2. J. Biolog. Chem. 283, 24617–24627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nielsen M. L., Vermeulen M., Bonaldi T., Cox J., Moroder L., and Mann M. (2008) Iodoacetamide-induced artifact mimics ubiquitination in mass spectrometry. Nat. Methods 5, 459–460 [DOI] [PubMed] [Google Scholar]
- 15. Elias J. E., and Gygi S .P. (2007) Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods 4, 207–214 [DOI] [PubMed] [Google Scholar]
- 16. Kirkpatrick D. S., Hathaway N. A., Hanna J., Elsasser S., Rush J., Finley D., King R. W., and Gygi S. P. (2006) Quantitative analysis of in vitro ubiquitinated cyclin B1 reveals complex chain topology. Nat Cell Biol. 8, 700–710 [DOI] [PubMed] [Google Scholar]
- 17. Huang D. W., Sherman B. T., and Lempicki R. A. (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 [DOI] [PubMed] [Google Scholar]
- 18. Marchler-Bauer A., Lu S., Anderson J. B., Chitsaz F., Derbyshire M. K., DeWeese-Scott C., Fong J. H., Geer L. Y., Geer R. C., Gonzales N. R., Gwadz M., Hurwitz D. I., Jackson J. D., Ke Z., Lanczycki C. J., Lu F., Marchler G. H., Mullokandov M., Omelchenko M. V., Robertson C. L., Song J. S., Thanki N., Yamashita R. A., Zhang D., Zhang N., Zheng C., and Bryant S. H. (2011) CDD: A conserved domain database for the functional annotation of proteins. Nucleic Acids Res. 39, D225–D229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhao B., Pisitkun T., Hoffert J. D., Knepper M. A., and Saeed F. (2012) CPhos: A program to calculate and visualize evolutionarily conserved functional phosphorylation sites. Proteomics 12, 3299–3303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hornbeck P. V., Kornhauser J. M., Tkachev S., Zhang B., Skrzypek E., Murray B., Latham V., and Sullivan M. (2012) PhosphoSitePlus: A comprehensive resource for investigating the structure and function of experimentally determined post-translational modifications in man and mouse. Nucleic Acids Res. 40, D261–D270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Schwartz D., and Gygi S. P. (2005) An iterative statistical approach to the identification of protein phosphorylation motifs from large-scale data sets. Nat. Biotechnol. 23, 1391–1398 [DOI] [PubMed] [Google Scholar]
- 22. Haglund K., and Dikic I. (2012) The role of ubiquitylation in receptor endocytosis and endosomal sorting. J. Cell Sci. 125, 265–275 [DOI] [PubMed] [Google Scholar]
- 23. Burke M. C., Oei M. S., Edwards N. J., Ostrand-Rosenberg S., and Fenselau C. (2014) Ubiquitinated proteins in exosomes secreted by myeloid-derived suppressor cells. J. Proteome Res. 13, 5965–5972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Buschow S. I., Liefhebber J. M., Wubbolts R., and Stoorvogel W. (2005) Exosomes contain ubiquitinated proteins. Blood Cells Molecules Diseases 35, 398–403 [DOI] [PubMed] [Google Scholar]
- 25. Mageswaran S. K., Dixon M. G., Curtiss M., Keener J. P., and Babst M. (2014) Binding to any ESCRT can mediate ubiquitin-independent cargo sorting. Traffic 15, 212–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mosesson Y., Shtiegman K., Katz M., Zwang Y., Vereb G., Szollosi J., and Yarden Y. (2003) Endocytosis of receptor tyrosine kinases is driven by monoubiquitylation, not polyubiquitylation. J. Biolog. Chem. 278, 21323–21326 [DOI] [PubMed] [Google Scholar]
- 27. Traub L. M., and Lukacs G. L. (2007) Decoding ubiquitin sorting signals for clathrin-dependent endocytosis by CLASPs. J. Cell Sci. 120, 543–553 [DOI] [PubMed] [Google Scholar]
- 28. Huang F., Zeng X., Kim W., Balasubramani M., Fortian A., Gygi S. P., Yates N. A., and Sorkin A. (2013) Lysine 63-linked polyubiquitination is required for EGF receptor degradation. Proc. Natl. Acad. Sci. U.S.A. 110, 15722–15727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Erpapazoglou Z., Dhaoui M., Pantazopoulou M., Giordano F., Mari M., Léon S., Raposo G., Reggiori F., and Haguenauer-Tsapis R. (2012) A dual role for K63-linked ubiquitin chains in multivesicular body biogenesis and cargo sorting. Mol. Biol. Cell 23, 2170–2183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wagner S. A., Beli P., Weinert B. T., Nielsen M. L., Cox J., Mann M., and Choudhary C. (2011) A proteome-wide, quantitative survey of in vivo ubiquitylation sites reveals widespread regulatory roles. Mol. Cell. Proteomics 10, M111 013284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Danielsen J. M., Sylvestersen K. B., Bekker-Jensen S., Szklarczyk D., Poulsen J. W., Horn H., Jensen L. J., Mailand N., and Nielsen M. L. (2011) Mass spectrometric analysis of lysine ubiquitylation reveals promiscuity at site level. Mol. Cell Proteomics 10, M110.003590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wagner S. A., Beli P., Weinert B. T., Schölz C., Kelstrup C. D., Young C., Nielsen M. L., Olsen J. V., Brakebusch C., and Choudhary C. (2012) Proteomic analyses reveal divergent ubiquitylation site patterns in murine tissues. Mol. Cell Proteomics 11, 1578–1585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Radivojac P., Vacic V., Haynes C., Cocklin R. R., Mohan A., Heyen J. W., Goebl M. G., and Iakoucheva L. M. (2010) Identification, analysis, and prediction of protein ubiquitination sites. Proteins 78, 365–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jadhav T., and Wooten M. W. (2009) Defining an Embedded Code for Protein Ubiquitination. J. Proteomics Bioinform. 2, 316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Danielsen J. M., Sylvestersen K. B., Bekker-Jensen S., Szklarczyk D., Poulsen J. W., Horn H., Jensen L. J., Mailand N., and Nielsen M. L. (2011) Mass spectrometric analysis of lysine ubiquitylation reveals promiscuity at site level. Mol. Cell. Proteomics 10, M110 003590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Denis N. J., Vasilescu J., Lambert J. P., Smith J. C., and Figeys D. (2007) Tryptic digestion of ubiquitin standards reveals an improved strategy for identifying ubiquitinated proteins by mass spectrometry. Proteomics 7, 868–874 [DOI] [PubMed] [Google Scholar]
- 37. Stuffers S., Sem Wegner C., Stenmark H., and Brech A. (2009) Multivesicular endosome biogenesis in the absence of ESCRTs. Traffic 10, 925–937 [DOI] [PubMed] [Google Scholar]
- 38. Woodman P. G., and Futter C. E. (2008) Multivesicular bodies: Co-ordinated progression to maturity. Curr. Opin. Cell Biol. 20, 408–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Edgar J. R., Eden E. R., and Futter C. E. (2014) Hrs- and CD63-dependent competing mechanisms make different sized endosomal intraluminal vesicles. Traffic 15, 197–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Miranda K. C., Bond D. T., McKee M., Skog J., Păunescu T. G., Da Silva N., Brown D., and Russo L. M. (2010) Nucleic acids within urinary exosomes/microvesicles are potential biomarkers for renal disease. Kidney Int. 78, 191–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gildea J. J., Carlson J. M., Schoeffel C. D., Carey R. M., and Felder R. A. (2013) Urinary exosome miRNome analysis and its applications to salt sensitivity of blood pressure. Clin. Biochem. 46, 1131–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Cheng L., Sun X., Scicluna B. J., Coleman B. M., and Hill A. F. (2014) Characterization and deep sequencing analysis of exosomal and non-exosomal miRNA in human urine. Kidney Int. 86, 433–444 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.