Abstract
Retinal vascular hyperpermeability causes macular edema, leading to visual deterioration in retinal diseases such as diabetic retinopathy and retinal vascular occlusion. Dysregulation of junction integrity between endothelial cells by vascular endothelial growth factor (VEGF) was shown to cause retinal vascular hyperpermeability. Accordingly, anti-VEGF agents have been used to treat retinal vascular hyperpermeability. However, they can confer potential toxicity through their deleterious effects on maintenance and survival of neuronal and endothelial cells in the retina. Thus, it is important to identify novel therapeutic targets for retinal vascular hyperpermeability other than VEGF. Here, we prepared murine retinas showing VEGF-induced vascular leakage from superficial retinal vascular plexus and prevention of VEGF-induced leakage by anti-VEGF antibody treatment. We then performed comprehensive proteome profiling of these samples and identified retinal proteins for which abundances were differentially expressed by VEGF, but such alterations were inhibited by anti-VEGF antibody. Functional enrichment and network analyses of these proteins revealed the β2 integrin pathway, which can prevent dysregulation of junction integrity between endothelial cells through cytoskeletal rearrangement, as a potential therapeutic target for retinal vascular hyperpermeability. Finally, we experimentally demonstrated that inhibition of the β2 integrin pathway salvaged VEGF-induced retinal vascular hyperpermeability, supporting its validity as an alternative therapeutic target to anti-VEGF agents.
Macular edema resulting from increased vascular permeability leads to visual deterioration in a broad spectrum of retinal diseases, including diabetic retinopathy and retinal vein occlusion (1, 2). In vitreous samples from patients with macular edema, the concentrations of vascular endothelial growth factor (VEGF) were reported to be higher than those of normal controls (3, 4). A number of in vitro and in vivo studies have shown that VEGF, also known as vascular permeability factor, caused loss of tightness between endothelial cells and resultant vascular leakage (5–7). In monkeys, intravitreal injection of bioactive VEGF led to fluorescein leakage from retinal vessels (5). Furthermore, VEGF was shown to affect the integrity of tight junction complexes along endothelial cell-cell interfaces (6, 7). These data indicate that VEGF plays a major role in the pathogenesis of retinal vascular hyperpermeability by dysregulating tight junction integrity between endothelial cells in the retinal vascular system (8, 9).
It has been observed in the clinic that anti-VEGF agents have therapeutic potential to treat retinal vascular hyperpermeability induced by VEGF (8, 9). However, VEGF not only promotes vascular permeability, but also plays an important role in the survival of normal endothelial and neuronal cells in the retina (10, 11). Furthermore, VEGF affects various downstream intracellular molecular pathways that are associated with the maintenance of retinal endothelial cells (9, 12). These data collectively suggest potential toxicity of anti-VEGF agents when they were used to treat retinal vascular hyperpermeability. In this context, it is important to identify novel therapeutic targets other than VEGF to treat VEGF-induced retinal vascular hyperpermeability.
Here, in search of new therapeutic targets, we first prepared mouse retina that exhibited VEGF-induced vascular leakage from the superficial vascular plexus and effective phenotypical prevention of vascular leakage by anti-VEGF antibody, as described previously (13, 14). We then performed comprehensive proteome profiling of the retinas treated with VEGF and with VEGF plus anti-VEGF antibody by liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis. Using the isobaric tag for relative and absolute quantitation (iTRAQ)1 data obtained from LC-MS/MS analyses, we identified retinal proteins whose abundances were altered by treatment of VEGF, but the alterations were inhibited by cotreatment of anti-VEGF antibody with VEGF. Functional enrichment and network analyses of these proteins suggested the β2 integrin pathway regulating cytoskeletal rearrangement of endothelial cells as a potential therapeutic target for VEGF-induced retinal vascular hyperpermeability. Both in vitro immunoassay and in vivo experiments confirmed that the modulation of the β2 integrin pathway inhibited VEGF-induced retinal vascular hyperpermeability, suggesting its validity as a therapeutic target other than VEGF.
EXPERIMENTAL PROCEDURES
Mice
6-Week-old male C57BL/6 mice (Central Laboratory Animal, Seoul, Republic of Korea) were used in this study. All animal procedures were approved by Institutional Animal Care and Use Committee of Seoul National University and conducted in agreement with the Association for Research in Vision and Ophthalmology statement for the use of animals in ophthalmic and vision research.
VEGF-induced Retinal Vascular Hyperpermeability
To induce retinal vascular hyperpermeability, we injected recombinant mouse VEGF164 (100 ng/1.5 μl; catalog no. 493-MV-005, R&D Systems, Minneapolis, MN) into the vitreous cavity of the right eyes of mice (VEGF condition). For control samples, phosphate-buffered saline (PBS) (1.5 μl) was injected (control condition). To show the effects of VEGF scavenging, we injected both recombinant mouse VEGF164 (100 ng) and affinity-purified polyclonal antibody against mouse VEGF164 (1 μg; catalog no. AF-493-NA, R&D Systems; source, goat IgG) in 1.5 μl of PBS (anti-VEGF condition). To analyze therapeutic effects of β2 integrin antagonism, we injected both recombinant mouse VEGF164 (100 ng) and monoclonal antibody against mouse β2 integrin with neutralizing effects of β2 integrin activity (1 μg; catalog no. MA1-10122, Thermo, Waltham, MA; source, rat IgG) in 1.5 μl of PBS. To evaluate the effect of the injection of VEGF, VEGF plus anti-VEGF antibody, or VEGF plus anti-β2 integrin antibody on retinal vascular hyperpermeability, and the levels of proteins measured, the retinas were prepared at 24 h after the injection. At 24 h after the injection, enucleation was performed after deep anesthesia and subsequent sacrifice with CO2 inhalation. To demonstrate vascular integrity, intracardiac injection of fluorescein isothiocyanate-dextran (FITC-dextran, catalog no. 46945, Sigma, St. Louis, MO) was performed 1 h prior to sacrifice with CO2 inhalation. Retinal flat mounts were observed under the fluorescence microscope (Eclipse 80i, Nikon, Tokyo, Japan). Quantitative analyses of mean green fluorescence intensity were performed using four randomly selected images of paracentral areas of retinas from ×200 magnification photographs per each eye with ImageJ software (National Institutes of Health, Bethesda, MD) (n = 3).
Sample Preparation
In each of the three conditions (control, VEGF, and anti-VEGF), we obtained four samples (n = 4). In VEGF or anti-VEGF condition, we obtained four samples after treatment of VEGF or VEGF plus anti-VEGF antibody, respectively. For each sample in the three conditions, we collected the retinas from six mice (six retinas per sample) into the microcentrifuge tube to minimize the effect of interindividual variability in mice. From each of the six mice, enucleation and subsequent sacrifice were performed as described above. Enucleated eyes were immediately put into cold PBS. Under the stereomicroscope (Leica, Wetzlar, Germany), the cornea was dissected, and the lens was removed to facilitate retinal preparation. By gentle pressure on the sclera with forceps, the retinas were isolated and put into clean microcentrifuge tubes, which were prepared in ice. In total, the 12 samples (n = 4 per condition) were stored at −80 °C before further preparation.
Protein Extraction and Digestion
20 mg of each sample in the three conditions (control, VEGF, and anti-VEGF) was cryo-pulverized by Covaris CP02 Prep (Covaris, Woburn, MA). Briefly, the retinas were transferred to a Covaris tissue bag (Covaris, TT1, 520007), and the tissue bag was placed in liquid nitrogen for 30 s. Immediately after the freezing, the tissue bag was placed in the impact chamber of the cryo-pulverizer and pulverized by using impact level 2 (tissue weight <50 mg, impact level 2). The cryo-pulverized retinas were transferred into a siliconized low-retention microcentrifuge tube (Thermo) and mixed with lysis buffer (4% SDS in 0.1 m Tris-HCl, pH 7.6, containing a Complete Mini Protease inhibitor tablet (Roche Applied Science, Basel, Switzerland)), followed by tissue lysis using a handheld sonicator (Q55, QSONICA, Newtown, CT) for 30 s (at 30 watts) on ice. The tissue lysate was centrifuged at 16,000 × g and 20 °C for 10 min, and the supernatant was then transferred to a new tube. Protein concentration of the supernatant was determined by BCA assay (Pierce, Waltham, MA).
For each sample in the three conditions, the retinal proteins were divided into 500-μg protein units, and each 500-μg protein unit was digested using a slightly modified version of filter-aided sample preparation digestion method (15). Briefly, the proteins were reduced with SDT buffer (4% SDS in 0.1 m Tris-HCl, pH 7.6, and 0.1 m DTT) for 45 min at 37 °C and then boiled for 10 min at 95 °C. Subsequently, protein samples were sonicated for 10 min in a bath sonicator (Power Sonic 505, Hwashin Technology, Seoul, Republic of Korea) and centrifuged at 16,000 × g for 5 min. The protein sample was transferred to a membrane filter device (YM-30, Millipore, Billerica, MA) and mixed with 200 μl of 8 m urea in 0.1 m Tris-HCl, pH 8.5. The device was centrifuged at 14,000 × g at 20 °C for 60 min to remove the SDS. This step was repeated three times. Subsequently, proteins were alkylated with 100 μl of 50 mm iodoacetamide in 8 m urea for 25 min at room temperature in the dark and followed by centrifugation at 14,000 × g for 30 min. The filter was washed with 200 μl of 8 m urea four times and then washed with 100 μl of 50 mm NH4HCO3 twice for buffer exchange. Trypsin (Promega, Madison, WI) was added to the proteins at an enzyme to protein ratio of 1:50 (w/w), and the filter device was placed in a thermomixer (Eppendorf, Hamburg, Germany) and incubated at 37 °C overnight. After the first digestion, the second digestion was carried out with additional trypsin (1:100 enzyme to protein ratio) at 37 °C for 6 h. After digestion, the tryptic peptides were eluted by centrifugation at 14,000 × g at 20 °C for 30 min. After collecting the tryptic peptides, the filter were rinsed with 60 μl of 50 mm NH4HCO3 and centrifuged at 14,000 × g at 20 °C for 20 min, and the eluent was combined with the first eluent. The combined eluent was dried by vacuum centrifugation, and the peptide concentration was determined by BCA assay. The peptide sample was divided into 100-μg units in Eppendorf tubes and kept in −80 °C until the subsequent iTRAQ labeling.
iTRAQ Labeling and Peptide Fractionation
We performed three 4-plex iTRAQ labeling experiments (Fig. 1A). For each experiment, we used one sample from the three conditions (control, VEGF, and anti-VEGF) for 114, 115, and 116 channels, respectively, and an additional sample from one of the three conditions for the 117 channel (i.e. experimental sets 1–3 with additional samples from control, VEGF, anti-VEGF conditions, respectively). A total of 800 μg of peptides (200 μg of peptides per channel) from each experimental set were labeled with two units of 4-plex iTRAQ reagent (AB Sciex, Framingham, MA) according to the manufacturer's instructions. After the iTRAQ labeling, all iTRAQ peptides were pooled and immediately subjected to a mid-pH reverse-phase fractionation, as described previously (16).
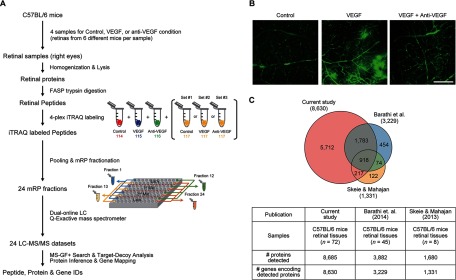
Fig. 1.
Comprehensive proteome profiling of retina tissues. A, overall scheme describing sample preparation, proteome profiling, and data analysis. See text for detailed description of each experimental step (“Experimental Procedures” and “Results”). B, VEGF-induced retinal vascular hyperpermeability. Intravitreally injected VEGF (100 ng)-induced vascular leakage of the superficial vascular plexus in the retina of C57BL/6 mice. Cotreatment of VEGF with anti-VEGF antibody (1 μg) effectively prevented the phenotype of increased permeability demonstrated by the leakage of FITC-dextran. Scale bar, 200 μm. C, comparison of our retinal proteome with two previously reported proteomes associated with VEGF-induced vascular permeability. The Venn diagram shows the relationships between the three retinal proteomes. The numbers of the detected proteins (see “Results” for the detection criteria) in the three studies and the genes encoding the proteins are shown in the table.
800 μg of iTRAQ-labeled tryptic peptides was separated using Agilent 1260 Infinity HPLC system (Agilent, Santa Clara, CA) equipped with an Xbridge C18 guard column (4.6 × 20 mm, 130 Å, 5 μm) and an analytical column (4.6 × 250 mm, 130 Å, 5 μm). Mid-pH reverse-phase liquid chromatography was performed at a flow rate of 0.5 ml/min during the 130-min gradient using solvent A (10 mm triethylammonium bicarbonate (TEAB) in water, pH 7.5) and solvent B (10 mm TEAB in 90% ACN, pH 7.5). The gradient used is as follows: 0% solvent B for 10 min, 0–5% solvent B in 10 min, 5–35% in 60 min, 35–70% in 15 min, 70% for 10 min, 70–0% in 10 min, and finally held at 0% over 15 min. 96 fractions were collected every minute from 15 to 110 min and were non-contiguously concatenated into 24 fractions by pooling four fractions from each of the early section (fractions 1–24), the first mid-section (fractions 25–48), the second mid-section (fractions 49–72), and the late section (fractions 73–96) of fractions (Fig. 1A). The 24 fractions were dried in a vacuum centrifuge concentrator and stored at −80 °C until LC-MS/MS experiments.
LC-MS/MS Experiments
10 μg of iTRAQ-labeled peptides from each of 24 fractions was individually analyzed by Q Exactive mass spectrometer (Thermo), which was coupled to a dual on-line LC system (17). The dual on-line LC system was equipped with two capillary columns (75-μm inner diameter × 360-μm outer diameter, 100 cm) and two solid-phase extraction columns (150-μm inner diameter × 360-μm outer diameter, 3 cm) that were prepared by slurry packing frit-ended fused silica capillaries with C18 resin (3 μm diameter, 300-Å pore size, Jupiter). A 180-min linear gradient (1–40% solvent B over 160 min, 40–80% over 5 min, 80% for 10 min and holding at 1% for 5 min) was used. Solvent A and B were 0.1% formic acid in water and 0.1% formic acid in ACN, respectively. The column flow rate was 300 nl/min.
The eluting peptides were ionized at the electric potential of 2.4 kV and the desolvation capillary temperature of 250 °C. MS precursor scans (m/z 400–2,000 thomson) were acquired at the resolution of 70,000 with an automated gain control target value of 1.0 × 106 and a maximum ion injection time of 20 ms. The MS/MS data for up to the 10 most abundant ions were acquired in a data-dependent mode using higher energy collisional dissociation at a normalized collision energy of 30 with fixed first mass of 100 thomson at the resolution of 17,500 with automated gain control target value of 1.0 × 106 and a maximum injection time of 60 ms.
LC-MS/MS Data Analysis
Each of the three iTRAQ datasets includes 24 MS/MS datasets for 24 fractions. For each MS/MS dataset, post-experiment monoisotopic mass refinement method was used to process the MS/MS data, which was previously demonstrated to accurately assign precursor mass to the tandem mass spectrometric data (18). The resultant MS/MS data from post-experiment monoisotopic mass refinement process (i.e. mgf files) were subjected to database search using the MS-GF+ search engine (version 9387) (19) against Swiss-Prot mouse reference database (released September, 2013; 24,544 entries). Search parameters were set to the following: precursor mass tolerance of 10 ppm, semi-tryptic, static modifications of carbamidomethylation (+57.0214 Da) to cysteine and iTRAQ (+144.102063 Da) to N termini and lysine, and variable modification of oxidation (+15.994915 Da) to methionine. The search results from the 24 MS/MS datasets were combined. The peptide-spectrum matches (PSMs) at the false discovery rate (FDR) of 1% were obtained by the target-decoy method built in the MS-GF+ search engine (19). The identified peptides with PSM-level FDR <0.01 from the three iTRAQ datasets were combined to generate an alignment table in which the identified peptides from different iTRAQ datasets with the same sequences were matched. The identified peptides in the alignment table were used to infer protein groups by a bipartite graph analysis (21) using an in-house software (22). Finally, we selected protein groups that have protein-level FDR ≤1% and more than two detected non-redundant peptides. Global protein-level FDR was estimated in the target-decoy setting as described previously (20). The representative protein of protein group was selected as the protein with the highest number of peptides. When more of two proteins in one group had the same number of peptides, the protein with the higher sequence coverage was selected as the representative protein, and the protein containing a unique peptide also was selected as the representative protein. Protein groups of two or more peptide hits were used for further analyses. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (23) partner repository with the dataset identifier PXD003656 and 10.6019/PXD003656.
Identification of Differentially Expressed Proteins (DEPs)
Of the identified peptides, we first selected the unique peptides that were detected in two or more of the three iTRAQ experiments. The reporter ion intensities of these selected unique peptides in the alignment table were normalized by the medians of control samples in the corresponding iTRAQ experiment to remove the batch effects in iTRAQ experiments. Using the normalized intensities, the previously reported integrative statistical method (24) was performed to identify differentially expressed peptides in the two comparisons: 1) VEGF versus control (VEGF/control) and 2) anti-VEGF versus VEGF (anti-VEGF/VEGF). Briefly, two sample t tests (e.g. four samples in VEGF versus four samples in control) and log2-median ratio test were applied to calculate T values and log2-median ratios for the selected peptides. To compute p values of selected peptides for the individual tests, empirical distributions of T values and log2-median ratios for the null hypothesis (i.e. a peptide is not differentially expressed) were estimated by performing all possible random permutations of the samples and then by applying the Gaussian kernel density estimation method to T values, and log2-median ratios resulted from the random permutations (25). For each peptide, the adjusted p values from the two tests were computed by the two-sided test using the corresponding empirical null distributions and then combined into an overall p value using Stouffer's method (26). Next, FDRs were estimated for the overall p values using the Storey method (27). The differentially expressed peptides were selected as the ones with FDR ≤0.05 and absolute log2 fold-changes ≥0.58 (1.5-fold). Finally, we selected a set of DEPs that have at least two differentially expressed peptides.
Enrichment Analysis of Gene Ontology Biological Processes (GOBPs)
Functional enrichment analysis was performed using DAVID software to identify the GOBPs represented by the DEPs in the two major clusters (clusters 1 and 2 in Fig. 2B) (28). The GOBPs represented by the DEPs were identified as the ones with p ≤ 0.1.
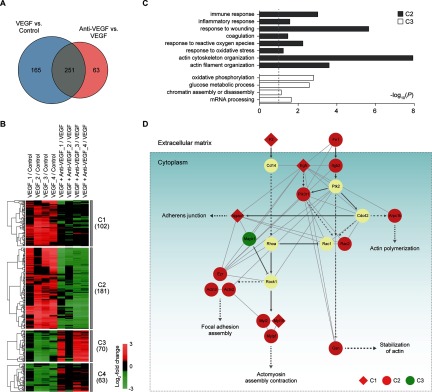
Fig. 2.
Retinal proteomes affected by VEGF and anti-VEGF antibody. A, relationships of the DEPs from the two comparisons (VEGF versus control and VEGF plus anti-VEGF versus VEGF). B, four major clusters (clusters 1–4) of the DEPs identified from the two comparisons. For each DEP in the two comparisons, the log2 fold-changes were calculated as the median ratios of the normalized reporter ion intensities of the differentially expressed peptides for the protein in individual samples (see also supplemental Fig. S2). The log2 fold-changes of the DEP were normalized by the median value of four replicates in control for VEGF versus control (VEGF/control, 1st to 4th columns) or in VEGF for anti-VEGF versus VEGF (anti-VEGF/VEGF, 5th to 8th columns). The underscored number followed by the condition indicates the replicate sample in the condition (e.g. VEGF_2 indicates the 2nd replicate sample under VEGF condition). The dendrogram shows how the DEPs in each cluster were clustered using a hierarchical clustering method (complete linkage and Euclidian distance). Color bar, gradient of log2 fold-changes between the two conditions indicated in each column. C, GOBPs represented by the DEPs in clusters 2 and 3 (gray and white, respectively). The bars represent −log10(P), where P is the significance of individual GOBPs being enriched by the DEPs, and the dotted line represents the cutoff of p value used (a default cutoff of DAVID). D, network model describing the DEPs associated with the regulation of actin cytoskeleton in the four major clusters (clusters 1–4). Node colors represent up-regulation (red) or down-regulation (green) of the corresponding proteins by VEGF treatment, and node shapes represent whether the alterations of the corresponding proteins by VEGF treatment were inhibited (circle, cluster 2) or not (diamond, cluster 1) by cotreatment of VEGF and anti-VEGF antibody. The arrow and inhibition symbol represent activation and suppression between the proteins obtained from the KEGG pathway database. Solid and dotted lines represent direct and indirect protein-protein interactions between the nodes, respectively, in cytoplasm (box) and extracellular matrix.
Fibronectin Enzyme-linked Immunosorbent Assay (ELISA)
The level of fibronectin in retinal proteins, which were extracted using RIPA buffer, was estimated using mouse fibronectin ELISA kit (catalog no. ab108849, Abcam, Cambridge, United Kingdom) according to the manufacturer's instructions (n = 3).
Western Blot Analysis
An equal amount (50 μg) of retinal proteins, which were extracted using RIPA buffer, was separated by SDS-PAGE and transferred to a nitrocellulose membrane. The membrane was incubated with primary antibodies overnight at 4 °C. The list of primary antibodies utilized in this study is as follows: anti-β2 integrin (catalog no. sc-8420, Santa Cruz Biotechnology, Dallas, TX); anti-gelsolin (catalog no. 12953, Cell Signaling, Danvers, MA); anti-CD14 (catalog no. MAB982-SP, R&D Systems); anti-RhoA (catalog no. 2117, Cell Signaling); and anti-β-actin (catalog no. A2668, Sigma). Species-specific secondary antibodies (Pierce) were treated on the membrane for 1 h at room temperature. The visualization of bands was performed using ECL solution (Daeillab, Seoul, Republic of Korea) and ImageQuant LAS4000 system and accompanying analysis software (GE Healthcare, Chicago, IL).
Immunocytochemistry
Human retinal microvascular endothelial cells (HRMECs) were cultured in EBM-2 Basal Medium supplemented with 1% penicillin/streptomycin (Gibco, Waltham, MA) and EGM-2 SingleQuotTM kit supplement and growth factors. The following treatments were performed using EBM-2 Basal Medium supplemented with 1% penicillin/streptomycin and 1% fetal bovine serum (FBS). Confluent HRMECs on fibronectin (50 μg/ml; catalog no. F2006, Sigma)-coated dish were treated with VEGF (20 ng/ml; catalog no. 8065, Cell Signaling) or VEGF plus affinity-purified polyclonal antibody against human β2 integrin (1 μg/ml; catalog no. AF1730, R&D Systems). The concentration of anti-β2 integrin was chosen not to induce changes in cell adhesiveness on dish. 30 min after the treatment, cells were fixed with 4% paraformaldehyde for 15 min at room temperature and then treated with 0.2% Triton X-100 for 10 min at room temperature. To minimize nonspecific binding, blocking with 3% bovine serum albumin was performed for 10 min at room temperature. After incubation with Alexa Fluor® 488 phalloidin (catalog no. A12379, Life Technologies, Inc., Waltham, MA) overnight at 4 °C, nuclear staining was performed with 4′,6-diamidino-2-phenylindole (catalog no. D1306, Invitrogen, Waltham, MA) for 10 min at room temperature. Cells were observed using the inverted fluorescence microscope (Leica).
Real Time Polymerase Chain Reaction (RT-PCR)
Confluent HRMECs on thrombin (4 IU/ml; catalog no. T9549, Sigma)- or fibronectin (50 μg/ml)-coated dish were treated with VEGF (20 ng/ml) in EBM-2 basal medium supplemented with 1% penicillin/streptomycin and 1% FBS after a 6-h starvation. Cells were harvested from the dish 6 h after the treatment, and total RNA was isolated from cells using TRI Reagent (Molecular Research Center, Cincinnati, OH) according to the manufacturer's instruction. The cDNA was then prepared with High Capacity RNA-to-cDNA kit (Life Technologies, Inc.). Real time PCR was performed with TaqMan® fast advanced master mix (Life Technologies, Inc.) and specific gene expression assays (catalog no. 4453320; Life Technologies, Inc.). Product IDs of Gene Expression Assays for genes are as follows: CD14, Hs02621496_s1; ITGB2, Hs00164957_m1; and GAPDH, Hs99999905_m1. All analyses were done using StepOnePlus RT-PCR System (Life Technologies, Inc.) and accompanying StepOne Software (version 2.2). All procedures were performed in accordance with the MIQE guidelines.
Phospho-focal Adhesion Kinase (FAK) and FAK ELISA
Confluent HRMECs on the fibronectin (50 μg/ml; catalog no. F2006, Sigma)-coated dish were treated with VEGF (20 ng/ml; catalog no. 8065, Cell Signaling) or VEGF plus affinity-purified polyclonal antibody against human β2 integrin (1 μg/ml; catalog no. AF1730, R&D Systems). 10 min after the treatment, cells were isolated, and extracted proteins were utilized to measure the levels of phospho-FAK and FAK with human phospho-FAK (Tyr397) and total FAK ELISA (catalog no. PEL-FAK-Y397-T, RayBiotech, Norcross, GA) according to the manufacturer's instructions.
Statistics
Differences among control and treatment conditions were assessed using Mann-Whitney U test or Kruskal-Wallis test (non-parametric analysis of variance) with post hoc Dunn's multiple comparison test. All statistical analyses were performed using GraphPad Prism 5 (version 5.01, GraphPad, La Jolla, CA). The mean ± S.E. is shown in the figures.
RESULTS
VEGF-induced Vascular Hyperpermeability
To investigate retinal proteins affected by VEGF or VEGF plus anti-VEGF antibody, we first prepared four samples (n = 4) in the three conditions with the following intravitreal injection (Fig. 1A, Retinal samples): 1) control condition with treatment of PBS (1.5 μl); 2) VEGF condition with treatment of recombinant mouse VEGF164 (100 ng per eye); and 3) anti-VEGF condition with treatment of both VEGF (100 ng per eye) and anti-VEGF antibody (1 μg per eye). For each of the four samples in the three conditions, the retinas were obtained from six different mice after the intravitreal injection and then combined (Fig. 1A, Retinal samples). We confirmed that VEGF treatment induced vascular leakage from the superficial vascular plexus in the retina, and treatment of VEGF plus anti-VEGF antibody effectively suppressed the effects of VEGF in whole mount retinas after systemic circulation of FITC-dextran (Fig. 1B), consistent with the previous findings (13, 14).
Comprehensive Quantitative Retinal Proteome Profiling
For each of four samples in the three conditions (control, VEGF, and anti-VEGF), we next extracted proteins and then applied the filter-aided sample preparation method (15) to the extracted proteins for protein digestion (Fig. 1A, Retinal Peptides). This resulted in a total of 12 peptide samples for the three conditions (four per condition). For the 12 peptide samples, we then performed three different sets of 4-plex iTRAQ labeling (Fig. 1A, and see “iTRAQ Labeling and Peptide Fractionation” under “Experimental Procedures”). For each set of the iTRAQ-labeled peptides, we next used a mid-pH reverse-phase liquid chromatography fractionation (mRP fractionation) from which the initial 96 fractions of iTRAQ-labeled peptides were non-contiguously pooled into 24 fractions (Fig. 1A, 24 mRP fractions; see under “Experimental Procedures”). LC-MS/MS analysis was performed on each of the 24 fractions, resulting in 24 MS/MS datasets (Fig. 1A, 24 LC-MS/MS datasets). For the three sets of iTRAQ-labeled peptides (12 samples for the three conditions), we generated a total of 72 MS/MS datasets. For each set of iTRAQ-labeled peptides (24 MS/MS datasets), we then identified the peptides using the Swiss-Prot mouse reference protein sequence database in the target-decoy setting by the MS-GF+ (version 9387) search engine (19).
From the three experimental sets, we identified a total of 205,730 non-redundant peptides with a PSM-level FDR <0.01 (supplemental Table S1) and 8,685 proteins that have protein-level FDR <0.01 and also more than two different non-redundant peptides (Fig. 1C and supplemental Table S2). The 8,685 proteins were mapped into 8,630 genes. Previously, a number of studies have reported retinal proteomes whose sizes were in the range of 896 to 3,882 (Fig. 1C) (29–32). For example, Skeie and Mahajan et al. (29) also analyzed retinal tissues of C57BL/6 mice and then identified 1,680 proteins (1,331 genes) with protein-level false positive rate <0.01. Also, Barathi et al. (31) analyzed retinal tissues of C57BL/6 mice and then identified 3,882 proteins (3,229 genes) with protein-level FDR <0.01 and more than two non-redundant peptides. Compared with these retinal proteomes using the same cutoffs (PSM and protein FDRs <0.01 and two distinct peptides), our profiling provided the most comprehensive retinal proteome that can be used to identify the proteins associated with increased vascular permeability. The large proteome size could be ascribed mainly to the effective mRP fractionation combined with the high resolution peptide separation by ultra-high pressure LC utilizing long gradients (i.e. 180 min for each fraction) on long capillary columns (i.e. 100 cm long; see supplemental Fig. S1).
Retinal proteins affected by VEGF and anti-VEGF antibody
To identify the proteins associated with VEGF-induced vascular hyperpermeability, we first quantified relative abundances of the identified proteins between control and VEGF-treated samples (VEGF versus control) and also between VEGF plus anti-VEGF antibody- and VEGF-treated samples (anti-VEGF versus VEGF) using the iTRAQ intensities of the peptides. We then identified a total of 479 DEPs from the two comparisons (Fig. 2A and supplemental Table S3) as described under “Experimental Procedures”: 416 DEPs (283 up-regulated and 133 down-regulated) in VEGF-treated samples, compared with the control samples (VEGF versus control); and 314 DEPs (86 up-regulated and 228 down-regulated) in the VEGF plus anti-VEGF-treated samples, compared with VEGF-treated samples (anti-VEGF versus VEGF).
Of the 479 DEPs, 251 (52.4%) were shared between the two comparisons, and the remaining DEPs were uniquely identified in the individual comparisons. To explore the relationships between the DEPs from the two comparisons, the DEPs were categorized into six clusters (clusters 1–6) based on their differential expression in the two comparisons (supplemental Table S4). Of them, we focused on four major clusters (clusters 1–4) that include more than 10% of the total number of the DEPs (Fig. 2B; supplemental Fig. S2). Cluster 2 (181 proteins) and cluster 3 (70 proteins) showed up- and down-regulation in their abundances by VEGF, respectively, but such alterations were inhibited by cotreatment of VEGF with anti-VEGF antibody. Clusters 1 and 4 showed up- and down-regulation by VEGF, respectively, but inhibition of the alterations by cotreatment with anti-VEGF antibody was not statistically significant (see “Discussion”).
In this study, we aimed to identify alternative therapeutic targets to anti-VEGF antibody for VEGF-induced retinal vascular hyperpermeability. Clusters 2 and 3 represent the inhibition of VEGF-induced retinal vascular hyperpermeability by anti-VEGF antibody. Thus, we focused on these two clusters because they are likely to include the molecules governing the inhibition by anti-VEGF antibody. To understand cellular processes represented by these two clusters, we performed the enrichment analysis of GOBPs for the DEPs in clusters 2 and 3 using DAVID software. Interestingly, cluster 2 was associated with the processes related to vascular permeability (response to reactive oxygen species and actin cytoskeleton/filament organization) and also to immune responses (immune response, inflammatory response, response to wounding, and coagulation) (Fig. 2C and supplemental Table S5). Cluster 3 was associated with the processes related to metabolic processes (oxidative phosphorylation and glucose metabolic process) and transcription-related processes (chromatin assembly or disassembly and mRNA processing) (Fig. 2C and supplemental Table S5). These data indicate that cluster 2 is more relevant to retinal vascular hyperpermeability or leakage that was induced by VEGF but was prevented by cotreatment with anti-VEGF antibody (Fig. 1B).
Retinal Proteome Profile Representing VEGF-induced Vascular Hyperpermeability
VEGF-induced vascular hyperpermeability and leakage have been shown to be associated with a loss of tight junction integrity between endothelial cells in the retina (33–35). The tight junction complexes consist of proteins spanning the intercellular cleft, such as occludin and claudin, and junctional adhesion molecules (33–35). Interestingly, among the processes represented by cluster 2 (Fig. 2C), actin cytoskeleton organization is closely linked to the tight junction integrity of endothelial cells because the actin cytoskeleton is connected with transmembrane proteins through zonula occludens proteins and cingulin in the cytoplasm (33, 34). Furthermore, actin cytoskeleton modulates adherens junction dynamics through its binding to the vascular endothelial cadherin (12). These data suggest that actin cytoskeleton organization is potentially linked to the VEGF-induced vascular hyperpermeability through its association with tight junction integrity of endothelial cells. The following 11 DEPs in cluster 2 can serve as a retinal proteome profile associated with the actin cytoskeleton or filament organization and thus VEGF-induced vascular hyperpermeability (supplemental Table S5): fibronectin 1 (FN1); β2 integrin (ITGB2); RAS-related C3 botulinum substrate 2 (RAC2); gelsolin (GSN); myosin light chain 2 and myosin light chain; phosphorylatable, fast skeletal muscle (MYL2/MYLPF); phosphatidylinositol 3-kinase, regulatory subunit, polypeptide 1 (PIK3R1); ezrin (EZR); actin related protein 2/3 complex, subunit 1B (ARPC1B), and actinin α/3 (ACTN2/3).
To understand functional associations among the DEPs involved in actin cytoskeleton and filament organization in clusters 1–4, which include the 11 DEPs in cluster 2, we reconstructed a network model describing the interactions among them based on the Kyoto Encyclopedia of Genes and Genomes pathway (Fig. 2D). The network model showed activation of F2-CD14 and FN1-β2 integrin pathways by VEGF, which could lead to the alteration in actin cytoskeleton and cellular junction through their links to actin cytoskeleton (actin polymerization, stabilization of actin, and actomyosin assembly contraction) and cellular junction (adherens junction and focal adhesion assembly). In the FN1-β2 integrin pathway, FN1, β2 integrin, and RAC2, a downstream molecule, were up-regulated by VEGF treatment, but the alterations were inhibited by cotreatment of VEGF and anti-VEGF antibody (cluster 2), indicating that the activation of the FN1-β2 integrin pathway by VEGF was inhibited by the anti-VEGF antibody. In contrast, in the F2-CD14 pathway, F2 was up-regulated by VEGF treatment (cluster 1), suggesting potential activation of the F2-CD14 pathway, but its downstream molecules (CD14 and ROHA) showed no significant changes by treatment of VEGF or VEGF plus anti-VEGF antibody, indicating that the activation of the F2-CD14 pathway was not inhibited by anti-VEGF antibody.
To confirm this finding, we examined the activities of the two pathways in HRMECs, which are the major players that undergo dysregulation of tight junction integrity by VEGF, leading to breakdown of inner blood-retinal barrier and eventually to retinal vascular hyperpermeability (34). To this end, we measured VEGF-induced mRNA expression of CD14 and β2 integrin genes in confluent HRMECs on F2 and FN1-coated dish, respectively, using RT-PCR analysis (see under “Experimental Procedures”). Consistent with the LC-MS/MS data, ITGB2 showed the significant (>2-fold) up-regulation in HRMECs, whereas CD14 showed no definite alteration in the expression (supplemental Fig. S3). Furthermore, we confirmed protein expression of two downstream proteins (CD14 and RhoA) of the F2-CD14 pathway using Western blotting (supplemental Figs. S4 and S5). Consistent with the LC-MS/MS data, these two proteins showed no significant alterations by treatments of VEGF and VEGF plus anti-VEGF antibody. All these data indicated that the FN1-β2 integrin pathway, rather than the F2-CD14 pathway, was activated as a primary pathway by VEGF to regulate the actin cytoskeleton organization in retinal microvascular endothelial cells, and its activation was inhibited by anti-VEGF antibody.
Inhibition of VEGF-induced Vascular Leakage by Targeting β2 Integrin
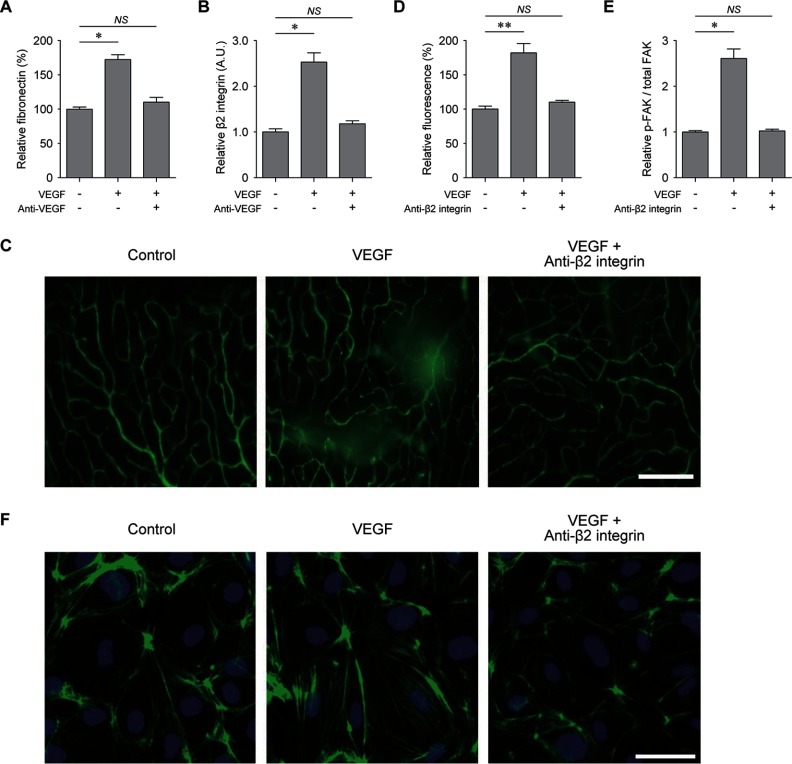
The above data suggest that the FN1-β2 integrin pathway can control VEGF-induced retinal vascular hyperpermeability through regulation of the actin cytoskeleton organization. To test this hypothesis, we first verified the differential expression of FN1 and β2 integrin using the ELISA and Western blotting, respectively. These independent assays showed the consistent changes of FN1 (Fig. 3A) and β2 integrin (Fig. 3B and supplemental Fig. S4) in their abundances after treatments of VEGF and VEGF plus anti-VEGF antibody to those measured by LC-MS/MS analyses. Also, we verified the differential expression of GSN, one of the downstream molecules of the FN1-β2 integrin pathway, and we found that it showed the consistent changes to those measured by LC-MS/MS analysis (supplemental Figs. S4 and S5). Next, we further examined the effect of β2 integrin on VEGF-induced retinal vascular hyperpermeability. To this end, we inhibited the function of β2 integrin by treating VEGF and anti-β2 integrin antibody together and analyzed the VEGF-induced vascular leakage from the superficial vascular plexus in the retina. Interestingly, anti-β2 integrin antibody effectively suppressed the VEGF-induced retinal vascular permeability, similar to anti-VEGF antibody (Fig. 1B), as indicated by reduced leakage of FITC-dextran (Fig. 3, C and D). Furthermore, we examined activation of FAK, one of key downstream molecules in the FN1-β2 integrin pathway. To this end, we cultured confluent HRMECs on fibronectin-coated dish and treated them with VEGF or VEGF plus anti-β2 integrin antibody. The data showed that phosphorylation of FAK at Tyr-397 was increased by VEGF, but such alteration was inhibited by anti-β2 integrin antibody (Fig. 3E). Also, we further found that the anti-β2 integrin antibody significantly reduced actin cytoskeleton rearrangement associated with tight junction integrity of retinal endothelial cells (Fig. 3F). Thus, all these data together suggest that β2 integrin antibody rescued the retinal vascular hyperpermeability induced by VEGF through the cytoskeleton rearrangement and also that β2 integrin can serve as an effective therapeutic target for VEGF-induced retinal vascular hyperpermeability.
Fig. 3.
Inhibition of VEGF-induced vascular leakage by anti-β2 integrin antibody. A, levels of fibronectin in the retina measured by ELISA. Anti-VEGF, VEGF plus anti-VEGF antibody (n = 4). B, levels of β2 integrin in the retina measured by Western blotting analysis (n = 3). C, qualitative assessment of retinal vascular integrity demonstrated by the leakage of FITC-dextran after treatments of PBS (control), VEGF (VEGF), and VEGF plus anti-β2 integrin antibody (anti-β2 integrin). D, quantitative analysis of relative fluorescence intensity after treatments of VEGF (VEGF) and VEGF plus anti-β2 integrin antibody (anti-β2 integrin) (n = 4). E, levels of phosphorylated FAK normalized by the total amount of FAK (p-FAK/total FAK) in confluent HRMECs on FN1-coated dish at 5 min after treatments of VEGF (VEGF) and VEGF plus anti-β2 integrin antibody (Anti-β2 integrin) (n = 4). F, actin cytoskeleton arrangement after treatments of PBS (Control), VEGF (VEGF), and VEGF plus anti-β2 integrin antibody (Anti-β2 integrin) demonstrated by immunofluorescence staining of F-actin (green). Nucleus was identified with DAPI staining. NS, p > 0.05; *, p < 0.05; **, p < 0.01 using Kruskal-Wallis test with post hoc Dunn's multiple comparison test.
DISCUSSION
VEGF is a trophic factor for neuronal and endothelial cells in the retina, as well as a driving factor for vascular hyperpermeability (10, 11, 36). Thus, therapeutic targets other than VEGF have been searched to circumvent potential toxicity of anti-VEGF agents. In this study, we identified the β2 integrin pathway as an alternative therapeutic target for VEGF-induced vascular hyperpermeability through the comparative proteome profiling followed by functional enrichment and network analyses of the proteins affected by VEGF and anti-VEGF antibody. We then experimentally verified that the inhibition of the β2 integrin pathway using anti-β2 integrin antibody effectively rescued the VEGF-induced vascular leakage, thereby indicating the validity of the β2 integrin pathway as a therapeutic target.
Vascular hyperpermeability in retinal vasculature is the consequence of breakdown of the inner blood-retinal barrier (BRB), which is composed of retinal endothelial cells (8). The integrity of the blood neural barriers is determined by the components of tight junction complexes in retinal endothelial cells (34). The proteins for which the expression was increased by VEGF treatment, but such alteration was inhibited by cotreatment of VEGF and anti-VEGF antibody (cluster 2 in Fig. 2B), were significantly associated with actin cytoskeleton or filament organization (Fig. 2C). The actin cytoskeleton can regulate the arrangement of the tight junction complexes through its interactions with proteins, such as zonula occludens and cingulin, which also have direct connection with transmembrane tight junction proteins (33, 34). Therefore, all these data together suggest that the rescue of VEGF-induced vascular hyperpermeability could be achieved by inhibiting the actin cytoskeleton dysregulation, preventing the breakdown of BRB formed by tight junction complexes between endothelial cells in the retina.
The network model (Fig. 2D) revealed FN1 as an upstream factor of the β2 integrin pathway associated with actin cytoskeleton. Our proteomic analysis showed up-regulation of FN1 by VEGF treatment, which was confirmed by ELISA, consistent with the results of the previous study on VEGF-induced changes in the murine retina (37). Interestingly, FN1 was previously shown to be overexpressed in retinal microvessels of patients with diabetic retinopathy (38, 39). Accordingly, these data suggest the possibility of targeting the FN1-mediated pathway to treat VEGF-mediated microvascular changes (40). As the downstream factors of FN1, previous studies, most of which are based on experiments with endothelial cells from large vessels, have identified the integrins αv, α3, α5, β1, and β3 (41, 42). In contrast, our proteomics analysis identified β2 integrin, which is known to be abundantly expressed in endothelial progenitor cells (43) and also microvascular endothelial cells (44, 45). For example, β2 integrin plays an important role in angiogenic properties of skin microvascular endothelial cells through its interaction with actin cytoskeleton (44). In addition to proteomic analyses of the whole retinal samples, we confirmed VEGF-induced expression of β2 integrin in retinal microvascular endothelial cells on FN1-coated dish. In contrast, we could not detect such a change of CD14 in HRMECs on F2-coated dish. F2-CD14 was the other upstream signaling pathway identified by the network analysis using the DEPs associated with actin cytoskeleton or filament organization. VEGF-induced recruitments of other cell types, such as mononuclear phagocytes, which express CD14 and exert less direct and significant impact on BRB, might affect the proteomes of the whole retina (46). All these data suggest the validity of the FN1-β2 integrin pathway in endothelial cells as a target pathway that can modulate actin cytoskeleton under the condition involving VEGF-induced retinal vascular hyperpermeability.
Our proteomic analysis detected 11 integrins (Itga1, Itga2, Itga2b, Itga3, Itga5, Itgav, Itgb1, Itgb1bp1, Itgb2, Itgb2l, and Itgb5), but of them, only β2 integrin (Itgb2) was altered in its abundance by VEGF treatment and showed the prevention of the alteration by cotreatment of VEGF and anti-VEGF antibody. Thus, we examined the possibility that β2 integrin can serve as an alternative therapeutic target to anti-VEGF antibody for retinal vascular hyperpermeability. Several studies have reported the association of the integrin signaling with functions of vascular endothelial cells. For example, Senger et al. (47) showed that Itga1/b1 and Itga2/b1 provide critical support for VEGF signaling and endothelial cell migration and tumor angiogenesis. Compared with these studies, however, our proteomic data provide a comprehensive proteome resource that can be used in various studies of retinal vascular hyperpermeability and also the network model (Fig. 2D) that delineates both upstream (FN1) and downstream (GSN, MYL2, MYLPF, MYH9, and ACTN2, ACTN3) molecules in the integrin signaling pathway. The network model can provide improved understanding of the integrin signaling and also serve as a molecular basis at the pathway/network level for those who study molecular mechanisms underlying retinal vascular hyperpermeability through cytoskeleton rearrangement, dysregulation of tight junction integrity, and breakdown of BRB.
In this study, we used the experimental protocol of VEGF-induced retinal hyperpermeability, which has been commonly used (13, 14). The retinal vascular hyperpermeability was prevented by the cotreatment of VEGF and anti-VEGF antibody. Despite the prevention of the phenotype, our analysis showed that the alterations by VEGF at the molecular level were not completely inhibited by the cotreatment of VEGF and anti-VEGF antibody; 283 proteins were up-regulated by VEGF, but for 181 of the 283 proteins (64.0%) such alterations were inhibited by anti-VEGF antibody; and 133 proteins were down-regulated by VEGF, but for 70 of the 133 proteins (52.6%) such alterations were inhibited by anti-VEGF antibody. The partial inhibition of the VEGF-induced DEPs by the cotreatment of VEGF and anti-VEGF antibody can be attributed to the differences of VEGF and anti-VEGF antibody in several factors: 1) injected amounts of VEGF (100 ng per eye) and anti-VEGF antibody (1 μg per eye); 2) binding affinity of VEGF and anti-VEGF antibody to VEGF receptors; and 3) accessibility of VEGF and anti-VEGF antibody to VEGF receptors. These differences can result in the changes unique to VEGF or anti-VEGF antibody as shown in clusters 1, 3, 4, and 6 (Fig. 2B; supplemental Table S4), which should be further verified in detailed studies that involve variation of these factors followed by LC-MS/MS analysis.
In conclusion, our quantitative proteomic analysis of retinal tissues mimicking pathological phenotypes identified a novel therapeutic target, β2 integrin, against VEGF-induced vascular hyperpermeability. Upon up-regulation of FN1 in VEGF-treated retina, β2 integrin might be implicated in cytoskeletal rearrangement in endothelial cells through the downstream signaling of the FN1-β2 integrin pathway, resulting in the increased permeability. Based on the experimental validation, we propose β2 integrin inhibition as an effective means to treat a broad spectrum of retinal diseases implicated with VEGF-induced vascular hyperpermeability, including diabetic retinopathy and retinal vascular occlusion.
Supplementary Material
Footnotes
Author contributions: D.J., D.H., S.L., and Jeong Hun Kim designed the research; D.J., J.B., and S.C. performed the research; D.J., J.B., S.C., Jin Hyoung Kim, D.H., S.L., and Jeong Hun Kim analyzed the data; D.J., S.C., D.H., and Jeong Hun Kim wrote the paper.
* This work was supported the Bio-Signal Analysis Technology Innovation Program 2009-0090895, Pioneer Research Program 2012-0009544, the Bio and Medical Technology Development Program (2015M3A9E6028949, 2015M3A9E6028947) from NRF/MEST, Republic of Korea, Seoul National University Research Grant 800-20140542, Multi-omics Research Program NRF-2012M3A9B9036675,National Research Foundation Grant NRF-2014M3C7A1046047, and Institute for Basic Science Grant IBS-R013-G1-2015-a00 funded by the Korean Ministry of Science, ICT & Future Planning.
 This article contains supplemental materials.
This article contains supplemental materials.
1 The abbreviations used are:
- iTRAQ
- isobaric tag for relative and absolute quantitation
- PSM
- peptide-spectrum match
- FDR
- false discovery rate
- DEP
- differentially expressed protein
- GOBP
- gene ontology biological process
- HRMEC
- human retinal microvascular endothelial cell
- FAK
- focal adhesion kinase
- mRP fractionation
- mid-pH reverse-phase liquid chromatography fractionation
- FN1
- fibronectin 1
- ITGB2
- β2 integrin
- F2
- coagulation factor II
- CD14
- CD14 antigen
- GSN
- gelsolin
- MYL2
- myosin light chain 2
- MyYLPF
- myosin light chain, phosphorylatable, fast skeletal muscle
- BRB
- blood-retinal barrier.
REFERENCES
- 1. Campochiaro P. A., Wykoff C. C., Singer M., Johnson R., Marcus D., Yau L., and Sternberg G. (2014) Monthly versus as-needed ranibizumab injections in patients with retinal vein occlusion: The SHORE study. Ophthalmology 121, 2432–2442 [DOI] [PubMed] [Google Scholar]
- 2. Campochiaro P. A., Wykoff C. C., Shapiro H., Rubio R. G., and Ehrlich J. S. (2014) Neutralization of vascular endothelial growth factor slows progression of retinal nonperfusion in patients with diabetic macular edema. Ophthalmology 121, 1783–1789 [DOI] [PubMed] [Google Scholar]
- 3. Ehlken C., Rennel E. S., Michels D., Grundel B., Pielen A., Junker B., Stahl A., Hansen L. L., Feltgen N., Agostini H. T., and Martin G. (2011) Levels of VEGF but not VEGF(165b) are increased in the vitreous of patients with retinal vein occlusion. Am. J. Ophthalmol. 152, 298–303 [DOI] [PubMed] [Google Scholar]
- 4. Shimada H., Akaza E., Yuzawa M., and Kawashima M. (2009) Concentration gradient of vascular endothelial growth factor in the vitreous of eyes with diabetic macular edema. Invest. Ophthalmol. Vis. Sci. 50, 2953–2955 [DOI] [PubMed] [Google Scholar]
- 5. Tolentino M. J., Miller J. W., Gragoudas E. S., Jakobiec F. A., Flynn E., Chatzistefanou K., Ferrara N., and Adamis A. P. (1996) Intravitreous injections of vascular endothelial growth factor produce retinal ischemia and microangiopathy in an adult primate. Ophthalmology 103, 1820–1828 [DOI] [PubMed] [Google Scholar]
- 6. Kim J. H., Kim J. H., Lee Y. M., Ahn E. M., Kim K. W., and Yu Y. S. (2009) Decursin inhibits VEGF-mediated inner blood-retinal barrier breakdown by suppression of VEGFR-2 activation. J. Cereb. Blood Flow Metab. 29, 1559–1567 [DOI] [PubMed] [Google Scholar]
- 7. Maeng Y. S., Maharjan S., Kim J. H., Park J. H., Suk Yu Y., Kim Y. M., and Kwon Y. G. (2013) Rk1, a ginsenoside, is a new blocker of vascular leakage acting through actin structure remodeling. PLoS ONE 8, e68659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jo D. H., Kim J. H., and Kim J. H. (2012) How to overcome diabetic retinopathy: focusing on blood-retinal barrier. Immunol. Endocr. Metab. Agents Med. Chem. 12, 110–117 [Google Scholar]
- 9. Miller J. W., Le Couter J., Strauss E. C., and Ferrara N. (2013) Vascular endothelial growth factor A in intraocular vascular disease. Ophthalmology 120, 106–114 [DOI] [PubMed] [Google Scholar]
- 10. Heo J. W., Kim J. H., Cho C. S., Jun H. O., Kim D. H., Yu Y. S., and Kim J. H. (2012) Inhibitory activity of bevacizumab to differentiation of retinoblastoma cells. PLoS ONE 7, e33456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kurihara T., Westenskow P. D., Bravo S., Aguilar E., and Friedlander M. (2012) Targeted deletion of Vegfa in adult mice induces vision loss. J. Clin. Invest. 122, 4213–4217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chen X. L., Nam J. O., Jean C., Lawson C., Walsh C. T., Goka E., Lim S. T., Tomar A., Tancioni I., Uryu S., Guan J. L., Acevedo L. M., Weis S. M., Cheresh D. A., and Schlaepfer D. D. (2012) VEGF-induced vascular permeability is mediated by FAK. Dev. Cell 22, 146–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jones C. A., London N. R., Chen H., Park K. W., Sauvaget D., Stockton R. A., Wythe J. D., Suh W., Larrieu-Lahargue F., Mukouyama Y. S., Lindblom P., Seth P., Frias A., Nishiya N., Ginsberg M. H., et al. (2008) Robo4 stabilizes the vascular network by inhibiting pathologic angiogenesis and endothelial hyperpermeability. Nat. Med. 14, 448–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Scheppke L., Aguilar E., Gariano R. F., Jacobson R., Hood J., Doukas J., Cao J., Noronha G., Yee S., Weis S., Martin M. B., Soll R., Cheresh D. A., and Friedlander M. (2008) Retinal vascular permeability suppression by topical application of a novel VEGFR2/Src kinase inhibitor in mice and rabbits. J. Clin. Invest. 118, 2337–2346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wiśniewski J. R., Zougman A., Nagaraj N., and Mann M. (2009) Universal sample preparation method for proteome analysis. Nat. Methods 6, 359–362 [DOI] [PubMed] [Google Scholar]
- 16. Park J. M., Park J. H., Mun D. G., Bae J., Jung J. H., Back S., Lee H., Kim H., Jung H. J., Kim H. K., Lee H., Kim K. P., Hwang D., and Lee S. W. (2015) Integrated analysis of global proteome, phosphoproteome, and glycoproteome enables complementary interpretation of disease-related protein networks. Sci. Rep. 5, 18189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lee H., Lee J. H., Kim H., Kim S. J., Bae J., Kim H. K., and Lee S. W. (2014) A fully automated dual-online multifunctional ultrahigh pressure liquid chromatography system for high-throughput proteomics analysis. J. Chromatogr. A 1329, 83–89 [DOI] [PubMed] [Google Scholar]
- 18. Shin B., Jung H. J., Hyung S. W., Kim H., Lee D., Lee C., Yu M. H., and Lee S. W. (2008) Postexperiment monoisotopic mass filtering and refinement (PE-MMR) of tandem mass spectrometric data increases accuracy of peptide identification in LC/MS/MS. Mol. Cell. Proteomics 7, 1124–1134 [DOI] [PubMed] [Google Scholar]
- 19. Kim S., Mischerikow N., Bandeira N., Navarro J. D., Wich L., Mohammed S., Heck A. J., and Pevzner P. A. (2010) The generating function of CID, ETD, and CID/ETD pairs of tandem mass spectra: applications to database search. Mol. Cell. Proteomics 9, 2840–2852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tang W. H., Shilov I. V., and Seymour S. L. (2008) Nonlinear fitting method for determining local false discovery rates from decoy database searches. J. Proteome Res. 7, 3661–3667 [DOI] [PubMed] [Google Scholar]
- 21. Zhang B., Chambers M. C., and Tabb D. L. (2007) Proteomic parsimony through bipartite graph analysis improves accuracy and transparency. J. Proteome Res. 6, 3549–3557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kim S. J., Chae S., Kim H., Mun D. G., Back S., Choi H. Y., Park K. S., Hwang D., Choi S. H., and Lee S. W. (2014) A protein profile of visceral adipose tissues linked to early pathogenesis of type 2 diabetes mellitus. Mol. Cell. Proteomics 13, 811–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Vizcaíno J. A., Csordas A., Del-Toro N., Dianes J. A., Griss J., Lavidas I., Mayer G., Perez-Riverol Y., Reisinger F., Ternent T., Xu Q. W., Wang R., and Hermjakob H. (2016) 2016 update of the PRIDE database and related tools. Nucleic Acids Res. 44, D447–D456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chae S., Ahn B. Y., Byun K., Cho Y. M., Yu M. H., Lee B., Hwang D., and Park K. S. (2013) A systems approach for decoding mitochondrial retrograde signaling pathways. Sci. Signal. 6, rs4. [DOI] [PubMed] [Google Scholar]
- 25. Bowman A. W., and Azzalini A. (1997) Applied Smoothing Techniques for Data Analysis: The Kernel Approach with S-Plus Illustrations, pp. 25–47, Oxford University Press, Oxford, NY [Google Scholar]
- 26. Hwang D., Rust A. G., Ramsey S., Smith J. J., Leslie D. M., Weston A. D., de Atauri P., Aitchison J. D., Hood L., Siegel A. F., and Bolouri H. (2005) A data integration methodology for systems biology. Proc. Natl. Acad. Sci. U.S.A. 102, 17296–17301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Storey J. D., and Tibshirani R. (2003) Statistical significance for genome-wide studies. Proc. Natl. Acad. Sci. U.S.A. 100, 9440–9445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Huang da W., Sherman B. T., and Lempicki R. A. (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 [DOI] [PubMed] [Google Scholar]
- 29. Skeie J. M., and Mahajan V. B. (2013) Proteomic interactions in the mouse vitreous-retina complex. PLoS ONE 8, e82140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kim S. J., Jin J., Kim Y. J., Kim Y., and Yu H. G. (2012) Retinal proteome analysis in a mouse model of oxygen-induced retinopathy. J. Proteome Res. 11, 5186–5203 [DOI] [PubMed] [Google Scholar]
- 31. Barathi V. A., Chaurasia S. S., Poidinger M., Koh S. K., Tian D., Ho C., Iuvone P. M., Beuerman R. W., and Zhou L. (2014) Involvement of GABA transporters in atropine-treated myopic retina as revealed by iTRAQ quantitative proteomics. J. Proteome Res. 13, 4647–4658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Reidel B., Thompson J. W., Farsiu S., Moseley M. A., Skiba N. P., and Arshavsky V. Y. (2011) Proteomic profiling of a layered tissue reveals unique glycolytic specializations of photoreceptor cells. Mol. Cell. Proteomics 10, M110.002469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Abbott N. J., Patabendige A. A., Dolman D. E., Yusof S. R., and Begley D. J. (2010) Structure and function of the blood-brain barrier. Neurobiol. Dis. 37, 13–25 [DOI] [PubMed] [Google Scholar]
- 34. Lai C. H., Kuo K. H., and Leo J. M. (2005) Critical role of actin in modulating BBB permeability. Brain Res. Brain Res. Rev. 50, 7–13 [DOI] [PubMed] [Google Scholar]
- 35. Kim J. H., Kim J. H., Park J. A., Lee S. W., Kim W. J., Yu Y. S., and Kim K. W. (2006) Blood-neural barrier: intercellular communication at glio-vascular interface. J. Biochem. Mol. Biol. 39, 339–345 [DOI] [PubMed] [Google Scholar]
- 36. Saint-Geniez M., Maharaj A. S., Walshe T. E., Tucker B. A., Sekiyama E., Kurihara T., Darland D. C., Young M. J., and D'Amore P. A. (2008) Endogenous VEGF is required for visual function: evidence for a survival role on Muller cells and photoreceptors. PLoS ONE 3, e3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kuiper E. J., Hughes J. M., Van Geest R. J., Vogels I. M., Goldschmeding R., Van Noorden C. J., Schlingemann R. O., and Klaassen I. (2007) Effect of VEGF-A on expression of profibrotic growth factor and extracellular matrix genes in the retina. Invest. Ophthalmol. Vis. Sci. 48, 4267–4276 [DOI] [PubMed] [Google Scholar]
- 38. Casaroli Marano R. P., Preissner K. T., and Vilaró S. (1995) Fibronectin, laminin, vitronectin and their receptors at newly-formed capillaries in proliferative diabetic retinopathy. Exp. Eye Res. 60, 5–17 [DOI] [PubMed] [Google Scholar]
- 39. Roy S., Cagliero E., and Lorenzi M. (1996) Fibronectin overexpression in retinal microvessels of patients with diabetes. Invest. Ophthalmol. Vis. Sci. 37, 258–266 [PubMed] [Google Scholar]
- 40. Lim Y., Jo D. H., Kim J. H., Ahn J. H., Hwang Y. K., Kang D. K., Chang S. I., Yu Y. S., Yoon Y., and Kim J. H. (2012) Human apolipoprotein(A) kringle V inhibits ischemia-induced retinal neovascularization via suppression of fibronectin-mediated angiogenesis. Diabetes 61, 1599–1608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Silva R., D'Amico G., Hodivala-Dilke K. M., and Reynolds L. E. (2008) Integrins: the keys to unlocking angiogenesis. Arterioscler. Thromb. Vasc. Biol. 28, 1703–1713 [DOI] [PubMed] [Google Scholar]
- 42. Avraamides C. J., Garmy-Susini B., and Varner J. A. (2008) Integrins in angiogenesis and lymphangiogenesis. Nat. Rev. Cancer 8, 604–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hayakawa K., Pham L. D., Arai K., and Lo E. H. (2014) Reactive astrocytes promote adhesive interactions between brain endothelium and endothelial progenitor cells via HMGB1 and β-2 integrin signaling. Stem Cell Res. 12, 531–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Margheri F., Manetti M., Serratì S., Nosi D., Pucci M., Matucci-Cerinic M., Kahaleh B., Bazzichi L., Fibbi G., Ibba-Manneschi L., and Del Rosso M. (2006) Domain 1 of the urokinase-type plasminogen activator receptor is required for its morphologic and functional β2 integrin-mediated connection with actin cytoskeleton in human microvascular endothelial cells: failure of association in systemic sclerosis endothelial cells. Arthritis Rheum. 54, 3926–3938 [DOI] [PubMed] [Google Scholar]
- 45. Wilson S. H., Ljubimov A. V., Morla A. O., Caballero S., Shaw L. C., Spoerri P. E., Tarnuzzer R. W., and Grant M. B. (2003) Fibronectin fragments promote human retinal endothelial cell adhesion and proliferation and ERK activation through α5β1 integrin and PI 3-kinase. Invest. Ophthalmol. Vis. Sci. 44, 1704–1715 [DOI] [PubMed] [Google Scholar]
- 46. Dejda A., Mawambo G., Cerani A., Miloudi K., Shao Z., Daudelin J. F., Boulet S., Oubaha M., Beaudoin F., Akla N., Henriques S., Menard C., Stahl A., Delisle J. S., Rezende F. A., et al. (2014) Neuropilin-1 mediates myeloid cell chemoattraction and influences retinal neuroimmune cross-talk. J. Clin. Invest. 124, 4807–4822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Senger D. R., Perruzzi C. A., Streit M., Koteliansky V. E., de Fougerolles A. R., and Detmar M. (2002) The α(1)β(1) and α(2)β(1) integrins provide critical support for vascular endothelial growth factor signaling, endothelial cell migration, and tumor angiogenesis. Am. J. Pathol. 160, 195–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.