Abstract
Co-chaperones containing tetratricopeptide repeat (TPR) domains enable cooperation between Hsp70 and Hsp90 to maintain cellular proteostasis. Although the details of the molecular interactions between some TPR domains and heat shock proteins are known, we describe a novel mechanism by which Tomm34 interacts with and coordinates Hsp70 activities. In contrast to the previously defined Hsp70/Hsp90-organizing protein (Hop), Tomm34 interaction is dependent on the Hsp70 chaperone cycle. Tomm34 binds Hsp70 in a complex process; anchorage of the Hsp70 C terminus by the TPR1 domain is accompanied by additional contacts formed exclusively in the ATP-bound state of Hsp70 resulting in a high affinity entropically driven interaction. Tomm34 induces structural changes in determinants within the Hsp70-lid subdomain and modulates Hsp70/Hsp40-mediated refolding and Hsp40-stimulated Hsp70 ATPase activity. Because Tomm34 recruits Hsp90 through its TPR2 domain, we propose a model in which Tomm34 enables Hsp70/Hsp90 scaffolding and influences the Hsp70 chaperone cycle, providing an additional role for co-chaperones that contain multiple TPR domains in regulating protein homeostasis.
The homeostasis and spatial organization of the cellular proteome is vitally dependent on coordinated action of the highly conserved Hsp70 and Hsp90 molecular chaperones (1, 2). Polypeptides processed by Hsp70/Hsp90 can either enter pathways leading to their final maturation (3, 4) and degradation (5, 6) or remain associated with molecular chaperones in a semifolded state required for their cytosolic transport to target sites (7, 8). The diversity of Hsp70/Hsp90-mediated actions is governed by a variety of co-chaperone proteins. These include proteins regulating chaperone ATPase cycles (9, 10), substrate recognition (11, 12), or proteins involved in scaffolding of the Hsp70/Hsp90 complexes (3, 13–15). Many co-chaperones contain specialized tetratricopeptide repeat (TPR)1 domains mediating their interaction with the evolutionarily conserved C-terminal EEVD motif of Hsp70/Hsp90 (16, 17). Hsp70/Hsp90-binding TPR domains form a so-called two-carboxylate clamp coordinating the terminal aspartate residue of the chaperones. Binding specificity to Hsp70 or Hsp90 is determined by the ability of TPR domains to accommodate neighboring Hsp70/Hsp90-specific hydrophobic residues (18–20). However, a more extensive interaction mode of TPR domain association with the C terminus of the chaperones has been described recently (5).
The Hsp70 family of molecular chaperones is essential for cellular proteostasis (21, 22). Hsp70 proteins consist of the following two structurally independent domains: an N-terminal nucleotide-binding domain (NBD) and a C-terminal substrate-binding domain (SBD). The structure of the SBD can be further subdivided into the SBDβ subdomain, representing the peptide-binding pocket, and the SBDα subdomain, which forms an α-helical lid that covers the SBDβ (23). The extreme C terminus of Hsp70 preceding the EEVD motif is largely unstructured (24, 25). NBD and SBD are connected by a highly conserved hydrophobic linker (26). ATP coordination with the NBD is allosterically transmitted to the SBD, leading to large conformational changes in the Hsp70 molecule in which the SBD docks onto the NBD (23, 27, 28). The SBD of Hsp70 in its ATP-bound state is characterized by high on/off rates of substrate binding (29–31). Simultaneous action of substrate and various Hsp40 co-chaperones delivering the substrate to Hsp70 promotes ATP hydrolysis, which, in turn, induces major reorganization in the SBD reducing substrate on/off rates and thus stabilizing the Hsp70/substrate interaction (31–34). Substrate release and subsequent chaperone cycles are enabled by ADP dissociation and binding of a new ATP molecule assisted by nucleotide exchange factor co-chaperones (35–37).
Tomm34 (34-kDa translocase of the outer mitochondrial membrane) containing two TPR domains has been recently described as an Hsp70/Hsp90 scaffolding co-chaperone (8, 13). Tomm34 TPR1/TPR2 domain binding specificities enable its simultaneous interaction with both Hsp70 and Hsp90 (13) in cytosolic complexes mediating preprotein transport (8). Although the TPR2 domain of Tomm34 binds Hsp90 mainly through a two-carboxylate clamp/EEVD interaction, the TPR1 association with the C terminus of Hsp70 was shown to be more complex involving additional contacts (13). The aim of this work was to disclose the details of the interaction between Tomm34 and Hsp70. We showed that Tomm34 extensively interacts with the C-terminal structures of Hsp70 selectively in its ATP-bound state and that this interaction is strongly entropically driven. Moreover, Tomm34 profoundly affected Hsp40-stimulated ATPase and Hsp70 refolding activity, indicating that Tomm34 plays a direct role in the Hsp70 chaperone cycle beyond mere scaffolding. Our results suggest the hypothesis of Tomm34 function in substrate shuttling between Hsp70 and Hsp90 during cytosolic preprotein transport. Because deregulation of Tomm34 levels is increasingly reported in various cancers (38–41), the structural and functional analysis of the Hsp70/Tomm34 interaction might provide initial insight into the role of Tomm34 in malignant transformation. Finally, for the purposes of this study we have prepared and partly characterized new human Hsp70 conformational mutants I164D and D529A. Our structural data describing these mutants answered several previous questions about Hsp70 mechanics and provided a useful tool for future investigation.
EXPERIMENTAL PROCEDURES
Cloning and Protein Preparation
All coding sequences were cloned by Gateway recombination technology (Invitrogen). The full coding sequences of the human TOMM34 (NM_006809.4), STIP1 (Hop, NM_001282652.1), DNAJB1 (Hsp40, NM_006145.2), and BAG1 (NM_001172415.1) genes, sequences coding for Tomm34 TPR domains (T1S, amino acids 1–137; T1L, amino acids 1–188; T2L, amino acids 137–309; and T2S, amino acids 188–309) and Tomm34 point mutants (W140A, K143A, P151A, and W158A) were cloned into a vector containing an N-terminal His6-GST tag cleavable by TEV protease. The full coding sequences of the human HSPA1A gene (Hsp70, NM_005345.5) and sequences coding for Hsp70 NBD (amino acids 1–383), SBD (amino acids 383–641), and Hsp70 mutant constructs (I164D, T204A, V438F, D529A, and Δ533–543) were cloned into a vector containing an N-terminal His6 or SBP tag; the coding sequence of the human HSP90AA1 gene (Hsp90α, NM_001017963.2) was cloned into a vector containing an N-terminal His6 tag cleavable by TEV protease. All cloned genes were expressed in BL21(DE3) RIPL cells.
The cells were grown in LB medium at 37 °C up to an A600 of 0.5. Induction of gene expression was achieved by adding isopropyl β-d-thiogalactopyranoside to the culture (final concentration 1 mm). The bacterial culture was grown at 30 °C for another 3–4 h and then pelleted by centrifugation. The cells producing Tomm34 and all its variants, Hop (where Hop is Hsp70/Hsp90 organizing protein) and Hsp40, were resuspended in GST binding buffer I (50 mm Tris, pH 7.8, 0.5 m NaCl); Bag-1-producing cells were resuspended in GST binding buffer II (50 mm Hepes, pH 7.4, 0.2 m NaCl, 0.1 m KAc); the cells expressing His6-tagged Hsp70 constructs were resuspended in His binding buffer I (50 mm Hepes, pH 7.6, 0.3 m KCl, 5 mm imidazole, 5% glycerol); the cells expressing SBP-tagged Hsp70 constructs were resuspended in SBP binding buffer (50 mm Hepes, pH 7.6, 0.1 m NaCl, 0.2 m KCl, 10% glycerol, 1 mm avidin); Hsp90α-expressing cells were resuspended in His binding buffer II (50 mm Tris, pH 8.0, 0.2 m NaCl, 2 mm MgCl2, 10% glycerol). Cell suspensions were enriched with lysozyme (1 mg/ml) and PMSF (1 mm) and then sonicated. Bacterial lysates were obtained by centrifugation for 30 min at 12,000 × g.
His6-GST-tagged proteins were captured on a GSTrap glutathione-agarose column (GE Healthcare), eluted with 20 mm glutathione, and subjected to TEV protease cleavage. To remove His6-GST with His6-TEV, proteins were applied to a HisTrap column (GE Healthcare). Finally, the flow-through fraction containing purified proteins was concentrated and further processed by preparative gel filtration using a HiPrep 16/60 Sephacryl S-100 HR column (GE Healthcare). His6-tagged Hsp70 constructs were purified using a HisTrap column and then applied to a HiPrep 16/60 Sephacryl S-100 HR column. His6-Hsp70 wild type (WT) and T204A proteins used for the malachite green ATPase assay were after immobilized metal affinity chromatography subjected to buffer exchange into Q buffer A (20 mm Tris, pH 7.4, 75 mm KCl), applied to a HiTrap Q column (GE Healthcare), and eluted by a linear gradient of Q buffer B (20 mm Tris, pH 7.4, 1 m KCl). Fractions containing purified proteins were concentrated and subjected to gel filtration using a HiPrep 16/60 Sephacryl S-100 HR column. SBP-tagged Hsp70 constructs were captured on streptavidin-agarose beads (Thermo Fischer Scientific, Waltham, MA), washed with SBP washing buffer (50 mm Hepes, pH 7.6, 0.1 m NaCl, 0.2 m KCl), and eluted with 2 mm biotin. His6-tagged Hsp90α was purified using a HisTrap column, and the His6 tag was cleaved by overnight incubation with TEV protease. The protein was subsequently exchanged into His binding buffer II and then subjected to a second immobilized metal affinity chromatograph to remove His6 tag and His6-TEV. The flow-through fractions were concentrated and further processed by gel filtration using a HiPrep 16/60 Sephacryl S-200 HR column (GE Healthcare). The purity of all isolated proteins was confirmed by SDS-PAGE/Coomassie staining. All proteins were finally exchanged into final assay buffers using 7-kDa molecular mass cutoff Zeba spin desalting column (Thermo Fischer Scientific). Tobacco etch virus protease His6-TEV(S219V)-Arg5 was prepared in-house following the modified method of Tropea et al. (42).
Site-directed Mutagenesis
The full-length Hsp70 and Tomm34 coding sequences were mutated to encode the desired mutations using QuikChange site-directed mutagenesis kit (Agilent Technologies, Santa Clara, CA) and mutagenic oligonucleotides according to the manufacturer's instructions.
SBP Pulldown Assays
Before performing pulldown experiments, all proteins were exchanged to pulldown assay buffer (50 mm Hepes, pH 7.5, 0.1 m KAc, 2 mm MgCl2). Next, 70 pmol of various SBP-tagged Hsp70 constructs was incubated with streptavidin-agarose beads at 4 °C for 30 min. After washing with assay buffer, 140 pmol of Tomm34 variants (WT, mutants, or TPR domains) or Hop protein in the buffer containing 0.2 mm ATP, ADP, or no nucleotide was added to the beads. After 1 h of incubation at 4 °C, beads were washed with buffers supplemented with matching nucleotides. The proteins were eluted with 2 mm biotin in assay buffer and analyzed by Western blotting. In Hsp40 or Bag-1 containing assays, 25 pmol of SBP-Hsp70 WT was immobilized on streptavidin-agarose beads, and equimolar amounts (25 pmol) of Tomm34 and Hsp40 were mixed in increasing ATP concentrations (0.01, 0.1, and 1 mm). Alternatively, 25 pmol of Tomm34 was mixed with increasing Bag-1 concentrations (10, 25, and 50 pmol) in the presence of 0.2 mm ATP. Prepared protein solutions were added to the beads and incubated for 1 h at 4 °C. After the washing steps, the proteins were eluted with 2 mm biotin and analyzed by Western blotting. In pulldown assay with HEK293 cell lysates, 70 pmol of bacterially produced SBP-Hsp70 WT or C-terminally truncated mutant lacking PTIEEVD motif (ΔC) was immobilized onto streptavidin-agarose beads. Before incubation, the HEK293 lysates were exchanged to buffer containing 50 mm Hepes, pH 7.5, 0.1 m KAc and then enriched with 2 mm MgCl2 and/or 1 mm ATP. Next, beads with immobilized SBP proteins were incubated with prepared lysates for 1 h at 4 °C. After the washing steps, the proteins were eluted with 2 mm biotin and analyzed by Western blotting.
Antibodies
Monoclonal and polyclonal anti-Tomm34, Hop, Hsp70, and Hsp40 antibodies used in this study were prepared in-house and characterized. For SBP-tagged protein detection, we used peroxidase-conjugated streptavidin (Sigma-Aldrich) diluted in 5% milk in the presence of avidin (10 ng/μl). Avidin was added to prevent streptavidin binding to biotinylated proteins present in the milk. The blots were developed with polyclonal anti-mouse rabbit IgG or anti-rabbit swine IgG secondary antibodies conjugated with horseradish peroxidase (Dako, Santa Clara, CA).
Isothermal Titration Calorimetry
ITC experiments were performed using a MicroCal Auto-iTC200 (Malvern Instruments, Worcestershire, UK). A total of 38 μl of 120 μm Tomm34 was injected into 200 μl of 12 μm His6-Hsp70 (monomer concentration) in 16 aliquots (1 × 0.5 and 15 × 2.5 μl). In experiments looking at the effect of ATP on Tomm34 binding, both His6-Hsp70 and Tomm34 were preincubated with 5 mm ATP (30 min at room temperature) before ITC measurement. Parallel titrations were performed in which the injectant was added to buffer without protein or buffer was injected into the protein. Titrations were carried out at 5, 15, 25, and 30 °C. Alternatively, 38 μl of 280 μm Tomm34 was injected into a cell containing 200 μl of 20 μm Hsp90α at 25 °C to measure the thermodynamics for Tomm34 interaction with Hsp90α. The heats of reaction were corrected for the heat of dilution and analyzed using the Origin software package supplied with the instrument (Malvern Instruments). All experiments were carried out in 50 mm Hepes, pH 7.5, 0.1 m KAc, 2 mm MgCl2.
Luciferase Refolding Assay
Firefly luciferase (8.2 μm) was chemically denatured (25 mm Hepes, pH 7.2, 50 mm KAc, 5 mm DTT, 2 mm MgCl2, 6 m guanidine hydrochloride) and incubated for 1 h at 25 °C. Similarly, native luciferase was diluted in buffer lacking guanidine hydrochloride. Next, the denatured/native luciferase was diluted into non-denaturing buffer to a final concentration of 0.2 μm. The refolding protein mixture contained 1 μm His6-Hsp70, 2 μm Hsp40, 8 nm denatured luciferase, and 0, 0.01, 0.1, 1, and 2 μm Tomm34/Hop in 25 mm Hepes, pH 7.2, 50 mm KAc, 5 mm DTT, 2 mm MgCl2. The reaction was started by addition of ATP at a 1 mm final concentration and incubated at 37 °C. At the given times (0, 10, 20, 40, 60, and 80 min), 2-μl aliquots of the reactions were taken and diluted into 48 μl of 50 mm glycylglycine, pH 7.8, 30 mm MgSO4, 4 mm DTT in 96-well white Nunc Plate (Thermo Fischer Scientific). Next, the luciferase activity was initiated by addition of d-luciferin at 0.2 mm final concentration in the presence of 1 mm ATP and measured at 21 °C using an Infinite M1000 Pro (Tecan, Männedorf, Switzerland) at emission wavelengths of 560 nm and 2000-ms integration time.
The signal from the sample with native luciferase was set as 100%. As negative controls, we measured the luciferase activity of denatured luciferase only, the refolding activity of chaperone mixture without Hsp40, and the refolding activity of individual Tomm34 and Hop proteins.
Malachite Green ATPase Assay
Experiments were performed according to previous protocols (43). Stock solutions of malachite green (0.081% w/v), polyvinyl alcohol (2.3% w/v), and sodium molybdate (5.7% w/v in 6 m HCl) were prepared, stored at 4 °C, and mixed with water at the ratio of 2:1:1:2 to prepare the malachite green reagent. His6-Hsp70 (2 μm) was titrated with Hsp40 protein with/without the addition of Tomm34 (0.1, 1, and 10 μm) in buffer containing 50 mm Hepes, pH 7.5, 0.1 m KAc, 2 mm MgCl2. As a negative control, 10 μm bovine serum albumin (BSA) was used. The ATPase reactions were initiated with the addition of ATP (2 mm) and proceeded for 3 h at 37 °C. After incubation, malachite green reagent was added into each well in clear 96-well plates. Immediately following this step, 34% sodium citrate was used to halt the non-enzymatic hydrolysis of ATP. The samples were incubated for another 15 min before measuring absorbance at 620 nm. A phosphate standard curve was used to calculate picomoles of ATP/μm Hsp70/min.
Peptide Binding
Fluorescein-NRLLLTG peptide binding experiments were performed in 50 mm Hepes, pH 7.5, 0.1 m KAc, 2 mm MgCl2, 0.01% Tween 20. To obtain equilibrium binding curves, mixtures containing 30 nm peptide were titrated with different concentrations of His6-Hsp70 constructs (WT, I164D, V438F, D529A, and Δ533–543) and incubated overnight at 4 °C. Peptide binding kinetics was analyzed using peptide and protein concentrations of 30 nm and 25 μm, respectively. To distinguish nonspecific binding, we used BSA with the same concentration as analyzed protein. All reactions were carried out in a total volume of 12 μl in a 384-well black Nunc Plate (Thermo Fischer Scientific). Fluorescence polarization was measured at 21 °C using an Infinite M1000 Pro (Tecan) with excitation and emission wavelengths of 470 and 520 nm, respectively. The analysis was performed using GraphPad Prism version 5.03 for Windows (GraphPad Software, San Diego).
H/D Exchange
Deuteration of Tomm34, Hop, and His6-Hsp70 was followed on peptides generated after pepsin digestion. His6-Hsp70 protein was analyzed with either free or with 2.5 m excess of Tomm34/Hop proteins. To assess the effect of ATP, protein mixtures were analyzed in nucleotide-free buffer or in the presence of 5 mm ATP. His6-Hsp70 was incubated with Tomm34/Hop for 1 h at 21 °C prior to the exchange. The exchange was initiated by a 10-fold dilution into D2O containing 50 mm Hepes, pD 7.5, 0.1 m KAc, 2 mm MgCl2 and was performed at 21 °C. After various time points (30 s to 3 h), exchange reaction was quenched by decreasing the pH with 1 m glycine (pH 2.3) and rapid freezing in liquid nitrogen. The amount of His6-Hsp70 added into H/D exchange reaction was 200 pmol.
Deuteration of Tomm34 and Hop proteins was followed in the presence of 2.5 m excess of His6-Hsp70 in the same experimental setup. The amount of Tomm34/Hop added into reaction was 200 pmol.
In experiments looking at the structural comparison of WT and mutant Hsp70s, nucleotide-free proteins were incubated with 5 mm ATP for 30 min at 21 °C prior to the exchange. The experimental setup was as before, but the exchange was measured only after a 1-h time interval.
Digestion and HPLC Separation
Each sample was quickly thawed and injected onto an immobilized pepsin column (66-μl bed volume). Digestion was driven by a flow of water solution containing 2% acetonitrile and 0.05% trifluoroacetic acid, and the rate was 20 μl/min. Peptides were trapped and desalted on line on a peptide microtrap (Michrom BioResources, Auburn, CA), eluted onto an analytical column (Jupiter C18, 1.0 × 50 mm, 5 μm, 300 Å; Phenomenex, Torrance, CA), and separated by a linear gradient elution of 10–40% B in 29 min at a flow rate of 20 μl/min. Solvents were as follows: A, 0.1% formic acid in water; and B, 80% acetonitrile, 0.08% formic acid. In all analyses, injection and switching valves, immobilized pepsin column, trap cartridge, and the analytical column were kept at 0 °C in a Peltier cooled insulated box to minimize back-exchange. The outlet of the LC system was interfaced to the electrospray ionization source of a mass spectrometer.
Mass Spectrometry and Data Analysis
Mass spectrometric analysis of deuterated samples used an ESI-FT-ICR with a 12-tesla superconducting magnet (Bruker Daltonics, Billerica, MA) or an ESI-Orbitrap Elite (Thermo Fischer Scientific). MS and MS/MS analysis using ESI-FT-ICR was done as described previously (13, 44). Data were searched using MASCOT against a custom-made database containing sequences of analyzed proteins. ESI-Orbitrap analysis was performed as follows. For peptide mapping (HPLC-MS/MS), the instrument was operated in a data-dependent mode. Each MS scan was followed by MS/MS scans of the top three most intense ions. Tandem mass spectra were searched using Sequest HT against a database containing sequences of analyzed proteins. Sequence coverage was visualized using Proteome Discoverer 1.4 software (Thermo Fischer Scientific). Data analysis of deuterated samples was done by HD Examiner version 1.4 (Sierra Analytics, Modesto, CA). No correction for back-exchange was done.
Software
Multiple sequence alignments were performed using DNASTAR Lasergene Suite. Molecular structures were rendered using PyMOL.
RESULTS
Tomm34 Interacts with Hsp70 in ATP-dependent Manner
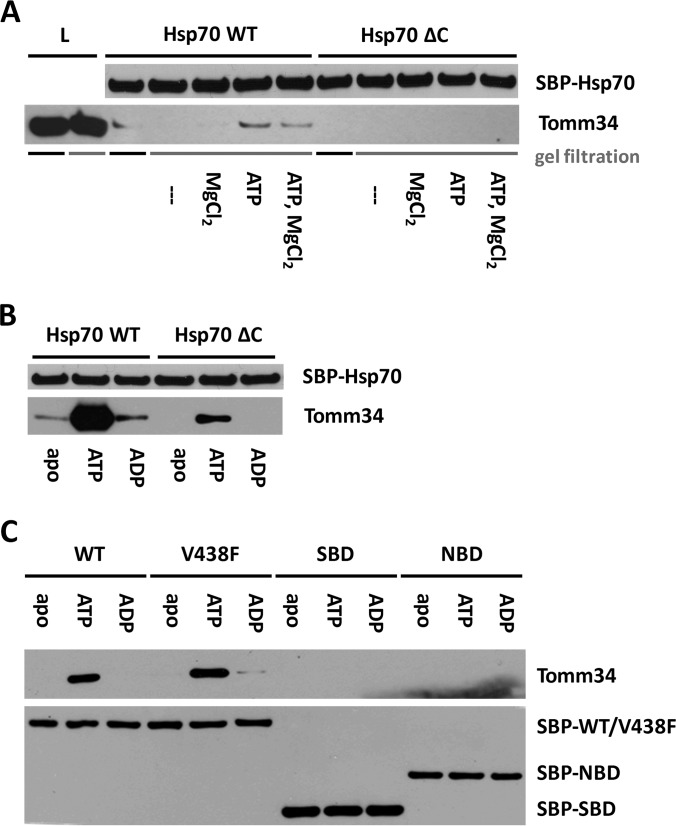
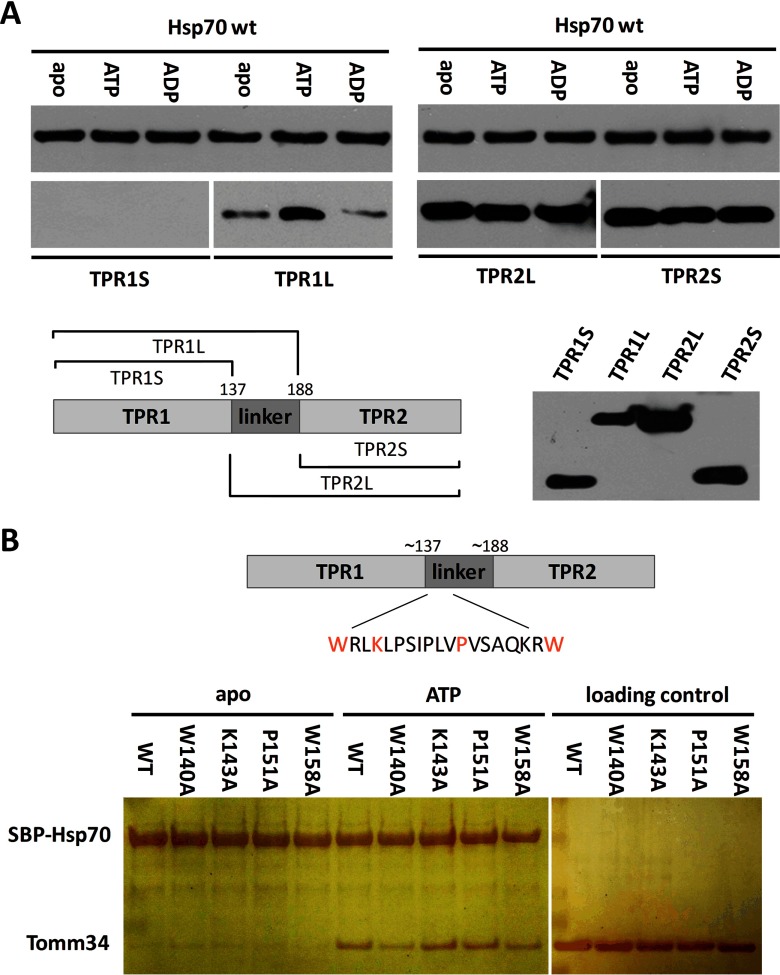
We have previously suggested that binding between Hsp70 chaperone and Tomm34 co-chaperone is not solely mediated through TPR1 two-carboxylate clamp recognition of Hsp70 C-terminal EEVD motif but involves other intermolecular contacts (13). Moreover, we observed apparent discrepancy in Hsp70/Tomm34 complex formation efficiency in vitro and in eukaryotic cell lysates, where Tomm34 forms stable assemblies with both molecular chaperones. Faou and Hoogenraad (8) analyzed the Hsp70/Tomm34 interaction using bovine cytosolic extracts and reported it as ATP/ADP-independent. Because intracellular concentration of ATP is in the millimolar range (45), we speculate that the putative nucleotide-dependent nature of Hsp70/Tomm34 interaction might be undetectable in unconditioned cell extracts. To assess the influence of ATP on Hsp70/Tomm34 binding, we performed pulldown of endogenous Tomm34 from HEK293 cell lysates by purified bacterially expressed SBP-Hsp70/SBP-Hsp70ΔEEVD (Fig. 1A). In this assay, the cell lysate was first processed by gel filtration to remove free low molecular weight compounds. Importantly, the level of Tomm34 protein in cell extracts was not reduced by gel filtration. Subsequently, indicated fractions of filtered lysates were supplemented with magnesium chloride and/or ATP and incubated with either SBP-Hsp70 or SBP-Hsp70ΔEEVD. Although Tomm34 protein co-precipitated with SBP-Hsp70 from unfiltered cell extracts, its level was undetectable after precipitation from gel-filtered lysates. The interaction of Tomm34 with SBP-Hsp70 in filtered lysates was restored by addition of ATP independently on Mg2+ presence. Conversely, the SBP-Hsp70ΔEEVD/Tomm34 complex was not detected in either filtered or unfiltered lysates, in concordance with the importance of TPR-EEVD contacts for Hsp70/Tomm34 interaction (13). Next, we confirmed the strong enhancing effect of ATP (but not ADP) on Hsp70/Tomm34 complex formation using purified proteins (Fig. 1B). Moreover, we observed a weak SBP-Hsp70ΔEEVD/Tomm34 interaction only in the presence of ATP, indicating the existence of additional nucleotide-dependent non-EEVD contacts. These results revised our current knowledge about Hsp70/Tomm34 interaction suggesting the important role of ATP during efficient complex assembly.
Fig. 1.
Tomm34 binding to Hsp70 is ATP-dependent and requires cooperation of both SBD and NBD. Bacterially purified SBP-Hsp70 constructs were immobilized on streptavidin-agarose beads and incubated with gel-filtered HEK293 cell lysates conditioned by the indicated compounds (2 mm MgCl2, 1 mm ATP) (A) or with purified Tomm34 protein in buffer containing 0.2 mm ATP/ADP or no nucleotide (B and C). After washing, bound proteins were eluted by biotin and analyzed by Western blotting. ΔC, deletion of PTIEEVD; V438F, mutant defective in substrate binding; L, total cell lysate.
Cooperation of NBD and SBD of Hsp70 Is Necessary for Tomm34 Binding
Hsp70 undergoes large allosterically coupled conformational changes of its domain organization triggered by ATP/substrate binding (28). Additionally, Tomm34 was reported to possess ATPase activity (46). To assign the key effect of ATP in Hsp70/Tomm34 interaction to one of the interacting proteins, we compared the binding of Tomm34 to full-length Hsp70, NBD(1–383), and SBD(383–641) in the presence of ATP, ADP, or nucleotide-free (Fig. 1C). To rule out the possibility that Tomm34 interacts with Hsp70 as its substrate during the ATP-triggered chaperone cycle, we tested Tomm34 interaction with the Hsp70 V438F mutant defective in substrate binding (31, 47). These experiments revealed that Tomm34 increasingly interacts only with full-length Hsp70/Hsp70 V438F proteins in the presence of ATP. This result suggests that the conformational change of allosterically coupled NBD-SBD domains is required for efficient Tomm34 binding and is independent of Hsp70 substrate binding capacity.
Tomm34 Has Nanomolar Affinity for Hsp70 in the Presence of ATP
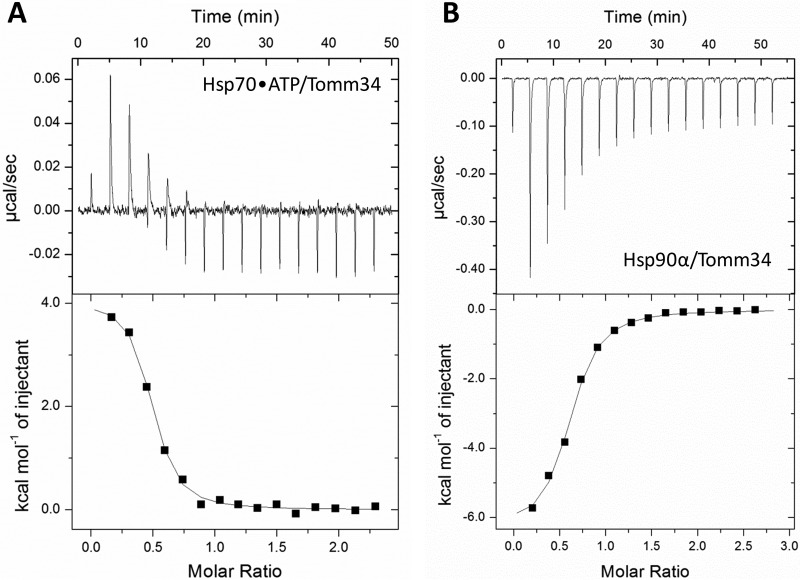
To investigate the thermodynamics of the Hsp70/Tomm34 interaction, we performed isothermal titration calorimetry. Tomm34 was titrated into a cell containing full-length Hsp70 protein pre-equilibrated with ATP (Fig. 2A). We did not detect any binding in the absence of ATP. All negative controls needed for assessment of possible nonspecific heat changes, e.g. ATP hydrolysis, were performed as shown in supplemental Fig. 1. The experiment was initially carried out at 25 °C. To our surprise, there was a very small enthalpy change for the interaction at this temperature, and it was not possible to analyze the data. Therefore, we repeated the experiment over a temperature range from 5 to 30 °C (supplemental Fig. 1). At 5 °C, the Hsp70/Tomm34 interaction is endothermic, with an enthalpic contribution, ΔH, of 5672 cal mol−1, an entropy change, ΔS, of 49.7 cal mol−1 K−1, and a KD of 0.4 μm (Table I). The Hsp70/Tomm34 interaction has a large positive entropic contribution at all temperatures measured. Although the increase of temperature causes lowering of entropic contribution to the binding, the affinity of the interaction seems to be slightly strengthened. This is provided by an increase in the enthalpic contribution to the Gibbs free energy. Nevertheless, ΔS still remains the main factor that drives the Hsp70/Tomm34 interaction. Interestingly, Tomm34 binding to the other major human molecular chaperone Hsp90 is an enthalpically driven exothermic interaction (Fig. 2B), which suggests a different binding mechanism. According to our previous study (13), the binding of Tomm34 to Hsp90 is mediated solely through electrostatic interaction of Tomm34-TPR2 with EEVD-Hsp90. On the contrary, the Hsp70/Tomm34 interaction involves additional non-EEVD contacts. Considering the unusually large entropic term, these additional contacts might be explained by hydrophobic effects and/or structural rearrangement initiated by Tomm34 binding.
Fig. 2.
Different thermodynamics of Tomm34 binding to Hsp70·ATP and Hsp90α. Tomm34 was injected into a cell containing Hsp70·ATP (A) or Hsp90α (B) for isothermal titration calorimetry. Both chaperones bound to Tomm34 with stoichiometries of ∼1:2 (Tomm34:Hsp70/Hsp90α) and dissociation constants of 0.2 μm (at 15 °C) and 0.96 μm (at 25 °C) for Hsp70·ATP and Hsp90α, respectively. See Table I for the full results.
Table I. ITC measurements of Hsp70/Hsp90-Tomm34 interactions.
ITC was used to investigate the ATP-dependent interaction between Tomm34 and Hsp70. At 25 °C, we observed only a very small enthalpy change for the interaction; thus we explored the interaction at different temperatures (for ITC plots see supplemental Fig. 1). To compare Tomm34 binding to another chaperone, we also measured the Hsp90α/Tomm34 interaction.
| Protein complex | Nucleotide | T | KD | ΔH | ΔS | TΔS | N |
|---|---|---|---|---|---|---|---|
| °C | μm | cal mol−1 | cal mol−1 K−1 | cal mol−1 | |||
| Hsp70/Tomm34 | ATP | 5 | 0.40 ± 0.06 | 5672 ± 168 | 49.7 | 13824 | 0.43 ± 0.01 |
| Hsp70/Tomm34 | ATP | 15 | 0.20 ± 0.05 | 3625 ± 126 | 43.3 | 12477 | 0.40 ± 0.01 |
| Hsp70/Tomm34a | ATP | 25 | NAb | NA | NA | NA | NA |
| Hsp70/Tomm34 | ATP | 30 | 0.10 ± 1.40 | −1647 ± 146 | 26.5 | 8033 | 0.60 ± 0.04 |
| Hsp90α/Tomm34 | 25 | 0.96 ± 0.10 | −6392 ± 146 | 6.1 | 1819 | 0.57 ± 0.01 |
a It was not possible to analyze the data due to a very small enthalpy change.
b NA means not applicable.
Tomm34 Binds Preferentially to Hsp70·ATP and Influences Hsp70 Chaperone Function
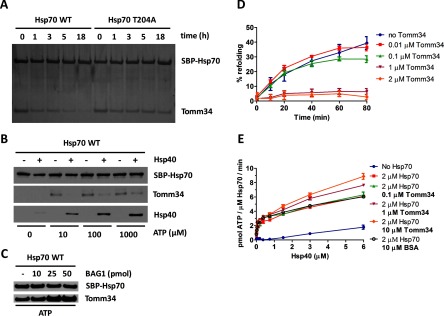
To investigate the nucleotide-bound state of Hsp70 in which it interacts with Tomm34, we evaluated the temporal stability of Hsp70/Tomm34 complexes preformed in the presence of ATP after washing out the free nucleotides using wild type Hsp70 and Hsp70 T204A mutant with compromised intrinsic ATPase activity but intact allosteric coupling (27, 48–50) (Fig. 3A and supplemental Fig. 2A). Compared with the Hsp70/Tomm34 assembly, we detected higher stability of preformed Hsp70 T204A/Tomm34 complexes. Next, we assessed the stability of the Hsp70/Tomm34 complex formed in the presence of ATP and Hsp40 (Fig. 3B). We observed rapid destabilization of Hsp70/Tomm34 assembly in the presence of Hsp40 and 10 μm ATP, indicating that accelerated ATP hydrolysis in the presence of Hsp40 leads to dissociation of Tomm34 from Hsp70. Interestingly, enhanced Hsp40 association with Hsp70 was accompanied by partial restoration of Hsp70/Tomm34 interaction at higher ATP concentrations (100 and 1000 μm, respectively) suggesting that Hsp40 is not competing with Tomm34 for binding to Hsp70. Conversely, we used increasing amounts of co-chaperone Bag-1-nucleotide exchange factor for Hsp70 (51) to enrich the population of Hsp70·ATP molecules and evaluated the level of Tomm34 bound to Hsp70 (Fig. 3C). As expected, the addition of Bag-1 enhanced the formation of Hsp70·ATP/Tomm34 complexes. Taken together, these experiments support the hypothesis that Tomm34 binds to full-length Hsp70 in its ATP rather than ADP·Pi-bound conformation. Furthermore, these results show that Tomm34 interaction with Hsp70·ATP is regulated by Hsp40/Bag-1 activity.
Fig. 3.
Tomm34 binding to Hsp70·ATP is dependent on ATP hydrolysis rate modulated by Hsp40 and Bag-1 co-chaperones and influences chaperone activity. A, SBP-Hsp70·ATP/Tomm34 complexes preformed in the presence of ATP (0.2 mm) were washed in nucleotide-free buffer and incubated at 4 °C for the indicated times before elution by biotin. The level of eluted proteins was analyzed by SDS-PAGE and silver staining. B, equimolar amounts (25 pmol) of SBP-Hsp70, Tomm34, and Hsp40 (as indicated) were mixed in increasing concentrations of ATP, eluted by biotin, and analyzed by Western blotting. C, equimolar amounts (25 pmol) of SBP-Hsp70 and Tomm34 were mixed with increasing amounts of Bag-1 in the presence of ATP (0.2 mm). Eluted proteins were analyzed by Western blotting. D, firefly luciferase was chemically denatured, mixed with Hsp70 (1 μm), Hsp40 (2 μm), ATP (1 mm), and varying Tomm34 concentrations, and recovered luminescence was measured. Negative controls described under “Experimental Procedures” did not exhibit luciferase activity and are not shown. E, ATPase activity of Hsp70 (2 μm) was tested at various Hsp40 and Tomm34 concentrations in malachite green assay. Refolding and ATPase experiments were performed in independent triplicates. Error bars represent S.E.
At the same time, the above-mentioned observations suggest that Tomm34 enters the Hsp40/Bag-1-directed ATPase cycle of Hsp70. To examine the effect of Tomm34 on Hsp70 chaperone function, we tested its influence on Hsp70/Hsp40-mediated luciferase refolding and ATPase activity (Fig. 3, D and E). We found that Tomm34 added at equimolar concentration to Hsp70 completely inhibits refolding of chemically denaturated luciferase (Fig. 3D). On the contrary, Tomm34 promotes Hsp40-stimulated Hsp70 ATPase activity in a concentration-dependent manner (Fig. 3E). These data imply that Tomm34 regulates Hsp70 chaperone activity. However, elucidation of the mechanism by which Tomm34 influences Hsp70 performance requires deeper structural insight into Hsp70/Tomm34 complex architecture.
Characterization of Human Hsp70 Structural Mutants
Most functional and structural studies have been done on the bacterial Hsp70 homologue DnaK, including the determination of DnaK·ADP and DnaK·ATP crystal/NMR structures (23, 27, 52). Because Hsp70s represent evolutionarily highly conserved proteins (53, 54), we decided to prepare human Hsp70 mutations homologous to previously described DnaK mutants with defective conformational properties to test their interaction with Tomm34. First, we prepared the Hsp70 I164D mutant that corresponds to DnaK I160D, Sse1 I163D, and Ssa1 I162D mutants (54). These proteins have impaired NBD-SBDα contacts in ATP-bound conformation. Second, we prepared the Hsp70 D529A mutant homologous to DnaK D526A (55), characterized by destabilization of the SBDβ subdomain and NBD in the ADP-bound state and by faster substrate association and dissociation rates. To evaluate the structure and conformational activity of WT, I164D, and D529A proteins, we measured the level of H/D exchange in these proteins in nucleotide-free conditions or in the presence of ATP by mass spectrometry (Fig. 4A and supplemental Fig. 3A). Additionally, we determined the ability of these mutants to bind peptide substrates (supplemental Fig. 3, B and C).
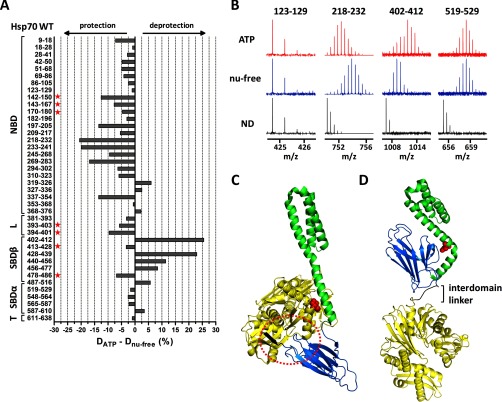
Fig. 4.
ATP-induced allosteric domain docking of Hsp70. A, deuteration level differences of Hsp70 peptides in ATP-bound and nucleotide-free state after 1 h of incubation in deuterated buffer. Numbers at left indicate the Hsp70 peptide fragments; schematic representation at left shows Hsp70 domain constitution; L, interdomain linker; T, C-terminal tail. B, original mass spectra of representative peptide fragments of Hsp70 incubated in non-deuterated buffer (ND) or for 1 h in deuterated buffer in the presence/absence of ATP. C and D, crystal structures of Hsp70 bacterial homologue DnaK in ATP-bound state (C, PDB code 4jne) and in nucleotide-free/ADP state (D, PDB code 2kho). NBD, interdomain linker, SBDβ, and SBDα are shown in yellow, black, marine, and green, respectively. Encircled region (C) shows tight association of IA subdomain of NBD, linker, and SBD loops L2,3/L6,7 (peptides indicated with a star in A) when SBDβ is docked onto NBD (compare with I164D mutant, Fig. 5A). Residues I160 and D526 (human numbering I164 and D529) are highlighted in red in ATP/ADP structures, respectively. The images were created in PyMOL.
In the case of WT protein (Fig. 4A), peptides covering NBD exchanged deuterium atoms more slowly in the ATP-bound state, indicating a tightening of the NBD domain upon ATP binding. On the contrary, SBDβ segments exhibited faster H/D exchange in the ATP-bound state, reflecting a destabilization of SBDβ induced by ATP. We also detected a different deuteration level of peptides localized in the linker region (amino acids 380–400) in the nucleotide-free and ATP-bound state. Peptides 393–403 and 394–401 showed higher solvent accessibility in the nucleotide-free state and protection induced by ATP binding. Similar structural changes upon ATP binding were observed for bacterial DnaK using a similar experimental approach (56) demonstrating that our data reliably reflect conserved conformational motions in human Hsp70.
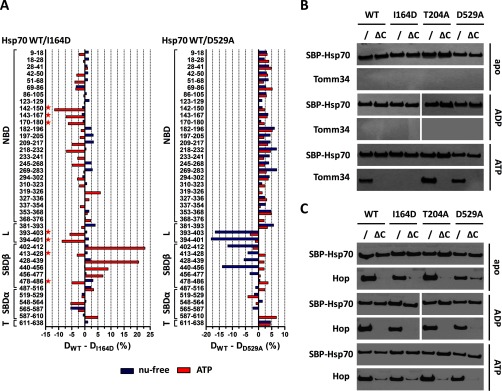
To assess the structural differences between WT and I164D/D529A mutants, we compared deuterium incorporation into WT and both mutants in the nucleotide-free and ATP-bound state (Fig. 5A). For the I164D mutant, we observed a similar deuteration profile as WT protein under nucleotide-free conditions, showing that I164D mutation does not affect the structural integrity of Hsp70 in its NBD-SBD undocked state. In line with this observation is the comparable substrate binding activity of this Hsp70 variant with WT protein (supplemental Fig. 3, B and C). On the contrary, I164D exhibited changed deuterium incorporation in the presence of ATP. Importantly, NBD regions covered by peptides (197–205, 218–232, 233–241, 269–283, and 337–354, see supplemental Fig. 3A) containing residues involved in ATP coordination (G202, E268, K271, S275, and G339) (57) demonstrated a similar trend of protection from deuteration as the WT protein, indicating that ATP binding and subsequent structural rearrangements in NBD are not perturbed by I164D mutation. Conversely, the SBDβ subdomain of I164D mutant remained largely inaccessible to solvent (peptides 402–412, 428–439, 440–456, and 456–477, see Fig. 5A, left), with the exception of segments covered by peptides 413–428 and 478–486 (corresponding to SBD loops L2,3 and L6,7, respectively (25)), that exhibit increased deuterium exchange. Concordantly, peptides derived from IA subdomain of NBD (142–150, 143–167, and 170–180) and the interdomain linker (393–403 and 394–401) also incorporated deuterium atoms more rapidly (Fig. 5A, left). These observations are in agreement with crystallographic data available for DnaK in the ATP-bound state revealing tight association of the interdomain linker, IA subdomain residues, and SBD loops L2,3/L6,7 when SBD is docked onto NBD (Fig. 4C) (23, 27). Taken together, our H/D exchange data show that although Hsp70 I164D mutant is able to bind ATP, its capacity to induce stable NBD-SBDβ docking is impaired. This is presumably caused by the abrogation of hydrophobic contacts between Ile-164 and conserved residues in the αA helix of SBDα (L510, M518, V519, and A522) (23, 25, 54).
Fig. 5.
Hsp70 I164D lacks the ability to induce NBD-SBDβ docking upon ATP binding, whereas D529A mutant exhibits destabilized SBDβ subdomain/linker conformation in nucleotide-free state. In contrast to Hsp70/Hop binding, the efficient Hsp70/Tomm34 interaction is dependent on NBD-SBDβ docking. A, deuteration level differences of Hsp70 WT and I164D (or D529A) peptides in ATP-bound (red) or nucleotide-free state (blue) after 1 h of incubation in deuterated buffer. Peptides indicated with a star (left panel) highlight the impaired NBD-SBDβ domain docking of I164D mutant (compare with WT, Fig. 4C). B and C, purified SBP-tagged WT or mutant Hsp70s immobilized on streptavidin-agarose beads were incubated with Tomm34 (B) or Hop (C) in buffer containing 0.2 mm ATP, ADP, or no nucleotide. After washing, eluted proteins were analyzed by Western blotting. ΔC, deletion of PTIEEVD.
In comparison with I164D, the D529A mutant exhibited reverse defects in the structure of nucleotide-free/ATP-bound conformations by being able to reach the wild type-like ATP-bound state but was unable to correctly establish the structure of the SBDβ subdomain in the absence of ATP (Fig. 5A, right). D529 present in the αB helix of the lid subdomain makes ionic contact with R447 localized in SBD loop L4,5, and thus it stabilizes the position of SBDβ and the lid subdomain in the nucleotide-free/ADP-bound state of Hsp70 (25, 55, 58, 59). In agreement with the mentioned structural role of D529, we detected an increased deuteration level of peptides 440–456 and 402–412 covering loops L4,5 and L1,2, respectively, accompanied by consistently elevated deuteration of the entire SBDβ subdomain in D529A protein. Surprisingly, the most solvent-exposed peptides of the D529A mutant in the nucleotide-free state, 393–403 and 394–401, were localized in the linker region of the protein. Correspondingly, to the observed rearrangements of SBDβ subdomain, the D529A mutant has impaired substrate binding activity (supplemental Fig. 3, B and C).
Efficient Hsp70/Tomm34 Interaction Requires Both Allosteric Coupling of NBD-SBD Domains and Accommodation of C-terminal EEVD Motif by TPR Domain
Next, we tested the interaction of I164D and D529A mutants with Tomm34 in the presence or absence of nucleotides (Fig. 5B). The T204A mutant was included to reflect the influence of ATP hydrolysis. We also analyzed the binding of Hop to compare the mode of interaction of these two Hsp70/Hsp90 organizing proteins with Hsp70 conformational mutants (Fig. 5C) (8, 13, 14, 60). Each Hsp70 variant was tested as the full-length protein or C-terminally truncated construct lacking the PTIEEVD motif. Expectedly, the PTIEEVD motif was indispensable for efficient Tomm34 and Hop binding to Hsp70 variants. Moreover, Hop binds the tested full-length Hsp70 proteins to the same extent under all nucleotide conditions, indicating that Hsp70/Hop interaction is not dependent on Hsp70 conformation and is mediated mostly by TPR domain recognition of the PTIEEVD motif (61). On the contrary, the ATP-strengthened Hsp70/Tomm34 interaction was lost for the I164D mutant independently of the PTIEEVD motif. Hsp70 T204A·ATP/Tomm34 complex formed more readily, corresponding to the decreased ATPase activity of the T204A mutant. These results support the hypothesis that efficient Hsp70/Tomm34 interaction is dependent on ATP-induced NBD-SBD docking (I164D lacks ATP-induced domain allostery, Fig. 5A, left) but does not require intact SBDβ subdomain/linker conformation of nucleotide-free Hsp70 (D529A, Fig. 5A, right). Furthermore, Tomm34 TPR1 domain accommodation of Hsp70 C-terminal PTIEEVD motif (13) is necessary to initiate or stabilize the ATP-induced Hsp70/Tomm34 interaction.
Tomm34 Binding to Hsp70·ATP Induces Structural Changesat the αA/αB Helices in Hsp70 SBDα
To determine the structural changes induced in Tomm34 and Hsp70 proteins upon their interaction, we performed H/D exchange measurement of the Hsp70·ATP/Tomm34 complex (Fig. 6). In parallel, we analyzed the structural impact of Hsp70/Hop complex formation (Fig. 7). Hsp70/Hop complex components were evaluated in nucleotide-free conditions because this assembly is not nucleotide-dependent (Fig. 5C).
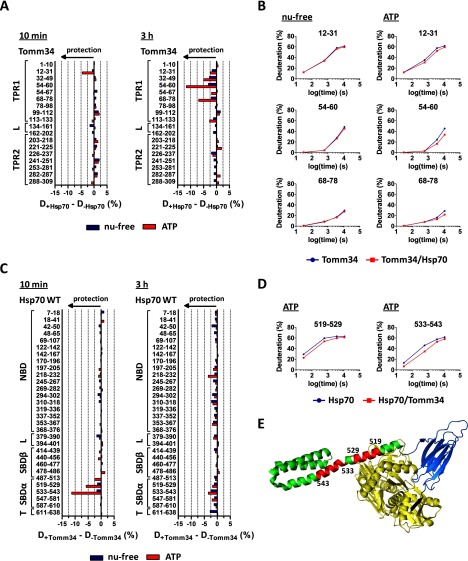
Fig. 6.
Although Hsp70·ATP protects Tomm34 peptides covering TPR1 domain, Tomm34 binding to Hsp70·ATP induces structural changes at the αA/αB helices in Hsp70 SBD. A, difference in deuteration of Tomm34 in the complex with Hsp70 and Tomm34 alone in the presence (red) or absence (blue) of ATP after 10 min (left) or 3 h (right) of incubation in deuterated buffer. B, deuteration kinetics of selected Tomm34 peptides protected by Hsp70 in the presence of ATP, but not in nucleotide-free state. C, difference in deuteration of Hsp70 in the complex with Tomm34 and Hsp70 alone in ATP-bound (red) or nucleotide-free state (blue) after 10 min or 3 h of incubation in deuterated buffer. Hsp70 peptides 519–529 and 533–543 exhibited consistent protection by Tomm34 in the presence of ATP. D, deuteration kinetics of 519–529/533–543 peptides of Hsp70·ATP protected from deuteration by the presence of Tomm34. E, peptides are indicated in red in the tertiary structure representation of DnaK·ATP (PDB code 4jne) with human Hsp70 numbering. Hsp70 domain constitution is colored similarly as in Fig. 4.
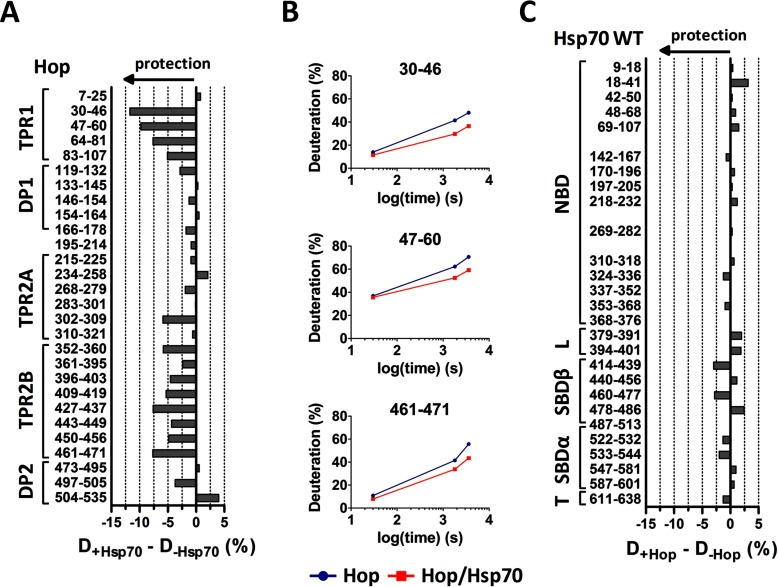
Fig. 7.
Peptides covering TPR1 and TPR2B domain of Hop are consistently protected by Hsp70. A, difference in deuteration of Hop in the complex with Hsp70 and Hop alone after 30 min of incubation in deuterated buffer. B, deuteration kinetics of selected Hop peptides shows stable protection by Hsp70. C, effect of Hop binding on deuteration of Hsp70 peptides after 30 min of incubation in deuterated buffer.
As we reported previously (13), the interaction of Tomm34 with Hsp70 under nucleotide-free conditions is weak and does not lead to significant differences in Tomm34/Hsp70 deuteration (Fig. 6, A and C). Before evaluating the deuteration changes of Tomm34/Hsp70 proteins in complex formed upon ATP addition, we first analyzed the influence of ATP on deuteration of Tomm34 protein only. ATP does not change the deuteration level of Tomm34 peptides determined under nucleotide-free conditions indicating that the structure of Tomm34 protein is not affected by ATP (supplemental Fig. 4). By measuring deuterium incorporation into Tomm34 protein in complex with Hsp70·ATP, we detected moderate but consistent protection of peptides 12–31, 54–60, and 68–78 (Fig. 6A). These peptides exhibited different deuteration kinetics (Fig. 6B), whereas the deuteration kinetics of peptide 12–31 implicate small and transient structural rearrangements, and structural changes induced in regions covered by peptides 54–60 and 68–78 are more stable. Importantly, these peptides cover residues of the TPR1 two-carboxylate clamp (R13, N17, and N56) that were shown to accommodate the Hsp70 C-terminal peptide (13). These results suggest that the two-carboxylate clamp of the Tomm34 TPR1 domain strengthens its contacts with the C-terminal EEVD motif of Hsp70 during ATP-induced Hsp70/Tomm34 complex formation. No other apparent changes in Tomm34 protein deuteration upon its interaction with Hsp70 were detected.
Importantly, measurement of deuterium incorporation into Hsp70·ATP protein in complex with Tomm34 revealed isolated protection of amino acid segment localized in the SBDα subdomain, highlighted by 519–529 and particularly 533–543 peptide covering the αA/αB-helices (Fig. 6, C–E). Deuteration kinetics of both peptides show a transient profile of the interaction (Fig. 6D). Other peptides of the Hsp70·ATP protein were not differentially deuterated in the presence of Tomm34, including peptide 611–638 covering the C terminus. The absence of protection of the extreme Hsp70 C terminus might be caused by the high flexibility of this region (24, 62) or by side chain-mediated interactions that cannot be trapped by H/D exchange. These data suggest that the identified Hsp70 segment spanning amino acids 519–529/533–543 represent the additional Hsp70/Tomm34 interaction interface formed exclusively in the ATP-bound conformation of Hsp70, or this region reflects allosteric changes induced by Tomm34 binding to different Hsp70·ATP sites.
Deuteration of Hop-derived peptides was selectively decreased in regions corresponding to its TPR1 and TPR2B domains upon Hsp70 binding (Fig. 7A). Deuteration kinetics of peptides belonging to these domains suggest stable rearrangements in this part of Hop (Fig. 7B). Because the Hsp70/Hop complex has a 1:1 stoichiometry (63) and TPR1/TPR2B domains were previously described as redundant (61) but differentially regulated (15, 63) binding sites for the C-terminal motif of Hsp70, our results provide structural evidence for random and individual Hsp70 binding to each one of these domains. Conversely, Hop binding does not influence the level of deuterium exchange in Hsp70 protein, including peptide 533–543 (Fig. 7C). The missing protection of the C terminus has been explained above. Together, these findings further support the notion that Hsp70/Hop interaction is mainly PTIEEVD:TPR1/TPR2B-dependent and is not affected by Hsp70 conformational status (Fig. 5C).
Deletion of 533–543 Segment in SBDα Diminishes Tomm34 Binding
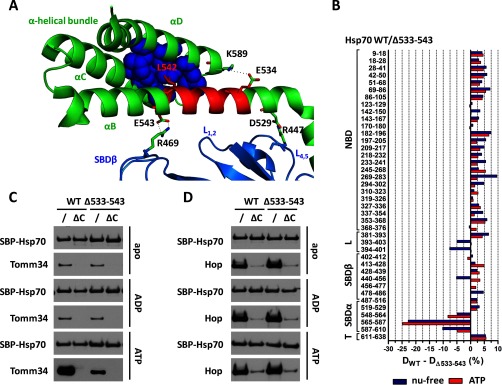
To assess the importance of αB helix for Tomm34 binding, we prepared the Hsp70 mutant with the deleted 533–543 amino acid segment. This region contains residues important for structural integrity of α-helical bundle by forming ionic contacts (E534-K589) and hydrophobic core (L542-V577, W580, L581, F592, K595, L599) (Fig. 8A) (25, 55). The lid-SBDβ positioning ionic contacts described for DnaK (human Hsp70 numbering E543–R469) seem to be preserved in human Hsp70 (25, 55). To analyze the structural differences between WT Hsp70 and Δ533–543 mutant, we compared deuterium incorporation into these proteins in nucleotide-free and ATP-bound states (Fig. 8B and supplemental Fig. 3A). The Δ533–543 mutant exhibited comparable structural changes as WT protein upon ATP binding demonstrated by consistent protection of NBD and linker region and deprotection of SBDβ indicating active allosterically coupled NBD-SBDβ docking of this deletion mutant (supplemental Fig. 3A). Unfortunately, peptide 478–486 from L6,7 SBD loop of the Δ533–543 mutant was not detected by MS analysis in samples with ATP. Strikingly, the deletion has led to destabilization of α-helical bundle folding unit as evidenced by enhanced solvent accessibility of peptides 548–564, 565–587, and 587–610 independent of the nucleotide state (Fig. 8B). The most pronounced difference was detected in peptide 565–587 covering the entire αC helix (25). Moreover, comparison of WT and Δ533–543 in the nucleotide-free states revealed increased deuteration of peptides covering linker (393–403 and 394–401) and L4,5 SBDβ loop (440–456) regions in the Δ533–543 protein, whereas the overall structure of its SBDβ subdomain remained intact (Fig. 8B). Additionally, Δ533–543 has impaired ability to bind substrate peptide (supplemental Fig. 3, B and C). Together, these data show that the 533–543 deletion leads to destabilization of the α-helical bundle followed by changes in linker/L4,5 region and abrogates stable peptide binding. However, Δ533–543 mutant retains the ability to induce ATP-triggered NBD-SBDβ docking.
Fig. 8.
Deletion of 533–543 region severely decreased EEVD/ATP-dependent Hsp70/Tomm34 interaction without abrogating allosteric domain docking. However, the deletion has led to destabilization of the α-helical bundle. A, crystal structure of human SBD (PDB code 4po2). SBDβ, SBDα, and 533–543 regions are shown in marine, green, and red, respectively. The lid-SBDβ positioning ionic contacts (D529–R447 and E543–R469) and residues important for structural integrity of α-helical bundle by forming ionic contacts (E534–K589) and hydrophobic core (L542 shown in red, V577, W580, L581, F592, K595, and L599 shown as blue spheres) are indicated. B, deuteration level differences of Hsp70 WT and Δ533–543 peptides in ATP-bound (red) or nucleotide-free state (blue) after 1 h of incubation in deuterated buffer. C and D, purified SBP-Hsp70 WT or Δ533–543 immobilized on streptavidin-agarose beads were incubated with Tomm34 (C) or Hop (D) in buffer containing 0.2 mm ATP, ADP, or no nucleotide. Eluted proteins were analyzed by Western blotting. ΔC, deletion of PTIEEVD.
Next, we analyzed the effect of 533–543 deletion on Tomm34 and Hop binding to Hsp70 (Fig. 8, C and D). The results show that TPR1:EEVD dependence of Tomm34 interaction with Hsp70 is not affected by 533–543 deletion. Conversely, Tomm34 binding to Δ533–543·ATP mutant was diminished compared with WT·ATP. These findings suggest the following: 1) 533–543 peptide represents part of or complete secondary interaction site of Hsp70·ATP/Tomm34 complex, or 2) the structural integrity of Hsp70·ATP α-helical bundle is required during efficient formation of Hsp70·ATP/Tomm34 interaction. Hsp70/Hop interaction is not affected by 533–543 deletion.
Interdomain Linker of Tomm34 Represents an Important Region for Conformation-specific Hsp70/Tomm34 Interaction
Finally, we delineated the binding interface of Hsp70-Tomm34 assembly in Tomm34 protein structure. H/D exchange experiments supported the importance of TPR1 two-carboxylate clamp for Tomm34 interaction with Hsp70·ATP; however, no other differentially deuterated regions of Tomm34 were detected (Fig. 6A). The prediction of Tomm34 protein secondary structure (supplemental Fig. 5A) revealed the presence of an interdomain segment (∼140–190) separating the TPR1 and TPR2 domains (these are formed by seven helices). To assess the role of this interdomain linker during Hsp70·ATP/Tomm34 interaction, we tested the binding of individual Tomm34 TPR domains with or without the interdomain linker sequence to Hsp70 (Fig. 9A). We observed ATP-strengthened binding only for Hsp70 interaction with the TPR1 domain containing the interdomain linker. Interestingly, absence of the linker from TPR1 completely abolished its Hsp70 binding. On the contrary, Hsp70 binds both TPR2 variants to the same extent in all nucleotide conditions indicating ATP-independent binding through TPR:EEVD contacts. This behavior of the TPR2 domain toward the Hsp70 C terminus was reported previously (13) and probably reflects non-physiological TPR2/Hsp70 association. Together, these results indicate that the interdomain linker is necessary for TPR1 domain capacity to accept the Hsp70 EEVD motif and contributes to ATP-induced Hsp70/Tomm34 interaction.
Fig. 9.
Interdomain linker of Tomm34 represents important region for conformation-specific Hsp70/Tomm34 interaction. The length of TPR domain constructs and mutated residues in the linker region are indicated. Purified SBP-Hsp70 WT was preincubated with streptavidin-agarose beads and mixed with TPR constructs (A) or Tomm34 mutants (B) in different nucleotide conditions. After washing, eluted proteins were subjected to SDS-PAGE and Western blot with mouse polyclonal sera against Tomm34 (A) or silver staining (B). The capacity of polyclonal sera to detect purified TPR domains was tested in A.
Furthermore, the interdomain linker contains a stretch of evolutionarily conserved residues predominantly of a hydrophobic nature (140–158, supplemental Fig. 5B). We hypothesized that these residues might participate in interaction with Hsp70 and examined the interaction of four substitution mutants (W140A, K143A, P151A, and W158A) with Hsp70 (Fig. 9B). Although K143A and P151A substitutions have no effect on Hsp70·ATP binding, substitutions of large hydrophobic residues W140 and W158 to alanines diminished Tomm34 binding to ATP-bound Hsp70. This suggests that W140/W158 tryptophan residues play a role in Hsp70·ATP/Tomm34 complex formation. The high flexibility (solvent exposure) of this conserved part of the interdomain linker was shown previously by measuring deuterium incorporation into Tomm34 (here and Ref. 13). Rapid deuteration in this region might have abrogated the detection of its structural changes induced upon Hsp70·ATP binding (Fig. 6A).
DISCUSSION
Our previous results suggested that Tomm34 TPR1 domain recognition of Hsp70 C-terminal EEVD motif is not sufficient for effective Hsp70/Tomm34 binding (13). Here, we have shown that ATP-induced allosteric domain docking and accompanying structural changes in Hsp70 chaperone are required for full Hsp70/Tomm34 interaction. Structural analyses revealed several regions implicated in the Hsp70/Tomm34 interaction. In Tomm34, TPR1 plus interdomain linker forms a structural unit mediating both Hsp70-EEVD motif accommodation and additional ATP-dependent Hsp70/Tomm34 contacts involving hydrophobic interactions (Figs. 6A and 9). In Hsp70·ATP, we identified a segment corresponding to residues 533–543 that is protected from deuteration by Tomm34 (Fig. 6C). This segment covers the central part of the αB helix that contains conserved amino acids involved in structural integrity of the α-helical bundle (E534 and L542) and lid-SBDβ positioning (E543) (Fig. 8, A and B) (25, 55). Two possible scenarios exist to explain the Tomm34-induced protection of 533–543 Hsp70·ATP region as follows. 1) The segment covering residues 533–543 directly represents the additional Hsp70·ATP/Tomm34 interaction interface. 2) The protection of 533–543 region is allosterically induced upon Tomm34 binding to a different site on Hsp70·ATP. In favor of the first scenario is that the 533–543 region closely contacts SBDβ subdomain in Hsp70·ADP (25) but becomes exposed in the ATP-induced conformation of Hsp70 (23, 27). Thus, the observed deprotection of 533–543 peptide upon ATP binding abrogated by Tomm34 might reflect direct contacts between these proteins enabled by accessibility of the central part of αB helix (Figs. 4 and 6C). The high affinity Hsp70·ATP/Tomm34 interaction is largely entropically driven (Fig. 2A). A positive entropic contribution to protein/protein interactions is commonly explained by the contribution of hydrophobic effects and/or structural rearrangements during binding (64, 65). We did not detect large changes in Hsp70·ATP and Tomm34 structures upon their interaction (Fig. 6, A and C) suggesting that hydrophobic contacts drive Hsp70·ATP/Tomm34 binding. This is in agreement with decreased association of Tomm34 W140A/W158A mutants with Hsp70·ATP (Fig. 9B). Because region 533–543 of Hsp70 contains mostly charged residues, it seems unlikely that this site represents a direct Hsp70·ATP/Tomm34 interaction interface. In addition to this notion is the observation that mainly electrostatic contacts between Tomm34-TPR2 domain and Hsp90-EEVD motif are characterized by large enthalpic contributions (Fig. 2B). Intriguingly, deletion of 533–543 severely decreased EEVD/ATP-dependent interaction of Tomm34 with Hsp70 without abrogating allosteric domain docking (Fig. 8, B and C). The Δ533–543 mutant exhibited a collapsed structure of the α-helical bundle in both nucleotide states (Fig. 8B) suggesting an allosteric mechanism of the Tomm34-induced 533–543 segment protection in which the putative Hsp70·ATP/Tomm34 interface is localized in the structure of the α-helical bundle and the binding event is allosterically reflected by stabilization of the 533–543 region. Indeed, a bipartite interaction between the CHIP TPR domain with accommodated EEVD motif and the residues of α-helical bundle of Hsc70 was described recently (5). Interestingly, the additional CHIP TPR/α-helical bundle contacts were disrupted by introducing charged residues (Asp) at non-polar positions V59 and L129 of CHIP implicating a hydrophobic contribution to this interaction. Moreover, the authors of this study suggested that this mode of CHIP TPR domain interaction with EEVD/α-helical bundle structures might be universal to several other TPR co-chaperones. Yet, another scenario explains Tomm34-induced protection of the 533–543 region and significant positive entropy measured during Hsp70·ATP/Tomm34 association. This scenario is based on recent results showing that conserved residues N540 and E543 (human Hsp70 numbering) form the interface of rare ATP-bound DnaK dimer with large buried surface area (33). Our ITC analysis of Hsp70·ATP/Tomm34 complex suggests a 2:1 stoichiometry (Table I). Thus, Tomm34 binding to Hsp70 might increase the population of Hsp70·ATP dimers, which would be reflected by decreased solvent accessibility of peptide 533–543 covering residues involved in Hsp70·ATP dimer interface. The reorganization of ordered water molecules upon dimer formation together with a hydrophobic Hsp70·ATP/Tomm34 interface might account for the high entropy increase, as described previously (65–67). Together, our H/D exchange and ITC analyses provided a number of testable hypotheses that will be addressed in future experiments to delineate the details of Hsp70·ATP/Tomm34 assembly topology. Our parallel experiments testing the Hsp70/Hop interaction (Fig. 5C) supported previous results showing that high affinity interaction of these proteins is mediated by TPR1/TPR2B accommodation of EEVD motif only, independent of Hsp70 conformation (15, 61, 63). Nevertheless, additional low affinity contacts of Hsp70 and Hop suggested elsewhere (3, 5, 18, 60, 68) might have remained undetected by our methods.
For the purposes of this work, we have prepared and partly characterized human Hsp70 structural mutants I164D, D529A, and Δ533–543 (Figs. 5A and 8B and supplemental Fig. 3). As discussed above, the Δ533–543 mutant has an unstructured α-helical bundle suggesting that residues in the deleted region are responsible for maintaining the α-helical subdomain (25). Conversely, the absence of E543-R469 ionic contacts stabilizing the so-called “latch” between the lid and the SBDβ subdomain in the nucleotide-free state of Hsp70 does not affect the structure of SBDβ (Fig. 8B) (55). This is consistent with the finding that the lid subdomain is not tightly bound to SBDβ and is largely mobile (69). However, Δ533–543 has impaired peptide binding activity (supplemental Fig. 3, B and C) supporting the role of the intact α-helical lid for stable substrate binding (30, 70, 71). Interestingly, disruption of the α-helical bundle in Δ533–543 was accompanied by destabilization of the interdomain linker and L4,5 SBDβ loop regions in the nucleotide-free state (Fig. 8B). This observation might reflect lid-linker allostery (72) or perturbations in Hsp70 dimerization mediated through the α-helical bundle-linker interface (73). D529A mutation caused reorganization of SBDβ structure in nucleotide-free Hsp70 and abrogated substrate binding (Fig. 5A and supplemental Fig. 3, B and C). The most prominent destabilization was in the interdomain linker. These results provide a structural explanation for increased on/off rates of peptide binding to Hsp70 mutants with disrupted network of ionic interactions between helix αA/B and the SBDβ subdomain reported previously (55, 59, 69) and highlight the stabilizing role of these ionic contacts on the linker region of Hsp70 in its nucleotide-free state. Interestingly, covalent fixation of D526-K446 contacts in DnaK (human Hsp70 residues D529-R447) disrupts the chaperone allostery (69). This finding together with our results indicates that free shuttling of interdomain linker between SBDβ and NBD is abrogated in this cross-linked variant (74).
The existence of Hsp70·ATP/Tomm34 complex is dependent on ATP hydrolysis rate (Fig. 3A) and is modulated throughout the Hsp70 ATPase cycle by Hsp40 and Bag-1 co-chaperones (Fig. 3, B and C). Tomm34 further accelerates Hsp40-stimulated Hsp70 ATPase activity (Fig. 3E). Conversely, its presence inhibits Hsp70/Hsp40-mediated refolding of denatured luciferase (Fig. 3D). The Hsp40 used in our assays is encoded by gene DNAJB1. The cooperation of this Hsp40 variant with Hsp70 in refolding assays was reported to be dependent on the presence of the EEVD motif (32, 75, 76). The association between Hsp40s and the C-terminal structures of Hsp70 seems to represent a more general phenomenon (77–81). Interestingly, the unstructured C-terminal sequences of Hsp70, including the EEVD motif, have been shown to play an inhibitory role in the regulation of the Hsp70 ATPase cycle (76, 79). Therefore, we hypothesize that the tight association of Tomm34 with both the EEVD motif and the α-helical lid region of Hsp70 may interfere with their inhibitory effect on Hsp70 ATPase activity. Furthermore, Tomm34 binding to Hsp70 might uncouple the Hsp40 ATPase-stimulating effect (Fig. 3E) from its ability to transfer denatured substrates to Hsp70 (Fig. 3D) (82). Concordant with this hypothesis, Hop protein exhibiting a different mode of interaction with the C terminus of Hsp70 (Figs. 5C and 7) inhibits Hsp70/Hsp40-mediated refolding only slightly and at high concentrations (supplemental Fig. 2B).
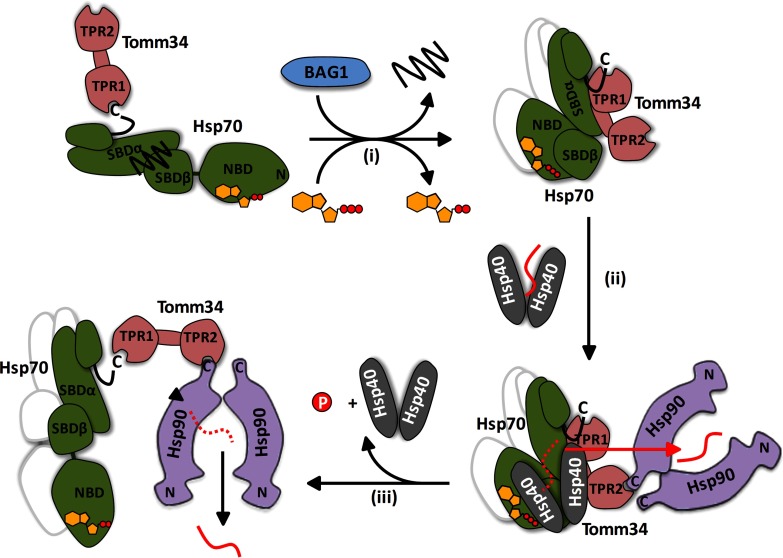
In cells, Tomm34 is mainly localized in the cytosol and functions as a component of large chaperone complexes that shuttle preproteins from ribosomes to mitochondria and other compartments (8, 83). During the transport process Tomm34 coordinates the action of Hsp70 and Hsp90 by their simultaneous binding (8, 13). The optimal chaperone/preprotein complexes are dynamically re-composed by recruitment of multiple co-chaperones with specialized properties, including various Hsp40s (84–87). Structural and functional data presented in this work suggest an hypothesis for the role of Tomm34 in chaperone-mediated preprotein transport (Fig. 10). Tomm34 binding to Hsp70·ATP through its TPR1/linker region interferes with the ability of certain Hsp40s (32, 84) to deliver polypeptide substrates to Hsp70. Instead, the substrates are directly transferred to Hsp90 in “open conformation” recruited to the Hsp40/substrate/Hsp70·ATP/Tomm34 assembly by Tomm34 TPR2 domain/Hsp90/EEVD interaction (13). Because Hsp90 is thought to recognize near-native conformation of its substrates (88, 89), the delivered polypeptide is not effectively processed by the Hsp90 chaperone cycle and is released (90). Interestingly, another two TPR domain co-chaperone, Tpr2, can mediate retrograde transfer of substrates from Hsp90 to Hsp70 in large chaperone complexes (7, 84). Excess of both Tomm34 and Tpr2 inhibits Hsp70/Hsp90-mediated substrate processing (7, 8). We hypothesize that the balanced action of Tomm34 (and Tpr2) maintains particular proteins in a transport-competent semifolded state by shuttling them between Hsp70 and Hsp90 and thus prevents their aggregation before they reach their destined compartment.
Fig. 10.
Proposed model of Tomm34 role in chaperone-mediated preprotein transport. ADP-bound Hsp70 with entrapped substrate interacts with Tomm34 weakly through EEVD motif. Bag-1 (NEF) accelerates ADP/ATP exchange and thus enhances formation of Hsp70·ATP/Tomm34 complexes (i). Tomm34 binding to Hsp70·ATP interferes with the ability of Hsp40 (32, 84) to deliver preproteins to Hsp70 (ii). Instead, the preproteins are directly transferred to Hsp90 in “open conformation” recruited to the Hsp40/preprotein/Hsp70·ATP/Tomm34 complex by Tomm34 TPR2 (13). Because Hsp90 is thought to recognize near-native conformation of its substrates (88, 89), the delivered preprotein is not effectively processed by Hsp90 chaperone cycle and is released (iii) (90). The released preprotein re-enters the cycle in a semifolded state until its delivery to the destined compartment. The consecutive association of the preprotein with Hsp40, Hsp70, and Hsp90 chaperones prevents its aggregation. The possible Hsp70 dimers are indicated by the shadow lines.
Clearly, much remains to be known about the cellular role of Tomm34. The data presented here provide important insight into the structural relationship between Hsp70 and Tomm34 and imply the significant influence of this TPR co-chaperone on Hsp70/Hsp40 chaperone machinery activity.
Supplementary Material
Acknowledgments
We gratefully thank Dr. P. J. Coates for a critical reading of the manuscript.
Footnotes
Author contributions: M.D., F.T., B.V., and P. Muller designed the research; M.D., F.T., P. Man, E.A.B., L.H., P.D., D.C., D.K., and P. Muller performed the research and analyzed the data; M.D., F.T., E.A.B., and P. Muller wrote the paper; and B.V. raised funding.
* This work was supported by Czech Science Foundation Grant P301/11/1678, by Ministry of Education, Youth and Sports of the Czech Republic Grant LO1413, by Ministry of Health of the Czech Republic-Conceptual Development of Research Organization Grant MMCI 00209805, and by Wellcome Trust Multi-User Equipment Grants 081287/Z/06/Z and 101527/Z/13/Z. The FT-ICR facility was supported by the Ministry of Education, Youth and Sports of the Czech Republic Grant LO1509 and by Operational Program Prague-Competitiveness Project CZ.2.16/3.1.00/24023.
 This article contains supplemental Figs. S1 to S5.
This article contains supplemental Figs. S1 to S5.
The authors declare no competing financial interests.
1 The abbreviations used are:
- TPR
- tetratricopeptide repeat
- Hsp
- heat shock protein
- Tomm
- translocase of outer mitochondrial membrane
- CHIP
- C terminus of Hsp70-interacting protein
- NBD
- nucleotide-binding domain
- SBD
- substrate-binding domain
- SBP
- streptavidin-binding peptide
- TEV
- tobacco etch virus
- H/D
- hydrogen/deuterium
- ITC
- isothermal titration calorimetry
- PDB
- Protein Data Bank.
REFERENCES
- 1. Young J. C., Agashe V. R., Siegers K., and Hartl F. U. (2004) Pathways of chaperone-mediated protein folding in the cytosol. Nat. Rev. Mol. Cell Biol. 5, 781–791 [DOI] [PubMed] [Google Scholar]
- 2. Mayer M. P. (2010) Gymnastics of molecular chaperones. Mol. Cell 39, 321–331 [DOI] [PubMed] [Google Scholar]
- 3. Alvira S., Cuéllar J., Röhl A., Yamamoto S., Itoh H., Alfonso C., Rivas G., Buchner J., and Valpuesta J. M. (2014) Structural characterization of the substrate transfer mechanism in Hsp70/Hsp90 folding machinery mediated by Hop. Nat. Commun. 5, 5484. [DOI] [PubMed] [Google Scholar]
- 4. Kirschke E., Goswami D., Southworth D., Griffin P. R., and Agard D. A. (2014) Glucocorticoid receptor function regulated by coordinated action of the Hsp90 and Hsp70 chaperone cycles. Cell 157, 1685–1697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zhang H., Amick J., Chakravarti R., Santarriaga S., Schlanger S., McGlone C., Dare M., Nix J. C., Scaglione K. M., Stuehr D. J., Misra S., and Page R. C. (2015) A bipartite interaction between Hsp70 and CHIP regulates ubiquitination of chaperoned client proteins. Structure 23, 472–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kundrat L., and Regan L. (2010) Balance between folding and degradation for Hsp90-dependent client proteins: a key role for CHIP. Biochemistry 49, 7428–7438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Brychzy A., Rein T., Winklhofer K. F., Hartl F. U., Young J. C., and Obermann W. M. (2003) Cofactor Tpr2 combines two TPR domains and a J domain to regulate the Hsp70/Hsp90 chaperone system. EMBO J. 22, 3613–3623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Faou P., and Hoogenraad N. J. (2012) Tom34: a cytosolic cochaperone of the Hsp90/Hsp70 protein complex involved in mitochondrial protein import. Biochim. Biophys. Acta 1823, 348–357 [DOI] [PubMed] [Google Scholar]
- 9. Cyr D. M., and Ramos C. H. (2015) Specification of Hsp70 function by Type I and Type II Hsp40. Subcell. Biochem. 78, 91–102 [DOI] [PubMed] [Google Scholar]
- 10. Rehn A. B., and Buchner J. (2015) p23 and Aha1. Subcell. Biochem. 78, 113–131 [DOI] [PubMed] [Google Scholar]
- 11. Calderwood S. K. (2015) Cdc37 as a co-chaperone to Hsp90. Subcell. Biochem. 78, 103–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dekker S. L., Kampinga H. H., and Bergink S. (2015) DNAJs: more than substrate delivery to HSPA. Front. Mol. Biosci. 2, 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Trcka F., Durech M., Man P., Hernychova L., Muller P., and Vojtesek B. (2014) The assembly and intermolecular properties of the Hsp70-Tomm34-Hsp90 molecular chaperone complex. J. Biol. Chem. 289, 9887–9901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Schmid A. B., Lagleder S., Gräwert M. A., Röhl A., Hagn F., Wandinger S. K., Cox M. B., Demmer O., Richter K., Groll M., Kessler H., and Buchner J. (2012) The architecture of functional modules in the Hsp90 co-chaperone Sti1/Hop. EMBO J. 31, 1506–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hernández M. P., Sullivan W. P., and Toft D. O. (2002) The assembly and intermolecular properties of the hsp70-Hop-hsp90 molecular chaperone complex. J. Biol. Chem. 277, 38294–38304 [DOI] [PubMed] [Google Scholar]
- 16. Allan R. K., and Ratajczak T. (2011) Versatile TPR domains accommodate different modes of target protein recognition and function. Cell Stress Chaperones 16, 353–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Scheufler C., Brinker A., Bourenkov G., Pegoraro S., Moroder L., Bartunik H., Hartl F. U., and Moarefi I. (2000) Structure of TPR domain-peptide complexes: critical elements in the assembly of the Hsp70-Hsp90 multichaperone machine. Cell 101, 199–210 [DOI] [PubMed] [Google Scholar]
- 18. Brinker A., Scheufler C., Von Der Mulbe F., Fleckenstein B., Herrmann C., Jung G., Moarefi I., and Hartl F. U. (2002) Ligand discrimination by TPR domains. Relevance and selectivity of EEVD-recognition in Hsp70 x Hop x Hsp90 complexes. J. Biol. Chem. 277, 19265–19275 [DOI] [PubMed] [Google Scholar]
- 19. Kajander T., Sachs J. N., Goldman A., and Regan L. (2009) Electrostatic interactions of Hsp-organizing protein tetratricopeptide domains with Hsp70 and Hsp90: computational analysis and protein engineering. J. Biol. Chem. 284, 25364–25374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ward B. K., Allan R. K., Mok D., Temple S. E., Taylor P., Dornan J., Mark P. J., Shaw D. J., Kumar P., Walkinshaw M. D., and Ratajczak T. (2002) A structure-based mutational analysis of cyclophilin 40 identifies key residues in the core tetratricopeptide repeat domain that mediate binding to Hsp90. J. Biol. Chem. 277, 40799–40809 [DOI] [PubMed] [Google Scholar]
- 21. Clerico E. M., Tilitsky J. M., Meng W., and Gierasch L. M. (2015) How hsp70 molecular machines interact with their substrates to mediate diverse physiological functions. J. Mol. Biol. 427, 1575–1588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Saibil H. (2013) Chaperone machines for protein folding, unfolding and disaggregation. Nat. Rev. Mol. Cell Biol. 14, 630–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Qi R., Sarbeng E. B., Liu Q., Le K. Q., Xu X., Xu H., Yang J., Wong J. L., Vorvis C., Hendrickson W. A., Zhou L., and Liu Q. (2013) Allosteric opening of the polypeptide-binding site when an Hsp70 binds ATP. Nat. Struct. Mol. Biol. 20, 900–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lee C. T., Graf C., Mayer F. J., Richter S. M., and Mayer M. P. (2012) Dynamics of the regulation of Hsp90 by the co-chaperone Sti1. EMBO J. 31, 1518–1528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhang P., Leu J. I., Murphy M. E., George D. L., and Marmorstein R. (2014) Crystal structure of the stress-inducible human heat shock protein 70 substrate-binding domain in complex with peptide substrate. PLoS ONE 9, e103518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhuravleva A., and Gierasch L. M. (2011) Allosteric signal transmission in the nucleotide-binding domain of 70-kDa heat shock protein (Hsp70) molecular chaperones. Proc. Natl. Acad. Sci. U.S.A. 108, 6987–6992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kityk R., Kopp J., Sinning I., and Mayer M. P. (2012) Structure and dynamics of the ATP-bound open conformation of Hsp70 chaperones. Mol. Cell 48, 863–874 [DOI] [PubMed] [Google Scholar]
- 28. Zhuravleva A., Clerico E. M., and Gierasch L. M. (2012) An interdomain energetic tug-of-war creates the allosterically active state in Hsp70 molecular chaperones. Cell 151, 1296–1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhuravleva A., and Gierasch L. M. (2015) Substrate-binding domain conformational dynamics mediate Hsp70 allostery. Proc. Natl. Acad. Sci. U.S.A. 112, E2865–2873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Buczynski G., Slepenkov S. V., Sehorn M. G., and Witt S. N. (2001) Characterization of a lidless form of the molecular chaperone DnaK: deletion of the lid increases peptide on- and off-rate constants. J. Biol. Chem. 276, 27231–27236 [DOI] [PubMed] [Google Scholar]
- 31. Mayer M. P., Schröder H., Rüdiger S., Paal K., Laufen T., and Bukau B. (2000) Multistep mechanism of substrate binding determines chaperone activity of Hsp70. Nat. Struct. Biol. 7, 586–593 [DOI] [PubMed] [Google Scholar]
- 32. Yu H. Y., Ziegelhoffer T., and Craig E. A. (2015) Functionality of Class A and Class B J-protein co-chaperones with Hsp70. FEBS Lett. 589, 2825–2830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sarbeng E. B., Liu Q., Tian X., Yang J., Li H., Wong J. L., Zhou L., and Liu Q. (2015) A functional DnaK dimer is essential for the efficient interaction with Hsp40 heat shock protein. J. Biol. Chem. 290, 8849–8862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Swain J. F., Schulz E. G., and Gierasch L. M. (2006) Direct comparison of a stable isolated Hsp70 substrate-binding domain in the empty and substrate-bound states. J. Biol. Chem. 281, 1605–1611 [DOI] [PubMed] [Google Scholar]
- 35. Sondermann H., Scheufler C., Schneider C., Hohfeld J., Hartl F. U., and Moarefi I. (2001) Structure of a Bag/Hsc70 complex: convergent functional evolution of Hsp70 nucleotide exchange factors. Science 291, 1553–1557 [DOI] [PubMed] [Google Scholar]
- 36. Xu Z., Page R. C., Gomes M. M., Kohli E., Nix J. C., Herr A. B., Patterson C., and Misra S. (2008) Structural basis of nucleotide exchange and client binding by the Hsp70 cochaperone Bag2. Nat. Struct. Mol. Biol. 15, 1309–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Melero R., Moro F., Pérez-Calvo M. Á., Perales-Calvo J., Quintana-Gallardo L., Llorca O., Muga A., and Valpuesta J. M. (2015) Modulation of the chaperone DnaK allosterism by the nucleotide exchange factor GrpE. J. Biol. Chem. 290, 10083–10092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhang B., Wang J., Wang X., Zhu J., Liu Q., Shi Z., Chambers M. C., Zimmerman L. J., Shaddox K. F., Kim S., Davies S. R., Wang S., Wang P., Kinsinger C. R., Rivers R. C., et al. (2014) Proteogenomic characterization of human colon and rectal cancer. Nature 513, 382–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Aleskandarany M. A., Soria D., Green A. R., Nolan C., Diez-Rodriguez M., Ellis I. O., and Rakha E. A. (2015) Markers of progression in early-stage invasive breast cancer: a predictive immunohistochemical panel algorithm for distant recurrence risk stratification. Breast Cancer Res. Treat. 151, 325–333 [DOI] [PubMed] [Google Scholar]
- 40. Shimokawa T., Matsushima S., Tsunoda T., Tahara H., Nakamura Y., and Furukawa Y. (2006) Identification of TOMM34, which shows elevated expression in the majority of human colon cancers, as a novel drug target. Int. J. Oncol. 29, 381–386 [PubMed] [Google Scholar]
- 41. Aleskandarany M. A., Negm O. H., Rakha E. A., Ahmed M. A., Nolan C. C., Ball G. R., Caldas C., Green A. R., Tighe P. J., and Ellis I. O. (2012) TOMM34 expression in early invasive breast cancer: a biomarker associated with poor outcome. Breast Cancer Res. Treat. 136, 419–427 [DOI] [PubMed] [Google Scholar]
- 42. Tropea J. E., Cherry S., and Waugh D. S. (2009) Expression and purification of soluble His(6)-tagged TEV protease. Methods Mol. Biol. 498, 297–307 [DOI] [PubMed] [Google Scholar]
- 43. Chang L., Bertelsen E. B., Wisén S., Larsen E. M., Zuiderweg E. R., and Gestwicki J. E. (2008) High-throughput screen for small molecules that modulate the ATPase activity of the molecular chaperone DnaK. Anal. Biochem. 372, 167–176 [DOI] [PubMed] [Google Scholar]
- 44. Kadek A., Mrazek H., Halada P., Rey M., Schriemer D. C., and Man P. (2014) Aspartic protease nepenthesin-1 as a tool for digestion in hydrogen/deuterium exchange mass spectrometry. Anal. Chem. 86, 4287–4294 [DOI] [PubMed] [Google Scholar]
- 45. Marcussen M., and Larsen P. J. (1996) Cell cycle-dependent regulation of cellular ATP concentration, and depolymerization of the interphase microtubular network induced by elevated cellular ATP concentration in whole fibroblasts. Cell. Motil. Cytoskeleton 35, 94–99 [DOI] [PubMed] [Google Scholar]
- 46. Yang C. S., and Weiner H. (2002) Yeast two-hybrid screening identifies binding partners of human Tom34 that have ATPase activity and form a complex with Tom34 in the cytosol. Arch. Biochem. Biophys. 400, 105–110 [DOI] [PubMed] [Google Scholar]
- 47. Morgner N., Schmidt C., Beilsten-Edmands V., Ebong I. O., Patel N. A., Clerico E. M., Kirschke E., Daturpalli S., Jackson S. E., Agard D., and Robinson C. V. (2015) Hsp70 forms antiparallel dimers stabilized by post-translational modifications to position clients for transfer to Hsp90. Cell Rep. 11, 759–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Barthel T. K., Zhang J., and Walker G. C. (2001) ATPase-defective derivatives of Escherichia coli DnaK that behave differently with respect to ATP-induced conformational change and peptide release. J. Bacteriol. 183, 5482–5490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Jiang J., Maes E. G., Taylor A. B., Wang L., Hinck A. P., Lafer E. M., and Sousa R. (2007) Structural basis of J cochaperone binding and regulation of Hsp70. Mol. Cell 28, 422–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Li Z., Hartl F. U., and Bracher A. (2013) Structure and function of Hip, an attenuator of the Hsp70 chaperone cycle. Nat. Struct. Mol. Biol. 20, 929–935 [DOI] [PubMed] [Google Scholar]
- 51. Höhfeld J., and Jentsch S. (1997) GrpE-like regulation of the hsc70 chaperone by the anti-apoptotic protein BAG-1. EMBO J. 16, 6209–6216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Bertelsen E. B., Chang L., Gestwicki J. E., and Zuiderweg E. R. (2009) Solution conformation of wild-type E. coli Hsp70 (DnaK) chaperone complexed with ADP and substrate. Proc. Natl. Acad. Sci. U.S.A. 106, 8471–8476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Gupta R. S. (1998) Protein phylogenies and signature sequences: A reappraisal of evolutionary relationships among archaebacteria, eubacteria, and eukaryotes. Microbiol. Mol. Biol. Rev. 62, 1435–1491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Liu Q., and Hendrickson W. A. (2007) Insights into Hsp70 chaperone activity from a crystal structure of the yeast Hsp110 Sse1. Cell 131, 106–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Fernández-Sáiz V., Moro F., Arizmendi J. M., Acebrón S. P., and Muga A. (2006) Ionic contacts at DnaK substrate binding domain involved in the allosteric regulation of lid dynamics. J. Biol. Chem. 281, 7479–7488 [DOI] [PubMed] [Google Scholar]
- 56. Rist W., Graf C., Bukau B., and Mayer M. P. (2006) Amide hydrogen exchange reveals conformational changes in hsp70 chaperones important for allosteric regulation. J. Biol. Chem. 281, 16493–16501 [DOI] [PubMed] [Google Scholar]
- 57. Arakawa A., Handa N., Shirouzu M., and Yokoyama S. (2011) Biochemical and structural studies on the high affinity of Hsp70 for ADP. Protein Sci. 20, 1367–1379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Hassan A. Q., Kirby C. A., Zhou W., Schuhmann T., Kityk R., Kipp D. R., Baird J., Chen J., Chen Y., Chung F., Hoepfner D., Movva N. R., Pagliarini R., Petersen F., Quinn C., et al. (2015) The novolactone natural product disrupts the allosteric regulation of Hsp70. Chem. Biol. 22, 87–97 [DOI] [PubMed] [Google Scholar]
- 59. Hu S. M., Liang P. H., Hsiao C. D., and Wang C. (2002) Characterization of the L399P and R447G mutants of hsc70: the decrease in refolding activity is correlated with an increase in the rate of substrate dissociation. Arch. Biochem. Biophys. 407, 135–141 [DOI] [PubMed] [Google Scholar]
- 60. Carrigan P. E., Nelson G. M., Roberts P. J., Stoffer J., Riggs D. L., and Smith D. F. (2004) Multiple domains of the co-chaperone Hop are important for Hsp70 binding. J. Biol. Chem. 279, 16185–16193 [DOI] [PubMed] [Google Scholar]
- 61. Flom G., Behal R. H., Rosen L., Cole D. G., and Johnson J. L. (2007) Definition of the minimal fragments of Sti1 required for dimerization, interaction with Hsp70 and Hsp90 and in vivo functions. Biochem. J. 404, 159–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Rezabkova L., Man P., Novak P., Herman P., Vecer J., Obsilova V., and Obsil T. (2011) Structural basis for the 14–3-3 protein-dependent inhibition of the regulator of G protein signaling 3 (RGS3) function. J. Biol. Chem. 286, 43527–43536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Röhl A., Wengler D., Madl T., Lagleder S., Tippel F., Herrmann M., Hendrix J., Richter K., Hack G., Schmid A. B., Kessler H., Lamb D. C., and Buchner J. (2015) Hsp90 regulates the dynamics of its cochaperone Sti1 and the transfer of Hsp70 between modules. Nat. Commun. 6, 6655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Tanford C. (1978) The hydrophobic effect and the organization of living matter. Science 200, 1012–1018 [DOI] [PubMed] [Google Scholar]
- 65. Luke K., Apiyo D., and Wittung-Stafshede P. (2005) Dissecting homo-heptamer thermodynamics by isothermal titration calorimetry: entropy-driven assembly of co-chaperonin protein 10. Biophys. J. 89, 3332–3336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Steinberg I. Z., and Scheraga H. A. (1963) Entropy changes accompanying association reactions of proteins. J. Biol. Chem. 238, 172–181 [PubMed] [Google Scholar]
- 67. Hong J., Hu Y., Li C., Jia Z., Xia B., and Jin C. (2010) NMR characterizations of the ice binding surface of an antifreeze protein. PLoS ONE 5, e15682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Onuoha S. C., Coulstock E. T., Grossmann J. G., and Jackson S. E. (2008) Structural studies on the co-chaperone Hop and its complexes with Hsp90. J. Mol. Biol. 379, 732–744 [DOI] [PubMed] [Google Scholar]
- 69. Schlecht R., Erbse A. H., Bukau B., and Mayer M. P. (2011) Mechanics of Hsp70 chaperones enables differential interaction with client proteins. Nat. Struct. Mol. Biol. 18, 345–351 [DOI] [PubMed] [Google Scholar]
- 70. Moro F., Fernández-Sáiz V., and Muga A. (2004) The lid subdomain of DnaK is required for the stabilization of the substrate-binding site. J. Biol. Chem. 279, 19600–19606 [DOI] [PubMed] [Google Scholar]
- 71. Slepenkov S. V., Patchen B., Peterson K. M., and Witt S. N. (2003) Importance of the D and E helices of the molecular chaperone DnaK for ATP binding and substrate release. Biochemistry 42, 5867–5876 [DOI] [PubMed] [Google Scholar]
- 72. Liebscher M., and Roujeinikova A. (2009) Allosteric coupling between the lid and interdomain linker in DnaK revealed by inhibitor binding studies. J. Bacteriol. 191, 1456–1462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Aprile F. A., Dhulesia A., Stengel F., Roodveldt C., Benesch J. L., Tortora P., Robinson C. V., Salvatella X., Dobson C. M., and Cremades N. (2013) Hsp70 oligomerization is mediated by an interaction between the interdomain linker and the substrate-binding domain. PLoS ONE 8, e67961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Swain J. F., Dinler G., Sivendran R., Montgomery D. L., Stotz M., and Gierasch L. M. (2007) Hsp70 chaperone ligands control domain association via an allosteric mechanism mediated by the interdomain linker. Mol. Cell 26, 27–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Yu H. Y., Ziegelhoffer T., Osipiuk J., Ciesielski S. J., Baranowski M., Zhou M., Joachimiak A., and Craig E. A. (2015) Roles of intramolecular and intermolecular interactions in functional regulation of the Hsp70 J-protein co-chaperone Sis1. J. Mol. Biol. 427, 1632–1643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Freeman B. C., Myers M. P., Schumacher R., and Morimoto R. I. (1995) Identification of a regulatory motif in Hsp70 that affects ATPase activity, substrate binding and interaction with HDJ-1. EMBO J. 14, 2281–2292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Suzuki H., Noguchi S., Arakawa H., Tokida T., Hashimoto M., and Satow Y. (2010) Peptide-binding sites as revealed by the crystal structures of the human Hsp40 Hdj1 C-terminal domain in complex with the octapeptide from human Hsp70. Biochemistry 49, 8577–8584 [DOI] [PubMed] [Google Scholar]
- 78. Li J., Wu Y., Qian X., and Sha B. (2006) Crystal structure of yeast Sis1 peptide-binding fragment and Hsp70 Ssa1 C-terminal complex. Biochem. J. 398, 353–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Gao X. C., Zhou C. J., Zhou Z. R., Wu M., Cao C. Y., and Hu H. Y. (2012) The C-terminal helices of heat shock protein 70 are essential for J-domain binding and ATPase activation. J. Biol. Chem. 287, 6044–6052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Sun L., Edelmann F. T., Kaiser C. J., Papsdorf K., Gaiser A. M., and Richter K. (2012) The lid domain of Caenorhabditis elegans Hsc70 influences ATP turnover, cofactor binding and protein folding activity. PLoS ONE 7, e33980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Michels A. A., Kanon B., Bensaude O., and Kampinga H. H. (1999) Heat shock protein (Hsp) 40 mutants inhibit Hsp70 in mammalian cells. J. Biol. Chem. 274, 36757–36763 [DOI] [PubMed] [Google Scholar]
- 82. Cuéllar J., Perales-Calvo J., Muga A., Valpuesta J. M., and Moro F. (2013) Structural insights into the chaperone activity of the 40-kDa heat shock protein DnaJ: binding and remodeling of a native substrate. J. Biol. Chem. 288, 15065–15074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Chewawiwat N., Yano M., Terada K., Hoogenraad N. J., and Mori M. (1999) Characterization of the novel mitochondrial protein import component, Tom34, in mammalian cells. J. Biochem. 125, 721–727 [DOI] [PubMed] [Google Scholar]
- 84. Bhangoo M. K., Tzankov S., Fan A. C., Dejgaard K., Thomas D. Y., and Young J. C. (2007) Multiple 40-kDa heat-shock protein chaperones function in Tom70-dependent mitochondrial import. Mol. Biol. Cell 18, 3414–3428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Pratt W. B., and Toft D. O. (2003) Regulation of signaling protein function and trafficking by the hsp90/hsp70-based chaperone machinery. Exp. Biol. Med. 228, 111–133 [DOI] [PubMed] [Google Scholar]
- 86. Wang X., Venable J., LaPointe P., Hutt D. M., Koulov A. V., Coppinger J., Gurkan C., Kellner W., Matteson J., Plutner H., Riordan J. R., Kelly J. W., Yates J. R. 3rd, and Balch W. E. (2006) Hsp90 cochaperone Aha1 downregulation rescues misfolding of CFTR in cystic fibrosis. Cell 127, 803–815 [DOI] [PubMed] [Google Scholar]
- 87. Sullivan W. P., Owen B. A., and Toft D. O. (2002) The influence of ATP and p23 on the conformation of hsp90. J. Biol. Chem. 277, 45942–45948 [DOI] [PubMed] [Google Scholar]
- 88. Jakob U., Lilie H., Meyer I., and Buchner J. (1995) Transient interaction of Hsp90 with early unfolding intermediates of citrate synthase. Implications for heat shock in vivo. J. Biol. Chem. 270, 7288–7294 [DOI] [PubMed] [Google Scholar]
- 89. Young J. C., Moarefi I., and Hartl F. U. (2001) Hsp90: a specialized but essential protein-folding tool. J. Cell Biol. 154, 267–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Li J., Richter K., Reinstein J., and Buchner J. (2013) Integration of the accelerator Aha1 in the Hsp90 co-chaperone cycle. Nat. Struct. Mol. Biol. 20, 326–331 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.