Abstract
The human primosome, a 340-kilodalton complex of primase and DNA polymerase α (Polα), synthesizes chimeric RNA-DNA primers to be extended by replicative DNA polymerases δ and ϵ. The intricate mechanism of concerted primer synthesis by two catalytic centers was an enigma for over three decades. Here we report the crystal structures of two key complexes, the human primosome and the C-terminal domain of the primase large subunit (p58C) with bound DNA/RNA duplex. These structures, along with analysis of primase/polymerase activities, provide a plausible mechanism for all transactions of the primosome including initiation, elongation, accurate counting of RNA primer length, primer transfer to Polα, and concerted autoregulation of alternate activation/inhibition of the catalytic centers. Our findings reveal a central role of p58C in the coordinated actions of two catalytic domains in the primosome and ultimately could impact the design of anticancer drugs.
Keywords: DNA polymerase, DNA primase, DNA-protein interaction, RNA synthesis, x-ray crystallography, 5′-triphosphate, DNA/RNA duplex, RNA primer length counting, human DNA replication, initiating NTP
Introduction
In eukaryotes, the primosome, a tight complex of DNA primase and DNA polymerase α (Polα),4 synthesizes primers for both leading and lagging strands in a highly coordinated fashion (1, 2). The primosome is indispensable for initiation of replication and has a large impact on genome stability (3–6). RNA primer synthesis by primase involves three steps: initiation, elongation, and termination (7, 8). During the rate-limiting initiation step, primase binds the DNA template and two ribonucleotide triphosphates (NTPs) and catalyzes the formation of a dinucleotide (9, 10). Further synthesis of the RNA primer is much faster but restricted, because of the intrinsic property of primases to count the primer length and terminate synthesis after incorporation of 8–10 nucleotides (7). Next, the mature so-called “unit length” RNA primer is intramolecularly translocated to Polα for the subsequent extension by dNTPs, and the primase became inhibited by an unknown mechanism (9, 11, 12). Orchestration of all these steps requires changes in primosome conformation (13).
Human Polα (Fig. 1A) is comprised of a large catalytic subunit (p180) and a smaller accessory subunit (p70), connected by the C-terminal domain of p180 (p180C) containing two conserved zinc-binding modules, Zn1 and Zn2 (14–16). p70 consists of an N-terminal (p70N), a phosphodiesterase, and oligonucleotide/oligosaccharide-binding (OB) domains (14, 17). The globular p70N is attached to the phosphodiesterase via a flexible linker (amino acid residues 79–156) (14, 18) and participates in interactions with other DNA replication proteins (19). The catalytic core of p180 (p180core) and p180C-p70 are connected by a 15-residue linker (1251–1265) (13). Human primase consists of catalytic (p49) and regulatory (p58) subunits (20). p58 has two distinct domains, N-terminal (p58N) and C-terminal (p58C), connected with an 18-residue linker (253–270) (21). p58N interacts with p49 and connects primase with Polα (22, 23), and an iron-sulfur cluster containing p58C plays an important role in substrate binding and primase activity (24–30).
FIGURE 1.
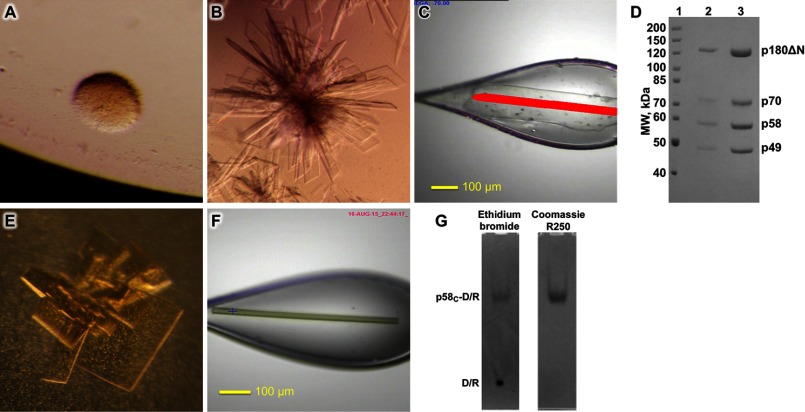
Crystallization of the primosome and p58C-D/R. A and B, photomicrographs of the primosome crystals before and after optimization, respectively. C, a single crystal of primosome scooped in a nylon-fiber loop. D, analysis of the content of primosome crystals by 7% SDS-PAGE. Lane 1, molecular weight markers; lanes 2 and 3 correspond to the crystals shown in A and B, respectively. E and F, photomicrographs of p58C-D/R crystals in the form of plates and needles, respectively. G, analysis of the content of a needle-like p58C-D/R crystal by 5% native PAGE. Gel was stained first with ethidium bromide (left panel) and then with Coomassie R-250 (right panel).
Structural characterization of eukaryotic primosome is limited to the low-resolution electronic microscopy data (13) and the x-ray structures of its parts. This includes the crystal structures of the intact human primase (21) and its truncated variant without p58C (22), as well as the high resolution structures of p180core (31), p180C-p70 (14), p58C (25, 30), p49 (33), and their yeast orthologs (15, 29, 32, 33). The interaction of eukaryotic primases with substrates has never been structurally characterized except for the binding site for an incoming nucleotide triphosphate (NTP) (22, 33). None of these data provide clear clues on the details of RNA-DNA primer synthesis including initiation, elongation, accurate counting of RNA primer length, primer transfer to Polα, and coordinated action of two catalytic centers. The comprehensive understanding of these mechanisms requires crystal structures of primosome in complex with a variety of substrates. Crystallization of such complexes is extremely challenging due to the significant mobility of the functional domains. We elected to obtain mechanistic insight into the primosome transactions by focusing on solving two key structures: the primosome in apo-form, and the C-terminal domain of the human primase large subunit (p58C) bound to the DNA template/RNA primer (D/R), p58C-D/R. The results of these efforts, in combination with functional analysis, lead to delineation of all the steps of RNA primer synthesis and its transfer to Polα.
Experimental Procedures
Protein Expression and Purification for X-ray Crystallography and Activity Analysis
Cloning, expression, and purification to homogeneity of the human primase heterodimer (p49-p58; p49 contains 420 amino acids, p58 contains 509), the C-terminal domain of p58 (p58C; residues 266–456), human Polα (p180-p70; p180 contains amino acids 335–1462, p70 contains 1–596), human primosome (p49-p58-p180-p70), and the primosome without Polα catalytic core (p49-p58-p180C-p70; p180C contains residues 1265–1462) have been described elsewhere (21, 34, 35). The N terminus of p180 (residues 1–334) is predicted to be poorly folded like that in yeast Polα (32) and has no conserved motifs necessary for DNA polymerizing activity or for interaction with other subunits in primosome complex, so it has been deleted to increase protein solubility and simplify its purification and crystallization.
Oligonucleotides for X-ray Crystallography and Activity Analysis
Sequences of all oligonucleotides are provided in Table 1. Oligonucleotides without 5′-triphosphate were obtained from IDT Inc. RNA primers containing the 5′-triphosphate (P1–P4) were obtained as described previously (26). DNA duplex T9-P5 and RNA polymerase of bacteriophage T7 were used for synthesis of the ribo-primer P1 (5′-pppGGCGGC). DNA duplexes T10-P5, T11-P5, and T12-P6 were used for synthesis of the ribo-primers P2 (5′-pppGGCUCGG), P3 (5′-pppGGCGUGCGG), and P4 (5′-pppGGCUGGUCGG), respectively. DNA/DNA and DNA/RNA duplexes were obtained at 0.1–0.2 mm concentration by 1 min heating to 80 °C followed by equilibration to room temperature for 15 min in buffer containing 20 mm Tris-HCl (pH 7.9) and 0.1 m NaCl.
TABLE 1.
Oligonucleotides used in the current study
| Name | Length | DNA/RNA | Sequence (template region involved in duplex formation is underlined) |
|---|---|---|---|
| T1 | 60 | DNA | 5′-gtgagagaaggagaaggagaggaagagaaggagaagaggagagaaacgccgccaaaaaaa |
| T2 | 59 | DNA | 5′-gtgagagaaggagaaggagaggaagagaaggagaagaggagagaacgccgccaaaaaaa |
| T3 | 58 | DNA | 5′-gtgagagaaggagaaggagaggaagagaaggagaagaggagagacgccgccaaaaaaa |
| P1 | 6 | RNA | 5′-pppGGCGGC |
| T4 | 20 | DNA | 5′-Cy3-AACTAACCCGAGCCAATACA |
| P2 | 7 | RNA | 5′-pppGGCUCGG |
| T5 | 22 | DNA | 5′-ATACAACCCGCACGCCAACATA |
| P3 | 9 | RNA | 5′-pppGGCGUGCGG |
| T6 | 60 | DNA | 5′-gcgagagaaggagaaggagaggaagagaaggagaagaggagacccgaccagccaaaaaaa |
| T7 | 25 | DNA | 5′-ATACTATCCCGACCAGCCAATACAA |
| P4 | 10 | RNA | 5′-pppGGCUGGUCGG |
| T8 | 12 | DNA | 5′-GCCGCCAACATA |
| T9 | 23 | DNA | 5′-GCCGCCTATAGTGAGTCGTATTA |
| T10 | 24 | DNA | 5′-CCGAGCCTATAGTGAGTCGTATTA |
| T11 | 26 | DNA | 5′-CCGCACGCCTATAGTGAGTCGTATTA |
| P5 | 18 | DNA | 5′-AATACGACTCACTATAGG |
| T12 | 27 | DNA | 5′-CCGACCAGCCTATAGTGAGTCGTATTA |
| P6 | 25 | DNA | 5′-TAATACGACTCACTATAGGCTGGTC |
Electrophoretic Mobility Gel Shift Assay (EMSA)
Reactions containing 0.2 μm primase-PolαΔcat and the 7-bp DNA/RNA duplex T4-P2 (T4 contains Cy3 dye at 5′-end) and varied concentrations of the 10-bp DNA/RNA duplex T7-P4 were incubated for 10 min at room temperature in a 10-μl reaction containing 10 mm Tris-HCl (pH 7.7), 50 mm KCl, 1% glycerol, and 1 mm DTT; then 5 μl was loaded on 5% native PAGE. Samples labeled with Cy3-dye were visualized using the Typhoon 9410 imager (GE Healthcare, emission of fluorescence at 580 nm).
Activity Assay
Activity of human primosome and primase-PolαΔcat in extension of riboprimers with the terminal 5′-triphosphate annealed to different DNA templates was tested in a 20-μl reaction containing 30 mm Hepes-KOH (pH 7.9), 50 mm KCl, 1 mm DTT, 2 mm MgCl2, 100 μm NTPs, 10 μm dNTPs, 0.25 μm [α-33P]GTP (3000 Ci/mmol; PerkinElmer, Inc.), 1 μm template/primer, and 50 nm enzyme. Reactions were incubated at 35 °C in a thermal cycler (Thermolyne Amplitron I) and stopped by mixing with an equal volume of formamide loading buffer (95% (v/v) formamide, 5 mm EDTA, 0.02% bromphenol blue, 0.02% xylene cyanol, and 0.025% SDS), heated at 65 °C for 10 min, and resolved by 20% urea-PAGE (UreaGel System (19:1 acrylamide/bisacrylamide), National Diagnostics) for 5 h at 2000 V. The gel was dried at 65 °C for 45 min using the Bio-Rad 583 gel dryer. The reaction products were visualized by phosphorimaging (Typhoon 9410, GE Healthcare). All activity gels were repeated at least two times.
Crystallization of Human Primosome
Primosome was concentrated to 10 mg/ml in buffer containing 10 mm Tris-HCl (pH 7.7), 0.1 m KCl, 1% glycerol, and 1 mm DTT and then flash-frozen in aliquots. The aliquots were defrosted and centrifuged at 40,000 × g to remove the aggregated molecules, and the sample monodispersity was verified with dynamic light scattering. Screening was performed with the sitting-drop vapor diffusion method at 295 K by mixing 1 μl of protein solution with 1 μl of reservoir solution. The reservoirs contained 2-fold diluted Crystal Screen, Crystal Screen 2, and Natrix (Hampton Research) solutions with the addition of 2 mm tris(2-carboxyethyl)phosphine (pH 7.5). Within 1–2 months the oil-like spherulites in the 17th condition of the Crystal Screen became covered by barely visible needles (Fig. 1A). After optimization and multiple screenings of additives, thin plates in the form of parallelograms appeared and grew on the precipitates in 3–5 days (Fig. 1B). The best reservoir solution contained 0.2 m lithium sulfate, 50 mm Tris-HCl (pH 8.5), 2 mm tris(2-carboxyethyl)phosphine (pH 7.5), 11.2% (w/v) PEG 4,000, 3% (v/v) ethanol, 0.5% (v/v) polypropylene glycol P400, and 0.2 mm EDTA. The crystals grew over 1 mm in length but were extremely thin and fragile, easily breaking into pieces due to the movement of solution around them. The resolution of the x-ray diffraction was time-dependent, indicating that crystals with larger volume (i.e. thicker) may have improved diffraction at shorter exposures. Multiple rounds of seeding produced crystals with a thickness of 2–4 μm that were diffracting anisotropically up to 3.2-Å resolution. However, the rate of success of seeding was too low; in addition to difficulties in peaking and washing the fragile single crystals, most seeds had a propensity to grow as layers and began to be covered by needle-shaped crystals within 30–40 h before completion of growth. Only single-appearing crystals were picked for the next round of seeding. We obtained over 50 crystals by seeding, but only two were diffracting as single crystals. One crystal diffracting up to 3.6 Å was used for the optimization of data collection conditions at the synchrotron, and the second, better diffracting crystal (Fig. 1C) was used for the complete data set collection. The washed and dissolved crystals revealed the presence of all subunits as in the original sample (Fig. 1D). We also crystallized the primosome with a longer p180 subunit (residues 292–1462). The growth conditions and shape of the crystals were similar as in primosome containing p180 residues 335–1462. However, the crystals were not diffracting.
Crystallization of p58C-D/R
Based on our recent biochemical results (26), we designed the optimal substrate for p58C with a 12-mer DNA template T8 annealed to a 6-mer RNA primer P1 containing a 5′-triphosphate. Purified p58C was exchanged into buffer containing 5 mm Tris-HCl (pH 7.9), 20 mm NaCl, 0.5% glycerol, and 1 mm DTT using a Superose 12 column (GE Healthcare). Next, the 6-bp DNA/RNA duplex T8-P1 was added at 10% molar excess; then the complex was concentrated to 12 mg/ml and flash-frozen in aliquots. Screening was performed as described for primosome, with the exception that the screen solutions were not diluted. Small thin plates appeared in the 15th condition of the Crystal Screen. The optimized condition (0.125 m ammonium sulfate, 0.1 m sodium cacodylate, pH 6.5, 0.02 m tris(2-carboxyethyl)phosphine, and 28.5% (w/v) PEG 8,000) produced slow-growing, well shaped rectangular plates in 3–4 weeks (Fig. 1E). These crystals diffracted poorly, with the diffused reflections extended up to a resolution of only 12–15 Å. In an attempt to obtain a new crystal form with better packing, we varied the type of salt and buffer and added a variety of compounds from the Hampton Research Additive screen. Eventually, we succeeded with a combination of 0.18 m lithium sulfate, 50 mm sodium citrate, 2 mm tris(2-carboxyethyl)phosphine, 0.1 m sodium malonate, 0.1 m Hepes-NaOH (pH 7.0), 19.7% PEG 8,000, and 0.02 m magnesium chloride that produced relatively fast-growing (within 2–4 days) needle-like prisms, with a square shape in cross-section (Fig. 1F). The crystals contained both p58C and DNA/RNA components (Fig. 1G) and diffracted to 2.2-Å resolution.
Data Collection
For diffraction data collection, the crystals were soaked in a cryoprotectant solution for a few seconds, scooped in a nylon-fiber loop, and flash-cooled in a dry nitrogen stream at 100 K. The cryoprotectant for the primosome crystals was prepared by increasing the PEG 4,000 concentration in the reservoir solution to 14% (w/v) and inclusion of 18% (v/v) ethylene glycol. In the case of p58C-D/R crystals, the PEG 8,000 and ethylene glycol concentrations were 21 and 18%, respectively. Because the latter crystals were grown in the presence of Mg2+, we refer to them as p58C-D/R-Mg. To exchange Mg2+ to Mn2+, we soaked crystals for 5 min in a reservoir solution containing 2 mm manganese chloride instead of magnesium chloride. The resulting crystals are referred to as p58C-D/R-Mn. The cryoprotectant was also prepared with 2 mm manganese chloride. Preliminary crystal characterizations and full dataset collection from p58C-D/R-Mn crystals were performed on a Rigaku R-AXIS IV imaging plate using Osmic VariMaxTM HR mirror-focused CuKα radiation from a Rigaku FR-E rotating anode operated at 45 kV and 45 mA. Complete diffraction data sets for primosome and p58C-D/R-Mg crystals were collected using synchrotron x-rays on the Argonne National Laboratory Advanced Photon source beamline 24ID-C with the Pilatus 6MF pixel-array detector. Continuous vector scan along the crystal length was applied to reduce the radiation damage (Fig. 1, C and F). All intensity data were indexed, integrated, and scaled with DENZO and SCALEPACK from the HKL-2000 program package (36). The crystal parameters and data processing statistics are summarized in Table 2.
TABLE 2.
Summary of data collection, phasing, and refinement statistics
| Molecule | Primosome | p58C-D/R-Mg | p58C-D/R-Mn |
|---|---|---|---|
| Data collection | |||
| Space group | P 21 | C 2221 | C 2221 |
| Cell dimensions | |||
| a, b, c (Å) | 113.096, 210.164, 172.565 | 122.507, 126.002, 83.942 | 122.753, 125.613, 83.690 |
| β (°) | 93.56 | 90 | 90 |
| Resolution (Å) | 50–3.6 (3.66–3.6)a | 50–2.2 (2.24–2.2) | 20 –3 (3.05–3) |
| Rmerge | 0.086 (0.363) | 0.062 (0.379) | 0.105 (0.435) |
| I/σI | 12.2 (1.9) | 31.2 (1.5) | 6.7 (1.8) |
| Completeness (%) | 80.5 (72.7) | 95.4 (58.3) | 89.9 (76.9) |
| Unique reflections | 74238 (3348) | 31361 (937) | 11834 (484) |
| Redundancy | 2.6 (1.9) | 5.3 (2.7) | 3.2 (1.9) |
| Refinement | |||
| Resolution (Å) | 39.94–3.6 (3.83–3.6) | 49.48–2.20 (2.34–2.2) | 19.87–3 (3.19–3) |
| No. reflections | 64105 (8030) | 30821 (3424) | 11818 (1650) |
| Rwork/Rfree | 0.268/0.326 (0.377/ 0.391) | 0.223/0.258 (0.409/0.455) | 0.232/0.269 (0.398/0.408) |
| No. atoms | |||
| Protein | 37642 | 3035 | 3035 |
| DNA/RNA | 0 | 721 | 721 |
| Ligand/ion | 22 | 18 | 18 |
| Water | 0 | 65 | 0 |
| Mean B-factors (Å2) | 61.2 | 63.8 | 52.5 |
| Root mean square deviations | |||
| Bond lengths (Å) | 0.012 | 0.007 | 0.01 |
| Bond angles (°) | 1.8 | 1.4 | 1.5 |
| Ramachandran plot (%) | |||
| Core | 73.8 | 88.7 | 88.4 |
| Allowed | 24.0 | 11.0 | 11.3 |
| Generously allowed | 1.7 | 0.3 | 0.3 |
| Disallowed | 0.5 | 0 | 0 |
a Numbers in parentheses refer to highest resolution shell.
Structure Determination
Only one complete native dataset from a single crystal was available for the phasing and refinement of the human primosome structure. Twinning and non-isomorphism among heavy-atom derivative crystals prevented merging data from several smaller crystals that were collected with a rotation of 10°. To apply the molecular replacement, we needed initial models; therefore, we started and completed structural studies of the three largest parts of primosome: p180core with bound substrates and the inhibitor aphidicolin, full-length primase, and p180C-p70 (PDB accession numbers 4QCL (31), 4Q5V (31), 4RR2 (21), and 4Y97 (14), respectively). An initial search was performed with the p180core coordinates from PDB entry 4Q5V, because, in complex with aphidicolin, the fingers were in an open conformation similar to apo-form. Clear solutions were obtained for both independent molecules only when we removed the thumb and palm domains. Both independent positions of p180C-p70 and their locations relative to p180core were also clearly resolved by molecular replacement. However, the search results were not clear with models containing the palm and thumb domains of p180core, or primase (with or without p58C), or any one of p49, p58N, and p58C. The density modifications and averaging using the phases from the model containing Polα without palm and thumb domains of p180 revealed positions of the latter domains and primase subunits that were manually placed into the electron density maps. Anomalous difference maps confirmed the locations of two zinc ions in p180C and one in p49, along with the location of the 4Fe-4S cluster in p58C for both independent primosome molecules. To avoid model bias, we verified the position of each domain by omitting its coordinates separately in each molecule as well as simultaneously from both molecules. After positioning of all the domains, we verified the structures of the loops and terminal residues. Zonal scaling (37) and bulk solvent corrections were applied to structure factors. Rigid-body refinement was performed after each cycle of model building. Positional and B-factor refinements were applied only after the completion of model building. Noncrystallographic 2-fold symmetry constraints were applied during the initial stages of refinement; these were replaced by restraints of backbone atoms during the final stages of refinement. The structure was refined at 3.6-Å resolution to an Rcryst of 26.9% and an Rfree of 32.8% and contains amino acids 1–283, 289–360, and 379–412 of p49; 22–455 of p58; 338–672, 680–808, 842–882, 898–1258, and 1266–1456 of p180; and 155–598 of p70.
The structure of p58C-D/R-Mg was solved by the molecular replacement method using the coordinates of p58C from the structure of human primase (PDB accession number 4RR2 (21)) as a search model. Clear solutions were found for two molecules in an asymmetric unit. Initial electron density maps revealed a 6-bp DNA/RNA duplex and part of the template 3′-overhang for both molecules. Their addition into the structure further clarified the packing of the remaining residues for the template 3′-overhang. One molecule contained all template residues that were stabilized by crystal packing, whereas no density was found for the dAde−6 and dThy−5 in another molecule. Residues 354 to 363 were rearranged in the p58C-D/R-Mg structure due to interaction with template DNA. No density was found for N-terminal residues 266–269. Non-crystallographic 2-fold symmetry restraints were applied during the initial stages of model refinement. After the addition of solvent molecules, the model was refined at 2.2-Å resolution to an Rcryst of 22.3% and an Rfree of 25.8%. The electron density peak close to the GTP phosphates of the primer was assigned as an Mg2+. For additional confirmation that the triphosphate moiety coordinates a divalent metal, the Mg2+ was exchanged with a heavier Mn2+. A difference Fo − Fc Fourier map revealed a significantly stronger peak at the same position with a contour level of over 8 (Fig. 7B), confirming the metal binding. The structure of p58C-D/R-Mn was refined at 3-Å resolution to an Rcryst of 22.5% and an Rfree of 27.6%.
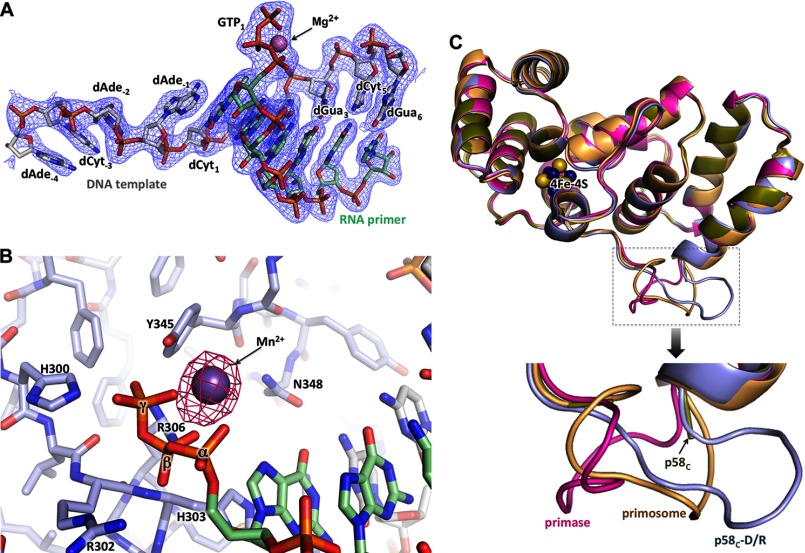
FIGURE 7.
Details of p58C interaction with a template/primer. A, electron density map for the DNA/RNA duplex and the triphosphate coordinating Mg2+. The carbons of DNA and RNA are colored green and gray, respectively. B, Fo − Fc Fourier map (contour level at 5σ) for Mn2+ coordinated by a 5′-triphosphate of RNA. C, alignment of p58C from different structures points to the flexibility of the DNA-interacting loop 354–366. The PDB accession numbers for the structures of the human primase and p58C are 4RR2 and 3Q36, respectively.
The final refinement statistics for all structures are provided in Table 2. Inspections of the electron density maps and the building/adjustment of the structures were performed with Turbo-Frodo. All crystallographic computing was performed with CNS version 1.1 (38). Supplemental Movie S1 and the figures containing molecular structures were prepared with PyMol (39).
Results
Crystal Structure of Human Primosome
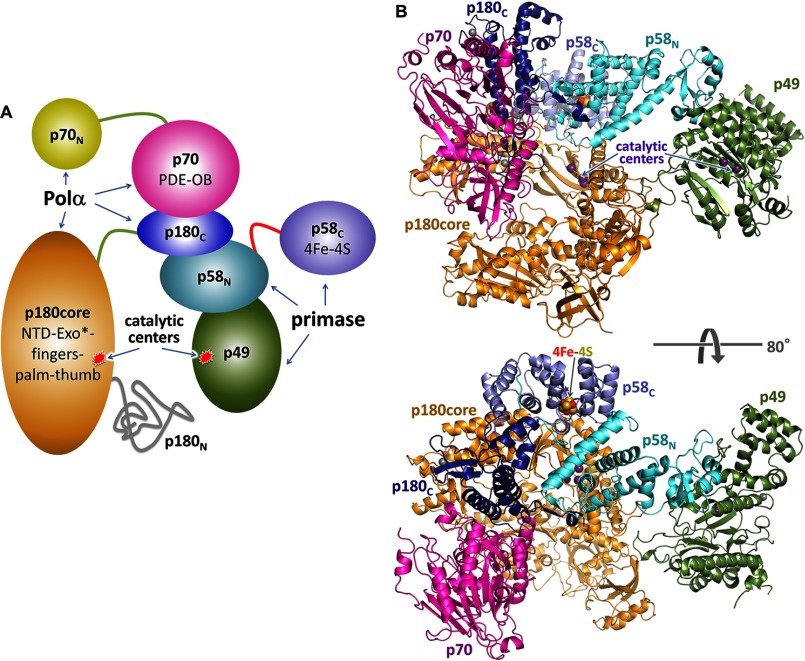
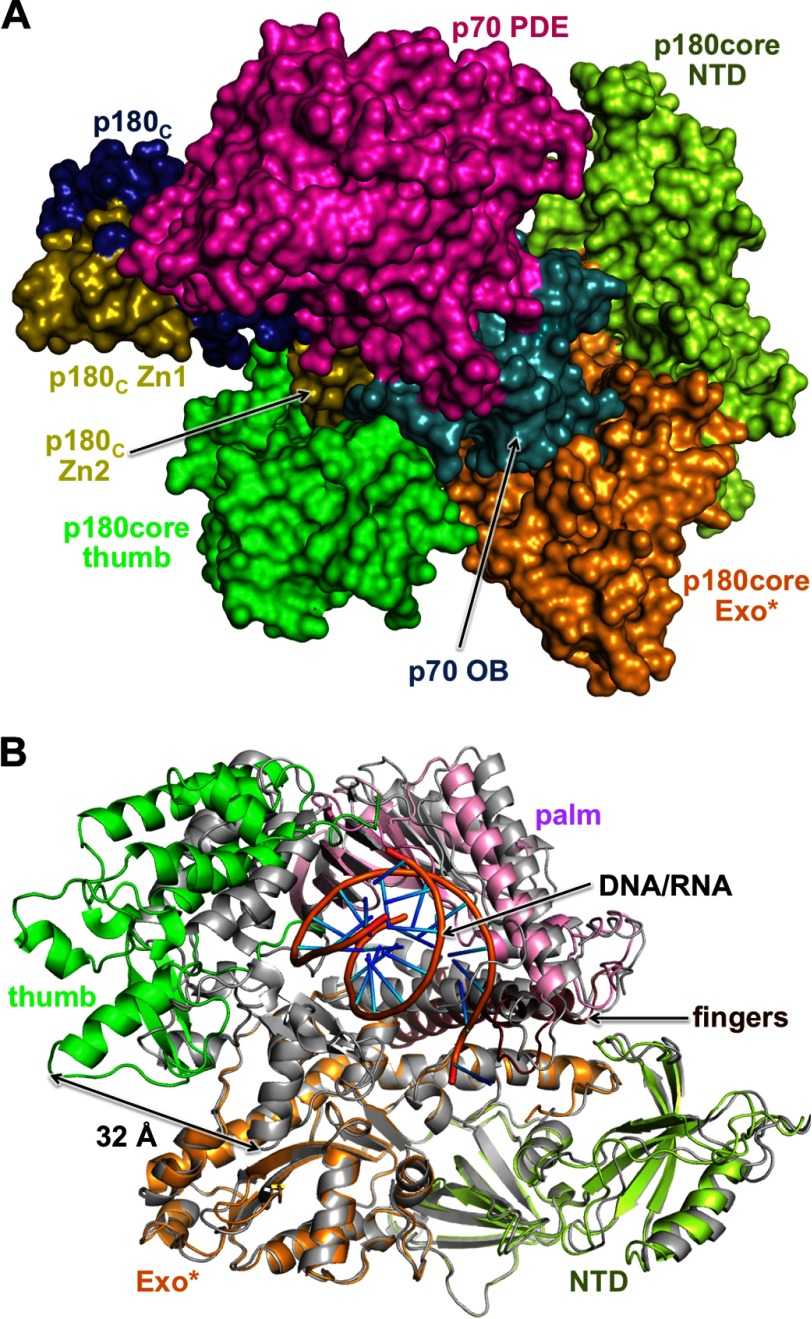
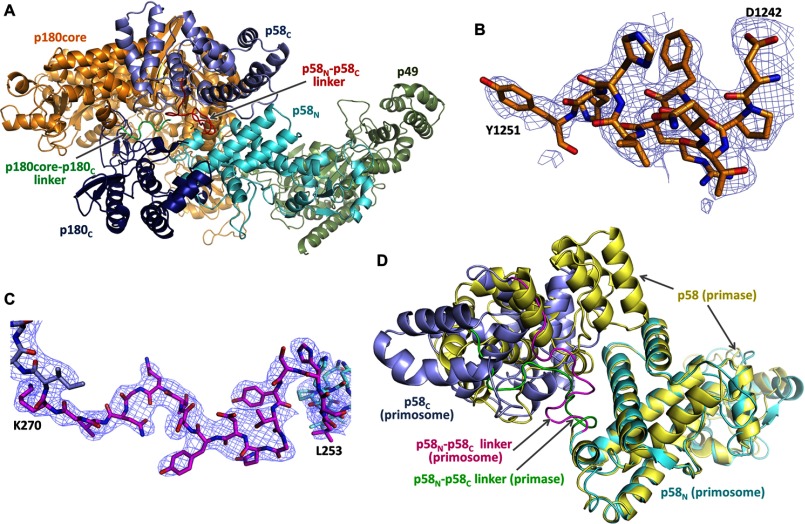
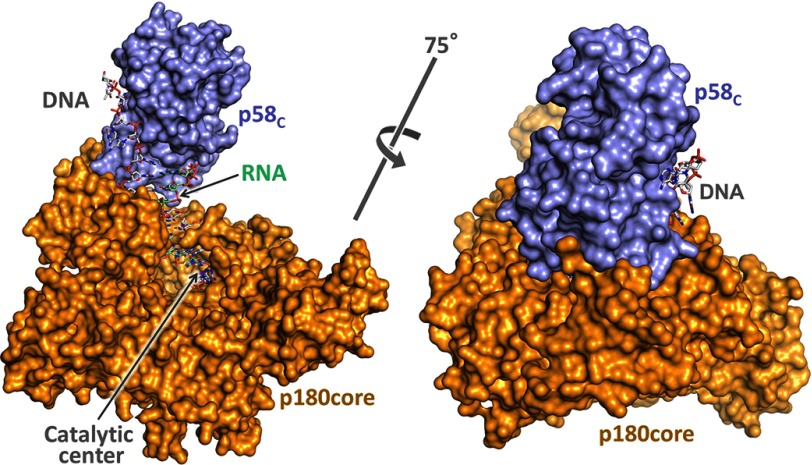
The structure of the apo-form of primosome has been determined at 3.6-Å resolution (Table 2). It acquires an extended shape with the dimensions of 148 × 119 × 107 Å3 and packs as three massive parts: p180core, p180C-p70, and primase (Fig. 2B). p180C-p70 associates with p180core by wedging the Zn2 module of p180C and OB domain of p70 into the substrate-binding area between the exonuclease and thumb domains of p180core and displacing the tip of the thumb up to 32 Å in comparison with a substrate-bound form (Fig. 3). As a result, the Polα active center is blocked, which explains the inhibitory role of p180C-p70 on Polα activity (35). An elongated primase locates on p180core and stretches from its thumb to the N-terminal domain. Total buried surface area between the p180core and the rest of the molecule is 6117 Å. The distance between the primase and Polα catalytic centers is 57 Å.
FIGURE 2.
Structure of the human primosome hetero-tetramer complex. A, schematic representation of the domain organization. Flexibly tethered domains are shown as separate parts. p58C coordinates the iron-sulfur cluster. Exo* is an exonuclease domain with no associated activity due to evolutionary substitution of the catalytic amino acid residues; alignment is as described in Ref. 57. PDE, phosphodiesterase. B, the crystal structure of primosome. Subunits are shown as schematics and colored as in A. The α-carbons of catalytic aspartates are shown as purple spheres.
FIGURE 3.
Mechanism of Polα autoinhibition in non-polymerizing primosome. A, Polα structure in apo-primosome. Zn2 module of p180C and OB domain of p70 block the entry to the Polα active site for a template/primer. PDE, phosphodiesterase. B, alignment of Polα catalytic cores in the ternary complex with substrates (colored gray; PDB code 4QCL) and in apo-primosome (colored as in A) shows a significant difference in positions of the palm and thumb relative to other subdomains.
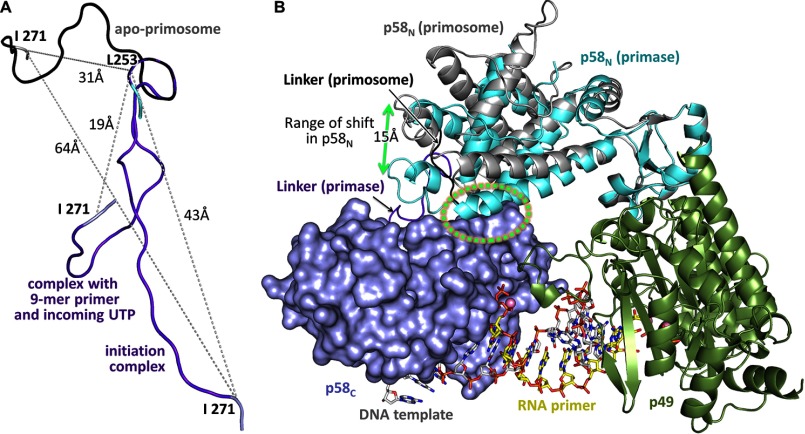
Substantial conformational changes are essential for the seamless accomplishment of all the steps of primer synthesis; therefore, the relative positioning of the moving domains and the linkers between them play the crucial role in primosome transactions (Fig. 4A). The 180core-p180C linker is only partially defined in the structure. Some electron density was found for linker residues 1251–1258 that are packed as a random coil, but no density was found for the remaining portion of the linker (1259–1265). The residues 1243–1250 preceding this linker form a well stabilized α-helix in the primosome structure (Fig. 4B). However, electron density for these residues was missing in the p180core structures (31), indicating that they could become a part of the 180core-p180C linker in the case of p180core dissociation from the rest of primosome. The p58N-p58C linker has a well defined density (Fig. 4C). The position of p58C relative to p58N is different in the primosome and primase (21), supporting the idea of flexibility of this linker (Fig. 4D).
FIGURE 4.
Domain and linkers organization in the primosome. A, primosome has two long linkers connecting p180core and p58C to the platform composed of p49-p58N-p180C-p70. Primase linker (colored red) connects p58C and p58N; Polα linker (colored green) connects p180core and p180C. B, analysis of the electron density map for p180 residues 1243–1250 confirms their packing as α-helix. Nitrogens, oxygens, and carbons are colored blue, red, and orange, respectively. C, the carbons in p58N, p58C, and the linker are colored cyan, gray, and magenta, respectively. D, the relative positions of p58C in the primase (PDB ID 4RR2) and the primosome structures (only p58N was used for alignment).
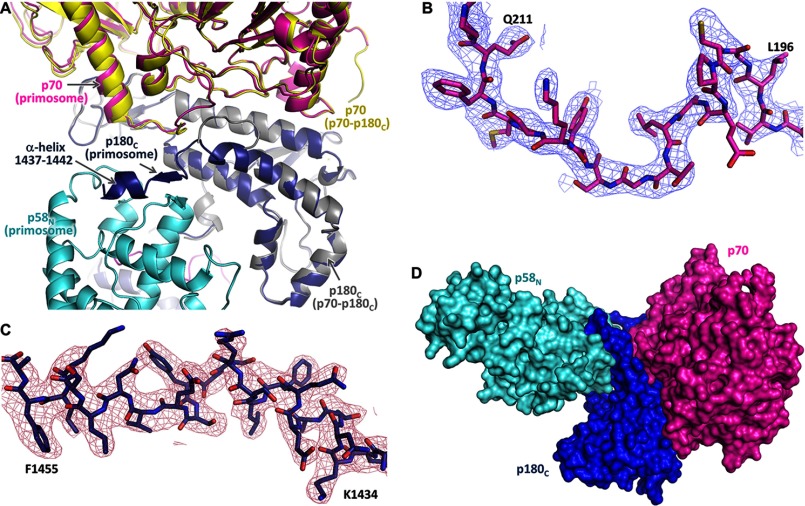
The third linker, which connects the p58N-interacting C-terminal peptide of p180 (1447–1456) with the rest of p180C, has been proposed to provide the flexible connection between p180C-p70 and primase (22). Interaction of p180C-p70 with primase rearranges the folding of p180 residues C-terminal to Thr-1436 and the surrounding residues 197–210 of p70, making this part of p180C-p70 much more stabilized and resolved in the primosome structure than in p180C-p70 (14) (Fig. 5, A-C). The folding of residues 1437–1442 into the α-helix reduces the size of the linker, placing p180C-p70 and primase in close proximity and restricting their movement relative to each other (Fig. 5D). As a result, the substructure p49-p58N-p180C-p70 of primosome has a defined elongated shape, whose basic architecture is not influenced by interactions with p180core or p58C and resembles the low-resolution electron microscopy images of the yeast ortholog (13). Therefore, p49-p58N-p180C-p70 can be considered as a platform that holds p180core and p58C stationary by docking in the inactive form or flexibly by linkers during various stages of primosome transactions. Both linkers are closely packed with a distance of 9 Å between the points of their attachment to the p49-p58N-p180C-p70 platform (Fig. 4A).
FIGURE 5.
The regions disordered in Polα are structured in primosome. A, superimposition of p180C-p70 (PDB code 4Y97) and primosome structures. In primosome, p180C and p70 are colored magenta and blue; in p180C-p70, they are colored yellow and gray. B and C, 2Fo − Fc Fourier map (contour level at 1σ) for p70 residues 195–212 and p180C residues 1433–1456, respectively. D, the relative position of p70 and p58N is stabilized by p180C.
Crystal Structure of p58C-D/R
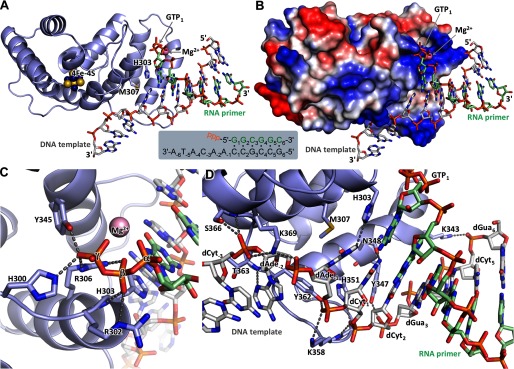
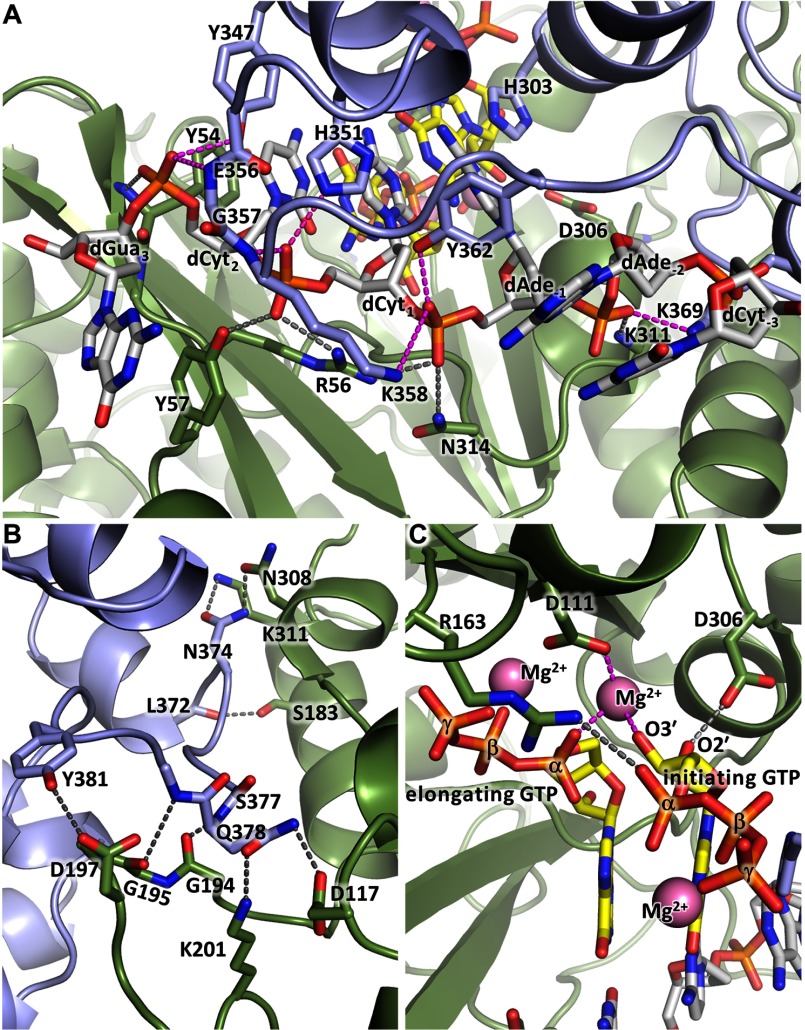
Currently, there is no structural data for any primase in the complex with a template-primer. We recently defined the elements of the DNA template and the RNA primer that are important for interaction with the human primase and found that all of them interact with p58C (26). These findings allowed us to design the optimal substrate and prepare a stable p58C-D/R complex for crystallographic studies. The structures of p58C-D/R in complex with Mg2+ or Mn2+ have been determined at 2.2- and 3.0-Å resolutions, respectively (Table 2). The only notable difference between these two structures is the nature of the divalent metal ion. The p58C-D/R-Mg complex has a bean shape where the RNA-primed DNA template binds to the positively charged depressions on the surface of p58C with a primer 5′-triphosphate on one side and a template 3′-overhang on other side (Fig. 6, A and B). Furthermore, the electrostatic interactions are extended by the DNA backbone of the DNA/RNA duplex. This structure explains why the elimination of 5′-triphosphate and/or 3′-overhang abolished the interaction of DNA/RNA with p58C as well as with full-length primase in our recent study (26).
FIGURE 6.
Structure of p58C in complex with the RNA-primed DNA template. A, specific recognition of the template/primer junction at the 5′ terminus of the primer by p58C. B, the surface electrostatic potential of p58C shows docking of the template and the 5′-triphosphate of the primer on two separate positively charged areas (colored blue). C, hydrogen bonds (dashed lines) between p58C and the β- and γ-phosphates of GTP1. D, hydrogen bonds between p58C and the template.
Only the β- and γ-phosphates of the triphosphate moiety are involved in interactions with p58C, via six hydrogen bonds with the side chains of His-300, Arg-302, Arg-306, Tyr-345, and the main chain of His-303 (Fig. 6C). Arg-306, which interacts with both the β- and γ-phosphates, has been shown to be important for RNA synthesis (22, 24, 27). The triphosphate coordinates the Mg2+ or Mn2+ ion that provides additional stability (Fig. 7, A and B). The template forms 13 hydrogen bonds with the protein: eight with the 3′-overhang and the rest with the duplex portion (Fig. 6D).
There are no dramatic conformational changes in p58C induced upon DNA/RNA binding apart from stabilization of the loop comprised by residues 354–366; it is partially disordered in the structure of apo-p58C (25) or acquires different crystal packing-induced conformations as seen in the structures of full-length primase (21) and primosome (Fig. 7C). However, p58C affects the conformation of the DNA/RNA substrate: it maintains the B-DNA conformation of the template deoxyriboses, except at positions 4–6 that are in the A-DNA conformation.
Primase Initiation Complex
p58C-D/R structure provides an intriguing and long-awaited mechanism elucidating the docking of the initiating GTP (GTP1) (Fig. 6). The well defined interactions of the template phosphates at positions −2, −1, 1, and 2 with p58C help to orient its bases at positions −1, 1, and 2 for a duplex formation. The dCyt1 base conformation is additionally stabilized via a hydrogen bond with a side chain of Asn-348. As a result, the dAde−1 base stacks between the hydrophobic side chain of Met-307 and dCyt1 base. Interestingly, the side chain of His-303 mimics the RNA base that is paired with the template dAde−1 via a hydrogen bond and supports the stacking of the initiating GTP.
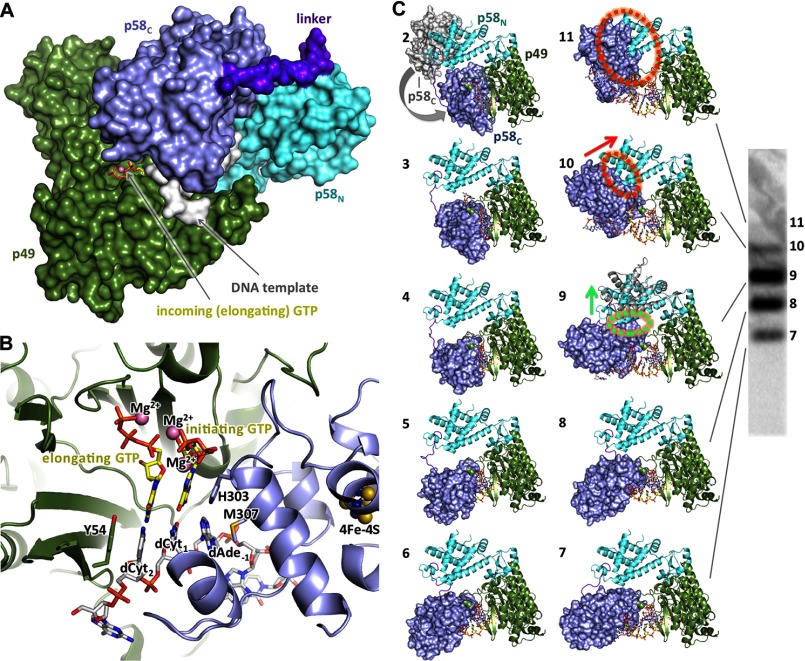
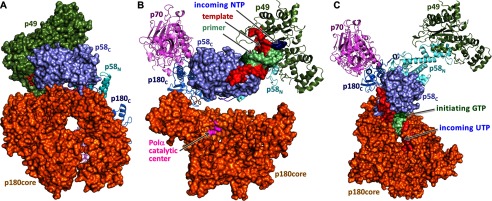
The structure of p58C-D/R together with the structures of the full-length human primase (21) and the primase-UTP complex (22) provides all necessary components for the modeling of the primase initiation and elongation complexes (Fig. 8). Superimposition of the second nucleotide of a primer in p58C-D/R with the incoming UTP in primase revealed a well defined shape complementarity between the catalytic p49 subunit and p58C domain (Fig. 8A). Unlike the elongated substrate-free primase (21), the initiating complex acquires a globular shape. It reveals the formation of a “mini duplex” (Fig. 8B) that is comprised of three template nucleotides at positions −1, 1, and 2 on one strand and His-303 of p58, initiating GTP, and incoming GTP on the other strand. The stacking of the DNA bases is extended with the side chains of p49 Tyr-54 from one side and p58 Met-307 from the other side. The duplex is partially buried in the middle of the complex, but the channel for loading an incoming NTP remains open (Fig. 8A). The formation of the initiation complex results in 16 additional hydrogen bonds: six between DNA phosphates and p49, and eight between p49 and p58C, one between p49 Arg-163 and the α-phosphate of initiating GTP, and one between p49 Asp-306 and O2′ of initiating GTP (Fig. 9). Moreover, the O3′ of the initiating GTP coordinates with an Mg2+ ion that is also bound to Asp-111 of p49 and the α-phosphate of the incoming GTP (Fig. 9C). The interaction between the O2′ of initiating GTP and Asp-306 of p49 stabilizes the ribose during dinucleotide synthesis, which explains the strict preference for ribonucleotides at this step. The abundance of additional interactions is supportive for the accuracy of the model and also explains why substitutions of the participating residues, Tyr-54, Arg-56, and Arg-163 of p49, resulted in the loss of primase activity (22, 40).
FIGURE 8.
Mechanism of RNA synthesis initiation, elongation, and termination. A, compact shape of primase in the initiation complex model. B, close-up view of the initiation mini duplex in the model. Position of the magnesium ion between the elongating and initiating GTPs was modeled to coordinate Asp-111, α-phosphate of elongating GTP, and O3′ of initiating GTP as in the active site of polymerases (58). C, models of the initiation and elongation complexes. The crystal structures of p58C-D/R, human primase (PDB code 4RR2), and p49-p58N–UTP complex (PDB code 4BPW) were used to build these models. Panel numbers correspond to the length of the growing RNA primer plus incoming NTP in the modeled complexes. Panel 2 corresponds to the initiation complex resulting in dinucleotide formation and also shows the position of p58C in apo-primase. The clash area is depicted by dotted circles (colored green or red to indicate avoidable or unavoidable steric hindrance, respectively), whereas the arrows show the direction at which p58C is pushing p58N. Close-up view of the complex in panel 9, explaining how steric hindrance is avoided, is provided in Fig. 10B. The image on the right shows the typical pattern of primase-catalyzed reaction products.
FIGURE 9.
p49-DNA and p49-p58C interaction interfaces in the model of initiation complex. A, interaction of the DNA template with p49 and p58C. The hydrogen bonds between DNA and p58C or p49 are depicted by pink or gray dashed lines, respectively. B, the potential p49-p58C interaction interface shows its plasticity due to the absence of hydrophobic contacts. C, p49 interacts with a ribose and α-phosphate of the initiating GTP. Second magnesium ion was modeled as described in the legend to Fig. 8B.
The Mechanism of Primer Length Counting by Primase
There are two alternative models for counting: one is based on the restriction of the distance between the two lobes of the large primase subunit by the linker (21, 41), and another is based on steric hindrance (7, 25). The obtained structures finally allow us to discover an exact mechanism of primer length counting.
The p58N-p58C linker acquires the most extended conformation in the initiation complex, with a distance of 43 Å between its N and C termini (Figs. 8C, panel 2, and 10A). Further RNA elongation up to a 9-mer gradually reduces this distance to 19 Å (Figs. 8C, panel 10, and 10A). Considering the capacity of the linker to be extended by over 60 Å, it can easily tolerate the extension of the primer well beyond the unit length, and hence cannot be the reason for primase counting. Indeed, when the linker was artificially shortened, the primase activity was reduced but the size of the products did not change (21).
FIGURE 10.
Conformational changes in human primase during RNA primer synthesis. A, position of the p58N-p58C linker in apo-primosome (Fig. 2B), in the initiation complex (Fig. 8A) and in the primase complex with a DNA template primed by a 9-mer primer and the incoming UTP (Fig. 8C, panel 10). The alignment is performed using p58N. B, close-up view of the primase-DNA/RNA-CTP elongation complexes containing an 8-mer primer. The complexes were modeled based on the structures of primosome (p58N is colored gray), primase (p58N is colored cyan; PDB code 4RR2), and p58C-D/R. The alignment was performed using p49 subunits. In the primosome, only p58N with the p58N-p58C linker is shown for clarity. Clash area between the two domains in the primase-based model is depicted by the green dotted circle; no clash is observed in the primosome-based model.
Inspection of elongation complexes with 2- to 7-bp DNA/RNA duplexes points to the absence of any steric hindrance between p58C and the helical bundle of p58N (Fig. 8C, panels 3–8). Some noticeable steric hindrance between them is observed for an 8-bp duplex (Figs. 8C, panel 9, and 10B). However, the helical bundle of p58N can move nearly perpendicular to its axis and acquire a range of positions that accommodate the structure with an 8-bp duplex and incoming NTP (Fig. 10B). Consequently, the reaction can proceed efficiently to form a 9-bp duplex. Unlike the case with an 8-mer primer, p58C with a 9-mer primer produces a steric hindrance at the top of the helical bundle in a direction that is parallel to its axis, and hence the clash cannot be relieved by flexibility in p58N (Fig. 8C, panel 10). As a result, the pre-catalytic alignment of the 3′-OH of a primer and the α-phosphate of incoming NTP is compromised, which explains the dramatically reduced efficiency of the 10th nucleotide incorporation. Addition of an 11th NTP would be impossible (panel 11). Therefore, interdomain steric hindrance is a cause of exact primer counting.
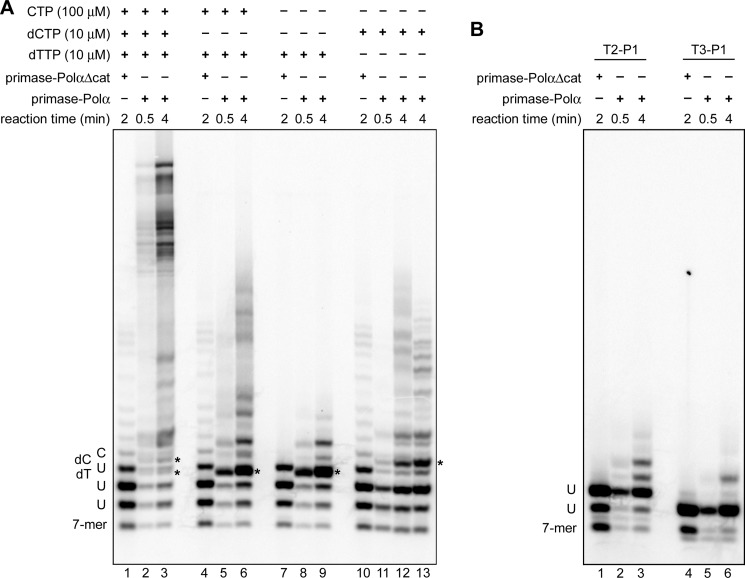
To verify the mechanism of counting, we examined the proficiency of primase in extension of 6-, 9-, and 10-mer primers (Fig. 11A). Primase extends a 6-mer primer very efficiently (lanes 2 and 3), whereas extension of 9-mer (lanes 8 and 9) and especially 10-mer primers (lanes 4–7) is dramatically decreased. The result means that primase not only terminates primer synthesis when its length is nine nucleotides, but also cannot efficiently extend the unit length primers.
FIGURE 11.
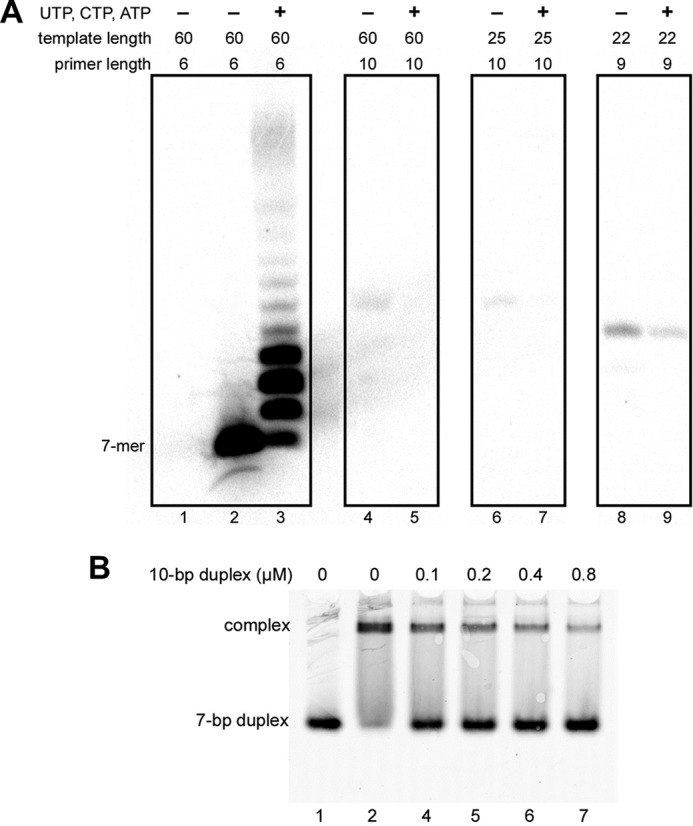
DNA/RNA duplex length affects primase activity but not affinity. A, the effect of RNA primer length on activity of primase-PolαΔcat. Lanes 1–3, T1-P1; lanes 4 and 5, T6-P4; lanes 6 and 7, T7-P4; lanes 8 and 9, T5-P3. Reactions corresponding to lanes 2, 4, 6, and 8 were conducted in the presence of [α-33P]GTP alone to show the position of the first product of primer extension. Reactions corresponding to lanes 3, 5, 7, and 9 were supplemented with 0.1 mm each UTP, CTP, and ATP. Reactions were run for 2 min at 35 °C, and products were resolved by 20% urea-PAGE. B, 10-bp DNA/RNA duplex (T7-P4) efficiently competes with a 7-bp duplex (T4-P2) for binding with primase-PolαΔcat. All reactions contained 0.2 μm T4-P2; reactions corresponding to lanes 2–7 contained 0.2 μm primase-PolαΔcat. Samples were resolved by 5% native PAGE and visualized using the Typhoon 9410 imager. T4 is labeled with Cy3 at the 5′-end.
The Mechanism of the Primase-Polα Switch
According to earlier data, the DNA/RNA duplex does not dissociate from primosome after synthesis of the unit length RNA primer, which allows for intramolecular substrate transfer between the two active sites (9, 11). However, the mechanism of switch and involved domains remained unknown. Recently we found that p58C makes a tight complex with a 7-bp DNA/RNA duplex with almost the same affinity for the template/primer as a whole primase (26). Moreover, the final and intermediate products of primase reaction have similar affinity for primase (Fig. 11B). Altogether, these data suggest that p58C holds the template/primer during its transition from primase to Polα, which results in concurrent binding of the unit length primer by p58C and Polα during the switch. We created a model of the switch complex, which contains p180core and p58C sharing the 9-bp DNA/RNA duplex, the optimal substrate for Polα recognition (31) (Fig. 12). This model illustrates good complementarity between surfaces of these two domains, indicating that the 3′-end of 8-mer or shorter primers will not reach the catalytic site.
FIGURE 12.
The model of the switch complex. The crystal structures of p58C-D/R and p180core-D/R-dCTP (PDB code 4QCL) were used for model building.
To verify the model and define the precise position of the switch (i.e. the length of the primer at which the 3′-end of RNA primer becomes accessible for the Polα catalytic site), we analyzed the products of primosome-catalyzed extension of a 6-mer primer in reactions with various composition of the rNTPs/dNTPs pool (Fig. 13A). The template sequence directs incorporation of UTP/dTTP at the 8–10th positions and CTP/dCTP at the 11th position of the primer. The incorporation of dNMPs (versus rNMPs) could be distinguished because of slightly higher mobility of the corresponding products on the gel. In the presence of ribo- and deoxy-NTPs, the primase incorporates only ribonucleotides (Fig. 13A, lane 1). Analysis of the products generated by the primosome reveals that the insertion of dNMPs begins from the 10th position (lanes 2 and 3). This means that Polα can gain access to the 3′-end of the primer when its length is 9 nucleotides or longer. The exclusion of dCTP (lanes 5 and 6) or both CTP/dCTP (lanes 8 and 9) from the reaction, which inhibits the processive primer extension by Polα, demonstrated that 10-mer primers mainly contain the 3′-dTMP. In the absence of dTTP, the primase to Polα switch with a 9-mer primer is abrogated and we can observe the switch with a 10-mer primer (lanes 11 and 12). These results clearly show that the switch mainly happens when the primer length is 9. If Polα is unable to pick up the 9-mer primer, it can easily start with a 10-mer. Incorporation of dTMP was not observed using templates T3 and T2 that were designed to stop the primosome after synthesis of the 8- and 9-mer primers, respectively (Fig. 13B). These results confirm that Polα cannot start extension of primers shorter than 9-mer (when p58C holds them) and support the structural model (Fig. 12).
FIGURE 13.
Determination of the Polα start position during RNA primer extension with primosome. A and B, analysis of the ribo- and deoxy-NTPs incorporation using the 6-mer RNA primer P1 annealed to the 60-mer DNA template T1 (A) or to the 59-mer and 58-mer DNA templates T2 and T3, respectively (B). All reactions with the T1-P1 duplex contained 100 μm UTP; all reactions with T2-P1 and T3-P1 duplexes contained 100 μm UTP and 10 μm dTTP. Primase-PolαΔcat is the primosome without p180core.
Discussion
Our study provides structural insight at the atomic level into eukaryotic primosome and DNA primase in complex with the DNA/RNA substrate that revealed the exact structure of the initiation site of the primase with bound GTP. The linkers connecting p180core and p58C to the p49-p58N-p180C-p70 platform provide significant freedom in their movement relative to each other and other subunits. Such flexibility allows p58C to be close to p49 during initiation, to move away together with the 5′-end of the primer during elongation, and, finally, to intramolecularly transfer and load the 9-mer primer to the Polα active site. To perform these duties, p58C conforms to several topological requirements: it is complementary to p49 during initiation and to p180core during switch, and makes a clash with p58N to terminate RNA synthesis at the defined position.
The p58C-D/R structure, elongation complex models, and biochemical data (24, 26, 42) show that p58C has a much stronger role in template/primer recognition than the catalytic p49 subunit. This is consistent with a distributive nature of RNA synthesis pointing to the high probability of p49 dissociation from the template/primer after incorporation of each nucleotide (Fig. 13A, lane 1). The unique mode of DNA/RNA substrate binding, with p58C holding the 5′-end of primer and the weakly bound p49, allows Polα to efficiently catch the 3′-end of RNA primer without dissociation of the template/primer from primosome. Biochemical data indicate that p58C holds the DNA/RNA duplex when Polα begins primer extension, thus providing the negative regulation for primase (11, 26).
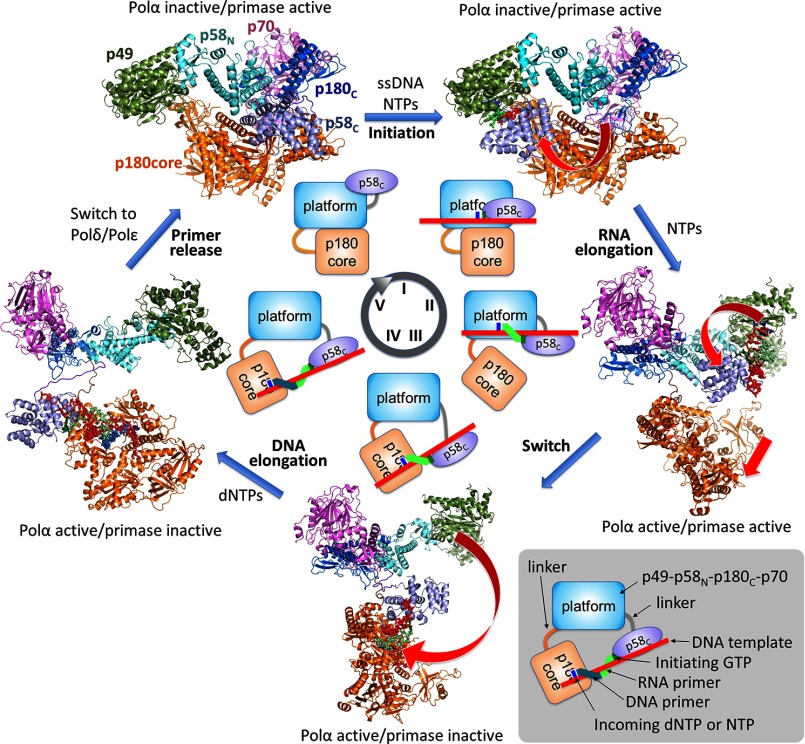
With the structures of apo-primosome and p58C-D/R, it became possible to visualize all key steps of the entire cycle of primer synthesis (Fig. 14 and supplemental Movie S1). The structure of apo-primosome suggests that Polα is in an autoinhibitory conformation, because entry to the DNA-binding site is blocked by the Zn2 module of p180C and OB-fold of p70; this is consistent with the increase of Polα activity when p70-p180C is absent (35). Interestingly, apo-primase has a space between p180core and p49 that is suitable for docking of p58C during initiation (Figs. 14 and 15A). Furthermore, upon RNA primer elongation, p58C moves toward p180core, pushing it to dissociate from the platform (Figs. 14 and 15B). As a result, the p180core became conveniently positioned to catch the p58C-bound 9-mer primer after its release from p49 and rotation by about 100° (Figs. 14 and 15C). The presented structures and models, together with biochemical data, showing that p58C remains bound to the template/primer during RNA primer elongation and switch (9, 11, 26), finally revealed the structural basis for coordination of the primase and Polα catalytic activities during all steps of primer synthesis.
FIGURE 14.
Mechanism of RNA-DNA primer synthesis by the human primosome. At the first step (steps are labeled by roman numerals at the circle in the center), p58C moves toward p49 (curved arrow) to initiate RNA synthesis. During the second step, p58C is moving toward p180core by following the helical path of the growing DNA/RNA duplex (curved arrow) and pushes the p180core to dissociate from the platform (straight arrow). Additionally, when RNA primer length is nine nucleotides, p58C makes a steric hindrance with p58N, which prevents primer extension by p49. At the third step, p58C rotates (curved arrow) and loads the template/primer to the Polα active site. At the fourth step, Polα extends the RNA primer with dNTPs, whereas p58C is holding the 5′ terminus of the primer. At the fifth step, primosome is replaced by Polϵ or Polδ. Models of primosome at different steps of primer synthesis are based on the apo-primosome structure (Fig. 2) and primase models (Fig. 8).
FIGURE 15.
Models of the human primosome during key steps of RNA primer synthesis. A, at the initiation step, p58C fits into a space between p180core and p49. B, at the RNA elongation step, p58C is moving toward p180core and displaces it from the p49-p58N-p180C-p70 platform. C, flexibly tethered p180core binds DNA template with a 9-mer RNA primer presented by p58C.
The mode of primase interaction with the template/primer is unique and differs from the mode employed by RNA polymerases where the initiating nucleotide does not interact permanently with the non-catalytic domain (43–46). This mode might be applicable to primases from other species where an accessory DNA(NTP)-binding domain is also present (47–51). The lack of interaction between the human primase and the emerging RNA strand (Fig. 8C) explains the significant ability of DNA primases to perform translesion synthesis (52–55). The data presented here have wide-ranging therapeutic implications, because they provide new perspectives for the design of specific primosome inhibitors ultimately aimed at arresting the onset of replication in targeted cancer cells. Moreover, these structures could be used as a reference for the detection of the differences between the human and bacterial or viral primases, thus facilitating the design of novel classes of antibiotics and antivirals.
Now that the mechanism of concerted primosome transactions in the synthesis of RNA-DNA primers has become clear, further study is needed to understand how primosome integrates into the replisome and how the primer is released and translocated to the main replicative DNA polymerases δ and ϵ (3, 6, 56). Polα switch to the autoinhibitory state would prevent its competition with Polδ and Polϵ for template/primer binding.
Author Contributions
A. G. B. and T. H. T. wrote the manuscript, with contributions and critical comments from the other authors. A. G. B. prepared all samples for crystallography and functional studies and designed the biochemical experiments. N. D. B. crystallized the complexes. N. D. B. and T. H. T. collected the diffraction data. Y. Z. performed and A. G. B. with Y. I. P. supervised the biochemical experiments. J. G. participated in expression and purification of protein complexes. Y. S. participated in optimization of p58C-D/R-Mg crystals. T. H. T. initiated the project, determined the crystal structures, and provided the structure-based models of primosome function.
Supplementary Material
Acknowledgments
We thank D. Temiakov (Rowan University) for kindly providing the expression vector for T7 RNA polymerase and A. Rizzino, D. Price, and K. Jordan for critical reading and editing of this manuscript. This work is also based upon research conducted at the APS on the NECAT beamlines, which are supported by NIGMS National Institutes of Health Grant P41 GM103403. The Pilatus 6M detector on 24-ID-C beamline is funded by National Institutes of Health-ORIP HEI Grant S10 RR029205. Use of the Advanced Photon Source, an Office of Science User Facility operated for the United States Department of Energy (DOE) Office of Science by Argonne National Laboratory, was supported by the U.S. DOE under Contract DE-AC02–06CH11357.
This work was supported, in whole or in part, by National Institutes of Health NIGMS Grant GM101167 (to T. H. T.). The Eppley Institute's X-ray Crystallography Core Facility is supported by National Institutes of Health NCI Grant P30CA036727. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The authors declare no competing financial interests.

This article contains supplemental Movie S1.
The atomic coordinates and structure factors (codes 5EXR, 5F0Q, and 5F0S) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- Polα
- DNA polymerase α
- p58N and p58C
- N- and C-terminal domains of the p58 subunit
- p180core
- catalytic core of Polα
- p180C
- C-terminal domain of the Polα catalytic subunit
- Zn1 and Zn2
- zinc-binding modules of p180C
- OB
- oligonucleotide/oligosaccharide-binding.
References
- 1. Pellegrini L. (2012) The Pol α-primase complex. Subcell. Biochem. 62, 157–169 [DOI] [PubMed] [Google Scholar]
- 2. Muzi-Falconi M., Giannattasio M., Foiani M., and Plevani P. (2003) The DNA polymerase α-primase complex: multiple functions and interactions. ScientificWorldJournal 3, 21–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Yeeles J. T., Deegan T. D., Janska A., Early A., and Diffley J. F. (2015) Regulated eukaryotic DNA replication origin firing with purified proteins. Nature 519, 431–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Reijns M. A., Kemp H., Ding J., de Procé S. M., Jackson A. P., and Taylor M. S. (2015) Lagging-strand replication shapes the mutational landscape of the genome. Nature 518, 502–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pavlov Y. I., Frahm C., Nick McElhinny S. A., Niimi A., Suzuki M., and Kunkel T. A. (2006) Evidence that errors made by DNA polymerase α are corrected by DNA polymerase δ. Curr. Biol. 16, 202–207 [DOI] [PubMed] [Google Scholar]
- 6. Sun J., Shi Y., Georgescu R. E., Yuan Z., Chait B. T., Li H., and O'Donnell M. E. (2015) The architecture of a eukaryotic replisome. Nat. Struct. Mol. Biol. 22, 976–982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kuchta R. D., and Stengel G. (2010) Mechanism and evolution of DNA primases. Biochim. Biophys. Acta 1804, 1180–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Frick D. N., and Richardson C. C. (2001) DNA primases. Annu. Rev. Biochem. 70, 39–80 [DOI] [PubMed] [Google Scholar]
- 9. Copeland W. C., and Wang T. S. (1993) Enzymatic characterization of the individual mammalian primase subunits reveals a biphasic mechanism for initiation of DNA replication. J. Biol. Chem. 268, 26179–26189 [PubMed] [Google Scholar]
- 10. Sheaff R. J., and Kuchta R. D. (1993) Mechanism of calf thymus DNA primase: slow initiation, rapid polymerization, and intelligent termination. Biochemistry 32, 3027–3037 [DOI] [PubMed] [Google Scholar]
- 11. Sheaff R. J., Kuchta R. D., and Ilsley D. (1994) Calf thymus DNA polymerase α-primase: “communication” and primer-template movement between the two active sites. Biochemistry 33, 2247–2254 [DOI] [PubMed] [Google Scholar]
- 12. Kuchta R. D., Reid B., and Chang L. M. (1990) DNA primase: processivity and the primase to polymerase α activity switch. J. Biol. Chem. 265, 16158–16165 [PubMed] [Google Scholar]
- 13. Núñez-Ramírez R., Klinge S., Sauguet L., Melero R., Recuero-Checa M. A., Kilkenny M., Perera R. L., García-Alvarez B., Hall R. J., Nogales E., Pellegrini L., and Llorca O. (2011) Flexible tethering of primase and DNA Polα in the eukaryotic primosome. Nucleic Acids Res. 39, 8187–8199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Suwa Y., Gu J., Baranovskiy A. G., Babayeva N. D., Pavlov Y. I., and Tahirov T. H. (2015) Crystal structure of the human Polα B subunit in complex with the C-terminal domain of the catalytic subunit. J. Biol. Chem. 290, 14328–14337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Klinge S., Núñez-Ramírez R., Llorca O., and Pellegrini L. (2009) 3D architecture of DNA Polα reveals the functional core of multi-subunit replicative polymerases. EMBO J. 28, 1978–1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mizuno T., Yamagishi K., Miyazawa H., and Hanaoka F. (1999) Molecular architecture of the mouse DNA polymerase α-primase complex. Mol. Cell. Biol. 19, 7886–7896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Baranovskiy A. G., Babayeva N. D., Liston V. G., Rogozin I. B., Koonin E. V., Pavlov Y. I., Vassylyev D. G., and Tahirov T. H. (2008) X-ray structure of the complex of regulatory subunits of human DNA polymerase δ. Cell Cycle 7, 3026–3036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhou B., Arnett D. R., Yu X., Brewster A., Sowd G. A., Xie C. L., Vila S., Gai D., Fanning E., and Chen X. S. (2012) Structural basis for the interaction of a hexameric replicative helicase with the regulatory subunit of human DNA polymerase α-primase. J. Biol. Chem. 287, 26854–26866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Huang H., Zhao K., Arnett D. R., and Fanning E. (2010) A specific docking site for DNA polymerase α-primase on the SV40 helicase is required for viral primosome activity, but helicase activity is dispensable. J. Biol. Chem. 285, 33475–33484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Copeland W. C. (1997) Expression, purification, and characterization of the two human primase subunits and truncated complexes from Escherichia coli. Protein Expr. Purif. 9, 1–9 [DOI] [PubMed] [Google Scholar]
- 21. Baranovskiy A. G., Zhang Y., Suwa Y., Babayeva N. D., Gu J., Pavlov Y. I., and Tahirov T. H. (2015) Crystal structure of the human primase. J. Biol. Chem. 290, 5635–5646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kilkenny M. L., Longo M. A., Perera R. L., and Pellegrini L. (2013) Structures of human primase reveal design of nucleotide elongation site and mode of Polα tethering. Proc. Natl. Acad. Sci. U.S.A. 110, 15961–15966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kilkenny M. L., De Piccoli G., Perera R. L., Labib K., and Pellegrini L. (2012) A conserved motif in the C-terminal tail of DNA polymerase α tethers primase to the eukaryotic replisome. J. Biol. Chem. 287, 23740–23747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zerbe L. K., and Kuchta R. D. (2002) The p58 subunit of human DNA primase is important for primer initiation, elongation, and counting. Biochemistry 41, 4891–4900 [DOI] [PubMed] [Google Scholar]
- 25. Agarkar V. B., Babayeva N. D., Pavlov Y. I., and Tahirov T. H. (2011) Crystal structure of the C-terminal domain of human DNA primase large subunit: implications for the mechanism of the primase-polymerase α switch. Cell Cycle 10, 926–931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Baranovskiy A. G., Zhang Y., Suwa Y., Gu J., Babayeva N. D., Pavlov Y. I., and Tahirov T. H. (2016) Insight into the human DNA primase interaction with template-primer. J. Biol. Chem. 291, 4793–4802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Klinge S., Hirst J., Maman J. D., Krude T., and Pellegrini L. (2007) An iron-sulfur domain of the eukaryotic primase is essential for RNA primer synthesis. Nat. Struct. Mol. Biol. 14, 875–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Weiner B. E., Huang H., Dattilo B. M., Nilges M. J., Fanning E., and Chazin W. J. (2007) An iron-sulfur cluster in the C-terminal domain of the p58 subunit of human DNA primase. J. Biol. Chem. 282, 33444–33451 [DOI] [PubMed] [Google Scholar]
- 29. Sauguet L., Klinge S., Perera R. L., Maman J. D., and Pellegrini L. (2010) Shared active site architecture between the large subunit of eukaryotic primase and DNA photolyase. PLoS ONE 5, e10083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Vaithiyalingam S., Warren E. M., Eichman B. F., and Chazin W. J. (2010) Insights into eukaryotic DNA priming from the structure and functional interactions of the 4Fe-4S cluster domain of human DNA primase. Proc. Natl. Acad. Sci. U.S.A. 107, 13684–13689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Baranovskiy A. G., Babayeva N. D., Suwa Y., Gu J., Pavlov Y. I., and Tahirov T. H. (2014) Structural basis for inhibition of DNA replication by aphidicolin. Nucleic Acids Res. 42, 14013–14021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Perera R. L., Torella R., Klinge S., Kilkenny M. L., Maman J. D., and Pellegrini L. (2013) Mechanism for priming DNA synthesis by yeast DNA polymerase α. Elife 2, e00482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Vaithiyalingam S., Arnett D. R., Aggarwal A., Eichman B. F., Fanning E., and Chazin W. J. (2014) Insights into eukaryotic primer synthesis from structures of the p48 subunit of human DNA primase. J. Mol. Biol. 426, 558–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Baranovskiy A. G., Gu J., Babayeva N. D., Agarkar V. B., Suwa Y., and Tahirov T. H. (2014) Crystallization and preliminary x-ray diffraction analysis of human DNA primase. Acta Crystallogr. F Struct. Biol. Commun. 70, 206–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhang Y., Baranovskiy A. G., Tahirov T. H., and Pavlov Y. I. (2014) The C-terminal domain of the DNA polymerase catalytic subunit regulates the primase and polymerase activities of the human DNA polymerase α-primase complex. J. Biol. Chem. 289, 22021–22034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Otwinowski Z., and Minor W. (1997) Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 37. Vassylyev D. G., Vassylyeva M. N., Perederina A., Tahirov T. H., and Artsimovitch I. (2007) Structural basis for transcription elongation by bacterial RNA polymerase. Nature 448, 157–162 [DOI] [PubMed] [Google Scholar]
- 38. Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., and Warren G. L. (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 39. DeLano W. L. (2002) The PyMOL Molecular Graphics System, DeLano Scientific, San Carlos, CA [Google Scholar]
- 40. Copeland W. C., and Tan X. (1995) Active site mapping of the catalytic mouse primase subunit by alanine scanning mutagenesis. J. Biol. Chem. 270, 3905–3913 [DOI] [PubMed] [Google Scholar]
- 41. Xie P. (2011) A model for dynamics of primer extension by eukaryotic DNA primase. Eur. Biophys. J. 40, 1157–1165 [DOI] [PubMed] [Google Scholar]
- 42. Arezi B., Kirk B. W., Copeland W. C., and Kuchta R. D. (1999) Interactions of DNA with human DNA primase monitored with photoactivatable cross-linking agents: implications for the role of the p58 subunit. Biochemistry 38, 12899–12907 [DOI] [PubMed] [Google Scholar]
- 43. Basu R. S., Warner B. A., Molodtsov V., Pupov D., Esyunina D., Fernández-Tornero C., Kulbachinskiy A., and Murakami K. S. (2014) Structural basis of transcription initiation by bacterial RNA polymerase holoenzyme. J. Biol. Chem. 289, 24549–24559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Cheetham G. M., and Steitz T. A. (1999) Structure of a transcribing T7 RNA polymerase initiation complex. Science 286, 2305–2309 [DOI] [PubMed] [Google Scholar]
- 45. Zhang Y., Feng Y., Chatterjee S., Tuske S., Ho M. X., Arnold E., and Ebright R. H. (2012) Structural basis of transcription initiation. Science 338, 1076–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Liu X., Bushnell D. A., Silva D. A., Huang X., and Kornberg R. D. (2011) Initiation complex structure and promoter proofreading. Science 333, 633–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Matsui E., Nishio M., Yokoyama H., Harata K., Darnis S., and Matsui I. (2003) Distinct domain functions regulating de novo DNA synthesis of thermostable DNA primase from hyperthermophile Pyrococcus horikoshii. Biochemistry 42, 14968–14976 [DOI] [PubMed] [Google Scholar]
- 48. Geibel S., Banchenko S., Engel M., Lanka E., and Saenger W. (2009) Structure and function of primase RepB′ encoded by broad-host-range plasmid RSF1010 that replicates exclusively in leading-strand mode. Proc. Natl. Acad. Sci. U.S.A. 106, 7810–7815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Beck K., Vannini A., Cramer P., and Lipps G. (2010) The archaeo-eukaryotic primase of plasmid pRN1 requires a helix bundle domain for faithful primer synthesis. Nucleic Acids Res. 38, 6707–6718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kato M., Ito T., Wagner G., and Ellenberger T. (2004) A molecular handoff between bacteriophage T7 DNA primase and T7 DNA polymerase initiates DNA synthesis. J. Biol. Chem. 279, 30554–30562 [DOI] [PubMed] [Google Scholar]
- 51. Corn J. E., Pease P. J., Hura G. L., and Berger J. M. (2005) Crosstalk between primase subunits can act to regulate primer synthesis in trans. Mol. Cell 20, 391–401 [DOI] [PubMed] [Google Scholar]
- 52. Urban M., Joubert N., Purse B. W., Hocek M., and Kuchta R. D. (2010) Mechanisms by which human DNA primase chooses to polymerize a nucleoside triphosphate. Biochemistry 49, 727–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Bianchi J., Rudd S. G., Jozwiakowski S. K., Bailey L. J., Soura V., Taylor E., Stevanovic I., Green A. J., Stracker T. H., Lindsay H. D., and Doherty A. J. (2013) PrimPol bypasses UV photoproducts during eukaryotic chromosomal DNA replication. Mol. Cell 52, 566–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. García-Gómez S., Reyes A., Martínez-Jiménez M. I., Chocrón E. S., Mourón S., Terrados G., Powell C., Salido E., Méndez J., Holt I. J., and Blanco L. (2013) PrimPol, an archaic primase/polymerase operating in human cells. Mol. Cell 52, 541–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Jozwiakowski S. K., Borazjani Gholami F., and Doherty A. J. (2015) Archaeal replicative primases can perform translesion DNA synthesis. Proc. Natl. Acad. Sci. U.S.A. 112, E633–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Simon A. C., Zhou J. C., Perera R. L., van Deursen F., Evrin C., Ivanova M. E., Kilkenny M. L., Renault L., Kjaer S., Matak-Vinković D., Labib K., Costa A., and Pellegrini L. (2014) A Ctf4 trimer couples the CMG helicase to DNA polymerase α in the eukaryotic replisome. Nature 510, 293–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Pavlov Y. I., Shcherbakova P. V., and Rogozin I. B. (2006) Roles of DNA polymerases in replication, repair, and recombination in eukaryotes. Int. Rev. Cytol. 255, 41–132 [DOI] [PubMed] [Google Scholar]
- 58. Yang W., Lee J. Y., and Nowotny M. (2006) Making and breaking nucleic acids: two-Mg2+-ion catalysis and substrate specificity. Mol. Cell 22, 5–13 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.