Abstract
The partially de-N-acetylated poly-β-1,6-N-acetyl-d-glucosamine (dPNAG) polymer serves as an intercellular biofilm adhesin that plays an essential role for the development and maintenance of integrity of biofilms of diverse bacterial species. Translocation of dPNAG across the bacterial outer membrane is mediated by a tetratricopeptide repeat-containing outer membrane protein, PgaA. To understand the molecular basis of dPNAG translocation, we determined the crystal structure of the C-terminal transmembrane domain of PgaA (residues 513–807). The structure reveals that PgaA forms a 16-strand transmembrane β-barrel, closed by four loops on the extracellular surface. Half of the interior surface of the barrel that lies parallel to the translocation pathway is electronegative, suggesting that the corresponding negatively charged residues may assist the secretion of the positively charged dPNAG polymer. In vivo complementation assays in a pgaA deletion bacterial strain showed that a cluster of negatively charged residues proximal to the periplasm is necessary for biofilm formation. Biochemical analyses further revealed that the tetratricopeptide repeat domain of PgaA binds directly to the N-deacetylase PgaB and is critical for biofilm formation. Our studies support a model in which the positively charged PgaB-bound dPNAG polymer is delivered to PgaA through the PgaA-PgaB interaction and is further targeted to the β-barrel lumen of PgaA potentially via a charge complementarity mechanism, thus priming the translocation of dPNAG across the bacterial outer membrane.
Keywords: bacterial adhesion; biofilm; extracellular matrix; membrane protein; outer membrane; polysaccharide; translocation; PgaA; poly-β-1,6-N-acetyl-d-glucosamine; outer membrane protein; biofilm; exopolysaccharide
Introduction
Bacterial biofilms are complex surface-attached microbial communities that cause severe medical problems because once established, they are notoriously difficult to eradicate (1). A hallmark of bacterial biofilm is that the bacteria are encased within a self-produced hydrated polymeric matrix that is typically composed of proteinaceous adhesion factors, exopolysaccharides and DNA (1–3). The extracellular matrix facilitates the adherence between bacteria and to surfaces and protects bacteria against certain antimicrobials, physical stresses, and the host immune system (4–7). A variety of exopolysaccharides, including poly-β-1,6-N-acetyl-d-glucosamine (PNAG),4 colonic acid, cellulose, alginate, and the Pel and Psl polysaccharides of Pseudomonas aeruginosa affect the development and structural integrity of various bacterial biofilms (8–13). Of these, the partially de-N-acetylated poly-β-1,6-N-acetyl-d-glucosamine (dPNAG) exopolysaccharides are required for the maintenance of biofilm structural stability in a diverse array of human pathogens, including Yersinia pestis, Staphylococcus epidermidis, Burkholderia spp., Acinetobacter baumannii, Aggregatibacter spp., Klebsiella pneumoniae, and pathogenic Escherichia coli strains, where it may provide protection against biocides and immune killing (11, 14–21).
In E. coli, four genes of the pgaABCD operon encode proteins required for the synthesis and translocation of dPNAG across the bacterial double-layer membrane (14, 17). PgaA, a tetratricopeptide repeat (TPR)-containing outer membrane protein, forms the secretion pore in the outer membrane and translocates the dPNAG polymer from the periplasm to the cell surface (14, 17). It is thought that the N-terminal TPR motifs of PgaA participate in protein-protein interactions in the periplasm, whereas the C terminus of PgaA forms a transmembrane β-barrel pore that is dedicated for the secretion of the partially dPNAG polymers across the outer membrane. De-N-acetylation of PNAG is achieved by the putative lipoprotein PgaB. The partially de-N-acetylated PNAG is the functional form of the exopolysaccharide (14, 17). Cell-bound PNAG, including the surface-exposed and periplasmic polymer, contains 3–5% de-N-acetylated glucosamine residues (14, 17), whereas the polymer that has been released from cells into the spent medium contains more than this (22). The positively charged glucosamine residues help to retain the polymer on the bacterial surface by interaction with the negatively charged surface component lipopolysaccharides (22). But in addition they are essential for secretion of the polymer as catalytically inactive PgaB proteins are unable to support biofilm formation and similar to the loss of PgaA, causing retention of PNAG polymers in the periplasm (14, 23, 24). PgaC, which is predicted to contain multiple transmembrane helices and a cytosolic domain homologous to family 2 glycosyltransferases, is necessary for the synthesis PNAG. Finally, PgaD, a small membrane protein with two predicted transmembrane helices, assists the glycosyltransferase in polymerizing PNAG and is involved in the regulation of PNAG synthesis. The two inner membrane proteins PgaC and PgaD form a complex that functions as a novel type of c-di-GMP receptor wherein ligand binding to the two proteins stabilizes their interaction and allosterically activates PNAG synthesis (25). The expression of the E. coli pgaABCD operon is tightly regulated. For example, the RNA-binding protein CsrA (carbon storage regulator A) negatively regulates pgaA translation, pgaABCD transcription, and mRNA stability (16, 26, 27).
Unlike various relatively well characterized bacterial protein secretion machineries, mechanisms of exopolysaccharide secretion by outer-membrane secretins remain largely elusive. Recently, structural studies have advanced our understanding of several types of exopolysaccharide secretion across the bacterial outer membrane. For instance, Keiski et al. 28 and Tan et al. 30 determined the crystal structure of AlgE, and Whitney et al. 29 solved the crystal structure of AlgK. In combination with functional analyses, they proposed a mechanism for alginate secretion across the bacterial outer membrane; Little et al. (23, 24) reported the crystal structures of PgaB and its complex with β-1,6-(GlcNAc)6 and disclosed the catalytic mechanism of the N-deacetylase PgaB on PNAG polymers and the manner of periplasmic translocation of PNAG. Despite this progress, many questions regarding how dPNAG, an essential biofilm-supporting exopolysaccharide, is polymerized and translocated across the bacterial outer membrane remain to be answered. Here we present the crystal structure of the PgaA C-terminal transmembrane domain. Structure-based functional studies indicate that a characteristic electronegative patch that lies inside of the secretion pore, in proximity to the periplasm, is important for the translocation of the positively charged dPNAG. In addition, the N-terminal TPR domain of PgaA directly interacts with expolysaccharide N-deacetylase PgaB, which may play an important role in bringing the PgaB-bound dPNAG polymers proximal to the secretion pore. These findings offer an explanation of how the PgaA protein is structured specifically for secreting dPNAG and why fully acetylated PNAG cannot exit from the periplasm to serve as a biofilm-supporting expolysaccharide.
Experimental Procedures
Protein Expression Purification and Crystallization
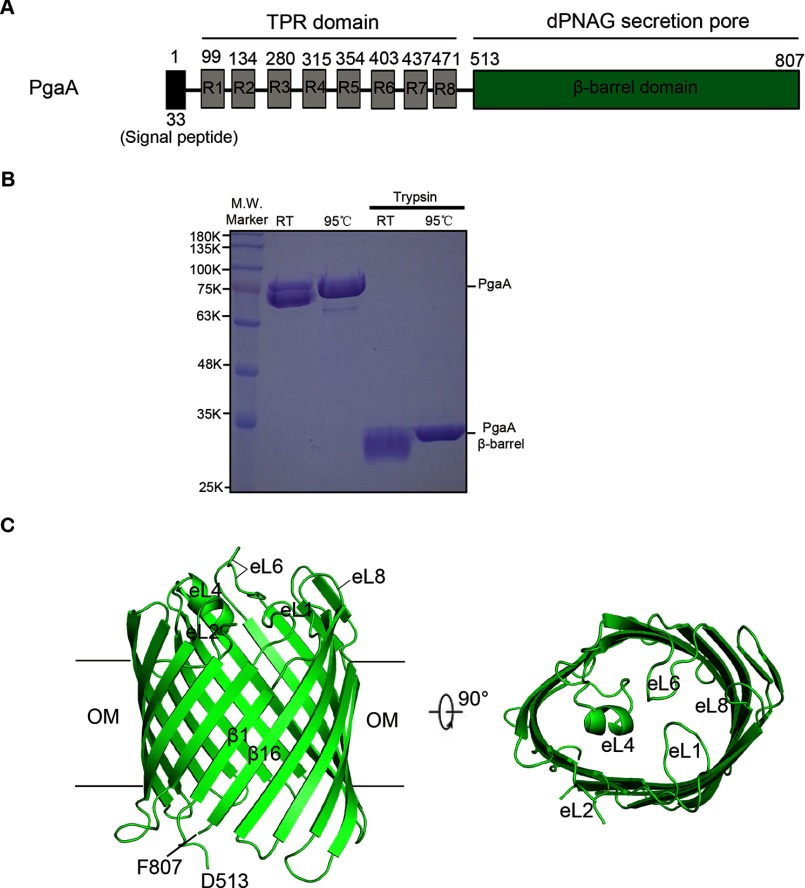
DNA coding sequence of the full-length PgaA protein was amplified by polymerase chain reaction (PCR) using E. coli strain K-12 gemonic DNA (E. coli strain MG1655, ATCC) as template, and the PCR fragments were subsequently cloned into pBAD22 vector under control of the arabinose promoter. The expressed PgaA protein contains a C-terminal six-histidine tag for affinity purification. Initially, we crystallized the full-length PgaA, but the crystals did not diffract beyond ∼10 Å. To obtain high quality crystals for structural determination, the purified full-length PgaA protein was subject to limited proteolysis with trypsin at a molar ratio of 100:1 (protein/enzyme) for 1 h at room temperature. Limited proteolysis produced a stable fragment that corresponds to the C-terminal transmembrane domain of PgaA with a molecular mass of 35 kDa (Fig. 1, A and B). N-terminal sequencing indicated that the N terminus of the fragment starts from residue Ala-511 of PgaA. Subsequently, the DNA coding sequence of this fragment in addition to an N-terminal signal peptide sequence of BamA from E. coli (signal peptide sequence of BamA: MAMKKLLIASLLFSSATVYG) was cloned into pBAD22 vector for overexpression.
FIGURE 1.
Molecular structure of PgaA-(513–807). A, schematic structure of the full-length PgaA. Residues 1–33 constitute a signal peptide. Eight TPR motifs are denoted as R1-R8. The C terminus of PgaA (residues 513–807) forms a transmembrane β barrel. B, the full-length PgaA protein purified from membrane fraction and its trypsin-digested product. Both the full-length PgaA and PgaA β-barrel exhibit heat-modifiable mobility to certain degree on 12% SDS-PAGE. RT, room temperature. C, ribbon representation of the structure of PgaA-(513–807). The PgaA β-barrel consists of 16 β-strands, closed on the extracellular surface by four long extracellular loops eL1, eL4, eL6. and eL8. OM, outer membrane.
For purification, cells of E. coli strain sf100 were induced to express the PgaA C-terminal domain by adding 0.2% arabinose for 3 h at 37 °C and lysed on ice in phosphate-buffered saline (PBS) buffer using a French press high pressure instrument (JN-02C, JNBio Ltd.) at 16,000 p.s.i. Membranes were collected by centrifugation for 40 min at 39,000 × g and solubilized in 1.0% (w/v) lauryldimethylamine oxide (Anatrace) in PBS buffer. After centrifugation for 30 min at 39,000 × g, the supernatant solution was loaded onto a 10-ml nickel-nitrilotriacetic acid column (metal-chelating-Sepharose, Amersham Biosciences). The column was washed with 10 volumes of 0.5% lauryldimethylamine oxide in PBS buffer in the presence of 20 mm imidazole. Detergent exchange was performed by washing the column with 5 volumes of 1.0% n-octyl-β-d-glucopyranoside (Anatrace) in a buffer containing 20 mm Tris, pH 8.0, and 150 mm NaCl. The PgaA β barrel protein was eluted with a buffer containing 20 mm Tris, pH 8.0, 150 mm NaCl, 250 mm imidazole, 5% glycerol, and 0.8% n-octyl-β-d-glucopyranoside. Further purification was carried out by gel filtration on Superdex-200 HR 10/30 column (GE Healthcare) in buffer containing 20 mm Tris, pH 8.0, 150 mm NaCl, and 0.8% n-octyl-β-d-glucopyranoside. The PgaA β-barrel protein was concentrated to 10 mg ml−1 with a Centricon-30 concentrator (Amicon; molecular mass cutoff, 30 kDa).
To improve the phasing power of selenomethionine (Se-Met)single-wavelength anomalous dispersion, two point mutations (L610M and I743M) were introduced into the PgaA β-barrel by overlapping PCR. Se-Met-substituted PgaA β-barrel protein was expressed in E. coli strain sf100 cells by inhibition of the methionine biosynthesis pathway. The cells were grown in M9 minimal medium with 1.5% (v/v) glycerol as carbon sources and induced with 1% arabinose. Purification procedures were as described for the native protein.
Crystallization of the PgaA barrel was performed by hanging-drop vapor diffusion at 16 °C by mixing 1 μl each of protein (10 mg ml−1) and reservoir solutions. Initial crystallization conditions were found using a broad screen. After optimization, the best crystals were obtained in a condition that contains 0.1 m NaOAc, pH 4.5, 31% 2-methyl-2,4-pentanediol, 0.4 m NaCl, and 0.3 m NDSB-195 (Hampton Research, Additive screenTM 63). The crystals appeared overnight and grew to maximum dimensions of 70 × 70 × 90 μm within 2 weeks.
Data Collection, Structure Determination, and Refinement
Crystals were flash-frozen in liquid nitrogen directly from the drop. Native datasets were collected at 100 K at a wavelength of 1.0000 Å at beamline BL17U of Shanghai Synchrotron Radiation Facility (SSRF, China), and Se-Met datasets were collected at the selenium peak (λ = 0.9795 Å) at beamline BL1A of Japanese KEK synchrotron. The data were indexed and scaled with HKL2000 package (31). The crystals belong to space group P6322 and diffract to 2.8 Å and 3.4 Å for native and Se-Met crystals, respectively. Five of six selenium sites were located using PHENIX (32). After solvent flattening, electron density maps were generated with useful phases to ∼3.5 Å. An initial model was manually built with the program COOT (33) using selenium sites and large amino acid side chains to determine the registry. The model was improved by an iterative process using solvent flattening and model building. This model was used in the subsequent refinement of the native dataset, giving an R factor of 21.5% (Rfree of 26.8%). Residues 567–572 of PgaA in the best-refined structure of the PgaA were not included, presumably because of conformational flexibility. The final refined model was analyzed with pro-check of CCP4 program suit (34), and detailed statistics of crystallographic analyses are shown in Table 1. All figures shown in the paper were rendered with PyMOL (35), and the coordinate file was deposited in PDB under ID code 4Y25.
TABLE 1.
Data collection and refinement statistics
Values in parentheses refer to the highest resolution shell. Rfree was calculated in identical manner to Rwork using a 5% test set of randomly selected reflections that were omitted from the refinement.
| Statistics | Values | |
|---|---|---|
| Data collection | ||
| Protein crystal | Native | Se-Met( L610M+I743M) |
| Beamline | BL17U (SSRF) | BL1A (KEK) |
| Wavelength (Å) | 1.00000 | 0.9795 |
| Space group | P6322 | P6322 |
| Cell dimensions | ||
| a, b, c (Å) | 144.0, 144.0, 90.6 | 144.4, 144.4, 88.4 |
| α, β, γ (°) | 90.0, 90.0, 120.0 | 90.0, 90.0, 120.0 |
| Resolution (Å) | 50-2.8 (2.9-2.8) | 50-3.4 (3.5-3.4) |
| Rsym (%) | 6.9 (>100) | 13.5 (83.8) |
| Completeness (%) | 94.4 (91.8) | 100.0 (100.0) |
| I/σ(I) | 28.9 (1.9) | 28.9 (4.5) |
| Unique reflections (n) | 13,051 (1,227) | 8,026 (771) |
| Redundancy | 10.9 (10.5) | 21.1 (21.8) |
| Refinement | ||
| Resolution (Å) | 32.3-2.8 | |
| Reflections (n) | 13,043 | |
| Rwork/Rfree (%) | 21.5/26.8 | |
| Number of atoms (n) | ||
| Protein | 2,402 | |
| Solvent | 12 | |
| B-factor (Å2) | 90.59 | |
| Root mean square deviation | ||
| Bond length (Å) | 0.010 | |
| Bond angles (°) | 1.418 | |
| Ramachandran plots | ||
| In preferred regions | 267 (93.36%) | |
| In allowed regions | 19 (6.64%) | |
| Outliers | 0 (0.00%) | |
Quantitative Biofilm Assay
The wild-type E. coli strain MG1655 (MG) produces little or no biofilm because under laboratory conditions, the RNA-binding protein negatively regulates pgaA translation, pgaABCD transcription, and mRNA stability (16, 26, 27). MG_csrA::kan_ΔpgaA strains (E. coli strain MG1655 with both genes csrA and pgaA disrupted) were transformed with the pgaA expression plasmid. For these experiments, the wild-type protein and the various mutants contain a C-terminal six-histidine tag. Overnight LB cultures (37 °C, 250 rpm) were inoculated 1:100 into LB medium containing 100 μg/ml ampicillin and 0.1% l-arabinose and incubated in a 96-well microtiter plate (total volume of 200 μl per each well) at 26 °C for 24 h without shaking. Growth of planktonic cells after 24 h was determined by absorbance at 600 nm. To measure biofilm formation, planktonic cells were removed by discarding the medium, and the wells were rinsed with water three times. Bound cells were stained with 1.25 volumes (250 μl) of crystal violet solution (Fisher; catalog #255-960B) for 2 min, and the wells were rinsed again three times with water to remove unbound and excess dye. Bound dye was solubilized with 1.25 volumes of 33% acetic acid, and absorbance at 630 nm was determined using a microtiter plate reader. Background staining was corrected by subtracting the crystal violet bound to un-inoculated controls. Each experiment was conducted at least twice in replicates of three.
Isolation and Immunoblotting of dPNAG
Cultures were grown in LB medium with or without 100 μg/ml ampicillin and 0.1% l-arabinose at 26 °C, 250 rpm for 24 h, cells were collected by centrifugation (10,000 rpm for 5 min), and dPNAG was isolated from cell-bound and spent medium fractions and detected following the procedures described earlier (14) with the modification that the aqueous extracts of dPNAG were concentrated using Amicon® Ultra Centrifugal filter (3000 Da cutoff, Millipore). dPNAG concentrates were applied onto a nitrocellulose membrane (PROTRAN®, Whatman GmbH) and allowed to dry at 37 °C for 1 h. The membrane was blocked for 1 h in 5% BSA in PBS, 0.5% Tween, washed twice for 15 min with PBS, 0.5% Tween, and treated for 1 h at room temperature with α-dPNAG monoclonal antibody (24 μg/ml in 1% BSA PBS, 0.5% Tween) that was raised against E. coli dPNAG (14). The membrane was washed five times (15, 5, 5, 5, 5 min) with PBS, 0.5% Tween and treated with horseradish peroxidase-conjugated anti-murine IgM antibody (Sigma A8786; 1:10,000 diluted in 1% BSA PBS, 0.5% Tween) for 1 h at room temperature. The membrane was washed again (15, 5, 5, 5, 5 min), and the signal was detected by chemiluminescence using Western Lighting Plus-ECL (PerkinElmer Life Sciences) and imaged using ChemiDoc.
Western Blotting Analysis of PgaA Protein Expression in Bacteria
To compare relative protein expression levels of pgaA mutants that have significantly reduced biofilm production to that of the wild-type strain, plasmids harboring wild-type plasmid or various pgaA mutants (all contain a C-terminal six-histidine tag) were transformed into E. coli MG_csrA::kan_ΔpgaA strain and spread on LB agar plates that contained 100 μg/ml ampicillin, 50 μg/ml kanamycin, and 0.1 mg/ml l-arabinose. After incubation at 26 °C for 24 h, colonies were scraped off of the plates, and the cell density was adjusted to A600 of 1.0 by the addition of 1 × PBS buffer. Resuspended cells were homogenized by sonication and then centrifuged at 40,000 × g for 1 h to collect total membrane fractions. Inner membranes were solubilized by 1.0% N-lauryl-sarcosine and discarded after a second round of centrifugation at 40,000 × g for 1 h. After that, outer membrane pellets were further solubilized using 1.0% lauryldimethylamine oxide. 20 μl of outer membranes for each PgaA mutant were withdrawn and mixed with 10 μl of SDS loading dye. Subsequently, the sample was equally allocated into two Eppendorf tubes (each 15 μl), kept at room temperature, or heated at 95 °C for 5 min before SDS-PAGE analysis. After electrophoresis, the proteins were transferred to a PVDF membrane and blocked using TBST buffer (20 mm Tris-HCl, pH 8.0, 150 mm NaCl, 0.05% Tween 20) that contained 8% skim milk for 2 h. The PVDF membrane was then incubated with anti-His antibody (1:3000) (TIANGEN) at room temperature for 2 h. PVDF membranes were subsequently washed with TBST buffer twice and further incubated with horseradish peroxidase-conjugated secondary antibody (1:5000) (TransTM) at room temperature for 2 h. PVDF membranes were exposed using enhanced chemiluminescence reagents (EasySee Western blot kit, TransTM).
Protein Co-expression and His-tag Pulldown Assay
DNA coding sequences of the E. coli full-length PgaA, the TPR domain of PgaA (residues 1–512, including the signal peptide and a C-terminal 6-histidine tag), and the full-length PgaB (with or without a C-terminal 6-histidine tag) were cloned in pQLink vector (36). To co-express PgaA and PgaB, plasmids (pQLink-PgaA and pQLink-PgaB-His) were utilized to construct plasmid pQLink-PgaA-PgaB-His that is capable of co-expressing both PgaA and PgaB-His proteins (36). Similarly, to co-express PgaB and TPR-His, plasmids (pQLink-PgaB and pQLink-TPR-His) were utilized to construct plasmid pQLink-PgaB-TPR-His.
To test whether PgaB binds PgaA, three plasmids (pQlink-PgaA, pQLink-PgaB-His, and pQLink-PgaA-PgaB-His) were individually transformed into E. coli BL21(DE3). Cells that carry each of the three plasmids were induced with 0.4 mm isopropyl 1-thio-β-d-galactopyranoside for 3 h at 37 °C and lysed on ice in PBS buffer by sonication. Membranes were collected by centrifugation for 40 min at 40,000 × g and solubilized in 1.0% (w/w) n-dodecyl-β-d-maltoside (Anatrace) in PBS buffer. After centrifugation for 30 min at 40,000 × g, the extract was loaded onto a 1-ml nickel column. The column was washed with 5 volumes of 0.5% n-dodecyl-β-d-maltoside in PBS buffer in the presence of 20 mm imidazole. The proteins bound to the nickel-nitrilotriacetic acid beads were eluted with a 1-ml buffer containing 20 mm Tris, pH 8.0, 150 mm NaCl, 250 mm imidazole, and 0.1% n-dodecyl-β-d-maltoside. The eluted samples (15 μl) were subject to 12% SDS-PAGE analysis. Detection of the interaction between TPR and PgaB proteins was performed in a similar procedure as described for PgaA and PgaB-His.
Results
Crystal Structure of PgaA-(513–807)
Based on prediction, the full-length PgaA contains eight periplasm-localized tetratricopeptide repeats at the N terminus and a C-terminal β-barrel domain located in the outer membrane (Fig. 1A). TPR domains are known primarily to be involved in protein-protein interactions, whereas the β-barrel domain of PgaA may serve as the portal and substrate specificity determinant for dPNAG secretion. Our initial attempts to crystallize the full-length PgaA failed to produce high resolution crystals for structural determination. To this end we performed limited proteolysis on the full-length PgaA protein. Limited proteolysis with trypsin produced a stable fragment that corresponds to the transmembrane β-barrel domain of PgaA (residues 513–807) (Fig. 1B). Subsequently, we cloned the coding sequence of PgaA (residues 513–807) in addition to a C-terminal six-histidine tag into pBAD22 vector with an N-terminal signal peptide coding sequence from E. coli BamA for efficiently targeting the PgaA transmembrane domain to the outer membrane for over-expression. PgaA-(513–807) was crystallized in space group P6322 with one molecule in the asymmetric unit. Both native and Se-Met-incorporated PgaA-(513–807) crystals were grown in the presence of the nonionic detergent n-octyl-β-d-glucoside, and the structure was solved by Se-Met single-wavelength anomalous dispersion by introducing two extra Met residues into the protein sequence (L610M and I743M). Refinement produced final models with good geometry and an R factor of 21.5% (Rfree factor of 26.8%) for the native data (Table 1). In the final refined model, residues 567–572 of PgaA (part of the extracellular loop 2) for the native structure were not included because there was no interpretable electron density present for these residues.
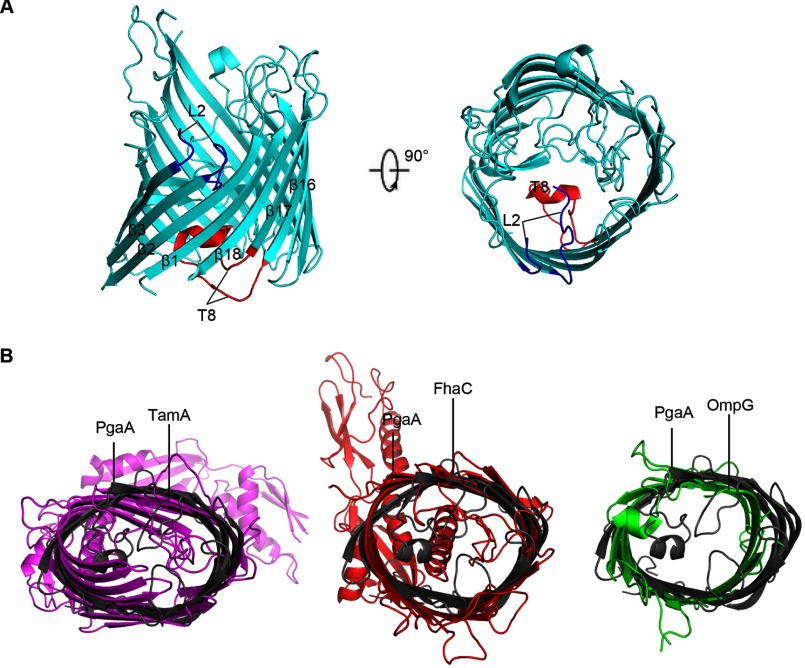
PgaA-(513–807) forms a 16-strand β-barrel that spans the outer membrane with antiparallel and interlocking β1 and β16 strands (Fig. 1C). Unlike AlgE, an 18-strand outer membrane protein that is responsible for exopolysaccharide alginate secretion, no narrow pore constriction is found in the β barrel of PgaA, and no extracellular or periplasmic loops occlude the lumen (Figs. 1C and 2A) (29). However, on the extracellular side, the PgaA β-barrel is mainly closed by four long extracellular loops, eL1, eL4, eL6, and eL8 (Fig. 1C). All of the periplasmic loops of the PgaA β-barrel are relatively short, a common feature found in most β-barrel outer membrane proteins. In search of similar structures using DALI (37), the β-barrel domain of PgaA was found to be related to a large number of outer membrane proteins such as TamA (38), FhaC (39), and some outer membrane porins (40) with top Z-scores ranging from 14 to 16.4. Superposition of PgaA β-barrel with that of TamA, FhaC, and outer membrane porin OmpG produced root mean square deviations of 3.2 Å for 278 Cα atoms, 3.2 Å for 280 Cα atoms, and 3.5 Å for 265 Cα atoms, respectively. Apparently, these three families of outer membrane proteins have significantly different overall β-barrel shape and extracellular loop conformations in comparison to those of the PgaA β-barrel (Fig. 2B).
FIGURE 2.
Structural comparison between PgaA-(513–807) and other representative β-barrel outer membrane proteins. A, ribbon representation of the structure of AlgE (PDB code 3RBH). AlgE β-barrel consists of 18 β-strands and the lumen is occluded by two functionally important loops: a long periplasmic loop 8 (T8, in red) and an extracellular loop 2 (L2, in blue). B, structural overlay between PgaA β-barrel and that of TamA (PDB code 4C00), FhaC (PDB code 3NJT), or OmpG (PDB code 2F1C).
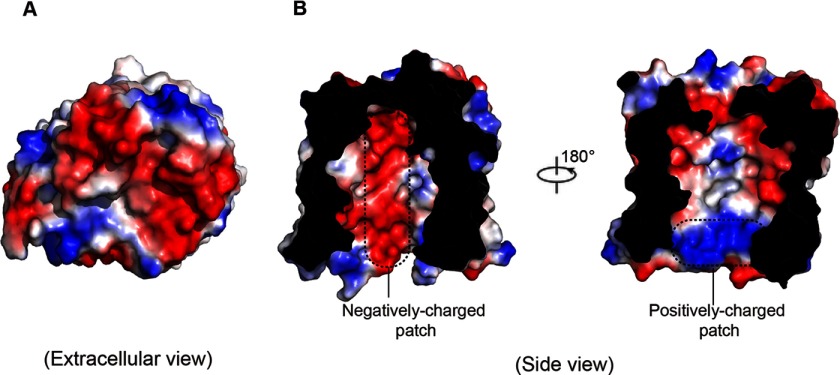
Electrostatic potential surface representation of PgaA β-barrel revealed that the extracellular surface of PgaA β-barrel is primarily electronegative (Fig. 3A). Remarkably, half of the interior surface of PgaA β-barrel along the substrate translocation pathway is electronegative, whereas the opposite half is mainly neutral with several dispersed positively charged residues located close to the periplasmic space (Fig. 3B).
FIGURE 3.
Electrostatic potential surface representation of PgaA-(513–807). A, electrostatic potential surface representation showing that the extracellular surface of PgaA is primarily electronegative. B, a slab view showing the electrostatic properties inside of the lumen of PgaA. The surface is colored according to electrostatic potential, ranging from blue (+70 kT/e) to red (−70 kT/e), where kT is thermal energy, and e is the elementary charge. The characteristic negatively charged patch and the positively charged patch inside of the β barrel are indicated with dotted lines.
A Cluster of Negatively Charged Residues, Inside of the β-Barrel Lumen and Proximal to the Periplasm, Is Important for Biofilm Formation
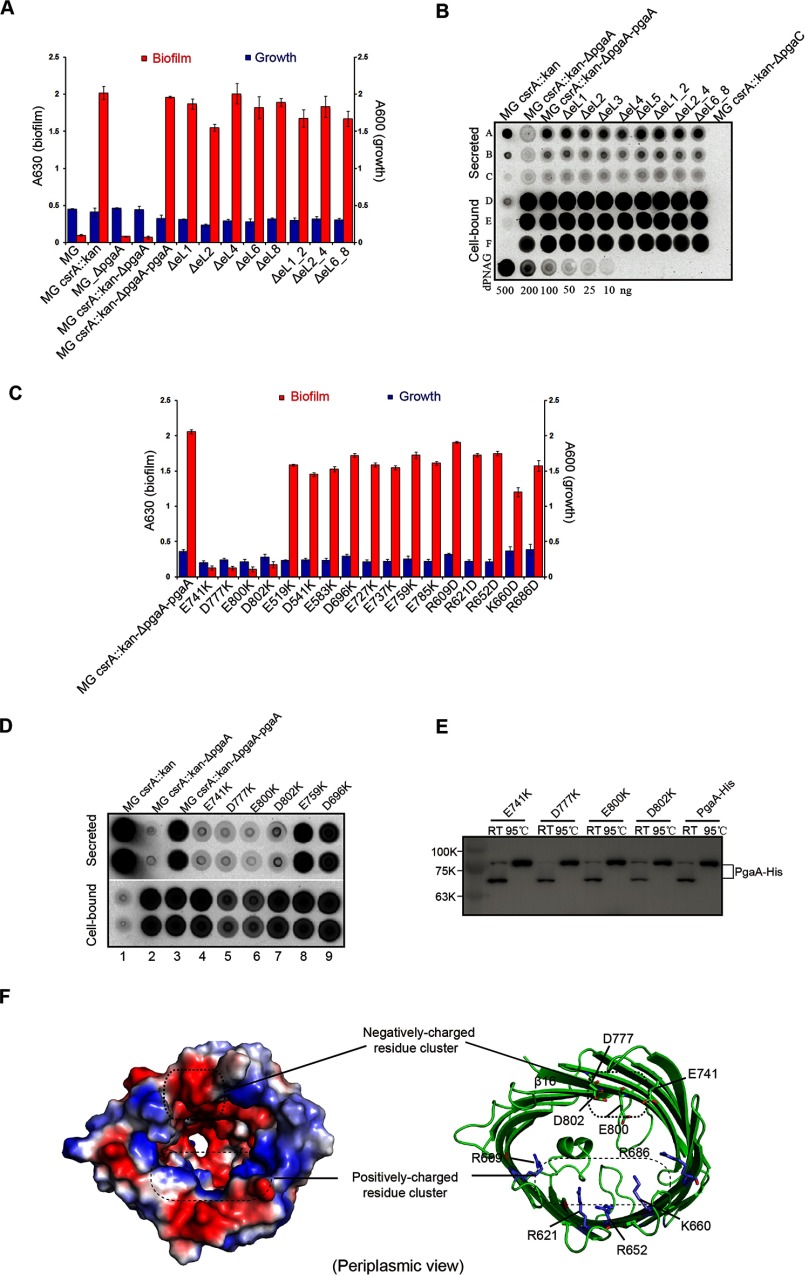
In many bacterial outer membrane proteins, long extracellular loops play important roles in regulating substrate translocation (29). To investigate the functional roles of the extracellular loops of PgaA in dPNAG secretion, a pgaA deletion mutant (E. coli. Strain MG_csrA::kan_ΔpgaA) was complemented with plasmids that express various mutant forms of PgaA. As reported previously, the pgaA deletion mutant does not secrete dPNAG and, therefore, produces very low levels of biofilm but was unaffected in its growth rate (14, 16). When the pgaA gene was introduced on a plasmid, high levels of biofilm production were restored (Fig. 4A). Using this complementation assay, we evaluated the production of biofilm by several loop deletion pgaA mutants. Surprisingly, when we tested for complementation by various extracellular loop deletion mutants pgaA_ΔeL1, pgaA_ΔeL2, pgaA_ΔeL4, pgaA_ΔeL6, and pgaA_ΔeL8, all were found to restore biofilm formation (Fig. 4A). We also tested biofilm production by several pgaA mutants, pgaA_ΔeL1_2, pgaA_ΔeL2_4, and pgaA_ΔeL6_8, that have two loops deleted. Similarly, these mutants also restored biofilm formation (Fig. 4A). Finally, we examined the ability of these complemented strains to secrete dPNAG using an established approach (14). In all cases we found that plasmids encoding the wild-type PgaA protein or proteins containing single or dual deletions of the extracellular loops were able to complement the pgaA deletion (Fig. 4, A and B). Taken together, these studies strongly suggest that, unlike alginate secretion, the extracellular loops of PgaA β-barrel seem not to be important for biofilm formation.
FIGURE 4.
A cluster of negatively charged residues inside of the barrel and in proximity to periplasm is important for dPNAG secretion. A, extracellular loops of PgaA are not important for biofilm production. Wild-type E. coli strain MG1655 (denoted as MG) produced low levels of biofilm due to repression by CsrA (A630 < 0.1); the csrA-disrupted E. coli strain (denoted as MG_csrA::kan) produced 20-fold more biofilm (A630 = 2.0); both the pgaA-deleted E. coli strain (denoted as MG_ΔpgaA) and E. coli strain MG1655 with both csrA- and pgaA-disrupted (denoted as MG_csrA::kan_ΔpgaA) produced low levels of biofilm (A630 < 0.1). When MG_csrA::kan_ΔpgaA was complemented with a plasmid that expresses PgaA (MG_csrA::kan_ΔpgaA_pgaA), biofilm production was restored. None of the loop deletion mutants affected biofilm production. B, extracellular loops of PgaA are not important for secretion of dPNA. Immunoblotting of secreted and cell-bound dPNAG from MG1655 csrA::kan_ΔpgaA strain was complemented with plasmids expressing wild-type pgaA or mutant pgaA proteins with extracellular loop deletions. Secreted and cell-bound dPNAG samples were blotted onto a nitrocellulose membrane and detected with α-PNAG monoclonal antibody as described (14). Rows A–C, dPNAG derived from 30, 6, and 2 μl of spent medium. Rows D-F, cell-bound dPNAG from 40, 4, and 1 ml of culture. Dilutions of purified dPNAG (bottom row) served as a standard for comparison. C, 12 negatively charged residues (Glu-519, Asp-541, Glu-583, Asp-696, Glu-727, Glu-737, Glu-741, Asp-759, Asp-777, Glu-785, Glu-800, and D802) and 5 positively charged residues (Arg-609, Arg-621, Arg-652, Lys-660, and Arg-686) inside the lumen were mutated to Lys and Asp, respectively, and their effects on biofilm production were assayed. Each experiment was conducted at least twice in replicates of three. Error bars denote S.D. D, immunoblotting of secreted and cell-bound dPNAG from MG_csrA:: kan_ΔpgaA strains expressing wild-type PgaA or single-point PgaA mutants. Duplicate samples represent dPNAG extracted from two independent cultures. E, nickel column purification and Western blot analysis of the expression levels of both the wild-type and mutant proteins from bacterial outer membrane. Both the wild-type and mutant proteins exhibited apparent heat-modifiable mobility, suggesting that they are all folded and resided in the outer membrane. RT, room temperature. F, residues Glu-741, Asp-777, Glu-800, and Asp-802 of PgaA form a negatively charged residue cluster in proximity to the periplasm.
In search of the elements of PgaA β barrel that are important for dPNAG secretion, we next tested whether the charged residues inside of the β-barrel lumen play roles in biofilm production. In the interior of the β-barrel, 12 negatively charged residues and 5 positively charged residues (not counting the charged resides located on the extracellular loops and short periplasmic loops) constitute a negatively charged patch and a positively charged patch, respectively (Fig. 3B). We mutated each of the negatively charged residues (Glu or Asp) to the positively charged residue Lys and mutated the positively charged residues (Arg or Lys) to negatively charged residue Asp. Subsequently, biofilm production by all of these single-point mutants was evaluated using the complementation assay. Of the 12 negative-to-positive mutations, E741K, D777K, E800K, and D802K significantly affected biofilm formation; each caused an ∼20-fold reduction of biofilm production in comparison to the control strain (MG_csrA::kan_ΔpgaA_pgaA), whereas the other 5 positive-to-negative mutation caused only modestly reduced biofilm production (Fig. 4C). To examine whether biofilm production correlates with dPNAG secretion, we grew a series of strains in cultures shaking at 26 °C, 250 rpm for 24 h, which allows secreted dPNAG to be released into the spent medium, and detected this polymer by immunoblotting (14). The cell-bound dPNAG was also isolated and detected to determine whether an effect on secretion of the polymer by a strain might be due to a defect in dPNAG synthesis. Consistent with previous observation (14), the parent strain (MG_csrA::kan) was efficient in secreting most of its dPNAG into the spent medium, whereas the MG_csrA::kan_ΔpgaA mutant failed to secrete this polymer (Fig. 4D, lanes 1 and 2). Expression of the wild-type pgaA gene from a plasmid was able to complement the observed ΔpgaA defect (Fig. 4D, lane 3), but the resulting strain retained a greater amount of the polymer in the cell-bound fraction than the parent strain, suggesting that the PgaA protein expressed from the pgaABCD genomic locus may function more effectively than the one from a multicopy plasmid. The pgaA mutant proteins with lysine substituted at the four critical residues of the electronegative patch of the PgaA channel (lanes 4–7) were all found to be defective for secretion, although they were capable of synthesizing and accumulating dPNAG in the cell-bound fraction. Two mutants with substitution of acidic residues that did not affect biofilm formation (Fig. 4C) were also able to secrete dPNAG (Fig. 4D, lanes 8 and 9). These studies suggest that biofilm production reflected the amount of dPNAG secretion into the spent medium for both wild-type strain and pgaA mutants. Furthermore, Western blot analysis revealed that all four point mutants of PgaA (E741K, D777K, E800K, and D802K) that significantly decreased biofilm production were expressed at levels similar to or higher than the wild-type protein (Fig. 4E). In addition, both the wild-type PgaA and PgaA mutant proteins exhibited heat-modifiable mobility difference on SDS-PAGE, suggesting that all of these proteins are folded and reside in the outer membrane (Fig. 4E). These experiments argue that the reduced biofilm production of these mutants is not caused by decreased expression of the mutant protein but by a decreased ability to secrete dPNAG across the outer membrane. Intriguingly, all of the negative-to-positive mutants that significantly reduced biofilm production were clustered at a position that is proximal to the periplasm, and they together form an electronegative patch inside the β-barrel (Fig. 4F). Thus, it is likely that this electronegative patch may make initial contacts with the positively charged dPNAG polymer and thereby help to usher the polymer into the secretion pore of PgaA.
Both the TPR Domain and β-Barrel Pore of PgaA Are Required and Specific for dPNAG Secretion
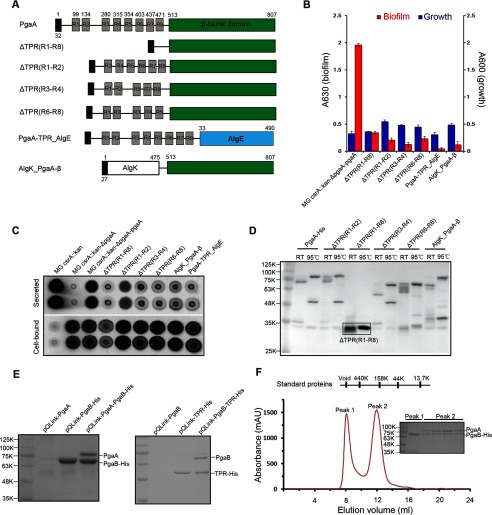
Of several types of exopolysaccharide outer-membrane secretins, the alginate secretin is among the best characterized. It mainly consists of two proteins: an 18-strand β-barrel outer-membrane protein, AlgE, which forms the secretion pore, and a TPR-containing lipoprotein, AlgK, which facilitates AlgE biogenesis and potentially interacts with other periplasmic components of the secretion machinery. In addition to AlgE, functional studies demonstrated that AlgK is also required for alginate secretion (28). Distinct from the alginate secretin, PgaA contains two functional domains with an N-terminal TPR domain that might have functions similar to AlgK and a C-terminal transmembrane secretion pore. To test whether the TPR domain of PgaA is required for dPNAG secretion, we studied the biofilm production of several TPR deletion PgaA mutants that have different helices of the TPR motifs deleted (Fig. 5A). As shown in Fig. 5, B and C, all of the tested TPR-deleted pgaA mutants significantly decreased biofilm production and dPNAG secretion in the spent medium, although their expression levels were close to or even higher than that of the wild-type protein (Fig. 5D). Furthermore, we investigated biofilm production by two fusion proteins by replacing the two domains of PgaA with either AlgK or AlgE. Likewise, AlgK_PgaA-β-barrel fusion protein that was expressed in the bacterial outer membrane also did not support biofilm production and dPNAG secretion (Fig. 5, B–D, PgaA-TPR_AlgE fusion protein seemed not to be expressed in the bacteria). Taken together, these studies show that the TPR domain of PgaA is important for biofilm production and suggest that both the TPR domain and the β-barrel domain of PgaA are required and specific for dPNAG secretion.
FIGURE 5.
The TPR domain of PgaA directly binds PgaB and is necessary for biofilm production. A, schematic structures of the full-length PgaA, TPR-deletion mutant PgaA proteins, and two fusion proteins (AlgK_PgaA-β and PgaA-TPR_AlgE). B, TPR domain of PgaA is required for biofilm production. TPR repeat deletion mutants ΔTPR(R1-R8) (Mr 35.1), ΔTPR(R1-R2) (Mr 76.3), ΔTPR(R3-R4) (Mr 73.5), and ΔTPR(R6-R8) (Mr 73.8) all significantly affected biofilm production; two fusion proteins AlgK_PgaA-β (Mr 76.3) and PgaA-TPR_AlgE (Mr 89.3) are unable to restore biofilm production. Each experiment was conducted at least twice in replicates of three. Error bars denote S.D. C, immunoblotting of secreted and cell-bound dPNAG from MG_csrA::kan_ΔpgaA strain expressing wild-type PgaA or PgaA with TPR deletions or PgaA fusion proteins. D, nickel-column purification and Western blot analysis of the expression levels of the wild-type, various TPR-deletion PgaA mutant proteins, and fusion protein AlgK_PgaA-β-barrel from bacterial outer membranes. All the proteins exhibited apparent heat-modifiable mobility, suggesting that they are all folded and resided in the outer membranes. RT, room temperature. E, the TPR domain of PgaA directly binds PgaB. When co-expressed in bacteria, PgaB-His is capable of pulling down PgaA (left panel), and TPR-His is capable of pulling down PgaB (right panel). Pulldown mixtures were resolved on 12% SDS-PAGE gels and stained with Coomassie Blue dye. F, size exclusion chromatographic profile of the pulldown mixture of PgaB-His and PgaA on a Superdex 200 10/30 column. PgaB-His (a lipoprotein) alone is not stable and forms aggregates in a gel filtration buffer that contains 20 mm Tris, pH 8.0, 150 mm NaCl, and 0.06% n-dodecyl-β-d-maltoside, so excess PgaB-His protein was eluted in a void volume (about 8.0 ml). PgaB-His and PgaA seem to form a 1:1 stable complex as resolved by 12% SDS-PAGE analysis. mAU, milliabsorption units.
The N-terminal TPR Domain of PgaA Directly Interacts with PgaB
Distinct from the outer-membrane alginate secretin that mainly consists of AlgK and AlgE, the dPNAG secretin includes an extra lipoprotein, PgaB, which is also required for dPNAG secretion and biofilm production (14, 23, 24). To further understand how the PNAG secretin is assembled and why the TPR domain of PgaA is required for biofilm production, we studied the interactions between PgaA and PgaB. To this end, DNA coding sequences of both PgaA and PgaB in addition to a C-terminal six-histidine tag were cloned in pQLink vector (36). Subsequently, a plasmid pQLink-PgaA-PgaB-His was constructed that was capable of co-expressing both PgaA and PgaB proteins upon induction in bacteria. Plasmids pQLink-PgaA, pQLink-PgaB-His, and pQLink-PgaA-PgaB-His were transformed into E. coli BL21(DE3) competent cells for protein overexpression. As shown in Fig. 5E, His-tagged PgaB protein was capable of pulling down a protein that has a molecular mass corresponding to that of PgaA as revealed by SDS-PAGE analysis. Protein identification using MALDI-TOF mass spectrometry (KRATOS Analytical, Shimadzu Corp.) confirmed that the suspected protein is PgaA. Furthermore, we found that the PgaB and PgaA proteins from the pulldown mixture co-migrate on size exclusion column (Fig. 5F), suggesting that PgaA and PgaB form a stable complex. As both the TPR domain of PgaA and the PgaB protein are localized in periplasm, we next tested whether the TPR domain of PgaA alone is responsible for the interaction. When the TPR domain of PgaA and PgaB was co-expressed in the bacteria, the TPR domain was capable of pulling down PgaB, indicating that the TPR domain of PgaA directly binds PgaB (Fig. 5E). Taken together, these experiments show the N-terminal TPR domain of PgaA interacts with PgaB and suggest that this interaction may play important roles in the assembly of the PNAG secretin by bringing the PgaB-bound dPNAG polymer proximal to the PgaA secretion pore.
Discussion
Similar to various bacterial protein secretion systems, outer-membrane polysaccharide secretins differ in composition of the components and the manner of polysaccharide translocation. For instance, the export of the tightly surface-associated capsular polysaccharides across the outer membrane occurs via a novel type of octameric α-helical pore formed by Wza and its otholog KpsD (41–43), whereas the exopolysaccharide alginate secretin AlgE forms an 18-strand β-barrel pore with an arginine-rich highly electropositive pore constriction acting as a selectivity filter for the negatively charged alginate polymer (29). Apart from this, the extracellular loop L2 and a long periplasmic loop T8 of the AlgE β-barrel seem to play important roles in regulating alginate secretion (Fig. 2A). Here, we showed that the transmembrane domain of the dPNAG secretin PgaA forms a 16-strand β barrel. Distinct from alginate secretion by AlgE, in which both extracellular L2 and long periplasmic loop T8 occlude the barrel lumen, the extracellular loops that close the lumen of PgaA do not appear to occlude the lumen or play an important role in substrate secretion. Rather, a cluster of negatively charged residues inside of the PgaA barrel lumen and in proximity to the periplasm is important for dPNAG secretion. This finding correlates well with the essential role of the lipoprotein PgaB in dPNAG secretion, which functions as a PNAG deacetylase, conferring a positive charge to the polymer by partial de-N-acetylation (14, 23, 24). We propose that complementary charge interactions between the secretion pore and the exopolysaccharide substrate are important for the secretion of both the negatively charged alginate and the positively charged dPNAG across the bacterial outer membrane. Our current hypothesis is also consistent with the observation that an electronegative patch on the N-terminal domain of PgaB interacts with the positively charged glucosamine as demonstrated by molecular simulation studies (23). Such unusual pore electrostatics has been previously observed in other outer membrane proteins and has been proposed to have consequences for solute translocation or substrate recognition. For example, Im and Roux (44) found that the electrostatic interactions between ions and the charge distribution of the OmpF channel play an important role in governing ion permeation and selectivity. An additional example of this type of pore feature that may support dPNAG translocation is the electropositive extracellular surface features of PorB from pathogenic Neisseria meningitides may mediate Toll-like receptor recognition through a nonspecific electrostatic attraction (45, 46).
In our structure, the PgaA barrel pore is closed on the extracellular surface by several loops, whereas the PgaA barrel is fully open on the periplasmic side, with a dimension of ∼25 Å × 35 Å in diameter. Given that the linear partially de-N-acetylated dPNAG polymer is ∼10 Å in width (23), the lumen of PgaA barrel is large enough to allow the passage of dPNAG polymer through the pore. However, it is unclear how the binding of dPNAG to PgaA elicits the dislocation of the extracellular loops that allows dPNAG to enter into the extracellular space. One possibility could be that the electronegative attributes of the PgaA extracellular surface and/or the lipopolysaccharides present in the outer leaflet of the outer membrane could facilitate this process (22).
We also provided evidence that the TPR motifs of PgaA are important for the secretion of dPNAG. In agreement with this, the TPR-containing protein AlgK is also required for exopolysaccharide alginate secretion (28). However, the β-barrel of PgaA alone or a fusion protein composed of the AlgK and the PgaA β-barrel was not functional, suggesting that both the TPR domain and the β-barrel of PgaA are necessary and specific for dPNAG secretion. The observed direct interaction between the TPR domain of PgaA and PgaB indicates that this interaction may position the PgaB-bound dPNAG in proximity to the PgaA secretion pore as a prerequisite for dPNAG interaction. However, it remains to be seen if the de-N-acetylated PNAG polymer directly binds to the PgaA β-barrel and/or the TPR domain of PgaA and how they exactly interact in the process of translocation. Further biochemical characterization of these interactions and structural studies of PgaA in complex with dPNAG and PgaB should help to clarify these issues.
In summary, our crystal structural studies and structure-based functional analysis provide important clues to explain how dPNAG is translocated across the bacterial outer membrane. Improving our understanding of the mechanism underlying dPNAG synthesis and translocation is of great biomedical interest given that it is critical for biofilm production by several important human pathogens (47, 48).
Author Contributions
Y. H. and T. R. conceived the study. Y. W. performed the protein purification, crystallization, structural determination, and biochemistry experiments. A. A. P. performed the biofilm assay and dot-blotting. D. N., H. Z., X. C. and X. L. collected x-ray diffraction data. Y. H., T. R., and Y. W. wrote the manuscript.
Acknowledgments
We thank Yan Zhao and staff members of beamline BL1A of Japanese KEK synchrotron and Shanghai Synchrotron Radiation Facility (China) synchrotron facilities for assistance in collecting diffraction data.
This work was supported by National Institutes of Health Grants GM059969 and GM066794 (to T. R.). This work was also supported by National Natural Science Foundation of China Grants 31470743 and 31170698 (to Y. H.), Ministry of Science and Technology (China) “973” Project Grants 2013CB910603 and 2012CB917302 (to Y. H.), Chinese Academy of Sciences Grant XDB08020302 (to Y. H.), and University of Florida CHRIS Project FLA-MCS-004949 (to T. R.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
The atomic coordinates and structure factors (code 4Y25) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- PNAG
- poly-β-1,6-N-acetyl-d-glucosamine
- dPNAG
- de-N-acetylated poly-β-1,6-N-acetyl-d-glucosamine
- TPR
- tetratricopeptide repeat
- CsrA
- carbon storage regulator A
- Se-Met
- selenomethionine.
References
- 1. López D., Vlamakis H., and Kolter R. (2010) Biofilms. Cold Spring Harb. Perspect. Biol. 2, a000398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Branda S. S., Vik S., Friedman L., and Kolter R. (2005) Biofilms: the matrix revisited. Trends Microbiol. 13, 20–26 [DOI] [PubMed] [Google Scholar]
- 3. Flemming H. C., and Wingender J. (2010) The biofilm matrix. Nat. Rev. Microbiol, 8, 623–633 [DOI] [PubMed] [Google Scholar]
- 4. Mah T. F., and O'Toole G. A. (2001) Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol. 9, 34–39 [DOI] [PubMed] [Google Scholar]
- 5. Donlan R. M., and Costerton J. W. (2002) Biofilms: survival mechanisms of clinically relevant microorganisms. Clin. Microbiol. Rev. 15, 167–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Messiaen A. S., Nelis H., and Coenye T. (2014) Investigating the role of matrix components in protection of Burkholderia cepacia complex biofilms against tobramycin. J. Cyst. Fibros. 13, 56–62 [DOI] [PubMed] [Google Scholar]
- 7. Bentancor L. V., O'Malley J. M., Bozkurt-Guzel C., Pier G. B., and Maira-Litrán T. (2012) Poly-N-acetyl-β-(1–6)-glucosamine is a target for protective immunity against Acinetobacter baumannii infections. Infect. Immun. 80, 651–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. O'Toole G., Kaplan H. B., and Kolter R. (2000) Biofilm formation as microbial development. Annu. Rev. Microbiol. 54, 49–79 [DOI] [PubMed] [Google Scholar]
- 9. Ryder C., Byrd M., and Wozniak D. J. (2007) Role of polysaccharides in Pseudomonas aeruginosa biofilm development. Curr. Opin. Microbiol 10, 644–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Danese P. N., Pratt L. A., and Kolter R. (2000) Exopolysaccharide production is required for development of Escherichia coli K-12 biofilm architecture. J. Bacteriol. 182, 3593–3596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cerca N., Maira-Litrán T., Jefferson K. K., Grout M., Goldmann D. A., and Pier G. B. (2007) Protection against Escherichia coli infection by antibody to the Staphylococcus aureus poly-N-acetylglucosamine surface polysaccharide. Proc. Natl. Acad. Sci. U.S.A. 104, 7528–7533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Irie Y., Borlee B. R., O'Connor J. R., Hill P. J., Harwood C. S., Wozniak D. J., and Parsek M. R. (2012) Self-produced exopolysaccharide is a signal that stimulates biofilm formation in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U.S.A. 109, 20632–20636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wan Z., Brown P. J., Elliott E. N., and Brun Y. V. (2013) The adhesive and cohesive properties of a bacterial polysaccharide adhesin are modulated by a deacetylase. Mol. Microbiol. 88, 486–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Itoh Y., Rice J. D., Goller C., Pannuri A., Taylor J., Meisner J., Beveridge T. J., Preston J. F 3rd, and Romeo T. (2008) Roles of pgaABCD genes in synthesis, modification, and export of the Escherichia coli biofilm adhesin poly-β-1,6-N-acetyl-d-glucosamine. J. Bacteriol. 190, 3670–3680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Itoh Y., Wang X., Hinnebusch B. J., Preston J. F. 3rd, Romeo T. (2005) Depolymerization of β-1,6-N-acetyl-d-glucosamine disrupts the integrity of diverse bacterial biofilms. J. Bacteriol. 187, 382–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang X., Dubey A. K., Suzuki K., Baker C. S., Babitzke P., and Romeo T. (2005) CsrA post-transcriptionally represses pgaABCD, responsible for synthesis of a biofilm polysaccharide adhesin of Escherichia coli. Mol. Microbiol. 56, 1648–1663 [DOI] [PubMed] [Google Scholar]
- 17. Wang X., Preston J. F. 3rd, and Romeo T. (2004) The pgaABCD locus of Escherichia coli promotes the synthesis of a polysaccharide adhesin required for biofilm formation. J. Bacteriol. 186, 2724–2734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yakandawala N., Gawande P. V., LoVetri K., Cardona S. T., Romeo T., Nitz M., and Madhyastha S. (2011) Characterization of the poly-β-1,6-N-acetylglucosamine polysaccharide component of Burkholderia biofilms. Appl. Environ. Microbiol. 77, 8303–8309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Choi A. H., Slamti L., Avci F. Y., Pier G. B., and Maira-Litrán T. (2009) The pgaABCD locus of Acinetobacter baumannii encodes the production of poly-β-1–6-N-acetylglucosamine, which is critical for biofilm formation. J. Bacteriol. 191, 5953–5963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Venketaraman V., Lin A. K, Le A., Kachlany S. C., Connell N. D., and Kaplan J. B. (2008) Both leukotoxin and poly-N-acetylglucosamine surface polysaccharide protect Aggregatibacter actinomycetemcomitans cells from macrophage killing. Microbial pathogenesis 45, 173–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chen K. M., Chiang M. K., Wang M., Ho H. C., Lu M. C., and Lai Y. C. (2014) The role of pgaC in Klebsiella pneumoniae virulence and biofilm formation. Microb. Pathog. 77, 89–99 [DOI] [PubMed] [Google Scholar]
- 22. Amini S., Goodarzi H., and Tavazoie S. (2009) Genetic dissection of an exogenously induced biofilm in laboratory and clinical isolates of E. coli. PLoS Pathog. 5, e1000432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Little D. J., Li G., Ing C., DiFrancesco B. R., Bamford N. C., Robinson H., Nitz M., Pomès R., and Howell P. L. (2014) Modification and periplasmic translocation of the biofilm exopolysaccharide poly-β-1,6-N-acetyl-d-glucosamine. Proc. Natl. Acad. Sci. U.S.A. 111, 11013–11018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Little D. J., Poloczek J., Whitney J. C., Robinson H., Nitz M., and Howell P. L. (2012) The structure- and metal-dependent activity of Escherichia coli PgaB provides insight into the partial de-N-acetylation of poly-β-1,6-N-acetyl-d-glucosamine. J. Biol. Chem. 287, 31126–31137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Steiner S., Lori C., Boehm A., and Jenal U. (2013) Allosteric activation of exopolysaccharide synthesis through cyclic di-GMP-stimulated protein-protein interaction. EMBO J. 32, 354–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pannuri A., Yakhnin H., Vakulskas C. A., Edwards A. N., Babitzke P., and Romeo T. (2012) Translational repression of NhaR, a novel pathway for multi-tier regulation of biofilm circuitry by CsrA. J. Bacteriol. 194, 79–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Figueroa-Bossi N., Schwartz A., Guillemardet B., D'Heygère F., Bossi L., and Boudvillain M. (2014) RNA remodeling by bacterial global regulator CsrA promotes Rho-dependent transcription termination. Genes Dev. 28, 1239–1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Keiski C. L., Harwich M., Jain S., Neculai A. M., Yip P., Robinson H., Whitney J. C., Riley L., Burrows L. L., Ohman D. E., and Howell P. L. (2010) AlgK is a TPR-containing protein and the periplasmic component of a novel exopolysaccharide secretin. Structure 18, 265–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Whitney J. C., Hay I. D., Li C., Eckford P. D., Robinson H., Amaya M. F., Wood L. F., Ohman D. E., Bear C. E., Rehm B. H., and Howell P. L. (2011) Structural basis for alginate secretion across the bacterial outer membrane. Proc. Natl. Acad. Sci. U.S.A. 108, 13083–13088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tan J., Rouse S. L., Li D., Pye V. E., Vogeley L., Brinth A. R., El Arnaout T., Whitney J. C., Howell P. L., Sansom M. S., and Caffrey M. (2014) A conformational landscape for alginate secretion across the outer membrane of Pseudomonas aeruginosa. Acta Crystallogr. D Biol. Crystallogr. 70, 2054–2068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Otwinowski Z., and Minor W. (1997) Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 32. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., and Zwart P. H. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Emsley P., and Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 34. Collaborative Computational Project, Number 4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 35. DeLano W. L. (2004) Use of PyMOL as a communications tool for molecular science. Abstr. Pap. Am. Chem. S 228, U313–U314 [Google Scholar]
- 36. Scheich C., Kümmel D., Soumailakakis D., Heinemann U., and Büssow K. (2007) Vectors for co-expression of an unrestricted number of proteins. Nucleic Acids Res. 35, e43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Holm L., and Sander C. (1995) a network tool for protein structure comparison. Trends Biochem. Sci. 20, 478–480 [DOI] [PubMed] [Google Scholar]
- 38. Gruss F., Zähringer F., Jakob R. P., Burmann B. M., Hiller S., and Maier T. (2013) The structural basis of autotransporter translocation by TamA. Nat. Struct. Mol. Biol. 20, 1318–1320 [DOI] [PubMed] [Google Scholar]
- 39. Clantin B., Delattre A. S., Rucktooa P., Saint N., Méli A. C., Locht C., Jacob-Dubuisson F., and Villeret V. (2007) Structure of the membrane protein FhaC: a member of the Omp85-TpsB transporter superfamily. Science 317, 957–961 [DOI] [PubMed] [Google Scholar]
- 40. Subbarao G. V., and van den Berg B. (2006) Crystal structure of the monomeric porin OmpG. J. Mol. Biol. 360, 750–759 [DOI] [PubMed] [Google Scholar]
- 41. Dong C., Beis K., Nesper J., Brunkan-Lamontagne A. L., Clarke B. R., Whitfield C., and Naismith J. H. (2006)Wza the translocon for E. coli capsular polysaccharides defines a new class of membrane protein. Nature 444, 226–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Russo T. A., Wenderoth S., Carlino U. B., Merrick J. M., and Lesse A. J. (1998) Identification, genomic organization, and analysis of the group III capsular polysaccharide genes kpsD, kpsM, kpsT, and kpsE from an extraintestinal isolate of Escherichia coli (CP9, O4/K54/H5). J. Bacteriol. 180, 338–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wunder D. E., Aaronson W., Hayes S. F., Bliss J. M., and Silver R. P. (1994) Nucleotide sequence and mutational analysis of the gene encoding KpsD, a periplasmic protein involved in transport of polysialic acid in Escherichia coli K1. J. Bacteriol. 176, 4025–4033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Im W., and Roux B. (2002) Ion permeation and selectivity of OmpF porin: a theoretical study based on molecular dynamics, Brownian dynamics, and continuum electrodiffusion theory. J. Mol. Biol. 322, 851–869 [DOI] [PubMed] [Google Scholar]
- 45. Kutzner C., Grubmüller H., de Groot B. L., and Zachariae U. (2011) Computational electrophysiology: the molecular dynamics of ion channel permeation and selectivity in atomistic detail. Biophys. J. 101, 809–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tanabe M., Nimigean C. M., and Iverson T. M. (2010) Structural basis for solute transport, nucleotide regulation, and immunological recognition of Neisseria meningitidis PorB. Proc. Natl. Acad. Sci. U.S.A. 107, 6811–6816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lu X., Skurnik D., Pozzi C., Roux D., Cywes-Bentley C., Ritchie J. M, Munera D., Gening M. L., Tsvetkov Y. E., Nifantiev N. E., Waldor M. K., and Pier G. B. (2014) A poly-N-acetylglucosamine-Shiga toxin broad-spectrum conjugate vaccine for Shiga toxin-producing Escherichia coli. mBio 5, e00974–00914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Gening M. L., Maira-Litrán T., Kropec A., Skurnik D., Grout M., Tsvetkov Y. E., Nifantiev N. E., and Pier G. B. (2010) Synthetic β-(1→6)-linked N-acetylated and nonacetylated oligoglucosamines used to produce conjugate vaccines for bacterial pathogens. Infect. Immun. 78, 764–772 [DOI] [PMC free article] [PubMed] [Google Scholar]