Abstract
Endothelial cell tumors are the most common soft tissue tumors in infants. Tumor-forming endothelial (EOMA) cells are able to escape cell death fate despite excessive nuclear oxidant burden. Our previous work recognized perinuclear Nox-4 as a key contributor to EOMA growth. The objective of this work was to characterize the mechanisms by which EOMA cells evade oxidant toxicity and thrive. In EOMA cells, compared with in the cytosol, the nuclear GSSG/GSH ratio was 5-fold higher. Compared to the ratio observed in healthy murine aortic endothelial (MAE) cells, GSSG/GSH was over twice as high in EOMA cells. Multidrug resistance-associated protein-1 (MRP-1), an active GSSG efflux mechanism, showed 2-fold increased activity in EOMA compared with MAE cells. Hyperactive YB-1 and Ape/Ref-1 were responsible for high MRP-1 expression in EOMA. Proximity ligand assay demonstrated MRP-1 and YB-1 binding. Such binding enabled the nuclear targeting of MRP-1 in EOMA in a leptomycin-B-sensitive manner. MRP-1 inhibition as well as knockdown trapped nuclear GSSG, causing cell death of EOMA. Disulfide loading of cells by inhibition of GSSG reductase (bischoloronitrosourea) or thioredoxin reductase (auranofin) was effective in causing EOMA death as well. In sum, EOMA cells survive a heavy oxidant burden by rapid efflux of GSSG, which is lethal if trapped within the cell. A hyperactive MRP-1 system for GSSG efflux acts as a critical survival factor for these cells, making it a potential target for EOMA therapeutics.
Keywords: endothelial cell, multidrug transporter, NADPH oxidase, thioredoxin, tumor, Ape/Ref-1, Nox-4, YB-1
Introduction
Endothelial cell tumors are the most common soft tissue tumor in infants, affecting 5–10% of all infants (1–3). These tumors result in residual deformity in 50% of affected children, threaten normal development in 10%, and threaten the life of 1% of affected children (4). Previous work using a validated in vivo murine endothelial (EOMA) cell tumor model has established that expression of oxidant-inducible monocyte chemoattractant protein-1 (MCP-1)3 is required for endothelial cell tumor formation and that Nox-4 is the primary source of oxidants inducing MCP-1 expression in EOMA cells (5–7). Elevated levels of Nox-4 and increased levels of oxidized nuclear DNA have been reported in human endothelial cell tumors (8). Elevated levels of Nox-4 have also been observed in melanoma, breast, and non-small cell lung cancer, and Nox-4 functions as a key mediator of epithelial to mesenchymal transition events. Thus, elevated levels of Nox-4 and downstream cellular oxidant loading may be viewed as a common etiologic finding between endothelial and other types of tumors.
A striking feature of the endothelial cell tumors is that Nox-4 is perinuclear and delivers H2O2 directly into the nucleus (7). In EOMA, Nox-4 is constitutively active and present in levels severalfold higher compared with non-tumor-forming endothelial cells (7). Given that the nuclear redox microenvironment must be maintained in an appropriate reduced state in order to defend cell survival and growth (8), it is intuitively clear that EOMA cells must depend on a pivotal mechanism to successfully handle the threat of nuclear oxidative stress. Given the high abundance of cellular GSH, rapid accumulation of cellular GSSG is one of the early responses to oxidant insult (9). Such excessive GSSG may be recycled to GSH in the presence of appropriate levels of NADPH (10–12). However, in EOMA compared with MAE, Nox-4 is substantially elevated, which may consume NADPH (7). Our previous work has shown that high intracellular GSSG is lethal in neurons (13). Thus, these cells must rely on rapid clearance of GSSG to ensure their survival in an oxidant-rich scenario (13). The objective of this work is to elucidate the mechanisms by which EOMA cells defend survival under conditions of high Nox-4-dependent oxidants, which are also necessary for tumor growth (7). We tested the hypothesis that EOMA cells rely on an extraordinary mechanism to efflux cellular GSSG, which acts as a critical survival factor. Inhibition of such a survival factor would make these cancer cells fall prey to excessive oxidants of their own making, which they otherwise use to support extraordinary growth.
Experimental Procedures
Materials
The following materials were obtained from the sources indicated. Hexadimethrine bromide, dimethyl sulfoxide, oxidized and reduced glutathione, l-buthionine-sulfoximine, and phenylarsine oxide were from Sigma. For cell culture, Dulbecco's modified Eagle's medium (DMEM), fetal calf serum (FCS), penicillin, and streptomycin were purchased from Invitrogen. Culture dishes were obtained from Nunc (Rochester, NY). EOMA cells (ATCC, Manassas, VA) and murine aortic endothelial (MAE) cells were a gift from Charles G. Orosz (Ohio State University).
Methods
Cell Culture
Murine endothelial (EOMA) cells were maintained under the same conditions as described previously (5). In brief, EOMA cells were maintained in DMEM supplemented with 10% FCS and 1% penicillin/streptomycin (complete medium) and incubated at 37 °C and 5% CO2.
Preparation of Nuclear Extracts
EOMA/MAE cells were seeded in 12-well plates at 1 × 105 cells/well. Vehicle and MK-571 treatments were carried out after 24 h of seeding. Nuclear and cytosolic extracts were isolated using a nuclear extraction kit (Active Motif Corp., Carlsbad, CA) per the manufacturer's instructions. Protein concentrations were measured by bicinchoninic acid protein assay reagent (BCA; Pierce), and the extracts were stored at −80 °C until analyzed. Purity of the nuclear and cytoplasmic fractions was confirmed by measuring nuclear and cytoplasmic specific protein markers, such as p84 (nuclear) and β-actin (cytoplasmic) with the immunoblotting method. In brief, following extraction, nuclear and cytosolic extracts (15 μg) were loaded in NuPAGETM NovexTM 4–12% BisTris protein gels (catalog no. NP0321BOX, lot 1508077, Thermo Fisher Scientific, Waltham, MA). After running, the gel was transferred to PVDF membrane (Amersham Biosciences Hybond P 0.45 PVDF) using NuPAGE® transfer buffer (20×). Transferred proteins in the nuclear fraction showed abundant p84 compared with cytosolic fraction, and cytosolic extract tested positive for the presence of cytosolic β-actin where the presence of nuclear p84 was markedly minimized (14, 15) .
Reduced (GSH) and Oxidized (GSSG) Glutathione Assay
GSH and GSSG were detected from EOMA/MAE cell lysates using an HPLC coulometric electrode array detector (CoulArray Detector, model 5600A, with 8 channels; ESA Inc., Chelmsford, MA) as described previously (13, 16). In brief, cells were seeded in 12-well plates at 1 × 105 cells/well for 72 h. Cells were harvested on day 3, combining three wells for each sample. For preparing whole cell lysate, cells were washed with cold PBS, scraped, and centrifuged (500 × g) for 5 min at 4 °C. Pellets were then treated with 5% (w/v) m-phosphoric acid (final concentration) and immediately snap-frozen and stored in liquid nitrogen until processing for HPLC (17). On the day of the HPLC assay, samples were quickly thawed on ice and centrifuged (12,000 × g, 5 min) at 4 °C for protein precipitation. Supernatants were collected and filtered through a 0.2-μm filter. The filtrate was immediately injected into HPLC. GSH and GSSH were separated using a C18 column and the following mobile phase: 50 mm sodium dihydrogen phosphate, 0.5 mm octanosulfonic acid, and 3% acetonitrile at pH 2.7 (17). Precipitated proteins were dissolved in 0.1 n NaOH, and protein was determined by the spectrophotometric quantitation method using BCA reagent (Pierce) (18).
Nuclear and cytosolic fractions were isolated according to the Active Motif extraction kit protocol (Active Motif, Carlsbad, CA). For GSH/GSSG measurements, cytosolic and nuclear fractions were treated with 5% (w/v) m-phosphoric acid and immediately snap-frozen and stored in liquid nitrogen until processing for HPLC (19, 20). Sample preparation, composition of the mobile phase, and specification of the column and electrochemical cell potentials used have been reported previously (17, 21). This system uses multiple channels set at different redox potentials. Data were collected using channels set at +500, +600, +700, +800, and +900 mV.
Flow Cytometric Determination of Thiols Using Bimane Probe
Chloro or bromo derivatives of bimane are nonfluorescent in their native forms but emit strong fluorescence when reacted with thiols (22). We used a monobromobimane (MBB) flow cytometric method to measure total cellular thiol pools (23–25) and glutathione (26) as described previously. Monochlorobimane (MCB) (27, 28), along with BSO (150 μm), was used to measure intracellular GSH, and phenylarsineoxide (PAO; 20 μm) was used to measure vicinal dithiol as described previously (29, 30).
To determine whether the oxidative status of tumor-forming EOMA cells was affected by elevated levels of Nox-4-derived oxidants, flow cytometry experiments were performed to compare the composition of the thiol pools in tumor-forming EOMA cells compared with non-tumor-forming MAE cells. EOMA/MAE cells were seeded in 12-well plates at 1 × 105 cells/well. After 18 h of seeding, cells were treated in three different groups: (a) control, untreated to measure total thiol content; (b) BSO treatment for 18 h to deplete GSH; and (c) 20 μm PAO pretreatment for 10 min prior to the addition of bimane (40 μm) to block vicinal dithiols. Bimane-loaded cells were excited using a 20-milliwatt powered UV line of an argon ion laser set at 350 nm in a BD FACS Aria flow cytometer. A morphometrically homogeneous cell population, typically representing <90% of the total population, was gated. Data were collected from 10,000 cells at a flow rate of 200–250 cells/s.
Determination of Cell Viability
The viability of cells in culture was assessed by measuring leakage of lactate dehydrogenase (LDH) from cells into medium using an in vitro toxicology assay kit from Sigma-Aldrich. This protocol has been described in detail in a previous report (13, 31). The percentage of cell viability was determined by cellular LDH content/total LDH content (LDH content in culture medium + detached cellular LDH content + attached cellular LDH content) × 100 (32, 33).
Calcein Clearance Assay
The calcein clearance assay was used to measure MRP-1 activity. After the indicated time point of treatment in EOMA and/or MAE cells, calcein-AM (25 nmol/liter) was loaded to the cells for 15 min at 37 °C. Cells were washed with PBS, collected, and analyzed using the AccuriTM C6 flow cytometer (Accuri Cytometers, Ann Arbor, MI). MRP-1 activity was measured on the basis of intracellular calcein retention (16).
Western Blotting
Immunoblotting was performed using EOMA/MAE cell lysates, and the protein concentration was determined using a BCA protein assay. Samples (15–30 μg of protein/lane) were separated using 4–12% SDS-polyacrylamide gel electrophoresis and probed with mouse monoclonal anti-MRP-1 antibody (1:750 dilution, catalog no. ab24108, lot GR203008-5, Abcam, Cambridge, MA) (34), rabbit polyclonal anti-YB-1 antibody (1:1000 dilution, catalog no. ab12148, lot GR162790-3, Abcam) (35), rabbit polyclonal anti-Apex-1 antibody (1:500 dilution, catalog no. ab 137708, lot GR109895-7, Abcam) (8, 36), anti-mouse β-actin (1:10,000 dilution, catalog no. A5441, lot 055K4854, Sigma) (8, 37), anti-mouse monoclonal p84 (1:5000 dilution, catalog no. GTX70220, lot 40953, GeneTex, Irvine, CA) (38), and normal rabbit IgG (catalog no. sc-2027, lot E1512, Santa Cruz Biotechnology, Inc., Dallas, TX) (39). Bands were visualized by using horseradish peroxidase-conjugated donkey anti-rabbit-IgG (1:2000, catalog no. NA934V, lot 9583369, Amersham Biosciences) and anti-mouse-IgG (1:2000, catalog no. NA931V, lot 6652622, Amersham Biosciences) (8, 37) and the enhanced chemiluminescence assay (Amersham Biosciences) according to the manufacturer's instructions. Pixel densitometry for individual bands was done using ImageJ software.
Measurement of Protein Carbonyl Levels and Lipid Peroxidation
Protein-bound carbonyls were measured via a protein carbonyl assay kit (catalog no. 1005020 Cayman Chemical, Ann Arbor, MI). The samples were isolated per the manufacturer's protocol. The utilized method was based on the covalent reaction of the carbonylated protein side chain with 2,4-dinitrophenylhydrazine and detection of the produced protein hydrazone at an absorbance of 280 nm. The results were calculated using the extinction coefficient of 11 mm−1 cm−1 for aliphatic hydrazones and were expressed as nmol/mg of protein. Lipid peroxidation was measured using the thiobarbituric acid-reactive substances assay kit (catalog no. 10009055, Cayman Chemical). Sample absorbance was measured at 532 nm, and concentration was determined based on the standard provided in the kit.
Measurement of Glutathione Reductase, Thioredoxin, and Thioredoxin Reductase Activity
Glutathione reductase activity was measured by using the glutathione reductase assay kit (catalog no. 703202, Cayman Chemical). The absorbance was measured at 340 nm, and the concentration was determined according to the manufacturer's protocol. Thioredoxin/thioredoxin reductase activity was measured using the thioredoxin/thioredoxin reductase mammalian assay kit (catalog no. 11526, Cayman Chemical).
Blue Native PAGE to Measure Oxidized Proteins
For protein extraction, EOMA/MAE cells were grown in T-75 flasks for 36 h. After 36 h, cells were washed twice with PBS, trypsinized, and harvested. The pelleted cells were lysed in the appropriate volume of lysis buffer (Cell Signaling, Danvers, MA). Crude extracts were centrifuged at 12,000 × g for 10 min to remove debris, and then protein in the supernatant was quantified using the BCA protein assay kit (Pierce). 50 μg of protein was mixed in 1× native PAGE sample buffer (Life Technologies) and loaded on a 4–16% Novex native PAGE BisTris gel system (Life Technologies). After electrophoretic separation, proteins were transferred onto PVDF membranes and probed with rabbit polyclonal anti-Trx-1 antibody (1:1000 dilution, Santa Cruz Biotechnology). Isotype-matched, horseradish peroxidase-conjugated secondary antibodies (Bio-Rad) were used, followed by detection by chemiluminescence (SuperSignal Pico, Pierce).
NADPH/NADP Ratio
NADPH/NADP ratio in EOMA and MAE cells was measured using the Sigma-Aldrich NADP/NADPH quantification kit according to the protocol of the manufacturer (MAK038, Sigma). In brief, EOMA/MAE cells were seeded in 12-well plates at 1 × 105 cells/well. After 36 h of seeding, cells were washed twice with cold PBS, trypsinized, and harvested using extraction buffer as per the protocol. Equal amounts of protein (150 μg) for each sample were loaded in 96-well plates along with kit standards. Sample absorbance was measured at 450 nm, and the NADPH/NADP ratio was determined according to kit protocol (40).
Fluorescence Detection of Cellular Glutathione (GSH)
Cellular GSH levels were analyzed using 5-chloromethylfluorescein diacetate (Molecular Probes, Inc., Eugene, OR). It is a thiol-reactive cell-permeable dye that is incorporated into glutathione by glutathione S-transferase. This fluorescent marker is retained within cells for >24 h and passed to daughter cells. EOMA/MAE cells were seeded at 0.05 × 105 cells/well, in 24-well plates containing sterile coverslips. After the indicated treatment, 24-well plates containing coverslips of EOMA/MAE cells were incubated with prewarmed (37 °C) chloromethylfluorescein diacetate-containing medium (10 μm). After 30 min at 37 °C incubation, cells were washed with PBS, and real-time images were collected using a Zeiss Axiovert 200M microscope (41).
Transfection Experiments and Analysis of Gene Expression
The cells were seeded in a 12-well plate at a density of 1 × 105 cells/well for 24 h before treatment. Delivery of siRNAs was achieved using DharmaFECTTM 1 transfection reagent (Thermo Fisher Scientific). All siRNA SMARTpool reagents were from Dharmacon RNA Technologies (Lafayette, CO). For controls, siControl non-targeting siRNA pool (mixture of four siRNAs, designed to have ≥4 mismatches with the corresponding gene) was used (42). DharmaFECT 1 transfection reagent (Dharmacon RNA Technologies) was used to transfect cells with the mentioned siRNAs (Dharmacon RNA Technologies) for 72 h per the manufacturer's instructions. Unless specified, the cells were lysed after 72 h of transfection, and RNA/protein were collected for the gene expression study (13, 31, 43, 44). For detection of mRNA, total RNA from cells was extracted using the miRVana miRNA isolation kit according to the manufacturer's protocol (Ambion, Life Technologies, Inc.). For mRNA expression studies, cDNA synthesis was achieved by a SuperScriptTM III first strand synthesis system (Applied Biosystems, Life Technologies, Inc.). The abundance of mRNA for genes of interest was quantified by using a real-time polymerase chain reaction with double-stranded DNA binding dye SYBR Green-I. The following primer sets (Invitrogen) were used: m_MRP-1_F, 5′-GGT CCT GTT TCC CCC TCT ACT TCT T-3′; m_MRP-1_R, 5′-GCA GTG TTG GGC TGA CCA GTA A-3′; m_GAPDH_F, 5′-ATG ACC ACA GTC CAT GCC ATC ACT-3′; m_GAPDH_R, 5′-TGT TGA AGT CGC AGG AGA CAA CCT-3′.
Capillary Electrophoresis Immunoassay
Capillary electrophoresis immunoassay or simple Western analyses were performed using the SimonTM machine (ProteinSimple Santa Clara, CA) according to the manufacturer's protocol. Simple Western analysis is carried out at room temperature, and instrument default settings were used as discussed earlier (37). MRP-1 (1:50 dilution), p84 (1:50 dilution), and β-actin (1:100 dilution) primary antibody were diluted with antibody diluent (ProteinSimple). The digital image was analyzed with Compass software (ProteinSimple), and the quantified data of the detected protein were reported as molecular weight.
Immunoprecipitation and Protein Expression
Cytosolic and nuclear fractions of EOMA cell lysate were collected using a nuclear extraction kit as described under “Preparation of Nuclear Extracts.” Whole cell lysate was collected using lysis buffer (150 mm KCl, 25 mm Tris-HCl, 5 mm EDTA, 0.5% IgePal, 1 mm PMSF, 1× protease inhibitor) as described previously (8, 37). Protein concentration was determined using the Coomassie Plus Assay (Thermo Scientific). 5–10 μg of anti-rabbit YB-1 and control IgG antibody were added to the different cell lysates (250 μg) and incubated overnight in a rotisserie shaker at 4 °C. After 18 h of incubation, cell lysates were centrifuged at 500 × g and 4 °C for 10 min. TrueBlot anti-rabbit Ig immunoprecipitation beads (eBiosciences Inc., San Diego, CA) were prewashed with lysis buffer for 40 min. Cell lysates (500 μg) were incubated with prewashed beads for 3 h at 4 °C on a rotisserie shaker (Barnstead/Thermolyne, Dubuque, IA). The beads were then washed three times with ice-cold lysis buffer (centrifugation at 2500 × g at 4 °C for 5 min). For Western blotting analysis, samples were subjected to SDS-PAGE after reduction with 1 m DTT as described previously (8, 37).
In Situ Proximity Ligation Assay (PLA)
The Duolink reagent kit (Olink Biosciences, Uppsala, Sweden) was used for these studies. EOMA cells were seeded at 5 × 103/well in a 24-well plate. After 24 h, cells were fixed in 1% paraformaldehyde for 15 min and rinsed twice with Wash Buffer A for 5 min. Cells were incubated with a blocking solution at 37 °C for 30 min and then rinsed with Wash Buffer A for 5 min twice. The procedures for administrating the primary antibodies (MRP-1 versus YB-1), PLA probes, hybridization, ligation, amplification, detection, and mounting followed the manufacturer's protocol and were as described (45). The images were captured by confocal microscopy, and fluorescent intensity was quantified using FV10-ASW version 3.0 software (Olympus, Tokyo, Japan).
Statistics
All experiments were conducted at least three times in duplicate unless otherwise specified in the figure legends. A two-sided two-sample t test was used to compare the differences between two groups. Residual data from every fitted model were investigated to ensure that the assumptions of the respective model were satisfied. Sensitivity analyses were also conducted using nonparametric procedures or with proper data transformation to ensure that the conclusions were robust to the selection of the statistical methods. A p value of <0.05 was considered statistically significant.
Results
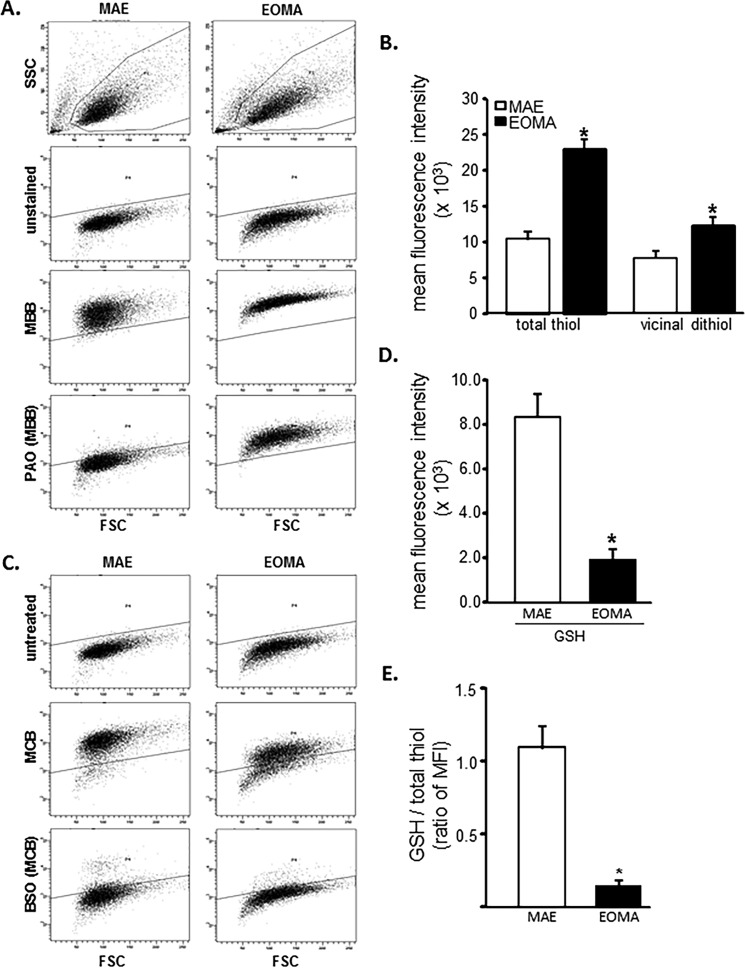
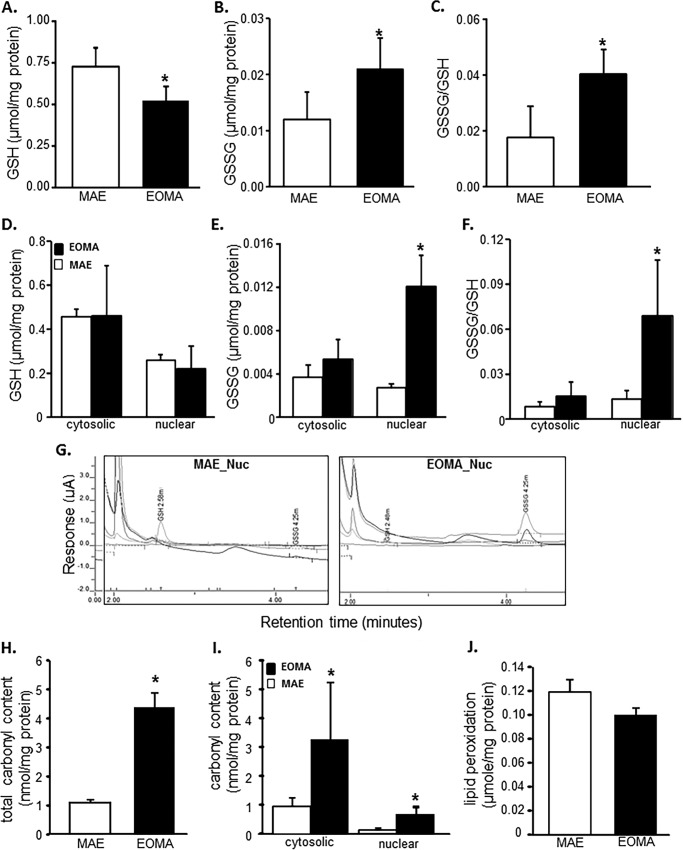
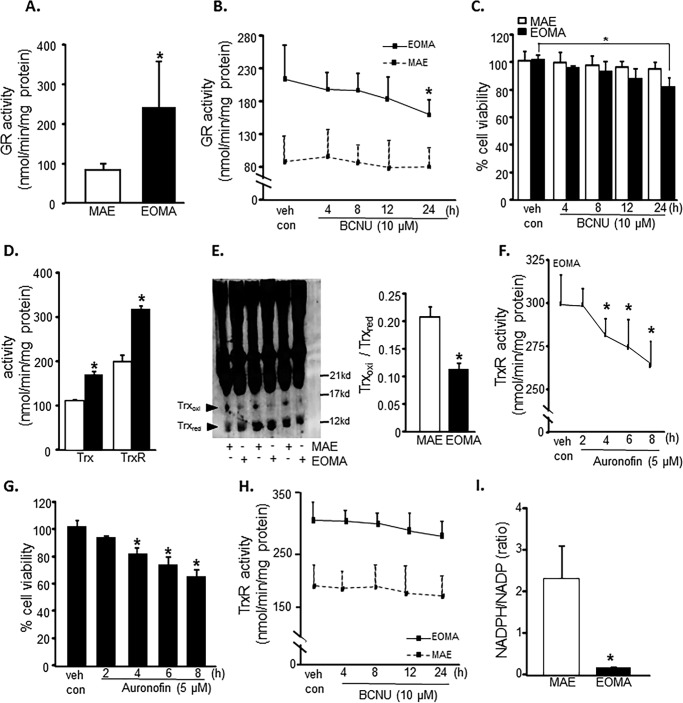
Flow cytometric analyses were used to look at the distribution of thiols across an entire population of cells to determine whether subsets of cells had different thiol levels. Results showed that, compared with healthy MAE cells, EOMA cells are uniformly high in cellular thiol content, as indicated by total thiol as well as vicinal dithiol status (Fig. 1, A and B). Interestingly, despite containing such a high cellular total thiol pool, GSH levels in EOMA are significantly lower than that in MAE (Fig. 1, C and D). Thus, compared with that in MAE, the proportion of cellular GSH compared with total thiol is strikingly low (Fig. 1E). Low cellular GSH is a hallmark of high oxidant burden (13, 46, 47). Consistently, EOMA cells showed low GSH (Fig. 2A), high GSSG (Fig. 2B), and high GSSG/GSH ratio (Fig. 2C). Interestingly, most of this imbalance was detected in the nuclear compartment demonstrating higher GSSG/GSH than that of MAE (Fig. 2, D–G). Study of other oxidative stress markers detected higher oxidative protein modification in EOMA than in MAE (Fig. 2H). Both cytosolic and nuclear compartments of EOMA demonstrated significantly elevated oxidative protein modification (Fig. 2I). Although this was the case, lipid peroxidation in EOMA was comparable with that in MAE, recognizing that thiols are a key target of oxidants generated in excess in EOMA cells (Fig. 2J). Survival of EOMA under conditions of elevated oxidant generation and high disulfides (Fig. 2) would require the presence of bolstered thiol reductase function. Indeed, compared with MAE, EOMA cells demonstrated elevated GSSG reductase and thioredoxin reductase defense systems (Fig. 3, A and D). Strengthened thioredoxin reductase was capable of keeping this critical thiol in a favorable redox state (Fig. 3E). As would be evident from the higher GSSG/GSH ratio of EOMA presented in Fig. 2, it is clear that high GSSG reductase in EOMA was ineffective in correcting the high GSSG/GSH noted in this cells. Although that was the case, it was also true that the GSSG reductase function in EOMA is critical for its survival. Pharmacologic inhibition of GSSG reductase (Fig. 3B) of EOMA resulted in loss of cell viability (Fig. 3C), which is not observed in MAE cells (Fig. 3, B and C). Likewise, inhibition of thioredoxin reductase also resulted in cell death (Fig. 3, E and F). Although there are reports that treatment with 1,3-bis(2-chloroethyl)-N-nitrosourea (BCNU) (inhibitor of GSSG reductase) inhibits thioredoxin reductase, in this case, there was no such observation (Fig. 3G). These observations highlight the critical importance of these thiol redox states in supporting cell survival. Of note, both of these thiol reductases rely on NADPH as their reducing equivalent. Interestingly, the NADPH content of EOMA was noted to be substantially low compared with that of MAE (Fig. 3H), as evidenced by NADPH/NADP values where the mean value for MAE (2.32 ± 0.48) was significantly higher than for EOMA (0.17 ± 0.02).
FIGURE 1.
EOMA cells have elevated total thiol pool and depleted GSH compared with MAE. A, flow cytometry was used on MAE and EOMA cells to detect the total thiol pool using (40 μm) MBB, and pretreatment with 20 μm PAO was used to identify the portion of total thiols consisting of vicinal dithiols. Threshold detection of autofluorescence from unstained cell population is indicated by the solid line. B, quantification of flow cytometry mean fluorescence intensity (MFI) demonstrates increased thiol pool in EOMA compared with MAE cells. Here total thiol intensity (MBB) was measured by subtracting the autofluorescence level (mean intensity of total MBB − intensity of unstained samples = total thiol signal), and vicinal thiol was measured by subtracting PAO-sensitive thiol (mean intensity of total MBB − PAO-sensitive MBB intensity = vicinal dithiol signal) from total thiol (MBB). C, flow cytometry detection of GSH pools in MAE and EOMA was done by using (40 μm) MCB and pretreating cells with 150 μm BSO for 18 h and then loading with MCB. BSO depletes GSH, so the residual MCB staining cells after pretreatment with BSO represent the low molecular weight thiols other than GSH. D, the bar graph represents flow cytometry quantification of mean fluorescence intensity (absolute values) generated in the presence of MCB. GSH is calculated as mean intensity of total MCB − BSO-sensitive MCB intensity = GSH signal, as described in our previous work (29). E, the fluorescence intensity ratio of total GSH pool to total thiol pool as described is significantly less in EOMA versus MAE cells. Results are expressed as mean ± S.D. (error bars); *, p < 0.05.
FIGURE 2.
Glutathione oxidation is elevated in EOMA cell nucleus. GSH and GSSG content in EOMA and MAE cells were measured by HPLC electrochemical detection. A–C, total GSH (A), total GSSG (B), and cellular GSSG/GSH level (C) in whole cell extract of both MAE and EOMA. D–F, nuclear and cytosolic fractions of GSH (D), GSSG (E), and GSSG/GSH level (F) in MAE and EOMA. C, chromatograms of HPLC CoulArray electrochemical detection of GSSG and GSH in EOMA and MAE cells. Measurement of protein oxidation was performed using the dinitrophenylhydrazine (DNPH) assay kit on whole cell (D) and nuclear and cytosolic fractions (E) with significantly increased levels of protein oxidation in EOMA cells for all measurements. Lipid peroxidation (malondialdehyde) was measured by a thiobarbituric acid-reactive substances assay on whole cell fractions (F) with no differences in total lipid peroxidation product in both EOMA and MAE cells. Results are expressed as mean ± S.D. (error bars); *, p < 0.05.
FIGURE 3.
Elevated thiol reductase activity is necessary for EOMA cell survival. A, glutathione reductase (GR) activity as measured by NADPH oxidation showed significantly elevated levels of glutathione reductase in EOMA versus MAE cells. B, the glutathione reductase activity in EOMA cells was significantly reduced after a 24-h treatment with 10 μm BCNU, whereas MAE did not have loss of GR activity during the 24-h treatment period. C, LDH leakage toxicity assay for cell viability correlated with glutathione reductase inhibition data. D, thioredoxin (Trx) and thioredoxin reductase (TrxR) activities were measured using a single assay system (Cayman Chemical), and both were increased in EOMA cells compared with MAE cells. E, immunoblotting of thioredoxin after native gel (native PAGE 4–16% BisTris gel) electrophoresis showing the oxidation status of thioredoxin protein in both EOMA and MAE cells. The formation of intermolecular disulfide bridges as a result of oxidation could impede migration of oxidized thioredoxin in MAE compared with EOMA cells. Both cell lines have single bands observed at 14 kDa. The protein densitometry calculation signifies that the ratio of oxidized to reduced thioredoxin is higher in MAE than EOMA. F, treatment of EOMA cells with 5 μm auranofin demonstrated a decrease in TrxR activity over time, which correlated with the LDH toxicity data (G) indicating that EOMA cells are highly susceptible to loss of TrxR activity. H, because BCNU treatment is reported to inhibit TrxR activity (84), we measured the TrxR activity level in the presence of BCNU (10 μm), which shows no significant inhibition at 24 h of treatment of both cell lines. I, NADPH/NADP measurements in normally cultured EOMA and MAE cells were obtained using a single assay system (Sigma). Results are expressed as mean ± S.D. (error bars); *, p < 0.05.
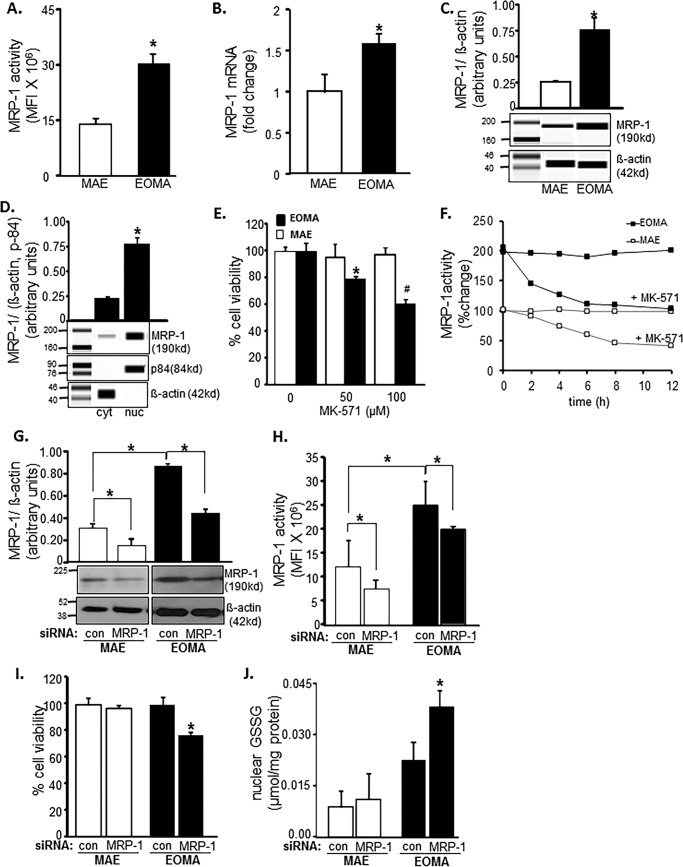
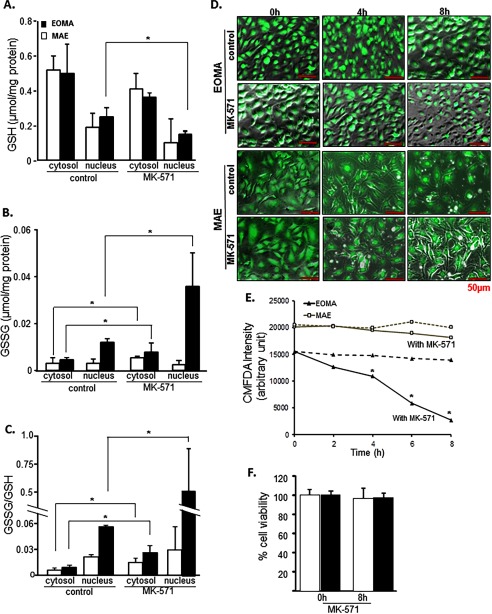
Survival of oxidant-rich EOMA would require that excessive cytotoxic GSSG generated in the cell be rapidly cleared. Compared with MAE, EOMA is supported by double MRP-1 activity (Fig. 4A). This observation was backed up by the presence of higher MRP-1 mRNA and protein in EOMA cells (Fig. 4, B and C). Interestingly, the excessive MRP-1 in EOMA was mostly present in the nucleus (Fig. 4D). Pharmacological inhibition of MRP-1 resulted in loss of EOMA cell viability, whereas MAE cells remained unaffected (Fig. 4E). This finding underscores the critical role of MRP-1 in supporting EOMA cell survival. Use of MK-571 as the pharmacological inhibitor of MRP-1 successfully inhibited MRP-1 function in both MAE and EOMA cells (Fig. 4F). However, such inhibition selectively killed EOMA cells (Fig. 4E). We also evaluated the knockdown approach by siRNA-dependent MRP-1 inhibition to show the decreased functional activity of MRP-1 (Fig. 4, F and G). Simultaneously, MRP-1 knockdown has a significant effect on cell survival (Fig. 4H) and also an effect on accumulation of GSSG inside the nucleus in EOMA, whereas there was no such effect observed in MAE cells (Fig. 4G).
FIGURE 4.
EOMA cell survival is MRP-1-dependent. A, MRP-1 activity was measured by calcein exclusion and was found to be significantly elevated in EOMA versus MAE cells. B and C, mRNA and capillary electrophoresis immunoblotting data show elevated MRP-1 expression in EOMA versus MAE cells. D, nuclear fractions have elevated MRP-1 protein expression compared with the cytosolic fraction of EOMA cells, as evidenced by capillary electrophoresis immunoblotting images. E, treatment of EOMA and MAE cells with MK-571, a pharmacologic inhibitor of MRP-1, shows a dose-dependent decrease in EOMA cell survival with no effect on MAE cells. F, treatment of both EOMA and MAE cells with 50 μm MK-571 showed progressive loss of activity over time as measured by calcein exclusion. G, transient transfection with MRP-1 siRNA showed decreased MRP-1 protein expression in EOMA and MAE cells by immunoblotting with an approximately 20% decrease of MRP-1 activity (H) in both cells 72 h after transfection. I, LDH assay shows that cell viability was significantly compromised in EOMA after MRP-1 knockdown but not in MAE. J, nuclear GSSG was significantly elevated in MRP-1 knockdown EOMA cells, but no such effect was observed in MAE. Results are expressed as mean ± S.D. (error bars); *, p < 0.05.
To test the influence of MRP-1 on GSSG levels in the cytosolic and nuclear compartments of the cell, EOMA cells were treated with MK-571. Inhibition of MRP-1 resulted in substantial accumulation of nuclear GSSG (Fig. 5, A–C). Inhibition of MK-571 resulted in a ∼10-fold increase in GSSG/GSH in EOMA cells (Fig. 5C). Such elevation of nuclear GSSG resulted in rapid depletion of reduced GSH in the EOMA cells but not in MAE (Fig. 5, D and E). These changes were recorded at a time when cell viability remained unaffected (Fig. 5F). Soon thereafter, however, cells started losing viability.
FIGURE 5.
MRP-1 regulates nuclear glutathione homeostasis. EOMA cells were treated with 50 μm MK-571 for 8 h. GSH (A), GSSG (B), and GSSG/GSH (C) were measured by HPLC electrochemical detection with a significant accumulation of GSSG in the cytosol and nucleus of MK-571-treated EOMA cells. D, 5-chloromethylfluorescein diacetate (CMFDA) detection of GSH at 4, 6, and 8 h showed progressive depletion of GSH in MK-571-treated cells compared with untreated EOMA cells with no significant changes observed in MAE cells, as quantified in E. F, the LDH assay shows that cell viability was not compromised for either cell type within the experimental time period. Results are expressed as mean ± S.D. (error bars); *, p < 0.05.
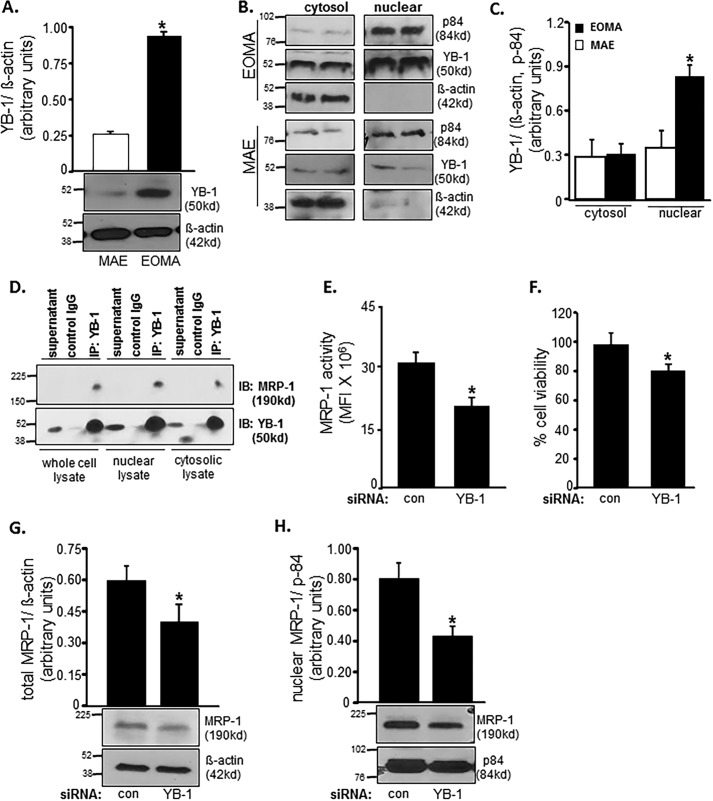
Because, in EOMA, MRP-1 function was noted to be critical in clearing nuclear GSSG and therefore in defending cell survival, our interest was directed toward understanding how MRP-1 is targeted to the nucleus in the absence of nuclear localization signal (NLS). Supported by NLS, YB-1 is a transcription factor that is known to translocate to the nucleus under conditions of genotoxic stress (48). In cooperation with Ape/Ref-1, YB-1 drives the expression of mdr-1, closely related to MRP-1. Thus, we studied the potential contribution of YB-1 on MRP-1 nuclear localization and function. Compared with MAE, YB-1 is more abundant in EOMA cells (Fig. 6A). In MAE, YB-1 was equally distributed across cytosolic and nuclear compartments (Fig. 6, A and C). However, in EOMA, YB-1 was more abundant in the nuclear fraction of cell extract (Fig. 6, B and C). Next, we sought to investigate whether MRP-1 physically interacts with YB-1 and utilizes the NLS of YB-1 to translocate to the nucleus. MRP-1 was detected in YB-1 immunoprecipitate, suggesting binding of MRP-1 with YB-1. Such physical interaction was most prominent in nuclear lysate, suggesting abundant interaction between these two proteins in the nucleus (Fig. 6D). Next, we asked whether because of such interaction or otherwise, does YB-1 influence MRP-1 function? Knockdown of YB-1 in EOMA significantly lowered MRP-1 function (Fig. 6E). YB-1 knockdown-induced compromise of MRP-1 function caused loss of cell viability (Fig. 6F). Further investigation on how YB-1 regulates MRP-1 function revealed that knockdown of YB-1 lowered MRP-1 protein expression (Fig. 6G). Interestingly, such YB-1 knockdown specifically compromised the nuclear abundance of MRP-1 protein (Fig. 6H). These observations support the notion that MRP-1 interacts with NLS-guided YB-1 such that nuclear localization of MRP-1 is dependent on YB-1.
FIGURE 6.
MRP-1 expression and activity are YB-1-dependent. A, Western blotting shows elevated YB-1 protein expression in whole cell lysates of EOMA versus MAE cells. B and C, Western blotting (B) was done on cytosolic and nuclear fractions of MAE and EOMA cells with densitometry (C) showing elevated levels of YB-1 protein localized to the nuclear compartment of EOMA cells. D, association of MRP-1 with YB-1 in cell nuclei was identified by co-immunoprecipitation (IP). Cell lysates (200 μg) were immunoprecipitated with YB-1 antibody or control IgG, and the immunoprecipitates were resolved with SDS-PAGE and immunoblotted (IB) for MRP-1 and YB-1. Transient transfection with YB-1 siRNA showed decreased MRP-1 activity (E) by means of a calcein clearance test and increased cell death measured by an LDH toxicity assay (F). Total (G) as well as nuclear (H) MRP-1 expression was decreased after YB-1 knockdown in EOMA cells. Results are expressed as mean ± S.D. (error bars); *, p < 0.05.
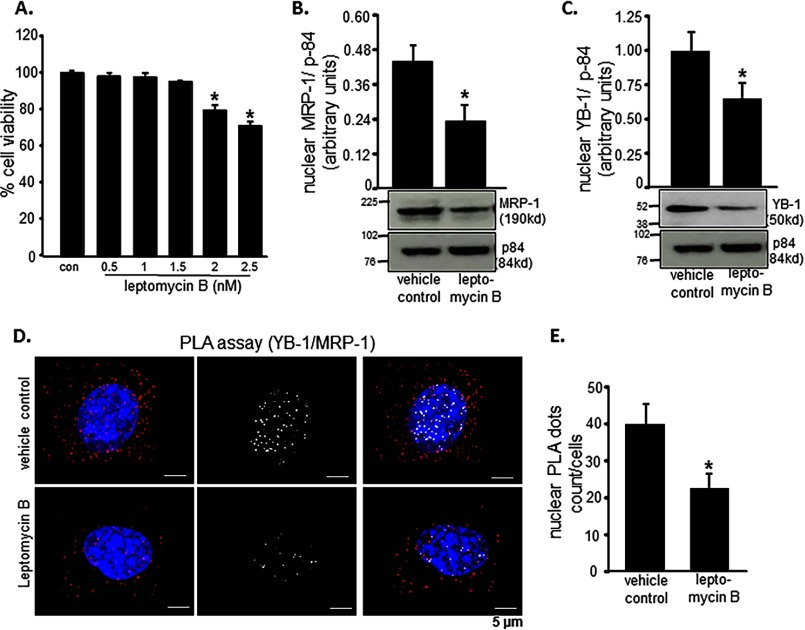
Leptomycin B is a pharmacological inhibitor of NLS function. If indeed YB-1-dependent nuclear translocation of MRP-1 is critical to the survival of EOMA cells, inhibition of NLS function should cause EOMA cell death. Treatment of EOMA cells with leptomycin B resulted in loss of cell viability (Fig. 7A). Indeed, in such leptomycin B-treated cells, nuclear abundance of MRP-1 as well as of YB-1 was significantly lowered (Fig. 7, B and C). Further interrogation of the physical interaction between MRP-1 and YB-1 was conducted employing the proximity ligation assay. Treatment of EOMA cells with leptomycin B minimized the abundance of YB-1-interacting MRP-1 in the nucleus (Fig. 7, D and E).
FIGURE 7.
MRP-1 nuclear translocation is YB-1-dependent. Leptomycin B was used to inhibit nuclear translocation. A, toxicity was evaluated by LDH assay, and 4-h treatment with 2 nm leptomycin B resulted in loss of cell survival. B and C, Western blotting images show that 4-h treatment with leptomycin B (2 nm) causes a significant reduction in MRP-1 (B) and YB-1 (C) protein expression. Protein-protein interaction between MRP-1 and YB-1 inside the EOMA cell nucleus was demonstrated using an in situ PLA. D, treatment with leptomycin B (2 nm, 4 h) resulted in a significant reduction of PLA-positive signals (red dots) with those interactions specifically occurring in the nucleus by co-localization of red dots and blue DAPI staining and represented by white dots. E, quantification of nuclear PLA events reveals that roughly 50% of the interaction between MRP-1 and YB-1 was inhibited by 2 nm leptomycin B treatment for 4 h. Results are expressed as mean ± S.D. (error bars); *, p < 0.05.
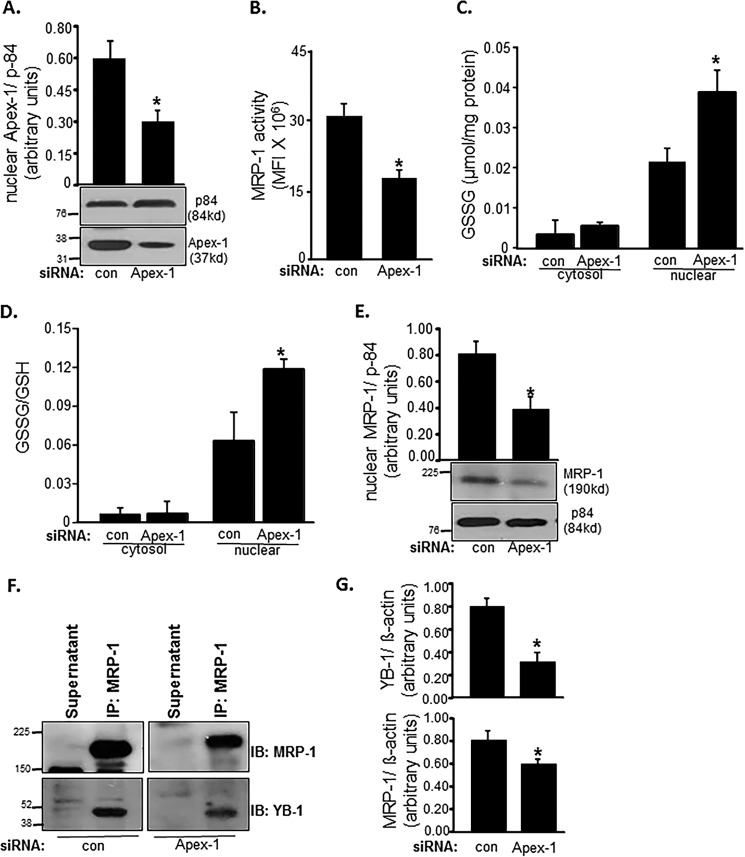
Ape-1/Ref-1 (Apex-1) regulates the expression of a number of proteins by promoting DNA binding of redox-sensitive transcription factors. Activation of the related mdr-1 gene is regulated by YB-1 in cooperation with Apex-1 (49, 50). Therefore, we were led to investigate whether Apex-1 is involved in regulating MRP-1 through its YB-1 regulatory function. Knockdown of Apex-1 in EOMA (Fig. 8A) resulted in lowering of MRP-1 function (Fig. 8B). Such compromised MRP-1 function in Apex-1 knockdown EOMA cells displayed a significant increase in nuclear GSSG build-up (Fig. 8, C and D). Interestingly, Apex-1 knockdown significantly lowered the abundance of MRP-1 protein in the nucleus (Fig. 8E). In Apex-1 knockdown EOMA cells, lower abundance of YB-1 was noted in MRP-1 immunoprecipitate (Fig. 8, F and G). These results implicate Apex-1 in the nuclear localization of YB-1 and MRP-1.
FIGURE 8.
MRP-1 function is Apex-1-dependent. apex-1 knockdown in EOMA cells resulted in a significant decrease of nuclear Apex-1 protein expression (A). MRP-1 activity as measured by calcein exclusion is also decreased in apex-1 knockdown cells (B). apex-1 knockdown resulted in a significantly elevated nuclear GSSG level (C) as well as ratio of oxidized to reduced glutathione (D) in the nucleus of EOMA cells compared with transfection controls. Western blotting demonstrates significantly decreased nuclear MRP-1 protein expression in apex-1 knockdown cells (E). F, association of MRP-1 with YB-1 was identified in apex-1 knockdown EOMA cells by the immunoprecipitation (IP) technique described previously. The cell lysates of control and apex-1 siRNA-treated EOMA cells were subjected to immunoprecipitation with MRP-1 antibody. The immunoprecipitates were separated by SDS-PAGE and immunoblotted (IB) for MRP-1 and YB-1 with densitometry analysis of both proteins showing an apex-1-dependent decrease in protein expression (G). Results are expressed as mean ± S.D. (error bars); *, p < 0.05.
Discussion
Previous works from our laboratory and others have underscored the significance of cellular GSSG in inducing cell death (13, 51, 52). Consistent with that notion, GSSG mimetics have been found to be productive in the treatment of cancer (53). Our previous work has reported that EOMA cells largely depend on Nox-4-derived oxidants for their rapid growth and survival (7–8, 37, 54). Reactive oxygen species are widely recognized to be directly implicated in tumor promotion. For example, the activation of oxidant-inducible transcription factors, such as AP-1 and NF-κB, stimulate cell proliferation and angiogenesis inherent to tumorigenesis (55, 56). Under such conditions, where oxidants play a central role in tumorigenesis, tumor survival depends on enhanced antioxidant defense mechanisms in cancer cells (57, 58). This work introduces the novel concept that efficient GSSG clearance from oxidant-rich EOMA cells represents the lifeline of these cells. The current prevalent notion is that the level of GSH in the cancer cell has a direct bearing on its survival (59). Thus, depletion of cellular GSH represents a common strategy to sensitize cancer cells to inducible death (60). In the current study, we observe for the first time that a cellular mechanism to clear excessive GSSG is of critical significance in EOMA cell survival.
Previously reported works by our laboratory and others have demonstrated that in neural cells and tissue, MRP-1 acts as a mechanism for the efflux of cellular GSSG (13, 61). MRP-1 efflux of GSSG is known to have favorable reaction kinetics with an approximate Km of 70 μm compared with 10 mm for GSH (62). However, this has never been upheld as a pathway by which cancer cells thrive in an oxidant-rich environment. Interestingly, agents that induce expression of γ-glutamylcysteine synthetase, the enzyme that synthesizes GSH, also induce MRP-1 expression (63, 64), indicating that the coordinated expression of these two proteins probably plays a role in redox homeostasis. The current work is the first of its kind recognizing that inhibition of MRP-1 may be utilized as a productive strategy to trap cytotoxic GSSG with the goal of killing the cell.
In tumor cells, MRP-1 is often found in the cell membrane and also in the cytosol (65, 66). Nuclear translocation of MRP-1 confers multidrug resistance to cancer cells (67). This work provides the first evidence suggesting that MRP-1 interacts with NLS-directed YB-1 to reach the nucleus. A non-canonical NLS is present in the C-terminal half of YB-1 (68). The gene transcription, mRNA processing, and translation regulatory functions of YB-1 justify the presence of an NLS. Although details on the transport of YB-1 in the cytoplasmic and nuclear compartments remain elusive, there is clear evidence that YB-1 interacts with other proteins to influence their subcellular localization (69).
Thiol-disulfide homeostasis is known to be of critical significance in cancer outcomes (70, 71). Arresting the glutathione and thioredoxin redox cycles has consistently provided productive returns in killing cancer cells (58, 72, 73). In oxidant-rich EOMA cells, intracellular GSSG levels are remarkably elevated. This GSSG burden within the cell is managed mostly by efflux processes in which MRP-1 plays a significant role as noted in this work. Another mechanism to lower cellular GSSG levels in EOMA is GSSG reductase-dependent recycling of GSSG to GSH. Contribution of this pathway in EOMA cells is evident from the observation that BCNU-dependent inhibition of GSSG reductase resulted in loss of cell viability. Under the trade name of Carmustine, BCNU has been used as a chemotherapy drug (74). More recent work points toward inhibition of thioredoxin reductase as a productive strategy to kill cancer cells (75). Cytosolic thioredoxin reductase 1 is auranofin-sensitive and reduces oxidized thioredoxin 1. In EOMA cells, auranofin-dependent inhibition of thioredoxin reductase was effective in causing cell death. This observation is consistent with the reported anticancer effects of auranofin (76). It is important to note in this context that inhibition of thioredoxin reductase is known to elevate cellular GSSG levels (77, 78). It is therefore plausible that inhibition of thioredoxin reductase pushes the oxidant burden in EOMA cells over the edge such that cytotoxicity is induced.
Certainly, MRP-1 is a major target for cancer therapeutics because MRP-1 levels are elevated in many different types of tumors (79–82), and inhibition of MRP-1 is aimed at limiting drug resistance (83). MRP-1 is known to efflux vinca alkaloids, such as vincristine, which is the chemotherapeutic agent used to treat the type of endothelial cell tumors (hemangioendothelioma) generated by EOMA. We have previously shown that Nox-4 induces Apex-1 expression in EOMA cells (8) and are now reporting that MRP-1 activity is dependent on Apex-1 expression. Thus, oxidative events in EOMA cells promote MRP-1 expression, and chemoresistance may be an undesired consequence of managing that oxidative stress.
In conclusion, this work provides the first evidence recognizing inhibition of MRP-1 to promote accumulation of cytotoxic GSSG as a productive strategy to kill cancer cells. Cancer cells, such as EOMA, that rely on ROS to thrive must be supported with extraordinary defense mechanisms that enable them to survive in the face of severe oxidant burden. In EOMA cells, a hyperactive MRP-1 system serves as a critical compensatory pathway to maintain cell survival. The remarkably depleted levels of GSH and NADPH indicate that the compensatory mechanism for managing oxidative stress is tenuous and vulnerable and merits further investigation of strategies to inhibit MRP-1 activity as a potential therapeutic target.
Author Contributions
G. M. G. and C. K. S. conceived the study, obtained funding, and wrote paper. S. K. coordinated and supervised the execution of experiments performed by A. B. and J. M. S., and A. B. also wrote some portions of the paper. X. P. provided critical review of the raw data, figures, and manuscript.
This work was supported by National Institutes of Health/NIGMS Grants R01 GM095657 (to G. M. G) and R01 GM108014 (to C. K. S.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- MCP-1
- monocyte chemoattractant protein-1
- BCNU
- 1,3-bis(2-chloroethyl)-N-nitrosourea
- MAE
- murine arterial endothelial
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- MBB
- monobromobimane
- MCB
- monochlorobimane
- BSO
- buthionine sulfoximine
- PAO
- phenylarsineoxide
- LDH
- lactate dehydrogenase
- PLA
- proximity ligation assay
- NLS
- nuclear localization sequence
- BCNU
- 1,3-bis(2-chloroethyl)-N-nitrosourea.
References
- 1. Haggstrom A. N., Drolet B. A., Baselga E., Chamlin S. L., Garzon M. C., Horii K. A., Lucky A. W., Mancini A. J., Metry D. W., Newell B., Nopper A. J., and Frieden I. J. (2006) Prospective study of infantile hemangiomas: clinical characteristics predicting complications and treatment. Pediatrics 118, 882–887 [DOI] [PubMed] [Google Scholar]
- 2. Jacobs A. H., and Walton R. G. (1976) The incidence of birthmarks in the neonate. Pediatrics 58, 218–222 [PubMed] [Google Scholar]
- 3. Mulliken J. B. (1988) in Vascular Birthmarks: Hemangiomas and Malformations (Mulliken J. B., and Young A. E., eds) pp. 41–62, W.B. Saunders, Philadelphia [Google Scholar]
- 4. Paller A. S. (2000) Responses to anti-angiogenic therapies. J. Investig. Dermatol. Symp. Proc. 5, 83–86 [DOI] [PubMed] [Google Scholar]
- 5. Gordillo G. M., Atalay M., Roy S., and Sen C. K. (2002) Hemangioma model for in vivo angiogenesis: inducible oxidative stress and MCP-1 expression in EOMA cells. Methods Enzymol. 352, 422–432 [DOI] [PubMed] [Google Scholar]
- 6. Gordillo G. M., Onat D., Stockinger M., Roy S., Atalay M., Beck F. M., and Sen C. K. (2004) A key angiogenic role of monocyte chemoattractant protein-1 in hemangioendothelioma proliferation. Am. J. Physiol. Cell Physiol 287, C866–C873 [DOI] [PubMed] [Google Scholar]
- 7. Gordillo G., Fang H., Park H., and Roy S. (2010) Nox-4-dependent nuclear H2O2 drives DNA oxidation resulting in 8-OHdG as urinary biomarker and hemangioendothelioma formation. Antioxid. Redox Signal. 12, 933–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Biswas A., Khanna S., Roy S., Pan X., Sen C. K., and Gordillo G. M. (2015) Endothelial cell tumor growth is ape/ref-1-dependent. Am. J. Physiol. Cell Physiol. 309, C296–C307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Janiszewski M., Pasqualucci C. A., Souza L. C., Pileggi F., da Luz P. L., and Laurindo F. R. (1998) Oxidized thiols markedly amplify the vascular response to balloon injury in rabbits through a redox active metal-dependent pathway. Cardiovasc. Res. 39, 327–338 [DOI] [PubMed] [Google Scholar]
- 10. Zhong L., Arnér E. S., and Holmgren A. (2000) Structure and mechanism of mammalian thioredoxin reductase: the active site is a redox-active selenolthiol/selenenylsulfide formed from the conserved cysteine-selenocysteine sequence. Proc. Natl. Acad. Sci. U.S.A. 97, 5854–5859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lu J., and Holmgren A. (2014) The thioredoxin antioxidant system. Free Radic. Biol. Med. 66, 75–87 [DOI] [PubMed] [Google Scholar]
- 12. Rahman I., Kode A., and Biswas S. K. (2006) Assay for quantitative determination of glutathione and glutathione disulfide levels using enzymatic recycling method. Nat. Protoc. 1, 3159–3165 [DOI] [PubMed] [Google Scholar]
- 13. Park H. A., Khanna S., Rink C., Gnyawali S., Roy S., and Sen C. K. (2009) Glutathione disulfide induces neural cell death via a 12-lipoxygenase pathway. Cell Death Differ. 16, 1167–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sharma-Walia N., Krishnan H. H., Naranatt P. P., Zeng L., Smith M. S., and Chandran B. (2005) ERK1/2 and MEK1/2 induced by Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) early during infection of target cells are essential for expression of viral genes and for establishment of infection. J. Virol. 79, 10308–10329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sharma-Walia N., George Paul A., Patel K., Chandran K., Ahmad W., and Chandran B. (2010) NFAT and CREB regulate Kaposi's sarcoma-associated herpesvirus-induced cyclooxygenase 2 (COX-2). J. Virol. 84, 12733–12753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Park H. A., Kubicki N., Gnyawali S., Chan Y. C., Roy S., Khanna S., and Sen C. K. (2011) Natural vitamin E α-tocotrienol protects against ischemic stroke by induction of multidrug resistance-associated protein 1. Stroke 42, 2308–2314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sen C. K., Khanna S., Babior B. M., Hunt T. K., Ellison E. C., and Roy S. (2002) Oxidant-induced vascular endothelial growth factor expression in human keratinocytes and cutaneous wound healing. J. Biol. Chem. 277, 33284–33290 [DOI] [PubMed] [Google Scholar]
- 18. Buşu C., Li W., Caldito G., and Aw T. Y. (2013) Inhibition of glutathione synthesis in brain endothelial cells lengthens S-phase transit time in the cell cycle: implications for proliferation in recovery from oxidative stress and endothelial cell damage. Redox Biol. 1, 131–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Radyuk S. N., Rebrin I., Luchak J. M., Michalak K., Klichko V. I., Sohal R. S., and Orr W. C. (2009) The catalytic subunit of Drosophila glutamate-cysteine ligase is a nucleocytoplasmic shuttling protein. J. Biol. Chem. 284, 2266–2274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rebrin I., Kamzalov S., and Sohal R. S. (2003) Effects of age and caloric restriction on glutathione redox state in mice. Free Radic. Biol. Med. 35, 626–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sen C. K., Roy S., Khanna S., and Packer L. (1999) Determination of oxidized and reduced lipoic acid using high-performance liquid chromatography and coulometric detection. Methods Enzymol. 299, 239–246 [DOI] [PubMed] [Google Scholar]
- 22. Kosower E. M., and Kosower N. S. (1995) Bromobimane probes for thiols. Methods Enzymol. 251, 133–148 [DOI] [PubMed] [Google Scholar]
- 23. Hedley D., and Chow S. (1994) Glutathione and cellular resistance to anti-cancer drugs. Methods Cell Biol. 42, 31–44 [DOI] [PubMed] [Google Scholar]
- 24. Ramos L., van der Heijden G. W., Derijck A., Berden J. H., Kremer J. A., van der Vlag J., and de Boer P. (2008) Incomplete nuclear transformation of human spermatozoa in oligo-astheno-teratospermia: characterization by indirect immunofluorescence of chromatin and thiol status. Hum. Reprod. 23, 259–270 [DOI] [PubMed] [Google Scholar]
- 25. Kosower N. S., and Kosower E. M. (1987) Thiol labeling with bromobimanes. Methods Enzymol. 143, 76–84 [DOI] [PubMed] [Google Scholar]
- 26. Hedley D. W., and Chow S. (1994) Evaluation of methods for measuring cellular glutathione content using flow cytometry. Cytometry 15, 349–358 [DOI] [PubMed] [Google Scholar]
- 27. Rice G. C., Bump E. A., Shrieve D. C., Lee W., and Kovacs M. (1986) Quantitative analysis of cellular glutathione by flow cytometry utilizing monochlorobimane: some applications to radiation and drug resistance in vitro and in vivo. Cancer Res. 46, 6105–6110 [PubMed] [Google Scholar]
- 28. Osseni R. A., Rat P., Bogdan A., Warnet J. M., and Touitou Y. (2000) Evidence of prooxidant and antioxidant action of melatonin on human liver cell line HepG2. Life Sci. 68, 387–399 [DOI] [PubMed] [Google Scholar]
- 29. Sen C. K., Roy S., Han D., and Packer L. (1997) Regulation of cellular thiols in human lymphocytes by α-lipoic acid: a flow cytometric analysis. Free Radic. Biol. Med. 22, 1241–1257 [DOI] [PubMed] [Google Scholar]
- 30. Friedrichs B., Müller C., and Brigelius-Flohé R. (1998) Inhibition of tumor necrosis factor-α- and interleukin-1-induced endothelial E-selectin expression by thiol-modifying agents. Arterioscler. Thromb. Vasc. Biol. 18, 1829–1837 [DOI] [PubMed] [Google Scholar]
- 31. Khanna S., Roy S., Slivka A., Craft T. K., Chaki S., Rink C., Notestine M. A., DeVries A. C., Parinandi N. L., and Sen C. K. (2005) Neuroprotective properties of the natural vitamin E α-tocotrienol. Stroke 36, 2258–2264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Khanna S., Roy S., Park H. A., and Sen C. K. (2007) Regulation of c-Src activity in glutamate-induced neurodegeneration. J. Biol. Chem. 282, 23482–23490 [DOI] [PubMed] [Google Scholar]
- 33. Han D., Sen C. K., Roy S., Kobayashi M. S., Tritschler H. J., and Packer L. (1997) Protection against glutamate-induced cytotoxicity in C6 glial cells by thiol antioxidants. Am. J. Physiol. 273, R1771–R1778 [DOI] [PubMed] [Google Scholar]
- 34. Scheffer G. L., Kool M., Heijn M., de Haas M., Pijnenborg A. C., Wijnholds J., van Helvoort A., de Jong M. C., Hooijberg J. H., Mol C. A., van der Linden M., de Vree J. M., van der Valk P., Elferink R. P., Borst P., and Scheper R. J. (2000) Specific detection of multidrug resistance proteins MRP1, MRP2, MRP3, MRP5, and MDR3 P-glycoprotein with a panel of monoclonal antibodies. Cancer Res. 60, 5269–5277 [PubMed] [Google Scholar]
- 35. Khaperskyy D. A., Emara M. M., Johnston B. P., Anderson P., Hatchette T. F., and McCormick C. (2014) Influenza a virus host shutoff disables antiviral stress-induced translation arrest. PLoS Pathog. 10, e1004217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lokanga R. A., Senejani A. G., Sweasy J. B., and Usdin K. (2015) Heterozygosity for a hypomorphic Polbeta mutation reduces the expansion frequency in a mouse model of the Fragile X-related disorders. PLoS Genet. 11, e1005181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gordillo G. M., Biswas A., Khanna S., Pan X., Sinha M., Roy S., and Sen C. K. (2014) Dicer knockdown inhibits endothelial cell tumor growth via microRNA 21a-3p targeting of Nox-4. J. Biol. Chem. 289, 9027–9038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tontonoz P., Cortez-Toledo O., Wroblewski K., Hong C., Lim L., Carranza R., Conneely O., Metzger D., and Chao L. C. (2015) The orphan nuclear receptor Nur77 is a determinant of myofiber size and muscle mass in mice. Mol. Cell. Biol. 35, 1125–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sundin T., Peffley D. M., and Hentosh P. (2013) Disruption of an hTERT-mTOR-RAPTOR protein complex by a phytochemical perillyl alcohol and rapamycin. Mol. Cell Biochem. 375, 97–104 [DOI] [PubMed] [Google Scholar]
- 40. Mims J., Bansal N., Bharadwaj M. S., Chen X., Molina A. J., Tsang A. W., and Furdui C. M. (2015) Energy metabolism in a matched model of radiation resistance for head and neck squamous cell cancer. Radiat. Res. 183, 291–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Seco-Cervera M., Spis M., García-Giménez J. L., Ibañez-Cabellos J. S., Velázquez-Ledesma A., Esmorís I., Bañuls S., Pérez-Machado G., and Pallardó F. V. (2014) Oxidative stress and antioxidant response in fibroblasts from Werner and atypical Werner syndromes. Aging 6, 231–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Tan L. P., Seinen E., Duns G., de Jong D., Sibon O. C., Poppema S., Kroesen B. J., Kok K., and van den Berg A. (2009) A high throughput experimental approach to identify miRNA targets in human cells. Nucleic Acids Res. 37, e137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Khanna S., Roy S., Ryu H., Bahadduri P., Swaan P. W., Ratan R. R., and Sen C. K. (2003) Molecular basis of vitamin E action: tocotrienol modulates 12-lipoxygenase, a key mediator of glutamate-induced neurodegeneration. J. Biol. Chem. 278, 43508–43515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Khanna S., Park H. A., Sen C. K., Golakoti T., Sengupta K., Venkateswarlu S., and Roy S. (2009) Neuroprotective and antiinflammatory properties of a novel demethylated curcuminoid. Antioxid. Redox. Signal. 11, 449–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Huang E. Y., Chen Y. F., Chen Y. M., Lin I. H., Wang C. C., Su W. H., Chuang P. C., and Yang K. D. (2012) A novel radioresistant mechanism of galectin-1 mediated by H-Ras-dependent pathways in cervical cancer cells. Cell Death Dis. 3, e251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ballatori N., Krance S. M., Notenboom S., Shi S., Tieu K., and Hammond C. L. (2009) Glutathione dysregulation and the etiology and progression of human diseases. Biol. Chem. 390, 191–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ciriolo M. R., Palamara A. T., Incerpi S., Lafavia E., Buè M. C., De Vito P., Garaci E., and Rotilio G. (1997) Loss of GSH, oxidative stress, and decrease of intracellular pH as sequential steps in viral infection. J. Biol. Chem. 272, 2700–2708 [DOI] [PubMed] [Google Scholar]
- 48. Cohen S. B., Ma W., Valova V. A., Algie M., Harfoot R., Woolley A. G., Robinson P. J., and Braithwaite A. W. (2010) Genotoxic stress-induced nuclear localization of oncoprotein YB-1 in the absence of proteolytic processing. Oncogene 29, 403–410 [DOI] [PubMed] [Google Scholar]
- 49. Sengupta S., Mantha A. K., Mitra S., and Bhakat K. K. (2011) Human AP endonuclease (APE1/Ref-1) and its acetylation regulate YB-1-p300 recruitment and RNA polymerase II loading in the drug-induced activation of multidrug resistance gene MDR1. Oncogene 30, 482–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Bhakat K. K., Mantha A. K., and Mitra S. (2009) Transcriptional regulatory functions of mammalian AP-endonuclease (APE1/Ref-1), an essential multifunctional protein. Antioxid. Redox Signal. 11, 621–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Filomeni G., Aquilano K., Civitareale P., Rotilio G., and Ciriolo M. R. (2005) Activation of c-Jun-N-terminal kinase is required for apoptosis triggered by glutathione disulfide in neuroblastoma cells. Free Radic. Biol. Med. 39, 345–354 [DOI] [PubMed] [Google Scholar]
- 52. Filomeni G., Rotilio G., and Ciriolo M. R. (2003) Glutathione disulfide induces apoptosis in U937 cells by a redox-mediated p38 MAP kinase pathway. FASEB J. 17, 64–66 [DOI] [PubMed] [Google Scholar]
- 53. Gumireddy K., Li A., Cao L., Yan J., Liu L., Xu X., Pazoles C., and Huang Q. (2013) NOV-002, A glutathione disulfide mimetic, suppresses tumor cell invasion and metastasis. J. Carcinog. Mutagen. 2013, S7–002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Gordillo G., Fang H., Khanna S., Harper J., Phillips G., and Sen C. K. (2009) Oral administration of blueberry inhibits angiogenic tumor growth and enhances survival of mice with endothelial cell neoplasm. Antioxid. Redox. Signal. 11, 47–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Hsu T. C., Young M. R., Cmarik J., and Colburn N. H. (2000) Activator protein 1 (AP-1)- and nuclear factor κB (NF-κB)-dependent transcriptional events in carcinogenesis. Free Radic. Biol. Med. 28, 1338–1348 [DOI] [PubMed] [Google Scholar]
- 56. Amiri K. I., and Richmond A. (2005) Role of nuclear factor-κB in melanoma. Cancer Metastasis Rev. 24, 301–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Dando I., Cordani M., Dalla Pozza E., Biondani G., Donadelli M., and Palmieri M. (2015) Antioxidant mechanisms and ROS-related microRNAs in cancer stem cells. Oxid. Med. Cell Longev. 2015, 425708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Raninga P. V., Trapani G. D., and Tonissen K. F. (2014) Cross talk between two antioxidant systems, thioredoxin and DJ-1: consequences for cancer. Oncoscience 1, 95–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Ortega A. L., Mena S., and Estrela J. M. (2011) Glutathione in cancer cell death. Cancers 3, 1285–1310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Liu C., Liu H., Li Y., Wu Z., Zhu Y., Wang T., Gao A. C., Chen J., and Zhou Q. (2012) Intracellular glutathione content influences the sensitivity of lung cancer cell lines to methylseleninic acid. Mol. Carcinog. 51, 303–314 [DOI] [PubMed] [Google Scholar]
- 61. Minich T., Riemer J., Schulz J. B., Wielinga P., Wijnholds J., and Dringen R. (2006) The multidrug resistance protein 1 (Mrp1), but not Mrp5, mediates export of glutathione and glutathione disulfide from brain astrocytes. J. Neurochem. 97, 373–384 [DOI] [PubMed] [Google Scholar]
- 62. Leier I., Jedlitschky G., Buchholz U., Center M., Cole S. P., Deeley R. G., and Keppler D. (1996) ATP-dependent glutathione disulphide transport mediated by the MRP gene-encoded conjugate export pump. Biochem. J. 314, 433–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Ishikawa T., Bao J. J., Yamane Y., Akimaru K., Frindrich K., Wright C. D., and Kuo M. T. (1996) Coordinated induction of MRP/GS-X pump and γ-glutamylcysteine synthetase by heavy metals in human leukemia cells. J. Biol. Chem. 271, 14981–14988 [DOI] [PubMed] [Google Scholar]
- 64. Yamane Y., Furuichi M., Song R., Van N. T., Mulcahy R. T., Ishikawa T., and Kuo M. T. (1998) Expression of multidrug resistance protein/GS-X pump and γ-glutamylcysteine synthetase genes is regulated by oxidative stress. J. Biol. Chem. 273, 31075–31085 [DOI] [PubMed] [Google Scholar]
- 65. Deeley R. G., Westlake C., and Cole S. P. (2006) Transmembrane transport of endo- and xenobiotics by mammalian ATP-binding cassette multidrug resistance proteins. Physiol. Rev. 86, 849–899 [DOI] [PubMed] [Google Scholar]
- 66. Van Luyn M. J., Müller M., Renes J., Meijer C., Scheper R. J., Nienhuis E. F., Mulder N. H., Jansen P. L., and De Vries E. G. (1998) Transport of glutathione conjugates into secretory vesicles is mediated by the multidrug-resistance protein 1. Int. J. Cancer 76, 55–62 [DOI] [PubMed] [Google Scholar]
- 67. Cai B. L., Xu X. F., Fu S. M., Shen L. L., Zhang J., Guan S. M., and Wu J. Z. (2011) Nuclear translocation of MRP1 contributes to multidrug resistance of mucoepidermoid carcinoma. Oral Oncol. 47, 1134–1140 [DOI] [PubMed] [Google Scholar]
- 68. Bader A. G., and Vogt P. K. (2005) Inhibition of protein synthesis by Y box-binding protein 1 blocks oncogenic cell transformation. Mol. Cell. Biol. 25, 2095–2106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Ohashi S., Atsumi M., and Kobayashi S. (2009) HSP60 interacts with YB-1 and affects its polysome association and subcellular localization. Biochem. Biophys. Res. Commun. 385, 545–550 [DOI] [PubMed] [Google Scholar]
- 70. Dirican N., Dirican A., Sen O., Aynali A., Atalay S., Bircan H. A., Ozturk O., Erdogan S., Cakir M., and Akkaya A. (2015) Thiol/disulfide homeostasis: a prognostic biomarker for patients with advanced non-small cell lung cancer? Redox Rep. 10.1179/1351000215Y.0000000027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Xu S., Sankar S., and Neamati N. (2014) Protein disulfide isomerase: a promising target for cancer therapy. Drug Discov. Today 19, 222–240 [DOI] [PubMed] [Google Scholar]
- 72. Traverso N., Ricciarelli R., Nitti M., Marengo B., Furfaro A. L., Pronzato M. A., Marinari U. M., and Domenicotti C. (2013) Role of glutathione in cancer progression and chemoresistance. Oxid. Med. Cell Longev. 2013, 972913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Arnér E. S., and Holmgren A. (2006) The thioredoxin system in cancer. Semin. Cancer Biol. 16, 420–426 [DOI] [PubMed] [Google Scholar]
- 74. Zhang Y. W., Zhang Y. L., Pan H., Wei F. X., Zhang Y. C., Shao Y., Han W., Liu H. P., Wang Z. Y., and Yang S. H. (2014) Chemotherapy for patients with gastric cancer after complete resection: a network meta-analysis. World J. Gastroenterol. 20, 584–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Tobe R., Yoo M. H., Fradejas N., Carlson B. A., Calvo S., Gladyshev V. N., and Hatfield D. L. (2012) Thioredoxin reductase 1 deficiency enhances selenite toxicity in cancer cells via a thioredoxin-independent mechanism. Biochem. J. 445, 423–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Fan C., Zheng W., Fu X., Li X., Wong Y. S., and Chen T. (2014) Enhancement of auranofin-induced lung cancer cell apoptosis by selenocystine, a natural inhibitor of TrxR1 in vitro and in vivo. Cell Death Dis. 5, e1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Martínez-González J. J., Guevara-Flores A., Rendón J. L., and del Arenal I. P. (2015) Auranofin-induced oxidative stress causes redistribution of the glutathione pool in Taenia crassiceps cysticerci. Mol. Biochem. Parasitol. 201, 16–25 [DOI] [PubMed] [Google Scholar]
- 78. Zhang H., Du Y., Zhang X., Lu J., and Holmgren A. (2014) Glutaredoxin 2 reduces both thioredoxin 2 and thioredoxin 1 and protects cells from apoptosis induced by auranofin and 4-hydroxynonenal. Antioxid. Redox Signal. 21, 669–681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Chow A. K., Ng L., Lam C. S., Wong S. K., Wan T. M., Cheng N. S., Yau T. C., Poon R. T., and Pang R. W. (2013) The enhanced metastatic potential of hepatocellular carcinoma (HCC) cells with sorafenib resistance. PLoS One 8, e78675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Ji L., Li H., Gao P., Shang G., Zhang D. D., Zhang N., and Jiang T. (2013) Nrf2 pathway regulates multidrug-resistance-associated protein 1 in small cell lung cancer. PLoS One 8, e63404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Hlaváč V., Brynychová V., Václavíková R., Ehrlichová M., Vrána D., Pecha V., Koževnikovová R., Trnková M., Gatěk J., Kopperová D., Gut I., and Souček P. (2013) The expression profile of ATP-binding cassette transporter genes in breast carcinoma. Pharmacogenomics 14, 515–529 [DOI] [PubMed] [Google Scholar]
- 82. Haber M., Smith J., Bordow S. B., Flemming C., Cohn S. L., London W. B., Marshall G. M., and Norris M. D. (2006) Association of high-level MRP1 expression with poor clinical outcome in a large prospective study of primary neuroblastoma. J. Clin. Oncol. 24, 1546–1553 [DOI] [PubMed] [Google Scholar]
- 83. Munoz M., Henderson M., Haber M., and Norris M. (2007) Role of the MRP1/ABCC1 multidrug transporter protein in cancer. IUBMB Life 59, 752–757 [DOI] [PubMed] [Google Scholar]
- 84. Kehrer J. P. (1983) The effect of BCNU (carmustine) on tissue glutathione reductase activity. Toxicol. Lett. 17, 63–68 [DOI] [PubMed] [Google Scholar]