Abstract
Intramembrane metalloproteases (IMMPs) are conserved from bacteria to humans and control many important signaling pathways, but little is known about how IMMPs interact with their substrates. SpoIVFB is an IMMP that cleaves Pro-σK during Bacillus subtilis endospore formation. When catalytically inactive SpoIVFB was coexpressed with C-terminally truncated Pro-σK(1–126) (which can be cleaved by active SpoIVFB) in Escherichia coli, the substrate dramatically improved solubilization of the enzyme from membranes with mild detergents. Both the Pro(1–20) and σK(21–126) parts contributed to improving SpoIVFB solubilization from membranes, but only the σK part was needed to form a stable complex with SpoIVFB in a pulldown assay. The last 10 residues of SpoIVFB were required for improved solubilization from membranes by Pro-σK(1–126) and for normal interaction with the substrate. The inactive SpoIVFB·Pro-σK(1–126)-His6 complex was stable during affinity purification and gel filtration chromatography. Disulfide cross-linking of the purified complex indicated that it resembled the complex formed in vivo. Ion mobility-mass spectrometry analysis resulted in an observed mass consistent with a 4:2 SpoIVFB·Pro-σK(1–126)-His6 complex. Stepwise photobleaching of SpoIVFB fused to a fluorescent protein supported the notion that the enzyme is tetrameric during B. subtilis sporulation. The results provide the first evidence that an IMMP acts as a tetramer, give new insights into how SpoIVFB interacts with its substrate, and lay the foundation for further biochemical analysis of the enzyme·substrate complex and future structural studies.
Keywords: Bacillus, bacterial signal transduction, enzyme purification, intramembrane proteolysis, mass spectrometry (MS), metalloenzyme, protein complex, sporulation, enzyme-substrate interaction, sigma factor
Introduction
Many critical cellular processes are regulated by intramembrane proteolysis (1). Intramembrane proteases (IPs)3 cleave their substrates within a transmembrane segment (TMS) or near the membrane surface. There are three classes of IPs: rhomboids, aspartyl IPs, and IMMPs (often called site-2 proteases or S2Ps). Rhomboids are serine IPs that promote animal cellular signaling, coordinate bacterial quorum sensing, regulate mitochondrial homeostasis, and control protozoan infection (2–5). Presenilin, an aspartyl IP, is the catalytic component of γ-secretase, which is involved in the processing of the amyloid precursor protein, Notch, and many other subtrates (6, 7). Dysfunction of γ-secretase contributes to the pathogenesis of Alzheimer disease (8) and many other diseases (7). Aspartyl IPs also include preflagellin and prepilin peptidases involved in bacterial pathogenesis (9), and signal peptide peptidases, which facilitate the clearance of signal peptides from membranes, participate in viral infection, and generate small peptides as signal molecules for immune systems (10, 11). IMMPs also play critical roles in a wide variety of biological functions. In eukaryotes, cholesterol metabolism, the unfolded protein response, and the acute-phase response are regulated by IMMPs (1, 12, 13). In bacteria, IMMPs control sporulation, envelope stress responses, mating signal production, polar morphogenesis, virulence, and signal peptide degradation from the membrane (14–16).
How IPs interact with their substrates remains a crucial knowledge gap. Crystal structures for representatives of all three IP types have been solved (9, 17–20), but no structure of an IP·substrate complex has been reported. Detailed knowledge of enzyme-substrate interactions can guide efforts to develop modulators of enzyme activity for therapeutic purposes. Here, we report the purification and characterization of a complex between the bacterial IMMP SpoIVFB and its substrate, Pro-σK.
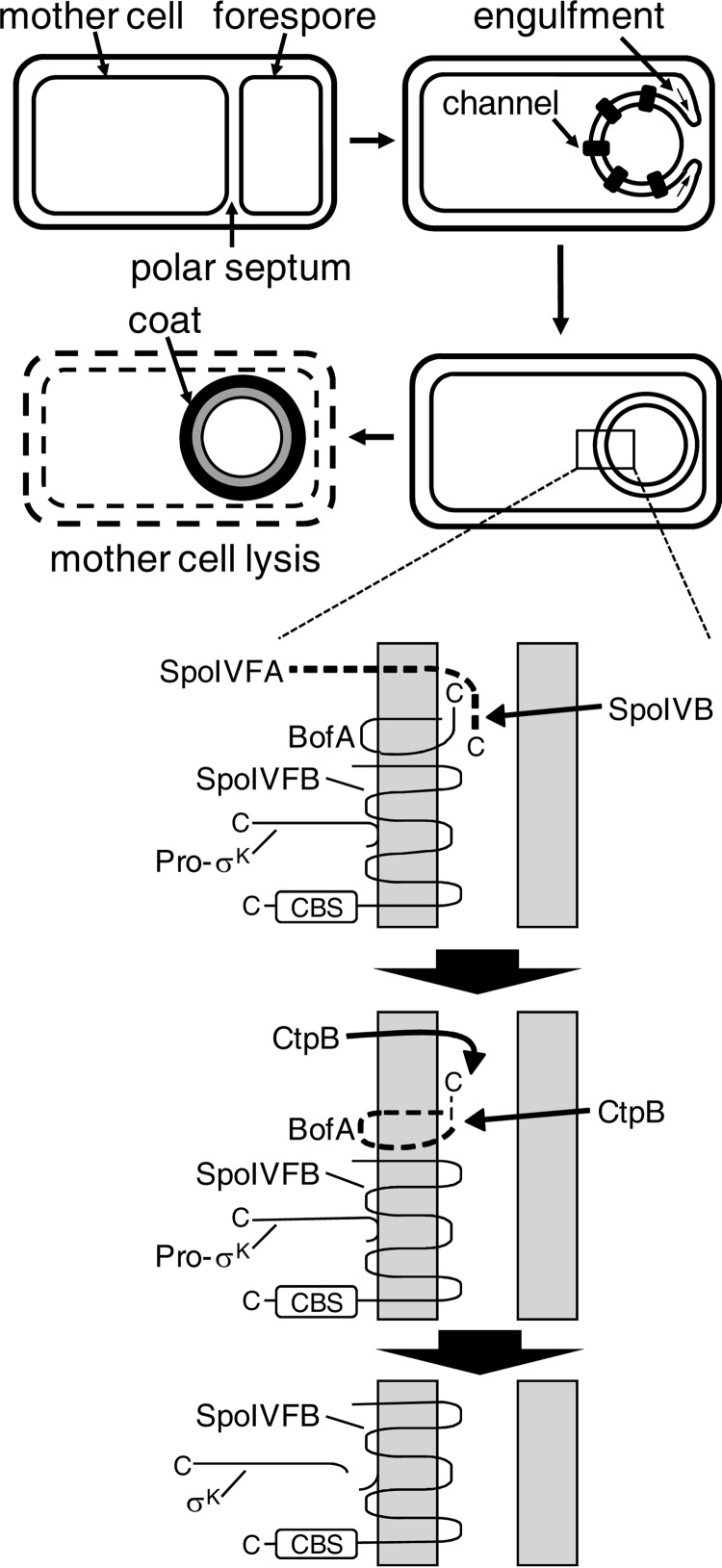
SpoIVFB is part of a signal transduction pathway that regulates the sporulation process of Bacillus subtilis (15). During endospore formation, an asymmetrically positioned septum divides the cell into forespore and mother cell compartments, and then the mother cell membrane of the septum engulfs the forespore, surrounding it with a second membrane and pinching it off within the mother cell (Fig. 1, top). SpoIVFB is expressed in the mother cell and localizes to the outermost membrane surrounding the forespore, with its active site facing toward the mother cell cytoplasm (Fig. 1, bottom). SpoIVFB is kept inactive by forming a complex with BofA and SpoIVFA (21, 22). A serine protease, SpoIVB, expressed in the forespore and secreted into the space between the membranes surrounding the forespore, initiates a proteolytic cascade that causes degradation of the SpoIVFB inhibitors (23–25) (Fig. 1, bottom). The activated SpoIVFB cleaves Pro-σK after residue 20 and releases σK into the mother cell (26–28). Genes transcribed by σK RNA polymerase are responsible for completing the sporulation process (29), including formation of the spore coat and lysis of the mother cell to release the spore (30) (Fig. 1, top).
FIGURE 1.
Morphological changes during B. subtilis sporulation and proteolytic cascade leading to cleavage of Pro-σK in B. subtilis. Top, morphological changes during sporulation of B. subtilis. Upon starvation, a polar septum forms, dividing the cell into the mother cell and forespore compartments. The mother cell membrane migrates around the forespore during the process of engulfment, and channels form connecting the two compartments. Completion of engulfment results in two membranes surrounding the forespore, triggering a proteolytic cascade detailed in the expanded view below. σK RNA polymerase activity in the mother cell results in expression of genes with products that form the spore coat and cause the mother cell to lyse. Bottom, proteolytic cascade that leads to cleavage of Pro-σK in the B. subtilis mother cell. This expanded view of the two membranes surrounding the forespore shows the signal transduction pathway that begins with SpoIVB in the forespore. SpoIVB crosses the first membrane and cleaves SpoIVFA. CtpB from both compartments cleaves BofA. Finally, SpoIVFB cleaves Pro-σK, releasing σK into the mother cell. See the text for references.
SpoIVFB has been studied as a model for a large subfamily of IMMPs that contain a CBS domain (31). Named after a domain found in the enzyme cystathionine β-synthase (32), CBS domains are found in thousands of proteins in all kingdoms of life (33). CBS domains undergo a conformational change upon binding a ligand and regulate protein activity in response to cellular energy levels or ion availability (34, 35). The C-terminal CBS domain of SpoIVFB is in the mother cell (Fig. 1, bottom) and has been shown to bind ATP and Pro-σK (28). ATP is required for purified SpoIVFB to cleave Pro-σK, so it has been proposed that CBS domains in IMMPs regulate substrate access to the active site in response to the cellular ATP level (28). Interestingly, channels form between the mother cell and the forespore during sporulation (36–38) (Fig. 1, top), and the channels have been proposed to be feeding tubes through which small molecules like ATP might pass from the mother cell to the forespore to allow late-stage gene expression in the forespore (39) and prevent its collapse (40). The SpoIVFB·BofA·SpoIVFA complex co-localizes with channel components (41, 42), and both undergo proteolytic modification upon completion of engulfment (23–25, 38, 42, 43), perhaps resulting in a rise in ATP in the mother cell that is sensed by the CBS domain of SpoIVFB as a signal that the channels have been closed and it is time to cleave Pro-σK (15, 44). In any case, it remains to be understood how the CBS domain of SpoIVFB regulates its activity in response to ATP and how this is coupled to physiological and morphological changes during sporulation.
Recently, considerable progress has been made toward understanding how SpoIVFB interacts with Pro-σK using mutational and cross-linking approaches. Mutational analysis revealed features of Pro-σK important for cleavage by SpoIVFB, including preferences and tolerances somewhat different from those of other IMMPs (45). Disulfide cross-linking between single-Cys versions of Pro-σK and catalytically inactive SpoIVFB have shown that residues in two conserved loops of SpoIVFB interact with residues near the cleavage site in Pro-σK (46). Both studies rely on coexpression of proteins in Escherichia coli, where it had been shown that active SpoIVFB can accurately and abundantly cleave C-terminally truncated Pro-σK(1–126) (22, 47) (Fig. 2A).
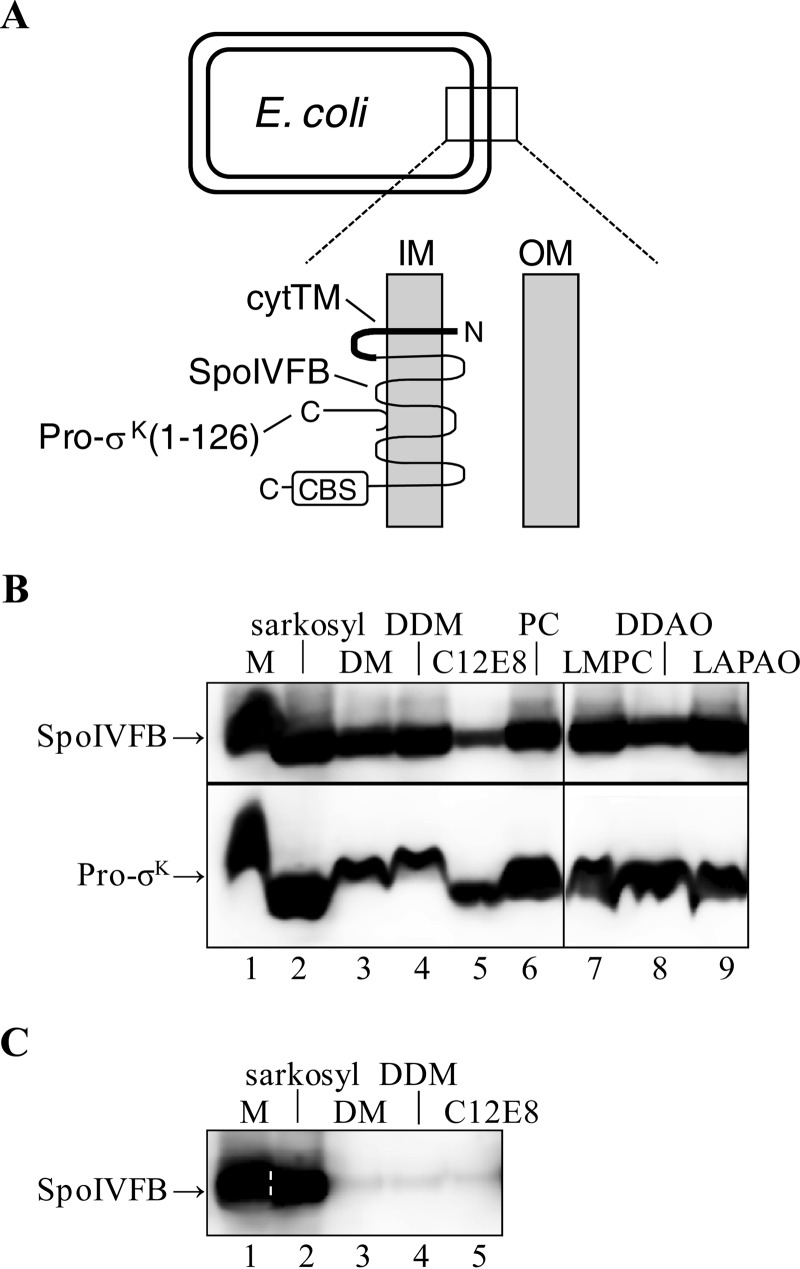
FIGURE 2.
Coexpression of Pro-σK(1–126)-His6 and SpoIVFB in E. coli and effect on solubilization of SpoIVFB from membranes. A, E. coli cell and an expanded view of the inner (IM) and outer membrane (OM). Pro-σK(1–126)-His6 associates peripherally with the inner membrane (45). The topology of SpoIVFB was determined previously (77). SpoIVFB has a C-terminal CBS domain, and in some experiments an extra TMS (cytTM) was added to its N-terminal end. B, solubilization of coexpressed proteins. E. coli bearing pYZ42 was grown in a fermentor and induced with IPTG to coexpress Pro-σK(1–126)-His6 and SpoIVFB-TEV-FLAG2 E44Q. The membrane (M) fraction from 5 g of cells was untreated or treated with the indicated detergent to solubilize proteins. After high-speed centrifugation, the supernatant was subjected to immunoblot analysis with anti-FLAG (top) or anti-His (bottom) antibodies. PC, n-dodecylphosphocholine; LMPC, 1-myristoyl-2-hydroxy-sn-glycero-3-phosphocholine; DDAO, n-dodecyl-N,N-dimethylamine-N-oxide; LAPAO, 3-dodecylamido-N,N′-dimethylpropyl amine oxide. C, solubilization of enzyme alone. This is the same as in A, except pYZ50 was used to express cytTM-SpoIVFB-TEV-FLAG2-His6 alone, and immunoblot analysis was done with only anti-FLAG antibodies. A dashed white line was added to help visualize the demarcation between lanes 1 and 2. In both panels, representative results from at least two biological replicates are shown.
Here, we report that catalytically inactive SpoIVFB forms a complex with Pro-σK(1–126)-His6 upon coexpression in E. coli. The complex can be solubilized from membranes with mild detergents, and it is stable during affinity purification and gel filtration chromatography. Parts of Pro-σK(1–126)-His6 and SpoIVFB that are necessary for solubilization and affinity purification were identified. Complexes appeared to contain four SpoIVFB and at least two Pro-σK(1–126)-His6, and evidence for tetrameric SpoIVFB in vivo was obtained. Our work deepens the knowledge of the SpoIVFB quaternary structure and interaction with its substrate and has important implications for further biochemical and structural studies of IMMP substrate recognition for the purpose of therapeutic design.
Experimental Procedures
Plasmids
Descriptions of the plasmids and primers used in this study are available upon request. DNA sequencing was used to confirm the desired sequences in cloned PCR products and in cloned genes that had been subjected to site-specific mutagenesis (QuikChange kit, Stratagene).
Cotransformation and Induction
Two plasmids with different antibiotic resistance genes and different B. subtilis genes fused to the T7 RNA polymerase promoter were used for cotransformation of E. coli BL21(DE3) and induction of gene expression as described (22), except 0.3 mm isopropyl β-d-thiogalactopyranoside (IPTG) was used for 2 h unless stated otherwise. Cells were grown in a 5-liter fermentor with vigorous aeration and pH control or in a flask with shaking, in both cases at 37 °C in Luria-Bertani medium (48) supplemented with antibiotics.
Cell Fractionation and Detergent Solubilization of Proteins
Cells (5 g if grown in a fermentor or about 2.5 g if harvested from 1 liter grown in a flask) were resuspended in 15 ml of lysis buffer (PBS containing 0.1 mg/ml lysozyme, 1 mg/ml RNase A, 2 mg/ml DNase I, 1 mm PMSF, 10 mm 2-mercaptoethanol, and 10% glycerol), incubated at 37 °C for 10 min, and passed three times through a French pressure cell (SLM Aminco) at 14,000 p.s.i. (96 megapascals). The cell lysate was centrifuged at low speed (15,000 × g for 15 min at 4 °C) to sediment cell debris and protein inclusion bodies. The supernatant was designated the low-speed supernatant and contains both cytoplasmic and membrane proteins. The low-speed supernatant was centrifuged at high speed (150,000 × g for 90 min at 4 °C) to sediment membrane vesicles. The supernatant was designated the high-speed supernatant and contains cytoplasmic proteins. The pellet was resuspended in 10 ml of resuspension buffer (PBS containing 1 mm Pefabloc SC, 5 mm 2-mercaptoethanol, and 10% glycerol) using a motorized Dounce homogenizer, resulting in the membrane fraction. To solubilize proteins, the membrane fraction was treated with the indicated detergent (1%; all detergents were from Anatrace, Maumee, OH) by rotating the mixture for 1 h at 4 °C. The mixture was then centrifuged at 150,000 × g for 75 min at 12 °C to sediment insoluble material. The supernatant contained detergent-solubilized proteins.
Immunoblot Analysis
To analyze cell fractions and detergent-solubilized proteins from the procedure described above or fractions from the cobalt affinity purification described below, samples were mixed with an equal volume of 2× sample buffer (50 mm Tris·HCl pH 6.8, 4% SDS, 20% glycerol, 200 mm DTT, and 0.03% bromphenol blue) and boiled for 3 min, and then samples from an equivalent amount of cells were subjected to SDS-PAGE (12% ProSieve polyacrylamide gel) (Lonza) and immunoblotting as described previously (49). After gel filtration chromatography, samples were treated in the same way, except equal volumes of samples were analyzed. To measure expression and cleavage of proteins, an equivalent amount of cells (based on the optical density of the culture at 600 nm) were collected from 0.5 to 1.0 ml of culture by centrifugation (12,000 × g), and cell extracts were prepared as described previously (22), mixed with an equal volume of 2× sample buffer, and boiled for 3 min; then equal volumes were subjected to SDS-PAGE and immunoblotting as described above. SeeBlue Plus2 prestained protein standard (Invitrogen) was used to judge the migration of protein species. Antibodies that recognize His6 (penta-His, Qiagen, catalogue No. 34460) or FLAG (M2 monoclonal antibody-peroxidase conjugate, Sigma, catalogue No. A8592) were used at 1:5000 or 1:10000 dilution, respectively. These antibodies specifically detected the proteins induced and exhibited very little cross-reaction with other proteins in extracts of E. coli. Signals were detected using a LAS-3000 imager (Fujifilm) with exposure times short enough to ensure the signals were not saturated. Signal intensities were quantified using MultiGauge (Fujifilm) software.
Cobalt Affinity Purification (Pulldown Assays)
Cells (about 2.5 g harvested from 1 liter grown in a flask) were fractionated as described above. The membrane fraction was treated with 1% n-decyl-β-d-maltoside (DM) to solubilize the proteins and then subjected to high-speed centrifugation as described above. The supernatant (10 ml) was mixed with 0.5 ml of Talon Superflow metal affinity resin (Clontech) that had been equilibrated with buffer (PBS containing 1% DM, 5 mm 2-mercaptoethanol, and 10% glycerol). The mixture was rotated for 1 h at 23 °C. The cobalt resin was sedimented by centrifugation at 708 × g for 2 min at 4 °C. The supernatant was the unbound sample. The cobalt resin was washed three times by rotating briefly and then sedimenting the resin as described above. The wash buffer base (WBB) was PBS containing 150 mm NaCl and 10% glycerol. The three washes were with 5 ml of WBB with 0.5% DM, 5 ml of WBB with 0.1% DM, and 3.5 ml of WBB with 0.1% DM and 80 mm imidazole. Finally, the resin was mixed with 9.5 ml of WBB with 0.1% DM, and then a 500-μl sample was added to 2× sample buffer and boiled for 3 min, resulting in the bound sample.
Disulfide Cross-linking in Vivo
A method described previously (50) was used with slight modifications as described recently (46). Based on the measurement of the optical density at 600 nm after induction with IPTG for 30 min, an equivalent amount of cells from 0.5 to 1.0 ml of culture was used to prepare the samples, and equal volumes were subjected to SDS-PAGE and immunoblotting as described above.
Purification of the SpoIVFB-TEV-FLAG2 E44Q·Pro-σK(1–126)-His6 Complex
Cells (10 g, grown in a fermentor) were fractionated as described above, except 40 ml of lysis buffer and 20 ml of resuspension buffer was used. The membrane fraction was treated with 1% DM to solubilize the proteins and then subjected to high-speed centrifugation as described above. The supernatant (20 ml) was cobalt affinity-purified as described above, except 1 ml of cobalt resin was used, and after removal of the unbound sample the resin was washed five times by rotating briefly and then sedimenting the resin as described above. The five washes were with 5 ml of WBB with 0.5% DM, 5 ml of WBB with 0.1% DM, 1 ml of WBB with 0.1% DM and 30 mm imidazole, 1 ml of WBB with 0.1% DM and 60 mm imidazole, and 1 ml of WBB with 0.1% DM and 100 mm imidazole. Finally, the resin was eluted with 2 ml of WBB with 0.1% DM and 400 mm imidazole. The eluted material was concentrated to 0.5 ml using an Amicon Ultra centrifugal device with a 10-kDa cut-off (Millipore), and the sample was loaded onto a 1.0 × 30 cm Superdex 200 gel filtration column equilibrated with PBS containing 150 mm NaCl, 5% glycerol, and 0.1% DM. The column was eluted with the same buffer at 0.4 ml/min, and 0.5-ml fractions were collected.
To purify the complex for disulfide cross-linking, cells (about 5 g harvested from 2 liters grown in a flask) were fractionated as described above, except 20 ml of lysis buffer, 10 ml of resuspension buffer, and 0.5 ml of cobalt resin was used. After removal of the unbound sample, the resin was washed three times by rotating briefly and then sedimenting the resin as described above. The three washes were with 5 ml of WBB with 0.5% DM, 5 ml of WBB with 0.1% DM, and 1 ml of WBB with 0.1% DM and 30 mm imidazole. Finally the resin was eluted with 1 ml of WBB with 0.1% DM and 400 mm imidazole. The eluted material was loaded directly onto the gel filtration column as described above. Equilibration and elution of the column were as described above.
Disulfide Cross-linking of Purified Proteins
Samples were treated with 1 mm Cu2+(phenanthroline)3 or 3 mm 2-phenanthroline for 10 min at 37 °C followed by incubation with 12.5 mm neocuproine for 5 min at 37 °C to terminate the oxidation reaction. Samples were mixed with an equal volume of 2× sample buffer with or without DTT and incubated for 5 min at 70 °C; then equal volumes were subjected to SDS-PAGE and immunoblotting as described above.
Ion Mobility-Mass Spectrometry (IM-MS)
To purify the SpoIVFB-TEV-FLAG2 E44Q·Pro-σK(1–126)-His6 complex for IM-MS analysis, cells (7.5 g, grown in a fermentor) were fractionated as described above, except 40 ml of lysis buffer and 40 ml of resuspension buffer was used. The membrane fraction was treated with 1% n-dodecyl-β-d-maltoside (DDM) by rotating the mixture for 1 h at room temperature and then subjected to high-speed centrifugation as described above. The supernatant (40 ml) was cobalt affinity-purified as described above, except 1 ml of cobalt resin was used and 0.1% DDM was substituted for 1% DM in the resin equilibration buffer. After removal of the unbound sample, the resin was washed twice with 5 ml of equilibration buffer and then with 5 ml of WBB with 0.1% DDM and 40 mm imidazole by rotating briefly and then sedimenting the resin as described above. Finally, the resin was eluted with 3 ml of WBB with 0.1% DDM and 400 mm imidazole. The eluted material was concentrated to 0.5 ml and loaded onto the gel filtration column as described above, except the column was equilibrated with buffer containing 150 mm ammonium acetate, pH 7.5, and 0.02% DDM. The column was eluted with the same buffer at 0.5 ml/min, and 0.5-ml fractions were collected. Samples were subjected to SDS-PAGE, the gel was stained with Coomassie Blue, and 3 or 4 fractions containing the most SpoIVFB-TEV-FLAG2 E44Q·Pro-σK(1–126)-His6 complex were concentrated to 0.1 ml using an Amicon Ultra centrifugal device with a 100-kDa cut-off (Millipore), flash-frozen, and stored at −80 °C.
For IM-MS analysis, the purified SpoIVFB-TEV-FLAG2 E44Q·Pro-σK(1–126)-His6 complex was thawed, and buffer was exchanged into 200 mm ammonium acetate, pH 8.1, using Amicon Ultra 0.5 ml centrifugal devices with a 100-kDa cut-off (Millipore). To try to maintain the native-like membrane-bound state of the protein complex in solution, the buffers contained either 0.02% DDM or 0.5% tetraethylene glycol monooctyl ether (C8E4), in a manner similar to previously described methods (51).
All IM-MS experiments were carried out on a Synapt G2 (Waters, Milford, MA). Samples were ionized using a nanoelectrospray source operated in positive ion mode. The capillary voltage was operated between 1.0 and 1.6 kV, with the sample cone run at 40 V. Trap collision energy was maintained at 100 V, with the transfer region operated between 50 and 240 V. Mass spectrometry instrumentation was operated at a backing pressure of 6.25 mbar, an ion mobility pressure of 3.5 mbar, and a time-of-flight pressure of 9.9 × 10−7 mbar. Mass spectra were calibrated externally using a solution of cesium iodide (24 mg/ml, 30% isopropanol). Ions were mobility-separated in the gas phase under the influence of a traveling wave maintained at a wave height of 35 V and velocity of 550 m/s. Data were analyzed using MassLynx 4.1 and DriftScope 2.0 (Waters). The masses of all species observed were assigned according to previously published work (52).
Stepwise Photobleaching
B. subtilis strain BDR347 containing enhanced green fluorescent protein (eGFP) fused to the C terminus of SpoIVFB, resulting in a functional SpoIVFB-eGFP fusion protein (21), was induced to undergo sporulation by the resuspension method and cells were collected at 3 h poststarvation for imaging, using total internal reflection fluorescence (TIRF) microscopy with a Nikon A1Rsi microscope and a ×100 PLANapo oil objective. Samples were mounted on poly-l-lysine-coated coverslips. Excitation of eGFP used a 488-nm Argon laser at 3% intensity. Fluorescence was filtered using a 525/550-nm band pass filter. Images were collected continuously for 30 s (to allow photobleaching) using 40-ms exposures on a 16-bit Andor DU-897 X-9795 camera set at the 10 MHz readout mode. The Nikon Perfect focus system was used to keep cells in focus during imaging. The objective was moved to a new field of view before excitation and the start of data collection to avoid missed photobleaching events.
Small foci believed to represent SpoIVFB-eGFP molecules were selected, and for each a 3 × 3 pixel region of interest (ROI) was drawn centered over the most intense pixel in the first image of the time lapse series. The total intensity of each ROI during each exposure was recorded and exported from ImageJ (53). A noise reduction algorithm (NoRSE) was used to extract an average intensity profile that aided in counting photobleaching steps (54). This algorithm was used with MATLAB (MathWorks, version R2014a 8.3.0.532). To count steps, the raw intensity and the noise-reduced profiles were plotted together. A step was defined as a decrease in intensity that was greater than noise and for which the decrease was approximately equal to other steps for that ROI. Steps were counted until the end of the time lapse series or until steps stopped occurring (steps were followed by continuous bleaching of autofluorescence). ROIs were discarded if noise exceeded steps in a major portion of the profile, if photobleaching appeared continuous rather than stepwise, or if there were inconsistent step sizes. Confidence and statistical tests (γ and θ) were computed using R, version 3.2.2, to perform a Baysian analysis designed to test support for the inferred subunit stoichiometry (55).
The dye FM 4-64 was used to stain the membranes of sporulating cells as described previously (56). Briefly, samples collected as described above were treated with FM 4-64 (1 μg/ml) on ice for 30 min prior to imaging. FM 4-64 fluorescence was imaged using TIRF microscopy as described above with the following changes. Excitation used a 514-nm laser diode at 6% intensity, and emission was captured using a 600–650-nm band pass filter. Fluorescence from eGFP and step counting was carried out as described above after imaging FM 4-64. To merge the fluorescence from FM 4-64 and eGFP, duplicate FM 4-64 images of each field of view were first subjected to a 12-pixel-size Gaussian blur, which was subtracted from each original image to remove background fluorescence due to the poly-l-lysine-coated coverslips. The ImageJ “find edges” feature was used to reduce the diffuse edges of the membrane to more defined edges. The duplicate images were then averaged to reduce noise. For eGFP the first 5 images of the time series were averaged to reduce noise, and the average was convolved using the default settings of ImageJ. The resulting images from these processing strategies were then merged to show the localization of SpoIVFB-eGFP with respect to membranes of sporulating cells.
Results
Effect of Pro-σK(1–126)-His6 Coexpression on Solubilization of SpoIVFB from Membranes
We discovered that when Pro-σK(1–126)-His6 was coexpressed with catalytically inactive SpoIVFB-TEV-FLAG2 E44Q in E. coli, both proteins were readily solubilized by a variety of detergents (all detergents were used at 1% for solubilization of proteins from membranes). The ionic detergent sodium dodecanoyl sarcosine (sarkosyl), the nonionic detergents DM and DDM, the lipid-like detergents n-dodecylphoscholine and 1-myristoyl-2-hydroxy-sn-glycero-3-phosphocholine, and the zwitterionic detergents n-dodecyl-N,N-dimethylamine-N-oxide and 3-dodecylamido-N,N′-dimethylpropyl amine oxide appeared to be equally effective at solubilization, whereas the nonionic detergent octaethylene glycol monododecyl ether (C12E8) appeared to be less effective (Fig. 2B). These results were surprising, because a previous study had shown that when cytTM-SpoIVFB-FLAG2-His6 (TM-SpoIVFB) alone was expressed in E. coli, sarkosyl was much more effective at solubilization than DM or DDM (28). The difference was not due to the tobacco etch virus (TEV) protease cleavage site in SpoIVFB-TEV-FLAG2 E44Q, because when cytTM-SpoIVFB-TEV-FLAG2-His6 alone was expressed in E. coli, it was readily solubilized by sarkosyl but poorly solubilized by nonionic detergents (Fig. 2C), like TM-SpoIVFB (28). Also, cytTM-SpoIVFB-FLAG2 E44Q lacking the TEV site but having an extra TMS from rabbit cytochrome P450 2B4 (cytTM) (57), shown previously to enhance SpoIVFB accumulation in E. coli (28), was readily solubilized by DM only if Pro-σK(1–126)-His6 was coexpressed (Fig. 3D, lanes 2 and 10). We concluded that coexpression of Pro-σK(1–126)-His6 with SpoIVFB E44Q variants dramatically improves solubilization of the catalytically inactive enzyme from membranes with mild, nonionic detergents and also allows it to be solubilized with lipid-like or zwitterionic detergents.
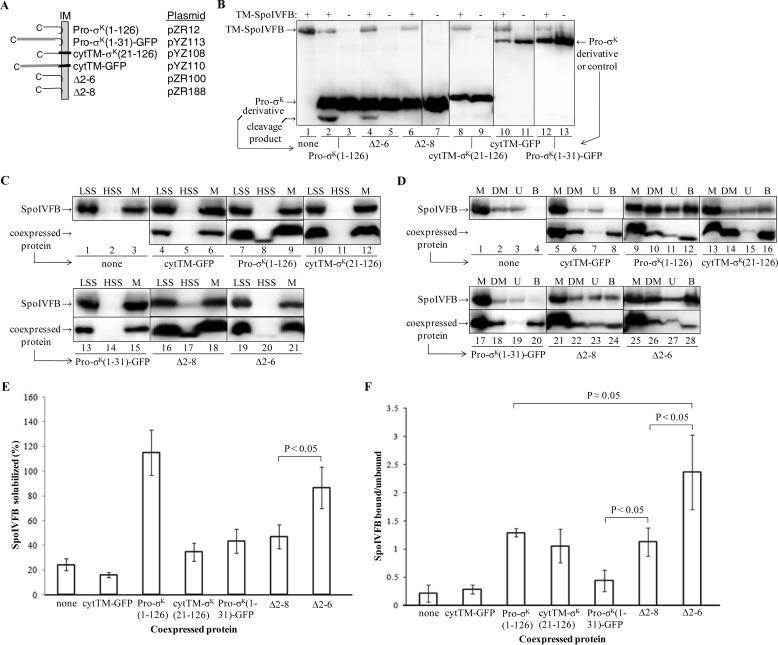
FIGURE 3.
Effect of Pro-σK(1–126)-His6 derivatives on solubilization of cytTM-SpoIVFB-FLAG2 E44Q and interaction with it. A, diagram of chimeras and deletion derivatives. Pro-σK(1–126)-His6 associates peripherally with the E. coli inner membrane (IM) (45). cytTM (thick black segment) is expected to insert into the inner membrane like a typical TMS. GFP is depicted as a gray segment. All proteins have a C-terminal His6 tag that is not shown. Chimeras and deletions are described in the text and are expressed from the listed plasmids. B, cleavage of Pro-σK(1–126)-His6 derivatives by TM-SpoIVFB. E. coli bearing pZR209 to express TM-SpoIVFB as indicated at the top and/or the plasmids listed in A to express Pro-σK(1–126)-His6 derivatives, indicated at the bottom, were cultured (10 ml) and induced with IPTG. Extracts were subjected to immunoblot analysis with anti-His antibodies. The cleavage product is indicated (lanes 2 and 4). Representative results from at least two biological replicates are shown. C, protein expression and initial fractionation. E. coli bearing pYZ68 to express cytTM-SpoIVFB-FLAG2 E44Q alone or in combination with the plasmids listed in A to coexpress Pro-σK(1–126)-His6 derivatives, indicated at the bottom, were cultured (1 liter) and induced with IPTG. Cell lysates were fractionated by centrifugation to produce low-speed supernatant (LSS), high-speed supernatant (HSS), and membrane fractions (M) containing cytoplasmic and membrane proteins, cytoplasmic proteins, and membrane proteins, respectively. Samples were subjected to immunoblot analysis with anti-FLAG (top) or anti-His (bottom) antibodies. D, solubilization of proteins and pulldown assays. This is the same as in C, except the membrane fraction was treated with DM to solubilize cytTM-SpoIVFB-FLAG2 E44Q and coexpressed protein followed by high-speed centrifugation and collection of a sample of the supernatant (DM). The rest of the supernatant was subjected to cobalt affinity purification of coexpressed protein and interacting cytTM-SpoIVFB-FLAG2 E44Q, resulting in unbound (U) and bound (B) samples, which were subjected to immunoblot analysis with anti-FLAG (top) or anti-His (bottom) antibodies. E, quantification of cytTM-SpoIVFB-FLAG2 E44Q solubilization by DM in the absence or presence of coexpressed proteins. The immunoblots shown in D plus two biological replicates were quantified, and the signal of the DM-solubilized sample (DM) was expressed as a percentage of the signal of the membrane fraction (M). Error bars show 1 S.D. The average with coexpressed Δ2–6 was significantly higher than with Δ2–8 (p < 0.05, Student's t test). F, quantification of cytTM-SpoIVFB-FLAG2 E44Q after cobalt affinity purification in the absence or presence of coexpressed proteins. The immunoblots shown in D plus two biological replicates were quantified, and the signals of the bound and unbound samples were expressed as a ratio. Error bars show 1 S.D. The indicated averages were compared using a Student's t test.
It is worth noting that when both Pro-σK(1–126)-His6 and SpoIVFB-TEV-FLAG2 E44Q were coexpressed from the same plasmid (pYZ42), cytTM was not necessary for the SpoIVFB E44Q variant to accumulate abundantly. This did not depend on the TEV site or the E44Q substitution, because SpoIVFB-FLAG2-His6 accumulated abundantly and cleaved Pro-σK(1–126)-His6 when both proteins were coexpressed from the same plasmid (pYZ6) (data not shown). In both plasmids, Pro-σK(1–126)-His6 is expressed from a T7 RNA polymerase promoter, and downstream a second such promoter drives expression of SpoIVFB. We did not investigate whether this arrangement of promoters or some other aspect of coexpression from the same plasmid would explain the abundant accumulation of SpoIVFB lacking cytTM. Also worth noting is that in the figures we labeled all SpoIVFB variants as “SpoIVFB” and all Pro-σK(1–126)-His6 variants as “Pro-σK” for simplicity and to conserve space, but in the figure legends and elsewhere in the text we have fully specified the variants used in the experiments for clarity.
Parts of Pro-σK(1–126)-His6 Required to Improve Solubilization of cytTM-SpoIVFB-FLAG2 E44Q from Membranes and to Form a Stable Complex
To determine whether parts of Pro-σK(1–126)-His6 would be sufficient to improve the solubilization of cytTM-SpoIVFB-FLAG2 E44Q from membranes with DM, we engineered chimeras containing part of Pro-σK(1–126)-His6 and either green fluorescent protein (GFP) or cytTM (Fig. 3A). We engineered Pro-σK(1–31)-GFP-His6 because previous studies indicated that the Pro part (residues 1–20) of Pro-σK is necessary for association with membranes (28, 47, 58) and that residues 1–27 fused to GFP allow membrane association upon expression in E. coli (47). Hence, this chimera was designed to test whether the Pro part plus a few residues of σK would be sufficient to improve the solubilization of SpoIVFB. Conversely, we engineered cytTM-σK(21–126)-His6 to test whether the σK part, associated with membrane via a generic TMS, would be sufficient to improve the solubilization of SpoIVFB. The TMS we used, cytTM, enhances the accumulation of a protein, as seen with TM-SpoIVFB (28), and/or promotes membrane association of a protein. As a negative control, we engineered cytTM-GFP-His6. We also tested two deletion derivatives of Pro-σK(1–126)-His6 reported recently to be membrane-associated when expressed in E. coli; one lacking residues 2–6 (Δ2–6-His6), which was cleaved by coexpressed TM-SpoIVFB in E. coli, and the other lacking residues 2–8 (Δ2–8-His6), which was not cleaved (45). As noted above, Δ2–6-His6, but not Δ2–8-His6, was reported to be cleaved by coexpressed TM-SpoIVFB in E. coli (45). We verified this finding and also found that none of the other His-tagged proteins were cleaved by coexpressed TM-SpoIVFB (Fig. 3B).
To measure solubilization from membranes and stable complex formation, each C-terminally His6-tagged protein described above was coexpressed in E. coli with cytTM-SpoIVFB-FLAG2 E44Q. As expected, cytTM-SpoIVFB-FLAG2 E44Q was present in the membrane fraction whether expressed alone or coexpressed with any of the His-tagged proteins, and each of the His-tagged proteins was predominantly in the membrane fraction, although small portions of Pro-σK(1–126)-His6 and its deletion derivatives remained in the cytosolic fraction after high-speed centrifugation (Fig. 3C). As noted above, cytTM-SpoIVFB-FLAG2 E44Q expressed alone was poorly solubilized from the membrane fraction by DM, and coexpressed Pro-σK(1–126)-His6 greatly improved its solubilization (Fig. 3D, lanes 2 and 10). In contrast, coexpressed cytTM-GFP-His6 did not improve the solubilization of cytTM-SpoIVFB-FLAG2 E44Q (Fig. 3D, lane 6), as expected, because cytTM-GFP-His6 does not contain any part of Pro-σK(1–126). Coexpressed cytTM-σK(21–126)-His6 (Fig. 3D, lane 14), Pro-σK(1–31)-GFP-His6 (lane 18), or Δ2–8-His6 (lane 22) did not appear to improve the solubilization of cytTM-SpoIVFB-FLAG2 E44Q as much as Pro-σK(1–126)-His6 (lane 10), but Δ2–6-His6 appeared to improve solubilization nearly as well (lane 26). Quantification from three experiments revealed that Δ2–6-His6 was significantly better than Δ2–8-His6 at improving the solubilization of cytTM-SpoIVFB-FLAG2 E44Q (Fig. 3E). Therefore, for all of the His-tagged proteins, including two proteins differing by just two residues, the ability to serve as substrate for the active enzyme (Fig. 3B) was positively correlated with the ability to improve the solubilization of the catalytically inactive enzyme (Fig. 3, D and E). We concluded that both the Pro(1–20) and σK(21–126) parts of Pro-σK(1–126)-His6 contribute to its ability to both serve as a substrate of TM-SpoIVFB and improve the solubilization of cytTM-SpoIVFB-FLAG2 E44Q from membranes.
It is worth noting that Pro-σK(1–126)-His6 itself was poorly solubilized by DM, and the other His-tagged proteins behaved similarly, except for cytTM-σK(21–126)-His6, which was readily solubilized (Fig. 3D, lane 14). Partial solubilization of Pro-σK(1–126)-His6 overproduced in E. coli was observed previously with the nonionic detergent Triton X-100 (1%), and this was attributed to interaction with itself and/or other cellular components resulting in detergent-insoluble aggregates too small to sediment upon low-speed centrifugation but large enough to sediment upon high-speed centrifugation (45).
Our experimental design allowed cobalt affinity purification of the His-tagged proteins to examine their interaction with cytTM-SpoIVFB-FLAG2 E44Q (i.e. pulldown assays). Proteins solubilized by DM were incubated with cobalt beads, and the unbound fraction was compared with the bound fraction using immunoblots. As expected, the major portion of each His-tagged protein was in the bound fraction (Fig. 3D), and cytTM-SpoIVFB-FLAG2 E44Q remained unbound when expressed alone (lanes 3 and 4) or in combination with cytTM-GFP-His6 (lanes 7 and 8). In contrast, the major portion of cytTM-SpoIVFB-FLAG2 E44Q was in the bound fraction upon coexpression with Pro-σK(1–126)-His6 (Fig. 3D, lanes 11 and 12), indicative of stable complex formation. Interestingly, cytTM-σK(21–126)-His6 appeared to interact with cytTM-SpoIVFB-FLAG2 E44Q, because a considerable portion was in the bound fraction (Fig. 3D, lanes 15 and 16), but this was not the case for Pro-σK(1–31)-GFP-His6 (lanes 19 and 20). Δ2–8-His6 (Fig. 3D, lanes 23 and 24) and especially Δ2–6-His6 (lanes 27 and 28) also appeared to interact with cytTM-SpoIVFB-FLAG2 E44Q. Quantification from three experiments (Fig. 3F) revealed that Δ2–6-His6 may be significantly better than Pro-σK(1–126)-His6 at forming a stable complex with cytTM-SpoIVFB-FLAG2 E44Q (p ≈ 0.05, Student's t test), suggesting that residues 2–6 inhibit the interaction of Pro-σK with SpoIVFB. Loss of just two more residues in Δ2–8-His6 or replacement of the Pro part with cytTM in cytTM-σK(21–126)-His6 resulted in similar levels of interaction, as observed for Pro-σK(1–126)-His6. As all the His-tagged proteins containing the σK part of Pro-σK(1–126)-His6 interacted with cytTM-SpoIVFB-FLAG2 E44Q better than did Pro-σK(1–31)-GFP-His6, we concluded that the σK part of Pro-σK(1–126)-His6 is required to form a stable complex with cytTM-SpoIVFB-FLAG2 E44Q upon solubilization from membranes with DM and that the Pro part is insufficient for stable complex formation under these conditions.
A Part of SpoIVFB Required for Pro-σK(1–126) to Improve Its Solubilization from Membranes and for Proximity between the SpoIVFB Active Site and the Cleavage Site in Pro-σK(1–126)
TM-SpoIVFB lacking the last 10 residues of SpoIVFB (TM-SpoIVFB CΔ10) failed to cleave coexpressed Pro-σK(1–126)-His6 in E. coli (28). As TM-SpoIVFB CΔ10 failed to interact productively with its substrate, we reasoned that Pro-σK(1–126) might be unable to improve the solubilization of TM-SpoIVFB CΔ10 from membranes, based on our results presented above. Indeed, we found that Pro-σK(1–126)-FLAG2 allowed solubilization of catalytically inactive TM-SpoIVFB E44Q from membranes with DM (Fig. 4A, lanes 1 and 2), but very little of its CΔ10 derivative was solubilized (lanes 3 and 4).
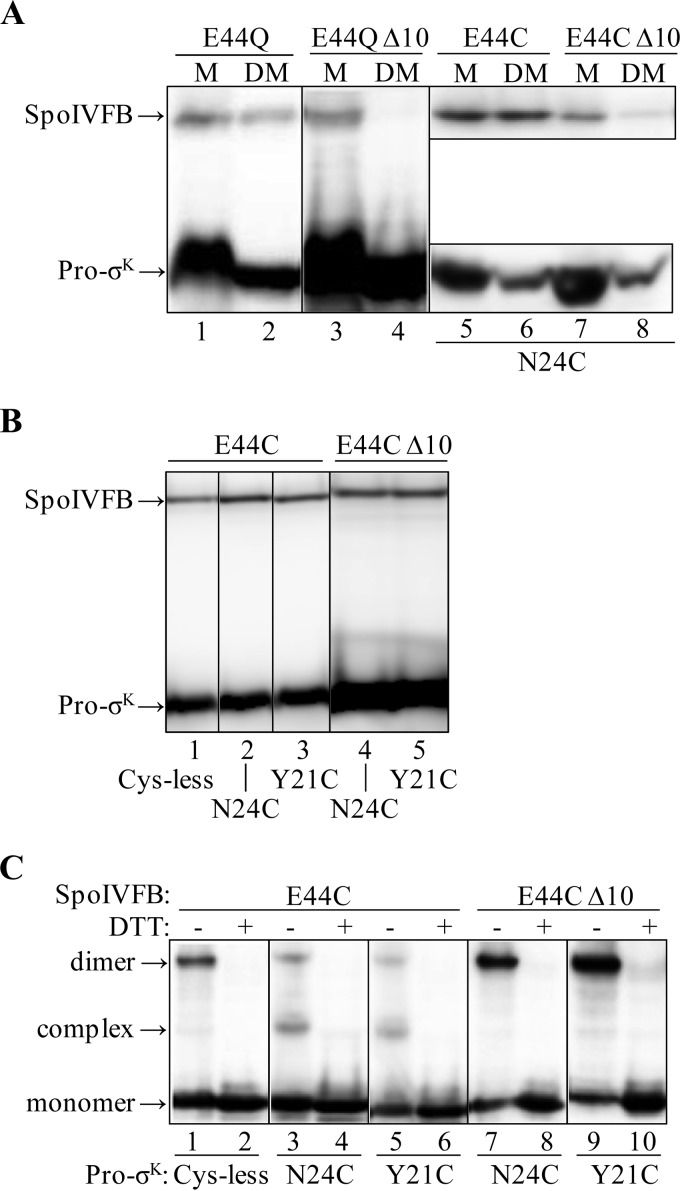
FIGURE 4.
Effects of deleting the last 10 residues of SpoIVFB. A, effects of Pro-σK(1–126) coexpression on the solubilization of TM-SpoIVFB from membranes. E. coli bearing pBS236 and pZR262 (lanes 1 and 2), pJZ7 and pZR262 (lanes 3 and 4), pYZ40 and pZR241 (lanes 5 and 6), or pYZ49 and pZR241 (lanes 7 and 8) were cultured (1 liter) and induced with IPTG to coexpress versions of TM-SpoIVFB, indicated at the top, and Pro-σK(1–126)-FLAG2 (lanes 1–4) or single-Cys Pro-σK(1–126)-His6 N24C (lanes 5–8). The membrane (M) fraction was treated with DM to solubilize the proteins followed by high-speed centrifugation and collection of a sample of the supernatant (DM). Samples were subjected to immunoblot analysis with anti-FLAG (lanes 1–4 and 5–8, top) or anti-His (lanes 5–8, bottom) antibodies. B, expression of proteins. E. coli bearing pYZ40 and pZR99 (lane 1), pYZ40 and pZR241 (lane 2), pYZ40 and pPL12 (lane 3), pYZ49 and pZR241 (lane 4), or pYZ49 and pPL12 (lane 5) were cultured (10 ml) and induced with IPTG to coexpress single-Cys versions of TM-SpoIVFB, indicated at the top, and Cys-less or single-Cys versions of Pro-σK(1–126)-His6, indicated at the bottom. Cell extracts were subjected to immunoblot analysis. Both proteins were detected with anti-His antibodies. C, disulfide cross-linking in vivo. E. coli bearing the same plasmid combinations and induced as in B were treated with 1 mm Cu2+(phenanthroline)3 for 10 min. Proteins were precipitated with TCA and resuspended in sample buffer with or without DTT as indicated. Antibodies against the FLAG tag on TM-SpoIVFB were used to detect monomer, complex with Pro-σK(1–126)-His6, and dimer. The results are presented in the same order as in B (two lanes for each plasmid combination). In A–C, representative results from at least two biological replicates are shown.
Because of the poor solubilization of the CΔ10 derivative, rather than attempting to implement a pulldown assay as described above, we employed an in vivo disulfide cross-linking assay that does not rely on the solubilization of proteins from membranes. Instead, it tests the ability of coexpressed proteins in E. coli to form a complex in which a residue at or near the active site of SpoIVFB is in close proximity to residues near the cleavage site in Pro-σK. because this assay used versions of the proteins that were engineered to each contain a single Cys residue, we first tested the ability of single-Cys Pro-σK(1–126)-His6 N24C, with a Cys residue near the cleavage site between residues 20 and 21, to allow solubilization of single-Cys TM-SpoIVFB E44C, which is inactive due to substitution of its catalytic Glu-44 residue with Cys (46). Consistent with the results described above, the full-length enzyme was solubilized from membranes with DM (Fig. 4A, lanes 5 and 6), but very little of its CΔ10 derivative was solubilized (lanes 7 and 8).
To ensure that single-Cys Pro-σK(1–126)-His6 N24C (or Y21C or a Cys-less version that served as a negative control) was not limiting for complex formation with single-Cys TM-SpoIVFB E44C or its CΔ10 derivative upon coexpression in E. coli, both His-tagged proteins were detected by immunoblot of whole-cell extracts with anti-His antibodies, and in each case Pro-σK(1–126)-His6 accumulated in excess of TM-SpoIVFB E44C or its CΔ10 derivative (Fig. 4B). Cells coexpressing the same combinations of enzyme and substrate were treated with the oxidant Cu2+(phenanthroline)3 to promote disulfide bond formation between Cys residues in proximity; then cells were lysed and proteins were precipitated by the addition of TCA, and finally samples were subjected to immunoblot analysis without or with DTT treatment to reverse disulfide cross-links. As shown previously (46), antibodies against the FLAG tags present in full-length TM-SpoIVFB E44C detected two species that migrated more slowly than the monomer in some of the samples (Fig. 4C), corresponding to a cross-linked complex with the N24C (lane 3) or Y21C (lane 5) versions of Pro-σK(1–126)-His6 and to a cross-linked dimer of TM-SpoIVFB E44C (lanes 1, 3, and 5), all of which were not present upon treatment with DTT, as expected (lanes 2, 4, and 6). Cys-less Pro-σK(1–126)-His6 cannot form a cross-linked complex, so the faint species migrating at a similar position as the cross-linked complex represents the background of the assay (Fig. 4C, lane 1). The CΔ10 derivative of TM-SpoIVFB E44C failed to form a cross-linked complex with the N24C (Fig. 4C, lane 7) or Y21C (lane 9) versions of Pro-σK(1–126)-His6, showing only the faint background species. We concluded that the CΔ10 derivative fails to interact with Pro-σK(1–126)-His6 in a way that brings the SpoIVFB active site into proximity with residues near the substrate cleavage site. The results do not distinguish between a lack of interaction and an altered interaction. In either case, the abnormal interaction presumably explains why TM-SpoIVFB CΔ10 fails to cleave Pro-σK(1–126)-FLAG2 (28) and why neither Pro-σK(1–126)-FLAG2 nor single-Cys Pro-σK(1–126)-His6 N24C coexpression facilitated solubilization of catalytically inactive TM-SpoIVFB from membranes with DM (Fig. 4A).
Purification and Characterization of SpoIVFB-FLAG2 E44Q·Pro-σK(1–126)-His6 Complexes
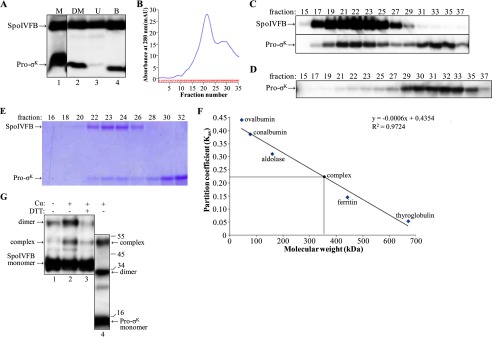
As noted above, Pro-σK(1–126)-His6 and SpoIVFB-TEV-FLAG2 E44Q both accumulate abundantly when coexpressed from pYZ42. We took advantage of this and used the cobalt affinity (pulldown) strategy described above for initial purification of a complex between the two proteins (Fig. 5A). After concentrating the bound fraction, it was subjected to gel filtration chromatography, monitoring the effluent absorbance at 280 nm and characterizing the fractions by immunoblot. The absorbance of the effluent reached a maximum in fractions 21–23, and this was followed by a smaller peak in fractions 29–31 (Fig. 5B). Immunoblotting showed that the larger peak contained both Pro-σK(1–126)-His6 and SpoIVFB-TEV-FLAG2 E44Q, whereas the smaller peak and subsequent fractions contained only Pro-σK(1–126)-His6 (Fig. 5C). Because Pro-σK(1–126)-His6 alone was retained for a longer time on the column, it may be monomeric and/or in relatively small multimeric complexes. Pro-σK(1–126)-His6 in complex with SpoIVFB-TEV-FLAG2 E44Q eluted earlier from the column, indicative of a relatively large complex. As a control, cells expressing only Pro-σK(1–126)-His6 were subjected to the same purification protocol, and Pro-σK(1–126)-His6 was not found in the early-eluting fractions from the gel filtration column (Fig. 5D). Taken together, the results suggest that Pro-σK(1–126)-His6 forms a complex with SpoIVFB-TEV-FLAG2 E44Q that is stable during gel filtration chromatography.
FIGURE 5.
Purification and characterization of the SpoIVFB-TEV-FLAG2 E44Q·Pro-σK(1–126)-His6 complex. A, cobalt affinity purification. E. coli bearing pYZ42 was grown in a fermentor and induced with IPTG to coexpress Pro-σK(1–126)-His6 and SpoIVFB-TEV-FLAG2 E44Q. The membrane (M) fraction was treated with DM to solubilize the complex followed by high-speed centrifugation and collection of a sample of the supernatant (DM), and the rest of the supernatant was subjected to cobalt affinity purification, resulting in unbound (U) and bound (B) samples, which were subjected to immunoblot analysis with anti-FLAG and anti-His antibodies. B, gel filtration chromatography. The bound fraction from the cobalt affinity purification was concentrated and applied to a size exclusion column. The absorbance at 280 nm was monitored as fractions were collected, generating the profile shown. C, immunoblot. Samples of fractions from the size exclusion column were subjected to immunoblot analysis with anti-FLAG (top) or anti-His (bottom) antibodies. D, purification of Pro-σK(1–126)-His6 alone. E. coli bearing pZR27 was grown in a fermentor and induced with IPTG to express Pro-σK(1–126)-His6. The membrane fraction was treated with DM, and the supernatant after high-speed centrifugation was subjected to cobalt affinity purification. The bound fraction was concentrated and applied to a size exclusion column, and samples of fractions were subjected to immunoblot analysis with anti-His antibodies. E, samples of fractions from a size exclusion column were subjected to SDS-PAGE as described for immunoblot analysis, but in this case gel electrophoresis was followed by Coomassie Blue staining of proteins. F, molecular weight estimate of the complex. The size exclusion column was calibrated with the indicated proteins. The partition coefficient was calculated as Kav = (elution volume − column void volume)/(geometric column volume − column void volume). The equation of the best fit line and the correlation coefficient are shown, as is the estimated molecular mass of the complex (360 kDa, which includes associated detergent) based on its Kav (0.22). G, disulfide cross-linking. Single-Cys versions of cytTM-SpoIVFB-FLAG2 E44C and Pro-σK(1–126)-His6 K23C were coexpressed in E. coli, and the complex was purified as described for A and B. Fractions 22–24 were pooled, and samples of the pooled fractions were treated with 1 mm Cu2+(phenanthroline)3 (Cu) for 10 min (+) or with 3 mm 2-phenanthroline for 10 min (−) as a control and then mixed with sample buffer with or without DTT as indicated and subjected to immunoblot analysis with antibodies against the FLAG tag on single-Cys cytTM-SpoIVFB-FLAG2 E44C (lanes 1–3) or against the His6 tag on single-Cys Pro-σK(1–126)-His6 K23C (lane 4). The immunoblot was cut between lanes 3 and 4 and realigned based on the migration of prestained marker proteins, for which sizes in kDa are indicated along the right side. Representative results from two biological replicates are shown.
We repeated the purification several times and found the absorbance pattern in the effluent of the gel filtration column to be reproducible (data not shown). Fractions from the column were subjected to SDS-PAGE followed by Coomassie Blue staining of proteins. The early-eluting peak fractions 22–24 contain predominantly two species that migrate as expected for Pro-σK(1–126)-His6 (15 kDa) and SpoIVFB-TEV-FLAG2 E44Q (37 kDa), whereas the late-eluting fractions 28–32 contain predominantly one species migrating as expected for Pro-σK(1–126)-His6 (Fig. 5E). Because the purification of the complex appeared to be quite effective, we quantified the two species in peak fraction 23 in this and another experiment. The upper species was 3-fold more intense than the lower species. If the upper species is exclusively SpoIVFB-TEV-FLAG2 E44Q and the lower species is exclusively Pro-σK(1–126)-His6, and if the two proteins stain equally well, the upper species should be 2.5-fold more intense than the lower species if the complex has 1:1 stoichiometry. Therefore, Pro-σK(1–126)-His6 appeared to be slightly substoichiometric compared with SpoIVFB-TEV-FLAG2 E44Q in the early-eluting peak fraction.
To estimate the size of the protein complex in the early-eluting peak fraction, the gel filtration column was calibrated. The partition coefficient (Kav) for each protein standard was calculated and plotted against its molecular weight, and then the Kav of the protein complex (0.22) was used to estimate its molecular mass, yielding 360 kDa (Fig. 5F). This estimate is slightly larger than the estimate of 320 kDa for TM-SpoIVFB solubilized from membranes with sarkosyl after expression in E. coli as found previously (28); in that study, it was assumed that detergent associated with protein doubles the apparent molecular weight, so it was inferred that TM-SpoIVFB is tetrameric. If the same assumption is made for the complex of Pro-σK(1–126)-His6 with SpoIVFB-TEV-FLAG2 E44Q, our data suggest that the protein complex is about 180 kDa, which is close to the size expected (208 kDa) for a complex containing four monomers of each protein. Some complexes may contain less than four monomers of Pro-σK(1–126)-His6, as it appeared to be slightly substoichiometric compared with SpoIVFB-TEV-FLAG2 E44Q in the early-eluting peak fraction described above.
To test whether a purified SpoIVFB-FLAG2 E44Q·Pro-σK(1–126)-His6 complex resembles the complex formed when the two proteins are coexpressed in E. coli, we employed single-Cys versions of the proteins so that purified complexes could be subjected to disulfide cross-linking to probe the proximity between the enzyme active site and the substrate cleavage site. We constructed single-Cys cytTM-SpoIVFB-FLAG2 E44C for this experiment and coexpressed it in E. coli with single-Cys Pro-σK(1–126)-His6 K23C, because E44C of the enzyme had been shown previously to cross-link abundantly with K23C of the substrate upon coexpression in vivo (46). Cobalt affinity purification followed by gel filtration chromatography and immunoblotting yielded results similar to those presented in Fig. 5, A–C (data not shown). Both single-Cys Pro-σK(1–126)-His6 K23C and single-Cys cytTM-SpoIVFB-FLAG2 E44C reached a maximum in early-eluting fractions 22–24; late-eluting fractions 29–31 contained predominantly single-Cys Pro-σK(1–126)-His6 K23C. Fractions 22–24, pooled and treated with Cu2+(phenanthroline)3 to promote disulfide bond formation, were then subjected to immunoblot analysis without or with DTT treatment to reverse cross-links. Single-Cys Pro-σK(1–126)-His6 K23C could be cross-linked to single-Cys cytTM-SpoIVFB-FLAG2 E44C, forming a complex that migrates more slowly than the monomer of single-Cys cytTM-SpoIVFB-FLAG2 E44C (Fig. 5G, lane 2). This species, as well as one that migrated at the position expected for a dimer of single-Cys cytTM-SpoIVFB-FLAG2 E44C, was much less abundant upon treatment with DTT, as expected for reversal of disulfide cross-links (Fig. 5G, lane 3). An immunoblot of the cross-linked sample was probed with anti-His antibodies to detect single-Cys Pro-σK(1–126)-His6 K23C, confirming its presence in the species migrating at the position expected for a cross-linked complex (Fig. 5G, lane 4). A species migrating at the position expected for cross-linked single-Cys Pro-σK(1–126)-His6 K23C dimer was also observed (Fig. 5G, lane 4). We concluded that in the purified complex, the active site of cytTM-SpoIVFB-FLAG2 (represented by E44C) is in close proximity to the cleavage site in Pro-σK(1–126)-His6 (represented by K23C), suggesting that the purified complex resembles the complex formed in vivo.
IM-MS Analysis of the SpoIVFB-TEV-FLAG2 E44Q·Pro-σK(1–126)-His6 Complex
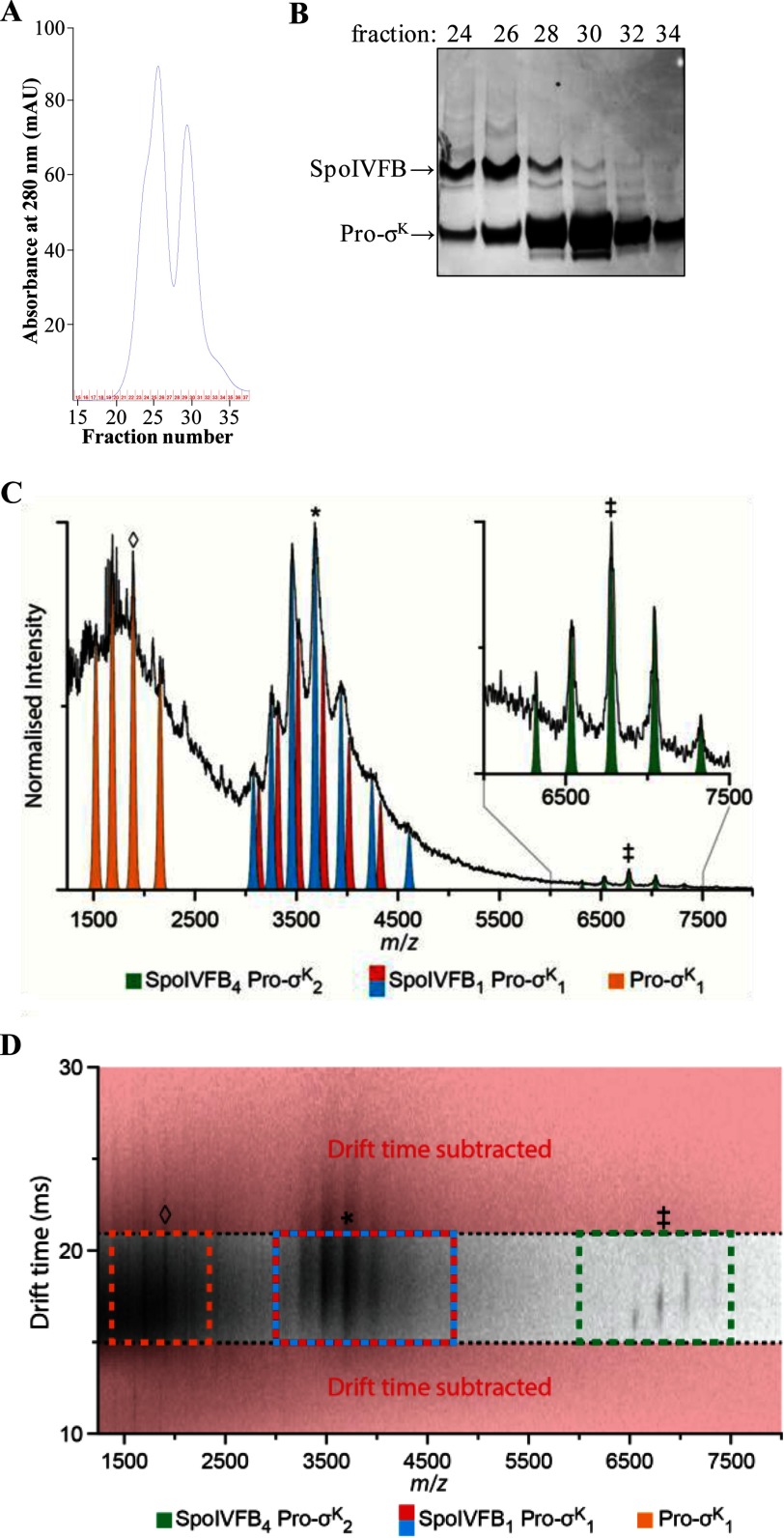
To purify the complex for IM-MS analysis, we modified the purification procedure to produce samples containing low levels of DDM or C8E4 detergent in ammonium acetate buffer (see “Experimental Procedures” for details). Briefly, we substituted DDM for DM during cobalt affinity purification, and we used ammonium acetate buffer containing a low concentration of DDM (0.02%) during gel filtration chromatography. The changes resulted in a higher yield of the complex with a slightly different elution profile in which the larger peak was preceded by a shoulder and the smaller peak was more prominent (compare Fig. 6, A and B). This appears to reflect more dissociation of Pro-σK(1–126)-His6 from SpoIVFB-TEV-FLAG2 E44Q during gel filtration chromatography (Fig. 6B). Fractions 25–27 containing the most SpoIVFB-TEV-FLAG2 E44Q·Pro-σK(1–126)-His6 complex were pooled, concentrated, and buffer-exchanged into ammonium acetate buffer containing low levels of DDM or C8E4 for IM-MS analysis.
FIGURE 6.
Purification of the SpoIVFB-TEV-FLAG2 E44Q·Pro-σK(1–126)-His6 complex and native IM-MS analysis. A, gel filtration chromatography. The bound fraction from the cobalt affinity purification was concentrated and applied to a size exclusion column. The absorbance at 280 nm was monitored as fractions were collected, generating the profile shown. B, samples of fractions from the size exclusion column were subjected to SDS-PAGE as described for immunoblot analysis, but in this case gel electrophoresis was followed by Coomassie Blue staining of proteins. C, analysis of the SpoIVFB-TEV-FLAG2 E44Q·Pro-σK(1–126)-His6 complex prepared in a buffer containing 0.2% DDM revealed a mass of 183.2 kDa (green; ‡ = +27), agreeing well with a stoichiometry of 4:2 (SpoIVFB4 Pro-σK2). Applying post-IM CID methods (drift time axis is presented in D), three mobility time-aligned dissociation products were observed: monomeric Pro-σK(1–126)-His6 (Pro-σK1, orange; ♢ = +8) and two masses closely agreeing with a 1:1 complex (SpoIVFB1 Pro-σK1, blue and red; * = +15). The mass difference between these two products (blue, 55.3 kDa; red, 56.6 kDa) suggests that some lipid or detergent molecules remain bound in the complex. D, post-IM CID analysis. To enable the stoichiometry assignment of the complex, drift time subtraction was used to remove those ions not aligning with the antecedent precursor ions (red). Drift time subtraction was performed post-process using software packages outlined under “Experimental Procedures.” SpoIVFB4 Pro-σK2 (green), ‡ = +27; SpoIVFB1 Pro-σK1 (blue and red), * = +15; Pro-σK1 (orange), ♢ = +8. In A–D, representative results from two technical replicates of the purification and IM-MS analysis are shown.
To identify the mass of the complex, we used gentle collision-induced dissociation (CID) conditions to remove lipid and detergent molecules bound to the surface of the protein complex. Analysis resolved an observed mass of 183.2 kDa (Fig. 6C), agreeing well with the expected mass of a 4:2 SpoIVFB-TEV-FLAG2 E44Q·Pro-σK(1–126)-His6 complex (176.4 kDa). The difference between the observed and expected masses (6.8 kDa) is likely due to lipid or detergent molecules that remained bound to the complex (51).
Samples prepared in DDM were further heated collisionally upon post-ion mobility (post-IM) separation to confirm our stoichiometry assignment. This approach exploited the fact that product ions produced by post-IM CID share the same arrival time as their antecedent precursor ions, enabling the mass assignment of precursor stoichiometry through the accurate correlation of precursor and product ions (59–62). This method was used because lipid and detergent adducts inhibited the quadrupole isolation of the 4:2 complex. Post-IM CID analysis of the data presented in Fig. 6C (transfer voltage 240 V, drift time dimension (Fig. 6D)) enabled the identification of three dissociation products from the 4:2 complex. Mass analysis of these dissociation products supported the identification of monomeric Pro-σK(1–126)-His6, along with two different masses in agreement with a 1:1 SpoIVFB-TEV-FLAG2 E44Q·Pro-σK(1–126)-His6 complex (55.3 and 56.6 kDa, respectively). These two closely related masses support the conclusion that some lipid or detergent molecules are retained within the complex (51).
Additional analyses using 0.5% C8E4 supported the identification of further masses corresponding to monomeric SpoIVFB-TEV-FLAG2 E44Q and 1:2 and 4:2 SpoIVFB-TEV-FLAG2 E44Q·Pro-σK(1–126)-His6 complexes (data not shown). A list of the observed masses and charge state distributions for these complexes, as well as those from Fig. 6, C and D, is available upon request. Although we do not understand why post-IM CID analysis detected different dissociation products when the complex was buffer-exchanged into two different detergents, in both cases the native IM-MS analysis resolved an observed mass that agreed well with the expected mass of a 4:2 SpoIVFB-TEV-FLAG2 E44Q·Pro-σK(1–126)-His6 complex.
Stepwise Photobleaching of SpoIVFB-eGFP in Vivo
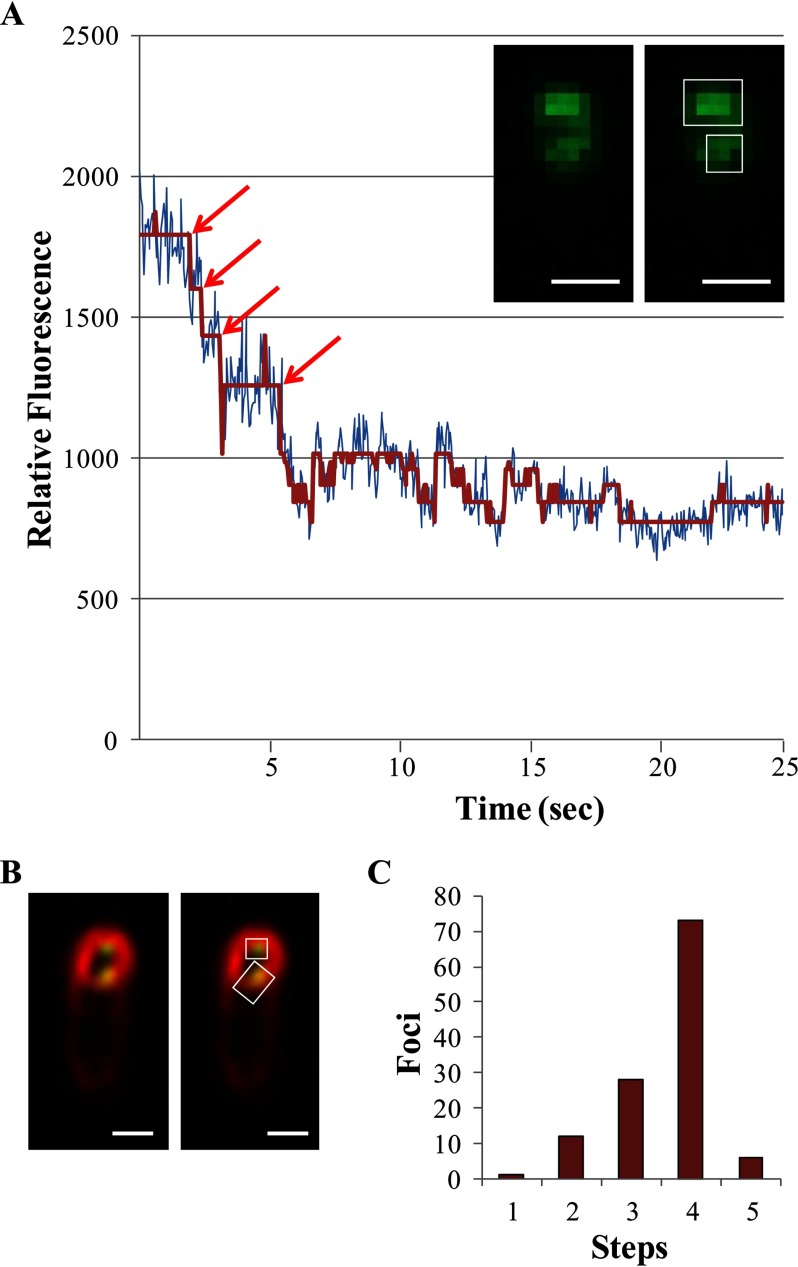
The in vitro analyses described above predicted that SpoIVFB is tetrameric. To test this prediction in vivo, stepwise photobleaching was performed on SpoIVFB fused at its C terminus to eGFP and expressed from the native spoIVF promoter during sporulation. Photobleaching of a fluorescent protein can decrease its fluorescence intensity in a stepwise fashion over time, revealing the number of subunits in a multisubunit protein (63–67). We used TIRF microscopy to limit excitation to the evanescent field and only excite SpoIVFB-eGFP near the coverslip. Small, fluorescent foci believed to represent individual molecules were analyzed for the number of photobleaching steps observed before continuous photobleaching of autofluorescence ensued. Most fluorescent foci were larger and likely represented multiple molecules (data not shown); these were not analyzed. The inset in Fig. 7A shows representative large and small foci in a single sporulating cell. For the small focus, the raw fluorescence intensity as a function of time during excitation and the noise-reduced profile were plotted together to illustrate the type of data used to count photobleaching steps (Fig. 7A). The small foci with countable photobleaching steps appeared to be localized to the mother cell membrane engulfing the forespore (Fig. 7B), as expected for sporulating cells at 3 h poststarvation based on standard fluorescence microscopy of SpoIVFB-GFP and membranes stained with FM 4-64 reported previously (42). The number of steps counted for many such foci is summarized in Fig. 7C. The majority of foci exhibited 4 steps, consistent with SpoIVFB-eGFP being tetrameric. Statistical analysis of the data supported this conclusion; using Bayesian inference, confidence was only slightly higher for 4 steps than 5 steps, but γ analysis suggested that a count of 5 steps was erroneous (Table 1). Also, the value of θ, representing the likelihood of observing every photobleaching event given the data, was highest for 4 steps and decreased with increasing step numbers (note that although 6-step profiles were not observed, the θ value for 6 steps was calculated to show that unobserved steps beyond step 5 would be unlikely). We concluded that stepwise photobleaching provides evidence that SpoIVFB is tetrameric in sporulating cells, in agreement with predictions based on in vitro analyses of the SpoIVFB-TEV-FLAG2 E44Q·Pro-σK(1–126)-His6 complex (Figs. 5 and 6) and of TM-SpoIVFB solubilized from membranes with the harsh, ionic detergent sarkosyl (28).
FIGURE 7.
Stepwise photobleaching of SpoIVFB-eGFP in sporulating B. subtilis. A, representative data showing relative fluorescence from a small focus over time. The inset shows the same TIRF microscopy image twice, that of a single sporulating cell at 3 h poststarvation (bar = 1 μm). The cell has distinct large and small fluorescent foci indicated by the rectangle and square, respectively, on the rightmost image. The large focus was excluded from the analysis. The graph shows the relative fluorescence of the small focus (i.e. the 3 × 3 pixel region of interest indicated by the square) at the indicated times after excitation. Both the raw intensity (blue) and noise-reduced (red) profiles are shown. The red arrows indicate the beginning of a photobleaching event. Four steps are observed followed by continuous photobleaching of autofluorescence. B, representative data showing fluorescence from SpoIVFB-eGFP (green) and membrane staining with FM 4-64 (red) of sporulating B. subtilis. The same merged TIRF microscopy image is shown twice, that of a single sporulating cell at 3 h poststarvation (bar = 1 μm). The cell has two foci of SpoIVFB-eGFP fluorescence associated with the membranes surrounding the forespore. The intense staining with FM 4-64 of the membranes surrounding the forespore indicates that engulfment was still in progress because the membranes surrounding the forespore become inaccessible to the dye upon completion of engulfment. As shown in A, the cell has distinct large and small fluorescent foci indicated by the rectangle and square, respectively, on the rightmost image. The large focus was excluded from the analysis. The small focus exhibited 4 steps of photobleaching (data not shown). C, summary showing the number of small foci exhibiting the indicated number of photobleaching steps. For statistical analysis, there were 1, 12, 28, 73, and 6 foci with 1–5 steps, respectively.
TABLE 1.
Statistical analysis of stepwise photobleaching data
a No foci with 6 photobleaching steps were observed, but θ could nevertheless be calculated, showing that the probability of having missed any 6-step profiles was relatively small.
b Not applicable.
Discussion
Our results include several important advances: 1) the discoveries that substrate improves the solubilization of inactive SpoIVFB from membranes with mild detergent and that the complex is stable during purification; 2) the identification of parts of SpoIVFB and Pro-σK important for solubilization and complex formation; 3) the use of IM-MS to determine the mass and composition of the complex, which agreed well with a 4:2 enzyme·substrate complex; and 4) the use of stepwise photobleaching to obtain evidence that SpoIVFB is tetrameric during sporulation. The implications of these advances are discussed below.
Solubilization of an IMMP·Substrate Complex from Membranes
Our discoveries that Pro-σK(1–126)-His6 improves the solubilization of catalytically inactive SpoIVFB from membranes with mild detergents and that the complex is stable during purification are important advances for several reasons. First, proteins solubilized with mild, nonionic detergents are more likely to retain their native structure and be amenable to crystallization efforts (68). The process of membrane protein solubilization by detergents is complex and not well understood. It appears typically to involve replacement of protein-lipid interactions with protein-detergent interactions in micelles, although some lipids often remain associated with the protein (68). Ionic detergents have charged head groups that in many cases facilitate membrane protein solubilization but may also alter protein structure and diminish activity. The ionic detergent sarkosyl was used previously for initial solubilization of overexpressed TM-SpoIVFB from E. coli membranes followed by a transition to nonionic detergent in an effort to preserve activity, which proved successful (28). However, partial loss of activity and alteration of structure could not be ruled out. It seemed possible that the interaction of the substrate with the enzyme might change its oligomeric state and/or its interactions with lipids in a way that facilitated solubilization from membranes. In recent studies of other types of IPs, small substrate mimics or inhibitors induced conformational changes ranging from modest shifts of TMSs and loops seen in rhomboid GlpG crystal structures (69–71) to widespread domain movements observed in presenilin-containing γ-secretase complexes using cyroelectron microscopy methods (72). Our results show that the SpoIVFB-TEV-FLAG2 E44Q·Pro-σK(1–126)-His6 complex is efficiently solubilized from membranes with nonionic detergents, as well as with lipid-like zwitterionic detergents (Fig. 2A). After affinity purification, the complex was fairly stable during gel filtration chromatography, although the large peak of absorbance at 280 nm was broad (Fig. 5B), suggesting some degree of heterogeneity in the composition of the complex. If the oligomeric complex dissociates slowly after affinity purification, a variety of smaller complexes could be produced and result in peak broadening. In some experiments, a very small peak preceded the large peak, and the small peak contained SpoIVFB-TEV-FLAG2 E44Q, based on immunoblotting, and eluted near the void volume of the column (data not shown), suggesting that protein aggregates had formed. In other experiments, the small peak was not observed, but the large peak was preceded by a shoulder (Fig. 5B) containing SpoIVFB-TEV-FLAG2 E44Q (Fig. 5C). SpoIVFB may have a tendency to aggregate after dissociation from Pro-σK(1–126)-His6. It might be necessary to optimize detergent and salt in the buffer to stabilize the SpoIVFB-TEV-FLAG2 E44Q·Pro-σK(1–126)-His6 complex to facilitate further biochemical analyses and future structural studies (e.g. x-ray crystallography and cryoelectron microscopy).
A second reason that our discoveries are important is that other IMMP·substrate complexes might exhibit similar solubilization and stability characteristics as the SpoIVFB-TEV-FLAG2 E44Q·Pro-σK(1–126)-His6 complex. To our knowledge, the only other IMMPs reported to be successfully solubilized from membranes are a fragment of a Methanocaldococcus jannaschii enzyme (referred to as mjS2P) lacking its CBS domain (17) and E. coli RseP (73). mjS2P produced in E. coli was solubilized from membranes with 1% DM, and it crystallized as a dimer; but the relevance of these observations to the full-length enzyme is unknown, as CBS domains typically promote oligomerization (34). RseP does not belong to the SpoIVFB subfamily, as it does not contain a CBS domain (31). Rather, RseP belongs to a large subfamily of IMMPs that contain one or more PDZ domains. RseP was readily solubilized from membranes with 1% DDM in the absence of its substrate (73). Also, catalytically inactive RseP could be solubilized from membranes with 1% DDM and co-immunoprecipitated with its substrate, RseA, suggesting that the two proteins form a fairly stable complex (50). This result, together with our discovery that the SpoIVFB-TEV-FLAG2 E44Q·Pro-σK(1–126)-His6 complex is fairly stable during purification, suggests that catalytically inactive IMMPs may form stable complexes with their substrates, allowing the substrates to be identified. For example, using a His-tagged IMMP with an inactivating substitution in place of the catalytic Glu residue of its easily recognizable HEXXH motif, it may be possible to solubilize complexes from membranes, affinity-purify them, and identify putative substrates by mass spectrometry after protease digestion. The conditions under which the substrate is associated with its cognate IMMP must be known for this approach to work. At least one IMMP in the RseP subfamily, Rip1 of Mycobacterium tuberculosis, requires a substrate-specific adapter protein, Ppr1, in order to interact with its substrate, RsmA (74). Such adapter proteins might also be identified by the approach just described.
A third reason that our discoveries are important is that they suggest a strategy to facilitate further biochemical analyses of active IMMPs that are difficult to solubilize from membranes, such as SpoIVFB. For example, coexpression of active SpoIVFB with uncleavable substrate in E. coli should result in a complex that can be solubilized from membranes with nonionic detergent and purified. If the uncleavable substrate can be exchanged for a cleavable one (e.g. upon the addition of ATP), this strategy might result in a more active enzyme than a previous purification strategy using the harsh ionic detergent sarkosyl for initial solubilization of TM-SpoIVFB from membranes (28). Recent progress toward identifying the features of Pro-σK important for cleavage by SpoIVFB revealed several single-residue substitutions in Pro-σK(1–126)-His6 (e.g. S20W, V22P, V22N, and V22G) that virtually eliminate cleavage by coexpressed TM-SpoIVFB (45). These uncleavable substrates are candidates for use in the purification strategy just described.
Parts of SpoIVFB and Pro-σK Important for Solubilization and Complex Formation
Both the Pro(1–20) and σK(21–126) parts contribute to improving the solubilization of cytTM-SpoIVFB-FLAG2 E44Q from membranes with 1% DM (Fig. 3, D and E). Analysis of two deletion derivatives of Pro-σK(1–126)-His6 provided further insight into the requirement for the Pro part. Δ2–6-His6 and Δ2–8-His6 differ by just two residues, yet Δ2–6-His6 improved enzyme solubilization significantly more than Δ2–8-His6. This implies that Ala7 and Leu8 of Pro-σK(1–126)-His6 change its interaction with the membrane and/or with SpoIVFB. In agreement, Δ2–6-His6 but not Δ2–8-His6 was observed previously to be cleaved by coexpressed TM-SpoIVFB in E. coli (45), as confirmed here (Fig. 3B, lanes 4 and 6). Slightly more Δ2–8-His6 than Δ2–6-His6 was in the supernatant after high-speed centrifugation (Fig. 3C, lanes 17 and 20), suggesting that slightly less Δ2–8-His6 was membrane-associated; but neither was wild-type Pro-σK(1–126) found exclusively in the membrane fraction (Fig. 3C, lane 8), so it seems unlikely that the slight difference in fractionation accounts for the observed differences in the solubilization of cytTM-SpoIVFB-FLAG2 E44Q and in cleavage by active TM-SpoIVFB. Rather, it seems likely that Δ2–6-His6 and Δ2–8-His6 differ subtly in their interaction with the membrane and/or with SpoIVFB. The Pro sequence, MTGVFAALGFVVKELVFLVS, has two hydrophobic regions separated by two adjacent, charged residues (Lys13 and Glu14). It was proposed that the charged residues interact with the membrane surface and the hydrophobic regions loop into the membrane, because Pro-σK(1–126)-His6 appeared to interact peripherally with the E. coli inner membrane (45). If so, the absence of Ala7 and Leu8 in the Pro part of Δ2–8-His6 might change the way its first hydrophobic region interacts with the membrane, perturbing its interaction with SpoIVFB.
The last 10 residues of SpoIVFB in catalytically inactive TM-SpoIVFB are also necessary for Pro-σK(1–126)-His6 to improve its solubilization from membranes with mild detergents (Fig. 4A). The single-Cys version of TM-SpoIVFB E44C lacking the 10 residues failed to interact normally with single-Cys versions of Pro-σK(1–126)-His6 in in vivo disulfide cross-linking assays (Fig. 4C). Although it is tempting to speculate that the C-terminal 10 residues of SpoIVFB interact directly with Pro-σK(1–126)-His6, we cannot rule out the possibility that the 10 residues are important for folding of the CBS domain and that other parts of the SpoIVFB CBS domain interact directly with the substrate.
The σK(21–126) part, but not the Pro(1–20) part, of Pro-σK(1–126) forms a stable complex with cytTM-SpoIVFB-FLAG2 E44Q when the parts of Pro-σK(1–126) are fused to heterologous domains. This conclusion is based on pulldown assays in which very little cytTM-SpoIVFB-FLAG2 E44Q bound to Pro-σK(1–31)-GFP-His6 and considerably more of the enzyme bound to cytTM-σK(21–126)-His6 (Fig. 3, D, lanes 16 and 20, and F). The inability of Pro-σK(1–31)-GFP-His6 to form a stable complex with the enzyme may reflect an inability to interact at all, which would explain the lack of cleavage (Fig. 3B, lane 12). On the other hand, despite the ability of cytTM-σK(21–126)-His6 to form a stable complex with the enzyme, it too is not cleaved (Fig. 3B, lane 8). cytTM-σK(21–126)-His6 may be unsuitable as a substrate for two reasons. First, cytTM likely interacts with membranes differently than the Pro part of Pro-σK(1–126)-His6. cytTM is expected to insert into membranes like a typical TMS (57), whereas the Pro sequence appears to interact peripherally with membranes (45), as discussed above. Second, certain residues near the cleavage site in Pro-σK(1–126)-His6 are important for cleavage to occur (45). cytTM-σK(21–126)-His6 may not contain a sequence of residues suitable for cleavage.
How does the σK part of Pro-σK(1–126)-His6 interact with SpoIVFB? Previously, it was shown that Pro-σK(1–126)-FLAG2 interacts with the CBS domain of SpoIVFB (28). It will be important to determine whether σK(21–126) interacts with the CBS domain and if so, how. Our purification of the SpoIVFB-TEV-FLAG2 E44Q·Pro-σK(1–126)-His6 complex lays the foundation for approaches like chemical cross-linking followed by protease digestion and mass spectrometry to identify residues of the two proteins that are in proximity. At least with respect to proximity between the active site of SpoIVFB (represented by E44C) and the cleavage site in Pro-σK(1–126)-His6 (represented by Y21C, K23C, or N24C), a purified complex resembled the complex formed in vivo, based on disulfide cross-linking results (Figs. 4C and 5G) (46).
Interestingly, five residues near the N terminus of Pro-σK(1–126)-His6 appear to inhibit its interaction with SpoIVFB slightly. Δ2–6-His6, lacking the five residues, appeared to pull down cytTM-SpoIVFB-FLAG2 E44Q more efficiently than did Pro-σK(1–126)-His6 (Fig. 3, D and F). The loss of just two more residues in Δ2–8-His6 restored the pulldown efficiency to that observed for Pro-σK(1–126)-His6 and cytTM-σK(21–126)-His6. Although Δ2–6-His6 appeared to interact more strongly than Pro-σK(1–126)-His6 with the enzyme in the pulldown assay, Pro-σK(1–126)-His6 appeared to be cleaved more efficiently (Fig. 3B, lanes 2 and 4). This suggests that a slightly weaker interaction benefits cleavage, perhaps by allowing better positioning of the substrate in the enzyme active site.
IM-MS Analysis of an IP·Substrate Complex
We used IM-MS to determine the composition of the SpoIVFB-TEV-FLAG2 E44Q·Pro-σK(1–126)-His6 complex. To our knowledge, this is the first time the composition of an IP·substrate complex has been determined. The detergent and buffer conditions used to prepare samples of the complex for IM-MS analysis appeared to cause more dissociation of Pro-σK(1–126)-His6 from SpoIVFB-TEV-FLAG2 E44Q during gel filtration chromatography, but analysis of pooled fractions containing the complex, after buffer exchange into two different detergents, resulted in an observed mass that agreed well with the expected mass of a 4:2 SpoIVFB-TEV-FLAG2 E44Q·Pro-σK(1–126)-His6 complex in both cases. Post-IM CID analysis of the samples detected some of the expected dissociation products, although the products were different depending on the detergent, as observed in other studies (75, 76). Notably, dissociation products in agreement with a 1:1 SpoIVFB-TEV-FLAG2 E44Q·Pro-σK(1–126)-His6 complex were observed in the 0.02% DDM sample (Fig. 6D), indicative of stable interaction between monomers of the two proteins. Interestingly, a dissociation product in agreement with a 1:2 SpoIVFB-TEV-FLAG2 E44Q·Pro-σK(1–126)-His6 complex was observed in the 0.5% C8E4 sample (data not shown). We speculated that this reflected SpoIVFB interacting with a dimer of Pro-σK(1–126)-His6, because a disulfide cross-linked dimer of Pro-σK(1–126)-His6 was detected upon treatment with Cu2+(phenanthroline)3 (Fig. 5G, lane 4). In any case, the IM-MS data strongly support the previous inference (28) that SpoIVFB is tetrameric and, together with our finding that Pro-σK(1–126)-His6 may be slightly substoichiometric compared with SpoIVFB-TEV-FLAG2 E44Q in the peak fraction during gel filtration in buffer containing 0.1% DM (Fig. 5E), at least two and perhaps as many as four Pro-σK(1–126)-His6 proteins may associate with the SpoIVFB tetramer.
Evidence That SpoIVFB Is Tetrameric in Vivo
Because previous analysis of purified TM-SpoIVFB (28) and our analyses of the purified SpoIVFB-TEV-FLAG2 E44Q·Pro-σK(1–126)-His6 complex (Figs. 5 and 6) suggested that SpoIVFB is tetrameric, we used TIRF microscopy and stepwise photobleaching to determine the number of subunits in SpoIVFB in sporulating B. subtilis. To our knowledge, this is the first time this methodology has been applied to an IP. The analysis was challenging because even though we used TIRF microscopy to only excite SpoIVFB-eGFP near the coverslip, we observed mostly large fluorescent foci likely representing multiple molecules and were unusable for counting photobleaching steps. We found that the small fluorescent foci had countable steps and such foci were localized to the mother cell membrane engulfing the forespore (Fig. 7, A and B). Most foci with countable steps exhibited 4 steps, but some had other numbers of steps (Fig. 7C). Potential sources of error in step counting are misfolded eGFP, quenching, partial overlap of foci, malformed complexes, and the subjective nature of counting steps. Hence, a distribution of the number of steps counted is expected, and our data are consistent with prior studies in this respect (63–67). The highest number of steps we counted was 5. Typically in stepwise photobleaching analysis, the highest number of steps is hypothesized to be the number of subunits in the protein. However, we rarely counted 5 steps, and statistical analysis of the γ function (55), which allows one to ask if a number of steps was counted erroneously, or more formally, tests the null hypothesis that the highest number of steps is not the number of subunits, yielded a very low γ value for 5 steps (Table 1), strongly suggesting that SpoIVFB is not pentameric. Moreover, analysis of the θ function yielded the highest value for 4 steps, indicating the lowest probability of missed photobeaching events. We concluded that the data provided strong evidence that SpoIVFB is tetrameric in vivo. Because purified TM-SpoIVFB appears to be tetrameric (28) and SpoIVFB in the purified SpoIVFB-TEV-FLAG2 E44Q·Pro-σK(1–126)-His6 complex (Figs. 5 and 6) appears to be tetrameric, we further conclude that the purified proteins resemble native SpoIVFB in terms of the number of subunits.
The C-terminal CBS domain of SpoIVFB may account for its formation of a tetramer. Four CBS domains form a conserved disk-like structure in a variety of proteins (34). The four CBS domains most commonly occur as pairs within two identical polypeptides of a homodimeric protein but may occur within a single polypeptide. In this respect, SpoIVFB, with its single CBS domain, is somewhat unusual among the CBS domain-containing proteins studied thus far. A pair of CBS domains forms a characteristic structure called a Bateman module, to which an adenine nucleotide typically binds in a cleft, although each CBS domain contains a potential nucleotide binding cavity (34). If the SpoIVFB tetramer follows this trend, it may bind two molecules of ATP. However, previously the recombinant CBS domain from SpoIVFB was shown to migrate during gel electrophoresis under mildly denaturing conditions as a mixture of predominantly dimers and some monomers, and radioactive ATP bound primarily to the monomers (28). ATP was required for the activity of purified TM-SpoIVFB, and it was speculated that ATP binding changes the conformation and/or oligomeric state of the enzyme. In most cases, ligand binding changes the conformation to relieve an autoinhibitory effect of the CBS domain (34). The tetrameric quaternary structure of SpoIVFB might allow cooperative binding of ATP and/or other adenine nucleotides, making the enzyme very sensitive to the energy level of the mother cell. Cooperative binding of the substrate is also possible. These questions and many others about the large subfamily of CBS domain-containing IMMPs represented by SpoIVFB can now be tackled by building on the foundation of insights and approaches reported here.
Author Contributions
Y. Z. and L. K. designed most of the study and wrote most of the paper, and Y. Z. performed most of the experiments. S. H. designed, performed, and analyzed the experiments shown in Fig. 6, A and B. R. A. K. and B. R. designed and analyzed the experiments shown in Fig. 6, C and D, and R. A. K. performed the experiments and wrote the corresponding parts of the paper. D. P. designed and performed the experiments shown in Fig. 7 and together with L. K. analyzed the results and wrote the corresponding parts of the paper. All authors reviewed the results and approved the final version of the manuscript.
Acknowledgments
We thank Bongjun Son, Jiajun Zhou, Paul Luethy, and Ruanbao Zhou for constructing plasmids. We thank David Rudner for B. subtilis strain BDR347.
This work was supported by National Institutes of Health Grants GM43585 (to L. K.) and GM105942 (to B. R.) and by Michigan State University AgBioResearch. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- IP
- intramembrane protease
- IMMP
- intramembrane metalloprotease
- TMS
- transmembrane segment
- IPTG
- isopropyl β-d-thiogalactopyranoside
- DM
- n-decyl-β-d-maltoside
- IM-MS
- ion mobility-mass spectrometry
- DDM
- n-dodecyl-β-d-maltoside
- C8E4
- tetraethylene glycol monooctyl ether
- eGFP
- enhanced green fluorescent protein
- TIRF
- total internal reflection fluorescence
- ROI
- region of interest
- sarkosyl
- sodium dodecanoyl sarcosine
- cytTM
- transmembrane segment from rabbit cytochrome P450 2B4
- TM-SpoIVFB
- cytTM-SpoIVFB-FLAG2-His6
- CID
- collision-induced dissociation
- post-IM
- post-ion mobility
- CBS
- cystathionine β-synthase
- TEV
- tobacco etch virus
- WBB
- wash buffer base.
References
- 1. Brown M. S., Ye J., Rawson R. B., and Goldstein J. L. (2000) Regulated intramembrane proteolysis: a control mechanism conserved from bacteria to humans. Cell 100, 391–398 [DOI] [PubMed] [Google Scholar]
- 2. Freeman M. (2008) Rhomboid proteases and their biological functions. Annu. Rev. Genet. 42, 191–210 [DOI] [PubMed] [Google Scholar]
- 3. Urban S., and Dickey S. W. (2011) The rhomboid protease family: a decade of progress on function and mechanism. Genome Biol. 12, 231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bergbold N., and Lemberg M. K. (2013) Emerging role of rhomboid family proteins in mammalian biology and disease. Biochim. Biophys. Acta 1828, 2840–2848 [DOI] [PubMed] [Google Scholar]
- 5. Rather P. (2013) Role of rhomboid proteases in bacteria. Biochim. Biophys. Acta 1828, 2849–2854 [DOI] [PubMed] [Google Scholar]
- 6. Wolfe M. S., and Kopan R. (2004) Intramembrane proteolysis: theme and variations. Science 305, 1119–1123 [DOI] [PubMed] [Google Scholar]
- 7. Jurisch-Yaksi N., Sannerud R., and Annaert W. (2013) A fast growing spectrum of biological functions of g-secretase in development and disease. Biochim. Biophys. Acta 1828, 2815–2827 [DOI] [PubMed] [Google Scholar]
- 8. Wolfe M. S. (2010) Structure, mechanism and inhibition of γ-secretase and presenilin-like proteases. Biol. Chem. 391, 839–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hu J., Xue Y., Lee S., and Ha Y. (2011) The crystal structure of GXGD membrane protease FlaK. Nature 475, 528–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Golde T. E., Wolfe M. S., and Greenbaum D. C. (2009) Signal peptide peptidases: a family of intramembrane-cleaving proteases that cleave type 2 transmembrane proteins. Semin. Cell Dev. Biol. 20, 225–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Voss M., Schröder B., and Fluhrer R. (2013) Mechanism, specificity, and physiology of signal peptide peptidase (SPP) and SPP-like proteases. Biochim. Biophys. Acta 1828, 2828–2839 [DOI] [PubMed] [Google Scholar]
- 12. Zhang K., Shen X., Wu J., Sakaki K., Saunders T., Rutkowski D. T., Back S. H., and Kaufman R. J. (2006) Endoplasmic reticulum stress activates cleavage of CREBH to induce a systemic inflammatory response. Cell 124, 587–599 [DOI] [PubMed] [Google Scholar]
- 13. Rawson R. B. (2013) The site-2 protease. Biochim. Biophys. Acta 1828, 2801–2807 [DOI] [PubMed] [Google Scholar]
- 14. Urban S. (2009) Making the cut: central roles of intramembrane proteolysis in pathogenic microorganisms. Nat. Rev. Microbiol. 7, 411–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kroos L., and Akiyama Y. (2013) Biochemical and structural insights into intramembrane metalloprotease mechanisms. Biochim. Biophys. Acta 1828, 2873–2885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Schneider J. S., and Glickman M. S. (2013) Function of site-2 proteases in bacteria and bacterial pathogens. Biochim. Biophys. Acta 1828, 2808–2814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Feng L., Yan H., Wu Z., Yan N., Wang Z., Jeffrey P. D., and Shi Y. (2007) Structure of a site-2 protease family intramembrane metalloprotease. Science 318, 1608–1612 [DOI] [PubMed] [Google Scholar]
- 18. Li X., Dang S., Yan C., Gong X., Wang J., and Shi Y. (2013) Structure of a presenilin family intramembrane aspartate protease. Nature 493, 56–61 [DOI] [PubMed] [Google Scholar]
- 19. Wang Y., Zhang Y., and Ha Y. (2006) Crystal structure of a rhomboid family intramembrane protease. Nature 444, 179–183 [DOI] [PubMed] [Google Scholar]
- 20. Wu Z., Yan N., Feng L., Oberstein A., Yan H., Baker R. P., Gu L., Jeffrey P. D., Urban S., and Shi Y. (2006) Structural analysis of a rhomboid family intramembrane protease reveals a gating mechanism for substrate entry. Nat. Struct. Mol. Biol. 13, 1084–1091 [DOI] [PubMed] [Google Scholar]
- 21. Rudner D. Z., and Losick R. (2002) A sporulation membrane protein tethers the pro-σK processing enzyme to its inhibitor and dictates its subcellular localization. Genes Dev. 16, 1007–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhou R., and Kroos L. (2004) BofA protein inhibits intramembrane proteolysis of pro-σK in an intercompartmental signaling pathway during Bacillus subtilis sporulation. Proc. Natl. Acad. Sci. U.S.A. 101, 6385–6390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Campo N., and Rudner D. Z. (2006) A branched pathway governing the activation of a developmental transcription factor by regulated intramembrane proteolysis. Mol. Cell 23, 25–35 [DOI] [PubMed] [Google Scholar]
- 24. Dong T. C., and Cutting S. M. (2003) SpoIVB-mediated cleavage of SpoIVFA could provide the intercellular signal to activate processing of Pro-σK in Bacillus subtilis. Mol. Microbiol. 49, 1425–1434 [DOI] [PubMed] [Google Scholar]
- 25. Zhou R., and Kroos L. (2005) Serine proteases from two cell types target different components of a complex that governs regulated intramembrane proteolysis of pro-σK during Bacillus subtilis development. Mol. Microbiol. 58, 835–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rudner D. Z., Fawcett P., and Losick R. (1999) A family of membrane-embedded metalloproteases involved in regulated proteolysis of membrane-associated transcription factors. Proc. Natl. Acad. Sci. U.S.A. 96, 14765–14770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yu Y. T., and Kroos L. (2000) Evidence that SpoIVFB is a novel type of membrane metalloprotease governing intercompartmental communication during Bacillus subtilis sporulation. J. Bacteriol. 182, 3305–3309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhou R., Cusumano C., Sui D., Garavito R. M., and Kroos L. (2009) Intramembrane proteolytic cleavage of a membrane-tethered transcription factor by a metalloprotease depends on ATP. Proc. Natl. Acad. Sci. U.S.A. 106, 16174–16179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kroos L., Kunkel B., and Losick R. (1989) Switch protein alters specificity of RNA polymerase containing a compartment-specific sigma factor. Science 243, 526–529 [DOI] [PubMed] [Google Scholar]
- 30. Eichenberger P., Fujita M., Jensen S. T., Conlon E. M., Rudner D. Z., Wang S. T., Ferguson C., Haga K., Sato T., Liu J. S., and Losick R. (2004) The program of gene transcription for a single differentiating cell type during sporulation in Bacillus subtilis. PLoS Biol. 2, e328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kinch L. N., Ginalski K., and Grishin N. V. (2006) Site-2 protease regulated intramembrane proteolysis: sequence homologs suggest an ancient signaling cascade. Protein Sci. 15, 84–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bateman A. (1997) The structure of a domain common to archaebacteria and the homocystinuria disease protein. Trends Biochem. Sci. 22, 12–13 [DOI] [PubMed] [Google Scholar]
- 33. Finn R. D., Mistry J., Tate J., Coggill P., Heger A., Pollington J. E., Gavin O. L., Gunasekaran P., Ceric G., Forslund K., Holm L., Sonnhammer E. L., Eddy S. R., and Bateman A. (2010) The Pfam protein families database. Nucleic Acids Res. 38, D211–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Baykov A. A., Tuominen H. K., and Lahti R. (2011) The CBS domain: a protein module with an emerging prominent role in regulation. ACS Chem. Biol. 6, 1156–1163 [DOI] [PubMed] [Google Scholar]
- 35. Scott J. W., Hawley S. A., Green K. A., Anis M., Stewart G., Scullion G. A., Norman D. G., and Hardie D. G. (2004) CBS domains form energy-sensing modules whose binding of adenosine ligands is disrupted by disease mutations. J. Clin. Invest. 113, 274–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Blaylock B., Jiang X., Rubio A., Moran C. P. Jr., and Pogliano K. (2004) Zipper-like interaction between proteins in adjacent daughter cells mediates protein localization. Genes Dev. 18, 2916–2928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Camp A. H., and Losick R. (2008) A novel pathway of intercellular signalling in Bacillus subtilis involves a protein with similarity to a component of type III secretion channels. Mol. Microbiol. 69, 402–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Meisner J., Wang X., Serrano M., Henriques A. O., and Moran C. P. Jr. (2008) A channel connecting the mother cell and forespore during bacterial endospore formation. Proc. Natl. Acad. Sci. U.S.A. 105, 15100–15105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Camp A. H., and Losick R. (2009) A feeding tube model for activation of a cell-specific transcription factor during sporulation in Bacillus subtilis. Genes Dev. 23, 1014–1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Doan T., Morlot C., Meisner J., Serrano M., Henriques A. O., Moran C. P. Jr., and Rudner D. Z. (2009) Novel secretion apparatus maintains spore integrity and developmental gene expression in Bacillus subtilis. PLoS Genet. 5, e1000566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Doan T., Marquis K. A., and Rudner D. Z. (2005) Subcellular localization of a sporulation membrane protein is achieved through a network of interactions along and across the septum. Mol. Microbiol. 55, 1767–1781 [DOI] [PubMed] [Google Scholar]
- 42. Jiang X., Rubio A., Chiba S., and Pogliano K. (2005) Engulfment-regulated proteolysis of SpoIIQ: evidence that dual checkpoints control sigma activity. Mol. Microbiol. 58, 102–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chiba S., Coleman K., and Pogliano K. (2007) Impact of membrane fusion and proteolysis on SpoIIQ dynamics and interaction with SpoIIIAH. J. Biol. Chem. 282, 2576–2586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Konovalova A., Søgaard-Andersen L., and Kroos L. (2014) Regulated proteolysis in bacterial development. FEMS Microbiol. Rev. 38, 493–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zhou R., Chen K., Xiang X., Gu L., and Kroos L. (2013) Features of Pro-σK important for cleavage by SpoIVFB, an intramembrane metalloprotease. J. Bacteriol. 195, 2793–2806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhang Y., Luethy P. M., Zhou R., and Kroos L. (2013) Residues in conserved loops of intramembrane metalloprotease SpoIVFB interact with residues near the cleavage site in Pro-σK. J. Bacteriol. 195, 4936–4946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Prince H., Zhou R., and Kroos L. (2005) Substrate requirements for regulated intramembrane proteolysis of Bacillus subtilis pro-σK. J. Bacteriol. 187, 961–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sambrook J., Fritsch E. F., and Maniatis T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Ed., p. A.1, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 49. Kroos L., Yu Y. T., Mills D., and Ferguson-Miller S. (2002) Forespore signaling is necessary for pro-σK processing during Bacillus subtilis sporulation despite the loss of SpoIVFA upon translational arrest. J. Bacteriol. 184, 5393–5401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Koide K., Ito K., and Akiyama Y. (2008) Substrate recognition and binding by RseP, an Escherichia coli intramembrane protease. J. Biol. Chem. 283, 9562–9570 [DOI] [PubMed] [Google Scholar]
- 51. Laganowsky A., Reading E., Hopper J. T., and Robinson C. V. (2013) Mass spectrometry of intact membrane protein complexes. Nat. Protoc. 8, 639–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. McKay A. R., Ruotolo B. T., Ilag L. L., and Robinson C. V. (2006) Mass measurements of increased accuracy resolve heterogeneous populations of intact ribosomes. J. Am. Chem. Soc. 128, 11433–11442 [DOI] [PubMed] [Google Scholar]
- 53. Rasband W. S. (1997–2016) ImageJ. U. S. National Institutes of Health, Bethesda, MD, http://imagej.nih.gov/ij/ [Google Scholar]
- 54. Reuel N. F., Bojo P., Zhang J., Boghossian A. A., Ahn J. H., Kim J. H., and Strano M. S. (2012) NoRSE: noise reduction and state evaluator for high-frequency single event traces. Bioinformatics 28, 296–297 [DOI] [PubMed] [Google Scholar]
- 55. Hines K. E. (2013) Inferring subunit stoichiometry from single molecule photobleaching. J. Gen. Physiol. 141, 737–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Pogliano J., Osborne N., Sharp M. D., Abanes-De Mello A., Perez A., Sun Y. L., and Pogliano K. (1999) A vital stain for studying membrane dynamics in bacteria: a novel mechanism controlling septation during Bacillus subtilis sporulation. Mol. Microbiol. 31, 1149–1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Saribas A. S., Gruenke L., and Waskell L. (2001) Overexpression and purification of the membrane-bound cytochrome P450 2B4. Protein Expr. Purif. 21, 303–309 [DOI] [PubMed] [Google Scholar]
- 58. Zhang B., Hofmeister A., and Kroos L. (1998) The pro-sequence of pro-σK promotes membrane association and inhibits RNA polymerase core binding. J. Bacteriol. 180, 2434–2441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Hoaglund-Hyzer C. S., Li J., and Clemmer D. E. (2000) Mobility labeling for parallel CID of ion mixtures. Anal. Chem. 72, 2737–2740 [DOI] [PubMed] [Google Scholar]
- 60. Hoaglund-Hyzer C. S., and Clemmer D. E. (2001) Ion trap/ion mobility/quadrupole/time-of-flight mass spectrometry for peptide mixture analysis. Anal. Chem. 73, 177–184 [DOI] [PubMed] [Google Scholar]
- 61. Stone E., Gillig K. J., Ruotolo B., Fuhrer K., Gonin M., Schultz A., and Russell D. H. (2001) Surface-induced dissociation on a MALDI-ion mobility-orthogonal time-of-flight mass spectrometer: sequencing peptides from an “in-solution” protein digest. Anal. Chem. 73, 2233–2238 [DOI] [PubMed] [Google Scholar]
- 62. Shepherd D. A., Marty M. T., Giles K., Baldwin A. J., and Benesch J. L. P. (2015) Combining tandem mass spectrometry with ion mobility separation to determine the architecture of polydisperse proteins. Int. J. Mass Spectrom. 377, 663–671 [Google Scholar]
- 63. Leake M. C., Chandler J. H., Wadhams G. H., Bai F., Berry R. M., and Armitage J. P. (2006) Stoichiometry and turnover in single, functioning membrane protein complexes. Nature 443, 355–358 [DOI] [PubMed] [Google Scholar]
- 64. Ulbrich M. H., and Isacoff E. Y. (2007) Subunit counting in membrane-bound proteins. Nat. Methods 4, 319–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Bai F., Morimoto Y. V., Yoshimura S. D., Hara N., Kami-Ike N., Namba K., and Minamino T. (2014) Assembly dynamics and the roles of FliI ATPase of the bacterial flagellar export apparatus. Sci. Rep. 4, 6528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Coste B., Xiao B., Santos J. S., Syeda R., Grandl J., Spencer K. S., Kim S. E., Schmidt M., Mathur J., Dubin A. E., Montal M., and Patapoutian A. (2012) Piezo proteins are pore-forming subunits of mechanically activated channels. Nature 483, 176–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Plant L. D., Xiong D., Dai H., and Goldstein S. A. (2014) Individual IKs channels at the surface of mammalian cells contain two KCNE1 accessory subunits. Proc. Natl. Acad. Sci. U.S.A. 111, E1438–E1446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Garavito R. M., and Ferguson-Miller S. (2001) Detergents as tools in membrane biochemistry. J. Biol. Chem. 276, 32403–32406 [DOI] [PubMed] [Google Scholar]
- 69. Freeman M. (2014) The rhomboid-like superfamily: molecular mechanisms and biological roles. Annu. Rev. Cell Dev. Biol. 30, 235–254 [DOI] [PubMed] [Google Scholar]
- 70. Zoll S., Stanchev S., Began J., Skerle J., Lepšík M., Peclinovská L., Majer P., and Strisovsky K. (2014) Substrate binding and specificity of rhomboid intramembrane protease revealed by substrate-peptide complex structures. EMBO J. 33, 2408–2421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Ha Y., Akiyama Y., and Xue Y. (2013) Structure and mechanism of rhomboid protease. J. Biol. Chem. 288, 15430–15436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Li Y., Lu S. H., Tsai C. J., Bohm C., Qamar S., Dodd R. B., Meadows W., Jeon A., McLeod A., Chen F., Arimon M., Berezovska O., Hyman B. T., Tomita T., Iwatsubo T., Johnson C. M., Farrer L. A., Schmitt-Ulms G., Fraser P. E., and St George-Hyslop P. H. (2014) Structural interactions between inhibitor and substrate docking sites give insight into mechanisms of human PS1 complexes. Structure 22, 125–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Akiyama Y., Kanehara K., and Ito K. (2004) RseP (YaeL), an Escherichia coli RIP protease, cleaves transmembrane sequences. EMBO J. 23, 4434–4442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Schneider J. S., Reddy S. P., Hock Y. E., Evans H. W., and Glickman M. S. (2013) Site-2 protease substrate specificity and coupling in trans by a PDZ-substrate adapter protein. Proc. Natl. Acad. Sci. U.S.A. 110, 19543–19548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Laganowsky A., Reading E., Allison T. M., Ulmschneider M. B., Degiacomi M. T., Baldwin A. J., and Robinson C. V. (2014) Membrane proteins bind lipids selectively to modulate their structure and function. Nature 510, 172–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Reading E., Walton T. A., Liko I., Marty M. T., Laganowsky A., Rees D. C., and Robinson C. V. (2015) The effect of detergent, temperature, and lipid on the oligomeric state of MscL constructs: insights from mass spectrometry. Chem. Biol. 22, 593–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Green D. H., and Cutting S. M. (2000) Membrane topology of the Bacillus subtilis Pro-σK processing complex. J. Bacteriol. 182, 278–285 [DOI] [PMC free article] [PubMed] [Google Scholar]