Abstract
Heme a is an essential cofactor for function of cytochrome c oxidase in the mitochondrial electron transport chain. Several evolutionarily conserved enzymes have been implicated in the biosynthesis of heme a, including the heme a synthase Cox15. However, the structure of Cox15 is unknown, its enzymatic mechanism and the role of active site residues remain debated, and recent discoveries suggest additional chaperone-like roles for this enzyme. Here, we investigated Cox15 in the model eukaryote Saccharomyces cerevisiae via several approaches to examine its oligomeric states and determine the effects of active site and human pathogenic mutations. Our results indicate that Cox15 exhibits homotypic interactions, forming highly stable complexes dependent upon hydrophobic interactions. This multimerization is evolutionarily conserved and independent of heme levels and heme a synthase catalytic activity. Four conserved histidine residues are demonstrated to be critical for eukaryotic heme a synthase activity and cannot be substituted with other heme-ligating amino acids. The 20-residue linker region connecting the two conserved domains of Cox15 is also important; removal of this linker impairs both Cox15 multimerization and enzymatic activity. Mutations of COX15 causing single amino acid conversions associated with fatal infantile hypertrophic cardiomyopathy and the neurological disorder Leigh syndrome result in impaired stability (S344P) or catalytic function (R217W), and the latter mutation affects oligomeric properties of the enzyme. Structural modeling of Cox15 suggests these two mutations affect protein folding and heme binding, respectively. We conclude that Cox15 multimerization is important for heme a biosynthesis and/or transfer to maturing cytochrome c oxidase.
Keywords: cytochrome c oxidase (complex IV), heme, mitochondria, mitochondrial disease, oligomerization, Cox10, Cox15
Introduction
Heme (iron protoporphyrin IX; heme b) is a vital cofactor in living cells, and the derivative heme a is critical for mitochondrial respiration. Cytochrome c oxidase (CcO),3 the heme-copper Complex IV of the mitochondrial electron transport chain, contains two heme a cofactors residing in its Cox1 subunit with different coordination environments, termed heme a and heme a3, that are essential for enzyme function (1).
In the biosynthesis of heme a, several evolutionarily conserved enzymes have been implicated, including the membrane proteins heme o synthase (Cox10) and heme a synthase (Cox15), which sequentially mediate the modification of heme b within mitochondria (1). Cox10 is responsible for farnesylation of heme b to yield the heme o intermediate, which Cox15 subsequently converts to heme a through oxidation of the C8 porphyrin methyl substituent to an aldehyde (1). This latter step is performed in conjunction with matrix-localized ferredoxin (Yah1) and ferredoxin reductase (Arh1) (2, 3).
The process by which heme a is delivered to CcO is incompletely understood and likely to be strictly controlled because free heme can be toxic to cells, and conditionally deleterious heme-populated intermediates have been observed to form during the process of CcO assembly (4, 5). The membrane protein Shy1 (homologous to human SURF1) and membrane-associated protein Coa2 have critical yet unclear roles in heme a delivery to Cox1 in yeast. Cox1 appears to receive heme a post-translationally within a Shy1-containing assembly intermediate, and Coa2 cooperates with Shy1 (6–12). Cells lacking Coa2 are impaired in Cox10 and Cox15 oligomerization and exhibit impaired hemylation and subsequent rapid degradation of newly synthesized Cox1 (6–8, 13).
Mutations resulting in alterations of conserved residues of COX10, COX15, and SURF1 have been linked to a variety of devastating diseases, including fatal infantile hypertrophic cardiomyopathy and the neurological disorder Leigh syndrome (LS) (14–25). These diseases underscore the essential role of each protein in CcO assembly and thus the function of the mitochondrial respiratory chain. In yeast, deletion of the COX10 or COX15 gene is lethal to cells grown under respiratory conditions, whereas cells are viable when grown on fermentable media, enabling studies in the absence of functional Cox10 or Cox15 (2, 3, 9, 10).
Limited information is available regarding the regulation, structure, and function of Cox10 and Cox15. Expression of Cox15, but not Cox10, is activated by the heme b-dependent transcription factor Hap1 (26). Both proteins are large, polytopic proteins residing in the mitochondrial inner membrane with seven to nine predicted transmembrane segments (1, 27). Cox10 can function in the absence of Cox15 (2, 14, 28). Experiments in bacteria led to a hypothesis that the heme o intermediate produced during heme a biosynthesis is directly channeled between Cox10 and Cox15 (29). However, topology modeling of eukaryotic Cox10 and Cox15 has revealed that the active sites of the enzymes face different sides of the inner mitochondrial membrane (1).
Cox10 and Cox15 exist as independent, oligomeric complexes (6, 10). Cox10 oligomerization is directly linked to the sophisticated process of Cox1 synthesis and maturation (for a review, see Ref. 30) wherein multimeric Cox10 appears to be the active state of the enzyme (7). The multimerization of Cox10 is in part mediated by the C-terminal portion of newly synthesized Cox1 and can be uncoupled from the catalytic activity of Cox10 (6, 7). A recent study has demonstrated that Cox15 associates with Shy1 and with Cox1-containing assembly intermediates (10). However, the significance of the latter associations remains unclear as Cox15 is neither a stoichiometric component of CcO assembly complexes nor is its association dependent on Cox1 or CcO assembly (10).
Many important questions about the eukaryotic heme a synthase remain unanswered, including the roles of oligomeric complexes formed by Cox15, whether the oligomerization process is regulated, and the molecular architecture of the Cox15 enzyme and its active site. In this study, we analyzed Cox15 high mass complexes, interrogated several aspects of Cox15 structure, tested the effects of the human COX15 mutations associated with mitochondrial disease on the stability and function of the Saccharomyces cerevisiae enzyme, and modeled the structure of Cox15 to analyze the roles of these structural features.
Our results suggest that Cox15 forms stable, evolutionarily conserved, oligomeric complexes containing more than one copy of Cox15. Oligomerization of Cox15 does not depend on heme or catalytic activity. Consistent with the bacterial heme a synthase, the four conserved histidine residues of Cox15 are each essential for function of the eukaryotic enzyme as their conversion to alanine renders the enzyme catalytically inactive, and even conversion to other potential heme-binding ligands disables enzyme function. The presence of a linker between the two heme-binding domains of Cox15 is crucial for both oligomerization and enzyme function, and the R217W single amino acid conversion found in patients with mitochondrial disease impairs the function and oligomerization of the yeast enzyme. Thus, Cox15 oligomers are important for heme a biosynthesis and/or subsequent delivery to maturing CcO and may serve as a platform for CcO assembly.
Experimental Procedures
Yeast Strains, Plasmids, and Media
Yeast strains used in this study were derivatives of the W303 genetic background and are listed in Table 1. LEU2 to URA3 selection marker swap was mediated by homologous recombination as described previously (32). For promoter swap experiments, the 450-bp region upstream of the ATG codon of COX15 was replaced with the TEF1 promoter sequence derived from the pUG6 vector (33) using primers TEF1-Cox15For (5′-ATGGAGCAAACGGATAGAGCAACGGT-3′) and TEF1-Cox15Rev (5′-TAGCTTAGCTGCCTGCCTGCCCACTTC-3′). The plasmid-borne variant of the 13xMyc-tagged Cox15 under control of its own promoter and the ADH1 terminator (Cox15-Myc) was obtained from the COX15::Myc strain by in vivo gap repair cloning using primers Cox15:MycFor (5′-GTATCGATAAGCTTGATATCGAATTCCTGCAGCCCGGGGTGGGTAAAGTCGACGTGTCTACAA-3′) and Cox15:MycRev (5′-CTGGAGCTCCACCGCGGTGGCGGCCGCTCTAGAACTAGTGTATATTACCCTGTTATCCCTAGCGGATCTG-3′) appended with vector sequence to facilitate insertion of the PCR product into BamHI-linearized pRS415 vector. The H169A, H245A, H368A, H431A, H169C, H245C, H368C, H431C, H169Y, H245Y, H368Y, H431Y, T236R, and T236W single amino acid conversions in pRS415-Cox15-Myc were each generated with use of a QuikChange (Agilent Technologies) or Q5 (New England BioLabs) site-directed mutagenesis kit. Linker region deletions of Cox15 were generated using the Q5 site-directed mutagenesis kit; the L-20 construct lacks the 20 amino acid residues (ECKWIKNPVQAISLFKKLDN) predicted to connect helices IV and V of Cox15, and the L-10 construct shortens the linker by half (via deletion of the residues KNPVQAISLF). The L+20 Cox15 linker region extension plasmid was generated by adding a sequence derived from the unstructured region of the Escherichia coli lacI gene as described previously (34) with use of the Q5 site-directed mutagenesis kit. This L+20 construct contains the following amino acids added to the middle of the predicted linker (at the plus sign in the linker sequence KNPVQ+AISLF): KPVTLYDVAEYAGVSYQTVS. A plasmid encoding Cox15 tagged with the FLAG epitope (Cox15-FLAG) was constructed from an existing pRS426-Cox15-His6 plasmid (Cox15-His) containing COX15 under the control of the MET25 promoter and CYC1 terminator via site-directed mutagenesis for use in co-immunoprecipitation experiments. All constructs were confirmed by DNA sequencing.
TABLE 1.
Yeast strains used in this work
| Strain | Genotype | Source/Ref. |
|---|---|---|
| W303-1B | MATα ade2-1 can1–100 his3–11,15 leu2–3,112 trp1-1 ura3–1 [rho+] | ATCC |
| DY5113 | MATa ade2-1 can1–100 his3–11,15 leu2–3,112 trp1Δ ura3-1 [rho+] | D. Winge |
| cox15Δ | MATa ade2-1 can1–100 his3–11,15 leu2–3,112 trp1-1 ura3–1 cox15Δ::KanMX4[rho+] | E. Hegg |
| hem1Δ | MATa ade2-1 can1–100 his3–11,15 leu2–3,112 trp1-1 ura3-1 hem1Δ::ADE2[rho+] | 31 |
| COX15::Myc | MATa ade2-1 can1–100 his3–11,15 leu2–3,112 trp1Δ ura3-1 COX15-13Myc::TRP1[rho+] | 9 |
| COX15::Myc hem1Δ | MATa ade2-1 can1–100 his3–11,15 leu2–3,112 trp1Δ ura3-1 COX15-13Myc::TRP1 hem1Δ::LEU2-Δ::URA3MX[rho+] | This study |
| COX15::Myc (TEF1P) | MATa ade2-1 can1–100 his3–11,15 leu2–3,112 trp1Δ ura3-1 TEF1P::KanMX4-COX15-13Myc::TRP1[rho+] | This study |
| COX15::Myc (TEF1P)hem1Δ | MATa ade2-1 can1–100 his3–11,15 leu2–3,112 trp1Δ ura3-1 TEF1P::KanMX4-COX15-13Myc::TRP1 hem1Δ::LEU2-Δ::URA3MX[rho+] | This study |
Cells were cultured and handled according to previously published protocols (35) using either yeast extract-peptone-dextrose (Amresco) or Brent Supplement Mixture synthetic selective medium (Sunrise Science Products) when necessary to maintain plasmid selection in transformed cells. Analysis of Cox15 protein levels and native complexes under heme-depleted conditions was performed by culturing cells in yeast extract-peptone-dextrose medium supplemented with either δ-aminolevulinic acid or Tween 80/ergosterol/methionine mixture as described by Crisp et al. (36).
Mammalian Cell Culture
Immortalized control and patient skin fibroblasts from Cox15-deficient patients (14, 15) (kind gifts from Drs. Eric Shoubridge and Massimo Zeviani) were cultured in high glucose DMEM supplemented with 10% (v/v) fetal bovine serum at 37 °C and 5% CO2 atmosphere. Cell culture reagents were from Thermo Scientific or Life Technologies. Cells (2 × 106) were released with trypsin, washed with PBS, and lysed in radioimmunoprecipitation assay buffer (150 mm NaCl, 10 mm Tris-HCl (pH 7.2), 0.1% (v/v) SDS, 0.1% (v/v) Triton X-100, 1% (w/v) deoxycholate, 5 mm EDTA (pH 8.0)) in the presence of protease inhibitor mixture (Thermo Scientific) and 2 mm PMSF (Sigma). Proteins from each lysate aliquot were precipitated with 10% TCA and subjected to SDS-PAGE.
Mitochondrial Isolation and Assays
Intact mitochondria were isolated from yeast cells according to published procedures (37). Porcine kidney mitochondria were isolated via published procedures (38) from organs obtained from a slaughterhouse. Blue native (BN)-PAGE separation of mitochondrial protein complexes was performed as described (9). Briefly, 30 μg of mitochondria were solubilized in lysis buffer (50 mm NaCl, 5 mm 6-aminocaproic acid, 50 mm imidazole (pH 7.0)) in the presence of 1% digitonin, 1 mm PMSF, and protease inhibitor mixture. After incubation for 15 min on ice and centrifugation (20,000 × g for 15 min at 2 °C), supernatants were mixed with sample buffer (5% Coomassie Brilliant Blue G-250, 0.5 m 6-aminocaproic acid (pH 7.0)) and separated on 5–13% gradient polyacrylamide gels. Analysis of Cox15 complex formation under different denaturing conditions was done via sucrose gradient fractionation by high velocity centrifugation as described previously (9, 13). Heme-pyridine redox difference spectra were determined from SDS-solubilized mitochondrial lysates according to the method of Berry and Trumpower (39). CcO enzymatic activity in isolated mitochondria was measured as described previously (40).
Co-immunoprecipitation
Immunoadsorption of Cox15-Myc from cox15Δ cells co-expressing Cox15-FLAG and either Cox15-Myc or vector control was carried out with mouse anti-c-Myc-agarose beads (sc-40 AC, Santa Cruz Biotechnology) as described previously (41) using 500 μg of mitochondrial protein solubilized in lysis buffer (1% digitonin, 1 mm PMSF in PBS at pH 7.4). Clarified lysates were incubated with the immunoreactive beads for 25 h at 4 °C with tube rotation. Following collection of the unbound fraction and wash fractions (using a wash buffer containing 0.5% digitonin, 1 mm PMSF in PBS at pH 7.4), the bound proteins were eluted with non-reducing Laemmli buffer, and fractions were subjected to SDS-PAGE and Western blotting analysis with rabbit (anti-FLAG and anti-Myc) antibodies (see below for antibody catalog numbers).
Immunoblotting
After gel electrophoresis, mitochondrial proteins or complexes were transferred to nitrocellulose or PVDF membranes. Membranes were blocked in 5% nonfat milk, PBS (with or without Tween 20) and decorated with the indicated primary antibodies and appropriate HRP-coupled secondary antibodies. The protein bands were visualized by incubation of immunodecorated membranes with chemiluminescence reagents (Millipore and Thermo Scientific) and subsequent exposure to x-ray film (BioExpress). The following commercially available antibodies were used: mouse anti-c-Myc (11667149001, Roche Diagnostics), rabbit anti-c-Myc (sc-789, Santa Cruz Biotechnology), anti-FLAG (F7425, Sigma), anti-porin (459500, Invitrogen), anti-His6 (MA1-21315 from Thermo Scientific and ab9108 from Abcam), anti-Cox1 (ab110270), -Cox2 (ab110271), and -Cox3 (ab110259) (Abcam), anti-human COX15 (ARP46442_T100, Aviva Systems Biology), anti-tubulin (T6074, Sigma), and goat anti-mouse and anti-rabbit IgG-HRP secondary antibodies (sc-2005 and sc-2030, Santa Cruz Biotechnology). In addition, we used antisera against Rip1 (kindly provided by Dr. Dennis Winge) and the Atp2 β subunit of F1 (a kind gift from Dr. Alexander Tzagoloff). All antibodies were validated to assure specificity and reliability in detection of the specified proteins at their respective migration distances in gel electrophoresis.
Structural Modeling
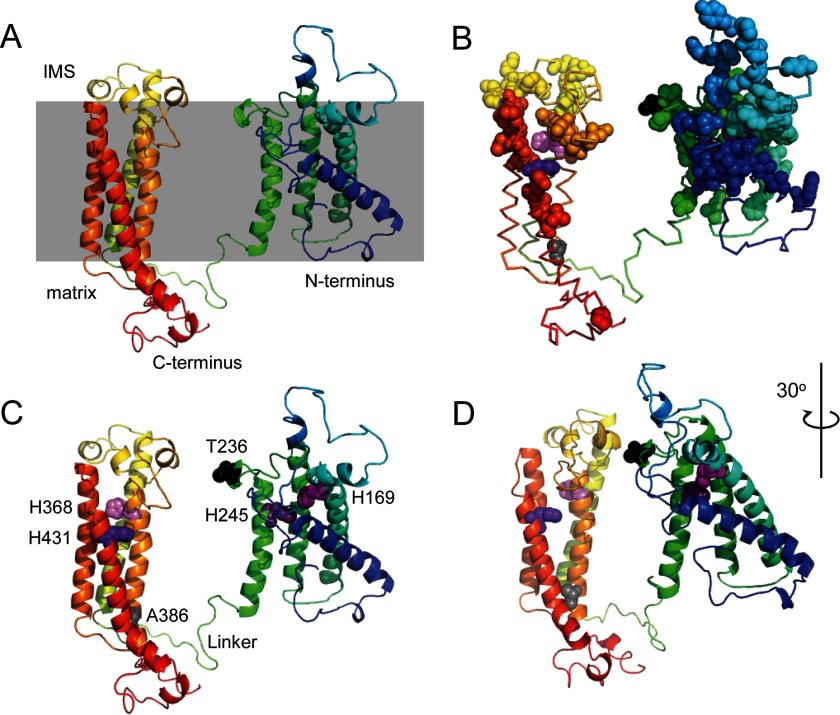
The Phyre2 protein fold recognition server (42) was used for structural prediction of the Cox15 protein in S. cerevisiae (residues 66–486) following removal of the MitoProt II-predicted (43) mitochondrial targeting sequence (residues 1–65). The contributors to this model were the heme-binding four-helical bundle fold of the cytochrome b subunit of the cytochrome bc1 complex (which was used in predicting structure for amino acid residues 281–316 and 332–417 with a reported 81% estimation of confidence), part of a structure for nitric-oxide reductase (which was used to model residues 241–339 and 364–454 with 59% confidence), the heme-binding four-helical bundle fold of the γ chain of respiratory nitrate reductase I (residues 74–108, 124–131, and 164–256 with 50% confidence), and a small turn from the structure of the cyclotide tricyclon A (residues 113–125 with 69% confidence). Fig. 7 was generated from this model using the PyMOL molecular graphics system (version 1.7.4.1).
FIGURE 7.
Structural modeling of yeast Cox15. A, model of yeast Cox15 generated using the Phyre2 protein fold recognition server shown with an indication of the predicted orientation relative to the mitochondrial inner membrane (gray box). The sequence progresses from the blue N terminus to the red C terminus. B, the yeast Cox15 model shown as a ribbon with conserved residues shown as colored spheres indicating their relative position in the sequence except for the four conserved histidines, which are in shades of purple. Also shown as spheres are the residues Thr-236 (aligns with human Arg-217; in black) and Ala-386 (aligns with human Ser-344; in gray). C, the model showing structural features tested in this study. Residues of interest are shown as spheres: the four conserved histidines are in shades of purple, Thr-236 is black, and Ala-386 is gray. The linker connecting the N- and C-terminal portions of the enzyme is the indicated green loop. D, alternate view of the structure shown in C rotated 30°. IMS, intermembrane space.
Growth Tests
Respiratory growth tests of cox15Δ cells expressing plasmid-borne variants of Cox15-Myc or vector control (and in Fig. 1A, WT and cox15Δ cells co-expressing vector control with an additional vector control or Cox15-Myc or Cox15-FLAG) were performed at 30 °C. Cells were precultured overnight in synthetic selective medium (lacking pertinent nutrients to maintain plasmid selection) containing 2% galactose and 0.1% glucose, then normalized to A600 = 1 in sterile water, serially diluted, and dropped onto synthetic selective medium plates containing 2% glucose or glycerol/lactate as a carbon source. The growth was assessed and documented after 1–2 (glucose plates) or 2–3 (glycerol/lactate plates) days of incubation at 30 °C.
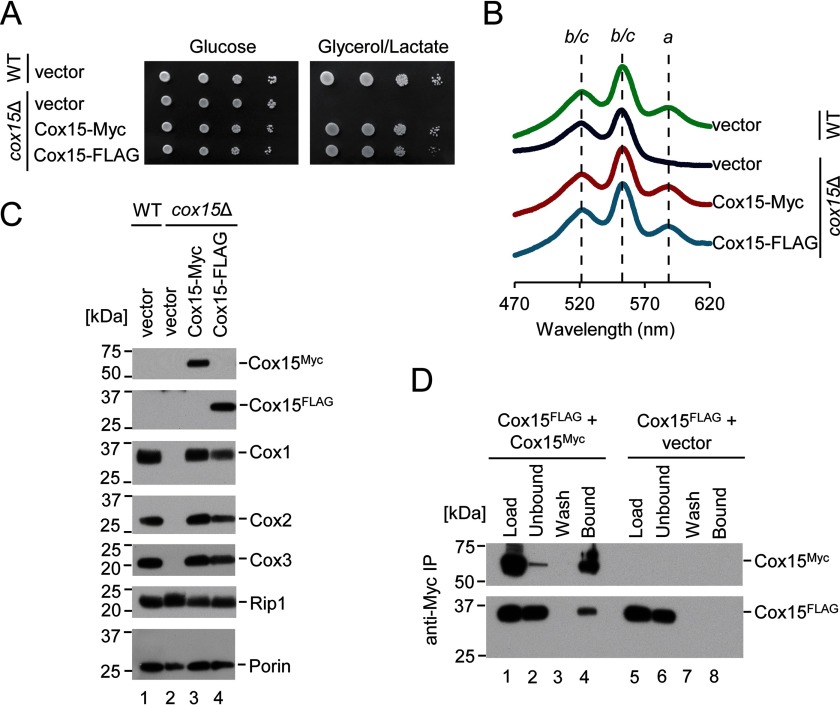
FIGURE 1.
Cox15 exhibits homotypic protein interaction. A, respiratory growth test at 30 °C of WT cells co-expressing two vector controls and cox15Δ cells co-expressing vector control with either another vector control, Cox15-Myc, or Cox15-FLAG. Cells were dropped onto synthetic selective medium containing either 2% glucose or 2% glycerol/lactate after normalization and serial dilution of cultures grown in 2% galactose and 0.1% glucose. Results are shown for one experiment, representative of three independent experiments (biological replicates). B, heme-pyridine redox difference spectra of mitochondria from the cells described in A. Spectra are offset in the y axis for clarity and are representative of three independent experiments (biological replicates). Absorption maxima for a-type hemes as well as the overlapping maxima for hemes b and c are marked with lines as a guide to the eye. C, steady-state levels of representative subunits of respiratory Complexes IV (Cox1–Cox3) and III (Rip1) along with porin (loading control) in mitochondria of the cells described in A analyzed by SDS-PAGE immunoblotting with appropriate antibodies. Positions of molecular mass markers are indicated on the left side of the blots. Results are shown for one experiment, representative of three independent experiments (biological replicates). D, co-immunoadsorption of differentially tagged variants of Cox15. Digitonin-solubilized mitochondrial lysates (500 μg) from cox15Δ cells co-expressing Cox15-FLAG and either Cox15-Myc or vector control were incubated with anti-Myc-agarose beads for 25 h at 4 °C. An SDS-PAGE immunoblot analyzed with anti-FLAG and anti-Myc antibodies shows 1% of mitochondrial lysates prior to (Load) and after (Unbound) the incubation with affinity resin, the whole fraction of protein precipitated from the final wash (Wash), and half of the eluate (Bound). Positions of molecular mass markers are indicated on the left side of the blots. Results are shown for one experiment, representative of three independent experiments (biological replicates). IP, immunoprecipitation.
Miscellaneous
Yeast cells were transformed using the lithium acetate procedure (44). Total mitochondrial protein concentrations were determined using a Coomassie Plus assay kit (Thermo Scientific) via the Bradford method. Whole-cell protein lysates were prepared as described previously (13). Amino acid sequence alignment was performed with use of the United States National Library of Medicine's National Center for Biotechnology Information online resources. Statistical significance was determined by one-way analysis of variance with Bonferroni's post hoc test in Kaleidagraph 4.1 (Synergy Software). All experiments reported in this study were performed in triplicate minimally.
Results
Yeast Heme a Synthase Complexes Contain Multiple Copies of Cox15
A previous report has indicated that Cox15 associates with Shy1 and with early CcO assembly intermediates but is a substoichiometric component of those intermediates (10). We therefore examined whether Cox15 may associate with other copies of itself to form homo-oligomeric complexes similarly to Cox10 (heme o synthase). To this end, we generated two plasmid-borne variants of Cox15 in which a 13xMyc or FLAG epitope tag is appended to the C terminus of the protein. The respiratory growth, mitochondrial heme a content, and steady-state levels of CcO core subunits Cox1, Cox2, and Cox3 of cox15Δ cells expressing the Cox15-Myc protein were comparable with those of WT cells, indicating that Cox15-Myc is expressed at adequate levels for function and that the 13xMyc tag does not interfere with enzyme function (Fig. 1, A–C). The expression of the Cox15-FLAG protein in cox15Δ cells also resulted in robust Cox15 function with slightly slower respiratory growth and slightly decreased levels of CcO core subunits (Fig. 1, A–C). We therefore used these two viable proteins for co-immunoprecipitation experiments. As there is no commercially available antibody to permit detection of untagged yeast WT Cox15, we chose to use the Cox15-Myc construct for all further work shown in the present study.
To test for Cox15-Cox15 interactions, mitochondria isolated from cox15Δ cells co-expressing Cox15-FLAG with either Cox15-Myc or vector control were used in co-immunoprecipitation experiments. Immunoadsorption of Cox15-Myc on anti-Myc-agarose beads resulted in specific co-purification of the Cox15-FLAG protein (Fig. 1D). Similar results were obtained with cells co-expressing Cox15-Myc and a Cox15-Yah1 fusion construct (2) that mimics the naturally occurring fusion protein found in Schizosaccharomyces pombe (data not shown). These results indicate that Cox15 likely functions as a homo-oligomer, which may form associative complexes with other known partner proteins such as Shy1.
Cox15 Forms a Highly Stable Complex via Hydrophobic Interactions
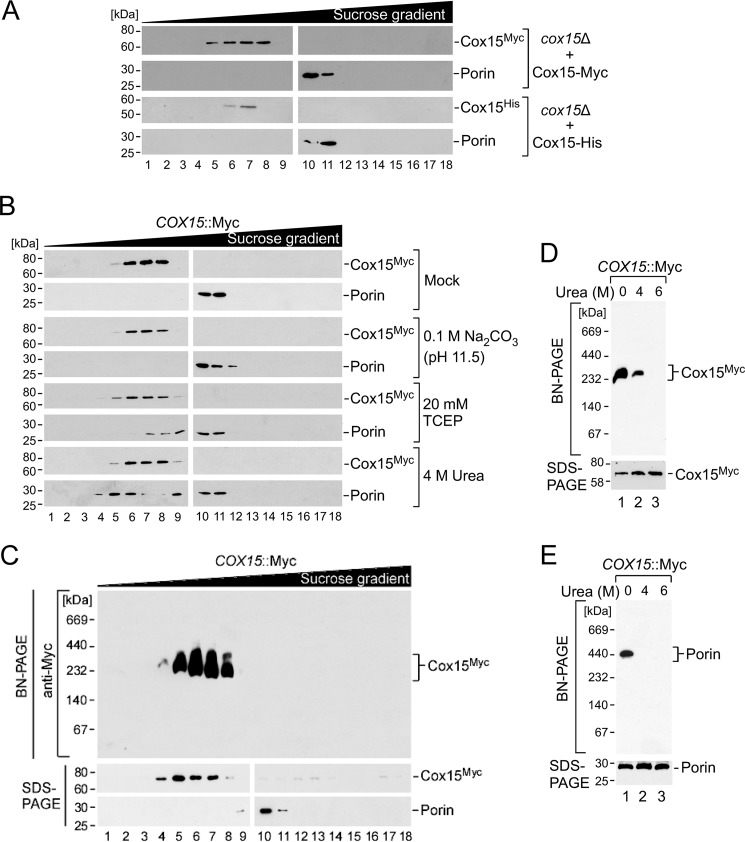
To gain additional insight into the oligomerization properties of Cox15, we interrogated the behavior of the Myc-tagged heme a synthase upon sucrose gradient ultracentrifugation under various physicochemical conditions. Cox15 oligomers fractionated in the molecular mass range of ∼200–350 kDa (Fig. 2). To verify that these complexes formed in the absence of the 13xMyc tag, we used Cox15 C-terminally modified with a minimal tag of seven additional residues (GHHHHHH; Cox15-His) and found it formed complexes of similar size (Fig. 2A).
FIGURE 2.
Analysis of Cox15 multimerization properties. A, mitochondria (1 mg) from cox15Δ cells expressing either Cox15-Myc or Cox15-His were lysed in a buffer containing 1% digitonin. Clarified lysates were loaded onto a continuous 7–50% sucrose gradient and subjected to ultracentrifugation. An SDS-PAGE immunoblot of collected gradient fractions shows the distribution of Cox15 oligomers (anti-Myc antibody or anti-His antibody), and the 440-kDa porin complex was used for size calibration (anti-porin antibody). Positions of molecular mass markers are indicated on the left side of the blots. Results are shown for one experiment, representative of three independent experiments (biological replicates). B, Cox15-Myc complexes from chromosomally tagged COX15::Myc cells were fractionated by sucrose gradient ultracentrifugation as in A after the indicated treatments. Results are shown for one experiment, representative of three biological replicates. C, Cox15-Myc complexes from COX15::Myc cells were fractionated by sucrose gradient ultracentrifugation as in A, and the collected fractions were analyzed by native electrophoresis (BN-PAGE using a 5–13% native gradient gel) with immunoblotting as in A. SDS-PAGE immunoblots are shown below the BN-PAGE immunoblot for each fraction. Results are shown for one experiment, representative of three independent experiments (biological replicates). D and E, gradient fractions from the experiment in C containing Cox15-Myc (fraction 7; D) or porin (fraction 11; E) complexes were incubated in the presence of 4 or 6 m urea or left untreated, and complex integrity was assessed by BN-PAGE immunoblotting using the antibodies described above. The bottom part of each panel shows steady-state levels of the respective protein in each sample tested, analyzed by SDS-PAGE immunoblotting as above. Results are shown for one experiment, representative of three biological replicates. TCEP, tris(2-carboxyethyl)phosphine.
The Cox15 complex remained stable in the presence of alkaline sodium carbonate and 20 mm tris(2-carboxyethyl)phosphine reductant, suggesting that ionic and disulfide bond interactions do not significantly contribute to oligomer stability. Importantly, the Cox15 oligomer exhibited marked resistance to treatment with 4 m urea, a condition sufficient for destabilization of the control porin complex (Fig. 2B). To verify this result, we confirmed the integrity and size of isolated Cox15 oligomers from sucrose gradient fractions enriched in Cox15 complexes by BN-PAGE (Fig. 2C). We also incubated isolated complexes with increasing concentrations of urea and tested their stability under native gel conditions. Unlike the control porin oligomers (Fig. 2E), Cox15 complexes displayed marked resistance to chemical denaturing, withstanding up to 4 m urea (Fig. 2D). These treatments did not affect steady-state levels of either protein (Fig. 2, D and E, bottom panels). Collectively, these results suggest that Cox15 forms highly stable assemblies that appear to be stabilized by strong hydrophobic interactions.
Cox15 Oligomerization is Independent of Heme and Is an Evolutionarily Conserved Feature
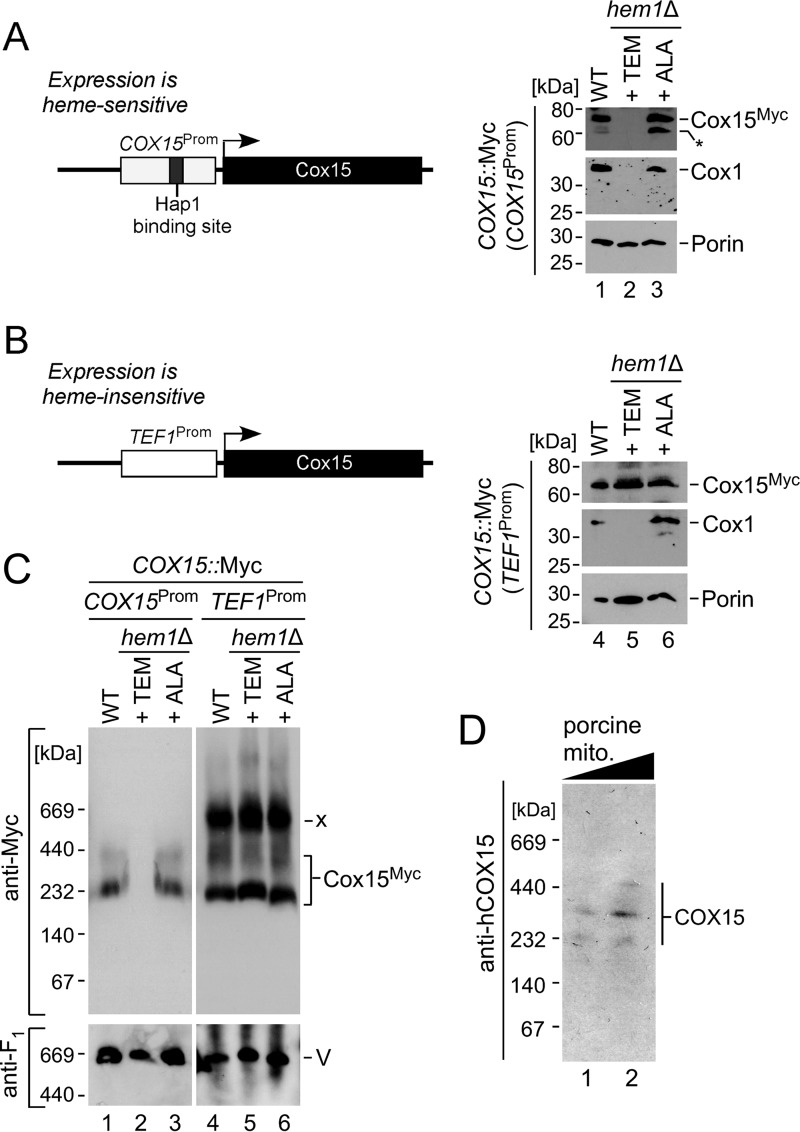
The lack of heme o substrate does not affect Cox15 complex stability as judged by the observation of normal levels of Cox15 oligomer in cox10Δ cells (1, 10). However, studies in bacteria have suggested an additional heme b molecule is stably bound to heme a synthase and might serve as a cofactor (27, 45). We therefore sought to determine whether such a cofactor heme might serve as a stabilizing factor for Cox15 oligomers. To test this hypothesis, we assessed the effect of cellular heme depletion on the stability of the Cox15 complexes. Heme-depleted cells are viable in a fermentable medium when supplemented with an oleic acid precursor, ergosterol, and methionine (46). One caveat of this experiment, however, is that the expression of yeast Cox15 is controlled by the Hap1 transcription factor and is therefore heme-dependent (26). Indeed, depletion of cellular heme via either deletion of 5-aminolevulinate synthase (Hem1, a key heme biosynthetic enzyme) or through pretreatment of the cells with the heme biosynthesis inhibitor 4,6-dioxoheptanoic acid ablated the expression of both Cox15 and the core CcO subunit Cox1, whereas supplementation of cells deficient in Hem1 with its reaction product, δ-aminolevulinic acid, restored the expression of both Cox15 and Cox1 proteins (Fig. 3A and data not shown). Therefore, to uncouple Cox15 expression from cellular heme concentration, we replaced the endogenous promoter region of the COX15::Myc locus with the heme-insensitive TEF1 promoter by homologous recombination. This genetic manipulation permitted the expression of Cox15-Myc (but not Cox1) under heme-deficient conditions (Fig. 3B). We then tested the effect of cellular heme depletion on the oligomerization of heme a synthase by BN-PAGE. Surprisingly, the heme-insensitive Cox15 demonstrated the ability to oligomerize irrespective of heme availability (Fig. 3C). Hence, oligomerization of Cox15 is not directly mediated by heme nor is it dependent upon the presence of the proposed heme b cofactor. In addition to the Cox15 complexes typically seen by native electrophoresis, we observed that the heme-insensitive Cox15 formed an additional high mass band of ∼660 kDa (Fig. 3C, labeled “x”). At present, the identity of this complex remains unknown and will be the subject of future analyses.
FIGURE 3.
Cox15 oligomerization is independent of heme binding and evolutionarily conserved. A and B, schematic depiction of the WT and modified COX15 loci. Expression of the WT Cox15 is heme-dependent due to the presence of a Hap1-binding site in the promoter region and is thus affected in heme-depleted cells (A). Replacement of the COX15 promoter (Prom) with the TEF1 promoter sequence renders Cox15 expression heme-independent (B). SDS-PAGE immunoblots show steady-state levels of the indicated proteins in WT and hem1Δ COX15::Myc cells cultured in the presence of Tween 80/ergosterol/methionine (TEM) mixture or δ-aminolevulinic acid (ALA) detected with anti-Myc, anti-Cox1, and anti-porin antibodies. Positions of molecular mass markers are indicated on the left side of the blots. The asterisk denotes a nonspecific cross-reacting band. Results are shown for one experiment, representative of three independent experiments (biological replicates). C, BN-PAGE analysis of Cox15-Myc complexes in mitochondria derived from the strains described above. Protein complexes were separated by BN-PAGE using a 5–13% native gradient gel and analyzed by anti-Myc immunoblotting to visualize Cox15 complexes. The monomeric form of respiratory Complex V visualized by immunodecoration with anti-Atp2 (anti-F1) serum served as a loading control. Results are shown for one experiment, representative of three independent experiments (biological replicates). Positions of molecular mass markers are indicated on the left side of the blots. x, unknown band. D, mitochondria (mito.) (40 and 80 μg) from porcine kidneys were analyzed by BN-PAGE. The mammalian heme a synthase complexes were detected by immunoblotting with an antibody against human COX15. Results are shown for one experiment, representative of three technical replicates.
To confirm evolutionary conservation of the oligomeric features of Cox15, we analyzed porcine kidney mitochondrial lysates by BN-PAGE immunoblotting using antibodies against human heme a synthase. This analysis revealed a series of discrete bands that resemble the Cox15 oligomers in yeast (Fig. 3D), suggesting that oligomerization of Cox15 is an evolutionarily conserved feature.
Each of the Four Conserved Histidine Residues of Cox15 Is Essential for Its Catalytic Function
Homology searches reveal heme a synthase orthologs in animals, plants, fungi, and bacteria. To identify and determine the roles of key active site residues of Cox15, we began with an investigation of potential heme-binding ligands that show strict conservation among members of these diverse kingdoms. Although there are no strictly conserved Cys, Met, Lys, or Tyr residues, there are four invariant histidine residues, which are each conserved from bacteria to humans. These residues correspond to the four conserved histidines previously identified in the bacterial heme a synthase ortholog CtaA (His-60, His-123, His-216, and His-278 in the Bacillus subtilis enzyme) (27). Although it is accepted that these residues are important for the catalytic function of the enzyme, their roles remain unclear. In S. cerevisiae, the invariant histidines are His-169, His-245, His-368, and His-431 with each residue positioned closer to the intermembrane space than the matrix side of the inner mitochondrial membrane (Fig. 4A). A recent study by Bareth et al. (10) has demonstrated that conversion of His-368 to methionine abolishes the function of Cox15 without affecting the stability or oligomerization of the protein. We sought to determine the importance of each of the invariant histidines for Cox15 multimerization and the significance of these residues in the function of the eukaryotic enzyme.
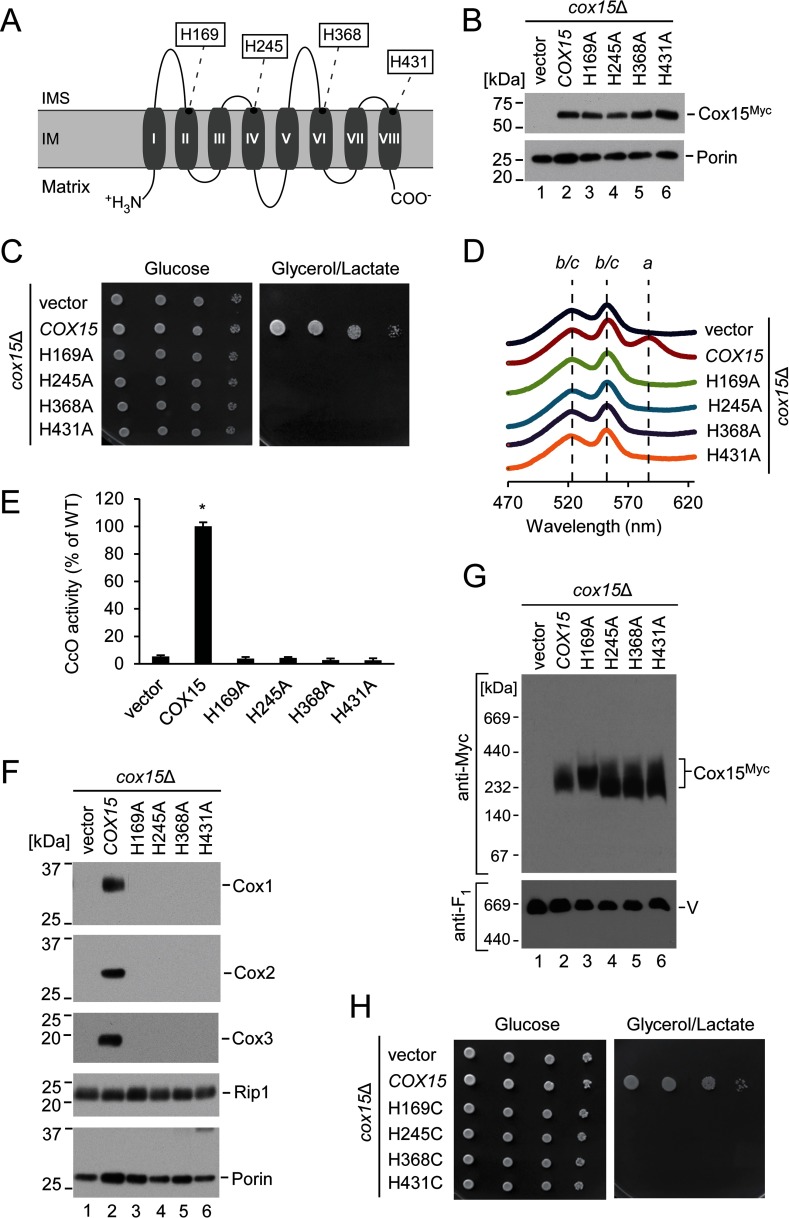
FIGURE 4.
Invariant histidine residues of Cox15 are critical for its function and cannot be functionally replaced with other heme-ligating residues. A, schematic representation of yeast Cox15, showing predicted topology and transmembrane domains of the protein. The four highly conserved histidine residues are indicated. B, steady-state mitochondrial protein levels of the wild type Cox15-Myc and its H169A, H245A, H368A, and H431A variants expressed in cox15Δ cells along with vector control. The indicated proteins were visualized by immunoblotting with anti-Myc and anti-porin (loading control) antibodies. Positions of molecular mass markers are indicated on the left side of the blots. Results are shown for one experiment, representative of three independent experiments (biological replicates). C, respiratory growth test of cox15Δ cells expressing the plasmids in B. Cells were handled and tested as described in Fig. 1A. Results are shown for one experiment, representative of four independent experiments (biological replicates). D, heme-pyridine redox difference spectra of mitochondria from the indicated cells presented as in Fig. 1B. Spectra are representative of three independent experiments (biological replicates). E, CcO enzymatic activity in mitochondria from the above cells determined by spectrophotometric cytochrome c oxidation assay and expressed as a percentage of WT activity. Bars indicate the average and S.D. of three technical replicates from one experiment representative of three independent experiments (biological replicates). Asterisks indicate a statistically significant difference from activity of cox15Δ cells expressing vector control (p < 0.0001). Error bars represent S.D. F, steady-state protein levels in mitochondria of the indicated cells as in Fig. 1C. Results are shown for one experiment, representative of three independent experiments (biological replicates). G, cox15 complexes from the above mitochondria analyzed by BN-PAGE as in Fig. 3C. Results are shown for one experiment, representative of three independent experiments (biological replicates). H, respiratory growth test as in C for the H169C, H245C, H368C, and H431C variants. Results are shown for one experiment, representative of four independent experiments (biological replicates). IM, inner mitochondrial membrane; IMS, intermembrane space.
Yeast Cox15 variants in which each of these histidines was converted to an alanine residue to disrupt any function of the imidazole side chain were stably expressed from plasmids in cox15Δ cells (Fig. 4B). None of these variants were able to support respiratory growth of cox15Δ yeast (Fig. 4C). Consistent with an absence of heme a synthase activity, analysis of heme redox difference spectra of mitochondria isolated from these cells revealed no detectable levels of the absorption peak at ∼588 nm characteristic of a-type heme (Fig. 4D). For each of the variants, we thus observed a loss of CcO enzymatic activity and a significant decrease in the steady-state concentrations of the core CcO subunits Cox1, Cox2, and Cox3 (Fig. 4, E and F). BN-PAGE analysis revealed that each of the mutant forms of heme a synthase retained its ability to oligomerize (Fig. 4G). In addition, we observed that proteins with the H245A, H368A, and H431A substitutions were predominantly organized into a slightly lower molecular weight species of Cox15. Thus, the conserved histidine residues are largely dispensable for Cox15 multimerization; however, their inactivation can exert modulatory effects on Cox15 oligomers.
Because the mechanism of Cox15-mediated catalysis necessitates binding of heme o to form an enzyme-substrate complex, one or more of the four invariant histidine residues may be axial ligands for either the heme o substrate or the proposed heme b cofactor. However, it is also possible that one or more of these residues do not bind to heme and instead serve a catalytic or structural role such as serving as a general acid/base catalyst, positioning the substrate for reaction, or stabilizing an active conformation of an oligomeric complex. To differentiate between heme binding and these alternative possibilities, we converted each conserved histidine to cysteine (or in a separate experiment to tyrosine). Cysteine and tyrosine residues can be potential heme axial ligands but are unlikely to provide an effective substitute for the histidine imidazole ring in catalysis (47). All eight variants (H169C, H245C, H368C, H431C, H169Y, H245Y, H368Y, and H431Y) were stably expressed in cox15Δ cells (data not shown), but surprisingly, none of these histidine to cysteine or histidine to tyrosine variants were able to rescue the heme a biosynthetic defect of the cox15Δ strain (Fig. 4H and data not shown). Altogether, these results suggest that all four invariant histidine residues are essential for and directly involved in either the heme binding or the catalytic mechanism of Cox15.
The Linker Connecting the N- and C-terminal Domains of Cox15 Is Important for Oligomerization and Function
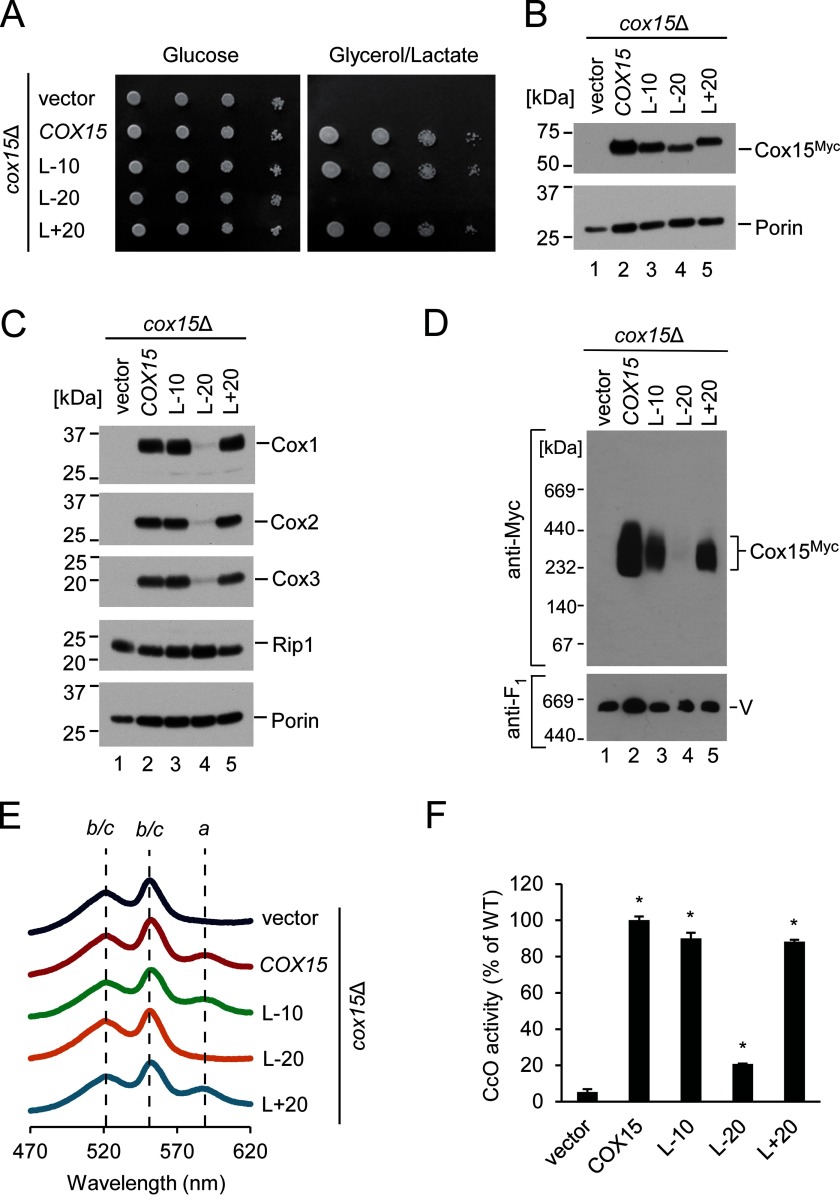
Studies in bacteria have indicated that the N- and C-terminal regions of heme a synthase display a significant degree of similarity (∼28% sequence identity in B. subtilis) and could be the result of a gene duplication and fusion (27, 48). The N- and C-terminal portions of Cox15 in eukaryotes appear to be similarly related to each other. These regions are joined by a ∼20-residue-long matrix-exposed linker region that connects helices IV and V. Although this linker exhibits a low degree of evolutionary conservation, it appears to be a common feature of heme a synthase in eukaryotes. To test whether this linker may be important for Cox15 function through maintenance of an optimal distance between conserved histidine-containing domains of the enzyme, we generated constructs wherein the length of the linker was either reduced or extended.
To test shortened linkers, truncated variants of Cox15 were generated that lacked 10 (L-10) or 20 (L-20) amino acid residues in the linker region. To interrogate whether extension of the linker region would modulate Cox15 function, we used a 20-amino acid spacer derived from the unstructured region of the E. coli lacI repressor protein (34) to increase the distance between the N- and C-terminal portions of Cox15-Myc, creating a construct designated L+20. This approach was taken because structural modeling predicts the linker region of Cox15 to be unstructured (see “Discussion”).
Neither shortening the linker by half its length (L-10) nor extending it to double its length (L+20) impaired the ability of plasmid-borne Cox15 to support respiratory growth of cox15Δ cells. However, loss of the linker (L-20) resulted in a significant respiratory growth defect (Fig. 5A) despite the stable expression of the construct (Fig. 5B). Consistent with these results, the functional L-10 and L+20 variants formed oligomeric complexes, and their mitochondrial lysates resembled the WT Cox15 in steady-state levels of core CcO subunits, heme a content, and CcO activity (Fig. 5, C–F). Interestingly, the L-20 variant lacking the linker region was severely compromised in its ability to form oligomeric complexes and exhibited defects in steady-state levels of core CcO subunits, mitochondrial heme a content, and CcO activity (Fig. 5, C–F), suggesting that the presence of this linker is critical for Cox15 oligomerization and function.
FIGURE 5.
Role of the linker region between N- and C-terminal domains of Cox15. A, respiratory growth test of cox15Δ cells expressing vector control or the indicated variants of Cox15-Myc with varying lengths of the linker region connecting helices IV and V (where L-10 and L-20 lack 10 and 20 amino acid residues, respectively, and L+20 contains an additional 20 residues). Cells were handled and tested as in Fig. 1A. Results are shown for one experiment, representative of four independent experiments (biological replicates). B, steady-state mitochondrial protein levels of the cells described in A assessed by SDS-PAGE immunoblotting as in Fig. 4B. Results are shown for one experiment, representative of three independent experiments (biological replicates). C, steady-state protein levels in mitochondria of the indicated cells as in Fig. 1C. Results are shown for one experiment, representative of three independent experiments (biological replicates). D, Cox15 complexes from the above mitochondria analyzed by BN-PAGE as in Fig. 3C. Results are shown for one experiment, representative of three independent experiments (biological replicates). E, heme-pyridine redox difference spectra of mitochondria from the indicated cells presented as in Fig. 1B. Spectra are representative of three independent experiments (biological replicates). F, CcO enzymatic activity in mitochondria from the above cells determined by spectrophotometric cytochrome c oxidation assay and expressed as a percentage of WT activity. Bars indicate the average and S.D. of three technical replicates from one experiment representative of three independent experiments (biological replicates). Asterisks indicate a statistically significant difference from activity of cox15Δ cells expressing vector control (p < 0.0001). Error bars represent S.D.
Effects of COX15 Patient Mutations on the Human and Yeast Enzymes
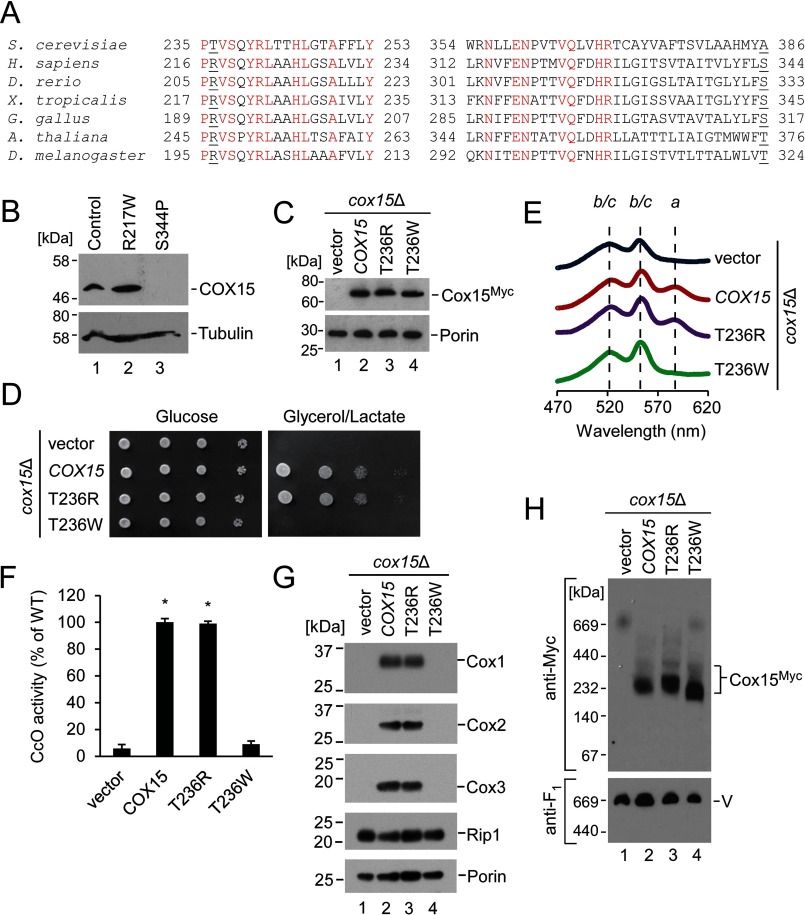
Mutations in human COX15 have been associated with fatal infantile hypertrophic cardiomyopathy and LS, and patients are variable in presentation (14–17). The mutations reported to date include the pathological substitutions S344P (found in a patient who was compound heterozygous with an S151X nonsense mutation introducing a premature stop codon) and R217W (found in a homozygous patient, a compound heterozygous patient with S151X, and a compound heterozygous patient with an intron splice site mutation, C447–3G, which introduces a frameshift and a premature stop codon) (Fig. 6A) (14–17). CcO activity was significantly reduced in at least one of the cell types examined for each of these patients (14–17). Heme levels in cardiac mitochondria from the R217W/C447–3G patient were consistent with COX15 deficiency; heme a content was drastically reduced, and the heme o intermediate accumulated (14).
FIGURE 6.
Effects of COX15 mutations associated with human mitochondrial disease on the human and yeast enzymes. A, amino acid sequence alignment (only partially depicted) with strictly conserved residues highlighted in red and the residues Thr-236 (human Arg-217) and Ala-386 (human Ser-344) underlined. B, SDS-PAGE immunoblot showing steady-state levels of COX15 and tubulin (loading control) in whole-cell extracts of skin fibroblasts from patients with hypertrophic cardiomyopathy (R217W/C447–3G-causing mutations) and LS (S344P/S151X). Because of the limited availability of these samples (cells from one patient for each mutation), results are shown for one experiment, representative of three technical replicates. Positions of molecular mass markers are indicated on the left side of the blots. C, steady-state mitochondrial protein levels of yeast Cox15-Myc, its T236R and T236W variants, and vector control expressed in cox15Δ cells. The respective proteins were visualized by SDS-PAGE immunoblotting as in Fig. 4B. Results are shown for one experiment, representative of four independent experiments (biological replicates). D, respiratory growth test of cox15Δ cells expressing the indicated variants of Cox15-Myc or vector control. Cells were handled and tested as described in Fig. 1A. Results are shown for one experiment, representative of four independent experiments (biological replicates). E, heme-pyridine redox difference spectra of mitochondria from the above cells presented as in Fig. 1B. Spectra are representative of three independent experiments (biological replicates). F, CcO enzymatic activity in mitochondria from the above cells determined by spectrophotometric cytochrome c oxidation assay and expressed as a percentage of WT activity. Bars indicate the average and S.D. of three technical replicates from one experiment representative of three independent experiments (biological replicates). Asterisks indicate a statistically significant difference from activity of cox15Δ cells expressing vector control (p < 0.0001). Error bars represent S.D. G, steady-state protein levels in mitochondria of the indicated cells as in Fig. 1C. Results are shown for one experiment, representative of three independent experiments (biological replicates). H, Cox15 complexes from the above mitochondria analyzed by BN-PAGE as in Fig. 3C. Results are shown for one experiment, representative of three independent experiments (biological replicates).
To investigate molecular consequences of these amino acid conversions, we evaluated the steady-state levels of heme a synthase in whole-cell lysates from cultured skin fibroblasts isolated from patients bearing the S344P/S151X-causing mutations (this patient presented with LS) and the R217W/C447–3G-causing mutations (this patient presented with fatal infantile hypertrophic cardiomyopathy). COX15 protein in the S344P/S151X cells was intrinsically unstable. However, steady-state levels of COX15 protein in cells with R217W/C447–3G were comparable with the control (Fig. 6B). Although differences in protein stability can potentially explain the diverse phenotypes associated with these substitutions, no additional useful information could be gained about the S344P/S151X variant due to its intrinsic instability. The observation of COX15 protein in the R217W/C447–3G variant lysates that is indistinguishable in size from the WT protein suggests that the R217W protein is stably expressed, whereas the allele bearing the C447–3G splicing mutation does not yield stable protein and may effectively be a null allele as suggested by Antonicka et al. (14). We thus focused our analysis on the R217W mutant protein. Because of sample limitations, we engineered this amino acid conversion at the corresponding residue in yeast Cox15.
Arg-217 is a conserved residue located in the intermembrane space-exposed loop of Cox15 between helices III and IV. Interestingly, baker's yeast heme a synthase is one of the few instances wherein this highly conserved arginine residue is replaced with threonine (Thr-236) (Fig. 6A). Therefore, two amino acid conversions were generated: 1) T236R, converting the yeast residue to the human residue at this location, and 2) T236W, mimicking the pathogenic substitution found in patients. Both variants were stably expressed in the cox15Δ strain (Fig. 6C). The T236R variant rescued respiratory growth similarly to the yeast WT; however, the T236W variant was unable to support respiratory growth of the heme a synthase-deficient strain (Fig. 6D). Consistent with this result, mitochondria from cells bearing the T236W variant exhibited impaired levels of heme a as determined by heme difference spectroscopy (Fig. 6E), were deficient in CcO activity (Fig. 6F), and had decreased steady-state levels of core CcO subunits (Fig. 6G). Native gel electrophoresis of these mitochondria revealed that, similarly to the H245A, H368A, and H431A substitutions, the ∼200-kDa Cox15 complex found in the T236W variant is shifted to a smaller mass than its functional counterparts (Fig. 6H). We thus concluded that the R217W substitution, which has been identified in two patients with hypertrophic cardiomyopathy and a patient with LS, impairs enzymatic activity and affects oligomerization of the Cox15 complex.
Discussion
The eukaryotic heme a synthase is essential for assembly of respiratory Complex IV in mitochondria. A deficiency in the formation of heme a and thus functional CcO has grievous pathological consequences and has been associated with severe clinical manifestations such as fatal infantile hypertrophic cardiomyopathy and LS (14–17). Despite its biomedical importance, very little information is available concerning the structural and functional properties of Cox15. In this study, we conducted a series of detailed analyses aiming to better understand the molecular properties of Cox15. We report several findings, discussed below, that provide new insight into the function of Cox15 in health and disease.
Cox15 Oligomeric Complexes
Heme a synthase complexes in yeast are shown to be highly stable and contain more than one copy of Cox15 protein. Additionally, multimerization of heme a synthase appears to be an evolutionarily conserved feature; COX15 protein was observed to form oligomeric complexes of comparable molecular weight in mammalian mitochondria. The multimer appears to be stabilized by strong hydrophobic interactions; as a consequence, the complex is rigid and cannot be easily dissociated even under typically denaturing conditions. A previous report has established that Cox15 can associate with Cox1 assembly intermediates, but heme a synthase is a substoichiometric component of these assembly modules (10). Consistently, oligomerization of Cox15 does not appear to be compromised in the absence of Cox1 synthesis or CcO assembly (7, 10). In light of these observations and our finding of the robust nature of Cox15 oligomers, it is plausible that Cox15 may have an additional function besides heme a formation, potentially serving as a scaffold for Cox1 maturation during heme a insertion.
We found that the oligomerization of Cox15 is not directly regulated by heme. The protein is able to oligomerize under heme-deficient conditions if its expression is rendered insensitive to heme levels. Therefore, if Cox15 contains a heme cofactor, as has been suggested by studies of the bacterial heme a synthase, the absence of that cofactor does not affect the ability of the enzyme to oligomerize. The oligomerization of Cox15 also contrasts with that of heme o synthase, Cox10, which is stimulated to multimerize primarily through heme-sensitive Cox1 translation (7). Cox15 complex formation appears to be regulated instead through transcription level control of expression by its heme-sensitive promoter (26) and its dependence upon the Coa2 assembly factor (7). This finding is consistent with previous reports demonstrating that loss of either heme o substrate-supplying Cox10 or heme a acceptor Cox1 has little to no effect on Cox15 oligomerization (7, 10). If Cox15 complexes serve as a scaffold for Cox1 maturation, their absence under heme-depleted conditions (due to Cox15 expression being regulated by Hap1) might represent another facet of an efficient feedback mechanism ensuring that no Cox1 assembly intermediates/scaffolds are formed when cellular heme levels are low. Indeed, Shy1-containing assembly intermediate complexes are impaired in the absence of Cox15 (10).
Oligomerization of Cox15 may be necessary but insufficient for enzymatic activity. Several catalytically inactive variants of Cox15 (H169A, H245A, H368A, H431A, and T236W) retained their ability to oligomerize, similarly to a catalytically inactive variant of Cox10 (6, 7) and consistent with the recent report by Bareth et al. (10), who showed that a H368M variant of Cox15 formed complexes and retained its associations with Cox1 assembly intermediates. We observed an apparent shift in the size of the Cox15 oligomer by BN-PAGE for some catalytically inactive variants (H245A, H368A, H431A, and T236W), suggesting oligomerization has been perturbed in some manner, whereas the catalytically inactive H169A oligomer resembled that of the WT and the functional variants (T236R, L-10, and L+20). Further studies are warranted to assess any implications of this observation for the role of His-169. Significantly, a variant of Cox15 (L-20) that had a significant defect in oligomerization also exhibited a significant defect in enzymatic activity, underscoring the importance of oligomerization for Cox15 function.
Key Cox15 Residues and Structural Features
This study reveals the essential role of four evolutionarily conserved histidine residues of Cox15 for heme a biosynthesis in eukaryotes. As demonstrated by a recent study, the prokaryotic variants of heme a synthase are markedly different from the eukaryotic enzyme as prokaryotic, but not eukaryotic, heme a synthase contains a disulfide bond essential for catalytic activity (49). Despite these differences, we have found that, similarly to the prokaryotic heme a synthase (27, 45), the four conserved histidine residues of eukaryotic Cox15 are each essential for proper assembly of CcO and electron transport chain function. The absence of heme a observed by difference spectroscopy in Δcox15 cells expressing Cox15 with the histidine to alanine substitutions suggests each histidine residue is essential for conversion of heme o to heme a. Therefore, although these residues might potentially be important in a possible chaperone function of Cox15 in CcO maturation, all four histidines appear to be primarily important for heme a biosynthesis.
Remarkably, Cox15 variants converting each histidine to alternative heme-binding ligands cysteine and tyrosine fail to rescue the respiratory growth defect of Δcox15 cells, demonstrating the essential role of histidine at these positions. In light of the structural modeling discussed below and the absence of alternative heme ligands conserved from bacteria to eukaryotes, it is likely that one or more of the conserved histidines do act as a heme-binding ligand, although in the case of Cox15, cysteine and tyrosine are evidently poor substitutes for this role. Additionally, an unsuccessful attempt to isolate any extragenic suppressors for the respiratory growth defect observed in Δcox15 cells expressing each of the Cox15 proteins with single histidine to alanine substitutions underscores the essential nature of His-169, His-245, His-368, and His-431.
The presence of the matrix-localized loop connecting the N- and C-terminal domains of Cox15 was also observed to be critical. Removal of this region resulted in a significant defect in both Cox15 oligomerization and enzyme function, whereas partial reduction or extension of the linker was not deleterious, indicating that the precise length of the linker is not critical and that the linker loop is unlikely to contain a site for interaction with other proteins.
Additional insight into eukaryotic heme a synthase was gained from analysis of patient mutations known to occur in human COX15. Although the LS-associated S344P conversion destabilizes the protein, the R217W substitution (corresponding to Thr-236 in the yeast enzyme), which has been associated with both fatal infantile hypertrophic cardiomyopathy and LS in different individuals, impairs catalytic activity and affects the assembly of the Cox15 oligomer in yeast. Our observations may shed light on the molecular basis of the diverse clinical manifestations associated with genetic disorders of COX15. Interestingly, the naturally occurring substitution of the highly conserved arginine to the polar but uncharged threonine residue seen in the yeast enzyme does not impair catalytic or oligomerization properties of Cox15.
Because very little is known about the structure of Cox15, we used the Phyre2 protein fold recognition server to model its structure based on homology to known protein folds (Fig. 7). The resulting model reveals that monomeric Cox15 comprises two heme-binding four-helical bundles, each containing a pair of evolutionarily conserved histidines that are positioned in proximity to one another. In the model, the His-169/His-245 pair within the N-terminal heme-binding domain is positioned with the two imidazole side chains pointed toward each other in a manner capable of binding heme by serving as two axial ligands for the iron ion. The His-368/His-431 pair is positioned similarly within the C-terminal heme-binding domain.
The enzymatic mechanism of Cox15 and whether or not it contains a heme cofactor involved in catalysis remain debated (26–28, 45). One possibility for the roles of the N- and C-terminal heme-binding domains is that each domain performs a different function. This idea is supported by the data demonstrating that all four conserved histidine residues (i.e. histidines in both domains) are essential for heme a biosynthesis. For example, one heme-binding domain of Cox15 could serve to ligate the heme o substrate with a pair of histidine axial ligands, whereas the other domain could form an active site wherein a heme cofactor is ligated by one histidine residue, leaving the other axial site of the iron ion available to catalyze the oxidation reaction with the remaining histidine serving a catalytic or structural function nearby (e.g. acting as a general acid/base catalyst for the reaction). The model does not suggest how the two domains might interact. One scenario, among many possibilities, is that a domain ligating substrate heme o could position the porphyrin C8 methyl group of the substrate so that it approaches the iron center of a cofactor heme bound in the other domain comprising the active site. Within a homo-oligomeric complex, it is possible that domains from different subunits could functionally interact.
We mapped the conserved residues of Cox15 onto the model (Fig. 7B), choosing strictly conserved residues from the sequence alignment partially depicted in Fig. 6A, and found that the residues surrounding the heme-binding pockets of each domain as well as the intermembrane space-exposed residues of each domain were highly conserved. In contrast, the matrix-exposed loops did not exhibit high sequence conservation. The human COX15 protein was also threaded onto the yeast Cox15 model, yielding a model with the same overall structure and histidine residue positioning as the yeast model (data not shown).
This Cox15 model was used to assess the relative positions of pathological substitutions reported in patients with different forms of mitochondrial disease, S344P and R217W. The position of residue Ser-344 (corresponding to yeast Ala-386) is within a transmembrane helix of the C-terminal heme-binding domain. The residue is located near the end of the helix toward the matrix side, and the helix is connected by only a short loop to the next transmembrane helix of the heme-binding four-helical bundle. Conversion to a proline at this location would be expected to have a helix-breaking effect, destabilizing the structure of the C-terminal heme-binding domain of the protein. This prediction is consistent with our observation of an absence of COX15 protein in patient fibroblasts with the S344P conversion, suggesting that the misfolded protein is degraded.
The Arg-217 residue (Thr-236 in yeast) is located in a highly conserved loop on the intermembrane space side of the N-terminal heme-binding domain. This loop is close to the heme-binding site of the His-169/His-245 pair and could potentially be involved in mediating heme o/a transfer into/out of this site. Alternatively, it could be positioned within the active site for a structural role or on the surface where it could be involved in interaction with the C-terminal domain or intersubunit contacts of the oligomer. The substitution of the hydrophobic, uncharged tryptophan residue at this position could disrupt any of these functions. Future structural studies are warranted to ascertain the roles of each of these important residues and refine the conclusions from our biochemical analyses.
Author Contributions
J. L. F. and O. K. designed and coordinated the study, performed experiments, analyzed results, and wrote the manuscript. S. S., A. C., N. J. H., and N. G. T. performed experiments and analyzed results. All authors reviewed the results and approved the final version of the manuscript.
Acknowledgments
We thank Drs. Dennis Winge (University of Utah), Eric Hegg (Michigan State University), Antoni Barrientos (University of Miami), Eric Shoubridge (McGill University), Massimo Zeviani (Medical Research Council-Mitochondrial Biology Unit), and Alexander Tzagoloff (Columbia University) for reagents. We acknowledge expert technical assistance of Nataliya Zahayko.
This work was supported by National Institutes of Health grants P30GM103335 (to the Nebraska Redox Biology Center) and R01GM108975 (to O. K.), Cottrell College Science Award 22643 (to J. L. F.) from Research Corporation for Science Advancement, National Science Foundation-Major Research Instrumentation Grant CHE-1229559 (to J. L. F. as co-principal investigator), College of Charleston Summer Undergraduate Research with Faculty grants (to J. L. F., N. J. H., and N. G. T.), and Howard Hughes Medical Institute Grant 52007537 through the Pre-college and Undergraduate Science Education Program (to the College of Charleston). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- CcO
- cytochrome c oxidase
- BN
- blue native
- LS
- Leigh syndrome.
References
- 1. Kim H. J., Khalimonchuk O., Smith P. M., and Winge D. R. (2012) Structure, function and assembly of heme centers in mitochondrial respiratory complexes. Biochim. Biophys. Acta 1823, 1604–1616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Barros M. H., Carlson C. G., Glerum D. M., and Tzagoloff A. (2001) Involvement of mitochondrial ferredoxin and Cox15p in hydroxylation of heme O. FEBS Lett. 492, 133–138 [DOI] [PubMed] [Google Scholar]
- 3. Barros M. H., Nobrega F. G., and Tzagoloff A. (2002) Mitochondrial ferredoxin is required for heme A synthesis in Saccharomyces cerevisiae. J. Biol. Chem. 277, 9997–10002 [DOI] [PubMed] [Google Scholar]
- 4. Khalimonchuk O., Bird A., and Winge D. R. (2007) Evidence for a pro-oxidant intermediate in the assembly of cytochrome oxidase. J. Biol. Chem. 282, 17442–17449 [DOI] [PubMed] [Google Scholar]
- 5. Bode M., Longen S., Morgan B., Peleh V., Dick T. P., Bihlmaier K., and Herrmann J. M. (2013) Inaccurately assembled cytochrome c oxidase can lead to oxidative stress-induced growth arrest. Antioxid. Redox Signal. 18, 1597–15612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bestwick M., Khalimonchuk O., Pierrel F., and Winge D. R. (2010) The role of Coa2 in hemylation of yeast Cox1 revealed by its genetic interaction with Cox10. Mol. Cell. Biol. 30, 172–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Khalimonchuk O., Kim H., Watts T., Perez-Martinez X., and Winge D. R. (2012) Oligomerization of heme o synthase in cytochrome oxidase biogenesis is mediated by cytochrome oxidase assembly factor Coa2. J. Biol. Chem. 287, 26715–26726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pierrel F., Khalimonchuk O., Cobine P. A., Bestwick M., and Winge D. R. (2008) Coa2 is an assembly factor for yeast cytochrome c oxidase biogenesis that facilitates the maturation of Cox1. Mol. Cell. Biol. 28, 4927–4939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Khalimonchuk O., Bestwick M., Meunier B., Watts T. C., and Winge D. R. (2010) Formation of the redox cofactor centers during Cox1 maturation in yeast cytochrome oxidase. Mol. Cell. Biol. 30, 1004–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bareth B., Dennerlein S., Mick D. U., Nikolov M., Urlaub H., and Rehling P. (2013) The heme a synthase Cox15 associates with cytochrome c oxidase assembly intermediates during Cox1 maturation. Mol. Cell. Biol. 33, 4128–4137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bundschuh F. A., Hannappel A., Anderka O., and Ludwig B. (2009) Surf1, associated with Leigh syndrome in humans is a heme-binding protein in bacterial oxidase biogenesis. J. Biol. Chem. 284, 25735–25741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bestwick M., Jeong M. Y., Khalimonchuk O., Kim H., and Winge D. R. (2010) Analysis of Leigh syndrome mutations in the yeast SURF1 homolog reveals a new member of the cytochrome oxidase assembly factor family. Mol. Cell. Biol. 30, 4480–4491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Khalimonchuk O., Jeong M. Y., Watts T., Ferris E., and Winge D. R. (2012) Selective Oma1 protease-mediated proteolysis of Cox1 subunit of cytochrome oxidase in assembly mutants. J. Biol. Chem. 287, 7289–7300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Antonicka H., Mattman A., Carlson C. G., Glerum D. M., Hoffbuhr K. C., Leary S. C., Kennaway N. G., and Shoubridge E. A. (2003) Mutations in COX15 produce a defect in the mitochondrial heme biosynthetic pathway, causing early-onset fatal hypertrophic cardiomyopathy. Am. J. Hum. Genet. 72, 101–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bugiani M., Tiranti V., Farina L., Uziel G., and Zeviani M. (2005) Novel mutations in COX15 in a long surviving Leigh syndrome patient with cytochrome c oxidase deficiency. J. Med. Genet. 42, e28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Alfadhel M., Lillquist Y. P., Waters P. J., Sinclair G., Struys E., McFadden D., Hendson G., Hyams L., Shoffner J., and Vallance H. D. (2011) Infantile cardioencephalopathy due to a COX15 gene defect. Am. J. Hum. Genet. 155A, 840–844 [DOI] [PubMed] [Google Scholar]
- 17. Oquendo C. E., Antonicka H., Shoubridge E. A., Reardon W., and Brown G. K. (2004) Functional and genetic studies demonstrate that mutation in the COX15 gene can cause Leigh syndrome. J. Med. Genet. 41, 540–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhu Z., Yao J., Johns T., Fu K., De Bie I., Macmillan C., Cuthbert A. P., Newbold R. F., Wang J., Chevrette M., Brown G. K., Brown R. M., and Shoubridge E. A. (1998) SURF1, encoding a factor involved in the biogenesis of cytochrome c oxidase, is mutated in Leigh syndrome. Nat. Genet. 20, 337–343 [DOI] [PubMed] [Google Scholar]
- 19. Tiranti V., Hoertnagel K., Carrozzo R., Galimberti C., Munaro M., Granatiero M., Zelante L., Gasparini P., Marzella R., Rocchi M., Bayona-Bafaluy M. P., Enriquez J. A., Uziel G., Bertini E., Dionisi-Vici C., Franco B., Meitinger T., and Zeviani M. (1998) Mutations of SURF1 in Leigh disease associated with cytochrome c oxidase deficiency. Am. J. Hum. Genet. 63, 1609–1621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Teraoka M., Yokoyama Y., Ninomiya S., Inoue C., Yamashita S., and Seino Y. (1999) Two novel mutations of SURF1 in Leigh syndrome with cytochrome c oxidase deficiency. Hum. Genet. 105, 560–563 [DOI] [PubMed] [Google Scholar]
- 21. Poyau A., Buchet K., Bouzidi M. F., Zabot M. T., Echenne B., Yao J., Shoubridge E. A., and Godinot C. (2000) Missense mutations in SURF1 associated with deficient cytochrome c oxidase assembly in Leigh syndrome patients. Hum. Genet. 106, 194–205 [DOI] [PubMed] [Google Scholar]
- 22. Piekutowska-Abramczuk D., Magner M., Popowska E., Pronicki M., Karczmarewicz E., Sykut-Cegielska J., Kmiec T., Jurkiewicz E., Szymanska-Debinska T., Bielecka L., Krajewska-Walasek M., Vesela K., Zeman J., and Pronicka E. (2009) SURF1 missense mutations promote a mild Leigh phenotype. Clin. Genet. 76, 195–204 [DOI] [PubMed] [Google Scholar]
- 23. Coenen M. J., van den Heuvel L. P., Ugalde C., Ten Brinke M., Nijtmans L. G., Trijbels F. J., Beblo S., Maier E. M., Muntau A. C., and Smeitink J. A. (2004) Cytochrome c oxidase biogenesis in a patient with a mutation in COX10 gene. Ann. Neurol. 56, 560–564 [DOI] [PubMed] [Google Scholar]
- 24. Antonicka H., Leary S. C., Guercin G. H., Agar J. N., Horvath R., Kennaway N. G., Harding C. O., Jaksch M., and Shoubridge E. A. (2003) Mutations in COX10 result in a defect in mitochondrial heme A biosynthesis and account for multiple, early-onset clinical phenotypes associated with isolated COX deficiency. Hum. Mol. Genet. 12, 2693–2702 [DOI] [PubMed] [Google Scholar]
- 25. Valnot I., von Kleist-Retzow J. C., Barrientos A., Gorbatyuk M., Taanman J. W., Mehaye B., Rustin P., Tzagoloff A., Munnich A., and Rötig A. (2000) A mutation in the human heme A:farnesyltransferase gene (COX10) causes cytochrome c oxidase deficiency. Hum. Mol. Genet. 9, 1245–1249 [DOI] [PubMed] [Google Scholar]
- 26. Wang Z., Wang Y., and Hegg E. L. (2009) Regulation of the heme A biosynthetic pathway: differential regulation of heme A synthase and heme O synthase in Saccharomyces cerevisiae. J. Biol. Chem. 284, 839–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hederstedt L. (2012) Heme A biosynthesis. Biochim. Biophys. Acta 1817, 920–927 [DOI] [PubMed] [Google Scholar]
- 28. Barros M. H., Tzagoloff A. (2002) Regulation of the heme A biosynthetic pathway in Saccharomyces cerevisiae. FEBS Lett. 516, 119–123 [DOI] [PubMed] [Google Scholar]
- 29. Brown B. M., Wang Z., Brown K. R., Cricco J. A., and Hegg E. L. (2004) Heme O synthase and heme A synthase from Bacillus subtilis and Rhodobacter sphaeroides interact in Escherichia coli. Biochemistry 43, 13541–13548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mick D. U., Fox T. D., and Rehling P. (2011) Inventory control: cytochrome c oxidase assembly regulates mitochondrial translation. Nat. Rev. Mol. Cell Biol. 12, 14–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lodi T., Fontanesi F., and Guiard B. (2002) Co-ordinate regulation of lactate metabolism genes in yeast: the role of the lactate permease gene JEN1. Mol. Genet. Genomics 266, 838–847 [DOI] [PubMed] [Google Scholar]
- 32. Pierrel F., Bestwick M. L., Cobine P. A., Khalimonchuk O., Cricco J. A., and Winge D. R. (2007) Coa1 links the Mss51 post-translational function to Cox1 cofactor insertion in cytochrome c oxidase assembly. EMBO J. 26, 4335–4346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nevoigt E., Kohnke J., Fischer C. R., Alper H., Stahl U., and Stephanopoulos G. (2006) Engineering of promoter replacement cassettes for fine-tuning of gene expression in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 72, 5266–5273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Prakash S., Tian L., Ratliff K. S., Lehotzky R. E., and Matouschek A. (2004) An unstructured initiation site is required for efficient proteasome-mediated degradation. Nat. Struct. Mol. Biol. 11, 830–837 [DOI] [PubMed] [Google Scholar]
- 35. Sherman F. (2002) Getting started with yeast. Methods Enzymol. 350, 3–41 [DOI] [PubMed] [Google Scholar]
- 36. Crisp R. J., Pollington A., Galea C., Jaron S., Yamaguchi-Iwai Y., and Kaplan J. (2003) Inhibition of heme biosynthesis prevents transcription of iron uptake genes in yeast. J. Biol. Chem. 278, 45499–45506 [DOI] [PubMed] [Google Scholar]
- 37. Diekert K., de Kroon A. I., Kispal G., and Lill R. (2001) Isolation and subfractionation of mitochondria from the yeast Saccharomyces cerevisiae. Methods Cell Biol. 65, 37–51 [DOI] [PubMed] [Google Scholar]
- 38. Boudina S., Bugger H., Sena S., O'Neill B. T., Zaha V. G., Ilkun O., Wright J. J., Mazumder P. K., Palfreyman E., Tidwell T. J., Theobald H., Khalimonchuk O., Wayment B., Sheng X., Rodnick K. J., Centini R., Chen D., Litwin S. E., Weimer B. E., and Abel E. D. (2009) Contribution of impaired myocardial insulin signaling to mitochondrial dysfunction and oxidative stress in the heart. Circulation 119, 1272–1283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Berry E. A., and Trumpower B. L. (1987) Simultaneous determination of hemes a, b and c from pyridine hemochrome spectra. Anal. Biochem. 161, 1–15 [DOI] [PubMed] [Google Scholar]
- 40. Barrientos A. (2002) In vivo and in organello assessment of OXPHOS activities. Methods 26, 307–316 [DOI] [PubMed] [Google Scholar]
- 41. Cui T. Z., Smith P. M., Fox J. L., Khalimonchuk O., and Winge D. R. (2012) Late-stage maturation of the Rieske Fe/S protein: Mzm1 stabilizes Rip1 but does not facilitate its translocation by the AAA ATPase Bcs1. Mol. Cell. Biol. 32, 4400–4409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kelley L. A., Mezulis S., Yates C. M., Wass M. N., and Sternberg M. J. (2015) The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10, 845–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Claros M. G., and Vincens P. (1996) Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur. J. Biochem. 241, 779–786 [DOI] [PubMed] [Google Scholar]
- 44. Gietz R. D., and Schiestl R. H. (2007) High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat. Protoc. 2, 31–34 [DOI] [PubMed] [Google Scholar]
- 45. Mogi T. (2009) Probing structure of heme A synthase from Bacillus subtilis by site-directed mutagenesis. J. Biochem. 145, 625–633 [DOI] [PubMed] [Google Scholar]
- 46. Gollub E. G., Liu K. P., Dayan J., Adlersberg M., and Sprinson D. B. (1977) Yeast mutants deficient in heme biosynthesis and a heme mutant additionally blocked in cyclization of 2,3-oxidosqualene. J. Biol. Chem. 252, 2846–2854 [PubMed] [Google Scholar]
- 47. Zoppellaro G., Bren K. L., Ensign A. A., Harbitz E., Kaur R., Hersleth H. P., Ryde U., Hederstedt L., and Andersson K. (2009) Studies of ferric heme proteins with highly anisotropic/highly axial low spin (s=1/2) electron paramagnetic resonance signals with bis-histidine and histidine-methionine axial iron coordination. Biopolymers 91, 1064–1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lewin A., and Hederstedt L. (2006) Compact archaeal variant of heme A synthase. FEBS Lett. 580, 5351–5356 [DOI] [PubMed] [Google Scholar]
- 49. Lewin A., and Hederstedt L. (2016) Heme A synthase in bacteria depends on one pair of cysteinyls for activity. Biochim. Biophys. Acta 1857, 160–168 [DOI] [PubMed] [Google Scholar]