Abstract
The intrinsic apoptosis pathway occurs through the release of mitochondrial cytochrome c to the cytosol, where it promotes activation of the caspase family of proteases. The observation that tRNA binds to cytochrome c revealed a previously unexpected mode of apoptotic regulation. However, the molecular characteristics of this interaction, and its impact on each interaction partner, are not well understood. Using a novel fluorescence assay, we show here that cytochrome c binds to tRNA with an affinity comparable with other tRNA-protein binding interactions and with a molecular ratio of ∼3:1. Cytochrome c recognizes the tertiary structural features of tRNA, particularly in the core region. This binding is independent of the charging state of tRNA but is regulated by the redox state of cytochrome c. Compared with reduced cytochrome c, oxidized cytochrome c binds to tRNA with a weaker affinity, which correlates with its stronger pro-apoptotic activity. tRNA binding both facilitates cytochrome c reduction and inhibits the peroxidase activity of cytochrome c, which is involved in its release from mitochondria. Together, these findings provide new insights into the cytochrome c-tRNA interaction and apoptotic regulation.
Keywords: apoptosis, caspase, cytochrome c, mitochondrial apoptosis, transfer RNA (tRNA)
Introduction
Cytochrome c is a heme-containing protein that normally resides in the mitochondrial inter-membrane space. It carries electrons from cytochrome c reductase (the cytochrome b-c1 complex) to cytochrome c oxidase as part of the electron transport chain that builds an electrochemical gradient driving the synthesis of ATP. This function of cytochrome c may be conserved over 1.5-billion years of eukaryotic evolution (1). In vertebrate cells, cytochrome c has taken on an additional role as a critical inducer of apoptosis or programmed cell death, which eliminates unwanted or harmful cells (1, 2). Apoptosis can occur through either of two major apoptotic pathways. The intrinsic apoptotic pathway is activated by intracellular stimuli such as DNA damage, oncogene activation, and developmental information. The extrinsic apoptotic pathway responds to extracellular stimuli via cell surface death receptors. The intrinsic pathway is evolutionarily more conserved than the extrinsic pathway, and it can be activated by the extrinsic pathway to amplify the apoptotic response. The defining event in the intrinsic pathway is the release of cytochrome c from mitochondria into the cytosol, where it binds to Apaf-1 (apoptotic protease activating factor-1) in the presence of ATP or dATP, facilitating the assembly of the oligomeric apoptosome (3–5). The apoptosome recruits and activates the initiator caspase, caspase-9 (6). Caspase-9 subsequently activates executioner caspases, leading to the cleavage of a large number of cellular proteins and eventually cellular death (7–9).
The activation of caspases by cytochrome c is intricately regulated. Release of cytochrome c is facilitated by the oxidation of cardiolipins, which anchor cytochrome c on the inner mitochondrial membrane, and by mitochondrial outer membrane permeabilization, a process that is regulated by members of the B-cell lymphoma protein-2 (Bcl2) family (1). In the cytoplasm, the ability of cytochrome c to activate caspases is modulated by its redox state, with the oxidized form showing a much more potent activity compared with the reduced form (10). Effective assembly of the apoptosome requires, in addition to cytochrome c and (d)ATP, the proteins HSP70, cellular apoptosis susceptibility protein, and the PHAPI tumor suppressor (11, 12). Apoptosome formation is inhibited by the oncoprotein prothymosin-α (11). Nucleic acid, specifically transfer RNA (tRNA), is also implicated in the regulation of cytochrome c-mediated caspase activation (13).
tRNA is responsible for the interpretation of nucleic acid sequences as amino acid sequences during protein synthesis in all known forms of life (14, 15). Mature tRNAs are 73–93 ribonucleotides in length and fold into a cloverleaf secondary and L-shaped tertiary structure. tRNA is “charged” by conjugation with an amino acid at the conserved 3′-CCA sequence, which resides at one end of the L-shaped structure. Opposite this end, a three-nucleotide anticodon sequence pairs with a specific mRNA codon and enables the translation of the codon into a specific amino acid. tRNA interacts with a number of proteins and other RNAs during its maturation, transport, aminoacylation (“charging”), and movement in and out of the ribosome. It also has a high degree of functional versatility in addition to protein synthesis. Non-canonical functions of tRNA include priming reverse transcription of specific viral genomes (16) and stimulating gene expression in response to amino acid deprivation (17).
We previously showed that tRNA binds directly to cytochrome c. The cytochrome c-tRNA binding prevents cytochrome c from interacting with Apaf-1 and activating apoptosis (13). This finding indicates that cytochrome c, in addition to supporting ATP production and to promoting apoptosis, is a tRNA-binding protein. However, although all cytosolic and mitochondrial tRNAs appear to participate in the interaction, the molecular basis remains unknown. Also mysterious is the effect of tRNA association on the redox state and peroxidase activity of cytochrome c, which have been implicated in the release of cytochrome c and the activation of the caspase cascade. Here, we further characterize the dynamics of the interaction between tRNA and cytochrome c, the influence of novel factors, and its consequences for each interaction partner. This study elucidates the basic tenets of an ancient molecular interaction that has important consequences for apoptosis.
Experimental Procedures
Reagents
A 78-nucleotide DNA encoding the sequence of human initiator tRNAMet was synthesized by Integrated DNA Technologies (IDT; Coralville, IA). Cy3 was purchased from AAT Bioquest (Sunnyvale, CA), Cy5 from Lumiprobe (Hannover, Germany), and 2-aminopurine (2AP)4 triphosphate from TriLink Biotech (San Diego). The following reagents were purchased from Sigma: bovine heart and yeast cytochrome c; total tRNA from bakers' yeast, ribosomal RNA, polyadenylic acid, NADH, NADPH, FAD, NaBH4, proflavine, ascorbic acid, and potassium ferricyanide. Onconase was provided by the Alfacell Corp. (Somerset, NJ).
Fluorophore-labeled tRNAs
A fluorescent tRNA based on the tRNACys sequence in Escherichia coli was prepared by in vitro reconstitution. E. coli tRNACys (etRNACys, Fig. 1A) has been well characterized (18, 19), and its crystal structure was determined (20). A 5′-fragment encoding nucleotides G1 to C16 was chemically synthesized by IDT with the Cy3 fluorophore (Fig. 1B) attached to the 5′-end (position 1) through a phosphodiester linkage. A 3′-fragment encoding G18 to A76 was synthesized by in vitro transcription, using T7 RNA polymerase, and was gel-purified. The two fragments were joined by T4 RNA ligase I in a 3:1 molar ratio of the short versus long fragment with a 70% yield (Fig. 1C). The ligated full-length tRNA (Cy3-etRNACys) was separated from individual fragments by a denaturing gel, heated at 85 °C, and re-annealed at 37 °C in the presence of Mg2+. Similar procedures were used to prepare a human elongator tRNAMet (htRNAMet) and a human tRNAPhe (htRNAPhe) (Fig. 1A) that were labeled with Cy3 and Cy5, respectively, at the 5′-end (Cy3-htRNAMet and Cy5-htRNAPhe) (Fig. 1B). To generate the 2AP-labeled E. coli tRNAVal (2AP-etRNAVal) (Fig. 1B), an E. coli tRNAVal transcript that terminated at nucleotide position 75 was generated. 2AP was added to nucleotide position 76 using the CCA-adding enzyme from E. coli and the triphosphate form of 2AP. The labeled tRNA was separated from unlabeled species on a denaturing PAGE, 7 m urea gel. The full-length tRNA was excited from gels, recovered by ethanol precipitation, and resuspended in the TE buffer. The proflavine-labeled E. coli tRNACys (Prf-etRNACys) was prepared by inserting proflavine (Fig. 1B) to the D-loop as described (Fig. 1D) (21, 22). Briefly, the transcript of E. coli tRNACys was modified by an insertion of U17, which was subsequently converted to dihydrouridine 17 (D17) by Dus1p and reduced to the ureidopropanal group by NaBH4. The ureidopropanal group was then reacted with proflavine to form adduct with the fluorophore (Fig. 1D).
FIGURE 1.
Generation of fluorescence-labeled tRNAs. A, sequence and cloverleaf structure of tRNAs used in this study. Species are E. coli tRNACys (etRNACys) and tRNAVal (etRNAVal) and human elongator tRNAMet (htRNAMet) and tRNAPhe (htRNAPhe). Arrows indicate the positions of joining between a synthetic Cy3- or Cy5-labeled 5′-fragment and an in vitro-transcribed 3′-fragment (for etRNACys, htRNAMet, and htRNAPhe), or the 3′-position labeled with 2AP (for etRNAVal). B, schematic representation of tRNAs labeled with Cy3 (maximal emission wavelength or λem = 570 nm) or Cy5 (λem = 670 nm) at the 5′-end, 2-AP (λem = 320 nm) at the 3′-end, and Prf (λem = 515 nm) in the D-loop. C, construction of tRNA labeled with Cy3 at the 5′-end. The 5′-Cy3 fragment and the 3′-tRNA transcript fragment were joined together using T4 RNA ligase in the presence of ATP. D, generation of tRNA labeled with proflavine attached to a ureidopropanal group at the D-loop.
Binding Affinity of Cytochrome c with Fluorescent tRNAs
Each fluorescent tRNA was titrated with bovine or yeast cytochrome c from 0.1 to 24.4 μm in the binding buffer (20 mm HEPES, pH 7.5, 35 mm KCl, 2.5 mm MgCl2, 0.5 mm EDTA, and 1 mm DTT) at room temperature, and the fluorescence emission was monitored. The peak intensity at 563 nm for Cy3 was corrected for the inner filter effect for each cytochrome c concentration, and the corrected data as a function of cytochrome c concentration were fit to a hyperbola equation to derive the Kd value.
Stoichiometry of Cytochrome c-tRNA Interaction
The stoichiometry of cytochrome c binding to tRNA was determined by monitoring the fluorescence quenching of Cy3-etRNACys and Cy5-htRNAPhe on a Photon Technology International instrument model QM-4 as described (23). The binding was performed at room temperature in a buffer containing 20 μm labeled tRNA, 200 mm HEPES, pH 7.5, 50 mm NaCl, 5% sucrose, and 5 mm DTT. The bovine cytochrome c was titrated from 0 to 454 μm, with the cytochrome c/tRNA molar ratio ranging from 0 to 22.7. The Cy3-etRNACys was excited at 550 nm, and the emission was monitored from 558 to 650 nm at room temperature. The Cy5-htRNAPhe was excited at 640 nm, and the emission was monitored from 655 to 720 nm. The emission peaks at 565 nm for Cy3-tRNA and at 662 nm for Cy5-tRNA were recorded and corrected for the inner filter effect, according to the formula, Fcorr = Fobs × anti-log((Aexcitation + Aemission)/2), where Fcorr is the corrected fluorescent signal at the peak wavelength, and Fobs is the observed fluorescent signal at the peak wavelength.
Surface Plasmon Resonance (SPR) Assay
The association of various nucleic acids with cytochrome c was assessed by surface plasmon resonance using a BIAcore 3000 system (GE Healthcare). Cytochrome c from bovine heart was immobilized on a CM5 sensor chip (BIAcore) by amine coupling. Each nucleic acid was individually injected onto the immobilized cytochrome c. Binding interaction was performed in a low salt buffer (20 mm HEPES, 20 mm KCl, 2.5 mm MgCl2, 0.5 mm EDTA) or in a buffer with physiologic ionic strength (by adding KCl to 135 mm and NaCl to 10 mm to the low salt buffer). After each binding experiment, the chips were washed with a 0.5 m NaCl regeneration solution.
CCA Addition and tRNA Charging Reactions
The transcript of E. coli tRNAVal was synthesized up to C75 and was internally labeled in the transcription reaction containing a trace amount of [α-32P]ATP. The addition of A76 to the gel-purified labeled tRNAVal (0.1 μm) was catalyzed by human CCA adding enzyme (2 μm), in the presence or absence of oxidized bovine cytochrome c (15 μm). The assay was performed at 37 °C for 0–1 s on a KinTek RQF-3 instrument in the reaction buffer previously described (24–26).
tRNA charging assay was performed using etRNACys as the substrate and E. coli cysteinyl-tRNA synthetase (CysRS) as the enzyme, as described previously (27–29). The efficiency of charging was monitored using [35S]cysteine. After the reaction, tRNA was acid-precipitated on filter pads, and the radioactivity of [35S]cysteine attached to each tRNA was quantified by scintillation counting (30).
Cytochrome c Peroxidase Activity
The peroxidase activity was determined by incubation of cytochrome c with enhanced chemiluminescence solution and measuring light emission using an illuminometer.
Cytochrome c Oxidation and Reduction
To generate the reduced protein, bulk cytochrome c was incubated with excess ascorbic acid and was then purified using a Sepharose column (GE Healthcare). To generate the oxidized protein, bulk cytochrome c was incubated with potassium ferricyanide and purified similarly. The oxidation and reduction were confirmed by measuring absorbance at 550 and 560 nm as well as by colorimetric inspection of the purified protein. In cytochrome c oxidation and reduction assays, the redox state was monitored by continuous measurement of absorption at 550 nm.
Results
Fluorescence-based Assay for the Cytochrome c-tRNA Interaction
We previously analyzed the cytochrome c-tRNA interaction using electrophoretic mobility shift assays (13). To provide an independent and quantitative evaluation of this interaction, we developed a fluorescence-based assay. Cytochrome c does not appear to strongly discriminate among various cytosolic and mitochondrial tRNA species (13), suggesting that prokaryotic tRNA may be just as capable of interacting with cytochrome c as eukaryotic tRNA. We used E. coli tRNACys (etRNACys) and tRNAVal (etRNAVal) and human elongator tRNAMet (htRNAMet) and tRNAPhe (htRNAPhe) as models (Fig. 1A). These tRNAs were labeled at the 5′-end with either Cy3 (for etRNACys and tRNAMet) or Cy5 (for htRNAPhe), at the 3′-end with 2AP (for etRNAVal), or in the D-loop with proflavine (Prf) (for etRNACys) (Fig. 1B).
To obtain Cy3-etRNACys, a chemically synthesized 5′-Cy3-attached fragment, encoding nucleotides G1 to C16, was joined with an in vitro-transcribed 3′-fragment, encoding nucleotides G18 to A76 (Fig. 1C) (26). Cy3-htRNAMet and Cy5-htRNAPhe were generated similarly. 2AP-etRNAVal was prepared by the addition of 2AP to 3′-end of an etRNAVal transcript containing nucleotides G1 to C75 (21). To produce Prf-etRNACys, etRNACys was modified by the insertion of a uridine at position 17, which was then converted to dihydrouridine and labeled with proflavine (Fig. 1D) (22).
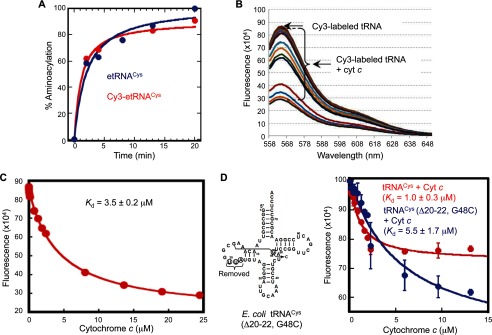
To assess whether the attachment of the fluorophores at the 5′-end affects the function of tRNA, we used etRNACys as an example and compared charging to the 3′-end of unlabeled and the 5′-end Cy3-labeled etRNACys. Using [35S]cysteine as the substrate and E. coli CysRS as the enzyme, we analyzed the attachment of [35S]cysteine to each tRNA over time. As shown in Fig. 2A, Cy3-etRNACys was charged to ∼85% of the unlabeled etRNACys. This indicates that the labeling at the 5′-end does not interfere with charging.
FIGURE 2.
Binding of cytochrome c to fluorophore-labeled tRNA in vitro. A, aminoacylation of unlabeled (in blue) and Cy3-labeled (in red) etRNACys (1 μm) with cysteine (25 μm) by E. coli CysRS (1 μm) over a time course. The Cy3-etRNACys is aminoacylated to 85% capacity relative to the unlabeled etRNACys. B, quenching of the fluorescence of Cy3-etRNACys (1 μm) upon binding with different concentrations of cytochrome c (0.1, 0.2, 0.4, 1.0, 1.5, 2.0, 2.5, 7.5, 12.5, 18, and 24.4 μm). The fluorescence peak intensity at 563 nm for Cy3 was monitored at room temperature and was corrected for the inner filter effect for each cytochrome c concentration. C, fitting the data of fluorescence change of Cy3-etRNACys as a function of cytochrome c concentration. The fluorescence intensity at 563 nm, corrected for the inner filter effect, was monitored as a function of the cytochrome c concentration and fit to a hyperbolic equation to derive the Kd value. The cytochrome c-tRNA interaction has a Kd of 1–3.5 μm (also see D). D, left, sequence and cloverleaf structure of the etRNACys mutant bearing a deletion of nucleotides 20–22 and a substitution of G48 with C. Right, fluorescence titration of the mutant and wild-type etRNACys as a function of cytochrome c concentration. Each data point was the average of three independent measurements. Error bars indicate standard deviation (S.D.).
Interaction of these tRNAs with cytochrome c was measured by fluorescence quenching of the labeled tRNAs following cytochrome c addition (Fig. 2B). Quenching data were fit to a hyperbolic equation to obtain the Kd value of the interaction, assuming a two-state (bound or unbound) model following correction for the inner filter effect and nonspecific binding.
Cytochrome c Binds to tRNA with an Affinity Comparable with Other tRNA-binding Proteins and Recognizes the Tertiary Structure of tRNA
We determined the affinity of bovine heart cytochrome c to Cy3-etRNACys and observed a Kd in the range of 1–3.5 μm (Fig. 2, C and D, and Table 1). Importantly, this affinity was comparable with those of known tRNA-binding proteins, including aminoacyl-tRNA synthetase (aaRS) (1–3 μm) (27, 31, 32) and CCA-adding enzyme (tRNA-nucleotidyltransferase) (0.8–3.3 μm) (25, 26). To corroborate this finding, we also analyzed cytochrome c binding to Cy3-htRNAMet and 2AP-etRNAVal. Cytochrome c bound to these two tRNAs with affinities similar to Cy3-etRNACys (Kd = 4.8 ± 0.7 and 5.1 ± 0.9 μm, respectively; Table 1). Therefore, analysis of three unrelated tRNA species labeled with distinct fluorophores at different ends showed that cytochrome c bound to tRNA species with an affinity akin to those of other tRNA-binding proteins.
TABLE 1.
Dissociation constant of bovine heart cytochrome c to different RNA
Measurement of the Kd (tRNA) for cytochrome c by fluorescence titration is shown. E. coli tRNACys and human tRNAMet labeled with Cy3 at the 5′-end, E. coli tRNAVal labeled with 2-AP on the 3′-end, or E. coli tRNACys labeled with Prf in the D-loop were used in the binding assay. The Cy3-oligonucleotide refers to the oligonucleotides corresponding to the sequence from positions 1 to 16 in E. coli tRNACys, carrying Cy3 at the 5′-end. Data shown are mean ± S.D.
| RNA | Kd |
|---|---|
| μm | |
| Cy3-etRNACys | 1–3.5 |
| Cy3-htRNAMet | 4.8 ± 0.7 |
| 2AP-etRNAVal | 5.1 ± 0.9 |
| Prf-etRNACys | 9.6 ± 2.8 |
| Cy3-oligonucleotide | >30 |
Compared with Cy3-etRNACys, cytochrome c bound to the D-loop-labeled Prf-etRNACys with a substantially reduced affinity (Kd of 9.6 ± 2.8 μm, Table 1). This might be due to proflavine in the D-loop interfering with cytochrome c binding directly or with folding of the tertiary structure of tRNA, both scenarios suggesting that the tRNA tertiary structure is required for high affinity interaction with cytochrome c. We also used a Cy3-labeled fragment of etRNACys encoding nucleotides 1–16 (Cy3-oligonucleotide), which resembled a microRNA or a tRNA-derived fragment and lacked the tertiary core structure (33). This fragment had greatly reduced affinity for cytochrome c (Kd >30 μm, Table 1).
To extend these analyses, we generated a mutant of etRNACys, in which three nucleotides in the D loop (U20, U21, and A22) were deleted, and the G48 nucleotide in the V-loop was changed to C (Fig. 2D, left). We have previously shown that the nucleotides in the D-loop and G48, which forms base pair with G15, are critical for the integrity of the core (18, 19, 34, 35). Thus, the resultant etRNACys mutant likely has an incomplete tertiary structure. Indeed, this mutant bound to cytochrome c with a Kd of 5.5 ± 1.7 μm, which was a 5-fold increase relative to the Kd of 1.0 ± 0.3 μm of the wild-type tRNA determined in the same experiment (Fig. 2D). Together, these results support the notion that cytochrome c recognizes the tertiary structural features of tRNA, particularly in the core region.
Surface Plasmon Resonance Analysis
To further independently assess the cytochrome c-tRNA interaction, we performed a surface plasmon resonance analysis using a BIAcore 3000 system. Bovine heart cytochrome c was immobilized on a CM5 sensor chip by amine coupling and tested for binding with total tRNA, ribosomal RNA (rRNA), a mixture of poly(A) RNAs, and a 78-nucleotide DNA oligonucleotide corresponding to a human initiator tRNAMet. In a low salt buffer, only tRNA had distinct association and dissociation phases (Fig. 3, A–C). In a buffer with physiologic ionic strength, only tRNA binding was detectable above the baseline (Fig. 3D). These results again indicate that cytochrome c is a specific tRNA-binding protein.
FIGURE 3.
Surface plasmon resonance analysis and stoichiometry of cytochrome c-tRNA binding. A–D, cytochrome c-RNA binding was determined by surface plasmon resonance (BIAcore) by titration of tRNA (A and D), a 78-nucleotide DNA oligonucleotide encoding the sequence of human initiator tRNAMet (B and D), polyadenylic acid matched to tRNA molecular weight (C and D), and rRNA (D). Each nucleic acid was injected at 100, 50, 25, 12.5, 6.25, and 0 μm (A–C) or at 100 μm (D) in low (A–C) or physiological salt (D) conditions. Real time graphs of response units (arbitrary units) over time are shown. E and F, stoichiometry of cytochrome c binding to tRNA was determined by monitoring the fluorescence quenching of 20 μm Cy3-etRNACys (E) or Cy5-htRNAPhe (F) by increasing amounts of bovine cytochrome c (0–454 μm).
Cytochrome c Associates with tRNA with a Stoichiometric Ratio of Three Cytochrome c Molecules for Each tRNA Molecule
We next interrogated the binding stoichiometry between tRNA and cytochrome c. We titrated a fixed amount of Cy3-etRNACys with increasing amounts of cytochrome c to shift the binding equilibrium in the direction of the bound complex. The titration produced a biphasic quenching of the Cy3-etRNACys fluorescence, with an initial steep phase followed by a second and much flatter phase (Fig. 3E). The two phases intercepted at a molar ratio of cytochrome c to tRNA at ∼3.0, indicating that three cytochrome c molecules bind to one tRNA molecule. Further fluorescence quenching was also observed at higher protein stoichiometries, likely indicating nonspecific formation of higher order complexes.
To confirm this binding stoichiometry, we tested Cy5-htRNAPhe. Despite the use of a different fluorophore and a different tRNA, the titration maintained the same two phases that intersected at the cytochrome c/tRNA molar ratio of ∼3.0 (Fig. 3F). The stoichiometry observed in these experiments is in agreement with the result of a recent study (36), and it further underscores the specificity of the cytochrome c-tRNA interaction.
Influence of the CCA-end of tRNA on Cytochrome c Binding
We next investigated whether some of the key features of tRNA and cytochrome c regulate their interaction. The addition of the CCA sequence is an essential step in the maturation of tRNA (14, 15). To determine whether the CCA sequence is required for cytochrome c binding, we used tRNA species with and without this sequence. An analysis of the interaction between Cy3-htRNAMet and either bovine or yeast cytochrome c showed no major difference regardless of the presence or absence of the CCA sequence (Table 2). Thus, the CCA sequence does not appear to affect cytochrome c binding.
TABLE 2.
Binding constant of CCA+ and CCA− tRNAs to cytochrome c
Human tRNAMet with (CCA+) or without (CCA−) the CCA sequence, and labeled with Cy3 at the 5′-end, were tested for the binding with bovine and yeast cytochrome c (cyt c). Data shown are mean ± S.D.
| tRNA | CCA end | Label |
Kd (μm) |
|
|---|---|---|---|---|
| Bovine cyt c | Yeast cyt c | |||
| htRNAMet | CCA+ | 5′-Cy3 | 4.8 ± 0.7 | 4.8 ± 0.9 |
| htRNAMet | CCA− | 5′-Cy3 | 3.7 ± 0.4 | 7.0 ± 2.0 |
Conversely, we assessed whether cytochrome c influences the CCA addition reaction. Using the human CCA-adding enzyme and under single-turnover conditions, we monitored the A76 addition to the transcript of E. coli tRNAVal that terminated with C75. Analysis of the time course of the reaction showed that the rate constant of the addition in the presence and absence of cytochrome c was virtually identical (14 versus 15 s−1) (Fig. 4A). Thus, the action of the CCA-adding enzyme is not perturbed by cytochrome c.
FIGURE 4.
Effects of CCA addition and charging on the interaction of tRNA with cytochrome c. A, kinetics of A76 addition to etRNAVal (1 μm) catalyzed by human CCA enzyme (2 μm) at 37 °C in the presence or absence of cytochrome c (15 μm). B, ability of free etRNACys and etRNACys in complex with CCA-adding enzyme to bind cytochrome c. Analysis of fluorescence quenching as a function of cytochrome c concentration in the fluorescence-based assay. Error bars indicate S.D. C, model of tRNA in complex with CCA-adding enzyme of Archaeoglobus fulgidus (green, Protein Data Bank code 1sz1) and CysRS of E. coli (gray, Protein Data Bank code 1u0b), showing the capacity to accommodate three molecules of cytochrome c (brown, purple, and pink, Protein Data Bank code 3ZCF) on the outside corner of the L-structure. D, dissociation constant (Kd) of cytochrome c with uncharged and charged Cy3-etRNACys. Error bars indicate S.D. E, ability of free etRNACys or tRNACys in complex with CysRS to bind cytochrome c. Analysis of fluorescence quenching as a function of cytochrome c concentration in the fluorescence-based assay. Error bars indicate S.D.
We also tested whether CCA-adding enzyme and cytochrome c compete for binding to tRNA. We pre-assembled an etRNACys-CCA-adding enzyme complex and performed the binding assay with a range of cytochrome c concentrations. The etRNA in the etRNACys-CCA-adding enzyme complex was able to bind cytochrome c with a Kd of 0.9 ± 0.3 μm, essentially the same as the binding of free tRNA with cytochrome c (Kd of 1.0 ± 0.3 μm), determined under the same condition (Fig. 4B). In the crystal structure of a tRNA-CCA-adding enzyme complex, the CCA-adding enzyme binds to the top half of the tRNA L-shaped tertiary structure near the 3′-end (Fig. 4C) (37). Thus, our data indicate that the CCA-adding enzyme does not block the access of cytochrome c to the tRNA structure.
Influence of the Charging State of tRNA on Cytochrome c Binding
tRNA charging by cognate aaRS is fundamental to the translation of mRNA into protein. The state of tRNA charging is also an indicator of the nutritional state, and amino acid deprivation leads to rapid accumulation of uncharged tRNAs (14, 15). The experiments described above were performed with uncharged tRNA. To assess the influence of the charging state of tRNA, we compared uncharged and charged Cy3-etRNACys for binding to cytochrome c. Bovine cytochrome c bound to these two forms of tRNA with similar affinities (Kd = 3.6 ± 0.8 and 3.3 ± 0.7 μm, respectively) (Fig. 4D). Thus, cytochrome c recognizes tRNA independent of its charging state.
To evaluate whether cytochrome c competes with aaRS for binding to tRNA, we compared the binding to cytochrome c of free etRNACys and etRNACys associated with CysRS. The etRNA molecules in these two states bound to cytochrome c with virtually the same affinities (Kd of 1.0 ± 0.3 and 1.2 ± 0.3 μm, respectively) (Fig. 4E). In the crystal structure of the etRNACys-CysRS complex, the CysRS enzyme binds to the inside of the tRNA L-shape (Fig. 4C) (38). The lack of competition between CysRS and cytochrome c is consistent with a model in which cytochrome c binds to the outside corner of the tRNA L-shape and hence is not in conflict with the binding by CysRS (Fig. 4C). It also suggests that labeling of tRNA at the 5′- or 3′-end unlikely interferes with its interaction with cytochrome c.
Binding of Oxidized and Reduced Cytochrome c to tRNA
Cytochrome c exists in either a reduced or an oxidized form, based on the oxidation state of the iron atom contained within the heme group. The oxidized form is much more potent than the reduced form in the activation of caspases (10). All experiments reported thus far employed oxidized cytochrome c. To test the influence of the redox state of cytochrome c, we compared binding of oxidized and reduced cytochrome c to tRNA directly. Oxidized bovine cytochrome c bound to Cy3-htRNAMet with a 2-fold weaker affinity compared with the reduced form (Kd = 4.8 ± 0.7 versus 2.5 ± 1.4 μm) (Fig. 5A, left). Interestingly, removal of the CCA sequence exacerbated the difference in binding between the two states (∼4-fold; Kd = 3.7 ± 0.4 versus 0.9 ± 0.2 μm) (Fig. 5A, right). We also tested cytochrome c from yeast, which, unlike its vertebrate counterpart, cannot induce caspase activation (39). Oxidized yeast cytochrome c displayed a weaker affinity to Cy3-htRNAMet relative to the reduced form, although this difference was less pronounced than for bovine cytochrome c regardless of the presence or absence of the CCA sequence (Kd = 4.8 ± 0.9 versus 3.4 ± 0.4 μm in the presence of CCA and Kd = 7.0 ± 2.0 versus 3.7 ± 0.7 μm in the absence of CCA) (Fig. 5B). The weaker binding of the oxidized form of mammalian cytochrome c to tRNA correlates with its stronger ability to activate apoptosis.
FIGURE 5.
Effects of the redox state of cytochrome c on its interaction with tRNA. A, Kd value of the oxidized and reduced form of bovine cytochrome (cyt) c with Cy3-etRNACys with or without the 3′-CCA. B, Kd value of the oxidized and reduced form of yeast cytochrome c with Cy3-etRNACys with or without the 3′-CCA.
Effect of tRNA Binding on the Activity of Cytochrome c
Cytochrome c possesses peroxidase activity, which promotes the oxidization of cardiolipins early in apoptosis, facilitating the detachment of cytochrome c from cardiolipins and its subsequent release into the cytosol (40). To further investigate how the cytochrome c-tRNA interaction may affect cytochrome c function, we tested whether tRNA binding modulates the peroxidase activity of cytochrome c. In a luminescence assay, tRNA inhibited the peroxidase activity of cytochrome c in a dose-dependent manner (Fig. 6A). This finding raises the possibility that tRNA, if present in the inter-membrane space upon mitochondrial outer membrane permeabilization, might impede the oxidation of cardiolipins and regulate apoptosis at the level of cytochrome c release.
FIGURE 6.
tRNA inhibits the peroxidase activity of cytochrome c and promotes the reduction of cytochrome c. A, tRNA inhibits cytochrome(cyt) c-peroxidase activity. Left, luminescence emission in an enhanced chemiluminescence assay, representing peroxidase activity, was linear with the amount of cytochrome c. Right, cytochrome c peroxidase activity, monitored by luminescence using Lumigen TMA-6 and hydrogen peroxide, in the presence or absence of tRNA. Concentrations of cytochrome c and tRNA (in μm) are shown. B, tRNA reduces oxidized cytochrome c. Left, oxidation of cytochrome c by peroxide as monitored by A550 nm. Right, incubation of tRNA in a buffer with physiologic ionic strength (150 mm) with oxidized cytochrome c at molar ratios of 1:2, 1:1, or 2:1 led to increases in A550 nm, indicating the reduction of cytochrome c over time. C, tRNA must be intact to reduce oxidized cytochrome c. tRNA-dependent reduction of cytochrome c is inhibited upon the degradation of tRNA by Onconase (ranpirnase). Error bars indicate S.D.
tRNA Binding Facilitates Cytochrome c Reduction
RNA is extensively oxidized during apoptosis (41). To test whether cytochrome c is capable of directly oxidizing tRNA, we incubated cytochrome c with tRNA and observed the changes in absorbance at 550 nm, which monitors the reduced state. In the absence of tRNA, oxidized cytochrome c was stable for days. However, upon incubation with tRNA, the oxidized state was gradually converted to the reduced state, as indicated by the increase in the absorption at 550 nm (Fig. 6B). This increase was dependent on the concentration of tRNA (Fig. 6B). Importantly, pre-digestion of tRNA by addition of the nuclease Onconase (ranpirnase) prevented cytochrome c reduction (Fig. 6C). These results suggest that tRNA, but not free NMPs, may serve as a substrate for oxidation by cytochrome c.
Discussion
Discovered by Keilin and co-workers (42, 43) in the 1920s as one of the color proteins involved in the respiratory chain, cytochrome c has an essential and evolutionarily conserved role in supporting aerobic eukaryotic life. More than 70 years later, a completely unanticipated role of cytochrome c in vertebrate cell death came to light, when Wang and co-workers (2) investigated the mechanism of caspase activation. The dichotomy of the dual roles of cytochrome c mechanistically links cell life and death and is fundamental to the evolutionary covenant required for multicellular life. Thus, virtually all vertebrate cells, by depending on cytochrome c for survival, carry this suicide pill for use when and where needed.
tRNA is even more evolutionarily ancient and fundamental to life (14, 15). Its function as the adaptor molecule in protein synthesis is based on the L-shaped tertiary structure that simultaneously recognizes a genetic codon and an amino acid. This tertiary structure allows for tRNA to interact with both general enzymes, such as the CCA-adding enzyme for 3′-end maturation, and specific enzymes, such as aaRS for charging. The L-shaped tertiary structure also affords tRNA non-conventional roles in cells, including priming reverse transcription (16) and, for uncharged tRNA, sensing nutrient deprivation (17).
The identification of the association between cytochrome c and tRNA revealed a previously unrecognized connection between two fundamental molecules in life and signified novel biochemical properties of each (13). The findings presented here show that cytochrome c binds to tRNA with an affinity comparable with other tRNA-binding proteins (Fig. 2 and Table 1). Interestingly, cytochrome c binds tRNA without sequence specificity, a property unlike aaRS and more similar to CCA-adding enzyme. Cytochrome c likely recognizes features of the tertiary structure of tRNA, particularly in the core region. The binding stoichiometry of three cytochrome c molecules to a single tRNA molecule (Fig. 3) additionally suggests that multiple cytochrome c molecules are coordinated to recognize a single tRNA. Interestingly, cytochrome c does not compete with either aaRS or CCA-adding enzyme. This could be accounted for in a model in which three cytochrome c molecules bind to the outside corner of the tRNA L-shape in a way that does not interfere with the binding of aaRS to the inside of the L-structure or with the binding of CCA-adding enzyme to the outside of the L-structure near the 3′-end (Fig. 4C).
In cells, tRNAs are extensively modified. These modifications can modulate the structure, function, and stability of tRNAs (15). The tRNAs used in this study lack the post-transcriptional modifications. Nevertheless, our data suggest that cytochrome c binds to the outside corner of the tRNA tertiary core, which is not extensively modified relative to the anticodon loop region. Post-transcriptional modifications to the outside corner of the tRNA tertiary core primarily consist of dihydrouridine residues, which have also been used extensively to introduce fluorophores to monitor tRNA dynamics on the ribosome (21, 22). This indicates that these residues themselves are not critical for the intra-molecular folding of the tRNA tertiary core. Therefore, we suggest that the use of unmodified tRNAs does not affect the binding with cytochrome c.
Apoptosis is tightly regulated at many levels (44, 45). The inhibition of the cytochrome c-Apaf-1 binding by tRNA may present an important cytosolic regulatory mechanism. Thus, tRNAs can bind to cytochrome c that is released into the cytoplasm, providing an inhibitory mechanism for apoptosis and linking cellular sensitivity to apoptotic stimuli with the state of protein synthesis (46–48). This study provides additional insights into the mechanism by which the apoptotic activity of cytochrome c is regulated. Oxidation of cytochrome c stimulates its apoptotic activity, whereas reduction of cytochrome c inhibits it (10). In apoptotic cells, cytochrome c released into the cytosol is likely maintained in the oxidized form by mitochondrial cytochrome c oxidase, which can act on the released cytochrome c due to the permeability of the mitochondrial outer membrane (10). By contrast, in cases where the release of cytochrome c fails to induce apoptosis, cytochrome c may be held in the reduced form by reduced glutathione (49). Our observation that tRNA binds to oxidized cytochrome c with a weaker affinity than to reduced cytochrome c (Fig. 5) provides a possible explanation for the different apoptotic activity of these two forms of cytochrome c.
tRNA can also convert cytochrome c from the oxidized to the reduced form (Fig. 5). The most likely explanation is that tRNA ribonucleotides contact with the heme group of cytochrome c and donate an electron to the ferric ion (Fe3+). This likely contributes to the reduction of the apoptotic activity of cytochrome c. The same redox reaction can also blunt the measured peroxidase activity of cytochrome c (Fig. 6). Because the peroxidase activity of cytochrome c is involved in the oxidation of cardiolipids and subsequent release of cytochrome c from the crista space (the space created by the invaginations of the inner membrane) to the cytosol (40), tRNA might also inhibit this function of cytochrome c if it has access to cardiolipid-bound cytochrome c. This scenario seems possible because of the permeability of mitochondrial outer membrane and the remodeling of the crista space during apoptosis. Thus, the redox reaction between cytochrome c and tRNA suggests a broad function by which tRNA impairs the pro-apoptotic activity of cytochrome c, beyond the disruption of the cytochrome c-Apaf-1 interaction. In contrast, cytochrome c-mediated oxidation could cause damage to tRNA and thus may impair translation, further promoting cell death.
In addition to representing new modes of regulation and function of cytochrome c in apoptosis, the discovery of the cytochrome c-tRNA interaction revealed a previously completely unanticipated role of tRNA. Although it had been appreciated that the three-dimensional structure of tRNA endows it with functions beyond gene expression, a direct function in cell fate decision is especially notable. The inhibitory role of tRNA in apoptosis may raise the threshold of apoptosis in cells that are highly active in protein synthesis, a sensible mechanism given the likely utility of these cells to the organism.
A recent study showed that tRNA halves, which are generated by endonucleolytic cleavage in the anticodon loop in response to oxidative and other stresses (50), bind to cytochrome c and confer resistance to apoptosis (51). We have shown that a pair of tRNA halves, separated by a nick in the anticodon loop, can nonetheless retain the L-shaped structure (26), which may account for their ability to bind to tRNA. Such tRNA halves represent an intriguing example whereby tRNA-mediated apoptotic inhibition is regulated physiologically. This mechanism may also be usurped under pathological conditions, including cancer. Mammalian cytoplasmic tRNAs are transcribed by RNA polymerase III, which is inhibited by the tumor suppressors p53 and the retinoblastoma protein (Rb) and activated by the oncoproteins, including c-Myc and Ras (52). Mutations in these tumor suppressors/oncogenes cause tRNA levels to rise in tumor cells, and high tRNA levels are required for proliferation and tumorigenesis (52). A better understanding of the molecular interaction between cytochrome c and tRNA and the regulation of this interaction should reveal evolutionarily conserved mechanisms that govern apoptosis, metabolism, and translation and the consequences of their deregulation in human diseases.
Author Contributions
Y. M. H. and X. Y. conceived and coordinated the study and wrote the paper. C. L. designed, performed, and analyzed the experiments shown in Figs. 2, B and C, 3, E and F, and 4. A. J. S. designed, performed, and analyzed the experiments shown in Figs. 3, A–D, and 5. C. L. and A. J. S. helped with manuscript preparation. T. C. designed, performed, and analyzed the experiments shown in Figs. 2, A and D, and 4, B and E. J. Y. provided advice and technical assistance. R. T. performed the tRNA-cytochrome c structural modeling in Fig. 4C.
Acknowledgments
We thank S. Seeholzer and H. Ding at the Children's Hospital of Philadelphia's Protein Core Facility for help with the surface plasmon resonance assay; K. Shogen and W. Ardelt for providing Onconase; Y. Mei, W. Prall, and N. Charan for technical assistance; and members of the Hou and Yang laboratories for helpful discussions.
This work was supported in part by National Institutes of Health Grants R01 GM081601, U01 GM108972, and R01 GM114343 (to Y. M. H.), R01 GM060911, and R21 CA178581 and United States Department of Defense Grant W81XWH-13-1-0446 (to X. Y.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- 2AP
- 2-aminopurine
- aaRS
- aminoacyl-tRNA synthetase
- Cy3-etRNACys
- etRNACys labeled with Cy3 at the 5′
- CysRS
- cysteinyl-tRNA synthetase
- etRNACys
- E. coli tRNACys
- etRNAVal
- E. coli tRNAVal
- htRNAPhe
- human tRNAPhe
- htRNAMet
- human elongator tRNAMet
- nt
- nucleotide
- Prf
- proflavine
- Prf-etRNACys
- etRNACys labeled with proflavine at the D-loop.
References
- 1. Ow Y. P., Green D. R., Hao Z., and Mak T. W. (2008) Cytochrome c: functions beyond respiration. Nat. Rev. Mol. Cell Biol. 9, 532–542 [DOI] [PubMed] [Google Scholar]
- 2. Liu X., Kim C. N., Yang J., Jemmerson R., and Wang X. (1996) Induction of apoptotic program in cell-free extracts: requirement for dATP and cytochrome c. Cell 86, 147–157 [DOI] [PubMed] [Google Scholar]
- 3. Zou H., Henzel W. J., Liu X., Lutschg A., and Wang X. (1997) Apaf-1, a human protein homologous to C. elegans CED-4, participates in cytochrome c-dependent activation of caspase-3. Cell 90, 405–413 [DOI] [PubMed] [Google Scholar]
- 4. Schafer Z. T., and Kornbluth S. (2006) The apoptosome: physiological, developmental, and pathological modes of regulation. Dev. Cell 10, 549–561 [DOI] [PubMed] [Google Scholar]
- 5. Riedl S. J., and Salvesen G. S. (2007) The apoptosome: signalling platform of cell death. Nat. Rev. Mol. Cell Biol. 8, 405–413 [DOI] [PubMed] [Google Scholar]
- 6. Li P., Nijhawan D., Budihardjo I., Srinivasula S. M., Ahmad M., Alnemri E. S., and Wang X. (1997) Cytochrome c and dATP-dependent formation of Apaf-1/caspase-9 complex initiates an apoptotic protease cascade. Cell 91, 479–489 [DOI] [PubMed] [Google Scholar]
- 7. Chang H. Y., and Yang X. (2000) Proteases for cell suicide: functions and regulation of caspases. Microbiol. Mol. Biol. Rev. 64, 821–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Li J., and Yuan J. (2008) Caspases in apoptosis and beyond. Oncogene 27, 6194–6206 [DOI] [PubMed] [Google Scholar]
- 9. Riedl S. J., and Shi Y. (2004) Molecular mechanisms of caspase regulation during apoptosis. Nat. Rev. Mol. Cell Biol. 5, 897–907 [DOI] [PubMed] [Google Scholar]
- 10. Borutaite V., and Brown G. C. (2007) Mitochondrial regulation of caspase activation by cytochrome oxidase and tetramethylphenylenediamine via cytosolic cytochrome c redox state. J. Biol. Chem. 282, 31124–31130 [DOI] [PubMed] [Google Scholar]
- 11. Jiang X., Kim H. E., Shu H., Zhao Y., Zhang H., Kofron J., Donnelly J., Burns D., Ng S. C., Rosenberg S., and Wang X. (2003) Distinctive roles of PHAP proteins and prothymosin-α in a death regulatory pathway. Science 299, 223–226 [DOI] [PubMed] [Google Scholar]
- 12. Kim H. E., Jiang X., Du F., and Wang X. (2008) PHAPI, CAS, and Hsp70 promote apoptosome formation by preventing Apaf-1 aggregation and enhancing nucleotide exchange on Apaf-1. Mol. Cell 30, 239–247 [DOI] [PubMed] [Google Scholar]
- 13. Mei Y., Yong J., Liu H., Shi Y., Meinkoth J., Dreyfuss G., and Yang X. (2010) tRNA binds to cytochrome c and inhibits caspase activation. Mol. Cell 37, 668–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Giegé R. (2008) Toward a more complete view of tRNA biology. Nat. Struct. Mol. Biol. 15, 1007–1014 [DOI] [PubMed] [Google Scholar]
- 15. Phizicky E. M., and Hopper A. K. (2010) tRNA biology charges to the front. Genes Dev. 24, 1832–1860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mak J., and Kleiman L. (1997) Primer tRNAs for reverse transcription. J. Virol. 71, 8087–8095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wek R. C., Jackson B. M., and Hinnebusch A. G. (1989) Juxtaposition of domains homologous to protein kinases and histidyl-tRNA synthetases in GCN2 protein suggests a mechanism for coupling GCN4 expression to amino acid availability. Proc. Natl. Acad. Sci. U.S.A. 86, 4579–4583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hou Y. M., Westhof E., and Giegé R. (1993) An unusual RNA tertiary interaction has a role for the specific aminoacylation of a transfer RNA. Proc. Natl. Acad. Sci. U.S.A. 90, 6776–6780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hou Y. M. (1994) Structural elements that contribute to an unusual tertiary interaction in a transfer RNA. Biochemistry 33, 4677–4681 [DOI] [PubMed] [Google Scholar]
- 20. Nissen P., Thirup S., Kjeldgaard M., and Nyborg J. (1999) The crystal structure of Cys-tRNACys-EF-Tu-GDPNP reveals general and specific features in the ternary complex and in tRNA. Structure 7, 143–156 [DOI] [PubMed] [Google Scholar]
- 21. Liu C., Betteridge T., and Hou Y. M. (2009) Fluorophore labeling to monitor tRNA dynamics. Methods Enzymol. 469, 69–93 [DOI] [PubMed] [Google Scholar]
- 22. Betteridge T., Liu H., Gamper H., Kirillov S., Cooperman B. S., and Hou Y. M. (2007) Fluorescent labeling of tRNAs for dynamics experiments. RNA 13, 1594–1601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hauenstein S. I., Hou Y. M., and Perona J. J. (2008) The homotetrameric phosphoseryl-tRNA synthetase from Methanosarcina mazei exhibits half-of-the-sites activity. J. Biol. Chem. 283, 21997–22006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Igarashi T., Liu C., Morinaga H., Kim S., and Hou Y. M. (2011) Pyrophosphorolysis of CCA addition: implication for fidelity. J. Mol. Biol. 414, 28–43 [DOI] [PubMed] [Google Scholar]
- 25. Kim S., Liu C., Halkidis K., Gamper H. B., and Hou Y. M. (2009) Distinct kinetic determinants for the stepwise CCA addition to tRNA. RNA 15, 1827–1836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dupasquier M., Kim S., Halkidis K., Gamper H., and Hou Y. M. (2008) tRNA integrity is a prerequisite for rapid CCA addition: implication for quality control. J. Mol. Biol. 379, 579–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Liu C., Gamper H., Liu H., Cooperman B. S., and Hou Y. M. (2011) Potential for interdependent development of tRNA determinants for aminoacylation and ribosome decoding. Nat. Commun. 2, 329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Christian T., Lipman R. S., Evilia C., and Hou Y. M. (2000) Alternative design of a tRNA core for aminoacylation. J. Mol. Biol. 303, 503–514 [DOI] [PubMed] [Google Scholar]
- 29. Liu C., Sanders J. M., Pascal J. M., and Hou Y. M. (2012) Adaptation to tRNA acceptor stem structure by flexible adjustment in the catalytic domain of class I tRNA synthetases. RNA 18, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhang C. M., and Hou Y. M. (2005) Domain-domain communication for tRNA aminoacylation: the importance of covalent connectivity. Biochemistry 44, 7240–7249 [DOI] [PubMed] [Google Scholar]
- 31. Zhang C. M., Perona J. J., Ryu K., Francklyn C., and Hou Y. M. (2006) Distinct kinetic mechanisms of the two classes of aminoacyl-tRNA synthetases. J. Mol. Biol. 361, 300–311 [DOI] [PubMed] [Google Scholar]
- 32. Liu C., Gamper H., Shtivelband S., Hauenstein S., Perona J. J., and Hou Y. M. (2007) Kinetic quality control of anticodon recognition by a eukaryotic aminoacyl-tRNA synthetase. J. Mol. Biol. 367, 1063–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lee Y. S., Shibata Y., Malhotra A., and Dutta A. (2009) A novel class of small RNAs: tRNA-derived RNA fragments (tRFs). Genes Dev. 23, 2639–2649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hamann C. S., and Hou Y. M. (2000) Probing a tRNA core that contributes to aminoacylation. J. Mol. Biol. 295, 777–789 [DOI] [PubMed] [Google Scholar]
- 35. Hamann C. S., and Hou Y. M. (1997) An RNA structural determinant for tRNA recognition. Biochemistry 36, 7967–7972 [DOI] [PubMed] [Google Scholar]
- 36. Suryanarayana T., Uppala J. K., and Garapati U. K. (2012) Interaction of cytochrome c with tRNA and other polynucleotides. Mol. Biol. Rep. 39, 9187–9191 [DOI] [PubMed] [Google Scholar]
- 37. Yamashita S., Takeshita D., and Tomita K. (2014) Translocation and rotation of tRNA during template-independent RNA polymerization by tRNA nucleotidyltransferase. Structure 22, 315–325 [DOI] [PubMed] [Google Scholar]
- 38. Hauenstein S., Zhang C. M., Hou Y. M., and Perona J. J. (2004) Shape-selective RNA recognition by cysteinyl-tRNA synthetase. Nat. Struct. Mol. Biol. 11, 1134–1141 [DOI] [PubMed] [Google Scholar]
- 39. Yuan J. (2006) Divergence from a dedicated cellular suicide mechanism: exploring the evolution of cell death. Mol. Cell 23, 1–12 [DOI] [PubMed] [Google Scholar]
- 40. Kagan V. E., Tyurin V. A., Jiang J., Tyurina Y. Y., Ritov V. B., Amoscato A. A., Osipov A. N., Belikova N. A., Kapralov A. A., Kini V., Vlasova I. I., Zhao Q., Zou M., Di P., Svistunenko D. A., et al. (2005) Cytochrome c acts as a cardiolipin oxygenase required for release of proapoptotic factors. Nat. Chem. Biol. 1, 223–232 [DOI] [PubMed] [Google Scholar]
- 41. Degen W. G., Pruijn G. J., Raats J. M., and van Venrooij W. J. (2000) Caspase-dependent cleavage of nucleic acids. Cell Death Differ. 7, 616–627 [DOI] [PubMed] [Google Scholar]
- 42. Keilin D. (1925) On cytochrome, a respiratory pigment, common to animals, yeast, and higher plants. Proc. R. Soc. Lond. B Biol. Sci. 98, 312–339 [Google Scholar]
- 43. Slater E. C. (2003) Keilin, cytochrome, and the respiratory chain. J. Biol. Chem. 278, 16455–16461 [DOI] [PubMed] [Google Scholar]
- 44. Jiang X., and Wang X. (2004) Cytochrome c-mediated apoptosis. Annu. Rev. Biochem. 73, 87–106 [DOI] [PubMed] [Google Scholar]
- 45. Tait S. W., and Green D. R. (2010) Mitochondria and cell death: outer membrane permeabilization and beyond. Nat. Rev. Mol. Cell Biol. 11, 621–632 [DOI] [PubMed] [Google Scholar]
- 46. Mei Y., Stonestrom A., Hou Y. M., and Yang X. (2010) Apoptotic regulation and tRNA. Protein Cell 1, 795–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Mei Y., Yong J., Stonestrom A., and Yang X. (2010) tRNA and cytochrome c in cell death and beyond. Cell Cycle 9, 2936–2939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hou Y. M., and Yang X. (2013) Regulation of cell death by transfer RNA. Antioxid. Redox Signal. 19, 583–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Vaughn A. E., and Deshmukh M. (2008) Glucose metabolism inhibits apoptosis in neurons and cancer cells by redox inactivation of cytochrome c. Nat. Cell Biol. 10, 1477–1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Thompson D. M., and Parker R. (2009) Stressing out over tRNA cleavage. Cell 138, 215–219 [DOI] [PubMed] [Google Scholar]
- 51. Saikia M., Jobava R., Parisien M., Putnam A., Krokowski D., Gao X. H., Guan B. J., Yuan Y., Jankowsky E., Feng Z., Hu G. F., Pusztai-Carey M., Gorla M., Sepuri N. B., Pan T., and Hatzoglou M. (2014) Angiogenin-cleaved tRNA halves interact with cytochrome c, protecting cells from apoptosis during osmotic stress. Mol. Cell. Biol. 34, 2450–2463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. White R. J. (2005) RNA polymerases I and III, growth control and cancer. Nat. Rev. Mol. Cell Biol. 6, 69–78 [DOI] [PubMed] [Google Scholar]