Abstract
In addition to the well-known light-driven outward proton pumps, novel ion-pumping rhodopsins functioning as outward Na+ and inward Cl− pumps have been recently found in eubacteria. They convert light energy into transmembrane electrochemical potential difference, similar to the prototypical archaeal H+ pump bacteriorhodopsin (BR) and Cl− pump halorhodopsin (HR). The H+, Na+, and Cl− pumps possess the conserved respective DTE, NDQ, and NTQ motifs in the helix C, which likely serve as their functional determinants. To verify this hypothesis, we attempted functional interconversion between selected pumps from each category by mutagenesis. Introduction of the proton-pumping motif resulted in successful Na+ → H+ functional conversion. Introduction of the respective characteristic motifs with several additional mutations leads to successful Na+ → Cl− and Cl− → H+ functional conversions, whereas remaining conversions (H+ → Na+, H+ → Cl−, Cl− → Na+) were unsuccessful when mutagenesis of 4–6 residues was used. Phylogenetic analysis suggests that a H+ pump is the common ancestor of all of these rhodopsins, from which Cl− pumps emerged followed by Na+ pumps. We propose that successful functional conversions of these ion pumps are achieved exclusively when mutagenesis reverses the evolutionary amino acid sequence changes. Dependence of the observed functional conversions on the direction of evolution strongly suggests that the essential structural mechanism of an ancestral function is retained even after the gain of a new function during natural evolution, which can be evoked by a few mutations. By contrast, the gain of a new function needs accumulation of multiple mutations, which may not be easily reproduced by limited mutagenesis in vitro.
Keywords: biophysics, chloride transport, mutant, photoreceptor, proton pump, rhodopsin, sodium transport
Introduction
Animal and microbial rhodopsins are photoreceptive proteins, which use retinal as their chromophore. The main function of several groups of microbial rhodopsins is light-driven ion transport across cellular membranes (1). In the last decade, these proteins became powerful tools in neuroscience, where they are used to control neural activity of animals by light (so-called “optogenetics”) (2, 3). Bacteriorhodopsin (BR)2 and HR, the first ion-transporting rhodopsins, were discovered in 1971 and 1977, respectively, from halophilic archaea (4, 5). Since 2000, genomic and metagenomic sequencing revealed that microbial rhodopsins are widely distributed among marine and freshwater bacteria, most of which were classified as light-driven H+ pumps (proteorhodopsins and xanthorhodopsins, PR and XR) (Fig. 1) (6, 7). In addition, eubacterial light-driven Na+ and Cl− pumps have been discovered recently (Fig. 1) (8–10), with the former forming a new functional class and the latter not being related to haloarchaeal HRs. These pumps are interesting not only from biophysical and biochemical point of view, but also for evolutionary and environmental microbiology and optogenetic applications. Recent structure determination of a light-driven Na+ pump Krokinobacter eikastus rhodopsin 2 (KR2) accelerated our understanding of ion selectivity, and K+ pump was engineered based on that structure (11, 12).
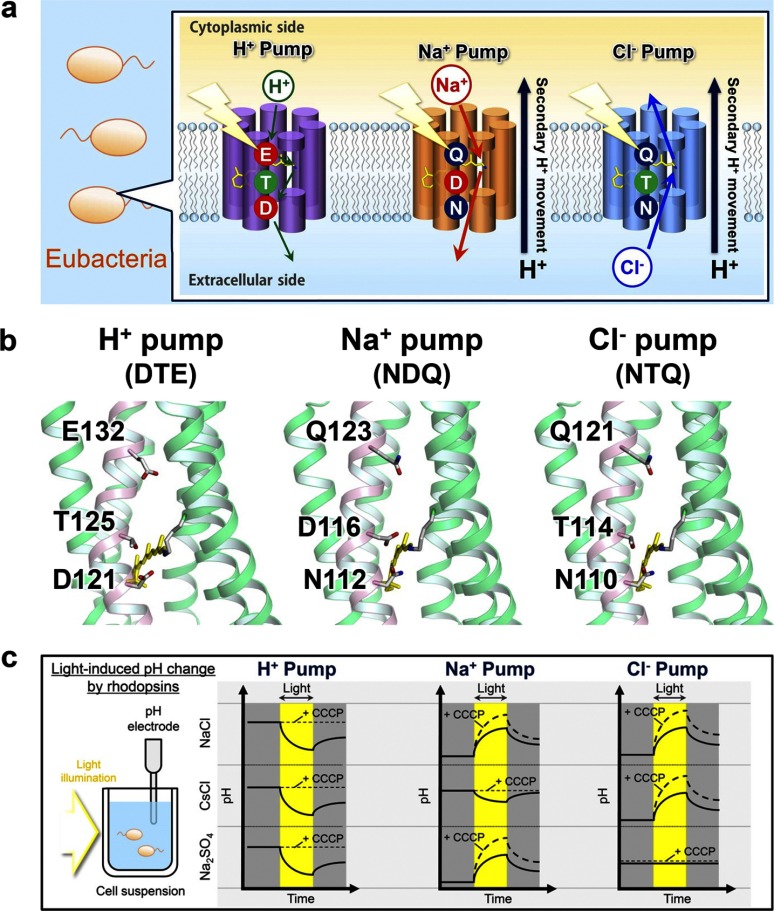
FIGURE 1.
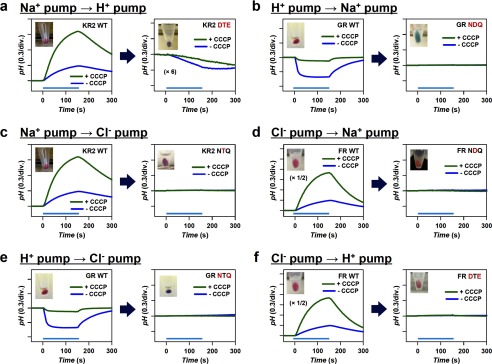
Light-driven ion-pumps in eubacteria and their transport activity. a, eubacterial rhodopsins contain DTE, NDQ, and NTQ motifs for H+, Na+, and Cl− pumps, respectively. b, motifs in GR, KR2, and FR based on the structure of XR (left, PDB ID: 3DDL) and KR2 (middle, right, PDB ID:3X3C). c, ion pumping activity assayed by pH changes. Light-driven H+ pump shows pH decrease in the presence of any ions, which is diminished by a protonophore CCCP. In contrast, Na+ and Cl− pumps cause pH increase, which is enhanced by CCCP. Light-driven Na+ and Cl− pumps are distinguished by their cation- and anion-dependent transport, respectively.
Functional interconversion of homologous proteins by site-directed mutagenesis is a powerful tool to understand molecular mechanisms of their functions. The first example of successful functional conversion of microbial rhodopsins is when BR was converted into an HR-like Cl− pump by a single amino acid replacement (13, 14). The conserved functional helix C motifs for BR and HR are DTD and TSA, respectively. In BR, Asp-85 (the first Asp in the DTD motif) is the H+ acceptor from the protonated Schiff base during H+ pumping (1), and when Asp-85 is mutated to Thr, the corresponding amino acid in HR, D85T BR pumps Cl− inwards, identical to HR (13, 14). This observation implied that BR and HR share a common transport mechanism, and its selectivity is determined at the position of primary proton acceptor (Asp-85 in BR). While this was a clear-cut result, the respective reverse mutation (Thr → Asp) did not convert HR into a H+ pump (15, 16). Ten additional mutations to make pharaonis HR resemble BR did not work either, indicating that functional conversion between archaeal H+ and Cl− pumps is asymmetric (17). The vibrational analysis suggested that protein-bound water molecules may be responsible for the asymmetric functional conversion (17). While two conversions (H+ ⇆ Cl−) were tested for archaeal pumps, eubacterial rhodopsins may undergo six functional conversions (H+ ⇆ Cl−, H+ ⇆ Na+, Na+ ⇆ Cl−). In the present study, we tested whether a limited number of mutations can achieve functional interconversion between eubacterial pumps.
Experimental Procedures
Gene Construction, Protein Expression, and Purification
KR2 and FR genes whose codons were optimized for Escherichia coli expression system were synthesized (Eurofins Genomics Inc.) and subcloned into a pET21a(+)-vector with C-terminal 6×His-tag (10). A plasmid vector of GR with the additional C-terminal 6×His-tag was constructed as described previously (18). For mutagenesis, a QuikChange site-directed mutagenesis kit (Stratagene) was used according to the standard protocol. Wild-type and mutant proteins were expressed in E. coli C41(DE3) strain. The protein expression was induced by 0.21 mm isopropyl β-d-thiogalactopyranoside (IPTG) for 4 h at 37 °C with the supplement of 10 μm all-trans-retinal (Sigma-Aldrich) to the culture. For the measurement of Cl− dependence of absorption spectra, the expressed proteins were purified from E. coli cells according to the previously reported protocols (8, 17). The cells were disrupted by French Press (Ohtake), and the membrane fraction was collected by ultracentrifugation (125,000 × g, 1 h). The protein was solubilized with 2% n-dodecyl-β-d-maltoside (DDM) (Anatrace) in the presence of 300 mm NaCl, 5 mm imidazole, and 50 mm MES (pH 6.5). After Co2+-NTA affinity chromatography, the collected fractions were dialyzed to the solution containing 100 mm NaCl, 50 mm Tris-HCl (pH 7.0), 0.02% DDM to remove imidazole used for elution from the column.
Assay of Light-driven Pump Activity of Rhodopsins in E. coli Cells
E. coli expressing rhodopsins were harvested by centrifugation (3,600 × g, 3 min), washed three times and resuspended in the aqueous solvent containing 100 mm salt (NaCl, CsCl, or Na2SO4). 7.5 ml of cell suspension at OD660 = 2 was placed in the dark and then illuminated at λ > 500 nm by a 1-kW tungsten-halogen projector lamp (Rikagaku, Japan) through a glass filter (Y-52, AGC Techno Glass, Japan). The light-induced pH changes were measured by a pH electrode (HORIBA, Japan).
Quantification of Rhodopsins Expressed in E. coli
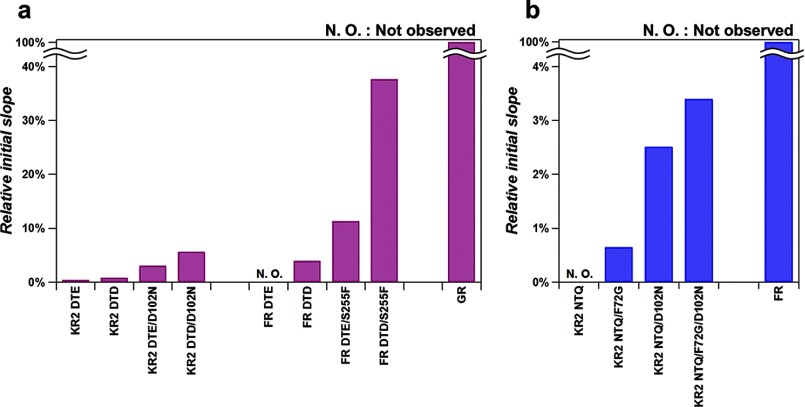
To quantify the expressed rhodopsin in E. coli cell used for pump activity assay, the same amount of cells used for pump activity assay was collected by low-speed centrifugation at 3,600 × g and 4 °C and suspended in the solution containing 100 mm NaCl, 50 mm Tris-HCl (pH 8.0), to a final volume of 3 ml. Then, 200 μl of 1 mm lysozyme was added to the suspension and it was gently stirred at room temperature for 1 h. The E. coli cells were disrupted by sonication (TAITEC, Japan) and solubilized in 3.0% DDM. The absorption change, representing the bleaching of rhodopsin by hydroxylamine (HA), was measured with a UV-vis spectrometer (Shimadzu, Japan) equipped with an integrating sphere after the addition of HA (100 mm NaCl, 50 mm Tris-HCl (pH 8.0)) to a final concentration of 500 mm HA and illumination at λ > 500 nm by a 1-kW tungsten-halogen projector lamp (Rikagaku, Japan) through a glass filter (Y-52, AGC Techno Glass, Japan). The molecular extinction coefficient of rhodopsin (ϵ) was calculated from the ratio between the absorbance of rhodopsin and retinal oxime (ϵ = 33,600 m−1 cm−1 at 360 nm) produced by the reaction between retinal Schiff base and HA (19). Molar extinction coefficients and absorption maxima wavelengths of each mutant are shown in Figs. 3, 4, and 5. The amount of rhodopsin expressed in E. coli cells was determined by the absorbance of the bleached rhodopsin and the value of ϵ, which is shown in Figs. 3, e, h; 4, e, h; 5, e and h. The transport activity of E. coli cells containing each rhodopsin was quantitatively determined from the initial slope of pH change after normalizing the expression level of protein, which is summarized in Fig. 6.
FIGURE 3.
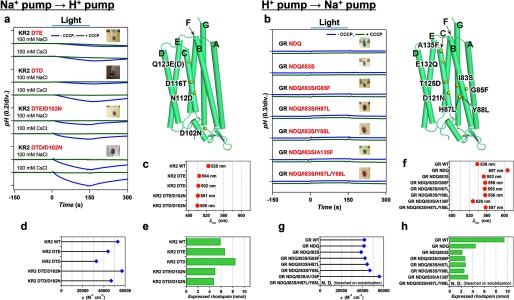
Optimization of functional conversions between light-driven Na+ and H+ pumps by additional mutations outside of the conserved helix C motifs. Improvement of functional conversion of Na+ → H+ pump in KR2 (a) in the presence of 100 mm NaCl or CsCl (pH ∼7.0) and unsuccessful functional conversion of H+ → Na+ pump in GR (b) in the presence of 100 mm NaCl (pH ∼7.0) in the absence (blue) and presence (green) of CCCP, respectively. The positions of mutations are indicated by yellow spheres in the modeled structure (right) and the names of helices are shown with green characters. The wavelength of absorption maxima (λmax) (c, f), molecular extinction coefficients (ϵ) (d, g), and the amount of expressed rhodopsin in the E. coli cell used for the pump activity assay (e, h) of wild type and mutant proteins.
FIGURE 4.
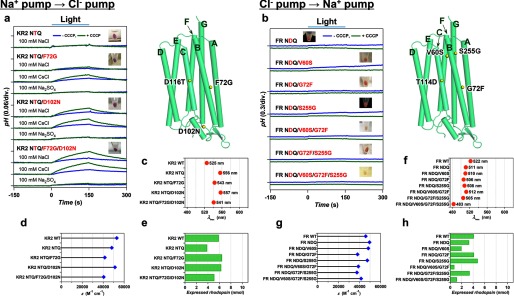
Optimization of functional conversions between light-driven Na+ and Cl− pumps by additional mutations outside of the conserved helix C motifs. Successful functional conversion of Na+ → Cl− pump in KR2 (a) in the presence of 100 mm NaCl, CsCl, or Na2SO4 (pH ∼7.0) and unsuccessful functional conversion of Cl− → Na+ pump in FR (b) in the presence of 100 mm NaCl (pH ∼7.0) in the absence (blue) and presence (green) of CCCP, respectively. The positions of mutations are indicated by yellow spheres in the modeled structure (right), and the names of helices are shown with green characters. The λmax (c, f), ϵ (d, g) and the amount of expressed rhodopsin in the E. coli cell used for the pump activity assay (e, h) of wild type and mutant proteins.
FIGURE 5.
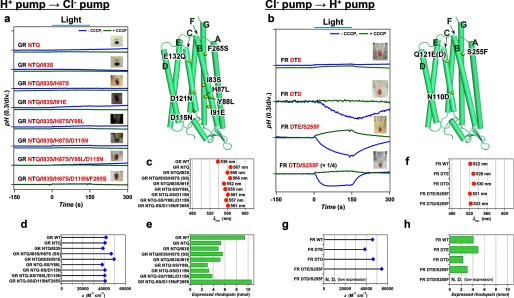
Optimization of functional conversions between light-driven H+ and Cl− pumps by additional mutations outside of the conserved helix C motifs. Unsuccessful functional conversion of H+ → Cl− pump in GR (a) and successful functional conversion of Cl− → H+ pump in FR (b) in the presence of 100 mm NaCl (pH ∼7.0) in the absence (blue) and presence (green) of CCCP, respectively. The positions of mutations are indicated by yellow spheres in the modeled structure (right), and the names of helices are shown with green characters. The λmax (c, f), ϵ (d, g) and the amount of expressed rhodopsin in the E. coli cell used for the pump activity assay (e, h) of wild type and mutant proteins.
FIGURE 6.
Ion pumping activities of the functionally-converted mutants. a, relative H+ pumping activities of KR2 and FR mutants compared with GR WT. b, relative Cl− pumping activities of KR2 mutants compared with FR WT.
Chloride Titrations
To measure the Cl−-concentration dependence of absorption spectra of rhodopsins, the proteins were dialyzed to the solution containing 0.1% DDM and 20 mm HEPES (pH 7.0). NaCl was added to the sample and UV-visible absorption spectra were measured with a UV-vis spectrometer (JASCO, Japan) equipped with an integrating sphere at various Cl−-concentrations.
Laser Flash Photolysis
The transient absorption change after the photo excitation of the rhodopsins were investigated by laser flash photolysis method (8, 11). The purified rhodopsin was reconstituted into the lipid bilayer of the mixture of 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (POPE) and 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-(1′-rac-glycerol) (POPG) (molecular ratio: POPE:POPG = 3:1) with the protein-to-lipid molecular ratio of 1:20 at 2–4 μm protein concentration. The reconstituted sample was suspended in 100 mm NaCl, 20 mm HEPES-NaOH (pH 7). The sample solution was illuminated with a second harmonics generation of nanosecond pulsed Nd3+-YAG laser (λ = 532 nm, INDI40, Spectra-Physics, CA) with the pulse energy of 3.8 mJ/cm2 pulse. The transient absorption spectrum of rhodopsin after the laser excitation was obtained by measuring the intensity of white light passed through the sample before and after laser excitation at λ = 350–750 nm with an ICCD linear array detector (C8808–01, Hamamatsu, Japan). To increase signal-to-noise ratio, 90 identical spectra were averaged and singular-value-decomposition (SVD) analysis was applied (8).
Phylogenic Analysis of Rhodopsin Genes
The amino acid sequences of rhodopsins were aligned using MUSCLE program (20) after the removal of weakly conserved interhelical loop, and N- and C-terminal extensions to increase the accuracy of alignment. The evolutionary history was inferred using the Neighbor-Joining method (21). The optimal tree with the sum of branch length = 23.00061278 was obtained. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) were calculated (22). The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Poisson correction method (23) and are in the units of the number of amino acid substitutions per site. The analysis involved 128 amino acid sequences. All positions containing gaps and missing data were eliminated. There were a total of 105 positions in the final dataset. Evolutionary analyses were conducted in MEGA6 (24).
Results and Discussion
We selected Gloeobacter rhodopsin (GR) (18), Krokinobacter eikastus rhodopsin 2 (KR2) (8), and Fulvimarina rhodopsin (FR) (10) as the representatives of H+, Na+, and Cl− pumps, respectively. The amino acid identity is 26% between GR and KR2, 33% between GR and FR, and 33% between KR2 and FR (see supplemental Table S1 for all amino acid sequences). It should be noted that the eubacterial H+, Na+, and Cl− pumps possess the highly conserved DTE, NDQ, and NTQ motifs in the helix C, respectively (Fig. 1, a and b), positions of which correspond to Asp-85, Thr-89, and Asp-96 in BR (25). As these motifs are likely to determine the function, we first examined functional conversions by altering them. Note that two, three, and one mutation(s) are needed for H+ ⇆ Cl−, H+ ⇆ Na+, and Na+ ⇆ Cl−, respectively. It should be also noted that a light-driven Na+ pump KR2 acts as a H+ pump in the presence of larger cations (8). Therefore, we define the Na+ → H+ pump conversion for the KR2 mutants as successful if it pumps H+ even in 100 mm NaCl.
To verify function of each rhodopsin mutant, the proteins were overexpressed in E. coli (C41 (DE3) strain) and light-induced pH changes of E. coli cell suspensions containing each protein were measured. Identification of H+, Na+, and Cl− pumps by the pH monitoring system is summarized in Fig. 1c. A cell suspension of E. coli expressing H+ pump shows acidification of external medium (pH decrease) upon illumination by the outward transport of H+ irrespective of the ionic species contained in the solvent, and the signals are diminished in the presence of a protonophore, carbonylcyanide m-chlorophenylhydrazone (CCCP) (Fig. 1c, left panel). In contrast, a cell suspension of E. coli expressing Na+ pump or Cl− pump shows alkalinization (pH increase) upon illumination, because of secondary H+-uptake occurs to compensate the increased negative membrane potential inside the cells. The signals are further enhanced in the presence of CCCP, because the observed pH changes originate from secondary H+ movement due to cation or anion transport. Analysis of ion dependence is used to distinguish cation and anion pumps. Na+ pump KR2 is converted to H+ pump in the solvent containing the salt of larger cations such as CsCl, while it pumps Na+ outwards in Na2SO4 (Fig. 1c, middle panel). On the other hand, while Cl− pump FR transports Cl− ion in the solution containing NaCl or CsCl, it does not transport larger anions such as SO42−, and no alkalization occurs in the solution of Na2SO4 (Fig. 1c, right panel). Thus, comparison of signals in NaCl, CsCl, and Na2SO4 allows distinguishing between the transported ions. Fig. 2 shows the results of functional conversion experiments performed by the replacement of the characteristic helix C motifs. Among the six functional conversions, only one conversion was achieved, from Na+ to H+ pump (Fig. 2a). This may be consistent with the fact that Na+-pumping KR2 transports H+ in the presence of larger cations (8), suggesting that KR2 retained the molecular machinery of H+ pump in its structure. On the other hand, none of the other motif-altering mutations achieved functional conversion (Fig. 2, b–f). Thus, we introduced additional plausible mutations based on the sequence comparisons outside of the conserved helix C motifs.
FIGURE 2.
Functional conversion of light-driven H+, Na+, and Cl− pumps by replacing the characteristic helix C motifs. a–f, three, one, and two amino acids in each motif are replaced for the functional conversions of H+ ⇆ Na+ (a, b), Na+ ⇆ Cl− (c, d), and H+ ⇆ Cl− (e, f), respectively. Blue and green traces represent pH changes upon illumination in the absence and presence of CCCP, respectively. All measurements are performed in the presence of 100 mm NaCl (pH ∼7.0). The KR2 DTE mutant shows H+ pumping activity, indicating a successful Na+ → H+ pump conversion by the motif mutation (a). All other mutants exhibit no pumping activities (b–f).
Figs. 3, 4, and 5 summarize the results of functional conversions between H+, Na+, and Cl− pumps which used such additional mutations. Fig. 3a shows the improvement of the Na+ → H+ pump conversion. Even though the DTE KR2 mutant pumps protons (Fig. 2a), the efficiency of transport estimated from the initial slope of pH change is very low (0.42% of that of GR, Fig. 6a). We found that the DTD KR2 mutant pumps better (0.79%, Fig. 6a), and the transport was more efficient in Cs+ than in Na+ (Fig. 3a). The latter may be related to the fact that KR2 possesses Na+ binding site at the extracellular surface (8), which inhibits outward H+ pumping only in Na+. As Asp-102 constitutes the Na+ binding site (11, 12), we replaced this residue, and the DTE/D102N and DTD/D102N KR2 mutants exhibited similar H+ pumping activities in NaCl and CsCl (3.0 and 5.6% of GR, respectively, Fig. 6a). The functional conversion of Na+→H+ pump is thus achieved by 4 mutations. In contrast, the reverse motif mutation for H+ → Na+ pump conversion (DTE to NDQ) has not worked (Fig. 2b), which may be related to the fact that the NDQ GR mutant looks blue (λmax = 607 nm, Fig. 3f) (i.e. exists in the non-transporting acidic form). This suggests that Asp in the NDQ motif of this mutant is protonated and does not act as the Schiff base counterion and proton acceptor as required for sodium transport (8, 26, 27). We introduced additional mutations to the NDQ GR mutant, to change the color from blue (λmax > 600 nm) to purple (λmax < 560 nm) and make the Schiff base proton acceptor functional. Fig. 3b shows clear color changes for the NDQ/I83S/H87L (λmax = 555 nm, Fig. 3f), NDQ/I83S/Y88L (λmax = 556 nm, Fig. 3f), and NDQ/I83S/H87L/Y88L (λmax = 557 nm, Fig. 3f) GR mutants, where mutations were introduced near the Schiff base region. However, despite the presence of the required ion-pair (Schiff base and counterion), these mutants do not pump Na+ (Fig. 3b). We conclude that functional interconversion for H+ ⇆ Na+ pair is asymmetric, where even 6 mutations are not sufficient for the H+ → Na+ pump conversion.
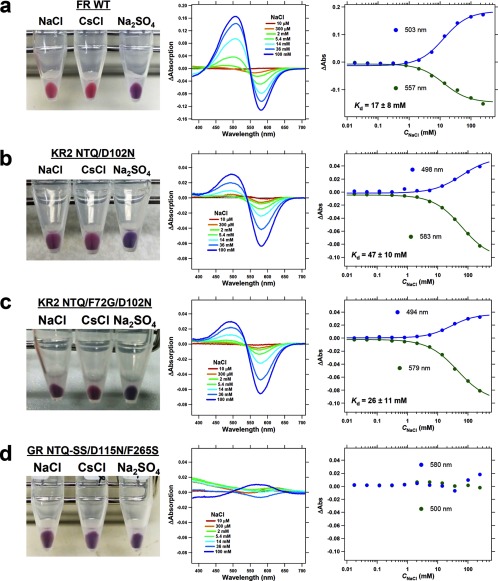
Next, we tried to improve functional interconversion of Na+ and Cl− pumps (KR2 and FR) with additional mutations. As the simple motif replacement resulted in unsuccessful Na+ → Cl− pump conversion, we suspected that the negatively charged Na+ binding site in KR2 (8, 11, 12) could prevent inward Cl− pumping (Fig. 2c). It is indeed the case, as the NTQ/D102N KR2 mutant exhibits light-induced pH increase in NaCl and CsCl, but not in Na2SO4 (Fig. 4a). This is characteristic for Cl− pumps, and in line with this, Fig. 7b shows absorption changes of the NTQ/D102N KR2 mutant upon Cl− binding. Thus, a double mutation (NTQ/D102N) is sufficient for converting KR2 into a Cl− pump. The pumping activity is further enhanced by introducing F72G mutation, which is highly conserved near the Schiff base region, and the NTQ/F72G/D102N KR2 mutant pumps Cl− with the 3.4% efficiency of that in FR (Figs. 4a and 6b). NTQ/F72G/D102N KR2 mutant also showed color change upon Cl− binding (Fig. 7c). To make a reverse Cl− → Na+ pump conversion possible, we searched for additional mutations outside of the conserved motif, based on the sequence differences (Fig. 4b) and structure of KR2. However, none of these mutants showed any ion transport (Fig. 4b). Thus, while Na+ → Cl− pump conversion was successful, the reverse Cl− → Na+ pump conversion could not be achieved, once again showing an asymmetric interconversion.
FIGURE 7.
Cl− concentration dependence of absorption spectra of rhodopsins having the NTQ motif reporting on chloride binding. The picture of E. coli cells in 100 mm NaCl, CsCl, and Na2SO4 (left), difference UV-visible absorption spectra upon the addition of NaCl of different concentrations (middle) and the titration curve for the absorption change at the peak wavelength shown in the difference spectra (right) for FR WT (a), KR2 NTQ/D102N (b), KR2 NTQ/F72G/D102N (c), GR NTQ-I83S/H87S(SS)/D115N/F265S (d). Solid lines in the titration curve indicate the fitting curves using a Hill equation and calculated dissociation constant of Cl− (Kd) is shown in the graph.
Finally, the H+ ⇆ Cl− interconversion was examined with additional mutations, as the motif replacement mutations (DTE ⇆ NTQ) were not sufficient for successful functional conversions (Fig. 2, e and f). The NTQ GR mutant looks blue (λmax = 567 nm) (Fig. 2e), suggesting that the protein does not bind Cl− in 100 mm NaCl. Therefore, we introduced additional mutations to the NTQ GR mutant to improve Cl− binding, as assayed by change of the color from blue (λmax > 560 nm) to purple (λmax < 560 nm) under these conditions. Fig. 5a shows that several mutants exhibit color changes, suggesting FR-like Cl− binding (10). Nevertheless, none of these mutants transported any ions (Fig. 5a). Using titrations of purified proteins, we found that Cl− binding does not take place, as shown for GR NTQ/I83S/H87S/D115N/F265S, which showed only small non-systematic absorption change probably representing unspecific interaction between the protein and ions (Na+ and/or Cl−) (Fig. 7d). Thus, the color changes were caused by the mutations themselves and not by Cl− binding, which explains the lack of ion transport. We then tried to optimize functional conversion from Cl− to H+ pump. Gly-263 of Na+ pump KR2 is a key residue for Na+ uptake, whose modification leads to K+ pumping (11, 12). The corresponding amino acids are S and F for FR and GR, respectively, which may be important for ion specificity of each pump. Indeed, Fig. 5b shows that the DTE/S255F FR mutant functions as an outward H+ pump whose relative activity is 11% of wild type GR (Fig. 6a). We thus conclude that functional interconversion for the H+ ⇆ Cl− pair is asymmetric as well, where an additional mutation outside of the conserved motif converted Cl− into H+ pump, but four additional mutations were not sufficient for the H+ → Cl− pump conversion.
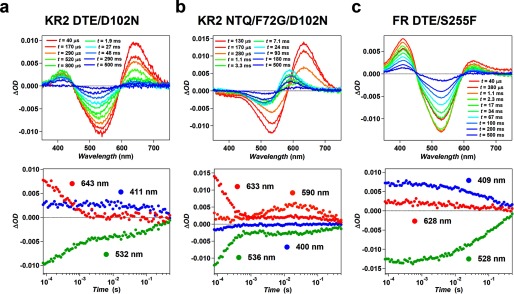
In this study, three types of functional conversion were succeeded in (Na+ → H+, Na+ → Cl−, and Cl− → H+) with an additional mutation to motif exchange. To investigate their transport mechanism and compare it with natural pumps, the photocycles of these functionally converted mutants were studied by laser flash photolysis. The transient absorption changes of KR2 DTE/D102N, KR2 NTQ/F72G/D102N, and FR DTE/S255F were shown in Fig. 8. While wild type KR2 shows small amount of accumulation of the M-intermediate at λ = 400 nm equilibrating with the L (8), KR2 DTE/D102N showed higher M-accumulation than wild type (Fig. 8a). This large accumulation of M is characteristic feature of the photocycle of many H+ pump rhodopsins including GR (18, 28). The similar high accumulation of M-intermediate was observed for FR DTE/S255F which also converted to H+ pump (Fig. 8c), in contrast to the photocycle of wildtype FR in which no M accumulation occurs (10). Therefore, the mutants converted to H+ pump transports H+ with the photocycle similar to that of natural H+ pump in which higher M accumulation occurs by the H+ transfer from retinal Schiff-base to the neighboring aspartate residues. On the other hand, the photocycle of KR2 NTQ/F72G/D102N showed no M-intermediate and red-shifted photo-intermediates (K and O) were dominantly observed (Fig. 8b). For natural Cl− pump, the photocycles of HR and FR are significantly different, that is, the former and latter mainly consist of blue-shifted L-intermediate (29, 30) and red-shifted K or O-intermediates (10), respectively. The photocycle of KR2 NTQ/F72G/D102N resembles to that of wild type FR which contains the same NTQ-motif. Therefore, KR2 NTQ/F72G/D102N transports Cl− with the identical mechanism to FR rather than HR with TSA-motif. The time constants of initial state recovery of these mutants (KR2 DTE/D102N: 251 ms, KR2 NTQ/F72G/D102N: 282 ms, FR DTE/S255F: 160 ms) were longer than wildtype GR (140 ms) (18) and FR (20.4 ms) (10). Therefore, a part of the reason of lower efficiencies of the mutants compared with natural pumps is considered to be its slower turnover rate (photocycle recovery) and more amino acid mutations would be required for the optimization of faster turnover rate.
FIGURE 8.
Transient absorption change of functionally converted mutants. Transient absorption spectra (upper) and the time-evolution of absorption changes (lower) at the wavelengths representing each photoreaction species (the K-intermediate (red), M-intermediate (blue), O-intermediate (orange), and initial state (green)) for KR2 DTE/D102N (a), KR2 NTQ/F72G/D102N (b), and FR DTE/S255F (c).
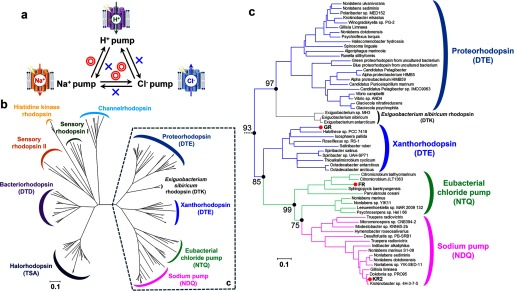
The asymmetric results of functional conversions among the light-driven retinal-binding pumps are summarized in Fig. 9a. In principle, mutations of key residues may destabilize proteins, leading to non-functional species which lost their original function but have not gained a new one. However, we achieved three functional conversions by a very limited number of mutations. Interestingly, only one functional conversion in each pair of proteins was possible, whereas the reverse conversions did not work. We discuss the observed asymmetry of the interconversions from the evolutionary viewpoint. Fig. 9b shows phylogenetic tree of microbial rhodopsins. Based on the conserved helix C motif, eubacterial rhodopsins can be classified into DTX (H+ pumps; X is mostly E), NTQ (Cl− pumps), and NDQ (Na+ pumps) rhodopsins, which are distinct from haloarchaeal DTD (BR; H+ pumps) and TSA (HR; Cl− pumps) rhodopsins. Fig. 9c focuses on eubacterial rhodopsins, whose phylogenetic subtree strongly suggests that the origin of eubacterial rhodopsins is an H+ pump, from which Cl− pumps emerged, followed by the appearance of Na+ pumps. Comparison of Fig. 9, a and c shows that successful functional conversions are attained exclusively when the mutagenesis tries to reverse the course of evolution, but not when it follows the evolutionary direction. Dependence of the observed functional conversions on the direction of evolution strongly suggests that the essential elements of an ancestral function are retained even after the gain of a new function, while the gain of a new function needs accumulation of multiple mutations, which may not be easily reproduced by limited mutagenesis in vitro.
FIGURE 9.
Functional conversions observed in the present study and their relationship to evolution of eubacterial ion-pumping rhodopsins. a, summary of functional conversions observed in the present study. Red double-circles and blue crosses represent successful and unsuccessful functional conversions, respectively. b, phylogenic tree of microbial rhodopsins. The bootstrap tests are calculated for 1000 replicates. The scale bar indicates 0.1 units of the number of amino acid substitutions per site. c, phylogenic subtree of eubacterial light-driven ion pumps. The corresponding branches are enclosed by dashed line in b. Bootstrap values of significantly higher score are indicated by the numbers at filled black circles.
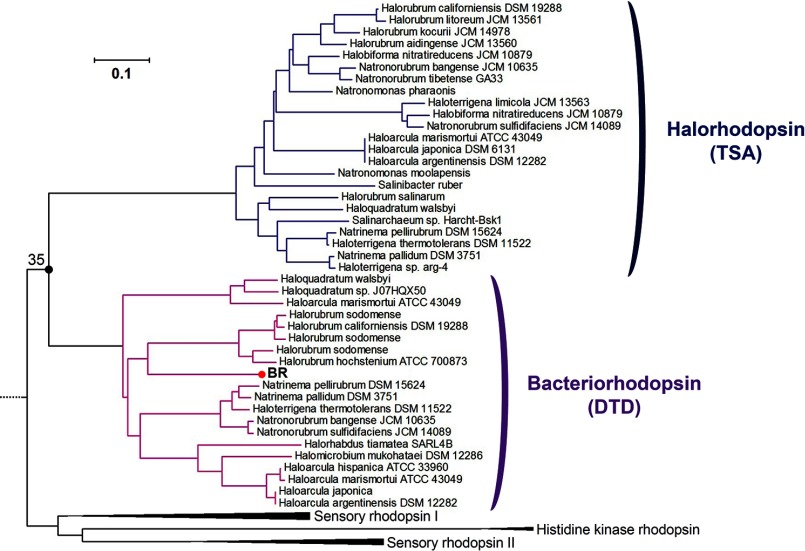
It should be noted that the asymmetry of functional interconversion between eubacterial H+ and Cl− pumps (GR and FR) is opposite to that between archaeal H+ and Cl− pumps (BR and HR), where the H+ → Cl− pump conversion was achieved by a single amino acid replacement (13, 14) but the reverse conversion was unsuccessful (15, 16). If success of functional conversion of ion-pumping rhodopsins depends on the direction of evolution, how can the results on the archaeal H+ and Cl− pumps be rationalized? Fig. 9b suggests that phylogenetic tree for BR and HR is less hierarchical than that for eubacterial pumps, with much older branching, as described in Fig 10. Thus, different molecular mechanism such as hydrogen-bonding strength of protein-bound water molecules (17) underlies asymmetric functional conversion in archaeal H+ and Cl− pumps. In this sense, each molecular mechanism of successful and unsuccessful functional conversions in the present study should be revealed by structural details in future.
FIGURE 10.
Phylogenic subtree of bacteriorhodopsins and halorhodopsins. Subtree of the phylogenic tree of microbial rhodopsins shown in Fig. 9b, including BR, HR, sensory rhodopsins, and histidine kinase rhodopsins. Subtrees of BR having DTD-motif and HR having TSA-motif are highlighted pink and indigo, respectively. The bootstrap tests are calculated for 1000 replicates. The scale bar indicates 0.1 units of the number of amino acid substitutions per site.
Author Contributions
K. I. and H. K. contributed to the study design. K. I. contributed to construction of mutant proteins and introduction to E. coli. K. I. and Y. N. conducted pumping activity measurement. Y. N. quantified the amount of protein expression in E. coli, purified proteins and measured their absorption spectra at different NaCl concentrations. K. I. and Y. N. carried out flash photolysis experiment of purified proteins. K. I. and H. K. wrote the paper. All authors discussed and commented on the manuscript.
Supplementary Material
This work was supported by grants from the Japanese Ministry of Education, Culture, Sports, Science, and Technology (to K. I.) (26708001, 26115706, 26620005) and to H.K. (25104009, 15H02391). The authors declare that they have no conflicts of interest with the contents of this article.

This article contains supplemental Table S1.
- BR
- bacteriorhodopsin
- HR
- halorhodopsin
- KR
- Krokinobacter eikastus rhodopsin
- DDM
- n-dodecyl-β-d-maltoside
- GR
- Gloeobacter rhodopsin
- FR
- Fulvimarina rhodopsin
- CCCP
- carbonylcyanide m-chlorophenylhydrazone.
References
- 1. Ernst O. P., Lodowski D. T., Elstner M., Hegemann P., Brown L. S., and Kandori H. (2014) Microbial and animal rhodopsins: structures, functions, and molecular mechanisms. Chem. Rev. 114, 126–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Deisseroth K. (2011) Optogenetics. Nat. Methods 8, 26–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hegemann P., and Möglich A. (2011) Channelrhodopsin engineering and exploration of new optogenetic tools. Nat. Methods 8, 39–42 [DOI] [PubMed] [Google Scholar]
- 4. Oesterhelt D., and Stoeckenius W. (1971) Rhodopsin-like protein from the purple membrane of Halobacterium halobium. Nat. New Biol. 233, 149–152 [DOI] [PubMed] [Google Scholar]
- 5. Matsuno-Yagi A., and Mukohata Y. (1977) Two possible roles of bacteriorhodopsin; a comparative study of strains of Halobacterium halobium differing in pigmentation. Biochem. Biophys. Res. Commun. 78, 237–243 [DOI] [PubMed] [Google Scholar]
- 6. Béjà O., Aravind L., Koonin E. V., Suzuki M. T., Hadd A., Nguyen L. P., Jovanovich S. B., Gates C. M., Feldman R. A., Spudich J. L., Spudich E. N., and DeLong E. F. (2000) Bacterial rhodopsin: evidence for a new type of phototrophy in the sea. Science 289, 1902–1906 [DOI] [PubMed] [Google Scholar]
- 7. Sharma A. K., Spudich J. L., and Doolittle W. F. (2006) Microbial rhodopsins: functional versatility and genetic mobility. Trends Microbiol. 14, 463–469 [DOI] [PubMed] [Google Scholar]
- 8. Inoue K., Ono H., Abe-Yoshizumi R., Yoshizawa S., Ito H., Kogure K., and Kandori H. (2013) A light-driven sodium ion pump in marine bacteria. Nat. Commun. 4, 1678. [DOI] [PubMed] [Google Scholar]
- 9. Yoshizawa S., Kumagai Y., Kim H., Ogura Y., Hayashi T., Iwasaki W., DeLong E. F., and Kogure K. (2014) Functional characterization of flavobacteria rhodopsins reveals a unique class of light-driven chloride pump in bacteria. Proc. Natl. Acad. Sci. U.S.A. 111, 6732–6737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Inoue K., Koua F. H., Kato Y., Abe-Yoshizumi R., and Kandori H. (2014) Spectroscopic study of a light-driven chloride ion pump from marine bacteria. J. Phys. Chem. B. 118, 11190–11199 [DOI] [PubMed] [Google Scholar]
- 11. Kato H. E., Inoue K., Abe-Yoshizumi R., Kato Y., Ono H., Konno M., Hososhima S., Ishizuka T., Hoque M. R., Kunitomo H., Ito J., Yoshizawa S., Yamashita K., Takemoto M., Nishizawa T., et al. (2015) Structural basis for Na+ transport mechanism by a light-driven Na+ pump. Nature 521, 48–53 [DOI] [PubMed] [Google Scholar]
- 12. Gushchin I., Shevchenko V., Polovinkin V., Kovalev K., Alekseev A., Round E., Borshchevskiy V., Balandin T., Popov A., Gensch T., Fahlke C., Bamann C., Willbold D., Büldt G., Bamberg E., and Gordeliy V. (2015) Crystal structure of a light-driven sodium pump. Nat. Struct. Mol. Biol. 22, 390–395 [DOI] [PubMed] [Google Scholar]
- 13. Sasaki J., Brown L. S., Chon Y. S., Kandori H., Maeda A., Needleman R., and Lanyi J. K. (1995) Conversion of bacteriorhodopsin into a chloride ion pump. Science 269, 73–75 [DOI] [PubMed] [Google Scholar]
- 14. Tittor J., Haupts U., Haupts C., Oesterhelt D., Becker A., and Bamberg E. (1997) Chloride and proton transport in bacteriorhodopsin mutant D85T: different modes of ion translocation in a retinal protein. J. Mol. Biol. 271, 405–416 [DOI] [PubMed] [Google Scholar]
- 15. Havelka W. A., Henderson R., and Oesterhelt D. (1995) Three-dimensional structure of halorhodopsin at 7 Å resolution. J. Mol. Biol. 247, 726–738 [DOI] [PubMed] [Google Scholar]
- 16. Váró G., Brown L. S., Needleman R., and Lanyi J. K. (1996) Proton transport by halorhodopsin. Biochemistry 35, 6604–6611 [DOI] [PubMed] [Google Scholar]
- 17. Muroda K., Nakashima K., Shibata M., Demura M., and Kandori H. (2012) Protein-bound water as the determinant of asymmetric functional conversion between light-driven proton and chloride pumps. Biochemistry 51, 4677–4684 [DOI] [PubMed] [Google Scholar]
- 18. Miranda M. R., Choi A. R., Shi L., Bezerra A. G. Jr., Jung K. H., and Brown L. S. (2009) The photocycle and proton translocation pathway in a cyanobacterial ion-pumping rhodopsin. Biophys. J. 96, 1471–1481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Scharf B., Hess B., and Engelhard M. (1992) Chromophore of sensory rhodopsin II from Halobacterium halobium. Biochemistry 31, 12486–12492 [DOI] [PubMed] [Google Scholar]
- 20. Edgar R. C. (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Saitou N., and Nei M. (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425 [DOI] [PubMed] [Google Scholar]
- 22. Felsenstein J. (1985) Confidence limits on phylogenies: An approach using the bootstrap. Evolution 39, 783–791 [DOI] [PubMed] [Google Scholar]
- 23. Zuckerkandl E., and Pauling L. (1965) Evolutionary divergence and convergence in proteins, in Evolving Genes and Proteins (Bryson V., and Vogel H. J., eds), pp, 97–166, Academic Press, New York [Google Scholar]
- 24. Tamura K., Peterson D., Peterson N., Stecher G., Nei M., and Kumar S. (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Inoue K., Kato Y., and Kandori H. (2015) Light-driven ion-translocating rhodopsins in marine bacteria. Trends. Microbiol. 23, 91–98 [DOI] [PubMed] [Google Scholar]
- 26. Balashov S. P., Imasheva E. S., Dioumaev A. K., Wang J. M., Jung K.-H., and Lanyi J. K. (2014) Light-driven Na+ pump from Gillisia limnaea: a high-affinity Na+ binding site is formed transiently in the photocycle. Biochemistry 53, 7549–7561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. da Silva G. F., Goblirsch B. R., Tsai A. L., and Spudich J. L. (2015) Cation-specific conformations in a dual-function ion-pumping microbial rhodopsin. Biochemistry 54, 3950–3959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Váró G., Brown L. S., Lakatos M., and Lanyi J. K. (2003) Characterization of the photochemical reaction cycle of proteorhodopsin. Biophys. J. 84, 1202–1207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Váró G., Zimányi L., Fan X., Sun L., Needleman R., and Lanyi J. K. (1995) Photocycle of halorhodopsin from Halobacterium salinarium. Biophys. J. 68, 2062–2072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sato M., Kubo M., Aizawa T., Kamo N., Kikukawa T., Nitta K., and Demura M. (2005) Role of putative anion-binding sites in cytoplasmic and extracellular channels of Natronomonas pharaonis halorhodopsin. Biochemistry 44, 4775–4784 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.