Abstract
Astrocytomas are the most common glial tumor of the central nervous system. Within this category, glioblastoma is the most prevalent and malignant primary brain tumor. Glioblastoma can arise de novo, or through progression from lower-grade lesions, but is uniformly associated with poor outcomes despite surgical resection, chemotherapy, and radiation therapy. Recent genomic discoveries have provided new insight into gliomagenesis and have identified key genetic alterations that have diagnostic, prognostic and predictive capacity. Numerous molecular classification schemes have been proposed to sort tumors into clinically meaningful categories to guide treatment. However, creating therapy targeted towards these alterations has been made challenging by the redundancy of essential signal transduction pathways affected in these tumors, intratumoral heterogeneity, and the hypermutated profiles of recurrent tumors. Future treatment strategies will require a personalized approach with consideration of the unique genetic profile of a specific tumor and the use of multimodality therapies.
Introduction
In 2007, the WHO proposed a classification system for tumors of the central nervous system (CNS), categorizing a heterogeneous group of cancers into discrete types based on histopathologic criteria. Grade I lesions included pilocytic astrocytoma, grade II consisted of diffuse astrocytoma; grade III of anaplastic astrocytoma, and grade IV of glioblastoma (GBM) [1]. GBMs are the most common primary malignant brain tumors; within GBMs, the majority arise de novo and are termed primary GBMs, and those that progress from lower-grade lesions and undergo transformation are secondary GBMs. Although this classification system has been used to guide prognostication and management, outcomes for GBM remain dismal, with median survival at 15–20 months [2]. Recently, however, our understanding of CNS tumors has been revolutionized by genomic studies. The identification of driver mutations in gliomas has already allowed for improved patient prognostication, and there is hope these new insights into gliomagenesis will generate novel and specific strategies for treatment [3].
Molecular alterations in astrocytomas
IDH1/2
Mutations of the isocitrate dehydrogenase (IDH1 and IDH2) gene are thought to occur early in gliomagenesis and drive cancer progression [4]. Although detected in 70–80% of grade II and III gliomas and secondary GBMs, IDH1 alterations rarely occur in primary GBM [5••]. The predominant causative single nucleotide polymorphism (SNP) results in an arginine to histidine substitution at codon 132 (R132H); IDH1 mutations are strongly associated with TP53 mutations in low-grade astrocytomas [4,6,7••]. The wild-type form catalyzes the reaction of isocitrate to α-ketoglutarate, but the mutated form results in the overproduction of 2-hydroxyglutarate (2HG) [8]. 2HG has been shown to induce DNA damage, prevent differentiation in hematopoietic cells, promote carcinogenesis. In addition, it also increases levels of and stabilizes hypoxia-inducible factor subunit HIF-1α, which is thought to promote tumor growth [9••,10,11,12]. In addition to effects on cellular metabolism, IDH1 mutations are associated with a global DNA hypermethylation phenotype, and result in blockade of histone demethylation and prevention of cell differentiation [13•,14,15,16••]. Even when accounting for tumor grade, patients with tumors that possess IDH1 mutations experience longer survival and have improved overall prognoses [17].
ATRX
Loss of expression mutations of the alpha-thalassemia/mental retardation syndrome X-linked (ATRX) gene have been described in the majority of grade II and III astrocytomas and secondary GBMs, but are rare in primary GBM [18•,19]. In addition to being a member of the SWI/SNF family of chromatin remodeling proteins, ATRX mutations are associated with an alternative lengthening of telomeres (ALT) phenotype [18•,20]. Indeed, the molecular basis of ALT appears to obligate inactivation of either ATRX or its binding partner DAXX. Jiao et al. also found that 99% of the tumors with ATRX mutations had co-occurring mutations of IDH1, and 94% with both ATRX and IDH1 alterations had TP53 mutations; alterations in ATRX were almost mutually exclusive with 1p/19q co-deletions, which are characteristic of oligodendrogliomas. Subsequent studies have reaffirmed these findings, and found that patients with tumors harboring ATRX mutations experienced longer times to treatment failure compared to those without such alterations [20].
EGFR
Amplification of the epidermal growth factor receptor (EGFR) gene has been reported in 40% of GBMs. Of these, 20–30% express a variant produced from the deletion of exons 2–7, EGFRvIII, a constitutively active receptor that is unable to bind ligand and results in continuous activation of cell growth and anti-apoptotic pathways [21]. Activation of EGFR in gliomas also occurs through gain-of-function mutations and double minute chromosomes [22]. There have been several reports noting glioma dependence on EGFR activation, citing that interruption of EGFR signaling results in short-term inhibition of glioma growth [23,24]. Recently, Frattini et al. described translocations of EGFR and in-frame fusion to either septin 14 (SEPT14) or phosphoserine phosphatase (PSPH) in 7% of GBMs [25•]. In 3% of GBMs, fibroblast growth factor receptor 1 (FGFR1) inversion and in-frame fusion to the coding domain of transforming acidic coiled-coil 1 (TACC1) results in a constitutively active protein (FGFR-TACC) [26••]. Although these fusion events appear to be rare, the resulting fusion proteins are promising therapeutic targets.
TERT
Mutations of the telomerase reverse transcriptase (TERT) promoter which result in increased telomerase expression have been observed in several human cancers; they have been found in the majority of primary GBM but are less common in lower-grade gliomas and secondary GBMs [27,28]. TERT promoter mutations appear to be mutually exclusive with ATRX mutations and activation of the ALT pathway, highlighting two distinct mechanisms for telomere maintenance in cancer cells. Recently, a new SNP near the telomerase RNA component (TERC) gene was found to be potentially associated with increased risk of glioma. This SNP and a previously identified risk loci near TERT demonstrated a correlation with longer telomeres [29,30]. The association between TERT promoter status and patient survival was not significant when IDH1 mutation status was accounted for [27].
Molecular classification systems
Although the WHO classification system has been universally used to guide diagnosis, treatment, and prognostication, the variability in the histologic appearance of gliomas has made uniform tumor grading challenging. By contrast, the above described genomic alterations appear to segregate consistently, providing insight into gliomagenesis and suggesting approaches for the molecular categorization of astrocytomas. For example, despite the fact that primary and secondary GBMs appear identical histologically, they are distinct in terms of their genetic signatures. Primary GBM is associated with EGFR amplifications and phosphatase and tensin homolog (PTEN) deletions, which are uncommon in lower-grade astrocytomas. These tumors also are associated with amplifications of platelet derived growth factor receptor a (PDGFRA), MET, CDK4, MDM2, and MDM4; mutations of phosphatidylinositol-3-OH kinase (PI3K); mutations and deletions of TP53, CDNK2A/ARF, CDKN2A/p16, RB1, and NF1 [5••,31]. By contrast, secondary GBMs are characterized by mutations in IDH1 and TP53. In one study, of the 80% anaplastic astrocytomas and secondary GBMs with IDH1 and IDH2 mutations, only 3% had mutations in EGFR, PTEN, CDKN2A and CDKN2B; in the 18% of tumors with wild-type IDH1 and IDH2, 74% exhibited alterations in EGFR, PTEN, CDKN2A and CDKN2B [7••,31]. As lower-grade astrocytomas also display mutations of IDH1 and TP53, these lesions likely progress to secondary GBM by accumulating new genetic alterations.
Genetic signature
Comprehensive genetic profiling studies have sought to use the divergent molecular profiles of gliomas to create new schema for tumor classification to be used in diagnosis, prognostication, and prediction [32]. In 2010, a study from The Cancer Genome Atlas described four distinct subgroups of GBM based on their molecular profile: (1) proneural, characterized by alterations in PDGFRA and IDH1; (2) classical, with mutations in EGFR, (3) mesenchymal, with mutations in NF1, and (4) neural, which did not exhibit a distinct genetic profile. The mesenchymal and classical subgroups displayed improved survival with aggressive therapy (temozolomide (TMZ) and radiation), but no benefit was seen in those with proneural tumors [33••]. Of gliomas classified as proneural, examination of DNA methylation patterns has led to further subcategorization, with some tumors displaying a glioma-CpG island methylator (G-CIMP) phenotype [13•,34]. G-CIMP are highly associated with IDH1 mutations, and resultingly, were associated with a more favorable prognosis. Subsequent studies found that IDH1 mutation alone was sufficient to induce the G-CIMP phenotype [35••].
Jiao et al. has since proposed another classification model in which tumors with IDH1 and ATRX mutations are ‘I-CF glioma;’ IDH1, CIC, and FUBP1 with 1p/19q loss are ‘I-A glioma;’ and gliomas without the previously noted mutations with multiple other genetic alterations are ‘I-X’ gliomas. This categorization not only aids in diagnosis and eliminating the histologically challenging diagnosis of oligoastrocytoma, but also has prognostic value, as those with I-CF gliomas experienced a median survival of 96 months, compared to 51 months in I-A gliomas and 13 months in I-X gliomas [18•]. A similar classification system using IDH1 and IDH2 and TERT has been proposed by Killela et al. They demonstrated that patients with gliomas with IDH1/2 mutations experienced a median survival of 57 months and those with TERT mutations and IDH1/2 mutations had a median survival of 125 months, in contrast to those with TERT mutations only, with a median survival of 11.5 months [28].
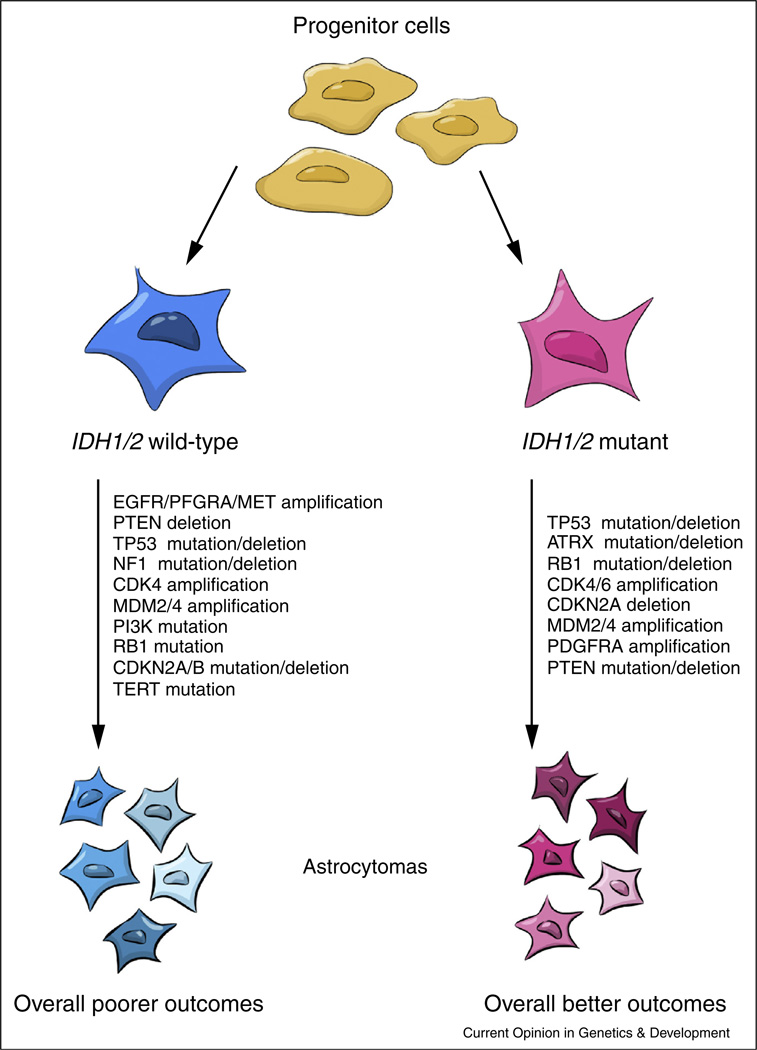
Although these proposed classification systems are valuable for diagnosis and prognostication, no one schema has emerged as more compelling than the others. Upon further examination, one predominant pattern appears: IDH1/2 mutations appear to be the fundamental genetic change by which other alterations segregate and is consistently predictive of a more favorable patient outcome. The use of other markers may allow for more nuanced prognostication, but they appear secondary to the effect driven by IDH1/2 (Figure 1).
Figure 1. IDH1/2 mutations occurs early in gliomagenesis and is the fundamental prognostic marker.
The molecular classification systems that have been proposed are based on a number of different markers, the most fundamental of which may involve IDH1/2. IDH1/2-mutant and IDH1/2-wild type tumors follow two distinct genetic pathways with divergent clinical outcomes.
Signal transduction pathways
The pattern of genetic alterations in astrocytomas also allows for tumor classification by the affected signal transduction pathway. TCGA described three core pathways altered in the majority of GBM: RTK/RAS/PI3K, p53, and Rb [36••]. Brennan et al., through targeted proteomic analyses of 27 glioma samples, found three distinct subclasses with mutually exclusive alterations: (1) EGFR activation, (2) PDGFR activation, and (3) loss of NF1 expression [37]. A subsequent study has proposed classifying gliomas by EGFR and PDGFRA expression and patterns of gene-coexpression upon demonstrating that gliomas that expressed EGFR and associated genes were associated with a significantly poorer prognosis, compared to tumors expressing PDGRFA and those with low expression of both [38]. Although the majority of gliomas have alterations in these pathways, recent studies have demonstrated the existence of alternate pathways that ultimately also result in tumor cell proliferation and escape from apoptosis. Heterozygous deletions of the NF-κB inhibitor-α (NFKBIA) gene were found in 25% of GBMs, which were mutually exclusive with EGFR amplifications. These two alterations appear to converge on the same downstream pathway, as patients with tumors that demonstrate either alteration experience shorter survival times compared to those who have neither [39]. In addition, Morris et al. recently identified inactivating mutations of FAT1, which binds β-catenin and antagonizes the Wnt signaling pathway, in 20% of GBMs [40]. The identification of these driver mutations and dysregulated core pathways has suggested potential therapeutic targets.
Strategies for targeted therapy
Although the standard of care for high-grade astrocytomas consists of surgical resection, chemotherapy (typically with TMZ), and radiotherapy, the treatment course for lower-grade lesions can vary. Most patients receive surgical resection, but use of additional therapies depends on both tumor and patient factors. Despite multimodality therapy, however, outcomes for GBM remain poor [3]. Given the prevalence of IDH1 alterations and its role as a driver mutation, there has been considerable interest in using inhibitors of mutant IDH1 as therapy. Rohle et al. identified a selective inhibitor of the R132H-IDH1 mutant, which was capable of inducing differentiation and impair tumor growth in vitro and in vivo with IDH1-mutant gliomas [41•]. They did not observe any effect of the inhibitor on DNA methylation; however, treatment of IDH1-mutant glioma cells with the DNA methyltransferase inhibitor decitabine resulted in reversal in mutant IDH1-induced methylation and caused similar effects on cell proliferation and differentiation [42]. There are ongoing phase I studies examining the effects of IDH inhibitors in IDH mutant gliomas (NCT02193347).
With the prevalence of alterations in the RTK/PI3K/Akt pathway in astrocytomas, specific therapy targeting EGFR signaling once held great promise. Erlotinib and gefitinib, first-generation EGFR inhibitors, as well as newer agents such as cetuximab and lapatinib, have not demonstrated a significant treatment benefit in trials [43–47]. Glioma resistance has been primarily attributed to redundancy in signaling pathway dysregulation in GBM, and the convergence of the effect of numerous alterations on the same downstream pathways [48]. Consistent with this idea, loss of PTEN predicts treatment failure with EGFR inhibitors, while patients with tumors without PTEN and TP53 alterations experience improved survival [49]. The importance of PTEN status in determining response to EGFR inhibitors has been reaffirmed in subsequent studies [23,50]. Effectors downstream of RTKs have also been targeted; the PI3K inhibitor PX-866 has been shown to attenuate glioma cell growth in vitro and in vivo, and is currently in clinical trials [2,51]. Mammalian mTOR inhibitors, including sirolimus and everolimus, have also been studied in clinical trials for GBM, but have also not demonstrated improvements in outcomes when alone or in combination with EGFR inhibitors [52].
Another treatment strategy that holds the promise is immunotherapy, which has seen success in the treatment of melanoma, renal cell carcinoma, and prostate carcinoma. Though the CNS was once thought to be a space of immune privilege, it is now clear that gliomas are not immunologically silent [53]. However, the interaction between the immune system and gliomas is a complex one, in which gliomas are able to create a state of immunosuppression to evade destruction by effector cells. The development of tumor-specific antigen vaccines is an area of active investigation in glioma, and new studies point to the potential of immune checkpoint therapies for use in tumors with high mutational burdens (Box 1).
Box 1. Immunotherapy: a promising treatment strategy.
Targeted therapy for glioblastoma remains an elusive goal, but there is hope that immunotherapeutic strategies may finally achieve treatment specificity. One category of immunotherapeutic agents, including peptide and dendritic cell (DC) vaccines, have been devised to target tumor-specific and tumor-associated antigens [65]. The most prominent example in gliomas is EGFRvIII; the earliest studies of rindopepimut (PEPvIII-KLH; CDX-110), a 14-amino acid peptide (PEPvIII) conjugated to keyhole limpet hemocyanin (KLH), demonstrated significantly improved patient outcomes in combination with radiation therapy and chemotherapy. A Phase II trial of this agent alone and a Phase III trial in combination with GM-CSF are currently underway (NCT00458601; NCT01480479). DC vaccines typically involve the administration of autologous DCs from patients previously pulsed with tumor lysates or tumor-specific antigens. ICT-107 and DCVax®-L are polyvalent DC vaccines currently in clinical trials (NCT01280552; NCT00045968). Another strategy that augments the host immune system response against tumor tissue is that of immune checkpoint blockade. Cytotoxic T lymphocyte antigen-4 prevents the co-stimulatory interaction between T-cells and antigen presenting cells, effectively inhibiting the immune response against certain antigens. This mechanism functions to prevent the development of autoimmunity, but is exploited by glioma cells, which overexpress CTLA-4. Ipilimumab, a monoclonal CTLA-4 antibody, was approved for the treatment of melanoma in 2010. The programmed cell death-1 (PD-1) pathway is another immune checkpoint which appears to negatively regulate T-cell responses. Nivolumab, a monoclonal antibody against PD-1, is currently in Phase III clinical trials for a variety of solid tumors. A Phase II trial comparing combination nivolumab and ipilimumab with bevacizumab in patients with recurrent glioblastoma is currently underway (NCT02017717). A recent report posited that the efficacy of immune checkpoint therapies depends in part on tumor mutational burden. Champiat and colleagues found that the mutational frequencies of solid tumors correlated with response to anti-PD-1 therapies; of the tumor types for which these therapies have been tested, melanoma and lung squamous cell carcinoma have the greatest frequencies of somatic mutations reported in the literature and also display the most robust responses to immune checkpoint therapies [66•]. The theory that the greater the mutational burden, and therefore, the more tumor neoantigens produced and more immunogenic the tumor, the better the response immune checkpoint therapies is particularly interesting to consider with glioblastoma, which is known for its intratumoral heterogeneity.
Treatment challenges
The difficulties in designing targeted therapy can be attributed to numerous factors. Originally named glioblastoma multiforme for its variable histologic appearance, GBM has become the exemplar of intratumoral heterogeneity in cancer. Not only are there cell subpopulations that display distinct phenotypes with tumors, with respect to self-renewal capabilities and response to treatment, but single-cell studies have demonstrated there can be tremendous variability at the genetic, transcriptional, and functional levels [54,55••]. Patel et al. and others have described mosaic expression of RTK and mutually exclusive expression of EGFR variants between cells [48,56]. Using the classification system proposed by Verhaak et al., they found that although tumors may overall demonstrate a predominant proneural, classical, or mesenchymal profile, all tumors had cells belonging to each of the proposed subtypes. Within the predominantly proneural tumors, more subtype heterogeneity was associated with decreased survival [33••,55••].
Several models have been proposed to explain both the initiation and the maintenance of intratumoral heterogeneity. Although their existence has not been definitively proven, cancer stem cells (CSCs) are one mechanism through which tumor heterogeneity can be continuously maintained. In this hierarchical model, a subpopulation of CSCs are capable of self-renewal and differentiation into cells with varying phenotypes [57–61]. Recent studies have suggested that there may not be a discrete subpopulation of CSCs, but the expression of stem cell-related genes may exist on a continuum in tumor cells [55••]. Earlier studies have also highlighted the necessary role of the tumor microenvironment and interactions between tumor cells in maintaining heterogeneity; cells with EGFR mutations are able to potentiate EGFR expression among EGFR-wild type GBM cells via paracrine cytokine signaling [62].
The mechanisms that generate striking intratumoral heterogeneity undoubtedly play a crucial role in the emergence of resistance to chemotherapy in recurrent tumors. A subpopulation of CSC-like cells were implicated in promoting tumor recurrence after treatment with TMZ in a mouse model of glioma [58]. This CSC model is not mutually exclusive with the hypothesis of clonal evolution, where cells accumulate varying mutations, and under the selective pressure of treatment, allow for the survival of certain cell populations. Novel somatic mutations in the mismatch repair gene MSH6 in recurrent GBM have also been identified, suggesting a mechanism underlying resistance to alkylating agents [63]. Additionally, exome sequencing of grade II gliomas at diagnosis and recurrence revealed that the majority of recurrences did not display driver mutations in genes including TP53 and ATRX, which were present in the initial tumor. Recurrences that arose after treatment with TMZ also revealed new mutations in the RB and PI3KAkt/mTOR pathways [64]. Both studies have described hypermutated genetic profiles after recurrences with TMZ treatment, demonstrating the ability of treatment to drive tumor evolution.
Conclusions and future directions
Our understanding of the genetic alterations driving astrocytoma initiation and progression has advanced dramatically with the advent of next-generation sequencing technologies. These alterations can have profound effects on cells, activating signal transduction pathways that result in uncontrolled proliferation, altering cell metabolism to promote cell growth, effecting global DNA methylation changes and chromatin remodeling, and activating mechanisms to maintain telomere length. Several molecular classification models have been proposed by examining the divergent molecular profiles of astrocytomas of different classes, all with prognostic significance. Insight into this complex genetic landscape is not only valuable from a diagnostic and prognostic perspective, as these genetic markers have been applied to provide molecular data to complement histopathologic data, but they have also revealed potential targets for rational therapeutic design. However, targeted therapies have so far had disappointing results in clinical trials; their limited success may be the result of several mechanisms and suggests areas for future investigation. First, the core dysregulated pathways in glioma appear to be cooperative and to converge on the same downstream effectors and there also exist alternative pathways that yield the same ultimate effect. Future therapeutic strategies cannot rely on the inhibition of one effector, but multiple targets at multiple signaling levels must be blocked to obtain a meaningful change in clinical outcomes. Furthermore, these therapies must be used on highly selected patient populations, which necessitates an understanding of the global genetic landscape of various tumor types, as well as patient-specific alterations that will enable matching the appropriate therapy to each individual. Finally, a deeper understanding of the marked intratumoral heterogeneity displayed by glioblastoma and judicious use of therapies is needed to prevent the evolution of recurrent tumors that are treatment-resistant.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, Jouvet A, Scheithauer BW, Kleihues P. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114:97–109. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grant R, Kolb L, Moliterno J. Molecular and genetic pathways in gliomas: the future of personalized therapeutics. CNS Oncol. 2014;3:123–136. doi: 10.2217/cns.14.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tanaka S, Louis DN, Curry WT, Batchelor TT, Dietrich J. Diagnostic and therapeutic avenues for glioblastoma: no longer a dead end? Nat Rev Clin Oncol. 2013;10:14–26. doi: 10.1038/nrclinonc.2012.204. [DOI] [PubMed] [Google Scholar]
- 4.Watanabe T, Nobusawa S, Kleihues P, Ohgaki H. IDH1 mutations are early events in the development of astrocytomas and oligodendrogliomas. Am J Pathol. 2009;174:1149–1153. doi: 10.2353/ajpath.2009.080958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Siu IM, Gallia GL, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. Landmark paper describing IDH1 mutations in secondary glioblastomas and their association with improved patient prognose.
- 6.Balss J, Meyer J, Mueller W, Korshunov A, Hartmann C, von Deimling A. Analysis of the IDH1 codon 132 mutation in brain tumors. Acta Neuropathol. 2008;116:597–602. doi: 10.1007/s00401-008-0455-2. [DOI] [PubMed] [Google Scholar]
- 7. Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, Kos I, Batinic-Haberle I, Jones S, Riggins GJ, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360:765–773. doi: 10.1056/NEJMoa0808710. Described mutations of IDH1/2 including the predominant R132H mutation in secondary glioblastom.
- 8.Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, Fantin VR, Jang HG, Jin S, Keenan MC, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462:739–744. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhao S, Lin Y, Xu W, Jiang W, Zha Z, Wang P, Yu W, Li Z, Gong L, Peng Y, et al. Glioma-derived mutations in IDH1 dominantly inhibit IDH1 catalytic activity and induce HIF-1alpha. Science. 2009;324:261–265. doi: 10.1126/science.1170944. Described the function of IDH1 as a tumor suppressor whose inactivation promotes tumorigenesis through activation of HIF-α.
- 10.Rakheja D, Medeiros LJ, Bevan S, Chen W. The emerging role of d-2-hydroxyglutarate as an oncometabolite in hematolymphoid and central nervous system neoplasms. Front Oncol. 2013;3:169. doi: 10.3389/fonc.2013.00169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Losman JA, Looper RE, Koivunen P, Lee S, Schneider RK, McMahon C, Cowley GS, Root DE, Ebert BL, Kaelin WG., Jr (R)-2-hydroxyglutarate is sufficient to promote leukemogenesis and its effects are reversible. Science. 2013;339:1621–1625. doi: 10.1126/science.1231677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koivunen P, Lee S, Duncan CG, Lopez G, Lu G, Ramkissoon S, Losman JA, Joensuu P, Bergmann U, Gross S, et al. Transformation by the (R)-enantiomer of 2-hydroxyglutarate linked to EGLN activation. Nature. 2012;483:484–488. doi: 10.1038/nature10898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Noushmehr H, Weisenberger DJ, Diefes K, Phillips HS, Pujara K, Berman BP, Pan F, Pelloski CE, Sulman EP, Bhat KP, et al. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17:510–522. doi: 10.1016/j.ccr.2010.03.017. Described the G-CIMP phenotype and its association with IDH1 mutations and improved outcome.
- 14.Lu C, Ward PS, Kapoor GS, Rohle D, Turcan S, Abdel-Wahab O, Edwards CR, Khanin R, Figueroa ME, Melnick A, et al. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature. 2012;483:474–478. doi: 10.1038/nature10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Figueroa ME, Abdel-Wahab O, Lu C, Ward PS, Patel J, Shih A, Li Y, Bhagwat N, Vasanthakumar A, Fernandez HF, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 2010;18:553–567. doi: 10.1016/j.ccr.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Xu W, Yang H, Liu Y, Yang Y, Wang P, Kim SH, Ito S, Yang C, Xiao MT, Liu LX, et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of alpha-ketoglutarate-dependent dioxygenases. Cancer Cell. 2011;19:17–30. doi: 10.1016/j.ccr.2010.12.014. Described the accumulation of 2-HG in tumors with IDH1/2 mutations and resultant hypermethylatio.
- 17.Hartmann C, Hentschel B, Wick W, Capper D, Felsberg J, Simon M, Westphal M, Schackert G, Meyermann R, Pietsch T, et al. Patients with IDH1 wild type anaplastic astrocytomas exhibit worse prognosis than IDH1-mutated glioblastomas, and IDH1 mutation status accounts for the unfavorable prognostic effect of higher age: implications for classification of gliomas. Acta Neuropathol. 2010;120:707–718. doi: 10.1007/s00401-010-0781-z. [DOI] [PubMed] [Google Scholar]
- 18. Jiao Y, Killela PJ, Reitman ZJ, Rasheed AB, Heaphy CM, de Wilde RF, Rodriguez FJ, Rosemberg S, Oba-Shinjo SM, Nagahashi Marie SK, et al. Frequent ATRX, CIC, FUBP1 and IDH1 mutations refine the classification of malignant gliomas. Oncotarget. 2012;3:709–722. doi: 10.18632/oncotarget.588. Described ATRX mutations in grade II and III astrocytomas and their association with IDH1 mutation.
- 19.Liu XY, Gerges N, Korshunov A, Sabha N, Khuong-Quang DA, Fontebasso AM, Fleming A, Hadjadj D, Schwartzentruber J, Majewski J, et al. Frequent ATRX mutations and loss of expression in adult diffuse astrocytic tumors carrying IDH1/IDH2 and TP53 mutations. Acta Neuropathol. 2012;124:615–625. doi: 10.1007/s00401-012-1031-3. [DOI] [PubMed] [Google Scholar]
- 20.Wiestler B, Capper D, Holland-Letz T, Korshunov A, von Deimling A, Pfister SM, Platten M, Weller M, Wick W. ATRX loss refines the classification of anaplastic gliomas and identifies a subgroup of IDH mutant astrocytic tumors with better prognosis. Acta Neuropathol. 2013;126:443–451. doi: 10.1007/s00401-013-1156-z. [DOI] [PubMed] [Google Scholar]
- 21.Gan HK, Kaye AH, Luwor RB. The EGFRvIII variant in glioblastoma multiforme. J Clin Neurosci. 2009;16:748–754. doi: 10.1016/j.jocn.2008.12.005. [DOI] [PubMed] [Google Scholar]
- 22.Vogt N, Lefevre SH, Apiou F, Dutrillaux AM, Cor A, Leuraud P, Poupon MF, Dutrillaux B, Debatisse M, Malfoy B. Molecular structure of double-minute chromosomes bearing amplified copies of the epidermal growth factor receptor gene in gliomas. Proc Natl Acad Sci U S A. 2004;101:11368–11373. doi: 10.1073/pnas.0402979101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vivanco I, Rohle D, Versele M, Iwanami A, Kuga D, Oldrini B, Tanaka K, Dang J, Kubek S, Palaskas N, et al. The phosphatase and tensin homolog regulates epidermal growth factor receptor (EGFR) inhibitor response by targeting EGFR for degradation. Proc Natl Acad Sci U S A. 2010;107:6459–6464. doi: 10.1073/pnas.0911188107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cloughesy TF, Cavenee WK, Mischel PS. Glioblastoma: from molecular pathology to targeted treatment. Annu Rev Pathol. 2014;9:1–25. doi: 10.1146/annurev-pathol-011110-130324. [DOI] [PubMed] [Google Scholar]
- 25. Frattini V, Trifonov V, Chan JM, Castano A, Lia M, Abate F, Keir ST, Ji AX, Zoppoli P, Niola F, et al. The integrated landscape of driver genomic alterations in glioblastoma. Nat Genet. 2013;45:1141–1149. doi: 10.1038/ng.2734. Described EGFR-SEPT14 fusion in glioblastom.
- 26. Singh D, Chan JM, Zoppoli P, Niola F, Sullivan R, Castano A, Liu EM, Reichel J, Porrati P, Pellegatta S, et al. Transforming fusions of FGFR and TACC genes in human glioblastoma. Science. 2012;337:1231–1235. doi: 10.1126/science.1220834. Described FGFR-TACC fusion in glioblastom.
- 27.Nonoguchi N, Ohta T, Oh JE, Kim YH, Kleihues P, Ohgaki H. TERT promoter mutations in primary and secondary glioblastomas. Acta Neuropathol. 2013;126:931–937. doi: 10.1007/s00401-013-1163-0. [DOI] [PubMed] [Google Scholar]
- 28.Killela PJ, Reitman ZJ, Jiao Y, Bettegowda C, Agrawal N, Diaz LA, Jr, Friedman AH, Friedman H, Gallia GL, Giovanella BC, et al. TERT promoter mutations occur frequently in gliomas and a subset of tumors derived from cells with low rates of self-renewal. Proc Natl Acad Sci U S A. 2013;110:6021–6026. doi: 10.1073/pnas.1303607110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Walsh KM, Codd V, Smirnov IV, Rice T, Decker PA, Hansen HM, Kollmeyer T, Kosel ML, Molinaro AM, McCoy LS, et al. Variants near TERT and TERC influencing telomere length are associated with high-grade glioma risk. Nat Genet. 2014;46:731–735. doi: 10.1038/ng.3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Codd V, Nelson CP, Albrecht E, Mangino M, Deelen J, Buxton JL, Hottenga JJ, Fischer K, Esko T, Surakka I, et al. Identification of seven loci affecting mean telomere length and their association with disease. Nat Genet. 2013;45:422–427. doi: 10.1038/ng.2528. 427e421–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Suva ML, Louis DN. Next-generation molecular genetics of brain tumours. Curr Opin Neurol. 2013;26:681–687. doi: 10.1097/WCO.0000000000000027. [DOI] [PubMed] [Google Scholar]
- 32.Gravendeel LA, Kouwenhoven MC, Gevaert O, de Rooi JJ, Stubbs AP, Duijm JE, Daemen A, Bleeker FE, Bralten LB, Kloosterhof NK, et al. Intrinsic gene expression profiles of gliomas are a better predictor of survival than histology. Cancer Res. 2009;69:9065–9072. doi: 10.1158/0008-5472.CAN-09-2307. [DOI] [PubMed] [Google Scholar]
- 33. Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, Miller CR, Ding L, Golub T, Mesirov JP, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. Categorization of glioblastoma into proneural, neural, classical, and mesenchymal subtypes by genetic signature.
- 34.Brennan CW, Verhaak RG, McKenna A, Campos B, Noushmehr H, Salama SR, Zheng S, Chakravarty D, Sanborn JZ, Berman SH, et al. The somatic genomic landscape of glioblastoma. Cell. 2013;155:462–477. doi: 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Turcan S, Rohle D, Goenka A, Walsh LA, Fang F, Yilmaz E, Campos C, Fabius AW, Lu C, Ward PS, et al. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature. 2012;483:479–483. doi: 10.1038/nature10866. The G-CIMP phenotype is mediated by IDH1 mutation.
- 36. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. Described alterations in NF1, TP53 ERB, PI3K and MGMT in glioblastoma.
- 37.Brennan C, Momota H, Hambardzumyan D, Ozawa T, Tandon A, Pedraza A, Holland E. Glioblastoma subclasses can be defined by activity among signal transduction pathways and associated genomic alterations. PLoS One. 2009;4:e7752. doi: 10.1371/journal.pone.0007752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun Y, Zhang W, Chen D, Lv Y, Zheng J, Lilljebjorn H, Ran L, Bao Z, Soneson C, Sjogren HO, et al. A glioma classification scheme based on coexpression modules of EGFR and PDGFRA. Proc Natl Acad Sci U S A. 2014;111:3538–3543. doi: 10.1073/pnas.1313814111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bredel M, Scholtens DM, Yadav AK, Alvarez AA, Renfrow JJ, Chandler JP, Yu IL, Carro MS, Dai F, Tagge MJ, et al. NFKBIA deletion in glioblastomas. N Engl J Med. 2011;364:627–637. doi: 10.1056/NEJMoa1006312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Morris LG, Kaufman AM, Gong Y, Ramaswami D, Walsh LA, Turcan S, Eng S, Kannan K, Zou Y, Peng L, et al. Recurrent somatic mutation of FAT1 in multiple human cancers leads to aberrant Wnt activation. Nat Genet. 2013;45:253–261. doi: 10.1038/ng.2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rohle D, Popovici-Muller J, Palaskas N, Turcan S, Grommes C, Campos C, Tsoi J, Clark O, Oldrini B, Komisopoulou E, et al. An inhibitor of mutant IDH1 delays growth and promotes differentiation of glioma cells. Science. 2013;340:626–630. doi: 10.1126/science.1236062. A selective inhibitor of R132-IDH1 inhibits growth and promotes differentiation of IDH1-mutant glioma cells in vitro and in vivo.
- 42.Turcan S, Fabius AW, Borodovsky A, Pedraza A, Brennan C, Huse J, Viale A, Riggins GJ, Chan TA. Efficient induction of differentiation and growth inhibition in IDH1 mutant glioma cells by the DNMT inhibitor decitabine. Oncotarget. 2013;4:1729–1736. doi: 10.18632/oncotarget.1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brown PD, Krishnan S, Sarkaria JN, Wu W, Jaeckle KA, Uhm JH, Geoffroy FJ, Arusell R, Kitange G, Jenkins RB, et al. Phase I/II trial of erlotinib and temozolomide with radiation therapy in the treatment of newly diagnosed glioblastoma multiforme: north central cancer treatment group study N0177. J Clin Oncol. 2008;26:5603–5609. doi: 10.1200/JCO.2008.18.0612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van den Bent MJ, Brandes AA, Rampling R, Kouwenhoven MC, Kros JM, Carpentier AF, Clement PM, Frenay M, Campone M, Baurain JF, et al. Randomized phase II trial of erlotinib versus temozolomide or carmustine in recurrent glioblastoma: EORTC brain tumor group study 26034. J Clin Oncol. 2009;27:1268–1274. doi: 10.1200/JCO.2008.17.5984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lassman AB, Rossi MR, Raizer JJ, Abrey LE, Lieberman FS, Grefe CN, Lamborn K, Pao W, Shih AH, Kuhn JG, et al. Molecular study of malignant gliomas treated with epidermal growth factor receptor inhibitors: tissue analysis from North American Brain Tumor Consortium Trials 01–03 and 00–01. Clin Cancer Res. 2005;11:7841–7850. doi: 10.1158/1078-0432.CCR-05-0421. [DOI] [PubMed] [Google Scholar]
- 46.Neyns B, Sadones J, Joosens E, Bouttens F, Verbeke L, Baurain JF, D’Hondt L, Strauven T, Chaskis C, In’t Veld P, et al. Stratified phase II trial of cetuximab in patients with recurrent high-grade glioma. Ann Oncol. 2009;20:1596–1603. doi: 10.1093/annonc/mdp032. [DOI] [PubMed] [Google Scholar]
- 47.Thiessen B, Stewart C, Tsao M, Kamel-Reid S, Schaiquevich P, Mason W, Easaw J, Belanger K, Forsyth P, McIntosh L, et al. A phase I/II trial of GW572016 (lapatinib) in recurrent glioblastoma multiforme: clinical outcomes, pharmacokinetics and molecular correlation. Cancer Chemother Pharmacol. 2010;65:353–361. doi: 10.1007/s00280-009-1041-6. [DOI] [PubMed] [Google Scholar]
- 48.Snuderl M, Fazlollahi L, Le LP, Nitta M, Zhelyazkova BH, Davidson CJ, Akhavanfard S, Cahill DP, Aldape KD, Betensky RA, et al. Mosaic amplification of multiple receptor tyrosine kinase genes in glioblastoma. Cancer Cell. 2011;20:810–817. doi: 10.1016/j.ccr.2011.11.005. [DOI] [PubMed] [Google Scholar]
- 49.Mellinghoff IK, Wang MY, Vivanco I, Haas-Kogan DA, Zhu S, Dia EQ, Lu KV, Yoshimoto K, Huang JH, Chute DJ, et al. Molecular determinants of the response of glioblastomas to EGFR kinase inhibitors. N Engl J Med. 2005;353:2012–2024. doi: 10.1056/NEJMoa051918. [DOI] [PubMed] [Google Scholar]
- 50.Fenton TR, Nathanson D, Ponte de Albuquerque C, Kuga D, Iwanami A, Dang J, Yang H, Tanaka K, Oba-Shinjo SM, Uno M, et al. Resistance to EGF receptor inhibitors in glioblastoma mediated by phosphorylation of the PTEN tumor suppressor at tyrosine 240. Proc Natl Acad Sci U S A. 2012;109:14164–14169. doi: 10.1073/pnas.1211962109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Koul D, Shen R, Kim YW, Kondo Y, Lu Y, Bankson J, Ronen SM, Kirkpatrick DL, Powis G, Yung WK. Cellular and in vivo activity of a novel PI3K inhibitor, PX-866, against human glioblastoma. Neuro Oncol. 2010;12:559–569. doi: 10.1093/neuonc/nop058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kreisl TN, Lassman AB, Mischel PS, Rosen N, Scher HI, Teruya-Feldstein J, Shaffer D, Lis E, Abrey LE. A pilot study of everolimus and gefitinib in the treatment of recurrent glioblastoma (GBM) J Neurooncol. 2009;92:99–105. doi: 10.1007/s11060-008-9741-z. [DOI] [PubMed] [Google Scholar]
- 53.Jackson CM, Lim M, Drake CG. Immunotherapy for brain cancer: recent progress and future promise. Clin Cancer Res. 2014;20:3651–3659. doi: 10.1158/1078-0432.CCR-13-2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shi Q, Qin L, Wei W, Geng F, Fan R, Shin YS, Guo D, Hood L, Mischel PS, Heath JR. Single-cell proteomic chip for profiling intracellular signaling pathways in single tumor cells. Proc Natl Acad Sci U S A. 2012;109:419–424. doi: 10.1073/pnas.1110865109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Patel AP, Tirosh I, Trombetta JJ, Shalek AK, Gillespie SM, Wakimoto H, Cahill DP, Nahed BV, Curry WT, Martuza RL, et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344:1396–1401. doi: 10.1126/science.1254257. Described the marked intratumoral heterogeneity of glioblastoma, including the mosaic expression of RTKs and presence of tumor cells of different genetic signatures within single tumor.
- 56.Szerlip NJ, Pedraza A, Chakravarty D, Azim M, McGuire J, Fang Y, Ozawa T, Holland EC, Huse JT, Jhanwar S, et al. Intratumoral heterogeneity of receptor tyrosine kinases EGFR and PDGFRA amplification in glioblastoma defines subpopulations with distinct growth factor response. Proc Natl Acad Sci U S A. 2012;109:3041–3046. doi: 10.1073/pnas.1114033109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, Henkelman RM, Cusimano MD, Dirks PB. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 58.Chen J, Li Y, Yu TS, McKay RM, Burns DK, Kernie SG, Parada LF. A restricted cell population propagates glioblastoma growth after chemotherapy. Nature. 2012;488:522–526. doi: 10.1038/nature11287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang J, Sakariassen PO, Tsinkalovsky O, Immervoll H, Boe SO, Svendsen A, Prestegarden L, Rosland G, Thorsen F, Stuhr L, et al. CD133 negative glioma cells form tumors in nude rats and give rise to CD133 positive cells. Int J Cancer. 2008;122:761–768. doi: 10.1002/ijc.23130. [DOI] [PubMed] [Google Scholar]
- 60.Bonavia R, Inda MM, Cavenee WK, Furnari FB. Heterogeneity maintenance in glioblastoma: a social network. Cancer Res. 2011;71:4055–4060. doi: 10.1158/0008-5472.CAN-11-0153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen R, Nishimura MC, Bumbaca SM, Kharbanda S, Forrest WF, Kasman IM, Greve JM, Soriano RH, Gilmour LL, Rivers CS, et al. A hierarchy of self-renewing tumor-initiating cell types in glioblastoma. Cancer Cell. 2010;17:362–375. doi: 10.1016/j.ccr.2009.12.049. [DOI] [PubMed] [Google Scholar]
- 62.Inda MM, Bonavia R, Mukasa A, Narita Y, Sah DW, Vandenberg S, Brennan C, Johns TG, Bachoo R, Hadwiger P, et al. Tumor heterogeneity is an active process maintained by a mutant EGFR-induced cytokine circuit in glioblastoma. Genes Dev. 2010;24:1731–1745. doi: 10.1101/gad.1890510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hunter C, Smith R, Cahill DP, Stephens P, Stevens C, Teague J, Greenman C, Edkins S, Bignell G, Davies H, et al. A hypermutation phenotype and somatic MSH6 mutations in recurrent human malignant gliomas after alkylator chemotherapy. Cancer Res. 2006;66:3987–3991. doi: 10.1158/0008-5472.CAN-06-0127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Johnson BE, Mazor T, Hong C, Barnes M, Aihara K, McLean CY, Fouse SD, Yamamoto S, Ueda H, Tatsuno K, et al. Mutational analysis reveals the origin and therapy-driven evolution of recurrent glioma. Science. 2014;343:189–193. doi: 10.1126/science.1239947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mitchell DA, Fecci PE, Sampson JH. Immunotherapy of malignant brain tumors. Immunol Rev. 2008;222:70–100. doi: 10.1111/j.1600-065X.2008.00603.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Champiat S, Ferte C, Lebel-Binay S, Eggermont A, Soria JC. Exomics and immunogenics: bridging mutational load and immune checkpoints efficacy. Oncoimmunology. 2014;3:e27817. doi: 10.4161/onci.27817. Described the relationship between tumor mutational burden, neoantigen production, and immunogenicity, with respect to response to immunotherap.