Abstract
Diffuse Intrinsic Pontine Glioma (DIPG) is an aggressive pediatric brain tumor with a median survival of one year after diagnosis. It has been reported recently that about 80% of DIPG cases and 70% of midline glioblastomas contain a mutation at one allele of the H3F3A gene (encoding histone H3 variant H3.3), replacing the lysine 27 with methionine (K27M). In order to facilitate diagnosis of DIPG patients, a quick and reliable method to identify the H3F3A K27M mutation is needed. Here, we describe a real-time PCR-based procedure involving a mutant-specific primer, a blocker oligonucleotide, and a reverse primer that can differentiate samples with H3F3A K27M mutation from those that do not. We first tested four different mutant-specific primers for their ability to selectively amplify H3F3A K27M-mutant allele and found that one primer amplified the mutant allele more efficiently than the rest. We then determined the optimal concentration of blocker oligo that significantly improved amplification of the H3F3A K27M-mutant allele. Using this optimized real-time PCR assay, we analyzed eleven samples, two of which containing H3F3A K27M mutation, and found that these two samples were differentially amplified from the nine others. In addition, we were able to discern the H3F3A K27M mutation in a newly obtained pediatric brainstem glioblastoma sample whose H3.3 status was not known previously, and in three other DIPG samples as well as paraffin embedded samples. These results demonstrate that we have developed a new reliable procedure for detecting the H3F3A K27M mutation in pediatric glioblastoma patient samples.
Keywords: Histone H3 variant, Diffuse Intrinsic Pontine Glioma, Real-Time PCR, pediatric glioblastoma
Introduction
Pediatric high grade gliomas, including Diffuse Intrinsic Pontine Glioma (DIPG) and glioblastoma multiforme (GBM), accounting for about 15% of all pediatric brain tumors, are the most aggressive brain tumors in children and is the leading cause of cancer-related death of children [1, 2]. Methods of diagnosis include MRI and biopsy, however, the deep and delicate locations of these tumors and the lack of targeted therapy often preclude them from a tissue diagnosis and the majority of DIPGs are diagnosed based on typical MRI imaging characteristics. DIPG has the worst prognosis of any malignancy in the central nervous system with a median time of survival of approximately 12-16 months after diagnosis [3, 4].
Mutations at the HIST1H3B and H3F3A genes have recently been detected in 15% and 60% of DIPG cases, respectively [5, 6]. HIST1H3B and H3F3A encode canonical histone H3.1 and H3 variant H3.3, respectively. Histone H3.1 is assembled into nucleosomes in a replication-dependent reaction. Histone H3 variant H3.3, differing from H3.1 by five amino acids, is assembled into nucleosomes via a replication-independent mechanism [7-9]. In all DIPG cases bearing a mutation at histone H3, lysine 27 of either histone H3.1 or H3.3 is mutated to methionine [5, 6]. Interestingly, it appears that mutations at HIST1H3B define a subgroup of younger aging DIPG samples. In addition, the H3F3A K27M mutation was also detected in other pediatric midline tumors including the thalamus (40%) [10-13]. Therefore, the H3F3A K27M mutation frequently occurs in high-grade pediatric brain tumors.
Histone H3 lysine 27 can be methylated by PRC2 complex and H3K27 di- and trimethylation (H3K27me2/me3) is associated with gene silencing [14-16]. H3K27 can also be acetylated (H3K27ac), and H3K27ac is associated with gene expression. Human genome has 14 genes encoding H3.1 and 2 genes encoding H3.3. Lysine 27 is conserved in all histone proteins expressed from these genes. We and others have shown previously that H3K27me3 was low in DIPG cells compared to reference cells such as neural stem cells or adult glioma cells. In cell lines expressing either the H3.1K27M or H3.3K27M mutant transgene, H3K27me3 levels of endogenous H3 including H3.3 and H3.1 are dramatically reduced. These results indicate that H3K27M mutant protein dominantly inhibit the levels of H3K27me3 in cells [17-21]. Because the H3F3A K27M mutation is found in over 60% of all DIPG cases, it is important to find a reliable method to detect this mutant allele in order to diagnose this class of brain tumor and guide future targeted therapy. Here, we report a method for the detection of the H3F3A K27M mutant allele specifically.
Materials and Methods
Sources of Nucleic Acids
Message RNA from neural stem cells and DIPG cells containing the H3F3A K27M mutation (SF7761) was isolated and converted to cDNA using published procedures [17]. During the initial test of various conditions, we used full-length wild type H3F3A cDNA and cDNA with the H3.3K27M mutation as the template. To isolate these cDNA templates, two PCR primers were used to amplify the cDNA libraries. Full-length H3F3A cDNA, one containing two wild type alleles (wild type template) and the other with one wild type and one mutant allele (mutant template) were purified using a QIAGEN PCR purification kit (QIAGEN) and used as templates for real-time PCR reactions. All the oligos except the blocker oligo (Life-technology) was synthesized by IDT. During the final test of our method, cDNA libraries from nine different tumor cell lines that did not have H3.3K27M mutation were collected and marked numerically. These cDNA libraries, along with cDNA isolated from two DIPG lines (SF7761 and SF8626), were then given to RZ for blind analysis. Finally, Dr. Baker kindly provided us three additional cDNA libraries (DIPG1, DIPG2 and DIPG4) for testing.
Real-Time PCR (qPCR) conditions
Standard 20 μl real-time PCR (qPCR) reactions were prepared, each containing 1 μl of cDNA template with concentration ranging from 0.1 to 10 ng/μl of DNA, 1 μl of mutant-specific primer (6.25 ng/μl), 1 μl of reverse primer (6.25 ng/μl), 10 μl of SYBR Green Supermix (BIO-RAD), and 7 μl H2O. To test the effect of blocker oligo, 1 μl of different concentrations of the blocker oligo ranging from 6.25 to 25 pmol/μl was added into each reaction; the amount of H2O was reduced to maintain the reaction volume of 20 μl. The real-time PCR amplification and detection were performed in a BIO-RAD Real-Time PCR machine under standard thermocycling conditions: 30 sec at 95°C, 42 cycles of (10 sec at 95°C, 30 sec at 55°C, 30 sec at 72°C), and 7 min at 72°C. All qPCR reactions were run in duplicate or triplicate when necessary.
Primary Tissue Culture
Primary pediatric human glioma cells were obtained from surgical biopsy of a brainstem tumor from a 12-year-old patient in accordance with an institutionally approved protocol (IRB: 12-003458). Informed consent was obtained from the patient's legal guardian prior to surgery. Minced tissue was placed in a centrifuge at 1200 rpm for 10 min and then passed through a 75-micron mesh into a T-75 flask. Primary stem cell culture was made by adding 50 mL of media hormone mix [composed of 500 ml DMEM/F12 (Gibco Life Technologies #14200-075), 5 ml 45 % glucose (Sigma #G8769), 6 ml 7.5% NaHCO3 (Gibco Life Technologies # 25080094), 5 ml 200 mM Glutamine (Gibco Life Technologies # 25030081), 2 ml 1 M HEPES (Gibco Life Technologies #15630080), 5 ml Pen Strep (Gibco Life Technologies #15140122) and 5 ml of N2 Supplement (Gibco Life Technologies #17502048) and the following growth factors were added at final concentrations of 20ng/ml: Human β-FGF stock (PeproTech # AF-100-18B), and Human EGF (PeproTech #AF-100-15). Neurosphere growth was confirmed within a couple of days and passaged in a serial fashion every one to two weeks. Tumor cells from the third passage were used for this study.
RNA extraction from FFPE tissues
Mouse xenograft tissues using SF8628 DIPG cells were first grossed and processed overnight in the Sakura Tissue Tek VIP 6 Tissue Processor. After processing, tissues were embedded in paraffin wax and labeled as 422 and 522 in Dr. Daniels's laboratory. To extract total RNA from FFPE tissue, 10 μm sections were cut from the paraffin-embedded 422 and 522 blocks using the microtome. Two sections from each sample were placed in 1.5 ml tubes. As a control, T/C 28a2 cell block was also processed. Then 1 ml xylene was added to each tube, mixed well, centrifuged 2 minutes at 16,100 × g. After removal of xylene, 1 ml 100% ethanol was added to each tube, mixed and centrifuged 2 minutes at 16,100 × g. After removal of ethanol, pellet was air-dried for approximately 5 minutes. Total RNA was extracted using the High Pure® FFPE RNA Micro Kit (Roche Diagnostics, Mannheim, Germany) according to manufacture protocols. cDNA was synthesized using total RNA and SuperScript® III First-Strand Synthesis System (Invitrogen, Carlsbad, CA). Oligo dT was used as the primer. cDNA libraries labeled as 422, 522 and T/C were given to RZ blindly for analysis of H3F3A K27M mutation shown in Figure 5d.
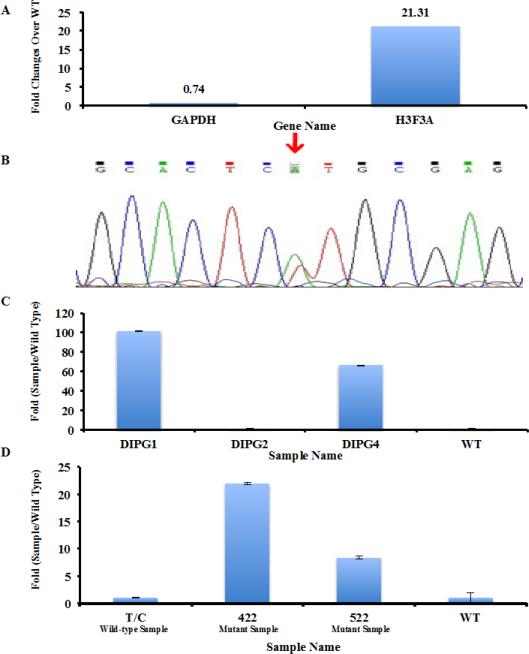
Fig. 5.
Detecting the H3F3A K27M mutation in DIPG samples with unknown H3F3A status. (a) Detection of H3F3A K27M mutation in a newly diagnosed DIPG case using qPCR. Amplification of a tumor sample with wild type H3F3A was used as control. The difference in amplification of house-keeping gene GAPDH and H3F3A between the wild type and test sample were shown. 1ng of cDNA library was used. (b) Sanger Sequencing of H3F3A cDNA isolated from the DIPG cancer was used to validate the presence of H3F3A K27M mutation. (c) Analysis of H3F3A K27M mutation in three additional DIPG samples. The mutation status of each sample was kept unknown to RZ. The PCR conditions established in Supplemental Fig. 2 and 10 ng of cDNA libraries were used. (d) Detecting the H3F3A K27M mutation in paraffin-embedded samples. 10 ng of cDNA libraries from three different paraffin samples were analyzed by real-time PCR.
Results
A description of PCR design
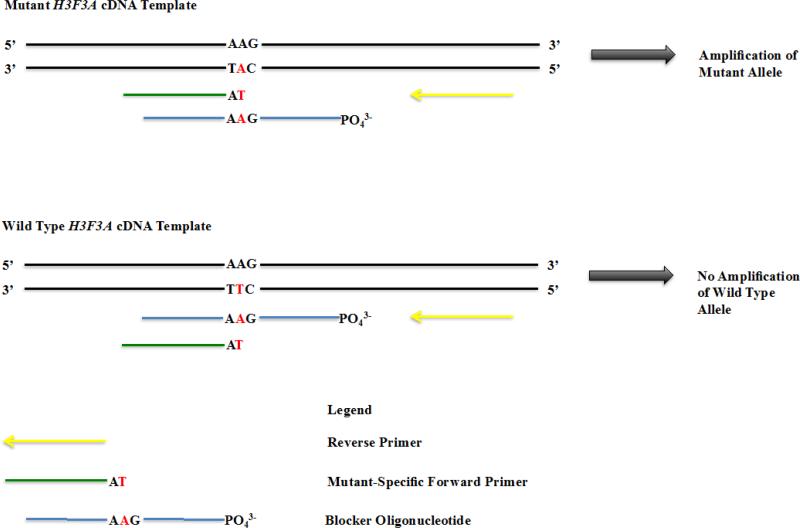
To develop a simple and reliable method for the detection of H3F3A K27M mutation allele in pediatric GBM patients, we decided to use a real-time PCR strategy that is similar to that described by Moran et al [22]. The success of this method relies on the use of mutant PCR primers; therefore, we designed four mutant-specific primers, a blocker oligo, and two reverse primers (Figure 1 and Supplemental Table 1). The 3’-end of each of the four mutant-specific primers was anchored to the variant-base of the H3F3A mutant allele. Each primer varied in length by a couple of nucleotides such that their melting temperatures (TM) ranges from 50 to 60 °C with 2-4°C incremental change. A blocker oligo was designed based on two principles. First, the variant-base corresponding to the wild type H3F3A allele was at the center of the sequence. Second, the TM of the blocker oligo, with its 3’ end of the sequence phosphorylated to prevent elongation, was about 4 to 10 °C higher than TM of mutant-specific primer. Two reverse primers complementary to the wild type sequence were also designed, amplifying 75bp and 200 bp cDNA of H3F3A gene, respectively. All PCR primers were optimized in silico using the open-source software “primer3” to eliminate primers forming hairpin and primer dimers. The unique features of each primer are described in Supplemental Table 1 and sequence of each primer is described in Supplemental Table 2.
Fig. 1.
An outline of the PCR design for detecting the H3.3K27M mutant allele. The codon for lysine 27 of the H3F3A gene is AAG and is mutated to ATG at one allele of the H3F3A gene. The 3’-end of each mutant-specific primer ends at mutant variant-base (T). The blocker oligo was phosphorylated at its 3’-end to prevent elongation, with the wild type base corresponding to the mutant allele placed in the center of the sequence. A reverse primer is also indicated.
Effect of mutant-specific primers on selective amplification of the mutant allele
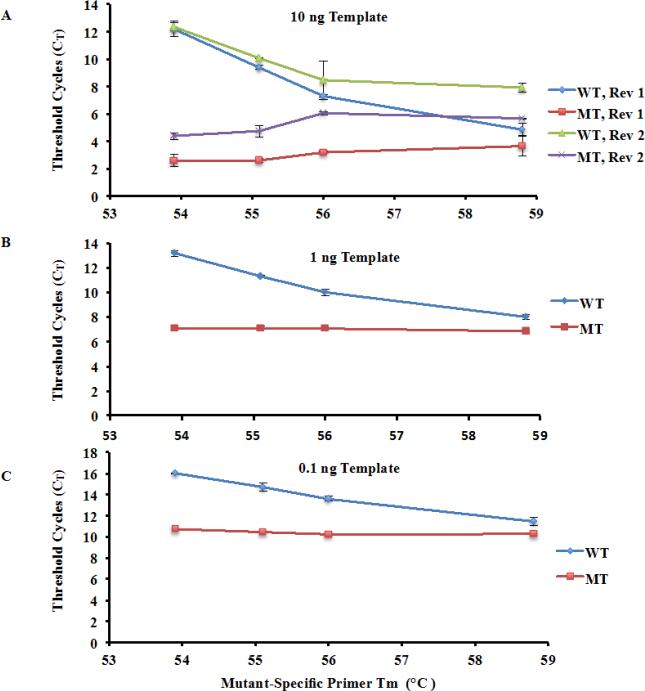
First, we tested how each of the two reverse primers and four mutant-specific primers amplified 10 ng of wild type and H3.3K27M mutant template. We used threshold cycles (CT) to measure amplification efficiency. Because each mutant-specific primer was directed towards the mutant allele, it generated a mismatch with wild type template. This drives up the threshold cycles for amplification of wild type template (CT). Therefore, a large CT value indicates a less efficient amplification. The difference in CT (ΔCT) between wild type and mutant templates was used to judge the ability of a primer and/or condition to amplification efficiency of the mutant allele. The larger the ΔCT was, the more selective for amplification of mutant allele over wild type one. As shown in Figure 2a, the threshold cycles (CT) for the mutant template were quite similar among the four mutant-specific primers combined with either reverse primer 1 or reverse primer 2. Moreover, ΔCT values for reverse primer 1 were generally larger than the corresponding ΔCT using reverse primer 2. These results indicate that reverse primer 1 gives rise to the larger amplification differences between wild type and mutant alleles than reverse primer 2. Therefore, we decided to use reverse primer 1 for all of the subsequent experiments.
Fig. 2.
Test the ability of four mutant-specific primers and two reverse primers for amplification of K27M mutant and wild type H3F3A. (a) Effects of four mutant-specific primers and two reverse primers on the amplification of wild type and K27M mutation-containing H3F3A. PCR reactions were set up using either Reverse Primer 1 (Rev 1) or Reverse Primer 2 (Rev 2). For subsequent experiments, Rev 1, which gave rise to the large ΔCT in amplification of wild type H3F3A (WT) and H3F3A K27M mutant (MT) allele, was used. (b-c) Amplification selectivity using two different amounts of H3F3A cDNA templates 1 ng (b) and 0.1 ng (c). Calculated TM of each mutant-specific primer is indicated on the X-axis.
Next, we tested how reduced template concentrations affected amplification specificity. The overall trends observed for lower template concentrations (1 and 0.1 ng) were similar. First, the CT for mutant template remained relatively constant among the four different mutant-specific primers, whereas CT for the wild type template decreased as TM of the mutant-specific primer increased. Second, mutant template was amplified significantly more efficiently than wild type. The mutant-specific primer with TM of 53.9°C attained the maximum ΔCT, which was 6.1 and 5.27 for 1 and 0.1 ng cDNA, respectively. These results suggest that mutant-specific primer 1 (TM=53.9°C) gives rise to the highest selectivity for amplification of mutant allele over wild type. Therefore, mutant-specific primer 1 was selected for the rest of experiments.
Effect of blocker oligo concentration on amplification selectivity
The differences in amplifying wild type and mutant alleles became smaller when the template concentration were reduced (Figure 2), indicating that the selectivity in amplification of mutant allele over wild type declines when template concentration is low. It is important to improve the selectivity in order to detect the mutant allele in clinical situations where tumor samples maybe limited. In principle, the mutant-specific primer should not anneal to the wild-type template because the primer:template mismatch at the variant-base causes immense instability in binding the two reagents together. However, because the remaining portion of the primer is complementary to the wild-type, there will be instances where the primer:template mismatch is overcome and the mutant-specific primer erroneously anneals and amplifies the wild-type template, consequently giving rise to non-specific amplification. To minimize this non-specific amplification of the wild-type and improve the selectivity of the PCR assay, the blocker oligonucleotide was designed with the variant-base positioned in the center of the blocker sequence, and its 3’-end was phosphorylated to prevent polymerase extension. In this way, the blocker will anneal to the wild-type template and obstruct the mutant-specific primer from binding to wild type allele, thereby selectively increasing the amplification of the mutant allele over the wild type.
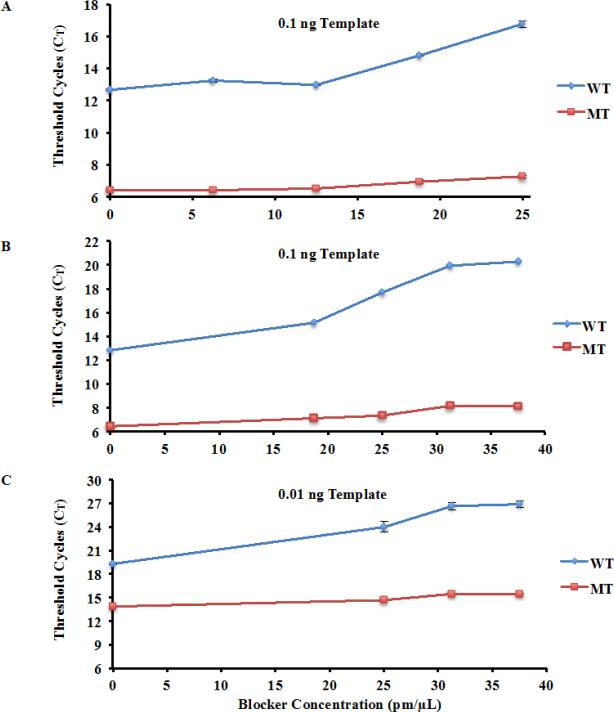
The effect of blocker concentration, ranging from 6.25 to 37.5 pm/μL (1 to 6-fold of PCR primer concentration), was tested. As shown in Figure 3a-b, with a template concentration of 0.1 ng, the mutant H3.3 template was amplified significantly more efficiently than wild type. Moreover, CT for wild type H3.3 cDNA increased with the increase of the concentration of blocker oligo, whereas CT for mutant allele barely changed, consistent with the idea that the blocker oligo inhibits amplification of wild type allele and thereby increases amplification specificity. The ΔCT between the wild type and mutant templates was largest (12.13) when the blocker oligo concentration was 37.5 pm/μL, indicating that the mutant allele was amplified 4882 (212.13) fold more efficiently than wild type (Student T-Test P<0.0001). Comparing to the same conditions without the blocker oligo, the blocker oligo increases amplification specificity of mutant allele over wild type by about 53-fold.
Fig. 3.
Effects of blocker oligo concentrations on the amplification efficiency of wild type and H3F3A K27M mutant allele. (a-b) Amplification selectivity increases with elevated blocker oligo concentrations. Blocker concentration ranging from 1X to 4X of the PCR primer concentration (a) and from 3X to 6X of the PCR primer concentration (b) was tested. (c) Amplification selectivity at a lower concentration of templates. Concentrations of blocker oligo are indicated in the figure.
Next, we tested whether the PCR primer with increasing concentration of blocker was effective at a lower concentration of template. As shown in Figure 3c, a similar trend was observed using 0.01 ng of template: while the CT for mutant template did not change dramatically with the increase in concentration of the blocker oligo, the Ct for wild type template was increased significantly. The largest ΔCT (11.44) between the wild type and mutant template was observed at blocker concentration of 37.5 pm/μL, which represents 2778 (211.44) fold more efficient amplification of the mutant allele over wild type (Student T-Test P<0.0001).
In all these tests, we noticed that when the blocker oligo concentration surpassed five- to six-fold of PCR primer concentration (31.25 and 37.5 pm/ml respectively), the ΔCT did not change dramatically, suggesting that a further increase in the blocker concentration will not have major effects on increasing amplification selectivity when cDNA of H3F3A gene was used as template. When cDNA libraries were used as a template, we found that eight- to nine-fold of PCR primer concentrations could still increase amplification specificity (Supplemental Figure 1). Therefore, we used blocker concentrations at either 6- or 8-fold of PCR primers for the rest of experiments.
Selectively amplify the H3.3K7M mutant allele using cDNA Libraries
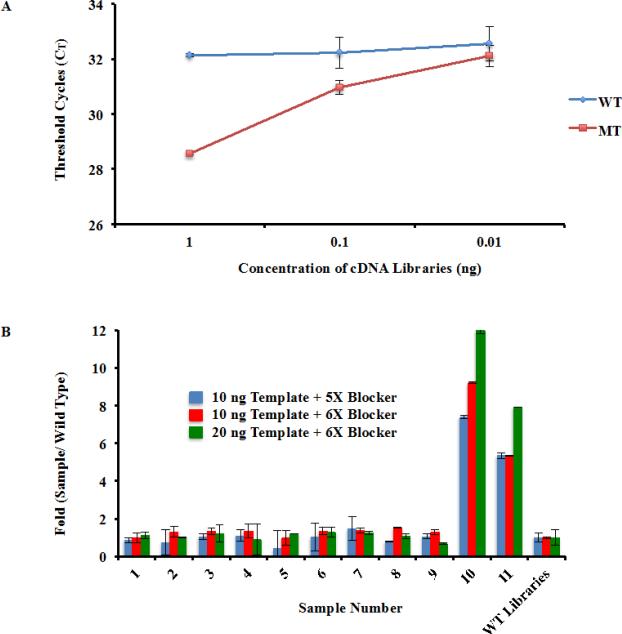
In the previous experiments, purified H3F3A cDNA was used to determine the optimal combination of a mutant-specific primer and the concentration of blocker oligonucleotide. To reduce the cost of the detection, it is desirable to detect the H3F3A mutation using cDNA libraries instead of pure H3F3A cDNA. Therefore, we tested whether the qPCR conditions could be used to detect the H3.3K27M mutation in whole human cDNA libraries. As shown in Figure 4A, there were significant differences in amplification of cDNA libraries prepared from cells with and without H3F3A K27M mutation, with a ΔCT value of 3.565 and 1.265 when 1 and 0.1 ng of cDNA libraries were used, respectively (Student T-Test P<0.0001, P=0.01 respectively). However, when 0.01 ng cDNA libraries were used, the difference in amplification of wild type and mutant libraries became insignificant. These results suggest that the PCR conditions could be used to identify H3F3A mutation in 0.1 ng or more cDNA libraries from tumor samples.
Fig. 4.
Identify the H3.3K27M mutation within cDNA libraries. (a) Amplification selectivity using cDNA libraries prepared from samples containing wild type and mutant H3F3A allele. The PCR conditions established in Fig. 3 was used to amplify three different amounts of cDNA libraries (1 ng, 0.1 ng, and 0.01 ng). (b) Identify samples containing H3F3A K27M mutation using the optimized procedure. To simulate actual medical situations, cDNA libraries from eleven different patient samples were gathered, two of which contained the H3F3A K27M mutation. This information was kept unknown from RZ during testing. The optimal qPCR conditions were used to blindly analyze all eleven samples using two different template and blocker oligo concentrations. The differences in amplification (fold changes) between a sample containing wild type H3F3A and each of the 11 patient samples were calculated.
Identify samples with the H3.3K27M mutation
In clinical situations, templates from patient samples will have unknown characteristics. To simulate this real world situation, we performed two separate experiments. First, cDNA libraries from eleven different samples were gathered, two of which were DIPG samples known to have the H3F3A K27M mutation. The contents of each sample were kept unknown to RZ, who performed the analysis on all eleven samples blindly using the optimal qPCR conditions (Figure 4b). The amplification efficiency of each sample was compared to the known wild type cDNA library. As shown in Figure 4b, patient samples 10 and 11 were amplified much more efficiently than either wild type or the nine other patient samples (all Student T-Tests P<0.0001), which is consistent with those samples containing the H3F3A K27M mutation after verification. This result confirms that our experimental procedures described here can be used to identify patient samples with the H3F3A K27M mutation.
To further validate our experimental procedure in the clinical setting, we tested a newly diagnosed brainstem GBM whose H3F3A mutational status was not known. We used 1 ng cDNA library for this experiment. As shown in Figure 5a, the housekeeping gene (GAPDH) was amplified similarly between tumor with wild type H3.3 and the brainstem GBM tumor, suggesting that similar amount of cDNA were used for amplification. In contrast, H3F3A was amplified much more efficiently in the newly diagnosed tumor sample compared to the sample with wild type H3F3A gene, supporting the idea that the newly diagnosed GBM having the H3F3A K27M mutation. Sanger Sequencing of H3F3A cDNA of this sample confirmed that this tumor had the H3F3A K27M mutation, with mutant base occurring at one allele of the H3F3A gene (Figure 5b). Together, these results indicate that the qPCR assay can identify pediatric high-grade gliomas with H3.3K27M mutation from those without this mutation.
Identifying H3F3A K27M in additional and paraffin samples
We also blindly analyzed three additional patient samples provided by Dr. Baker. As shown in Figure 5c, samples DIPG 1 and 4, but not DIPG2, were amplified much more efficiently than the control sample with wild type H3.3, indicating that these two samples harbor the H3F3A K27M mutation and DIPG2 appears not have the mutation.
To further test whether DIPG2 harbors H3F3A K27M mutation, we performed several experiments. First, we tested DIPG2 using a higher concentration of blocker oligos (8-fold of PCR primers) and found DIPG2 was amplified to a similar degree as samples containing wild type H3F3A despite the fact that under this condition the amplification selectivity increases significantly (Supplemental Figure 1). Second, we isolated H3F3A cDNA by amplifying DIPG2 cDNA libraries using two PCR primers and used H3.3 cDNA as the template to determine whether DIPG2 had H3F3A K27M mutation. As shown in Supplemental Figure 2a, DIPG2 was amplified more efficiently than control samples with wild type H3F3A (Student T-Test P<0.0001), indicating that DIPG2 contained the H3F3A K27M mutation. Third, we performed Sanger sequencing of H3.3 cDNA isolated from the DIPG2 sample and observed that the H3F3A mutant allele was barely detectable (Supplemental Figure 2b), providing an explanation for our inability to detect the H3F3A K27M mutation in this samples using cDNA library as templates. These results indicate the sensitivity and selectivity of our method to detect H3F3A K27M mutation will be enhanced dramatically when H3.3 cDNA is used as templates.
In clinical situations, most patient samples are preserved after paraffin fixation. To determine whether our method can detect H3F3A K27M mutation in paraffin embedded samples. We analyzed cDNA libraries from 3 different mouse xenograft samples and the amplification efficiency of each sample was compared to the known cDNA library with wild type H3.3. Samples marked with 422 and 522 were amplified much more efficiently than the sample labeled as T/C (Figure 5d), indicating that two samples marked with 422 and 522 harbored the H3F3A K27M mutation and T/C does not. This result demonstrates that the experimental procedures described here can be used to identify H3F3A K27M mutation in paraffin-embedded xenograft samples. While xenograft tissues were used in this test, it is quite likely that our method can detect H3F3A K27M mutation in paraffin embedded primary human tissues.
Discussion
In order to facilitate the diagnosis of pediatric patients with GBMs that harbor the H3.3K27M mutation, we have developed a PCR-based procedure to detect the frequently mutated allele of H3F3A in this high-grade pediatric brain tumor. This procedure involves a mutant-specific primer, a blocker oligo, and a reverse primer. We made the following attempts to identify the PCR conditions. First, using full-length human wild type H3F3A and mutant cDNA with the H3F3A K27M allele, we tested four-mutant-specific primers and concentration of a blocker oligo for their ability to differentially amplify H3F3A wild type and mutant allele. We found that the optimal mutant-specific primer has a TM of 53.9°C, and the blocker oligo concentration about 5- to 6-fold of PCR primer concentration. Under these conditions, the qPCR reaction gave rise to about 2778-fold amplification of mutant H3F3A cDNA allele over wild type one when 0.01 ng of purified H3F3A cDNA was used. We then tested whether the PCR assay could detect the H3F3A K27M mutation in whole human cDNA libraries instead of purified H3.3 cDNA. We found that the mutant allele was amplified 2.7-fold more efficiently than wild type allele when using 0.1 ng of cDNA libraries. In addition, a further increase in the concentration of blocker oligo (8-fold that of the PCR primer concentration) would increase amplification selectivity when cDNA libraries are used. To further validate the procedure, we simulated the test under clinical conditions in multiple ways. First, we blindly analyzed 11 different cDNA libraries using the qPCR procedure, and we successfully differentiated two samples harboring the H3F3A K27M mutation from nine other samples that do not. Second, we analyzed a newly diagnosed brainstem GMB in which the mutation status of H3F3A gene was not known and showed that this tumor has the H3F3A K27M mutation, a result that was confirmed using Sanger Sequencing. Third, we analyzed three DIPG samples and found two of them with H3F3A mutation using cDNA libraries and one had the mutation using H3.3 cDNA as templates. These results show a strategy to further validate a sample without H3F3A K27M mutation using cDNA library in the future. Finally, we analyzed cDNA libraries from 2 different paraffin embedded samples, and we successfully determined the two that have the H3F3A K27M mutation. Together, these results show that the qPCR procedure using the mutant-specific primer and blocker oligo can be used in real situations to identify tumor samples with the H3F3A K27M mutation. While we have simulated several situations for detecting the H3F3A K27M mutation at clinic settings, this protocol should be tested in a large sample size before used as clinical test. Because our assay is highly specific to detect H3F3A K27M mutation, we expect that our procedure will stand-up the test when a large sample size is applied.
Sanger sequencing is capable of detecting the H3F3A K27M mutation in tumor samples. The advantage of Sanger sequencing is that it can detect mutation with precision without usage of negative controls. However, Sanger sequencing requires pure genomic DNA fragments of H3F3A. In contrast, our qPCR method can use whole cDNA libraries as templates. Moreover, it is noted that Sanger sequencing could not definitively detect the H3F3A K27M mutant allele of DIPG2 samples because the wild-type allele was the predominant one in the sample. In contrast, our qPCR method can detect H3F3A K27M mutation when H3F3A cDNA was used as templates.
Two independent groups recently published their data showing that a polyclonal antibody that recognizes H3K27M for the detection of the H3K27M mutation in tumor samples using immunohistochemistry techniques [23, 24]. Furthermore, they show that the antibody for the H3K27M mutation is more sensitive than measuring global reduction of H3K27me3. Immunohistochemistry is a technique that is readily available to most tumor pathology practices making their application widely available and useful clinically. However, this is a burgeoning field, and additional procedures that confirm the diagnosis will be necessary. In addition, antibodies against H3K27M cannot differentiate whether H3F3A (encoding H3.3) and HIST1H3B (encoding H3.1) are mutated in tumors. Mutations at H3.1 and H3.3, which both results in lysine to methionine changes, are associated with different age group and may define different sub-group of DIPG tumors, therefore, it is important to differentiate mutations at either HIST1H3B and H3F3A. Our method will complement the IHC staining for detecting H3F3A K27M mutations.
The advantage of our procedure is its simplicity. Similar PCR methods utilizing a mutant-specific primer and blocker oligonucleotide to detect single nucleotide mutations and polymorphisms have been described before [22]. However, Morlan et al included a common Taqman Probe as part of their assay in order to increase selectivity and sensitivity. In our method, the Taqman Probe, which is expensive to synthesize, is unnecessary. Therefore, our qPCR design is likely to be cheaper in detecting the target mutation.
In addition to the detection of H3.3K27M mutation found in high-grade glioma samples, we expect that this procedure can be adapted to detect H3.3 mutations found in other tumors. For instance, G34 of H3.3 was mutated in another subgroup of brain tumor as well as giant cell tumor [5, 6, 25]. In addition, H3.3 lysine 36 is mutated in chondroblastoma [25]. We anticipate that by redesigning the mutant-specific PCR primers and blocker oligos while following similar procedures tested in this study, we will be able to detect other mutations on histone H3.3 found in these tumors as well. As more experimental therapies that target the writers and erasers of histone post-translational modifications become available there will be a great need for identifying the H3K27M mutation in tumors in an every day clinical practice.
Supplementary Material
Acknowledgements
We thank Dr. Dominick Sinicropi for providing advice to RZ during the primer and blocker oligonucleotide designs; Dr. Chan Kui-Ming for providing H3F3A wild type and K27M mutant cDNA libraries; and Dr. Yu Chao and Mrs. Hui Zhou for assistance with qPCR reactions. We also thank Dr. Suzanne Baker for cDNA from DIPG1, DIPG2 and DIPG4.
Funding: This work is supported by a NIH grant (CA157489) to ZZ. This paper is subject to the NIH Public Access Policy.
Footnotes
Conflict of Interest: The authors declare no competing interests
References
- 1.Jones C, Baker SJ. Unique genetic and epigenetic mechanisms driving paediatric diffuse high-grade glioma. Nat Rev Cancer. 2014;14 doi: 10.1038/nrc3811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Warren KE. Diffuse intrinsic pontine glioma: poised for progress. Front Oncol. 2012;2:205. doi: 10.3389/fonc.2012.00205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grimm SA, Chamberlain MC. Brainstem glioma: a review. Current neurology and neuroscience reports. 2013;13:346. doi: 10.1007/s11910-013-0346-3. [DOI] [PubMed] [Google Scholar]
- 4.Kebudi R, Cakir FB, Agaoglu FY, Gorgun O, Ayan I, Darendeliler E. Pediatric diffuse intrinsic pontine glioma patients from a single center. Child's nervous system : ChNS : official journal of the International Society for Pediatric Neurosurgery. 2013;29:583–588. doi: 10.1007/s00381-012-1986-3. [DOI] [PubMed] [Google Scholar]
- 5.Schwartzentruber J, Korshunov A, Liu XY, Jones DT, Pfaff E, Jacob K, Sturm D, Fontebasso AM, Quang DA, Tonjes M, Hovestadt V, Albrecht S, Kool M, Nantel A, Konermann C, Lindroth A, Jager N, Rausch T, Ryzhova M, Korbel JO, Hielscher T, Hauser P, Garami M, Klekner A, Bognar L, Ebinger M, Schuhmann MU, Scheurlen W, Pekrun A, Fruhwald MC, Roggendorf W, Kramm C, Durken M, Atkinson J, Lepage P, Montpetit A, Zakrzewska M, Zakrzewski K, Liberski PP, Dong Z, Siegel P, Kulozik AE, Zapatka M, Guha A, Malkin D, Felsberg J, Reifenberger G, von Deimling A, Ichimura K, Collins VP, Witt H, Milde T, Witt O, Zhang C, Castelo-Branco P, Lichter P, Faury D, Tabori U, Plass C, Majewski J, Pfister SM, Jabado N. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature. 2012;482:226–231. doi: 10.1038/nature10833. [DOI] [PubMed] [Google Scholar]
- 6.Wu G, Broniscer A, McEachron TA, Lu C, Paugh BS, Becksfort J, Qu C, Ding L, Huether R, Parker M, Zhang J, Gajjar A, Dyer MA, Mullighan CG, Gilbertson RJ, Mardis ER, Wilson RK, Downing JR, Ellison DW, Baker SJ. Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem glioblastomas. Nature genetics. 2012;44:251–253. doi: 10.1038/ng.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goldberg AD, Banaszynski LA, Noh KM, Lewis PW, Elsaesser SJ, Stadler S, Dewell S, Law M, Guo X, Li X, Wen D, Chapgier A, DeKelver RC, Miller JC, Lee YL, Boydston EA, Holmes MC, Gregory PD, Greally JM, Rafii S, Yang C, Scambler PJ, Garrick D, Gibbons RJ, Higgs DR, Cristea IM, Urnov FD, Zheng D, Allis CD. Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell. 2010;140:678–691. doi: 10.1016/j.cell.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Szenker E, Ray-Gallet D, Almouzni G. The double face of the histone variant H3.3. Cell Res. 2011;21:421–434. doi: 10.1038/cr.2011.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burgess RJ, Zhang Z. Histone chaperones in nucleosome assembly and human disease. Nature structural & molecular biology. 2013;20:14–22. doi: 10.1038/nsmb.2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buczkowicz P, Hoeman C, Rakopoulos P, Pajovic S, Letourneau L, Dzamba M, Morrison A, Lewis P, Bouffet E, Bartels U, Zuccaro J, Agnihotri S, Ryall S, Barszczyk M, Chornenkyy Y, Bourgey M, Bourque G, Montpetit A, Cordero F, Castelo-Branco P, Mangerel J, Tabori U, Ho KC, Huang A, Taylor KR, Mackay A, Bendel AE, Nazarian J, Fangusaro JR, Karajannis MA, Zagzag D, Foreman NK, Donson A, Hegert JV, Smith A, Chan J, Lafay-Cousin L, Dunn S, Hukin J, Dunham C, Scheinemann K, Michaud J, Zelcer S, Ramsay D, Cain J, Brennan C, Souweidane MM, Jones C, Allis CD, Brudno M, Becher O, Hawkins C. Genomic analysis of diffuse intrinsic pontine gliomas identifies three molecular subgroups and recurrent activating ACVR1 mutations. Nature genetics. 2014;46:451–456. doi: 10.1038/ng.2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fontebasso AM, Papillon-Cavanagh S, Schwartzentruber J, Nikbakht H, Gerges N, Fiset PO, Bechet D, Faury D, De Jay N, Ramkissoon LA, Corcoran A, Jones DT, Sturm D, Johann P, Tomita T, Goldman S, Nagib M, Bendel A, Goumnerova L, Bowers DC, Leonard JR, Rubin JB, Alden T, Browd S, Geyer JR, Leary S, Jallo G, Cohen K, Gupta N, Prados MD, Carret AS, Ellezam B, Crevier L, Klekner A, Bognar L, Hauser P, Garami M, Myseros J, Dong Z, Siegel PM, Malkin H, Ligon AH, Albrecht S, Pfister SM, Ligon KL, Majewski J, Jabado N, Kieran MW. Recurrent somatic mutations in ACVR1 in pediatric midline high-grade astrocytoma. Nature genetics. 2014;46:462–466. doi: 10.1038/ng.2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Taylor KR, Mackay A, Truffaux N, Butterfield YS, Morozova O, Philippe C, Castel D, Grasso CS, Vinci M, Carvalho D, Carcaboso AM, de Torres C, Cruz O, Mora J, Entz-Werle N, Ingram WJ, Monje M, Hargrave D, Bullock AN, Puget S, Yip S, Jones C, Grill J. Recurrent activating ACVR1 mutations in diffuse intrinsic pontine glioma. Nature genetics. 2014;46:457–461. doi: 10.1038/ng.2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zadeh G, Aldape K. ACVR1 mutations and the genomic landscape of pediatric diffuse glioma. Nature genetics. 2014;46:421–422. doi: 10.1038/ng.2970. [DOI] [PubMed] [Google Scholar]
- 14.Burgess R, Jenkins R, Zhang Z. Epigenetic changes in gliomas. Cancer Biol Ther. 2008;7:1326–1334. doi: 10.4161/cbt.7.9.6992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Plath K, Fang J, Mlynarczyk-Evans SK, Cao R, Worringer KA, Wang H, de la Cruz CC, Otte AP, Panning B, Zhang Y. Role of histone H3 lysine 27 methylation in X inactivation. Science. 2003;300:131–135. doi: 10.1126/science.1084274. [DOI] [PubMed] [Google Scholar]
- 16.Zhang Y, Reinberg D. Transcription regulation by histone methylation: interplay between different covalent modifications of the core histone tails. Genes & development. 2001;15:2343–2360. doi: 10.1101/gad.927301. [DOI] [PubMed] [Google Scholar]
- 17.Chan KM, Fang D, Gan H, Hashizume R, Yu C, Schroeder M, Gupta N, Mueller S, James CD, Jenkins R, Sarkaria J, Zhang Z. The histone H3.3K27M mutation in pediatric glioma reprograms H3K27 methylation and gene expression. Genes & development. 2013;27:985–990. doi: 10.1101/gad.217778.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chan KM, Han J, Fang D, Gan H, Zhang Z. A lesson learned from the H3.3K27M mutation found in pediatric glioma: a new approach to the study of the function of histone modifications in vivo? Cell cycle. 2013;12:2546–2552. doi: 10.4161/cc.25625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lewis PW, Muller MM, Koletsky MS, Cordero F, Lin S, Banaszynski LA, Garcia BA, Muir TW, Becher OJ, Allis CD. Inhibition of PRC2 activity by a gain-of-function H3 mutation found in pediatric glioblastoma. Science. 2013;340:857–861. doi: 10.1126/science.1232245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bender S, Tang Y, Lindroth AM, Hovestadt V, Jones DT, Kool M, Zapatka M, Northcott PA, Sturm D, Wang W, Radlwimmer B, Hojfeldt JW, Truffaux N, Castel D, Schubert S, Ryzhova M, Seker-Cin H, Gronych J, Johann PD, Stark S, Meyer J, Milde T, Schuhmann M, Ebinger M, Monoranu CM, Ponnuswami A, Chen S, Jones C, Witt O, Collins VP, von Deimling A, Jabado N, Puget S, Grill J, Helin K, Korshunov A, Lichter P, Monje M, Plass C, Cho YJ, Pfister SM. Reduced H3K27me3 and DNA hypomethylation are major drivers of gene expression in K27M mutant pediatric high-grade gliomas. Cancer Cell. 2013;24:660–672. doi: 10.1016/j.ccr.2013.10.006. [DOI] [PubMed] [Google Scholar]
- 21.Venneti S, Garimella MT, Sullivan LM, Martinez D, Huse JT, Heguy A, Santi M, Thompson CB, Judkins AR. Evaluation of histone 3 lysine 27 trimethylation (H3K27me3) and enhancer of Zest 2 (EZH2) in pediatric glial and glioneuronal tumors shows decreased H3K27me3 in H3F3A K27M mutant glioblastomas. Brain Pathol. 2013;23:558–564. doi: 10.1111/bpa.12042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morlan J, Baker J, Sinicropi D. Mutation detection by real-time PCR: a simple, robust and highly selective method. PloS one. 2009;4:e4584. doi: 10.1371/journal.pone.0004584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bechet D, Gielen GG, Korshunov A, Pfister SM, Rousso C, Faury D, Fiset PO, Benlimane N, Lewis PW, Lu C, David Allis C, Kieran MW, Ligon KL, Pietsch T, Ellezam B, Albrecht S, Jabado N. Specific detection of methionine 27 mutation in histone 3 variants (H3K27M) in fixed tissue from high-grade astrocytomas. Acta Neuropathol. 2014;128:733–741. doi: 10.1007/s00401-014-1337-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Venneti S, Santi M, Felicella MM, Yarilin D, Phillips JJ, Sullivan LM, Martinez D, Perry A, Lewis PW, Thompson CB, Judkins AR. A sensitive and specific histopathologic prognostic marker for H3F3A K27M mutant pediatric glioblastomas. Acta Neuropathol. 2014;128:743–753. doi: 10.1007/s00401-014-1338-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Behjati S, Tarpey PS, Presneau N, Scheipl S, Pillay N, Van Loo P, Wedge DC, Cooke SL, Gundem G, Davies H, Nik-Zainal S, Martin S, McLaren S, Goodie V, Robinson B, Butler A, Teague JW, Halai D, Khatri B, Myklebost O, Baumhoer D, Jundt G, Hamoudi R, Tirabosco R, Amary MF, Futreal PA, Stratton MR, Campbell PJ, Flanagan AM. Distinct H3F3A and H3F3B driver mutations define chondroblastoma and giant cell tumor of bone. Nature genetics. 2013;45:1479–1482. doi: 10.1038/ng.2814. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.