Abstract
Pseudomonas aeruginosa is an opportunistic pathogen that induces severe lung infections such as ventilator-associated pneumonia and acute lung injury. Under these conditions, the bacterium diminishes epithelial integrity and inhibits tissue repair mechanisms, leading to persistent infections. Understanding the involved bacterial virulence factors and their mode of action is essential for the development of new therapeutic approaches.
In our study we discovered a so far unknown effect of the P. aeruginosa lectin LecB on host cell physiology. LecB alone was sufficient to attenuate migration and proliferation of human lung epithelial cells and to induce transcriptional activity of NF-κB. These effects are characteristic of impaired tissue repair. Moreover, we found a strong degradation of β-catenin, which was partially recovered by the proteasome inhibitor lactacystin. In addition, LecB induced loss of cell–cell contacts and reduced expression of the β-catenin targets c-myc and cyclin D1. Blocking of LecB binding to host cell plasma membrane receptors by soluble l-fucose prevented these changes in host cell behavior and signaling, and thereby provides a powerful strategy to suppress LecB function.
Our findings suggest that P. aeruginosa employs LecB as a virulence factor to induce β-catenin degradation, which then represses processes that are directly linked to tissue recovery.
Keywords: NF-κB, Wnt, Acute lung injury, Migration, Proliferation, Bacterial pathogenesis
Highlights
-
•
The Pseudomonas aeruginosa lectin LecB reduces lung cell migration and proliferation
-
•
LecB also induces NF-kB-mediated inflammation in lung epithelial cells
-
•
ß-catenin degradation and reduction of c-myc and cyclin D1 protein levels inhibit cell proliferation and migration
-
•
Blocking of LecB binding to host cell receptors with L-fucose suppresses all effects caused by LecB
1. Introduction
Hospital-acquired infections are serious issues in human health and therapy. One of the most common nosocomial pathogens is Pseudomonas aeruginosa [1], [2]. Infections with this bacterium result in epithelial damage and affected tissues regenerate only slowly. For instance, P. aeruginosa causes ventilator-associated pneumonia leading to acute lung injury (ALI), which is characterized by loss of epithelial and endothelial integrity inside the alveoli, and strong induction of inflammation [3], [4]. During normal tissue repair, inflammation markers are reduced facilitating cell proliferation and proper tissue regeneration [5]. However, upon bacterial infection, high and persistent levels of inflammation markers (e.g. TNF-α) can undermine repair mechanisms [6].
To date, several P. aeruginosa virulence factors are known to influence host cell processes that are implicated in tissue repair [4], [7]. The P. aeruginosa lectin LecB was previously described as adhesion factor with a high binding affinity for l-fucose and its derivatives [8]. It occurs as a tetramer, is mainly found at the outer bacterial membrane and is involved in biofilm formation [9], [10]. Moreover, LecB agglutinates human peripheral lymphocytes, decreases the ciliary beating frequency of nasal epithelial cells and binds and stimulates follicular lymphoma cells [11], [12], [13]. Interestingly, a LecB-deficient mutant of P. aeruginosa and the LecB-inhibition with soluble carbohydrate ligands decreased the bacterial burden and dissemination in an in vivo murine model of ALI [14], [15].
Beta-catenin signaling is critically implicated in tissue homeostasis and repair [16], [17], [18]. As part of adherens junctions β-catenin stabilizes cell–cell contacts by connecting cadherins to the actin cytoskeleton. These junctional complexes can be further promoted and stabilized through interaction with α3β1-integrins and CD151-tetraspanins [19]. In addition, β-catenin is the central player of the canonical Wnt signaling, in which it translocates to the nucleus upon Wnt activation resulting in transcriptional regulation of target genes [20], [21]. Thereby, it promotes proliferation and migration during development, tissue homeostasis and repair [22], [23]. Without stimulation of Wnt ligands, the cytosolic β-catenin pool is continuously marked for proteasomal degradation by a destruction complex containing GSK-3β and Axin [20], [21]. Several studies also reported a regulatory function of β-catenin during inflammation. For instance, during infection with Salmonella typhimurium or Mycobacterium tuberculosis β-catenin was described as antagonist of NF-κB, which itself is a strong inducer of inflammatory processes [24], [25], [26]. In an in vivo keratitis model β-catenin degradation has also been demonstrated for P. aeruginosa infection [27]. The thereby induced strong inflammation diminished recovery and therapeutic success. However, no P. aeruginosa factors able to manipulate cellular β-catenin protein levels have been identified so far.
In this study, we show that soluble LecB caused β-catenin degradation together with attenuation of cell migration and proliferation in lung epithelial cells. At the same time, we observed an activation of NF-κB signaling and an increase in TNF-α expression indicating an induction of inflammation. Based on these results we propose that LecB has a role in disturbing cellular repair processes in order to facilitate robust and enduring colonization and infection of injured tissue.
2. Results
2.1. P. aeruginosa lectin LecB attenuates cell proliferation and migration and activates NF-κB p65
P. aeruginosa is one of the most common pathogens found in ALI and infected wounds [1], [4]. In order to assay if LecB is sufficient to inhibit tissue repair, we initially investigated three crucial host cell processes: migration, inflammation and proliferation. We chose the lung epithelial cell line H1299 as a model system because P. aeruginosa frequently infects lung tissue. For stimulation we used recombinantly produced, soluble LecB.
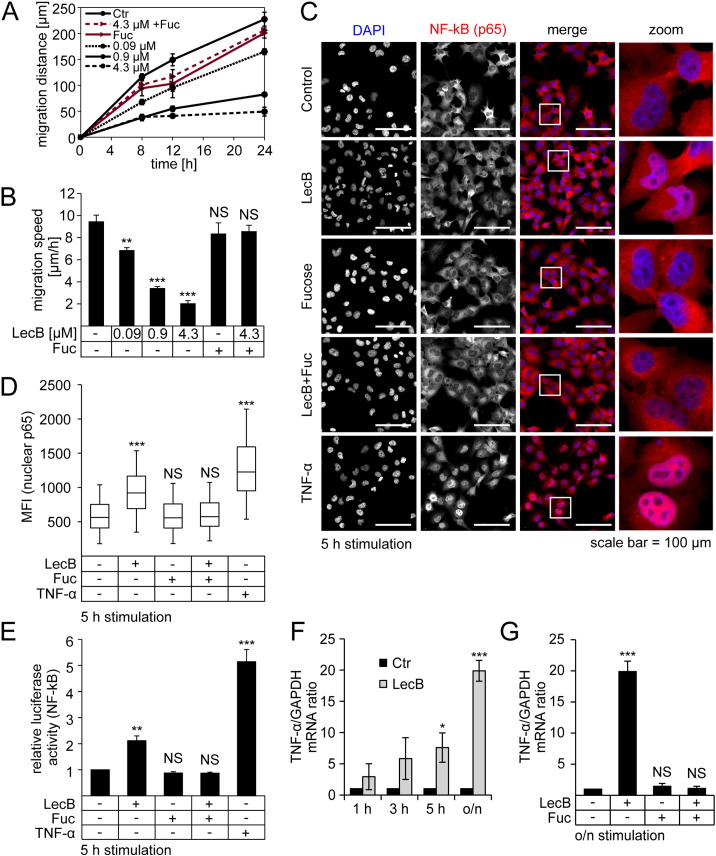
To measure the migratory activity of cells we performed conventional scratch assays. We observed a pronounced decrease in migration distance and speed during LecB treatment in a dose-dependent manner, whereas used concentrations ranged from 0.09 μM to 4.3 μM of LecB (Fig. 1A and B). Since the inhibitory effect was most pronounced with 4.3 μM LecB, the following experiments were conducted with this concentration. As control, co-incubation with 43 mM l-fucose was performed in order to saturate the fucose binding sites at LecB, which, in turn, inhibited the interaction between the lectin and host cell receptors. As shown in Fig. 1A (red lines) and Fig. 1B co-incubation with l-fucose rescued cell migration.
Fig. 1.
LecB attenuates migration and induces NF-κB nuclear translocation, transcriptional activity and TNF-α expression.
(A) H1299 cells were grown to confluence, scratched and stimulated with LecB (0.09 μM to 4.3 μM) in the presence or absence of l-fucose (43 mM). Distances between the cell fronts were monitored after 8, 12 and 24 h. The absolute migrated distance after different time periods as well as (B) the mean migration speed after indicated treatments is depicted. (C) Cells were treated as indicated and analyzed by confocal fluorescence microscopy with immunostaining for NF-κB-p65 (red) and counterstaining for DNA (DAPI, blue). White squares highlight areas that are further magnified in the right column. (D) Microscopy samples were prepared as described in (C) and nuclear location of p65 was quantified by estimating the nuclear mean fluorescence intensity (MFI) of p65. (E) Cells were transfected with NF-κB luciferase reporter plasmid and treated as indicated for 5 h. Luciferase activity was normalized to the protein concentration. (F) Cells were treated as indicated and mRNA levels of TNF-α were estimated by qPCR, normalized to GAPDH mRNA levels and are presented in relation to the corresponding control. TNF-α expression level after different time periods as well as (G) the expression after 24 h in the presence and absence of l-fucose is depicted. Except for (D) all values represent the mean of at least three independent experiments ± SEM. For (D) median values of three independent experiments with lower and higher quartile are shown. Error bars represent 1.5 interquartile range (IQR) values. Asterisks in all graphs indicate the statistical significance compared to the untreated control.
In order to investigate inflammation we focused on NF-κB signaling, which has been described to be induced by P. aeruginosa [28]. During its activation, NF-κB is translocated from the cytosol to the nucleus. To determine nuclear translocation of NF-κB, cells were co-stained for p65, the most common NF-κB protein in inflammation, and DAPI to visualize nuclei (Fig. 1C). Mean fluorescence intensity (MFI) values of p65 inside the nuclei were measured to quantify nuclear translocation (Fig. 1D). As seen in Fig. 1C and D, LecB and TNF-α, as positive control, resulted in significantly higher nuclear MFI values of p65. We also analyzed if LecB treatment influences the phosphorylation status of p65 (Suppl. Fig. 1A and 1B). Upon LecB treatment, phosphorylation at Ser536 was measurably elevated after 30 min and continuously increased for longer treatment times. Ser536 phosphorylation of p65 has been described to be important for increased transcriptional activity and acetylation of NF-κB p65 [29], [30]. In contrast, the phosphorylation site at Ser468 was unaffected during LecB treatment. Phosphorylation at Ser468 has been shown to usually have suppressive effects on NF-κB activity [31]. Therefore, the constant phosphorylation at Ser468 argues neither for nor against an activation of p65. However, compared to TNF-α, which is able to induce NF-κB activation within minutes [32], LecB-mediated NF-κB responses appear to start later (after approximately 30 min, Suppl. Fig. 1A and 1B). In order to verify if the observed phosphorylation and nuclear translocation of p65 also results in increased transcriptional activity, we transfected cells with a NF-κB luciferase reporter plasmid and analyzed the resulting luciferase expression [33]. Indeed, we measured a two-fold increase of NF-κB reporter activity upon 5 h of LecB stimulation, whereas TNF-α incubation elevated the reporter activity to a five times higher level (Fig. 1E). Again, co-incubation with LecB and l-fucose blocked both nuclear translocation and luciferase activity (Fig. 1C–E). Finally, we examined whether the increased transcriptional activity results in an enhanced expression of NF-κB targets. To this end, we analyzed mRNA levels of TNF-α, since it is a common target of NF-κB and is frequently associated with delayed tissue recovery [3], [6]. As depicted in Fig. 1F, LecB stimulation continuously elevated TNF-α mRNA levels resulting in a 20-fold induction after overnight incubation (16 h). In the presence of l-fucose mRNA levels of TNF-α remained at control levels (Fig. 1G).
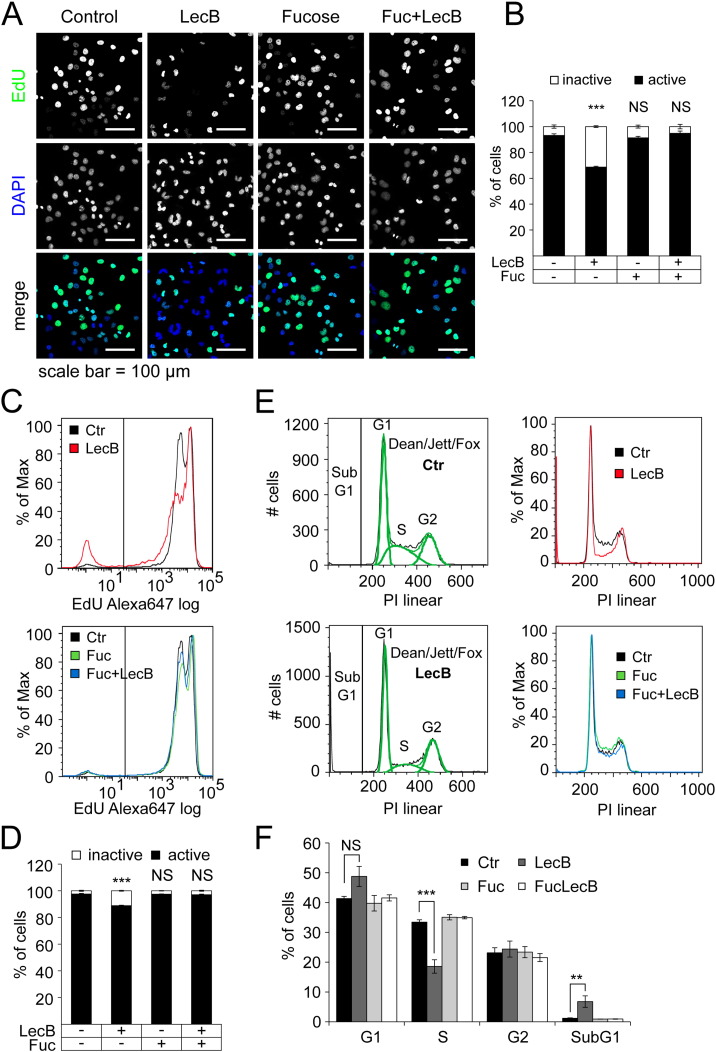
For analysis of the proliferative activity of LecB-treated cells, we quantified the incorporation of a thymidine derivative (EdU), which enables to determine DNA synthesis during proliferation. EdU was labeled with Alexa488 for confocal fluorescence microscopy and with Alexa647 for FACS analysis. For analysis of microscopy data nuclei were identified using DAPI co-staining. The nuclear MFI of EdU-Alexa488 was measured and used for classification of the proliferative activity (Fig. 2A and B). After 16 h of incubation with LecB, the amount of proliferative active cells was decreased from 93% to 69%, whereas co-incubation with l-fucose prevented this effect (Fig. 2B). In FACS analysis, we were able to identify clearly EdU-positive and -negative populations (Fig. 2C). Again, EdU-positive cells decline from 98% to 88% upon LecB treatment (Fig. 2D). The slight differences between the basic levels and differences of EdU-positive cells in microscopy and FACS approaches are probably due to varying procedures of staining and analysis. Interestingly, the FACS data showed two peaks for EdU-positive cells, whereas only the lower intensity peak was decreased upon LecB treatment. This suggests that cells exhibit different susceptibility to LecB depending on the phase of cell cycle. In order to test this hypothesis, we used a propidium iodide (PI) staining to determine the phase of cell cycle by measuring cellular DNA content. For quantification, the Dean/Jett/Fox algorithm (FlowJo) was used (Fig. 2E, left). Cells with less than diploid DNA content (SubG1) represent apoptotic cells with degraded DNA and were quantified separately. LecB treatment resulted in a significant decrease of the fraction of S phase cells from 33% to 19% together with an increase of cells with SubG1 DNA content from 1% to 6% (Fig. 2E, right and Fig. 2F). In addition, a clear, but not significant, increase of cells in G1 phase from 41% to 49% was observed. This may indicate a deregulation of G1 to S phase transition, partially leading to DNA degradation, which is typically followed by cell death. Again, co-incubation with l-fucose prevented LecB-induced effects (Fig. 2C–F). In order to elucidate whether the effects on proliferation are accompanied by induction of apoptosis, a luminescent caspase assay was performed. The results showed that activity of caspase 3 and 7 upon LecB treatment were similar to the untreated negative control, whereas incubation with 1 μM staurosporine significantly increased caspase 3 and 7 activity by 2.5- to 3-fold after 5 h and 16 h, respectively (Suppl. Fig. 1C). Thus, decreased proliferation is not caused by induction of apoptosis during LecB stimulation.
Fig. 2.
LecB attenuates proliferation and reduces number of cells in the S-phase.
All stimulations were performed with 4.3 μM LecB and 43 mM l-fucose. (A) Cells were treated with indicated stimuli in the presence of EdU (10 μM) for 16 h and analyzed by confocal fluorescence microscopy. EdU was linked to Alexa488 (green) and cells were counterstained with DAPI (blue). Representative images as well as (B) quantification of proliferative activity are depicted. The nuclear MFI of EdU was used as a measure for the proliferative activity. (C) Cells were treated with indicated stimuli in the presence of EdU (10 μM) for 16 h. EdU was linked to Alexa647 and cells were analyzed by FACS. Representative histograms as well as (D) quantification of proliferative activity are shown. Cells above the threshold that is delineated in (C) as vertical line were regarded as proliferative active. (E) Cells were treated with indicated stimuli for 16 h and stained for their DNA content with Propidium iodide (PI). Percentages of cells in each cell cycle phase were determined by Dean/Jett/Fox algorithm (FlowJo). Representative histograms with algorithm (left), different stimuli (right) as well as (F) percentage of cells in each cycle phase are shown. As SubG1 cells are not considered in Dean/Jett/Fox algorithm they were quantified separately by gating for cells with less DNA content than for G1 cells. All values represent the mean of at least three independent experiments ± SEM. Asterisks indicate the statistical significance compared to the untreated control.
To exclude that the described effects are unique to H1299 cells, key experiments were also performed in other cell lines. LecB inhibited cell migration and induced NF-κB nuclear translocation to a comparable extent in another lung epithelial cell line (H1975, Suppl. Fig. 2A–C). Furthermore, the repressing effects of LecB on proliferative activity have been also observed in the acute myeloid leukemia cell line THP-1 [34].
Taken together, our experiments demonstrate that soluble LecB is sufficient to strongly attenuate cell migration and proliferation and to induce NF-κB signaling.
2.2. LecB induces proteasomal β-catenin degradation depending on GSK-3β-activity
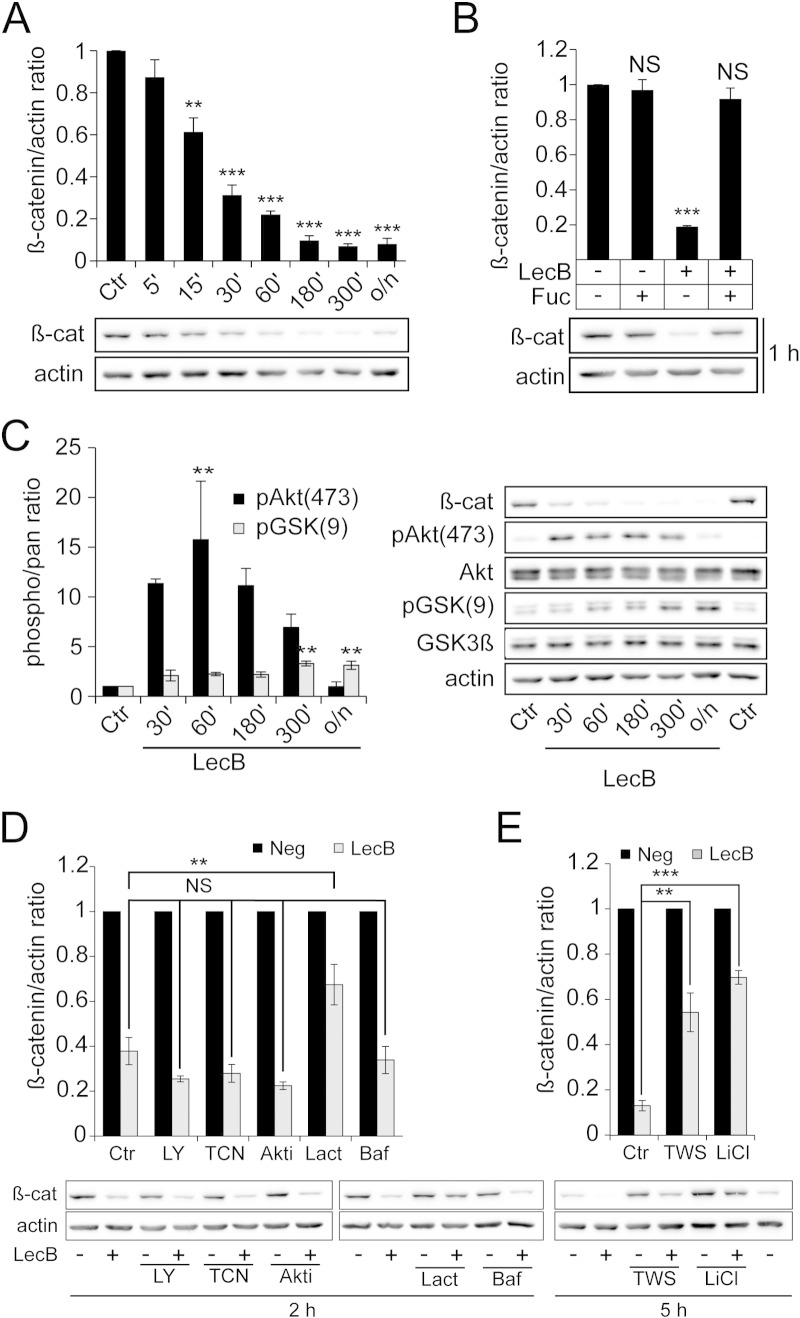
Since β-catenin is a key regulator of proliferation and migratory processes the total protein amount of β-catenin was estimated by Western blot analysis. LecB stimulation decreased β-catenin protein levels within 60 min to 20% of the initial protein amount (Fig. 3A). For longer incubation periods the protein levels remained below 10% of the initial expression level, whereas addition of l-fucose prevented β-catenin reduction by inhibiting LecB binding to host cells (Fig. 3B). Comparable LecB-mediated β-catenin degradation was also observed in H1975 (Suppl. Fig. 2D) and in THP-1 cells [34]. Degradation of β-catenin is induced by GSK-3β-dependent phosphorylation, which is negatively regulated by Akt through Ser9 phosphorylation. Since Akt activity has been reported to be induced by P. aeruginosa [35] we assayed the activating phosphorylation of Akt (Ser473) and the inhibitory phosphorylation of GSK-3β (Ser9) in response to LecB treatment. Indeed, LecB was able to induce Akt activation, as well as inhibitory phosphorylation of GSK-3β (Fig. 3C). Interestingly, the kinetics of both phosphorylations were different. Whereas Akt phosphorylation showed a peak after 60 min and reached control level after overnight incubations (16 h), GSK-3β phosphorylation continuously increased until 5 h and remained at high level during overnight experiments. The LecB-induced inhibitory phosphorylation of GSK-3β would be expected to cause an accumulation of β-catenin. However, our previous experiments (Fig. 3A) showed that LecB stimulation resulted in β-catenin degradation. In order to resolve this contradiction, we carried out experiments with different inhibitors of Akt (and its upstream kinase PI3K) and GSK-3β upon stimulation with LecB. Since some of the inhibitors alone already changed basic β-catenin levels, all LecB stimulation results were related to the corresponding control containing only the inhibitor. As seen in Fig. 3D, inhibition of PI3K and Akt signaling by LY294002 (LY: 100 μM) and Akt inhibitors (triciribine, TCN: 10 μM and Akt1/2 inhibitor, Akti: 10 μM), respectively, did not influence the LecB-induced β-catenin degradation indicating an independency from PI3K/Akt signaling. In contrast, an inhibition of GSK-3β activity using lithium chloride (LiCl: 20 mM) and TWS119 (TWS: 20 μM) partially blocked LecB-induced β-catenin degradation by elevating the protein level from 10% to 50% and 70% of the initial protein level, respectively (Fig. 3E). These experiments argue for an importance of GSK-3β activity for LecB-induced β-catenin degradation, whereas the inhibitory effect of Akt on GSK-3β may be blocked or circumvented by LecB.
Fig. 3.
LecB induces proteasomal β-catenin degradation, which requires GSK-3β activity but is independent from Akt signaling.
All stimulations were performed with 4.3 μM LecB and 43 mM l-fucose. All graphs present densitometric quantification of protein levels performed with ImageJ and corresponding representative blots. Protein and phosphorylation levels were normalized to actin or pan staining and actin, respectively. (A) H1299 cells were treated as indicated for different time periods and β-catenin protein level is depicted. (B) Cells were treated as indicated for 1 h and β-catenin protein level is depicted. (C) Cells were stimulated as indicated for different time periods and phosphorylation levels of Akt and GSK-3β are shown. (D) Cells were stimulated for 2 h with LecB and inhibitors of the proteasome (lactacystin, Lact, 10 μM), lysosomes/vacuolar H+-ATPase (bafilomycin, Baf, 100 μM), PI3K (LY294002, LY, 100 μM) and Akt (triciribine, TCN, 10 μM; Akt1/2 inhibitor, Akti, 10 μM). Inhibitors were pre-incubated for 30 min at 37 °C and maintained during stimulation. As inhibitors changed the basic levels of β-catenin, LecB samples are depicted in relation to controls containing only the inhibitor. (E) Cells were stimulated for 5 h with LecB and inhibitors of GSK-3β (TWS119, TWS, 20 μM and lithium chloride, LiCl, 20 mM). Inhibitors were incubated and data are presented as described in (D). All values represent the mean of at least three independent experiments ± SEM. Asterisks indicate the statistical significance compared to control samples.
In addition, we applied inhibitors of proteasomal and lysosomal degradation to elucidate the pathway of β-catenin depletion. Lysosomal inhibition by bafilomycin (Baf: 0.2 μM) had no effect on β-catenin degradation, whereas proteasomal inhibition using lactacystin (Lact: 10 μM) increased the protein level from 40% to 70% (Fig. 3D). This suggests that β-catenin is degraded by proteasomes during LecB stimulation.
The cellular amount of β-catenin is regulated by an equilibrium between continuous de-novo synthesis and degradation of protein excess. In order to elucidate whether suppression of de-novo protein synthesis modulates LecB-induced β-catenin degradation, cells were co-incubated with cycloheximide (Cyclo: 100 μg/ml), a protein synthesis inhibitor. The presence of cycloheximide did not significantly influence LecB-induced β-catenin degradation after 3 h and 5 h of treatment (Suppl. Fig. 3A and B). These data show that de-novo protein synthesis only marginally influences cellular β-catenin levels during LecB treatment. In summary, LecB induced proteasomal degradation of β-catenin, which was independent from Akt signaling, whereas GSK-3β activity was required to reach full degradation.
2.3. LecB stimulation reduces cell–cell contacts and promotes degradation of β1-integrin
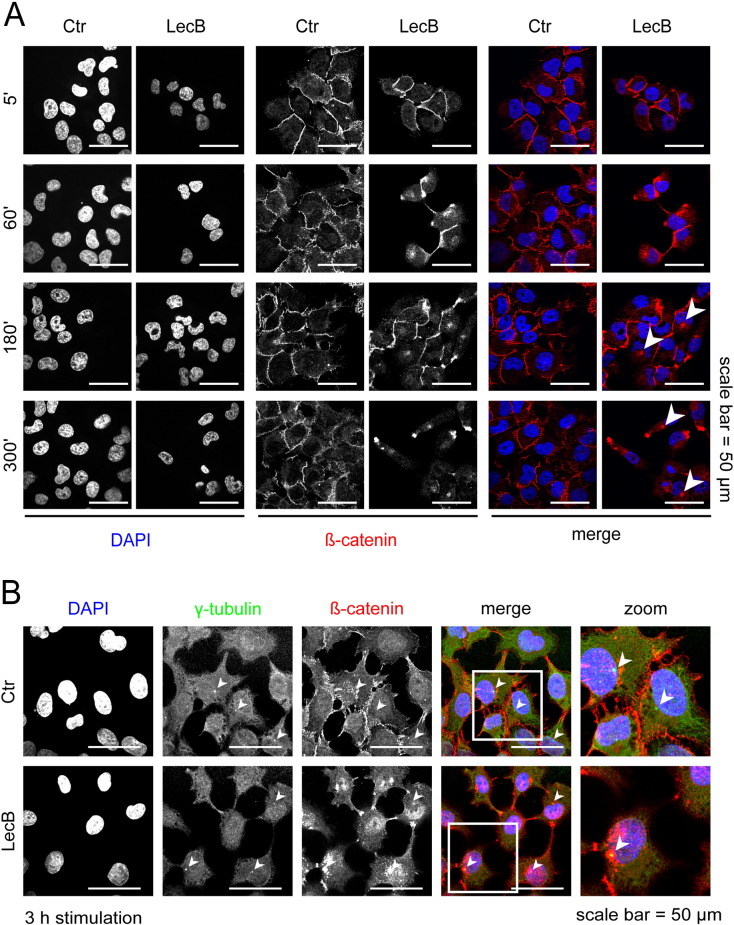
To elucidate if the membrane-bound pool of β-catenin is affected by LecB, we followed the localization of β-catenin upon LecB stimulation using confocal fluorescence microscopy. We observed a reduction of β-catenin staining at cell–cell contacts and an accumulation of β-catenin close to nuclei after 3 to 5 h of LecB stimulation, indicating that β-catenin was removed from the plasma membrane (Fig. 4A, arrowheads). Since cytosolic proteasomes are mainly localized at centrosome [36] samples were co-stained for γ-tubulin as centrosomal marker. We observed a clear accumulation of β-catenin around centrosomes (Fig. 4B), which further supports the role of proteasomes in β-catenin degradation.
Fig. 4.
LecB reduces cell–cell contacts and β-catenin locates to the centrosome.
All experiments were performed with 4.3 μM LecB. (A) Cells were stimulated as indicated for different time periods and analyzed by confocal fluorescence microscopy with immunostaining for β-catenin (red) and counterstaining for DNA (DAPI, blue). Arrowheads highlight the perinuclear accumulation of β-catenin. (B) Cells were treated as indicated for 3 h and analyzed by confocal fluorescence microscopy with immunostainings for γ-tubulin (green), β-catenin (red) and counterstaining for DNA (DAPI, blue). In order to visualize centrosomes of multiple focal planes, maximum intensity projections of 4 Z-stacks are depicted. White squares highlight areas that are further magnified in the right column. Arrowheads highlight the localization of centrosomes.
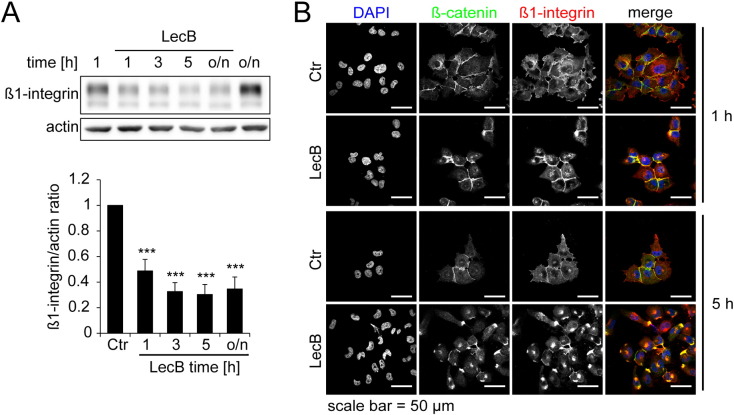
Interestingly, Western blot experiments showed that LecB also induces a reduction of the cell adhesion factor β1-integrin to 50% after 1 h, which remained at 30% afterwards (Fig. 5A and B). As depicted in Fig. 5B, we observed a strong internalization of β1-integrin to the same location as β-catenin after LecB treatment.
Fig. 5.
Beta1-integrin is degraded and locates to the perinuclear region.
All treatments were performed with 4.3 μM LecB. (A) Cells were treated as indicated for different time periods and β1-integrin protein levels were determined by Western blot analysis and densitometric quantification using ImageJ. Protein levels were normalized to actin. Representative blots (above) and quantification data (below) are depicted. Values represent the mean of at least three independent experiments ± SEM. Asterisks indicate the statistical significance. (B) Cells were treated as indicated for 1 h and 5 h and analyzed by confocal fluorescence microscopy with immunostainings for β-catenin (green), β1-integrin (red) and counterstaining for DNA (DAPI, blue).
In summary, LecB reduces β-catenin at cell–cell adhesion sites and induces an accumulation of the protein close to centrosomes, where a high abundance of proteasomes is described [36]. In addition, β1-integrin is internalized to the same location as β-catenin and also degraded.
2.4. LecB acts antagonistically to Wnt signaling and reduces protein levels of Wnt targets
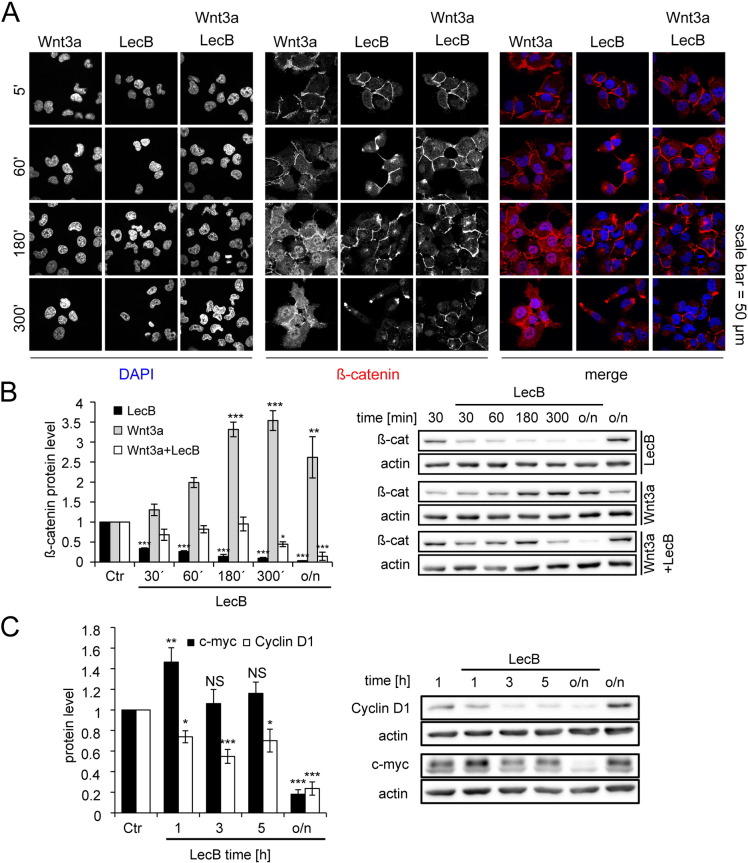
The cytosolic and the nuclear pool of β-catenin mediate its major effects on migration, proliferation and inflammation. Thus, we also examined effects of LecB on the free, cytosolic pool of β-catenin. For this, we co-stimulated the cells with Wnt3a (400 ng/ml), a prototypical inducer of β-catenin accumulation and its nuclear translocation. In immunofluorescence experiments, Wnt3a-stimulated cells showed a higher cytosolic signal of β-catenin, as well as an increased nuclear translocation compared to control cells (Fig. 6A). This accumulation of β-catenin was also confirmed in Western blot analysis, where the protein level was continuously increased until a maximum of 3.5 fold induction was achieved after 5 h (Fig. 6B). Again, LecB treatment reduced the β-catenin protein level, as seen in immunofluorescence experiments (Fig. 6A) as well as in Western blot analyses (Fig. 6B). Interestingly, co-incubation of Wnt3a and LecB completely prevented the Wnt3a-induced accumulation and nuclear translocation of β-catenin, but also slightly reduced the LecB-mediated loss of cell–cell contacts (Fig. 6A). Furthermore, Western blot experiments showed that co-incubation prevented β-catenin accumulation but also delayed β-catenin degradation (Fig. 6B). These results point to a crosstalk between Wnt3a and LecB effects and suggest that LecB-induced β-catenin degradation is also manipulating the cytosolic pool of the protein. Since this may influence the transcriptional activity of β-catenin, we additionally performed luciferase assays with the β-catenin-responsive reporter plasmid 7TFP [37]. The assays showed a continuous decrease of transcriptional activity to around 50% after 5 h of LecB stimulation (Suppl. Fig. 3C). This reduction supports the hypothesis that the strong degradation of β-catenin also negatively affects its transcriptional activity. In order to further confirm these findings, we directly analyzed the protein levels of the β-catenin targets cyclin D1 and c-myc, which are also implicated in the regulation of migration and proliferation. Indeed, the cyclin D1 protein level was significantly decreased to 60% and 70% after 1 and 5 h and to 20% upon overnight incubation for 16 h (Fig. 6C). In contrast, c-myc showed different kinetics with a significant 1.4 fold elevation of the protein level after 1 h of LecB stimulation, which was reduced back to control levels after 3 and 5 h. However, c-myc levels were finally reduced to the same extent as cyclin D1 after overnight incubations with LecB (Fig. 6C). The analysis of other cyclins showed that the protein level of cyclin E2, whose expression directly depends on cyclin D1 accumulation and interaction with corresponding Cdks [38], was also decreased during LecB stimulation (Suppl. Fig. 3D and E). In contrast, cyclin B1, which is important during progression of M phase and is not directly influenced by cyclin D1, remained mainly unaffected (Suppl. Fig. 3D and E). During cell cycle the cyclin D1/Cdk4/6 complex can phosphorylate the Rb protein at Ser795 in order to inhibit its suppressive effect on transcription [39]. Interestingly, this phosphorylation was also strongly reduced upon LecB treatment (Suppl. Fig. 3D and E). Furthermore, the pan staining of the Rb protein showed a shift to a slightly lower molecular weight suggesting the loss of numerous modifications of this temporarily highly phosphorylated protein. The LecB-induced regulation of cyclin D1, cyclin E2 and Rb phosphorylation, which are all proteins involved in G1–S-phase transition, is in accordance with the data from Fig. 2E and F showing a reduction of cells in S-phase upon LecB treatment.
Fig. 6.
LecB antagonizes Wnt3a-induced β-catenin nuclear translocation and reduces protein levels of cyclin D1 and c-myc.
All treatments were performed with 4.3 μM LecB and 400 ng/ml Wnt3a. (A) Cells were treated as indicated for different time periods and analyzed by confocal fluorescence microscopy with immunostaining for β-catenin (red) and staining of DNA (DAPI, blue). (B) and (C) Cells were stimulated as indicated for different time periods and protein levels of β-catenin and cyclin D1/c-myc, respectively, were determined by Western blot analysis and densitometric quantification using ImageJ. Protein levels were normalized to actin. Representative blots (right) and quantification data (left) are depicted. All values represent the mean of at least three independent experiments ± SEM. Asterisks indicate the statistical significance compared to the untreated control.
Taken together, LecB is capable of repressing Wnt3a-induced β-catenin accumulation and nuclear translocation. Furthermore, overnight stimulation with LecB diminished protein levels of c-myc and cyclin D1, which are typical transcriptional targets of β-catenin. The reduction of cyclin D1 also affected the protein level of cyclin E2 and the phosphorylation state of the Rb protein.
3. Discussion
P. aeruginosa is a common opportunistic pathogen and causes acute lung injury in ventilator-associated pneumonia [4]. Upon these infections, the tissue integrity is destroyed and a strong inflammation is generated [3]. The re-establishment of the tissue structure requires a reduction of inflammation as well as an induction of cell proliferation and migration [5]. P. aeruginosa is able to repress these mechanisms, in order to facilitate bacterial dissemination into blood circulation, which may lead to fatal sepsis [4].
In this study, we investigated the effects of the P. aeruginosa lectin LecB on these repression mechanisms in a lung cell model and observed a pronounced attenuation of host cell migration and proliferation together with an induction of NF-κB-mediated signaling. According to these results, we propose LecB as a player in P. aeruginosa-induced delay of tissue repair. Furthermore, we observed a strong degradation of the cytosolic and the membrane pool of β-catenin during LecB stimulation, which may represent the molecular basis for the inhibition of tissue recovery.
An antagonistic regulation between β-catenin and inflammation during bacterial infection has already been described for P. aeruginosa and S. typhimurium [24], [27]. However, the responsible bacterial factors were not identified. In particular, in a murine P. aeruginosa keratitis model it was shown that the bacterium induces β-catenin degradation and a strong inflammation, leading to enduring tissue destruction even after complete bacterial clearance [27]. In addition, a prevention of β-catenin degradation by transfecting a non-degradable mutant diminished the induction of inflammation. Interestingly, the induced inflammatory genes were TNF-α, IL-6 and IL-1β, which are all under the control of NF-κB. In accordance, mice challenged with S. typhimurium showed β-catenin degradation in intestine samples, which was accompanied by an induction of NF-κB activity [24]. Again, a stabilization of β-catenin decreased the inflammatory induction during infection. Both studies support our hypothesis that the loss of β-catenin during LecB stimulation can induce the activation of NF-κB signaling (Fig. 1, Fig. 3). An induction of NF-κB activity through β-catenin degradation could also explain the late onset of LecB-mediated effects on NF-κB as compared to the immediate NF-κB activation by other molecules like TNF-α. Furthermore, fucose-binding lectins from other lung pathogens, like Burkholderia cenocepacia and Aspergillus fumigatus, which are not related in sequence to LecB, were also described to display pro-inflammatory effects [40], [41].
For the effects of LecB on cell migration β-catenin degradation may be a suitable explanation as well. Migration is a precise and dynamic interplay between a variety of processes, including cell–matrix attachment and detachment, cell–cell adhesion, cell polarity as well as cytoskeletal rearrangement [42]. Several studies identified GSK-3β as key regulator of migration, whereas its exact effect was dependent on the cellular model system. In particular, it was described that inhibition of GSK-3β directly reduces formation of extended lamellipodia and migration of keratinocytes, which supports a promoting effect of GSK-3β on cell migration [43]. In contrast, Karrasch et al. described an activation of Akt signaling with following inhibitory phosphorylation of GSK-3β, leading to increased transcriptional activity of β-catenin and by this enhanced migration of intestinal epithelial cells [44]. Interestingly, we also observed Akt activation and inhibitory GSK-3β phosphorylation, but without the following increase in β-catenin protein level and migration (Fig. 3). Therefore, it is possible that the P. aeruginosa lectin LecB induces parallel pathways in order to evade the induction of migration, which can facilitate the establishment and stabilization of bacterial infection. In support of this, it was proposed that, depending on the interaction partner of GSK-3β, a β-catenin binding and phosphorylation is structurally possible, even if the inhibitory phosphorylation of GSK-3β is present [45]. Furthermore, it was hypothesized that GSK-3β phosphorylation is prevented if the protein is bound to Axin within the destruction complex [46].
Also integrins were reported to play a crucial role in cell migration [47]. For instance, β1-integrin was described to stabilize the primary lamellipodia and by this promoting migration in a complex with α3-integrin [48]. Besides the main localization at cell–matrix contacts, α3β1-integrins were also found to stimulate and localize to cadherin-catenin complexes together with tetraspanin CD151 [19]. CD151 itself was reported to suppress cell migration, when it is released from integrins [49]. Hence, the described depletion of β1-integrin (Fig. 5) and its removal from cell–cell and cell–matrix contacts, may contribute to decreased migration.
Cell proliferation constitutes a central mechanism in cell maintenance, tissue homeostasis and repair. Our observation of fewer cells in S phase and more cells in the G1 and SubG1 population upon LecB stimulation indicate a normal cell cycle progression of S phase cells, but a lack of new cells entering this phase (Fig. 2). This effect argues for an arrest at the G1-S transition point. As cyclin D1 is a central inducer of this transition, the observed down-regulation of this and other related proteins like cyclin E2 and Rb supports the hypothesis of a G1-S phase arrest upon LecB treatment, which leads to attenuated proliferation and minor cell death (SubG1) (Fig. 2, Fig. 6, Suppl. Fig. 3). Indeed, several studies showed that a downregulation of β-catenin by upstream regulators or shRNA decreases cyclin D1 expression [50], [51], which supports the hypothesis that β-catenin depletion directly contributes to reduced proliferation.
We propose that LecB may contribute to ALI and can work together with other P. aeruginosa cytotoxins, like ExoU, ExoT and LepA, which may also play roles in ALI and inflammation [52], [53], [54]. Our hypothesis of an impact of LecB on ALI is further supported by a study, which describes a decrease in bacterial burden and dissemination in an in vivo murine model of ALI during infections with a LecB-deficient mutant or LecB-inhibition with specific glycoconjugates [14], [15]. In accordance, it was shown that LecB and other bacterial lectins can efficiently be blocked by synthetic glycoconjugates [14], [55], [56], [57].
Our study not only broadens the knowledge how the catalytically inactive lectin LecB severely changes host cell signaling and may contribute to pathogenicity, but also provides additional rationale for using synthetic LecB-binding glycoconjugates for alternative therapies of P. aeruginosa infections.
4. Materials and methods
4.1. Cell cultivation, stimulation and inhibitors
H1299 and H1975 lung epithelial cells were grown at 37 °C and 5% CO2 in RPMI-1640 (Gibco) with 10% FCS and 2 mM l-glutamine (Gibco).
Lyophilized, recombinant LecB (Dr. Anne Imberty) was dissolved in PBS (with Ca/Mg) and used at concentrations of 0.09 μM, 0.9 μM and 4.3 μM for indicated time periods. Starvation in serum-free medium was performed, when indicated, for 2 h before treatment and maintained during stimulation. In order to block fucose-binding sites of LecB, l-fucose (Sigma-Aldrich) was directly dissolved in medium, sterile filtered and used at a final concentration of 43 mM for the same time periods as LecB. Other proteins for stimulation purposes were used at the following concentrations: Wnt3a (400 ng/ml, R&D Systems), TNF-α (10 ng/ml, Life Technologies) and staurosporine (Stauro, 1 μM, Sigma-Aldrich). The used inhibitors were pre-incubated for 30 min at 37 °C and maintained during stimulation at following concentrations: lactacystin (Lact, 10 μM, Biomol), bafilomycin (Baf, 0.2 μM, Invivogen), LY294002 (LY, 100 μM, Sigma-Aldrich), triciribine (TCN, 10 μM, Sigma-Aldrich), Akt1/2 inhibitor (Akti, 10 μM, Sigma-Aldrich), TWS119 (TWS, 20 μM, Selleckchem), lithium chloride (LiCl, 20 mM, Roth) and cycloheximide (Cyclo, 100 μg/ml, Sigma-Aldrich).
4.2. Migration assay
H1299 or H1975 cells were grown to confluence and scraped using a sterile pipet tip. Same positions of cell gaps with an approximate width of 500 μm were monitored using EVOS microscope (peqlab) after indicated time points. To minimize the effects of proliferation during this assay, it was performed under serum starvation. Distances between both cell fronts were measured using the vector graphic software Inkscape (Open Source, version 0.48).
4.3. PI cell cycle test
Cells were grown to 50% confluence and stimulated as indicated for 16 h. For staining, cells were trypsinized, nuclei were isolated and stained with the Cycletest™ Plus DNA Reagent Kit (BD Biosciences) according to the manufacturer's instructions. The PI fluorescence of the nuclei was measured using the Beckman Coulter Gallios™ Flow Cytometer. Analysis was performed with FlowJo software (version X 10.0.7r2). Dot plots displaying peak versus area values of forward scatter (FS) and sideward scatter (SS) were used to gate for single nuclei in order to exclude aggregates from analysis. Subsets of G1, S and G2 phase were estimated by Dean/Jett/Fox algorithm of FlowJo software. Not considered SubG1 subsets were assessed separately by gating for cells with less DNA content than for G1. Per experiment and condition 20.000 events were measured.
4.4. EdU proliferation assay
For investigating EdU incorporation, Click-iT® EdU assays (Life Technologies) for imaging and flow cytometry were used.
For microscopy experiments cells were seeded on glass coverslips, grown to 50% confluence and were incubated in the presence of 10 μM EdU and the distinct stimulus for 16 h. Afterwards, cells were trypsinized, fixed with 4% paraformaldehyde (PFA), stained according to the manufacturer's instruction and additionally counterstained with DAPI (1 μg/ml, Roth). Fluorescence images were recorded with a Nikon confocal laser scanning microscope as described below. Analysis was done using Nikon NIS analysis software (NIS-Elements 4.20). Regions of interest (ROIs) were identified corresponding to the DAPI signal and subsequently analyzed for mean fluorescence intensity (MFI) of EdU-Alexa488. The nuclear MFI of EdU-Alexa488 was measured and normalized by the DAPI signal. Cells with a Alexa488/DAPI ratio above a threshold, which was determined from the cumulative distribution function of all values, were classified as proliferative active. Per experiment and condition, at least 200 cells were analyzed.
For FACS analysis cells were seeded to 50% confluence and incubated with EdU (10 μM) and the corresponding stimulus for 16 h. Afterwards, cells were trypsinized, fixed with 4% PFA and stained following the manufacturer's instruction. Flow cytometry and analysis was performed using Beckman Coulter Gallios™ Cytometer and FlowJo software (version X 10.0.7r2), respectively. Dot plots displaying peak versus area values of FS and SS were used to gate for single cells in order to exclude doublets from analysis. For quantification cells above a threshold (shown in Fig. 2C) were classified as proliferative active. Per experiment and condition 20,000 events were measured.
4.5. Apoptosis assay
For analysis of apoptosis the Caspase-Glo 3/7 assay (Promega) was used following manufacturer's instruction. Briefly, 2.5 ∗ 104 H1299 cells were seeded into a well of a 96 well plate and grown over night. The next day cells were either incubated with staurosporine, LecB or left untreated for 5 and 16 h. Afterwards, supernatant was removed and 100 μl Caspase Glow reagent per well was added. Subsequently the plate was vortexed at 300 rpm for 30 s and incubated for 1.5 h at room temperature. Luminescence was detected with a micro plate reader (SynergyH4 Hybrid Reader, BioTek). Blank-corrected luminescence values were normalized to values of untreated, control cells. Statistical significance was assessed by one-way analysis of variance (ANOVA) followed by Dunnett's post-test.
4.6. Western blot performance and analysis
Cells were seeded to 90% confluence, starved for 2 h and stimulated as indicated for different time periods. Starvation was performed to exclude possible effects of growth factors and cytokines of serum, which may interfere with LecB effects. For better comparability, experiments described below were also performed under starving conditions. Afterwards, cells were lysed in Ripa buffer containing protease and phosphatase inhibitors (0.8 μM aprotinin, 11 μM leupeptin, 200 μM pefablock, 1 mM sodium orthovanadate, 1% (v/v) phosphatase inhibitor cocktail 3, all compounds were purchased from Sigma-Aldrich) for 30 min at 4 °C and centrifuged to remove cell debris. Determination of protein concentration was performed using BCA assay (Pierce) according to the manufacturer's protocol. Equal amounts of protein were separated on 8% polyacrylamide gels and transferred on nitrocellulose membranes. Blots were blocked in 3% BSA and incubated with primary antibodies at 4 °C overnight. Next day, blots were incubated with HRP-conjugated secondary antibodies. Luminescence was detected using Vilber Lourmat Fusion FX chemiluminescence imager. Following antibodies were used for Western blots in this study: anti-β-catenin (Abcam, #ab32572), anti-β-actin (Sigma, #A5316), anti-pAkt (Ser473) (CST, #4060), anti-Akt (pan) (CST, #A4691), anti-pGSK-3β (Ser9) (CST, #5558), anti-GSK (pan) (CST, #9315), anti-β1-integrin (Millipore, #MAB2000), anti-p65 pSer536 (CST, #3033), anti-p65 pSer 468 (CST, #3039), anti-cyclin E2 (CST, #4132), anti-cyclin B1 (CST, #12,231), anti-Rb pan (CST, #9309), anti-Rb pSer795 (CST, #9301), anti-rabbit-HRP (CST, #7074), anti-mouse-HRP (CST, #7076). Densitometric quantification of blots was performed using ImageJ (1.45b). Protein and phosphorylation levels were normalized to actin or corresponding pan staining and actin, respectively. As exception, Rb pSer795 signals were normalized to actin, in order to avoid a bias from the quantification of shifted Rb pan bands.
4.7. NF-κB reporter assay
To evaluate the transcriptional activity of NF-κB, a luciferase reporter assay was performed with the p1242 3 ×-KB-L plasmid (gift from Bill Sugden, Addgene #26699) containing three MHC I KB elements [33].
For this, cells were grown to 70% confluence and transfected with the mentioned plasmid with polyethyleneimine (PEI, Polysciences) as described elsewhere [58]. After 24 h cells were starved for 2 h and stimulated for 5 h with indicated stimuli. In the following, cells were lysed for at least 10 min in luciferase buffer (25 mM Tris–HCl pH 7.8, 15 mM MgSO4, 4 mM EGTA, 1% Triton X-100, 1 mM DTT). Lysates were transferred into a white plate and firefly luciferase-substrate (20 mM tricine, 2.67 mM MgSO4, 0.1 mM EDTA, 33.3 mM DTT, 524 μM ATP, 218 μg/ml Acetyl-CoA, 131 μg/ml Luciferin, 5 mM NaOH, 264 μM MgCO3) was added and luminescence was measured every 2 min for 30 min using the microplate reader Tecan Infinite 200Pro. The three highest values were used for quantification and normalized to the protein concentration of the lysates. At least three independent experiments were performed with three replicates per experiment and condition.
4.8. Beta-catenin reporter assay
To evaluate the transcriptional activity of β-catenin, a luciferase reporter assay was performed with the plasmid 7TFP (gift from Roel Nusse, Addgene #24,308 [37]). For the assay H1299 cells were seeded in 24 well plates (1 ∗ 105 cells per well) and grown for 24 h. Then, cells were transfected with the 7TFP plasmid and a plasmid encoding Renilla luciferase under control of a CMV promoter (gift from Wilfried Weber, University of Freiburg, Germany) using Lipofectamine 2000 (Thermo Fisher) for 12 h. After treatment with LecB for indicated time periods, cells were lysed with luciferase buffer, and two aliquots of each lysate were loaded on a white 96 well plate. One aliquot was supplemented with firefly luciferase substrate; the other aliquot was supplemented with Renilla luciferase substrate. The luminescence was measured with a plate reader as described before. For quantification, the firefly luciferase readings were normalized to the Renilla luciferase readings for each sample. Three independent experiments were performed with three replicates per experiment and condition.
4.9. Quantitative RT-PCR
Cells were seeded to 90% confluence, starved for 2 h and stimulated for indicated time periods. Cells were starved to estimate results that are comparable to Western blot experiments. Afterwards, cells were lysed with TRIzol (Sigma-Aldrich) and RNA was extracted following the manufacturer's instructions. 1 μg RNA was transcribed into cDNA after DNase digest using First Strand cDNA synthesis Kit with dsDNase (Thermo Scientific). For qPCR itself SYBR Select qPCR Kit (Applied Biosystems, Life Technologies) was used as recommended by the manufacturer with the following primers at a concentration of 10 μM: TNF-α (fwd: CTTCTCCTTCCTGATCGTGG, rev: GCTGGTTATCTCTCAGCTCCA), GAPDH (fwd: CCTCCAAAATCAAGTGGGGCGAT, rev: CAAATGAGCCCCAGCCTTCTCC). Samples were measured with Biorad CFX384 qPCR system and analyzed using the BioRad Analysis software (version 3.0). Relative mRNA levels of TNF-α were normalized to GAPDH levels.
4.10. Immunofluorescence, confocal microscopy and image analysis
Cells were grown on glass coverslips to 60% confluence, starved for 2 h and stimulated for indicated time periods at 37 °C. Starvation was performed to estimate results that are comparable to Western blot experiments. After fixation with 4% and saturation with 50 mM NH4Cl, cells were permeabilized in blocking buffer containing 5% BSA, 0.3% Triton X-100 in PBS for 1 h at RT. Primary antibodies were incubated overnight at 4 °C in a humidity chamber, secondary antibodies for 1 h at RT and finally cells were counterstained with DAPI (1 μg/ml) and mounted with Mowiol containing DABCO (Roth, antioxidant). As only exception, samples for γ-tubulin staining were fixed and permeabilized with acetone/methanol (1:1) at − 20 °C and processed as described above with 1 h incubation in blocking buffer. Following antibodies were used for immunofluorescence staining in this study: anti-β-catenin (Abcam, #ab32572), anti-β1-integrin (Millipore, #MAB2000), NF-κB-p65 (CST, #8242), anti-γ-tubulin (Sigma-Aldrich, #T5192), anti-rabbit-Cy3 (Jackson ImmunoResearch, #711-166-152), anti-rabbit-Alexa488 (Invitrogen, #A21206) and anti-mouse-Alexa647 (Invitrogen, #21,236).
For confocal microscopy the Nikon Eclipse Ti-E inverted microscope equipped with a Nikon A1R confocal laser scanning system was used. EdU assays were imaged with a 20 × multi-immersion objective (NA = 0.75, used with oil), NF-κB images with a 40 × air objective (NA = 0.6) and all remaining experiments with a 60 × oil immersion objective (NA = 1.49). Nuclear MFI of EdU was quantified with NIS-Elements software (Nikon, version 4.20), whereas nuclear localization of NF-κB was analyzed with ImageJ (1.45b). In both cases DAPI signal was used to determine nuclear regions and the corresponding MFI values were measured within these regions of interest.
4.11. Statistical analysis
Statistical analysis was performed with GraphPad Prism software (version 5.04). All data (except NF-κB nuclear translocation, Fig. 1D, Suppl. Fig. 2D) are presented as mean ± standard error of the mean (SEM) values that have been calculated from the results of independent experiments. Statistical significance compared to the untreated control was assessed by one-way analysis of variance (ANOVA) followed by Dunnett's post-hoc test. Data from NF-κB nuclear translocation origin from three independent experiments (Fig. 1D and Suppl. Fig. 2D). As the values were not normally distributed, data are presented as boxplot with median and the upper and lower quartile, whereas whiskers indicate 1.5 interquartile range (IQR) values. Statistical significance was assessed by Kruskal–Wallis test with Dunn's post-hoc test. All results are considered as statistically significant with a p-value lower than 0.05. Asterisks indicate significance levels in figures with NS = p ≥ 0.05, * = p ≤ 0.05, ** = p ≤ 0.01 and *** = p ≤ 0.001.
Transparency document
Transparency document.
Acknowledgments
This work was supported in part by the Excellence Initiative of the German Research Foundation (GSC-4, Spemann Graduate School and EXC 294, BIOSS). W.R. acknowledges the support by a grant from the Ministry of Science, Research and the Arts of Baden-Württemberg (Az: 33-7532.20), by a German Research Foundation grant (RO 4341/2-1), and by a starting grant of the European Research Council (Program “Ideas” — call identifier: ERC-2011-StG 282105). The authors declare no conflict of interest.
Footnotes
The Transparency document associated with this article can be found, in online version.
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.bbamcr.2016.02.004.
Appendix A. Supplementary data
Supplementary figures.
References
- 1.Bessa L.J., Fazii P., DI Giulio M., Cellini L. Bacterial isolates from infected wounds and their antibiotic susceptibility pattern: some remarks about wound infection. Int. Wound J. 2015;12:47–52. doi: 10.1111/iwj.12049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Emori T.G., Gaynes R.P. An overview of nosocomial infections, including the role of the microbiology laboratory. Clin. Microbiol. Rev. 1993;6:428–442. doi: 10.1128/cmr.6.4.428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Johnson E.R., Matthay M.A. Acute lung injury: epidemiology, pathogenesis, and treatment. J. Aerosol Med. Pulm. Drug Deliv. 2010;23:243–252. doi: 10.1089/jamp.2009.0775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sawa T. The molecular mechanism of acute lung injury caused by Pseudomonas aeruginosa: from bacterial pathogenesis to host response. J. Intensive Care. 2014;2:10. doi: 10.1186/2052-0492-2-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.González-López A., Astudillo A., García-Prieto E., Fernández-García M.S., López-Vázquez A., Batalla-Solís E. Inflammation and matrix remodeling during repair of ventilator-induced lung injury. Am. J. Physiol. — Lung Cell. Mol. Physiol. 2011;301:L500–L509. doi: 10.1152/ajplung.00010.2011. [DOI] [PubMed] [Google Scholar]
- 6.Meduri G.U., Headley S., Kohler G., Stentz F., Tolley E., Umberger R. Persistent elevation of inflammatory cytokines predicts a poor outcome in ARDS. Plasma IL-1 beta and IL-6 levels are consistent and efficient predictors of outcome over time. Chest. 1995;107:1062–1073. doi: 10.1378/chest.107.4.1062. [DOI] [PubMed] [Google Scholar]
- 7.Bjarnsholt T., Kirketerp-Møller K., Jensen P.Ø., Madsen K.G., Phipps R., Krogfelt K. Why chronic wounds will not heal: a novel hypothesis. Wound Repair Regen. Off. Publ. Wound Heal. Soc. Eur. Tissue Repair Soc. 2008;16:2–10. doi: 10.1111/j.1524-475X.2007.00283.x. [DOI] [PubMed] [Google Scholar]
- 8.Garber N., Guempel U., Gilboa-Garber N., Doyle R.J. Specificity of the fucose-binding lectin of Pseudomonas aeruginosa. FEMS Microbiol. Lett. 1987;48:331–334. [Google Scholar]
- 9.Tielker D., Hacker S., Loris R., Strathmann M., Wingender J., Wilhelm S. Pseudomonas aeruginosa lectin LecB is located in the outer membrane and is involved in biofilm formation. Microbiology. 2005;151:1313–1323. doi: 10.1099/mic.0.27701-0. [DOI] [PubMed] [Google Scholar]
- 10.Funken H., Bartels K.-M., Wilhelm S., Brocker M., Bott M., Bains M. Specific association of lectin LecB with the surface of Pseudomonas aeruginosa: role of outer membrane protein OprF. PLOS ONE. 2012;7 doi: 10.1371/journal.pone.0046857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schneider D., Dühren-von Minden M., Alkhatib A., Setz C., VAN Bergen C.A.M., Benkißer-Petersen M. Lectins from opportunistic bacteria interact with acquired variable region glycans of surface Ig in follicular lymphoma. Blood. 2015;125:3287–3296. doi: 10.1182/blood-2014-11-609404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Adam E.C., Mitchell B.S., Schumacher D.U., Grant G., Schumacher U. Pseudomonas aeruginosa II lectin stops human ciliary beating: therapeutic implications of fucose. Am. J. Respir. Crit. Care Med. 1997;155:2102–2104. doi: 10.1164/ajrccm.155.6.9196121. [DOI] [PubMed] [Google Scholar]
- 13.Avichezer D., Gilboa-Garber N. PA-II, the l-fucose and d-mannose binding lectin of Pseudomonas aeruginosa stimulates human peripheral lymphocytes and murine splenocytes. FEBS Lett. 1987;216:62–66. doi: 10.1016/0014-5793(87)80757-3. [DOI] [PubMed] [Google Scholar]
- 14.Chemani C., Imberty A., DE Bentzmann S., Pierre M., Wimmerová M., Guery B.P. Role of LecA and LecB lectins in Pseudomonas aeruginosa-induced lung injury and effect of carbohydrate ligands. Infect. Immun. 2009;77:2065–2075. doi: 10.1128/IAI.01204-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boukerb A.M., Rousset A., Galanos N., Méar J.-B., Thépaut M., Grandjean T. Antiadhesive properties of glycoclusters against Pseudomonas aeruginosa lung infection. J. Med. Chem. 2014;57:10275–10289. doi: 10.1021/jm500038p. [DOI] [PubMed] [Google Scholar]
- 16.Whyte J.L., Smith A.A., Helms J.A. Wnt signaling and injury repair. Cold Spring Harb. Perspect. Biol. 2012;4:a008078. doi: 10.1101/cshperspect.a008078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheon S.S., Wei Q., Gurung A., Youn A., Bright T., Poon R. Beta-catenin regulates wound size and mediates the effect of TGF-beta in cutaneous healing. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2006;20:692–701. doi: 10.1096/fj.05-4759com. [DOI] [PubMed] [Google Scholar]
- 18.Amini-Nik S., Cambridge E., Yu W., Guo A., Whetstone H., Nadesan P. β-Catenin-regulated myeloid cell adhesion and migration determine wound healing. J. Clin. Invest. 2014;124:2599–2610. doi: 10.1172/JCI62059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chattopadhyay N., Wang Z., Ashman L.K., Brady-Kalnay S.M., Kreidberg J.A. alpha3beta1 integrin-CD151, a component of the cadherin–catenin complex, regulates PTPmu expression and cell–cell adhesion. J. Cell Biol. 2003;163:1351–1362. doi: 10.1083/jcb.200306067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Logan C.Y., Nusse R. The Wnt signaling pathway in development and disease. Annu. Rev. Cell Dev. Biol. 2004;20:781–810. doi: 10.1146/annurev.cellbio.20.010403.113126. [DOI] [PubMed] [Google Scholar]
- 21.MacDonald B.T., Tamai K., He X. Wnt/β-catenin signaling: components, mechanisms, and diseases. Dev. Cell. 2009;17:9–26. doi: 10.1016/j.devcel.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vlad-Fiegen A., Langerak A., Eberth S., Müller O. The Wnt pathway destabilizes adherens junctions and promotes cell migration via β-catenin and its target gene cyclin D1. FEBS Open Bio. 2012;2:26–31. doi: 10.1016/j.fob.2012.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li H., Lee H.J., Ahn Y.H., Kwon H.J., Jang C.-Y., Kim W.-Y. Tussilagone suppresses colon cancer cell proliferation by promoting the degradation of β-catenin. Biochem. Biophys. Res. Commun. 2014;443:132–137. doi: 10.1016/j.bbrc.2013.11.062. [DOI] [PubMed] [Google Scholar]
- 24.Duan Y., Liao A.P., Kuppireddi S., Ye Z., Ciancio M.J., Sun J. Beta-catenin activity negatively regulates bacteria-induced inflammation. Lab. Investig. J. Tech. Methods Pathol. 2007;87:613–624. doi: 10.1038/labinvest.3700545. [DOI] [PubMed] [Google Scholar]
- 25.Schaale K., Neumann J., Schneider D., Ehlers S., Reiling N. Wnt signaling in macrophages: augmenting and inhibiting mycobacteria-induced inflammatory responses. Eur. J. Cell Biol. 2011;90:553–559. doi: 10.1016/j.ejcb.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 26.Silva-García O.A., Valdez-Alarcón J.J., Baizabal-Aguirre V.M. The Wnt/ß-catenin signaling pathway controls the inflammatory response in infections caused by pathogenic bacteria. Mediators Inflamm. 2014;2014:e310183. doi: 10.1155/2014/310183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen K., Yin L., Nie X., Deng Q., Wu Y., Zhu M. β-Catenin promotes host resistance against Pseudomonas aeruginosa keratitis. J. Infect. 2013;67:584–594. doi: 10.1016/j.jinf.2013.07.025. [DOI] [PubMed] [Google Scholar]
- 28.Tran C.S., Eran Y., Ruch T.R., Bryant D.M., Datta A., Brakeman P. Host cell polarity proteins participate in innate immunity to Pseudomonas aeruginosa infection. Cell Host Microbe. 2014;15:636–643. doi: 10.1016/j.chom.2014.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mattioli I., Sebald A., Bucher C., Charles R.-P., Nakano H., Doi T. Transient and selective NF-kappa B p65 serine 536 phosphorylation induced by T cell costimulation is mediated by I kappa B kinase beta and controls the kinetics of p65 nuclear import. J. Immunol. Baltim. Md. 2004;1950(172):6336–6344. doi: 10.4049/jimmunol.172.10.6336. [DOI] [PubMed] [Google Scholar]
- 30.Chen L.-F., Williams S.A., Mu Y., Nakano H., Duerr J.M., Buckbinder L. NF-κB RelA phosphorylation regulates RelA acetylation. Mol. Cell. Biol. 2005;25:7966–7975. doi: 10.1128/MCB.25.18.7966-7975.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Buss H., Dörrie A., Schmitz M.L., Frank R., Livingstone M., Resch K. Phosphorylation of serine 468 by GSK-3β negatively regulates basal p65 NF-κB activity. J. Biol. Chem. 2004;279:49571–49574. doi: 10.1074/jbc.C400442200. [DOI] [PubMed] [Google Scholar]
- 32.Karin M. How NF-kappaB is activated: the role of the IkappaB kinase (IKK) complex. Oncogene. 1999;18:6867–6874. doi: 10.1038/sj.onc.1203219. [DOI] [PubMed] [Google Scholar]
- 33.Mitchell T., Sugden B. Stimulation of NF-kappa B-mediated transcription by mutant derivatives of the latent membrane protein of Epstein–Barr virus. J. Virol. 1995;69:2968–2976. doi: 10.1128/jvi.69.5.2968-2976.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kühn K., Cott C., Bohler S., Aigal S., Zheng S., Villringer S. The interplay of autophagy and β-catenin signaling regulates differentiation in acute myeloid leukemia. Cell Death Discov. 2015;1:15031. doi: 10.1038/cddiscovery.2015.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kierbel A., Gassama-Diagne A., Mostov K., Engel J.N. The phosphoinositol-3-kinase-protein kinase B/Akt pathway is critical for Pseudomonas aeruginosa strain PAK internalization. Mol. Biol. Cell. 2005;16:2577–2585. doi: 10.1091/mbc.E04-08-0717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wójcik C., DeMartino G.N. Intracellular localization of proteasomes. Int. J. Biochem. Cell Biol. 2003;35:579–589. doi: 10.1016/s1357-2725(02)00380-1. [DOI] [PubMed] [Google Scholar]
- 37.Fuerer C., Nusse R. Lentiviral vectors to probe and manipulate the Wnt signaling pathway. PLoS ONE. 2010;5 doi: 10.1371/journal.pone.0009370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bertoli C., Skotheim J.M., DE Bruin R.A.M. Control of cell cycle transcription during G1 and S phases. Nat. Rev. Mol. Cell Biol. 2013;14:518–528. doi: 10.1038/nrm3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Burke J.R., Liban T.J., Restrepo T., Lee H.-W., Rubin S.M. Multiple mechanisms for E2F binding inhibition by phosphorylation of the retinoblastoma protein C-terminal domain. J. Mol. Biol. 2014;426:245–255. doi: 10.1016/j.jmb.2013.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Houser J., Komarek J., Kostlanova N., Cioci G., Varrot A., Kerr S.C. A soluble fucose-specific lectin from Aspergillus fumigatus conidia—structure, specificity and possible role in fungal pathogenicity. PLoS ONE. 2013;8 doi: 10.1371/journal.pone.0083077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sulák O., Cioci G., Lameignère E., Balloy V., Round A., Gutsche I. Burkholderia cenocepacia BC2L-C is a super lectin with dual specificity and proinflammatory activity. PLoS Pathog. 2011;7 doi: 10.1371/journal.ppat.1002238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sheetz M.P., Felsenfeld D., Galbraith C.G., Choquet D. Cell migration as a five-step cycle. Biochem. Soc. Symp. 1999;65:233–243. [PubMed] [Google Scholar]
- 43.Koivisto L., Alavian K., Hakkinen L., Pelech S., McCulloch C.A., Larjava H. Glycogen synthase kinase-3 regulates formation of long lamellipodia in human keratinocytes. J. Cell Sci. 2003;116:3749–3760. doi: 10.1242/jcs.00693. [DOI] [PubMed] [Google Scholar]
- 44.Karrasch T., Spaeth T., Allard B., Jobin C. PI3K-dependent GSK3ß(Ser9)-phosphorylation is implicated in the intestinal epithelial cell wound-healing response. PLoS ONE. 2011;6 doi: 10.1371/journal.pone.0026340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dajani R., Fraser E., Roe S.M., Yeo M., Good V.M., Thompson V. Structural basis for recruitment of glycogen synthase kinase 3β to the axin–APC scaffold complex. EMBO J. 2003;22:494–501. doi: 10.1093/emboj/cdg068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ding V.W., Chen R.H., McCormick F. Differential regulation of glycogen synthase kinase 3beta by insulin and Wnt signaling. J. Biol. Chem. 2000;275:32475–32481. doi: 10.1074/jbc.M005342200. [DOI] [PubMed] [Google Scholar]
- 47.Huttenlocher A., Horwitz A.R. Integrins in cell migration. Cold Spring Harb. Perspect. Biol. 2011;3:a005074. doi: 10.1101/cshperspect.a005074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Choma D.P., Pumiglia K., DiPersio C.M. Integrin alpha3beta1 directs the stabilization of a polarized lamellipodium in epithelial cells through activation of Rac1. J. Cell Sci. 2004;117:3947–3959. doi: 10.1242/jcs.01251. [DOI] [PubMed] [Google Scholar]
- 49.Palmer T.D., Martínez C.H., Vasquez C., Hebron K.E., Jones-Paris C., Arnold S.A. Integrin-free tetraspanin CD151 can inhibit tumor cell motility upon clustering and is a clinical indicator of prostate cancer progression. Cancer Res. 2014;74:173–187. doi: 10.1158/0008-5472.CAN-13-0275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chatterjee I., Humtsoe J.O., Kohler E.E., Sorio C., Wary K.K. Lipid phosphate phosphatase-3 regulates tumor growth via β-catenin and cyclin-D1 signaling. Mol. Cancer. 2011;10:51. doi: 10.1186/1476-4598-10-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lau M.-T., Klausen C., Leung P.C.K. E-cadherin inhibits tumor cell growth by suppressing PI3K/Akt signaling via β-catenin-Egr1-mediated PTEN expression. Oncogene. 2011;30:2753–2766. doi: 10.1038/onc.2011.6. [DOI] [PubMed] [Google Scholar]
- 52.Finck-Barbançon V., Goranson J., Zhu L., Sawa T., Wiener-Kronish J.P., Fleiszig S.M. ExoU expression by Pseudomonas aeruginosa correlates with acute cytotoxicity and epithelial injury. Mol. Microbiol. 1997;25:547–557. doi: 10.1046/j.1365-2958.1997.4891851.x. [DOI] [PubMed] [Google Scholar]
- 53.Geiser T.K., Kazmierczak B.I., Garrity-Ryan L.K., Matthay M.A., Engel J.N. Pseudomonas aeruginosa ExoT inhibits in vitro lung epithelial wound repair. Cell. Microbiol. 2001;3:223–236. doi: 10.1046/j.1462-5822.2001.00107.x. [DOI] [PubMed] [Google Scholar]
- 54.Kida Y., Higashimoto Y., Inoue H., Shimizu T., Kuwano K. A novel secreted protease from Pseudomonas aeruginosa activates NF-kappaB through protease-activated receptors. Cell. Microbiol. 2008;10:1491–1504. doi: 10.1111/j.1462-5822.2008.01142.x. [DOI] [PubMed] [Google Scholar]
- 55.Hauber H.-P., Schulz M., Pforte A., Mack D., Zabel P., Schumacher U. Inhalation with fucose and galactose for treatment of Pseudomonas aeruginosa in cystic fibrosis patients. Int. J. Med. Sci. 2008;5:371–376. doi: 10.7150/ijms.5.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Novoa A., Eierhoff T., Topin J., Varrot A., Barluenga S., Imberty A. A LecA ligand identified from a galactoside-conjugate array inhibits host cell invasion by Pseudomonas aeruginosa. Angew. Chem. Int. Ed. Engl. 2014;53:8885–8889. doi: 10.1002/anie.201402831. [DOI] [PubMed] [Google Scholar]
- 57.Eierhoff T., Bastian B., Thuenauer R., Madl J., Audfray A., Aigal S. A lipid zipper triggers bacterial invasion. Proc. Natl. Acad. Sci. U. S. A. 2014;111:12895–12900. doi: 10.1073/pnas.1402637111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Müller K., Engesser R., Schulz S., Steinberg T., Tomakidi P., Weber C.C. Multi-chromatic control of mammalian gene expression and signaling. Nucleic Acids Res. 2013;41 doi: 10.1093/nar/gkt340. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Transparency document.
Supplementary figures.