Summary
Clostridium difficile is a leading cause of antibiotic-associated diarrhea. The mechanisms underlying C. difficile expansion after microbiota disturbance are just emerging. We assessed the gene expression profile of C. difficile within the intestine of gnotobiotic mice to identify genes regulated in response to either dietary or microbiota compositional changes. In the presence of the gut symbiont Bacteroides thetaiotaomicron, C. difficile induces a pathway that metabolizes the microbiota fermentation end-product succinate to butyrate. The low concentration of succinate in the microbiota of conventional mice is transiently elevated upon antibiotic treatment or chemically-induced intestinal motility disturbance, and C. difficile exploits this succinate spike to expand in the perturbed intestine. A C. difficile mutant compromised in succinate utilization is at a competitive disadvantage during these perturbations. Understanding the metabolic mechanisms involved in microbiota-C. difficile interactions may help to identify approaches for the treatment and prevention of C. difficile-associated diseases.
Introduction
One of the most common causes of infectious antibiotic-associated diarrhea is the Gram-positive, spore-forming anaerobe Clostridium difficile (He et al. 2013). While C. difficile resides asymptomatically in the GI tract of 2–5% of the population, it can also cause pathology ranging from mild diarrhea to severe colitis, toxic megacolon, and other C. difficile-associated diseases (CDAD) (Rivera EV and Woods S 2003). Even after successful treatment with antibiotics such as vancomycin or metronidazole, roughly 20–30% of individuals experience disease recurrence (Pepin J et al. 2006, Johnson S 2009). Fecal transplants have been found to be effective in curing recurrent CDAD, consistent with the importance of a healthy gut ecosystem in outcompeting C. difficile (Khoruts et al. 2010; van Nood et al. 2013). The successful use of six bacterial species to cure persistent infection in mice, but failure of other cocktails of six or fewer species underscores the current poor understanding of ecological relationships that underlie C. difficile disease (Lawley et al. 2012). Additional mechanistic insight into how C. difficile exploits the perturbed gut ecosystem will facilitate prevention strategies to hold C. difficile at bay during the period of post-antibiotic vulnerability (Wilson and Perini, 1988; Ng et al. 2013).
The gut microbiota metabolizes dietary and host-derived nutrients and produces end products including heat, gases, organic acids (OAs) and short-chain fatty acids (SCFAs). Of all metabolites in the colon, dietary carbohydrates play the largest role in the formation of SCFAs and OAs (Macfarlane and Macfarlane 2003). The SCFAs acetate, propionate and butyrate are the most common end products of fermentation within the gut microbiota (Wong et al., 2006). The OAs succinate and lactate are also common end-products of primary fermenters that are taken up and metabolized by other microbes. Several recent studies have highlighted the role of the microbiota-produced metabolites in pathogen colonization (Thiennimitr et al., 2011; Ng. et al., 2013; Maier et al. 2013), however the contribution of SCFAs and OAs on pathogenesis remains understudied. While antibiotics contribute to the majority of cases of hospital-acquired C. difficile, dietary and motility changes represent two unexplored perturbations that may contribute to cases of community-acquired C. difficile infection, which occur independently of antibiotic-exposure (Wilcox et al. 2008; Khanna et al. 2012). Shifts in diet have been shown to affect the composition and diversity of the microbiota (Turnbaugh et al., 2009; Wu et al., 2011; McNulty et al., 2013), and the extent to which diet-induced transitions in microbiota membership share ecosystem disturbance-characteristics with antibiotic use is unknown. C. difficile and other opportunistic members of the microbiota have evolved mechanisms to capitalize upon such ecosystem alterations (Lozupone et al., 2013; Ng et al. 2013). In our previous study, microbiota-liberation of the host mucosal monosaccharide sialic acid was identified as a nutrient source used by C. difficile and Salmonella during post-antibiotic expansion (Ng et al. 2013). Further characterization of how microbiota perturbations affect availability of metabolites that contribute to pathogen niches is needed.
In this study, we aimed to understand the mechanisms underlying C. difficile expansion after microbiota disturbance, with a focus on microbiota-associated metabolites. We utilized germ-free mice colonized with a well-characterized representative of the prominent Bacteroides genus, B. thetaiotaomicron (Bt). We profiled the transcriptional response elicited by C. difficile in the presence and absence of Bt in vivo using standard, polysaccharide-rich dietary conditions versus a polysaccharide-deficient diet. We identified key metabolic strategies that C. difficile differentially regulates in these environmental conditions. Bt produces high levels of succinate during fermentation of dietary carbohydrates, and C. difficile couples reduction of succinate to butyrate with fermentation of remaining dietary carbohydrates. C. difficile relies on succinate in a conventional, antibiotic-treated setting as well as after gut motility disturbance to efficiently expand. This work sheds light on the important roles that microbiota-associated metabolites play in C. difficile expansion, which has implications for potential therapeutic strategies for enteric pathogens.
Results
Identification of key metabolic strategies for C. difficile colonization in vivo
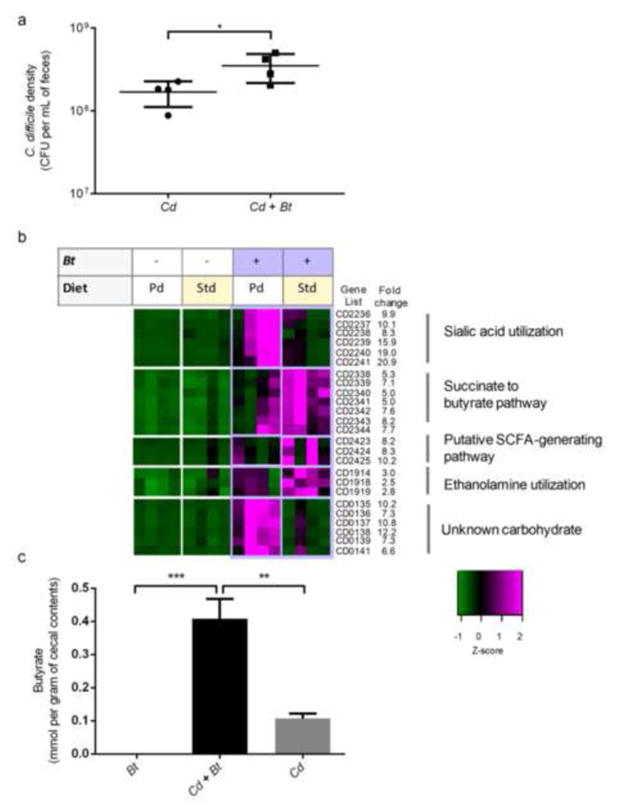
We sought to identify colonization strategies employed by C. difficile within the gut. When C. difficile infects mice colonized with Bt, it attains higher population densities on day 1 post-infection when the mice were fed a polysaccharide-rich standard diet (2.1-fold). Previous work has shown that C. difficile obtains higher population densities in the presence of Bt in polysaccharide-deficient chow (Ng et al., 2013) (Fig. 1a). We performed transcriptional profiling of C. difficile residing within the distal gut of gnotobiotic mice in these four conditions, to identify potential metabolic strategies employed by C. difficile in the presence of Bt that could explain its ability to attain higher densities (Fig. 1b). Five COG functional categories, including “carbohydrate transport and metabolism” are significantly differentially regulated (p<0.01; hypergeometric test) in response to the presence of Bt and three categories changed in response to polysaccharide availability in the diet (Supplemental Fig. S1a, S1b).
Figure 1. Metabolic strategies employed by C. difficile in response to a human gut commensal.
a. C. difficile density in feces at 1 day post-infection of germ free (Cd) or Bt-associated mice (Cd + Bt) and fed standard diet (Std diet) or polysaccharide-deficient diet (Pd diet) (n=4/group).
b. C. difficile catabolic pathways with significant differences in gene expression levels in vivo in the presence and absence of Bt. Mice were fed standard diet (Std) or polysaccharide-deficient diet (Pd). Colors indicate the deviation of each gene’s signal above (purple) and below (green) its mean (black) expression value across all sixteen in vivo samples (n=4/group) at 5 days post-infection in cecal contents. Fold change values correspond to gene expression in the Bt-biassociation relative to the monoassociation state.
c. Concentration of the short-chain fatty acid butyrate in cecal contents of standard diet-fed mice colonized with Bt, C. difficile and Bt (Cd + Bt), or C. difficile (Cd) at 5 days post-infection (n=4/group).
Several putative operons involved in nutrient acquisition and metabolism are responsive to changes in the gut environment. C. difficile expresses the sialic acid utilization pathway more highly in the Bt-bicolonized condition when mice are fed a polysaccharide-deficient diet (14.8–88-fold), consistent with our previous report (Ng et al. 2013) (Fig. 1b and Supplemental Table S1). In some cases, the changes are specific to a colonization state in a single dietary condition. For instance utilization pathways of sorbitol and fructose, both common dietary sugars found in many plants including fruit, are more highly expressed in the Bt-bicolonized condition when mice are fed standard diet, whereas the glucose utilization operon is more highly expressed in all other conditions (Supplemental Fig. S1c). Dietary change also affects C. difficile gene expression, and in standard diet, genes involved in sorbitol/glucitol utilization and cell-surface modification increase, whereas in polysaccharide-deficient diet, genes involved in acquiring cofactors such as cobalt and iron as well as cell-surface proteins and sugar ABC transporters increase (Supplemental Table S1). Because the genes involved in the conversion of succinate to butyrate were highly expressed in conditions where C. difficile achieves higher density (4.9–8.2-fold increase in expression in the Bt-biassociation relative to monoassociation) (Fig. 1b, Supplemental Table S1, Supplemental Fig. S1d, S1e), we pursued the importance of this pathway for gut colonization by C. difficile.
C. difficile relies upon microbiota-produced succinate to expand in the intestine
A similar succinate-to-butyrate pathway has been described for Clostridium kluyveri (Kenealy and Waselefsky, 1985; Wolff et al. 1993), which enables regeneration of NAD+ from NADH. Succinate metabolism does not directly yield ATP, however it acts as an electron sink allowing for the oxidation of electron carriers (Wolff et al. 1993). Notably, C. difficile’s induction of this NAD+ regenerating pathway is coincidently expressed with genes that enable consumption of the sugar alcohol, sorbitol, as well as other dietary sugars, including fructose, which require NAD+ for catabolism (Supplemental Fig. S2). We quantified the levels of SCFAs and OAs in the four in vivo conditions (Supplemental figure S3a, S3b). In monoassociation, Bt produces high levels of succinate (Supplemental Fig. S3a). Succinate is a common by-product of primary fermenters such as Bt, and is typically consumed by various secondary fermenters in the gut (Fischbach and Sonnenburg, 2011). In Bt-biassociation, C. difficile produced higher levels of butyrate compared to its monoassociation (3.8-fold, p<0.01) (Fig. 1c), consistent with its use of Bt-generated succinate to produce butyrate.
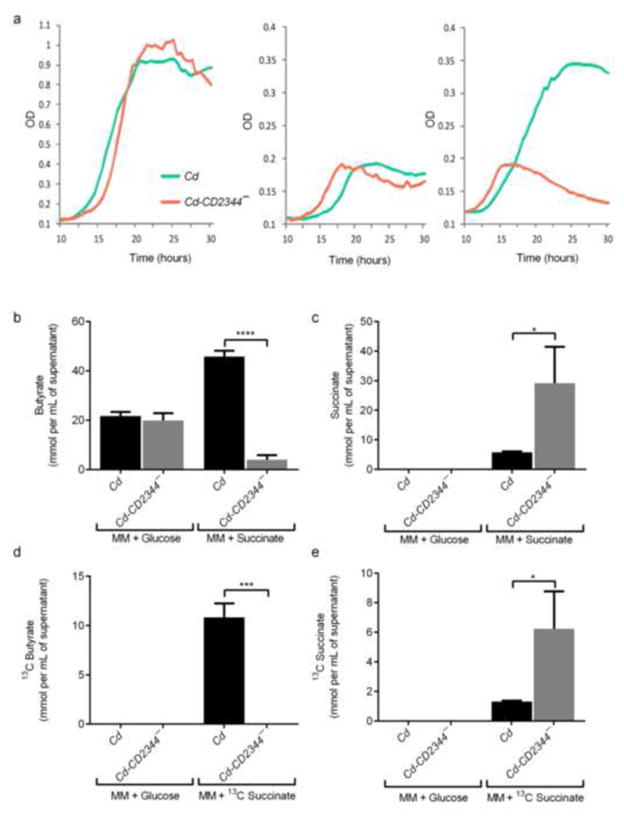
We next tested the metabolic role of the putative C. difficile succinate-to-butyrate pathway. Using ClosTron targeted mutagenesis (Heap et al. 2010) we mutated CD2344, a putative succinate transporter, of C. difficile, creating a new strain Cd-CD2344−. Although both wild-type and the mutant strain grow equally well in minimal medium (MM) containing glucose or water (Fig. 2a), Cd-CD2344− is unable to use succinate for growth (Fig. 2a). GC-MS measurement of SCFAs within the medium from the cultures at the maximum optical density revealed C. difficile wild-type and Cd-CD2344− show no differences in SCFA production when grown in MM-glucose (Figs. 2b, Supplemental Fig. S3c). The mutant Cd-CD2344− produces significantly less butyrate than wild type C. difficile grown in minimal medium containing succinate (11.4-fold, p<0.0001) (Fig. 2b) and does not deplete succinate from the medium (5-fold, p<0.05) (Fig. 2c). When C. difficile and Cd-CD2344− are grown in minimal medium containing 13C-succinate, only wild-type C. difficile is able to produce 13C-butyrate (p<0.001 and Fig. 2e, p<0.05)(Fig. 2d), and no 13C-propionate or 13C-acetate are found as byproducts, demonstrating that this pathway is specific for the conversion of succinate to butyrate and is not functional in the mutant strain.
Figure 2. A C. difficile operon encodes succinate-to-butyrate conversion.
a. Growth of wild-type C. difficile (Cd) and CD2344 mutant C. difficile (Cd-CD2344−) in minimal media (MM) containing 1% glucose (left), water (center), or 0.1% succinate (right). OD600 measurements were taken every 30 minutes (n=3/group). Representative of at least 3 replicates.
b. Concentrations of butyrate produced after growth of Cd or Cd-CD2344− in minimal media containing 0.5% glucose (MM + glucose) or 0.1% succinate (MM + succinate) (n=3/group).
c. Concentrations of succinate remaining after growth of Cd or Cd-CD2344− in minimal media containing 0.5% glucose (MM + glucose) or 0.1% succinate (MM + succinate) (n=3/group).
d. Levels of 13C-butyrate produced by Cd or Cd-CD2344− in minimal media containing 0.5% glucose (MM + glucose) or 0.1% 13C-succinate (MM + succinate) (n=3/group).
e. Levels of 13C-succinate remaining after growth of Cd or Cd-CD2344− in minimal media containing 0.5% glucose (MM + glucose) or 0.1% 13C-succinate (MM + succinate) (n=3/group).
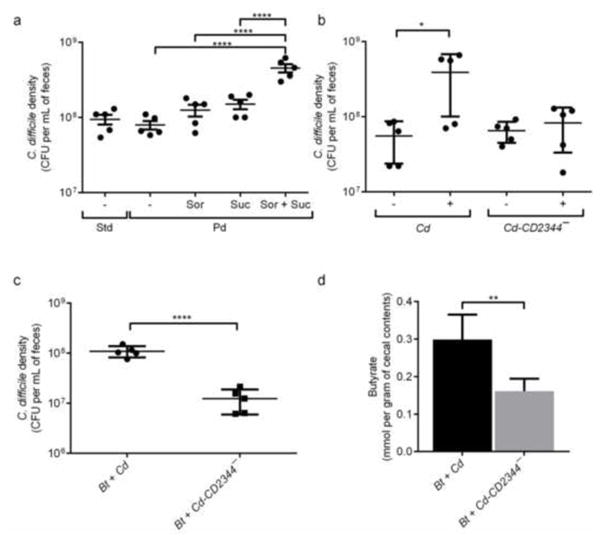
We conducted several experiments to assess whether the succinate-to-butyrate pathway is necessary for C. difficile to expand in the presence of Bt. C. difficile-monoassociated mice were fed a polysaccharide-deficient diet, which unlike the standard diet, is known to not to include sorbitol as a component. C. difficile is able to achieve marginally increased densities with succinate addition in a monoassociation, however a more dramatic benefit is observed when succinate is administered in the presence of the fermentable sugar alcohol, sorbitol, a common carbohydrate present in many fruits (Fig. 3a). In contrast, Cd-CD2344− does not reach high densities when colonizing GF mice supplemented with succinate and sorbitol-containing water (7.1-fold, p<0.05) (Fig. 3b), consistent with the inability of the mutant to consume succinate effectively. In mice colonized with succinate-producing Bt, Cd-CD2344− is unable to reach the same densities as wild type (8.8-fold, p<0.0001) (Fig. 3c and Supplemental Fig. S3d) and produces lower levels of butyrate compared to wild-type C. difficile (1.9-fold, p<0.001) (Fig. 3d), demonstrating that the succinate-to-butyrate pathway is important for C. difficile’s expansion in the presence of Bt. There are no significant differences in any other SCFAs or OAs quantified from the cecal contents of bi-associated mice (Supplemental Fig. S3e). Cd-CD2344− also induces less severe inflammation compared to wild-type C. difficile in Bt-associated germ-free mice (Supplemental Fig. S4).
Figure 3. C. difficile consumes the Bt fermentation product succinate to expand in the intestinal ecosystem.
a. C. difficile density in feces at 2 days post-infection of germ-free mice standard diet (Std) or polysaccharide deficient diet (Pd), supplemented with water containing 1% sorbitol (Sor), 1% succinate (Suc) or both 1% sorbitol and 1% succinate (Sor + Suc) (n=5/group). **** indicates statistical significance at an alpha value of <0.05 by one-way ANOVA with Bonferroni multiple comparisons test.
b. C. difficile density in feces of germ-free mice consuming polysaccharide-deficient diet and infected with wild-type C. difficile (Cd) or Cd-CD2344− mutant C. difficile (Cd-CD2344−) and administered water (−) or water containing 1% sorbitol and 1% succinate (+) at 1 day post-infection (n=5/group).
c. C. difficile density in feces of Bt-associated mice infected with wild-type C. difficile (Bt + Cd) or Cd-CD2344− mutant C. difficile (Bt + Cd-CD2344−) at day 1 post-infection and under standard dietary conditions (n=5/group).
d. Concentration of butyrate in cecal contents of Bt-associated mice infected with wild-type C. difficile (Cd) or Cd-CD2344− mutant C. difficile (Cd-CD2344−) at day 3 post-infection and under standard dietary conditions (n=5/group).
Post-antibiotic succinate accumulation is a key nutrient for C. difficile expansion
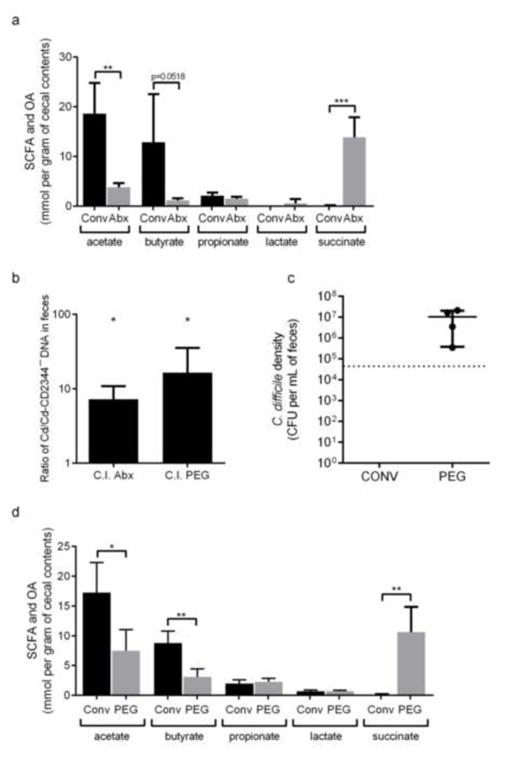
We next used antibiotic-treated conventional mice to determine the importance of succinate for C. difficile expansion in the context of a complex microbiota. Antibiotic treatment of conventional mice results in a significant reduction in the levels of the SCFAs acetate and butyrate and a concomitant significant increase in succinate levels (87-fold, p<0.001) (Fig. 4a). In a competition experiment, wild-type C. difficile outcompetes Cd-CD2344− (7.3-fold, p<0.05) (Fig. 4b). These data demonstrate that succinate is one of several key nutrients promoting C. difficile expansion after antibiotic treatment.
Figure 4. C. difficile exploits a spike in succinate availability after microbiota perturbations.
a. Concentrations of acetate, butyrate, propionate, lactate and succinate detected in cecal contents of streptomycin-treated conventional mice before treatment (Conv) and 3 days after antibiotic treatment (Abx) (n=5/group).
b. DNA of wild-type C. difficile (Cd) versus Cd-CD2344− mutant C. difficile (Cd-CD2344−) in feces of conventional mice treated with antibiotics (Abx) or PEG 3 days after infection with a 1:1 mixture of the two strains (n=5/group). *, p<0.05, one-sample t-test vs. a theoretical mean of 1.0.
c. C. difficile density in feces of conventional (Conv) or PEG-treated mice infected with wild-type C. difficile at day 3 post-infection (n=5 in Conv and n=4 in PEG). Dotted line represents the detection limit.
d. Concentrations of acetate, butyrate, propionate, lactate and succinate detected in cecal contents of PEG-treated conventional mice before treatment (Conv) and 3 days after treatment (PEG) (n=5/group).
Microbiota-derived succinate is utilized by C. difficile upon non-antibiotic perturbations
We tested whether perturbations to the gut microbiota other than antibiotics could enable C. difficile to gain access to succinate and expand to cause disease. A significant proportion (22–48%) of community-acquired C. difficile infections are not associated with recent antibiotic use (Wilcox et al. 2008; Khanna et al. 2012). Our previous work established that changes in gut motility induced by oral polyethylene glycol (PEG) treatment decreases diversity of the gut microbiota and significantly increases the presence of members of the family Peptostreptococcaceae, the same family that includes C. difficile (Kashyap et al. 2013). Given that members of the same family often share general metabolic characteristics, we wondered whether PEG-induced motility disturbance could share molecular features with antibiotic disturbance and result in C. difficile infection. Mice were administered a 10% PEG solution in drinking water, resulting in mild diarrhea and reduced gastrointestinal transit time (Kashyap et al. 2013). After five days of PEG treatment, mice were infected with C. difficile. C. difficile efficiently expands within the gut of mice with PEG-induced diarrhea (Fig. 4c) with the pathogen reaching densities comparable to post-antibiotic treatment (Ng et al. 2013). Diarrhea reduces the levels of butyrate and acetate and increases succinate (80-fold, p<0.01) (Fig. 4d), mirroring the changes that occur after antibiotic treatment. In competition, wild-type C. difficile outcompetes Cd-CD2344− in the presence of PEG (16.4-fold, p<0.05) (Fig. 4b), although in single infections of antibiotic-treated mice, Cd-CD2344− reached similar density levels as wild-type C. difficile (Supplemental Fig. S4c). These results demonstrate that succinate and other nutrients promote the expansion of C. difficile in the presence of PEG-induced diarrhea.
Discussion
Our results demonstrate that C. difficile relies upon the microbiota-produced metabolite succinate after antibiotic treatment as well as after the induction of diarrhea to efficiently expand and cause disease. The ability of C. difficile to reach high densities in the gut via pools of microbiota-produced metabolites is consistent with our previous work showing that both C. difficile and Salmonella typhimurium rely on microbiota-liberation of the host mucosal monosaccharide, sialic acid, as a nutrient source during post-antibiotic expansion (Ng et al. 2013). The succinate utilization pathway that C. difficile upregulates regenerates NAD+. In other studies, genes from both the nan operon (CD2240 and CD2241) and the succinate-to-butyrate pathway (CD2338, CD2339) were found to be present in the C. difficile spore proteome (Lawley et al. 2009), consistent with the utility of these pathways soon after germination. Our transcriptional profiling screen also identified C. difficile genes specifying an ethanolamine utilization pathway. The previous report of ethanolamine metabolism by Salmonella within the gut suggests that metabolic strategies common to the antibiotic-associated pathogens likely extend beyond the ones identified (Thiennimitr et al. 2011). Building an understanding of the hierarchy and relative importance of nutritional substrates used by antibiotic-associated pathogens is an important goal of future work.
Metabolomics studies on antibiotic-treated communities have shown that there are dramatic differences in host responses, bile acid composition, SCFA levels, and carbohydrate availability after antibiotic-treatment (Antunes et al. 2011; Theriot et al. 2014). Our work reveals that C. difficile responds to these changes by adjusting its metabolic pathways. The well-described role of diet in dictating fermentation products of the gut microbes, such as SCFAs and OAs, implies a potential additional link between our food and pathogen susceptibility. The OA succinate is not typically detected in the large intestine of healthy humans. However, succinate has been shown to accumulate in the large intestine of pigs with antibiotic-associated diarrhea (Tsukahara and Ushida, 2002). Combined with the elevated succinate we observe after PEG- or antibiotic-treatment, these data suggest that metabolic cross-feeding between gut resident microbes typically depletes succinate, and such interactions are disrupted by microbiota perturbations. It is well known that diet can have a profound impact on gut motility changes, and we have previously shown in a humanized mouse model that diet-induced motility changes are accompanied by large changes in SCFA profiles within the microbiota (Kashyap et al. 2013). How dietary interventions may promote or ameliorate CDAD is currently unknown, though dietary strategies that alter the composition and metabolic capabilities of the microbiota have proven successful against other enteric pathogens (Cummings et al. 1996; Ramakrishna et al. 2000; Muir et al. 2004; Alam et al. 2005; Alvarez-Acosta et al. 2009; Rabbani et al. 2009). Our data demonstrate that dietary change can rewire crossfeeding relationships relevant to pathogen expansion. Upon ecosystem disturbance the identity of available nutrients depends upon diet: C. difficile is able to capitalize upon available mucus-derived sialic acids in polysaccharide-deficient dietary conditions and high levels of succinate in the presence of a polysaccharide-rich diet.
This work points to common molecular features of gut microbiota disturbances, which create vulnerability to C. difficile expansion. Important questions include which alterations within the dynamic environment of the gut qualify as a “disturbance” and what are the common and specific molecular correlates of different disturbances (Antunes et al. 2011; Theriot et al. 2014). Microbiota-encoded metabolic pathways and the small molecules they produce represent therapeutic targets to prevent or combat C. difficile diseases.
Experimental Procedures
Animal diets and treatments
Germ-free (GF) Swiss-Webster mice of 6–8 weeks of age were utilized for experiments. All experiments were in accordance with A-PLAC, the Stanford IACUC. Mice were fed either an autoclaved standard diet (Purina LabDiet 5K67) or custom polysaccharide-deficient diet (BioServ) (Sonnenburg ED et al. 2010). All breeder and experimental isolators were periodically assessed for germ-free status by both culture-based methods (aerobic and anaerobic) as well as 16S-based PCR screens using universal primers. Sterility of multiple irradiated custom diets from this vendor (Bio-Serv) has been monitored in the lab over a five-year span with no samples or gnotobiotic mice fed the diets testing positive for contamination. For water supplementation experiments, succinic acid (Acros Organics) and sorbitol (Alfa Aesar) were administered at 1% (w/v) concentration in water after filtration through 0.22 μm Millipore filters. All mouse experiments presented in the manuscript are representative of at least two independent replicates of the experiment.
For antibiotic treatment, Streptomycin (Sigma) was administered at a concentration of 20 mg in 200 μl of PBS by gavage per mouse at 24 hours pre-infection, and mice were starved at 18 hours pre-infection with C. difficile. For PEG treatments, PEG 3350 (Miralax™) was administered at a 10% concentration in the drinking water continuously for 5 days prior to infection and removed at the time of infection.
Bacterial strains and culture conditions
B. thetaiotaomicron (VPI-5482; ATCC 29148), was grown anaerobically (6% H2, 20% CO2, 74% N2) overnight in TYG medium as previously described (Sonnenburg JL et al. 2005).
C. difficile strain 630, JIR8094 and Cd-CD2344 were cultured in Brain Heart Infusion (Becton Dickinson, MD) supplemented with 5 mg/ml of Yeast Extract (Remel) (BHIS) anaerobically (6% H2, 20% CO2, 74% N2). C. difficile growth curves were generated using minimal medium (MM) composed of Farmtech Tryptone (peptone) (EMD), beef extract (Becton Dickinson), yeast extract (Remel), monosodium phosphate (EMD), and disodium phosphate (EMD). OD600 was monitored using a BioTek PowerWave 340 plate reader (BioTek, Winooski, VT) every 30 minutes, at 37°C anaerobically (6% H2, 20% CO2, 74% N2). Fecal densities (CFU) of wild-type C. difficile after colonization of germ-free or Bt-monoassociated mice were quantified by duplicate sampling with 1 μl loops and subsequent dilution and spot plating on brain heart infusion agar (Becton Dickinson) with 10% v/v of defibrinated horse blood (Lampire Biological Laboratories) supplemented with 25 mg/L of erythromycin. To selectively quantify C. difficile fecal densities in antibiotic-treated conventional mice and in Bt-biassociation experiments involving C. difficile mutants, CDMN plates were utilized as previously described (Ng et al. 2013). Because Erm is not preset in CDMN plates, there is no difference in the recovery of Cd-CD2344− and wild-type C. difficile on CDMN plates.
ClosTron generation of Cd-CD2344− followed detailed protocols for targeted gene disruption in C. difficile (Heap JT et al. 2010). SOEing PCRs with primers IBS, EBS1d, EBS2 and EBS (Supplemental Table S2) were used to assemble and amplify the product for intron targeting to the 365/366 nucleotide region of the CD2344 gene, as outlined in the TargeTron users’ manual (Sigma Aldrich). The retargeting sequence was digested with BsrGI/HindIII and cloned into pMTL007C-E2. The resulting plasmid was transformed into HB101/pRK24 for conjugation into JIR8094 (O’Connor JR et al. 2006) (a generous gift from Aimee Shen) to generate Cd-CD2344−. Integration was confirmed by PCR. For all experiments involving Cd-CD2344−, the wild-type isogenic background strain, JIR8094, was used as the wild-type control.
Expression analysis
Genome-wide transcriptional profiling of C. difficile was conducted using custom-made Affymetrix GeneChips, which contain probes for all annotated coding sequences for C. difficile strain 630 as well as intergenic regions. RNA was purified from cecal contents and in vitro culture and cDNA was prepared, fragmented and labeled as described (Sonnenburg JL et al. 2005). GeneChip data were RMA-MS normalized as described and log2 transformed (Ng et al. 2013). Statistical significance for differential gene expression was determined using Significance Analysis of Microarrays (SAM) (Ng et al. 2013). The delta parameter was adjusted to achieve a FDR less than 1%, and this delta value was used to select significantly-regulated genes.
Quantification of fermentation metabolites
Frozen cecal contents (100–300mg) or frozen supernatants from in vitro growth curves were resuspended in 0.1% formic acid in water (Honeywell) and blended with a pellet pestle (Kimble chase). Acetate, butyrate, lactate, propionate and succinate standards were included in each run to generate standard curves for quantification. The samples were acidified (37% HCl), and SCFAs were extracted (500μl diethyl ether/extraction; 2 cycles). Each sample was derivatized with N-tert-butyldimethylsilyl-N-methyltrifluoracetamide (MTBSTFA; Sigma Aldrich) and quantified using a gas chromatograph (Model 7890A; Agilent Technologies, Santa Clara, CA) coupled to mass spectrometer detector (Model 5975C; Agilent Technologies). Analyses were carried out in a split mode (1:100) on a DB-5MSUI capillary column (30 m×0.25 mm, 0.25 μm film thickness, Agilent Technologies) using electronic impact (70 eV) as ionization mode and scan in m/z 50–550 mass range. The column head-pressure was 12 p.s.i. Injector, source and quadrupole temperatures were 250, 280 and 150 °C, respectively. The GC oven was programmed as follow: 75 °C held for 2 min, increased to 120 °C at 40 °C min−1, 120 °C held for 5 min, increased to 320 °C at 20 °C min−1 and held at 320 °C for 7 min.
Competitive index analysis
DNA was extracted from feces using the Powersoil DNA Isolation kit (MoBio). DNA quantification was conducted by Quantitative PCR using SYBR Green (ABgene) in a MX3000P thermocycler (Strategene, San Diego, CA). Levels of wild-type C. difficile were quantified by amplifying a 208 base-pair region of the CD2344 gene with the 2344F and 2344R primers (Supplemental Table S2), which included the area targeted for mutagenesis in the Cd-CD2344− mutant. Genomic DNA from Cd-CD2344− did not produce a band when amplified with these primers due to the insertion of the transposon within the region. The CDEP692 transposon specific sequencing primer and the 2344F primer (Supplemental Table S2) were used to determine the relative concentrations of the mutant, and did not amplify a band against wild-type C. difficile genomic DNA. The ratio of wild-type to mutant bacteria was normalized to the inoculum to determine the competitive index.
Statistical analyses
The Student’s t-test was used for statistical calculations involving two group comparisons, and * indicates p < 0.05, ** indicates p < 0.01 and *** indicates p < 0.001. For comparisons involving multiple experimental groups, one-way ANOVA with Bonferroni multiple comparisons test was employed, with **** indicating statistical significance at an alpha value of <0.05. Outliers were excluded from by the Grubbs’ statistical test. Error bars indicate standard deviation. n indicates the number of mice used per condition.
Supplementary Material
Supplemental Table S1, Related to Figure 1. Genes upregulated in Bt-biassociation vs. monoassociation
Fold change is provided when the gene’s expression provided a false discovery rate of <1% for the indicated comparison. Blocks of genes highlighted in red, green, and yellow indicate the succinate-to-butyrate pathway, sialic acid utilization, and ethanolamine pathway, respectively.
Supplemental Table S2, Related to Experimental Procedures. List of primers.
a., b., Each graph shows the number of genes in each COG category that were significantly upregulated in the comparison between (a.) germ-free mice mono-associated with C. difficile (mono) versus Bt-biassociated mice colonized with C. difficile (bi) and (b.) polysaccharide deficient diet versus standard diet. Letters represent the following COG categories: chromatin structure and dynamics (B); energy production and conversion (C); cell cycle, cell division and chromosome partitioning (D); amino acid transport and metabolism (E); nucleotide transport and metabolism (F); carbohydrate transport and metabolism (G); coenzyme transport and metabolism (H); lipid transport and metabolism (I); translation, ribosomal structure and biogenesis (J); transcription (K); replication, recombination and repair (L); cell wall, membrane and envelope biogenesis (M); cell motility (N); post-translational modification, protein turnover and chaperones (O); inorganic ion transport and metabolism (P); secondary metabolites biosynthesis, transport and catabolism (Q); general function prediction only (R); function unknown (S); signal transduction mechanisms (T); and intracellular trafficking, secretion and vesicular transport (U); and defense mechanisms (V). Bars labeled with * indicate a significant increase in gene abundance relative to representation in the genome (hypergeometric probability function p < 0.01).
c., Additional C. difficile operons with significant differences in expression between colonization state and diet. Colors indicate the deviation of each gene’s signal above (green) and below (purple) its mean expression value across all sixteen in vivo samples (n=4/group) at 5 days post-infection in the cecal contents.
d., e., Induction of C. difficile genes from the succinate to butyrate pathway, including cat1 (CD2343) (d) and abfT (CD2339) (e) in cecal contents at 5 days post-infection, normalized to growth in minimal media with glucose.
a. Schematic of the gene cluster comprising the succinate-to-butyrate pathway in Clostridium difficile strain 630.
b. Schematic of the metabolic pathway for the conversion of succinate to butyrate and fermentation of sorbitol. Potential complementarity of electron flow is noted between pathways.
a. Concentration of short-chain fatty acids acetate, succinate and propionate in cecal contents of mice consuming standard diet and colonized with C. difficile (Cd), Bt (Bt) or C. difficile and Bt (Cd + Bt) at 5 days post-infection (n=4/group).
b. Concentration of short-chain fatty acids acetate, butyrate, succinate and propionate in cecal contents of mice consuming polysaccharide-deficient diet and colonized with C. difficile (Cd), Bt (Bt) or C. difficile and Bt (Cd + Bt) and at 5 days post-infection (n=4/group).
c. Concentrations of acetate, propionate or lactate produced by Cd or Cd-CD2344− in minimal media containing 0.5% glucose (filled bars, MM + G) or 0.1% succinate (checkered bars, MM + S) (n=3/group).
d. Cd-CD2344− mutant C. difficile (Cd-CD2344−) density in feces of germ-free mice consuming polysaccharide-deficient diet and administered water (−) or water containing 1% sorbitol and 1% succinate (+) at 1 day post-infection (n=5/group).
e. C. difficile density in feces of Bt-biassociated mice infected with wild-type C. difficile (Bt + Cd) or Cd-CD2344− mutant C. difficile (Bt + Cd-CD2344−) at day 2 and 3 post-infection (n=5/group).
f. Levels of acetate, succinate and propionate detected in cecal contents of Bt-biassociated mice infected with wild-type C. difficile (Cd) or Cd-CD2344− mutant C. difficile (Cd-CD2344−) at day 3 post-infection (n=5/group).
a. Mean pathology score of H&E stained formalin fixed sections from mouse cecum of mice in monoassociation with C. difficile (Cd) or Bt-biassociation with C. difficile (Cd + Bt) at day 5 post-infection (n=4/group). Statistical analysis conducted by Mann Whitney nonparametric test. * indicates p<0.05.
b. Mean pathology scores from the lamina propria of the colon or cecum of mice colonized with Bt-biassociated mice infected with wild-type C. difficile (Bt + Cd) or Cd-CD2344− mutant C. difficile (Bt + Cd-CD2344−) at day 3 post-infection (n=4/group). Statistical analysis conducted by Mann Whitney nonparametric test. * indicates p<0.05.
c. Density of C. difficile in feces of conventional, streptomycin-treated mice infected with wild-type C. difficile (Cd) or Cd-CD2344− mutant C. difficile (Cd-CD2344−) (n=10/group).
Acknowledgments
The authors would like to give a special thanks to Steven Higginbottom for help with mouse experiments, Michelle St. Onge for the preparation of reagents, Purna Kashyap for valuable discussions, and Kristen Earle for protocol development. We would like to thank Allis Chien and the Stanford Mass Spectrometry facility for help with GC-MS. This research was supported by R01-DK085025 (to J.L.S.) and an NSF graduate fellowships (to K.J.W. and J.A.F). Justin L. Sonnenburg, Ph.D., holds an Investigators in the Pathogenesis of Infectious Disease Award from the Burroughs Wellcome Fund.
Footnotes
Author Contributions
J.A.F and J.L.S designed experiments, analyzed data and wrote the paper. J.A.F and K.J.W. performed experiments. D.M.B. conducted pathology scoring and imaging. B.C.W. contributed reagents.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alam NH, Meier R, Sarker SA, Bardhan PK, Schneider H, Gyr N. Partially hydrolysed guar gum supplemented comminuted chicken diet in persistent diarrhoea: a randomised controlled trial. Arch Dis Child. 2005;90:195–199. doi: 10.1136/adc.2003.040089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Acosta T, Leon C, Acosta-Gonzalez S, Parra-Soto H, Cluet-Rodriguez I, Rossell MR, Colina-Chourio JA. Beneficial role of green plantain [Musa paradisiaca] in the management of persistent diarrhea: a prospective randomized trial. J Am Coll Nutr. 2009;28:169–176. doi: 10.1080/07315724.2009.10719768. [DOI] [PubMed] [Google Scholar]
- Antunes LC, Han J, Ferreira RB, Lolic P, Borchers CH, Finlay BB. Effect of antibiotic treatment on the intestinal metabolome. Antimicrob Agents Chemother. 2011;55:1494–1503. doi: 10.1128/AAC.01664-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings JH, Beatty ER, Kingman SM, Bingham SA, Englyst HN. Digestion and physiological properties of resistant starch in the human large bowel. Br J Nutr. 1996;75:733–747. doi: 10.1079/bjn19960177. [DOI] [PubMed] [Google Scholar]
- Fischbach MA, Sonnenburg JL. Eating for two: how metabolism establishes interspecies interactions in the gut. Cell Host Microbe. 2011;10:336–347. doi: 10.1016/j.chom.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He M, Miyajima F, Roberts P, Ellison L, Pickard DJ, Martin MJ, Connor TR, Harris SR, Fairley D, Bamford KB, et al. Emergence and global spread of epidemic healthcare-associated Clostridium difficile. Nat Genet. 2013;45:109–113. doi: 10.1038/ng.2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heap JT, Cartman ST, Kuehne SA, Cooksley C, Minton NP. ClosTron-targeted mutagenesis. Methods Mol Biol. 2010;646:165–182. doi: 10.1007/978-1-60327-365-7_11. [DOI] [PubMed] [Google Scholar]
- Johnson S. Recurrent Clostridium difficile infection: a review of risk factors, treatments, and outcomes. J Infect. 2009;58:403–410. doi: 10.1016/j.jinf.2009.03.010. [DOI] [PubMed] [Google Scholar]
- Kashyap PC, Marcobal A, Ursell LK, Larauche M, Duboc H, Earle KA, Sonnenburg ED, Ferreyra JA, Higginbottom SK, Million M, et al. Complex interactions among diet, gastrointestinal transit, and gut microbiota in humanized mice. Gastroenterology. 2013;144:967–977. doi: 10.1053/j.gastro.2013.01.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenealy WR, Waselefsky DM. Studies on the substrate range of Clostridium kluyveri: The use of propanol and succinate. Arch Microbiol. 1985;141:187–194. [Google Scholar]
- Khoruts A, Dicksved J, Jansson JK, Sadowsky MJ. Changes in the composition of the human fecal microbiome after bacteriotherapy for recurrent Clostridium difficile-associated diarrhea. J Clin Gastroenterol. 2010;44:354–360. doi: 10.1097/MCG.0b013e3181c87e02. [DOI] [PubMed] [Google Scholar]
- Lawley TD, Clare S, Walker AW, Stares MD, Connor TR, Raisen C, Goulding D, Rad R, Schreiber F, Brandt C, et al. Targeted restoration of the intestinal microbiota with a simple, defined bacteriotherapy resolves relapsing Clostridium difficile disease in mice. PLoS Pathog. 2012;8:e1002995. doi: 10.1371/journal.ppat.1002995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawley TD, Croucher NJ, Yu L, Clare S, Sebaihia M, Goulding D, Pickard DJ, Parkhill J, Choudhary J, Dougan G. Proteomic and genomic characterization of highly infectious Clostridium difficile 630 spores. J Bacteriol. 2009;191:5377–5386. doi: 10.1128/JB.00597-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone CA, Stombaugh J, Gonzalez A, Ackermann G, Wendel D, Vázquez-Baeza Y, Jansson JK, Gordon JI, Knight R. Meta-analyses of studies of the human microbiota. Genome Res. 2013;23:1704–1714. doi: 10.1101/gr.151803.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macfarlane S, Macfarlane GT. Regulation of short-chain fatty acid production. Proc Nutr Soc. 2003;62:67–72. doi: 10.1079/PNS2002207. [DOI] [PubMed] [Google Scholar]
- Maier L, Vyas R, Cordova CD, Lindsay H, Schmidt TS, Brugiroux S, Periaswamy B, Bauer R, Sturm A, Schreiber F, et al. Microbiota-derived hydrogen fuels salmonella typhimurium invasion of the gut ecosystem. Cell Host Microbe. 2013;14:641–651. doi: 10.1016/j.chom.2013.11.002. [DOI] [PubMed] [Google Scholar]
- McNulty NP, Wu M, Erickson AR, Pan C, Erickson BK, Martens EC, Pudlo NA, Muegge BD, Henrissat B, Hettich RL, et al. Effects of diet on resource utilization by a model human gut microbiota containing Bacteroides cellulosilyticus WH2, a symbiont with an extensive glycobiome. PLoS Biol. 2013;11:e1001637. doi: 10.1371/journal.pbio.1001637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller BA, Chen LF, Sexton DJ, Anderson DJ. Comparison of the burdens of hospital-onset, healthcare facility-associated Clostridium difficile Infection and of healthcare-associated infection due to methicillin-resistant Staphylococcus aureus in community hospitals. Infect Control Hosp Epidemiol. 2011;32:387–390. doi: 10.1086/659156. [DOI] [PubMed] [Google Scholar]
- Muir JG, Yeow EG, Keogh J, Pizzey C, Bird AR, Sharpe K, O’Dea K, Macrae FA. Combining wheat bran with resistant starch has more beneficial effects on fecal indexes than does wheat bran alone. Am J Clin Nutr. 2004;79:1020–1028. doi: 10.1093/ajcn/79.6.1020. [DOI] [PubMed] [Google Scholar]
- Ng KM, Ferreyra JA, Higginbottom SK, Lynch JB, Kashyap PC, Gopinath S, Naidu N, Choudhury B, Weimer BC, Monack DM, et al. Microbiota-liberated host sugars facilitate post-antibiotic expansion of enteric pathogens. Nature. 2013;502:96–99. doi: 10.1038/nature12503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connor JR, Lyras D, Farrow KA, Adams V, Powell DR, Hinds J, Cheung JK, Rood JI. Construction and analysis of chromosomal Clostridium difficile mutants. Mol Microbiol. 2006;61:1335–1351. doi: 10.1111/j.1365-2958.2006.05315.x. [DOI] [PubMed] [Google Scholar]
- Pepin J. Improving the treatment of Clostridium difficile-associated disease: where should we start? Clin Infect Dis. 2006;43:553–555. doi: 10.1086/506357. [DOI] [PubMed] [Google Scholar]
- Pettit LJ, Browne HP, Yu L, Smits WK, Fagan RP, Barquist L, Martin MJ, Goulding D, Duncan SH, Flint HJ, et al. Functional genomics reveals that Clostridium difficile Spo0A coordinates sporulation, virulence and metabolism. BMC Genomics. 2014;15:160. doi: 10.1186/1471-2164-15-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabbani GH, Ahmed S, Hossain I, Islam R, Marni F, Akhtar M, Majid N. Green banana reduces clinical severity of childhood shigellosis: a double-blind, randomized, controlled clinical trial. Pediatr Infect Dis J. 2009;28:420–425. doi: 10.1097/INF.0b013e31819510b5. [DOI] [PubMed] [Google Scholar]
- Ramakrishna BS, Venkataraman S, Srinivasan P, Dash P, Young GP, Binder HJ. Amylase-resistant starch plus oral rehydration solution for cholera. N Engl J Med. 2000;342:308–313. doi: 10.1056/NEJM200002033420502. [DOI] [PubMed] [Google Scholar]
- Rivera EV, Woods S. Prevalence of asymptomatic Clostridium difficile colonization in a nursing home population: a cross-sectional study. J Gend Specif Med. 2003;6:27–30. [PubMed] [Google Scholar]
- Rivera-Chávez F, Winter SE, Lopez CA, Xavier MN, Winter MG, Nuccio SP, Russell JM, Laughlin RC, Lawhon SD, Sterzenbach T, et al. Salmonella uses energy taxis to benefit from intestinal inflammation. PLoS Pathog. 2013;9:e1003267. doi: 10.1371/journal.ppat.1003267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenburg ED, Zheng H, Joglekar P, Higginbottom SK, Firbank SJ, Bolam DN, Sonnenburg JL. Specificity of polysaccharide use in intestinal bacteroides species determines diet-induced microbiota alterations. Cell. 2010;141:1241–1252. doi: 10.1016/j.cell.2010.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenburg JL, Xu J, Leip DD, Chen CH, Westover BP, Weatherford J, Buhler JD, Gordon JI. Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science. 2005;307:1955–1959. doi: 10.1126/science.1109051. [DOI] [PubMed] [Google Scholar]
- Theriot CM, Koenigsknecht MJ, Carlson PE, Hatton GE, Nelson AM, Li B, Huffnagle GB, Li ZJ, Young VB. Antibiotic-induced shifts in the mouse gut microbiome and metabolome increase susceptibility to Clostridium difficile infection. Nat Commun. 2014;5:3114. doi: 10.1038/ncomms4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiennimitr P, Winter SE, Winter MG, Xavier MN, Tolstikov V, Huseby DL, Sterzenbach T, Tsolis RM, Roth JR, Bäumler AJ. Intestinal inflammation allows Salmonella to use ethanolamine to compete with the microbiota. Proc Natl Acad Sci U S A. 2011;108:17480–17485. doi: 10.1073/pnas.1107857108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukahara T, Ushida K. Succinate accumulation in pig large intestine during antibiotic-associated diarrhea and the constitution of succinate-producing flora. J Gen Appl Microbiol. 2002;48:143–154. doi: 10.2323/jgam.48.143. [DOI] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ridaura VK, Faith JJ, Rey FE, Knight R, Gordon JI. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci Transl Med. 2009;1:6ra14. doi: 10.1126/scitranslmed.3000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Nood E, Vrieze A, Nieuwdorp M, Fuentes S, Zoetendal EG, de Vos WM, Visser CE, Kuijper EJ, Bartelsman JF, Tijssen JG, et al. Duodenal infusion of donor feces for recurrent Clostridium difficile. N Engl J Med. 2013;368:407–415. doi: 10.1056/NEJMoa1205037. [DOI] [PubMed] [Google Scholar]
- Wilson KH, Perini F. Role of competition for nutrients in suppression of Clostridium difficile by the colonic microflora. Infection and immunity. 1988;56:2610–2614. doi: 10.1128/iai.56.10.2610-2614.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilcox MH, Mooney L, Bendall R, Settle CD, Fawley WN. A case-control study of community-associated Clostridium difficile infection. J Antimicrob Chemother. 2008;62:388–396. doi: 10.1093/jac/dkn163. [DOI] [PubMed] [Google Scholar]
- Wolff RA, Urben GW, O’Herrin SM, Kenealy WR. Dehydrogenases involved in the conversion of succinate to 4-hydroxybutanoate by Clostridium kluyveri. Appl Environ Microbiol. 1993;59:1876–1882. doi: 10.1128/aem.59.6.1876-1882.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong JM, de Souza R, Kendall CW, Emam A, Jenkins DJ. Colonic health: fermentation and short chain fatty acids. J Clin Gastroenterol. 2006;40:235–243. doi: 10.1097/00004836-200603000-00015. [DOI] [PubMed] [Google Scholar]
- Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA, Bewtra M, Knights D, Walters WA, Knight R, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334:105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Table S1, Related to Figure 1. Genes upregulated in Bt-biassociation vs. monoassociation
Fold change is provided when the gene’s expression provided a false discovery rate of <1% for the indicated comparison. Blocks of genes highlighted in red, green, and yellow indicate the succinate-to-butyrate pathway, sialic acid utilization, and ethanolamine pathway, respectively.
Supplemental Table S2, Related to Experimental Procedures. List of primers.
a., b., Each graph shows the number of genes in each COG category that were significantly upregulated in the comparison between (a.) germ-free mice mono-associated with C. difficile (mono) versus Bt-biassociated mice colonized with C. difficile (bi) and (b.) polysaccharide deficient diet versus standard diet. Letters represent the following COG categories: chromatin structure and dynamics (B); energy production and conversion (C); cell cycle, cell division and chromosome partitioning (D); amino acid transport and metabolism (E); nucleotide transport and metabolism (F); carbohydrate transport and metabolism (G); coenzyme transport and metabolism (H); lipid transport and metabolism (I); translation, ribosomal structure and biogenesis (J); transcription (K); replication, recombination and repair (L); cell wall, membrane and envelope biogenesis (M); cell motility (N); post-translational modification, protein turnover and chaperones (O); inorganic ion transport and metabolism (P); secondary metabolites biosynthesis, transport and catabolism (Q); general function prediction only (R); function unknown (S); signal transduction mechanisms (T); and intracellular trafficking, secretion and vesicular transport (U); and defense mechanisms (V). Bars labeled with * indicate a significant increase in gene abundance relative to representation in the genome (hypergeometric probability function p < 0.01).
c., Additional C. difficile operons with significant differences in expression between colonization state and diet. Colors indicate the deviation of each gene’s signal above (green) and below (purple) its mean expression value across all sixteen in vivo samples (n=4/group) at 5 days post-infection in the cecal contents.
d., e., Induction of C. difficile genes from the succinate to butyrate pathway, including cat1 (CD2343) (d) and abfT (CD2339) (e) in cecal contents at 5 days post-infection, normalized to growth in minimal media with glucose.
a. Schematic of the gene cluster comprising the succinate-to-butyrate pathway in Clostridium difficile strain 630.
b. Schematic of the metabolic pathway for the conversion of succinate to butyrate and fermentation of sorbitol. Potential complementarity of electron flow is noted between pathways.
a. Concentration of short-chain fatty acids acetate, succinate and propionate in cecal contents of mice consuming standard diet and colonized with C. difficile (Cd), Bt (Bt) or C. difficile and Bt (Cd + Bt) at 5 days post-infection (n=4/group).
b. Concentration of short-chain fatty acids acetate, butyrate, succinate and propionate in cecal contents of mice consuming polysaccharide-deficient diet and colonized with C. difficile (Cd), Bt (Bt) or C. difficile and Bt (Cd + Bt) and at 5 days post-infection (n=4/group).
c. Concentrations of acetate, propionate or lactate produced by Cd or Cd-CD2344− in minimal media containing 0.5% glucose (filled bars, MM + G) or 0.1% succinate (checkered bars, MM + S) (n=3/group).
d. Cd-CD2344− mutant C. difficile (Cd-CD2344−) density in feces of germ-free mice consuming polysaccharide-deficient diet and administered water (−) or water containing 1% sorbitol and 1% succinate (+) at 1 day post-infection (n=5/group).
e. C. difficile density in feces of Bt-biassociated mice infected with wild-type C. difficile (Bt + Cd) or Cd-CD2344− mutant C. difficile (Bt + Cd-CD2344−) at day 2 and 3 post-infection (n=5/group).
f. Levels of acetate, succinate and propionate detected in cecal contents of Bt-biassociated mice infected with wild-type C. difficile (Cd) or Cd-CD2344− mutant C. difficile (Cd-CD2344−) at day 3 post-infection (n=5/group).
a. Mean pathology score of H&E stained formalin fixed sections from mouse cecum of mice in monoassociation with C. difficile (Cd) or Bt-biassociation with C. difficile (Cd + Bt) at day 5 post-infection (n=4/group). Statistical analysis conducted by Mann Whitney nonparametric test. * indicates p<0.05.
b. Mean pathology scores from the lamina propria of the colon or cecum of mice colonized with Bt-biassociated mice infected with wild-type C. difficile (Bt + Cd) or Cd-CD2344− mutant C. difficile (Bt + Cd-CD2344−) at day 3 post-infection (n=4/group). Statistical analysis conducted by Mann Whitney nonparametric test. * indicates p<0.05.
c. Density of C. difficile in feces of conventional, streptomycin-treated mice infected with wild-type C. difficile (Cd) or Cd-CD2344− mutant C. difficile (Cd-CD2344−) (n=10/group).