Abstract
Geminiviruses are ssDNA plant viruses that cause significant agricultural losses worldwide. The viruses do not encode a polymerase protein and must reprogram differentiated host cells to re-enter the S-phase of the cell cycle for the virus to gain access to the host-replication machinery for propagation. To date, 3 Beet curly top virus (BCTV) encoded proteins have been shown to restore DNA replication competency: the replication-initiator protein (Rep), the C2 protein, and the C4 protein. Ectopic expression of the BCTV C4 protein leads to a severe developmental phenotype characterized by extensive hyperplasia. We recently demonstrated that C4 interacts with 7 of the 10 members of the Arabidopsis thaliana SHAGGY-like protein kinase gene family and characterized the interactions of C4 and C4 mutants with AtSKs. Herein, we propose a model of how C4 functions.
Keywords: AtSK, BCTV, C4 protein, curtovirus, cell cycle, geminivirus, hyperplasia, replication
Geminiviruses (family Geminiviridae) are small plant DNA viruses with circular single-stranded genomes that cause significant losses in food, fiber, and cash crops worldwide.1 The family consists of 7 genera: Becurtovirus, Curtovirus, Eragrovirus, Mastrevirus, Topocuvirus, and Turncurtovirus have monopartite genomes, while the Begomovirus can have mono- or bipartite genomes.2 The absence of a polymerase gene in geminiviruses requires that the S-phase of the cell cycle be reactivated in terminally differentiated cells following virus infection to gain access to cellular DNA replication machinery for virus propagation. The replication-initiator protein (Rep) is present in all geminiviruses and is the only viral protein essential for replication. It is required to restore DNA replication competency to terminally differentiated host cells.3,4 In addition, in the curtovirus genus, the Beet curly top virus (BCTV) C2 protein is involved in enhancing replication competency, although the molecular mechanism is unclear,5 and the BCTV C4 protein has been shown to restore DNA replication competency.6
Most geminiviruses do not induce cell proliferation, but some curtovirus C4 proteins are very proficient at regulating cell cycle progression and promoting mitosis.6-9 The BCTV C4 protein induces vein swelling and enations in BCTV-infected hosts and hyperplasia in transgenic Arabidopsis plants.6-10 Curtovirus C4 genes, like all geminivirus C4 and AC4 genes (the C4 gene in bipartite begomoviruses is designated AC4), are nested within the Rep genes and encode small proteins of approximately 10 kDa, depending on the virus. The BCTV C4 protein interacts with members of the Arabidopsis SHAGGY-like protein kinase (AtSK) family.7,11,12 AtSKs are homologues of the glycogen synthase kinase 3 (GSK3) family of serine/threonine kinases (GSK3α and GSK3β) in animals.13 However, in plants the AtSK gene family has evolved into 10 members to address diverse plant-specific functions.14,15 Seven AtSK members have been implicated in brassinosteroid (BR) signaling.16 The BRs are steroid hormones that regulate plant growth and development. We and others showed that expression of C4 in transgenic Arabidopsis disrupts the BR pathway, indicating a direct role for C4 in regulating BR signaling.7,11,12
C4 protein from Beet curly top virus interacts with multiple AtSKs
We recently showed that C4 interacts with 7 of the 10 AtSKs (clade 1 AtSKs-11,-12,-13; clade 2 AtSKs-21,-22,-23; and clade 3 AtSK32),12 the same 7 AtSKs involved in regulating BR signaling (Table 1). Bikinin, a selective inhibitor of the 7 AtSKs implicated in BR signaling, induced hyperplasia in Arabidopsis similar to that induced by C4 (Table 1).12 An array of functions are attributed to AtSKs,16 especially AtSK21, which is the most well studied AtSK. The numerous cellular functions regulated by AtSK21 suggests that other AtSK family members may also regulate a wide variety of overlapping and individual AtSK-specific functions.15,16,17 Even within redundant functions, the degree of substrate specificity can vary between AtSKs.18 In some instances, preferential tissue-specific AtSK expression is observed,15 and it is likely that expression levels of individual AtSKs vary at different stages of development. Therefore, C4 likely impacts a large number of diverse functions regulated by clade 1 AtSKs, clade 2 AtSKs and AtSK32.
Table 1.
Arabidopsis thaliana SHAGGY-like protein kinase family.
| Clade | Name | Locus Identifier | C4 Interaction/BR Signaling/Bikinin Inhibition |
|---|---|---|---|
| 1 | AtSK11 | AT5G26751 | + |
| AtSK12 | AT3G05840 | + | |
| AtSK13 | AT5G14640 | + | |
| 2 | AtSK21 | AT4G18710 | + |
| AtSK22 | AT1G06390 | + | |
| AtSK23 | AT2G30980 | + | |
| 3 | AtSK31 | AT3G61140 | − |
| AtSK32 | AT4G00720 | + | |
| 4 | AtSK41 | AT1G09840 | − |
| AtSK42 | AT1G57870 | − |
Requirements for a functional C4/AtSK interaction
The C4/AtSK interactions and C4 function require the presence of a phosphorylated Ser/Thr residue at amino acid position 49.12 In addition, AtSKs must be catalytically active to interact with C4, supporting their role in phosphorylating C4.12 Residue 49 is part of a proline-directed Ser/Thr kinase phosphorylation motif (Ser/Thr-Pro). Ser/Thr-Pro motifs are phosphorylated by members of a large family of proline-directed Ser/Thr kinases in eukaryotes, including AtSKs, and regulate a diverse array of cellular processes.19 While most peptide bonds have the more energetically favored trans isomer, proline can exist in either a cis or trans conformation. Phosphorylation of Ser/Thr-Pro motifs limits the rate of cis/trans isomerization, which plays an important role in regulating protein structure,19 and may be essential in regulating C4 function.
C4 requires plasma membrane localization for function
Geminivirus C4/AC4 proteins contain a conserved N-myristoylation motif required for localization to the plasma membrane (PM).11,20 We recently showed that BCTV C4/AtSK complexes localize primarily to the PM and nucleus. Disruption of the C4 N-myristoylation motif resulted in a nonfunctional C4 mutant, C4G2A, that failed to induce hyperplasia. The mutant retained the ability to bind AtSKs, but localized to the cytosol and nucleus, indicating that the formation of C4/AtSK complexes does not require association with the PM, but that functional C4/AtSK complexes do require PM localization.12 The targeting of a portion of both C4G2A/AtSK and C4/AtSK complexes to the nucleus suggests that nuclear localization is not necessary to induce hyperplasia. Similarly, the AC4 protein of the East African cassava mosaic Cameroon virus (EACMCV) required an N-myristoylation motif and PM localization for function. Interestingly, the AC4 protein suppresses the systemic phase of RNA silencing and has not been shown to induce hyperplasia.20
N-myristoylation alone is likely not sufficient to stably anchor a protein to the PM.21,22 A second signal in the myristoylated protein, such as another fatty acyl chain, a polybasic group of amino acids that binds the negatively charged phospholipids within the PM, or a domain that interacts with another membrane protein is required for stability. Indeed, PM localization of the AC4 protein of EACMCV was shown to require palmitoylation of Cys 3 in addition to myristoylation of Gly 2 in the N-myristoylation motif.20 While BCTV C4 requires Gly2 for myristoylation,11 a second signal has not been identified. Two likely possibilities are the sole Cys9 residue in C4, which is a putative site for palmitoylation, and/or the 3 basic amino acids Lys13, Lys15, and Arg17. The latter would be similar to the Src protein of Rous sarcoma virus, where 3 basic amino acids at the N-terminus of the protein are necessary to stabilize localization of the myristoylated protein to the PM.23 Indeed, a mutation in C4 of Lys13 to Ala13 resulted in a very mild C4-like phenotype.12
How does the BCTV C4 protein induce hyperplasia?
While extensive details of the mechanism underlying the function of the C4 protein remain unknown, our recent findings,12 and previously published work,7,11 provide a basis to propose a tentative mechanistic model of how the BCTV C4 protein functions (Fig. 1). As indicated above, the ability of the C4 protein to restore replicational competency, as manifested by extensive hyperplasia, requires the formation of functional C4/AtSK complexes at the PM. Therefore, inhibition or modulation of AtSK function(s) by C4 likely occurs at the PM. This is supported by the fact that the presence or absence of C4 influences specific AtSK-PM interactions. In the absence of C4, clade 2 AtSKs localized to the cytosol, nucleus, and punctate regions on the PM.12 This is in line with previous reports of AtSKs localizing to the PM; AtSK21 was localized to the nucleus, cytosol, and PM and AtSK41 was localized to the cytosol and PM.24-26 When C4 is present, C4/clade 2 AtSK complexes are distributed evenly along the PM membrane. This shift in localization is C4-dependent, since the PM localization of AtSK41, which does not interact with C4, is unchanged in the presence of C4.12 More importantly, recent evidence suggests that PM localization is critical for some AtSK function, AtSKs (putatively clade 1 and clade2) were shown to interact with the tracheary element differentiation inhibitory factor receptor (TDR) at the PM and play a crucial role in regulating xylem cell differentiation.26 Furthermore, in Drosophila, a PM-proximal GSK-3 activates the Wnt signaling pathway by phosphorylating LRP6, a transmembrane protein, that is critical for signal transduction.27,28 Similarly, the bikinin-induced hyperplastic phenotype could be explained by bikinin inhibiting the function of AtSKs that function at the PM.
Figure 1.
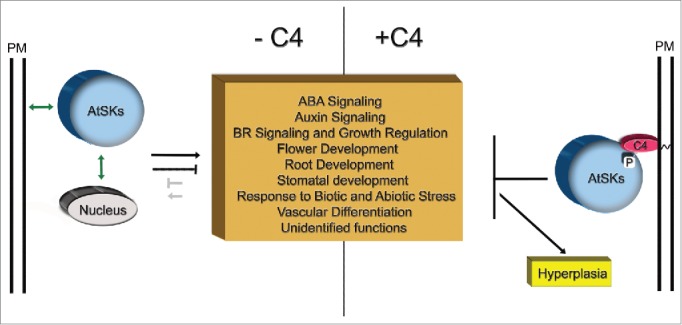
C4/AtSK interaction model. In the absence of C4, AtSKs (clade 1 AtSKs, clade 2 AtSKs and AtSK32) activate (black arrow) or inhibit (black bar) proteins involved in the various cellular processes indicated (brown box). In a few instances, cellular proteins activate (gray arrow) or inhibit (gray bar) AtSKs. In the absence of C4, AtSKs may be localized to the cytosol, nucleus and/or PM. Double-head arrows indicate AtSKs association/disassociation with the nucleus and PM. In the presence of C4, AtSK activities are inhibited (black bar). Detailed description of activation or inhibitory functions attributed to specific members of clade 1 and clade2 AtSKs, as well as AtSK32 and cellular proteins are described.16.
While current experimental data support the model presented, additional information is needed to elucidate details on how the C4 protein modulates the host cell cycle. Future experiments to identify specific C4/AtSK complexes involved in the induction of hyperplasia, possible additional C4-interacting host proteins, and possible host proteins that interact with C4/AtSK complexes or modified C4/AtSK complexes will provide additional insights into how the BCTV C4 protein usurps the host cell cycle to promote mitosis.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
The authors thank John Sherwood and Brian Kvitko for critically reviewing the article and Kisha Shelton for assistance with Figure. 1
Funding
This research was funded by the Georgia Agricultural Experiment Station.
References
- 1.Varma A, Malathi VG. Emerging geminivirus problem: a serious threat to crop production. Ann Appl Biol 2003; 142:145-64; http://dx.doi.org/ 10.1111/j.1744-7348.2003.tb00240.x [DOI] [Google Scholar]
- 2.Varsani A, Navas-Castillo J, Moriones E, Hernandez-Zepeda C, Idris A, Brown JK, Murilo Zerbini F, Martin DP. Establishment of three new genera in the family Geminiviridae: Becurtovirus, Eragrovirus and Turncurtovirus. Arch Virol 2014; 159:2193-203; PMID:24658781; http://dx.doi.org/ 10.1007/s00705-014-2050-2 [DOI] [PubMed] [Google Scholar]
- 3.Hanley-Bowdoin L, Bejarano ER, Robertson D, Mansoor S. Geminiviruses: masters at redirecting and programming plant processes. Nature Rev Microbiol 2013; 11:777-88; http://dx.doi.org/ 10.1038/nrmicro3117 [DOI] [PubMed] [Google Scholar]
- 4.Gutzat R, Borghi L, Gruissem W. Emerging roles of RETINOBLASTOMA-RELATED proteins in evolution and plant development. Trends Plant Sci 2012; 17:139-48; PMID:22240181; http://dx.doi.org/ 10.1016/j.tplants.2011.12.001 [DOI] [PubMed] [Google Scholar]
- 5.Caracuel Z, Lozano-Duran R, Huguet S, Arroyo-Mateos M, Rodriguez-Negrete EA, Bejarano ER. C2 from Beet curly top virus promotes a cell environment suitable for efficient replication of geminiviruses, providing a novel mechanism of viral synergism. New Phytol 2012; 194:846-858; PMID:22404507 http://dx.doi.org/ 10.1111/j.1469-8137.2012.04080.x [DOI] [PubMed] [Google Scholar]
- 6.Latham JR, Saunders K, Pinner MS, Stanley J. Induction of plant cell division by beet curly top virus gene C4. Plant J 1997; 11:1273-1283; http://dx.doi.org/ 10.1046/j.1365-313X.1997.11061273.x [DOI] [Google Scholar]
- 7.Mills-Lujan K, Deom CM. Geminivirus C4 protein alters Arabidopsis development. Protoplasma 2010; 239:95-110; PMID:20091067; http://dx.doi.org/ 10.1007/s00709-009-0086-z [DOI] [PubMed] [Google Scholar]
- 8.Park J, Hwang HS, Buckley KJ, Park JB, Auh CK, et al.. C4 protein of Beet severe curly top virus is a pathomorphogenetic factor in Arabidopsis. Plant Cell Rep 2010; 29:1377-1389; PMID:20960205; http://dx.doi.org/ 10.1007/s00299-010-0923-8 [DOI] [PubMed] [Google Scholar]
- 9.Lai J, Chen H, Teng K, Zhao Q, Zhang Z, et al.. RKP, a RING finger E9 ligase induced by BSCTV C4 protein, affects geminivirus infection by regulation of the plant cell cycle. Plant J 2009; 57:905-917; PMID:19000158; http://dx.doi.org/ 10.1111/j.1365-313X.2008.03737.x [DOI] [PubMed] [Google Scholar]
- 10.Esau K, Hoefert LL. Hyperplastic phloem in sugarbeet leaves infected with the beet curly top virus. Amer J Bot 1978; 65:772-783; http://dx.doi.org/ 10.2307/2442153 [DOI] [Google Scholar]
- 11.Piroux N, Saunders K, Page A, Stanley J. Geminivirus pathogenicity protein C4 interacts with Arabidopsis thaliana shaggy-related protein kinase AtSKη, a component of the brassinosteroid signaling pathway. Virol 2007; 362:428-440; PMID:17280695; http://dx.doi.org/ 10.1016/j.virol.2006.12.034 [DOI] [PubMed] [Google Scholar]
- 12.Mills-Lujan K, Andrews DL, Chou C-W, Deom CM. The roles of phosphorylation and SHAGGY-like protein kinases in geminivirus C4 protein induced hyperplasia. PLoS One 2015; 10(3):e0122356; PMID:25815729; http://dx.doi.org/ 10.1371/journal.pone.0122356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Doble BW, Woodgett JR. GSK-3: tricks of the trade for a multi-tasking kinase. J Cell Sci 2003; 116:1175-86; PMID:12615961; http://dx.doi.org/ 10.1242/jcs.00384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saidi Y, Hearn TJ, Coates JC. Function and evolution of “green” GSK3/Shaggy-like kinases. Trends in Plant Sci 2012; 17:39-46; http://dx.doi.org/ 10.1016/j.tplants.2011.10.002 [DOI] [PubMed] [Google Scholar]
- 15.Qi X, Chanderbali AS, Wong GK-S, Soltis DE, Soltis PS. Phylogeny and evolutionary history of glycogen synthase kinase 3/SHAGGY-like kinase genes in land plants. BMC Evol Biol 2013; 13:143PMID:23834366; http://dx.doi.org/ 10.1186/1471-2148-13-143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Youn J-H, Kim TW. Functional insights of plant GSK-like kinases: multi-taskers in diverse cellular signal transduction pathways. Mol Plant 2015; 8:552-65; PMID:25655825; http://dx.doi.org/ 10.1016/j.molp.2014.12.006 [DOI] [PubMed] [Google Scholar]
- 17.Youn JH, Kim TW, Kim EJ, Bu S, Kim SK, Wang ZY, Kim TW. Structural and functional characterization of Arabidopsis GSK3-like kinase AtSK12. Mol Cells 2013; 36:564-70; PMID:24292946; http://dx.doi.org/ 10.1007/s10059-013-0266-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rozhon W, Mayerhofer J, Petutschnig E, Fujioka S, Jonak C. ASKθ, a group-III Arabidopsis GSK3, functions in the brassinosteroid signaling pathway. Plant J 2010; 62:215-23; PMID:20128883; http://dx.doi.org/ 10.1111/j.1365-313X.2010.04145.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ubersax JA and Ferrell JE Jr. Mechanism of specificity in protein phosphorylation. Nat Rev Mol Cell Biol. 2007; 8:530-41; http://dx.doi.org/ 10.1038/nrm2203 [DOI] [PubMed] [Google Scholar]
- 20.Fondong VN, Reddy RV, Lu C, Hankoua B, Felton C, Czymmek K, Achenjang F. The consensus N-myristoylation motif of a geminivirus AC4 protein is required for membrane binding and pathogenicity. Mol Plant-Microbe Interact 2007; 20:380-91; PMID:17427808; http://dx.doi.org/ 10.1094/MPMI-20-4-0380 [DOI] [PubMed] [Google Scholar]
- 21.Resh MD. Trafficking and signaling by fatty-acylated and prenylated proteins. Nature Chem Biol 2006; 2:584-90; http://dx.doi.org/ 10.1038/nchembio834 [DOI] [PubMed] [Google Scholar]
- 22.Wright MH, Heal WP, Mann DJ, Tate EW. Protein myristoylation in health and disease. J Chem Biol 2010; 3:19-35; PMID:19898886; http://dx.doi.org/ 10.1007/s12154-009-0032-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Buser CA, Sigal CT, Resh MD, McLaughlin S. Membrane binding of myristoylated peptides corresponding to the NH2 terminus of Src. Biochem 1994; 33:13093-101; http://dx.doi.org/ 10.1021/bi00248a019 [DOI] [PubMed] [Google Scholar]
- 24.Vert G, Chory J. Downstream nuclear events in brassinosteroid signaling. Nature 2006; 441:96-100; PMID:16672972; http://dx.doi.org/ 10.1038/nature04681 [DOI] [PubMed] [Google Scholar]
- 25.Bayer RG, Stael S, Rocha AG, Mair A, Vothknecht UC, Teige M. Chloroplast-localized protein kinases: a step forward towards a complete inventory. J Exp Biol 2012; 63 1713-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kondo Y, Ito T, Nakagami H, Hirakawa Y, Saito M, Tamaki T, Shirasu K, Fukuda H. Plant GSK3 proteins regulate xylem cell differentiation downstream of TDIF-TDR signaling. Nature Comms 2013; 5:3504. [DOI] [PubMed] [Google Scholar]
- 27.Zeng X, Tamai K, Doble B, Li S, Huang H, Habas R, Okamura H, Woodgett J, He X. A dual-kinase mechanism for Wnt co-receptor phosphorylation and activation. Nature 2005; 438:873-7; PMID:16341017; http://dx.doi.org/ 10.1038/nature04185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mannava AG, Tolwinski NS. Membrane bound GSK-3 activates Wnt signaling through Disheveled and Arrow. PLoS One 2015; 10:e0121879; PMID:25848770; http://dx.doi.org/ 10.1371/journal.pone.0121879 [DOI] [PMC free article] [PubMed] [Google Scholar]


