Abstract
Background:
A genome-wide association study (GWAS) suggested inherited genetic single-nucleotide polymorphisms (SNPs) affecting overall survival (OS) in advanced pancreatic cancer. To identify robust clinical biomarkers, we tested the strongest reported candidate loci in an independent patient cohort, assessed cellular drug sensitivity, and evaluated molecular effects.
Methods:
This study comprised 381 patients with histologically verified pancreatic ductal adenocarcinoma treated with gemcitabine-based chemotherapy. The primary outcome was the relationship between germline polymorphisms and OS. Functional assays addressed pharmacological dose-response effects in lymphoblastoid cell lines (LCLs) and pancreatic cancer cell lines (including upon RNAi), gene expression analyses, and allele-specific transcription factor binding. All statistical tests were two-sided.
Results:
The A allele (26% in Caucasians) at SNP rs11644322 in the putative tumor suppressor gene WWOX conferred worse prognosis. Median OS was 14 months (95% confidence interval [CI] = 12 to 15 months), 13 months (95% CI = 11 to 15 months), and nine months (95% CI = 7 to 12 months) for the GG, GA, and AA genotypes, respectively (P trend < .001 for trend in univariate log-rank assuming a codominant mode of inheritance; advanced disease subgroup P trend < .001). Mean OS was 25 months (95% CI = 21 to 29 months), 19 months (95% CI = 15 to 22 months), and 13 months (95% CI = 10 to 16 months), respectively. This effect held true after adjustment for age, performance status according to Eastern Cooperative Oncology Group classification, TNM, grading, and resection status and was comparable with the strongest established prognostic factors in multivariable analysis. Consistently, reduced responsiveness to gemcitabine, but not 5-fluorouracil, along with lower WWOX expression was demonstrated in LCLs harboring the AA genotype. Likewise, RNAi-mediated WWOX knockdown in pancreatic cancer cells confirmed differential cytostatic drug sensitivity. In electrophoretic mobility shift assays, the A allele exhibited weaker binding of Sp family members Sp1/Sp3.
Conclusions:
WWOX rs11644322 represents a major predictive factor in gemcitabine-treated pancreatic cancer. Decreased WWOX expression may interfere with gemcitabine sensitivity, and allele-specific binding at rs11644332 might be a causative molecular mechanism behind the observed clinical associations.
Pancreatic ductal adenocarcinoma (PDAC) represents the predominant phenotype of pancreatic cancer. In Western countries, the current cumulative life risk adds up to 1.5%, with no marked sex preference. Among all malignancies, its incidence amounts to 3.5%, and it is the fourth leading cause of death from cancer (1). The overall five-year survival rate is around 4% to 8% and barely exceeds 20% to 25%, even when surgical resection for curative intention is feasible (2). The nucleoside analogue gemcitabine constitutes a current standard first-line therapy. The advantage in overall survival (OS) in comparison with 5-fluorouracil is very moderate; however, gemcitabine has some benefits regarding symptom control, with fewer side effects (3,4). Even though the overall therapeutic response toward gemcitabine is rather small, a subset of patients demonstrates substantial benefit. The reasons for these differences are not yet understood and reveal a need for robust predictive biomarkers.
Innocenti et al. recently reported a genome-wide association study (GWAS) for single-nucleotide polymorphisms (SNPs) in regard to OS in patients treated with gemcitabine for advanced pancreatic cancer (5,6). That GWAS suggested a single-nucleotide polymorphism in IL17F (rs763780) as a predictive biomarker (5).
We set out to evaluate the top five ranking associations in the referenced GWAS data in an independent and similarly scaled patient cohort and to assess cellular and molecular functionality. As the SNP rs763780 (1st in GWAS) is in high linkage disequilibrium (LD) with rs7771466 (2nd in GWAS), we decided to replace the latter in our analysis with another SNP among the top twenty in the GWAS list. We selected rs10883617 in BTRC (ranked on 13th) because this gene was reported to show oncogenic activity in pancreatic cancer cell lines (7) and was related to gemcitabine effectiveness (8). Thus, we investigated five genetic markers in 381 patients with adenoductal pancreatic cancer in relation to OS following gemcitabine-based chemotherapy. For a reproducibly clinical association, functional elucidations were intended to understand underlying mechanistic actions. A robust biomarker supported by molecular effects and proof-of-principle investigations is expected to assist for therapy stratification in gemcitabine-treated PDAC.
Methods
Patient Cohorts
Patients with histopathologically confirmed PDAC (without ampullary carcinoma) with adjuvant or palliative gemcitabine-containing chemotherapy were included in this study. Written informed consent was obtained from each patient. In total, 381 patients were recruited at three university medical centers in Germany (Göttingen, n = 142; Hamburg, n = 159; Heidelberg, n = 80), with investigations approved by local institutional ethic review boards at these three sites. Table 1 summarizes patient and clinical baseline data. Staging and grading were employed according to current standard classifications (9,10).
Table 1.
Distribution of patient baseline parameters, tumor stages, chemotherapy regimens, as well as time of follow-up and overall survival in the three study cohorts
| Variable | Cohort 1 Göttingen (n = 142) |
Cohort 2 Heidelberg (n = 80) |
Cohort 3 Hamburg (n = 159) |
|---|---|---|---|
| Age, median (IQD, range), y | 68 (61–73, 44–88) | 62 (55–67, 34–77) | 65 (58–72, 28–88) |
| Sex, No. (%) | |||
| Female | 68 (47.9) | 38 (47.5) | 64 (40.3) |
| Male | 74 (52.1) | 42 (52.5) | 95 (59.7) |
| Performance status, No. (%) | |||
| Classified | 142 | 80 | 151 |
| ECOG 0 | 5 (3.5) | 43 (53.7) | 26 (17.2) |
| ECOG 1 | 87 (61.3) | 28 (35.0) | 66 (43.7) |
| ECOG 2 | 45 (31.7) | 8 (10.0) | 53 (35.1) |
| ECOG 3 | 5 (3.5) | 1 (1.3) | 6 (4.0) |
| T stage, No. (%)* | |||
| Classified | 142 | 80 | 159 |
| 1 | 2 (1.4) | 0 (0.0) | 8 (5.0) |
| 2 | 7 (4.9) | 0 (0.0) | 21 (13.2) |
| 3 | 108 (76.1) | 76 (95.0) | 106 (66.7) |
| 4 | 25 (17.6) | 4 (5.0) | 24 (15.1) |
| N stage, No. (%) | |||
| Classified | 142 | 79 | 159 |
| 0 | 22 (15.5) | 5 (6.3) | 50 (31.4) |
| 1† | 120 (84.5) | 74 (93.7) | 109 (68.6) |
| M stage, No. (%) | |||
| Classified | 142 | 80 | 159 |
| 0 | 113 (79.6) | 73 (91.2) | 105 (66.0) |
| 1 | 29 (20.4) | 7 (8.8) | 54 (34.0) |
| Resection status, No. (%) | |||
| Classified | 142 | 80 | 159 |
| 0 | 45 (31.7) | 37 (46.3) | 98 (61.6) |
| 1 | 52 (36.6) | 39 (48.7) | 42 (26.4) |
| 2 | 2 (1.4) | 4 (5.0) | 19 (12.0) |
| Not resected | 43 (30.3) | 0 (0) | 0 (0) |
| Grading, No. (%) | |||
| Classified | 142 | 80 | 159 |
| G1 | 0 (0.0) | 2 (2.5) | 21 (13.2) |
| G2 | 101 (71.1) | 55 (68.8) | 92 (57.9) |
| G3 | 41 (28.9) | 23 (28.7) | 46 (28.9) |
| Chemotherapy regimen, No. (%) | |||
| Classified | 142 | 80 | 159 |
| Gemcitabine mono | 90 (63.4) | 60 (75.0) | 90 (56.6) |
| Gemcitabine combination | 52 (36.6) | 20 (25.0) | 69 (43.4) |
| Follow-up, median (IQD, range), mo | 11.0 (6–18, 1–124) | 22.7 (13–48, 1–115) | 11.5 (8–20, 2–69)‡ |
| OS, median (IQD, range), mo | 12.0 (7–23, 1–124) | 28.1 (14–49, 3–115) | 11.5 (8–20, 2–69)‡ |
* T stage substratified by resection status is shown in Supplementary Table 3 (available online). ECOG = Eastern Cooperative Oncology Group (classification for general patient performance status); IQD = interquartile distance.
† In pancreatic cancer classification, there is only one category (N1) for tumor spread to local lymph nodes.
‡ In the Hamburg cohort, all patients were deceased at time of data acquisition, ie, follow-up time was identical to overall survival in this case.
Cell Lines
Cellular drug sensitivity and gene expression were analyzed in 89 lymphoblastoid cell lines (LCLs) obtained from the National Human Genome Research Institute Sample Repository for Human Genetic Research at the Coriell Institute for Medical Research (www.coriell.org, cell line identifiers in Supplementary Methods, available online). The human adenoductal pancreatic carcinoma cell lines MIA-PaCa-2, PaTu-8988t, and L3.6 were obtained from ATCC (Manassas, VA).
Technical Procedures
All applied technical procedures are described in detail in the Supplementary Methods (available online). The five selected SNPs (Supplementary Table 1, available online) were genotyped by primer extension method (SNaPshot, Applied Biosystems, Foster City, CA). Variability in toxicity of gemcitabine was addressed in LCLs by dose-response relationships of serial gemcitabine dilutions. WWOX expression in PaTu-8988t and L3.6 was depleted via siRNA transfect, successful knockdown was verified by Western blotting (Supplementary Figure 3, available online), and functional consequences on gemcitabine sensitivity were evaluated. Transcript numbers of the WWOX gene were measured by quantitative real-time polymerase chain reaction. Genetic sites with suggested allelic distinctions in protein binding were analyzed by electrophoretic mobility shift assay (EMSA) using 32P-labelled probes for interaction with nuclear protein extracts.
Bioinformatic Analysis
Bioinformatic analyses refer to the assessment of linkage disequilibrium (LD) between genetic polymorphisms, analysis of database entries concerning expression regulating features, and calculations to predict transcription factor binding (Supplementary Methods, available online).
Statistical Analysis
OS in dependence on nongenetic and genetic variables is illustrated by Kaplan-Meier plots, with statistical assessment performed by log-rank test associated with median values. Respective mean values are also reported. Hazard ratios (HRs) in relation to OS were examined for genetic and nongenetic variables by univariate and multivariable Cox regression analyses. Genotype effects on functional parameters were assessed by Mann-Whitney U test for two-group and by Jonckheere-Terpstra for three-group comparisons. For the initial clinical testing of five independent genetic variables, the threshold for statistical significance was set at a P value of .01, further functional testing of one candidate polymorphism at a P value of .05. All reported P values refer to two-sided testing. Calculations were carried out using SPSS 12.0 (IBM, Chicago, IL).
Results
Univariate Genetic Analysis
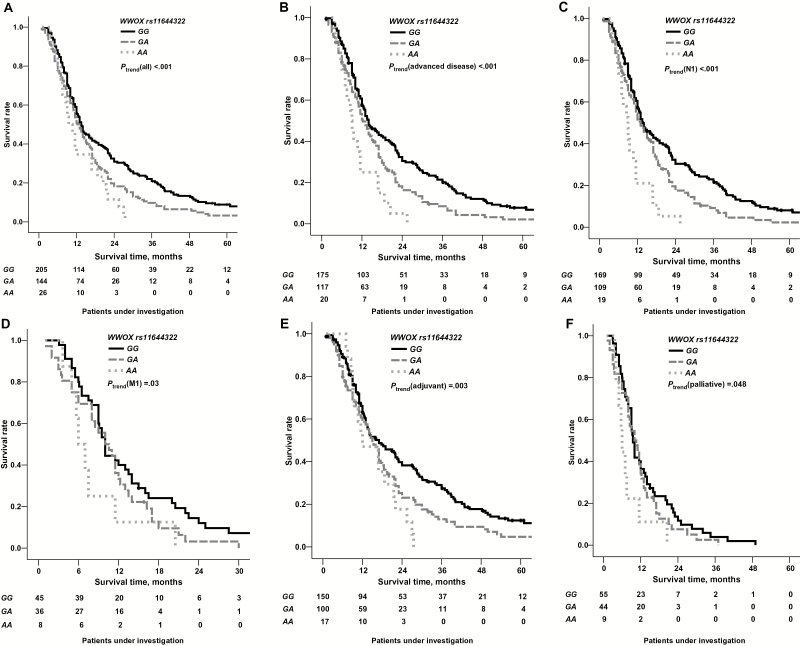
We identified the genetic polymorphism WWOX rs11644322 G>A to be strongly linked to the clinical outcome of patients treated with a gemcitabine-containing regimen for PDAC. The variant A allele at WWOX rs11644322, which features a frequency of 26% in Caucasians, conferred worse OS. For the three genotypes of WWOX rs11644322, median OS was 14 months (95% confidence interval [CI] = 12 to 15 months) for GG, 13 months (95% CI = 11 to 15 months) for GA, and 9 months (95% CI = 7 to 12 months) for AA (P trend < .001 for trend in univariate log-rank assuming a codominant mode of inheritance) (Figure 1A). Respective mean OS was 25 months (95% CI = 21 to 29 months) for GG, 19 months (95% CI = 15 to 22 months) for GA, and 13 months (95% CI = 10 to 16 months) for AA.
Figure 1.
Impact of WWOX rs11644322 on clinical outcome. Overall survival of 381 pancreatic cancer patients treated with a gemcitabine-containing regimen in dependence on the genotype configuration of WWOX rs11644322. A) The entire patient cohort. B) The subgroup of advanced disease stage (T4 and/or N1 and/or M1). C) Lymph node-positive patients. D) Patients with proven distant metastasis. E) Patients with adjuvant treatment intent. F) Patients with palliative treatment intent. The given P values indicate univariate testing by unadjusted log-rank test. Patient numbers under investigation are specified in 12-month intervals, for distant metastasis in six-month intervals. Genotyping for six patients at this position failed, leaving 375 assessable (see Supplementary Table 2, available online). All statistical tests were two-sided.
For the IL17F rs763780 SNP, we observed a trend in the opposite direction, as formerly suggested by GWAS (5). Regarding the variant allele of PRB2 rs2900174, we could reproduce a worse prognosis. However, this did not reach statistical significance. The other two investigated candidate polymorphisms DCP1B rs11062040 and BTRC rs10883617 did not reveal any link to OS in our study cohorts (Supplementary Figure 1, available online).
Combined Genetic and Nongenetic Analysis
Apart from rs11644322, univariate analysis of nongenetic factors revealed M stage, resection status, Easter Cooperative Oncology Group performance status (Supplementary Figure 2, available online), and age at therapy start as strong (P < .001)—and histopathologic grading and T stage as weak (.001 ≤ P < .2)—predictors for OS (Table 2). Distribution of these variables was not affected by rs11644322 (Supplementary Table 4, available online). Sex, N stage, and mode of gemcitabine chemotherapy (single or combined) were not associated with OS in univariate analysis and thus were not considered in the multivariable Cox model. This model, corrected for the three study sites, revealed age at therapy start, WWOX rs11644322, M stage, histopathologic grading, and resection status as predictors for OS (Table 3). In this comprehensively adjusted analysis, rs11644322 retained its strong impact on OS. Referred to GG, the hazard ratio for patients with GA genotype was 1.34 (95% CI = 1.12 to 1.60) and 1.80 (95% CI = 1.51 to 2.15) for those with AA, with the latter exceeding any considered single nongenetic parameter.
Table 2.
Univariate Cox regression analysis for nongenetic factors and WWOX rs11644322 with respect to overall survival
| Variable | HR (95% CI)* | P† |
|---|---|---|
| Age (per y)‡ | 1.04 (1.02 to 1.06) | <.001 |
| Sex | ||
| Female vs male | 0.95 (0.76 to 1.17) | .62 |
| Performance status | ||
| ECOG 1 vs 0 | 1.59 (1.19 to 2.13) | .002 |
| ECOG 2/3 vs 0 | 2.12 (1.53 to 2.92) | <.001 |
| ECOG 1/2/3 vs 0 | 1.73 (1.32 to 2.28) | <.001 |
| T stage | ||
| T3 vs T1/T2 | 0.87 (0.61 to 1.25) | .44 |
| T4 vs T1/T2 T4 vs T1/T2/T3 |
1.45 (0.93 to 2.26) 1.56 (1.15 to 2.12) |
.10 .004 |
| N stage | ||
| N1 vs N0 | 1.00 (0.77 to 1.30) | 1.0 |
| M stage | ||
| M1 vs M0 | 2.05 (1.59 to 2.64) | <.001 |
| Resection status | ||
| R1 vs R0‡ | 1.47 (0.98 to 2.21) | .06 |
| R1/R2 vs R0‡ | 1.54 (1.06 to 2.26) | .02 |
| R2/not resected vs R0 R2/not resected vs R0/R1 |
1.90 (1.42 to 2.54) 1.85 (1.41 to 2.42) |
<.001 <.001 |
| Grading | ||
| G2 vs G1 | 1.27 (0.81 to 1.99) | .30 |
| G3 vs G1 G3 vs G1/G2 |
1.46 (0.91 to 2.34) 1.20 (0.95 to 1.51) |
.12 .13 |
| Chemotherapy regimen | ||
| Gemcitabine combination vs mono | 1.06 (0.85 to 1.32) | .60 |
| WWOX rs11644322 | ||
| GA vs GG | 1.33 (1.06 to 1.67) | .01 |
| GA+AA vs GG
Trend (number of A alleles) |
1.40 (1.13 to 1.74) 1.36 (1.14 to 1.62) |
.002 <.001 |
* For the variable “patient age at therapy start,” the hazard ratio (HR) refers to each additional year. For the other variables, the HR of each category with respect to the denoted reference is indicated. When a linear trend was assumed, the HR refers to the effect size of one unit increase in the variable category. Proportionality over time was tested for all variables. CI = confidence interval; ECOG = Eastern Cooperative Oncology Group (classification for general patient performance status); HR = hazard ratio.
† The reported P values refer to Cox regression analysis and are two-sided.
‡ An interaction with time at P < .05 for the respective variable was observed, and hence time-dependent adjusted values are reported (for details, see “Survival analysis” in the Supplementary Methods, available online).
Table 3.
Impact of WWOX rs11644322 on overall survival adjusted for nongenetic factors*
| Variable | HR (95% CI) | P† |
|---|---|---|
| Age (per y)‡ | 1.04 (1.02 to 1.06) | <.001 |
| Performance status (ECOG 1/2/3 vs 0) | 1.16 (0.86 to 1.57) | .34 |
| T stage (T4 vs T1/T2/T3) | 1.01 (0.72 to 1.42) | .94 |
| M stage (M1 vs M0) | 1.49 (1.12 to 1.98) | .006 |
| Resection status (R2/not-resected vs R0/R1) | 1.42 (1.04 to 1.95) | .03 |
| Grading (G3 vs G1/G2) | 1.23 (0.96 to 1.56) | .10 |
| WWOX rs11644322 (per A allele) | 1.34 (1.12 to 1.60) | .001 |
* This multivariable analysis was carried out using a Cox proportional hazard regression model with the denoted variables as independent factors. Variables with P < .2 according to the univariate analysis reported in Table 2 were included. Adjustment for study site was performed in the model to account for variations at the three locations. CI = confidence interval; ECOG = Eastern Cooperative Oncology Group (classification for general patient performance status); HR = hazard ratio.
† The reported P values refer to Cox regression analysis and are two-sided.
‡ Because of the interaction with time at P < .05, time-dependent adjusted values are reported for this variable (for details, see “Survival analysis” in the Supplementary Methods, available online).
Subgroup Analysis According to Clinical Features
Detailed subgroup analyses revealed the effect of rs11644322 being present in advanced disease stage (T4 and/or N1 and/or M1, P trend < .001) (Figure 1B), lymph node (Figure 1C), or distant metastasis (Figure 1D). Dichotomization into one group with resected PDAC (R0, R1, or R2) and M0 status (regarded as “adjuvant” treatment intent, 71% of all cases) and a second group with nonresectability and/or M1 status (“palliative”) proved the strongest combined discriminator for OS identified in this cohort (P < .001) (Supplementary Figure 2D, available online). The WWOX index SNP affected OS both in the “adjuvant” (P = .003) and in the “palliative” (P = .048) group (Figure 1, E and F). The effect of this SNP on OS also remained virtually unaltered when adjusted for adjuvant vs palliative treatment intent in a multivariable Cox model.
Cellular Drug Sensitivity
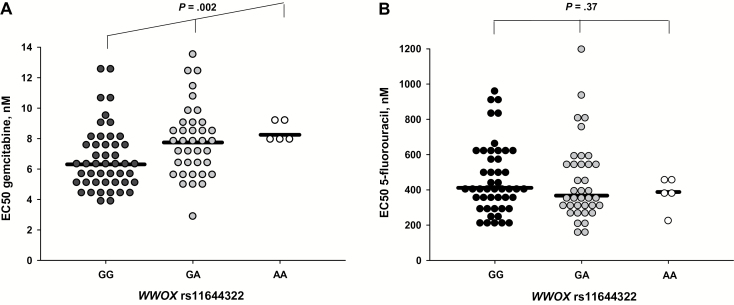
In order to determine whether cellular gemcitabine sensitivity is dependent on WWOX rs11644322, we examined the dose-response effects of serial drug dilutions on cytotoxicity in 89 LCLs. Consistent with the clinical findings, the A allele was associated with increased resistance (P = .002) (Figure 2A). In contrast, cytotoxicity of 5-fluorouracil (5-FU) assessed in the same 89 LCLs was not modulated by rs11644322 (P = .37) (Figure 2B).
Figure 2.
Impact of WWOX rs11644322 on cellular drug sensitivity of lymphoblastoid cell lines. A) EC50 values representing cellular sensitivity towards gemcitabine in relation to the three genotype configurations at rs11466322. Out of 89 LCLs, 47 harbored GG genotype, 37 GA, and five AA. EC50 data for proliferation inhibition were calculated from seven serial gemcitabine dilutions (1.9–76.0nM) with respect to a drug-free control by a three-parameter Gompertz model (details in the Supplementary Methods, available online). Statistical differences were assessed by the nonparametric Jonckheere-Terpstra trend test. B) Respective data for 5-fluorouracil. The samples used were exactly identical to those assessed for gemcitabine drug sensitivity (A). Eight concentrations of 5-fluorouracil, ranging from 75 to 385 000nM, were analyzed with respect to a drug-free control. The median value for each group is highlighted by a horizontal black line. All statistical tests were two-sided.
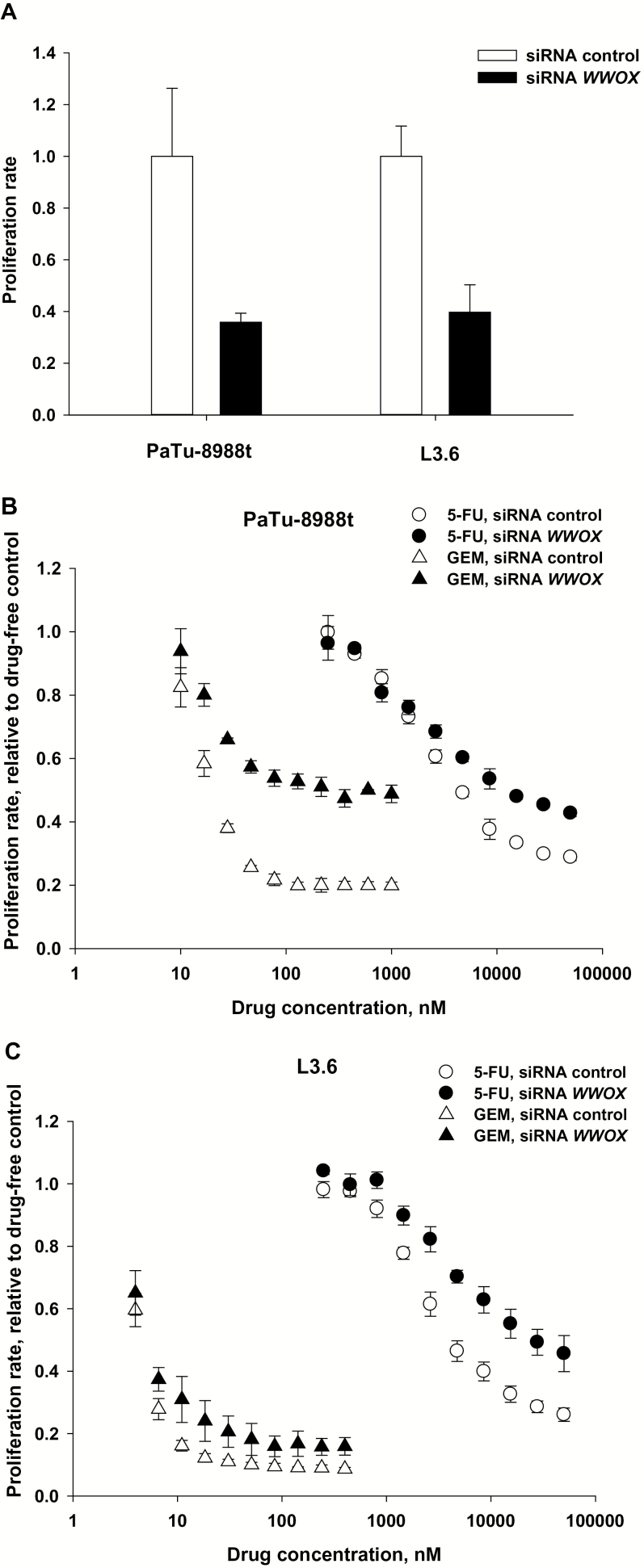
Drug Sensitivity Upon WWOX Knockdown
We next tested the hypothesis that WWOX directly influences gemcitabine sensitivity by performing siRNA-mediated knockdown in the two pancreatic cancer cell lines, PaTu-8988t and L3.6. Successful abrogation of WWOX protein expression was verified by western blot (Supplementary Figure 3, available online). We noted a marked decrease in basal proliferation of both cell lines following transfection of WWOX siRNA in comparison with control siRNA (Figure 3A). The effects of WWOX knockdown on 5-FU responsiveness were moderate and similar in the two investigated cell lines. In contrast, while WWOX depletion moderately decreased gemcitabine sensitivity in L3.6 cells, PaTu-8988t cells displayed a reproducibly profound resistance to gemcitabine following WWOX depletion (Figure 3, B and C). Thus, the dependence of gemcitabine sensitivity on WWOX expression might be a feature that varies between different pancreatic cancer cells. Cell-specific interactions between WWOX and gemcitabine, but not 5-FU sensitivity, are supposed, consistent with our findings in LCLs.
Figure 3.
WWOX knockdown. A) Effects of WWOX knockdown by siRNA on basal proliferation rates of the two adenoductal pancreatic cancer cell lines PaTu-8988t and L3.6. Cells were transfected either with a panel of four siRNAs intended to target WWOX or with a scrambled panel of unspecific siRNAs as control. Technical details are described in the Supplementary Methods (available online). Data of this panel refer to drug-free conditions. The bars represent means of three independent experiments, with the errors indicating one standard deviation. B and C) Consequences of WWOX knockdown on cytostatic drug sensitivity. B) Displays data for the PaTu-8988t, and (C) for the L3.6 cell line. Drug concentrations are denoted in a log10-scale. Data for gemcitabine are shown as triangles (open ones for control siRNA, filled ones for siRNA against WWOX), for 5-FU analogously as circles. For each transfection condition and each drug, the proliferation rate for a drug-free control was set to 1.0, to which each drug concentration was referred. Data represent means of three independent experimental series, with one standard deviation indicated as error symbols. Within each series, each single condition was assayed in quadruplicates, of which median values were taken for analysis.
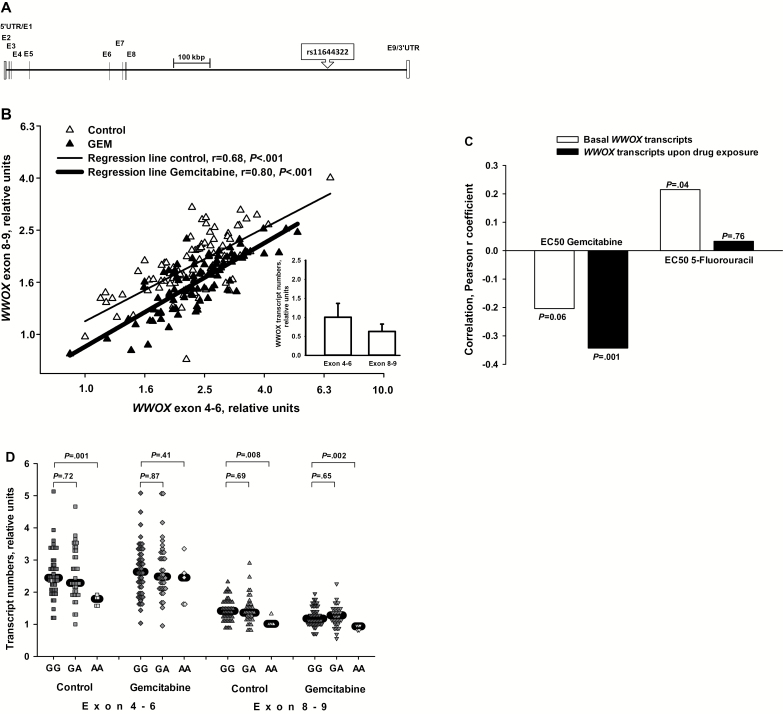
WWOX Transcription: Region-Specific Features, Implications for Drug Sensitivity, and Impact of the WWOX Index SNP
Rs11644322 is located in the 778856bp long intron 8, which separates exons 8 and 9 (Figure 4A). Updated GeneBank entries (http://www.ncbi.nlm.nih.gov/gene/) indicate several alternative WWOX transcripts, terminating within this intron 8. Using cloned entire WWOX cDNA as a reference, we could demonstrate 67% mean transcription rate of exon 8–9 in relation to the core coding region in 88 LCLs (insert in Figure 4B). Together with the high intra-cell line expression correlation (r = 0.68, P < .001) between these two WWOX regions it is suggested that the majority of WWOX transcripts contain the last exon. As this correlation was found to have even increased upon exposure to gemcitabine (r = 0.80, P < .001), a link between this genotoxic stress and transcription of the entire WWOX gene is assumed. Moreover, high WWOX transcript numbers were accompanied by low EC50 values for gemcitabine in LCLs, indicative of increased sensitivity toward this drug. This relationship was further increased if WWOX transcription upon gemcitabine exposure was considered (Figure 4C). In sharp contrast, higher WWOX transcription rather enhanced resistance toward 5-FU. These data suggest a specific gemcitabine-sensitizing effect of WWOX. In case of the AA genotype at rs11644322, which rendered both the worst prognosis (Figure 1) and the highest cellular resistance toward gemcitabine (Figure 2A), statistically significant effects on WWOX transcription were observed: Transcripts concerning both the core and the last exon region were reduced under baseline conditions whereas upon gemcitabine exposure these two regions were differentially affected by this genotype. (Figure 4D). In this short-term incubation time of 24 hours, a statistically significant difference between the GA and GG genotypes in relation to WWOX expression could not be delineated.
Figure 4.
Gene expression analyses. A) Genetic architecture at the WWOX locus. The coding region encompasses nine exons, with the first and the last one flanked by a 5′- and 3′-untranslated region (UTR), respectively. The exons are depicted as vertical lines. In the scheme, the relationships of physical sizes and distances are retained. The position of the index SNP rs11644322 is denoted. B) Expression of the last exon in relation to that of the core WWOX coding region. The mRNA expression of the terminal exon 9 (captured by an exon 8/exon 9-spanning primer pair) was compared with the major part of the coding region (represented by a primer pair spanning exons 4–6). The graph summarizes the data obtained in 88 lymphoblastoid cell lines (for one cell line, reverse transcription failed) treated either with cell culture medium only (baseline) or with 30nM gemcitabine at 37°C for 24 hours. The scatter plot illustrates expression correlation between regions 4–6 and 8–9. Both axes are displayed in log10-scale, for which normal distributions of the data could be assumed. The respective regression lines with the Pearson correlation coefficient r are indicated. All expression data were referred to the cell line with the lowest transcript numbers for exon 4–6 under basal conditions (set to “1”). The lower right insert illustrates, under baseline conditions and using cloned full-length WWOX cDNA as reference, the transcript numbers of the last WWOX exon in relation to those of the core coding region, of which the mean of the entire LCL cohort was set to “1” (error symbols denote one standard deviation). C) Correlation of WWOX transcripts with EC50 values of gemcitabine and 5-fluorouracil. Data are based on the same 88 LCLs as in Figure 4B and refer to WWOX transcripts of the exon 4–6 region (very similar for exon 8–9, not shown). Transcript numbers were normalized to the weighted mean of five reference genes (36b4, B2MG, GAPDH, HPRT1, and UBC). P values are according to the Pearson correlation coefficient r, which is displayed on the y-axis. Note that a negative correlation indicates higher WWOX expression along with lower EC50 values, ie, increased sensitivity toward cytotoxic effects. Gemcitabine was administered at 30nM and 5-fluorouracil at 3 µM for 24 hours at 37°C prior to RNA harvesting. These drug concentrations were chosen about five-fold higher than mean EC50 values observed upon 72 hours of drug exposure (Figure 2). D) Impact of rs11644322 SNP on WWOX regional transcription (exon 4–6/8–9). The left side of the image refers to the central coding region (exon 4–6), the right to that of exon 8–9, each for baseline conditions and upon 30nM gemcitabine incubation for 24 hours at 37°C. Shown data refer to the same panel of 89 LCLs as in Figure 2A. The median value for each group is highlighted by a horizontal black line. Statistical differences between two groups were assessed by the nonparametric Mann-Whitney U test. The lower line of P values refers to testing between GG and GA genotype, the upper one between GG and AA configuration. All statistical tests were two-sided.
Allele-Specific Protein Binding at the WWOX Index SNP
Based on the differential mRNA levels in cells possessing different rs11644322 alleles, we hypothesized that this region may contain a specific regulatory sequence that regulates WWOX gene expression. Prior to conducting EMSA, we evaluated the genetic linkage pattern of rs11644322 because markers in high linkage disequilibrium (eg, r2 > 0.8) associate similarly with clinical and functional traits. The tagger algorithm implemented in Haploview (see “Bioinformatic analysis” in the Supplementary Methods, available online) uncovered three polymorphisms at r2 > 0.8 with rs11644322: rs2062903, rs34310485, and rs12598700. Among those, only rs34310485 was highlighted by a screen for regulatory elements employing the University of California, Santa Cruz browser (https://genome.ucsc.edu/). We therefore performed EMSA using sequences containing rs34310485 in addition to the clinically associated rs11466322. Interestingly, probes for rs11644322, but not rs34310485, revealed decreased mobility in EMSA, indicating that this region can be bound by one or more transcription factors. Importantly, a reproducibly stronger binding was observed for the G in comparison with the A allele and confirmed by competition experiments using nonradiolabeled probes (Figure 5B, lanes 2–3).
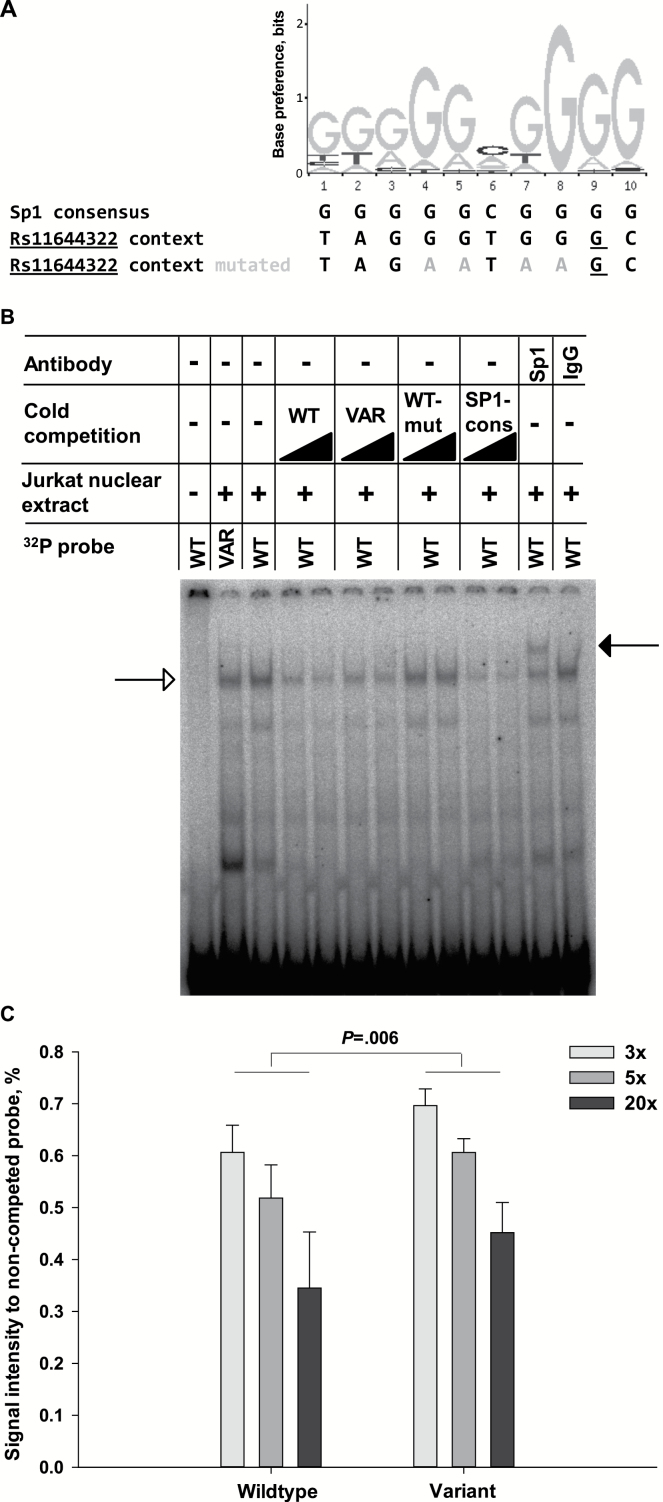
Figure 5.
Transcription factor binding motif and identity of binding protein. A) Composition of the SP1_Q6 (systematic name M9525) binding motif. The plot was derived from JASPAR database (29). Out of the two models annotated for Sp1 in JASPAR, SP1_Q6 was identified corresponding to the sequence context of rs11644322 according to a matrix-derived consensus sequence (www.telis.ucla.edu/TFBM). The y-axis highlights the preference of a specific base in bits ranging from 0 (no base preferred) to 2 (only one base preferred). The base positions in the motif are ordered from 1 to 10 on the x-axis. Beneath the plot, the consensus sequence of SP1_Q6 is displayed in the first row. The second row represents the sequence context of WWOX rs11644322 with wild-type G allele configuration (underlined). In the third line, this sequence context was mutated at four bases (shaded in gray), particularly prominent in the SP1_Q6 motif. B) Representative EMSA plot for assessing transcription factor binding of nuclear protein extracts from Jurkat cells at WWOX rs11644322. Lane 1 indicates the negative control without nuclear proteins. Lanes 2 and 3 illustrate 32P-labeled probes containing the variant A and the wild-type G allele, respectively. In lanes 4–7, the interaction between nuclear proteins and the G wild-type allele-containing probe was competed with each three-fold and five-fold excess of nonradioactive wild-type and variant allele-containing probes. Analogously, competition with the wild-type rs11644322 probe mutated at the four most crucial bases of the Sp1 binding motif, last row in (A), is shown in lanes 8–9 and with the consensus sequence for Sp1, first row in (A), in lanes 10–11. Supershift with the Sp1 antibody 1C6 is illustrated in lane 12, with IgG control in lane 13. The arrow with open head at the left indicates the bands of interaction between nuclear proteins and the radio-labeled probes, and that with filled head at the right the supershifted complex. Technical details are provided in the Supplementary Methods (available online). C) Quantification of cold competition using nuclear extracts of the pancreatic cancer cell line MIA-PaCa-2. Signal intensities of the noncompeted 32P-labeled probe with wild-type G allele at the WWOX rs11644322 site were set at 1.0. The nonlabeled probes (containing the wild-type G or variant A allele) were applied with three-, five-, and 20-fold excess with respect to the 32P-labeled probe with the G allele. For each bar, the mean value of three independent binding reactions is depicted, with the errors indicating one standard deviation. Statistical significance was tested by a linear regression model, with the excess of the nonradioactive probes and the allelic configuration at rs11644322 as independent variables. The indicated P value denotes the impact of rs11644322 in this model. All statistical tests were two-sided.
We next tested whether these findings with standardized Jurkat nuclear extracts also hold true in pancreatic carcinoma. Using the human MIA-PaCa-2 cell line, again allele-specific distinctions at rs11644322 for Sp1 binding affinities were observed (summarized in Figure 5C). A linear regression model considering the excess of cold competitor and the allelic configuration as independent variables elicited a statistically significant impact of the allelic configuration at rs11644322 (P = .006). To rule out any bias on the linear regression model, we also performed an ordinal regression, which confirmed the significant allelic impact on this probe-protein interaction (P = .002).
Identity of Protein Binding at the WWOX Index SNP
A bioinformatic analysis was conducted to identify candidate proteins that may bind to rs11644322 in an allele-specific manner. For the most statistically significant results of this in silico screen, double-stranded probes representing the consensus sequences were tested in EMSA for competition. The probe corresponding to the SP1_Q6 binding pattern (Figure 5A) abolished the interaction between the nuclear protein extract and the probe with the G wild-type allele (Figure 5B, lanes 10–11) suggesting that the binding protein might be Sp1 or a structural homologue like Sp3. Moreover, mutation of the four most pivotal positions in the Sp1 consensus motif (third row in Figure 5A) abrogated its ability to compete for binding (lanes 8–9 in Figure 5B). The identity of Sp1 as a protein capable of binding to the rs11644322-containing sequence was verified via super shift analysis with a specific antibody and was not seen with a control IgG (Figure 5B, lanes 12–13). Members of the Sp transcription factor family, including Sp1-4, display high homology in their protein domain structure (11). In particular, Sp1 and Sp3 exhibit the most similar features in terms of ubiquitous expression and binding affinity to the same DNA elements (12). Consistently, we also observed a supershift of the binding to the WWOX rs11466322 site when using an Sp3-specific antibody (data not shown).
These data establish an allele-specific affinity, rendering differential transcription factor recruitment of the Sp family and subsequent gene expression differences a plausible explanation for the observed clinical findings.
Discussion
An impact of the WWOX rs11644322 SNP on OS in gemcitabine-treated adenoductal pancreatic cancer could be demonstrated for the first time in a highly statistically significant fashion. This association supports earlier findings obtained in an exclusively palliative setting (5), in which the statistical significance threshold was not passed. In our study, the effect of this genetic marker held true when adjusting for all common, established prognostic features in pancreatic cancer and was proven for both adjuvant and palliative treatment intent. Subgroup analysis revealed that the effect of rs11644322 was because of advanced disease stage, for which most patients with pancreatic cancer are usually diagnosed and as was the case in our study. Specifically, the “predictive cut” of this SNP appears to be between N-negative and N-positive conditions, suggesting a role for WWOX in metastatic processes. For patients with distant metastasis, superiority of the chemotherapeutic combination FOLFIRINOX vs gemcitabine has been reported (13), with a benefit comparable with what we detected between carriers of GG vs AA at rs11644322. Thus, it should be evaluated if patients with the GG genotype might be spared from the more toxic FOLFIRINOX regimen.
A recent report in endometrial cancer cells implicated WWOX in the suppression of mesenchymal markers (14). The processes of epithelial to mesenchymal transition (EMT) at the primary tumor site and backward in remote tissue (MET) are believed to be important in tumor metastasis. Furthermore, acquisition of a mesenchymal phenotype is associated with chemotherapeutic resistance in pancreatic cancer (15,16). This is consistent with our observation linking decreased WWOX expression to inferior cellular responsiveness toward gemcitabine. In this manner, lower WWOX levels probably impacted by the rs11644322 variant allele might favor a shift toward EMT.
In 88 LCLs, increased cellular resistance toward gemcitabine but not 5-fluorouracil was observed in presence of the variant A allele at rs11644322. These findings of drug-specific distinctions in individual cellular cytotoxicity dependent on WWOX are underscored by our investigations in pancreatic cancer cells, in which WWOX was depleted.
We demonstrate, for the first time, that rs11644322 interacts with protein binding of the SP family. Depending on the context and post-translational modifications, Sp1 and Sp3 can either activate or suppress gene transcription (12,17). Current literature does not contain any information about rs11644322 beyond the reported GWAS (5), and to the best of our knowledge no functional assessments considering this SNP have been undertaken so far.
There are two particular limitations of this study. First, we could not assess the clinical effect of rs11644322 for treatments other than gemcitabine. Thus, it is debatable whether the observed impact of this genetic polymorphism is a general feature of cytostatic treatment or related to gemcitabine as a specific drug. The latter hypothesis is supported by rs11644322-dependent modulation of cellular sensitivity toward gemcitabine, but not 5-FU in LCLs, and by dramatic variations in pancreatic cancer cell line responsiveness toward gemcitabine, but not 5-FU upon WWOX knockdown. Although unlikely, we cannot rule out that rs11644322 modifies the natural disease course rather than treatment effects. However, this objection is of minor relevance, as standard of care usually implicates any chemotherapy. The second limitation refers to the use of LCLs instead of pancreatic cancer cells to evaluate cytotoxic effects of gemcitabine in dependence on rs11644322. We decided for LCLs in this issue as the number of genetically diverse pancreatic cancer cell lines is limited. Because LCLs display EC50 values for gemcitabine in a similar range as determined for the pancreatic cancer cell lines PaTu-8988t and L3.6 (compare Figure 2A and Figure 3, B and C), it is conceivable that modulation of gemcitabine sensitivity by rs11644322 might be similar in pancreatic cancer cells and LCLs.
WWOX encodes for a protein of 46kDa involved in a variety of cellular functions including transcription, RNA splicing, and protein degradation. Mapping to chromosome 16q, it spans FRA16D, the second most fragile site in the human genome (18,19). The physical position of rs11644322 is more than 300 kbp distant from the downstream end of the FRA16D region, making an interaction unlikely. Likewise, no marker in high LD with rs11644322 touches the FRA16D region.
WWOX was found to be a potent tumor suppressor gene, affecting multiple malignancies including pancreatic cancer (18,20–23); however, it may differ from a classical tumor suppressor (24). It encodes a protein that interacts with p53 and its homologue p73 via the WW-containing domains and enhances stress response–induced cell death when translocated to the nucleus (25–27). However, an interaction with JNK1 was described, possibly counteracting WWOX-mediated apoptosis (27,28). Our data support an apoptosis-promoting role for WWOX following gemcitabine exposure.
In conclusion, WWOX rs11644322 represents a promising biomarker for gemcitabine-treated pancreatic cancer, with the perspective of tailoring future treatment. The impact of rs11644322 on OS was comparable with the strongest established prognostic markers in PDAC. This effect may be particularly pronounced in homozygous AA carriers concerning 7% of our Caucasian study cohort, but about 22% in Asian populations. The identified cellular and molecular mechanisms for WWOX rs11644322 convey a potential functional mechanistic basis for the observed clinical association. Beyond that, our study suggests that enhanced WWOX expression or activity might be relevant for drug-specific actions in cancer treatment.
Funding
This work was supported by the German Research Foundation (DFG, GRK1034/2 to JB, BMG, and SAJ) and by the Research Program, University Medical Center, University of Göttingen (to MAS).
Supplementary Material
The funders had no role in the design of the study; the collection, analysis, or interpretation of the data; the writing of the manuscript; or the decision to submit the manuscript for publication.
The authors state that there are no conflicts of interest concerning this study.
References
- 1. Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63 (1):11–30. [DOI] [PubMed] [Google Scholar]
- 2. Vincent A, Herman J, Schulick R, Hruban RH, Goggins M. Pancreatic cancer. Lancet. 2011;378(9791):607–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Burris HA, 3rd, Moore MJ, Andersen J, et al. Improvements in survival and clinical benefit with gemcitabine as first-line therapy for patients with advanced pancreas cancer: a randomized trial. J Clin Oncol. 1997;15 (6):2403–2413. [DOI] [PubMed] [Google Scholar]
- 4. Neoptolemos JP, Stocken DD, Bassi C, et al. Adjuvant chemotherapy with fluorouracil plus folinic acid vs gemcitabine following pancreatic cancer resection: a randomized controlled trial. JAMA. 2010;304 (10):1073–1081. [DOI] [PubMed] [Google Scholar]
- 5. Innocenti F, Owzar K, Cox NL, et al. A genome-wide association study of overall survival in pancreatic cancer patients treated with gemcitabine in CALGB 80303. Clin Cancer Res. 2012;18 (2):577–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kindler HL, Niedzwiecki D, Hollis D, et al. Gemcitabine plus bevacizumab compared with gemcitabine plus placebo in patients with advanced pancreatic cancer: phase III trial of the Cancer and Leukemia Group B (CALGB 80303). J Clin Oncol. 2010;28 (22):3617–3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Muerkoster S, Arlt A, Sipos B, et al. Increased expression of the E3-ubiquitin ligase receptor subunit betaTRCP1 relates to constitutive nuclear factor-kappaB activation and chemoresistance in pancreatic carcinoma cells. Cancer Res. 2005;65 (4):1316–1324. [DOI] [PubMed] [Google Scholar]
- 8. Pan X, Arumugam T, Yamamoto T, et al. Nuclear factor-kappaB p65/relA silencing induces apoptosis and increases gemcitabine effectiveness in a subset of pancreatic cancer cells. Clin Cancer Res. 2008;14 (24):8143–8151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hrubran RH, Bishop Pitman M, Klimstra DS. Tumors of the pancreas. AFIP Atlas of Tumor Pathology. 4th Series, Fascicle 6. Washington, DC: American Registry of Pathology; 2007. [Google Scholar]
- 10. Sobin LH, Gospodarowicz MK, Wittekind C, eds. TNM Classification of Malignant Tumours. 7th ed New York: John Wiley & Sons; 2009. [Google Scholar]
- 11. Schaeper ND, Prpic NM, Wimmer EA. A clustered set of three Sp-family genes is ancestral in the Metazoa: evidence from sequence analysis, protein domain structure, developmental expression patterns and chromosomal location. BMC Evol Biol. 2010;10:88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Li L, Davie JR. The role of Sp1 and Sp3 in normal and cancer cell biology. Ann Anat. 2010;192 (5):275–283. [DOI] [PubMed] [Google Scholar]
- 13. Conroy T, Desseigne F, Ychou M, et al. FOLFIRINOX versus gemcitabine for metastatic pancreatic cancer. N Engl J Med. 2011;364 (19):1817–1825. [DOI] [PubMed] [Google Scholar]
- 14. Pluciennik E, Nowakowska M, Pospiech K, et al. The role of WWOX tumor suppressor gene in the regulation of EMT process via regulation of CDH1-ZEB1-VIM expression in endometrial cancer. Int J Oncol. 2015;46 (6):2639–2648. [DOI] [PubMed] [Google Scholar]
- 15. Arumugam T, Ramachandran V, Fournier KF, et al. Epithelial to mesenchymal transition contributes to drug resistance in pancreatic cancer. Cancer Res. 2009;69 (14):5820–5828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Meidhof S, Brabletz S, Lehmann W, et al. ZEB1-associated drug resistance in cancer cells is reversed by the class I HDAC inhibitor mocetinostat. EMBO Mol Med. 2015;7(6):831–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lee HS, Park CK, Oh E, et al. Low SP1 expression differentially affects intestinal-type compared with diffuse-type gastric adenocarcinoma. PLoS One. 2013;8 (2):e55522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bednarek AK, Keck-Waggoner CL, Daniel RL, et al. WWOX, the FRA16D gene, behaves as a suppressor of tumor growth. Cancer Res. 2001;61 (22):8068–8073. [PubMed] [Google Scholar]
- 19. Ried K, Finnis M, Hobson L, et al. Common chromosomal fragile site FRA16D sequence: identification of the FOR gene spanning FRA16D and homozygous deletions and translocation breakpoints in cancer cells. Hum Mol Genet. 2000;9 (11):1651–1663. [DOI] [PubMed] [Google Scholar]
- 20. Aqeilan RI, Trapasso F, Hussain S, et al. Targeted deletion of Wwox reveals a tumor suppressor function. Proc Natl Acad Sci U S A. 2007;104 (10):3949–3954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kuroki T, Yendamuri S, Trapasso F, et al. The tumor suppressor gene WWOX at FRA16D is involved in pancreatic carcinogenesis. Clin Cancer Res. 2004;10 (7):2459–2465. [DOI] [PubMed] [Google Scholar]
- 22. Nakayama S, Semba S, Maeda N, Aqeilan RI, Huebner K, Yokozaki H. Role of the WWOX gene, encompassing fragile region FRA16D, in suppression of pancreatic carcinoma cells. Cancer Sci. 2008;99 (7):1370–1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Paige AJ, Taylor KJ, Taylor C, et al. WWOX: a candidate tumor suppressor gene involved in multiple tumor types. Proc Natl Acad Sci U S A. 2001;98 (20):11417–11422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Watanabe A, Hippo Y, Taniguchi H, et al. An opposing view on WWOX protein function as a tumor suppressor. Cancer Res. 2003;63 (24):8629–8633. [PubMed] [Google Scholar]
- 25. Abu-Odeh M, Bar-Mag T, Huang H, et al. Characterizing WW domain interactions of tumor suppressor WWOX reveals its association with multiprotein networks. J Biol Chem. 2014;289 (13):8865–8880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Aqeilan RI, Pekarsky Y, Herrero JJ, et al. Functional association between Wwox tumor suppressor protein and p73, a p53 homolog. Proc Natl Acad Sci U S A. 2004;101 (13):4401–4406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chang NS, Doherty J, Ensign A, et al. Molecular mechanisms underlying WOX1 activation during apoptotic and stress responses. Biochem Pharmacol. 2003;66 (8):1347–1354. [DOI] [PubMed] [Google Scholar]
- 28. Chang NS, Doherty J, Ensign A. JNK1 physically interacts with WW domain-containing oxidoreductase (WOX1) and inhibits WOX1-mediated apoptosis. J Biol Chem. 2003;278 (11):9195–9202. [DOI] [PubMed] [Google Scholar]
- 29. Bryne JC, Valen E, Tang MH, et al. JASPAR, the open access database of transcription factor-binding profiles: new content and tools in the 2008 update. Nucleic Acids Res. 2008;36(Database issue):D102-D106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.